Abstract
In eukaryotes, the C-terminal domain (CTD) of Rpb1 contains a heptapeptide repeat sequence of (Y1S2P3T4S5P6S7)n that undergoes reversible phosphorylation through the opposing action of kinases and phosphatases. Rtr1 is a conserved protein that colocalizes with RNA polymerase II (RNAPII) and has been shown to be important for the transition from elongation to termination during transcription by removing RNAPII CTD serine 5 phosphorylation (Ser5-P) at a selection of target genes. In this study, we show that Rtr1 is a global regulator of the CTD code with deletion of RTR1 causing genome-wide changes in Ser5-P CTD phosphorylation and cotranscriptional histone H3 lysine 36 trimethylation (H3K36me3). Using chromatin immunoprecipitation and high-resolution microarrays, we show that RTR1 deletion results in global changes in RNAPII Ser5-P levels on genes with different lengths and transcription rates consistent with its role as a CTD phosphatase. Although Ser5-P levels increase, the overall occupancy of RNAPII either decreases or stays the same in the absence of RTR1. Additionally, the loss of Rtr1 in vivo leads to increases in H3K36me3 levels genome-wide, while total histone H3 levels remain relatively constant within coding regions. Overall, these findings suggest that Rtr1 regulates H3K36me3 levels through changes in the number of binding sites for the histone methyltransferase Set2, thereby influencing both the CTD and histone codes.
INTRODUCTION
Reversible phosphorylation of the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (RNAPII) controls transcription elongation and termination through the orderly recruitment of transcription regulatory factors. These factors are involved in various processes including mRNA processing and cotranscriptional histone modifications (1–3). The CTD is comprised of the heptapeptide repeat of Y1S2P3T4S5P6S7 that is highly conserved with 26 repeats in Saccharomyces cerevisiae, 42 repeats in Drosophila melanogaster, and 52 repeats in humans and is essential for viability (4–10). The CTD is an unstructured domain that is unique to RNAPII and contains some degenerative repeats that deviate from the canonical sequence (11). Multiple residues within the CTD repeats are phosphorylated (P) at different stages of the transcription cycle; these residues include Ser2, Ser5, Ser7, Tyr1, and Tyr4, which thereby serve as a signaling scaffold for RNAPII transcription machinery (12). The phosphorylation of Ser2, Ser5, and Ser7 in S. cerevisiae has been well characterized by several laboratories (13, 14). Phosphorylation of Tyr1 and Thr4 are less characterized but have also been implicated in the regulation of RNAPII transcription (12, 15). Additionally, the proline at position six in the CTD repeats is subject to cis-trans isomerization by Ess1 (16, 17). Despite a great deal of research into these modifications, the extent to which these modifications coexist in a phosphorylated and nonphosphorylated state on a single heptapeptide repeat in vivo remains enigmatic.
The reversible phosphorylation of the CTD through the action of kinases and phosphatases generates a “CTD code” that has been shown to correlate with RNAPII location on the gene (13, 18–20). At the beginning of transcription, RNAPII is in a hypophosphorylated state as it is recruited to the preinitiation complex at the promoter of a target gene (21). Promoter escape correlates with a rise in the levels of Ser5 and Ser7 phosphorylation (Ser5-P and Ser7-P, respectively), which are modified through the action of the transcription factor IIH (TFIIH) subunit Kin28 (14, 18, 19, 22, 23). Once RNAPII enters into productive transcript elongation, Ser5-P levels decrease while Ser7-P and Ser2-P levels continue to accumulate (13, 19). These two modifications are significant for the recruitment of factors involved in transcription elongation, termination, and 3′-end processing (24–26). Chromatin immunoprecipitation (ChIP) studies investigating Ser5-P using the H14 antibody (shown to detect CTD sequences phosphorylated at both Ser5 and Ser2 [27]) suggest that Ser5 is dephosphorylated within the first few hundred base pairs of transcription elongation at a small number of RNAPII target genes (22). However, a more-recent global study with the 3E8 antibody found that Ser5-P levels begin to decrease within 500 bp after the transcription start site (TSS), but approximately half of the detectable signal remains throughout the gene body (18). Concurrent with Ser5 dephosphorylation, Ser2-P increases through the action of the kinase complex CTDK-I and remains phosphorylated through the transcription termination site (TTS), during which this modification has been shown to recruit transcription termination factors (25, 28). The phosphatase Fcp1 removes the Ser2-P mark at the end of transcription, thereby making hypophosphorylated RNAPII available for a subsequent round of transcription (29, 30).
The transition of RNAPII from Ser5-P to Ser2-P is an important step in the transcription cycle and is regulated by multiple layers of regulation. The dual-specificity phosphatase family member Ssu72 has been shown to remove Ser5-P from the CTD (31). Interestingly, Ssu72 localizes primarily at the 3′ end of the gene as a subunit of the cleavage and polyadenylation factor (CPF) and is involved in mRNA processing events and control of RNAPII termination (32–36). Ssu72 also participates in gene looping and the suppression of divergent transcription (37, 38). Rtr1 is a conserved protein that has been reported by both in vitro and in vivo experimental studies to be a phosphatase responsible for the removal of Ser5-P from the CTD (14, 39–42). ChIP assays found that the localization of Rtr1 is predominant between the peaks of Ser5-P and Ser2-P. Analysis of RNAPII in RTR1 deletion cells found that Ser5-P RNAPII accumulates in whole-cell extracts in the absence of Rtr1 activity (39). In addition, the human homologue of Rtr1, RPAP2, also displays phosphatase activity that is specific for Ser5-P (40, 43). Overall, these studies have implicated both Ssu72 and Rtr1 in the removal of Ser5-P during the RNAPII transcription cycle.
Set2 is a histone lysine methyltransferase that modifies coding region histones on histone H3 at K36 and is the sole histone H3 lysine 36 (H3K36) methyltransferase in Saccharomyces cerevisiae. Set2 has been shown to interact with the Ser2 and Ser5 diphosphorylated form of transcribing RNAPII through the Set2-Rpb1-interacting (SRI) domain in a Ctk1-dependent manner (44–48). We have previously found that the Rtr1 interaction with RNAPII is dependent on Ctk1 activity and that Rtr1 can interact with RNAPII phosphorylated at Ser2, Ser5, and/or Ser7 (49). In fact, multiple lines of evidence suggest that Rtr1 binds to a multiply phosphorylated form of the RNAPII CTD (40–42, 49). These data suggest that Rtr1 can act as both a reader and an eraser for the CTD code and could function as a potential regulator of Set2 targeting to RNAPII target genes through regulation of Ser5-P RNAPII levels. The recruitment of Set2 by Ser2 and Ser5 CTD phosphorylation directs H3K36 methylation across transcribed RNAPII genes (47, 50). However, the role of Rtr1-dependent Ser5-P CTD dephosphorylation in Set2 recruitment and histone H3K36 methylation has not been investigated.
In this study, we show that S. cerevisiae Rtr1 is a regulator of Ser5 phosphorylation in the RNAPII CTD in agreement with previous findings (14, 39, 41, 49, 51, 52). We show that deletion of RTR1 leads to global increases in Ser5-P CTD occupancy at RNAPII target genes, suggesting that Rtr1 is a general regulator of RNAPII phosphorylation during transcription elongation. Chromatin immunoprecipitation experiments in an RTR1 deletion strain (rtr1Δ) show global accumulation of Ser5-P RNAPII primarily at the 3′ ends of target genes regardless of gene length. This finding is specific for Rtr1, since the RTR2 deletion strain (rtr2Δ) does not appear to affect Ser5-P levels at PMA1. Furthermore, we provide genome-wide evidence that Rtr1 activity regulates H3K36 trimethylation (H3K36me3) levels across RNAPII target genes. Overall, this study clearly shows that Rtr1 is an important global regulator of the CTD code during RNAPII transcription elongation. Changes in Rtr1-mediated dephosphorylation of RNAPII lead to increased Ser5-P levels at the 3′ ends of mRNA-encoding genes, which causes a coordinate increase in histone H3K36me3 levels within the coding region.
MATERIALS AND METHODS
Chromatin immunoprecipitation.
For a detailed description of chromatin immunoprecipitation, see Mosley et al. (39) and Fox et al. (53). Briefly, 200-ml cultures were grown at 30°C until they reached an optical density at 600 nm (OD600) of ∼0.8 to 1.0. The cultures were cross-linked with formaldehyde (1% vol/vol) for 15 min, and the reaction was quenched by adding glycine to a final concentration of 125 mM. The cells were spun down, washed, and stored at −80°C. The cells were thawed and lysed by bead beating. The chromatin pellet was sonicated to yield fragments between 200 and 500 bp using a Branson 450 instrument (at a setting of 4, duty cycle 90%, 12 pulses for 7 cycles) while cooling samples on ice between cycles. Following an overnight incubation with the H14 antibody against Ser5-P RNAPII (Covance), a histone H3 lysine 36 trimethylation antibody (Abcam) or an antibody directed against the C terminus of histone H3 (Abcam) was used. The antibodies were subsequently pulled down using protein G-Sepharose. The beads were washed extensively, and DNA was eluted as previously described.
Chromatin immunoprecipitation with microarray technology (ChIP-chip) sample preparation and hybridization.
A mass of 5 μg Cy-labeled samples with specific activity of greater than 15 pmol/μg were combined for each channel and hybridized according to the Agilent Yeast ChIP-on-chip Analysis Protocol (54). Hybridization occurred in an Agilent rotisserie hybridization oven set at 20 rpm and 65°C for 40 h.
ChIP-chip data analysis.
Agilent feature text files were read into R (3.1.2) using the limma package (3.22.1). Data were normalized within arrays using median normalization. Each gene in the yeast genome was divided into six equally sized bins. Five hundred base pairs of the upstream and downstream intergenic regions were divided into two bins on each side of the gene. The normalized average microarray log2 (immunoprecipitated [IP]/input) value for each probe was assigned to the closest bin (whichever bin midpoint was closest to the probe midpoint). In this way, a matrix was generated with 10 columns and a row for each gene in the yeast genome. Each column was then averaged and plotted to display average expression over the gene region. Clustering was performed as previously described by k-means clustering using the multiexperiment viewer, part of the TM4 microarray software suite (55, 56).
GO term analysis.
GOstat analysis was performed to determine significantly overrepresented genes from our five clusters (57). The 7,112 yeast genes, divided into their respective clusters, were entered into the GOstat web interface (http://gostat.wehi.edu.au/cgi-bin/goStat.pl) to determine the most overrepresented gene ontology (GO) terms and to obtain P values to indicate the significance of enrichment (see Table S1 in the supplemental material).
Accession number.
All data have been deposited into the NCBI Gene Expression Omnibus (GEO) under accession number GSE68181.
RESULTS
RTR1 deletion causes a shift in the distribution of Ser5-P.
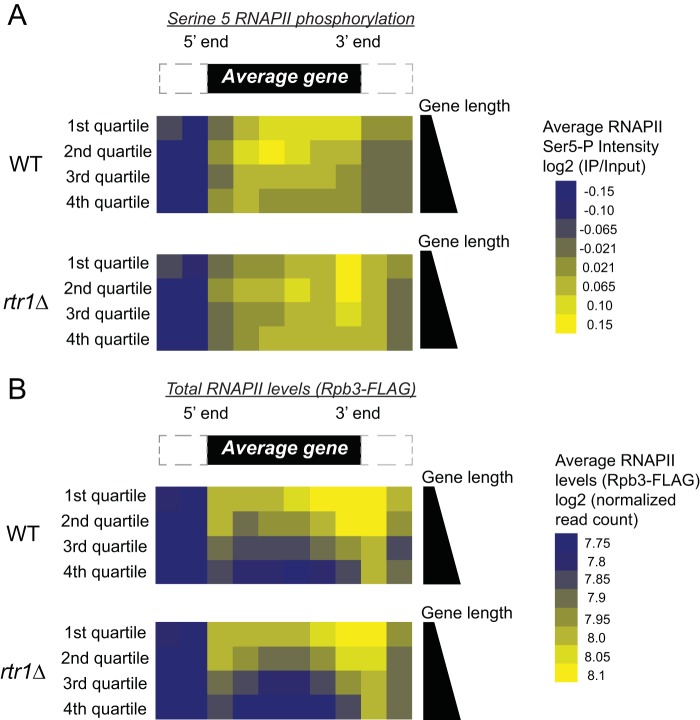
To capture a global picture of the distribution changes of Ser5-P RNAPII in wild-type (WT) and rtr1Δ cells, we performed average gene analysis from biological replicate ChIP-chip studies (Fig. 1A, n = 2). For illustration purposes, target genes were separated into quartiles according to gene length, since shorter genes (such as RPS5), show high levels of Ser5-P RNAPII occupancy across their entire coding regions (Fig. 1A, WT 1st quartile). In the first quartile that contains the shortest set of RNAPII target genes, we observe a 3′ shift in Ser5-P RNAPII intensity in strains lacking Rtr1 activity (Fig. 1A, compare the 1st quartiles in WT and rtr1Δ cells). A similar 3′ shift in the Ser5-P intensity can be seen for each subsequent quartile, with the 1st and 2nd quartile in rtr1Δ cells displaying the highest intensity of Ser5-P at the 3′ end of an average gene. In addition, we observed a higher intensity of Ser5-P downstream of the 3′ end in rtr1Δ cells than in WT cells. The increase occurs in each of the four quartiles. These results, in conjunction with our previous findings, show that Rtr1 maintains the distribution of RNAPII Ser5-P levels in WT cells. Ser5-P levels peak at the 5′ ends of RNAPII target genes in yeast, and this mark has been shown to play a critical role in early transcription events, namely, mRNA capping (58). Changes in the Ser5-P distribution throughout the genome could lead to changes in specific CTD-interacting proteins that act as readers of the CTD code. To illustrate that the changes we observe in Ser5-P distribution following deletion of RTR1 are not caused by global changes in total RNAPII occupancy across target genes, we have measured total RNAPII occupancy using Rpb3-FLAG ChIP-exonuclease (exo) (Fig. 1B). For ease of comparison, gene averaging was performed as for the Ser5-P RNAPII data set and is shown as quartiles of gene length (see Supplemental Methods in the supplemental material for more details). These data clearly show that the average total RNAPII occupancy across the genome either decreases (quartiles 1, 2, and 3) or stays relatively unchanged (quartile 4 in Fig. 1B). Overall, these data indicate that the increased levels of Ser5-P RNAPII are due to decreased Ser5-P CTD dephosphorylation in rtr1Δ cells and not due to increases in overall RNAPII occupancy at the 3′ ends of genes.
FIG 1.
Rtr1 affects the global distribution of Ser5-P RNAPII. (A) Average gene analysis from ChIP-chip experiments is shown in WT and rtr1Δ strains. Each heat map is divided into quartiles based on gene length, with the 1st quartile representing the shortest 25% of RNAPII target genes and the 4th quartile representing the longest 25% of RNAPII target genes. The color scale of Ser5-P RNAPII intensity is shown to the right. A schematic representation of an average gene is shown at the top. (B) Average gene analysis from median normalized Rpb3-FLAG ChIP-exo experiments is shown from WT and rtr1Δ strains. Each heat map is divided into quartiles based on gene length, with the 1st quartile representing the shortest 25% of RNAPII target genes and the 4th quartile representing the longest 25% of RNAPII target genes. The color scale of total RNAPII occupancy is shown to the right.
Deletion of RTR1 causes a 3′ prime shift in the distribution of Ser5-P, which was independent of gene length (Fig. 1). To determine the general classes of RNAPII Ser5-P distribution changes in RTR1 deletion cells, we performed unsupervised k-means clustering analysis of differential enrichment data [(rtr1Δ IP − WT IP)/input values] for annotated RNAPII target genes. We obtained five clusters from the k-means clustering, the majority of which show increased Ser5-P in the 3′ end consistent with our length analysis (Fig. 2, clusters 2, 3, and 4). In Fig. 2A, cluster 1 genes display an increase in Ser5-P RNAPII enrichment at the 3′ ends of genes in the absence of Rtr1. Although cluster 3 genes show a pattern similar to the pattern of cluster 1 genes, on average, they show a higher enrichment of Ser5-P levels in WT samples relative to data from rtr1Δ strains. The average differential Ser5-P RNAPII enrichment across cluster 4 genes is elevated in rtr1Δ cells than in WT cells throughout the transcribed region (Fig. 2B), whereas the genes in cluster 2 show Ser5-P in the rtr1Δ enrichment primarily at the 3′ end of the gene. Cluster 5 genes show a distinct differential enrichment pattern of Ser5-P RNAPII in rtr1Δ cells primarily at the 5′ ends of genes (Fig. 2C). We next performed GO term enrichment analysis and determined the top overrepresented terms (P < 0.01) within our clusters in order to better understand the classes of cellular functions that are represented by the individual gene clusters (see Table S1 in the supplemental material). Interestingly, cluster 4, in which Ser5-P RNAPII is enriched in rtr1Δ cells throughout the gene body, had GO term enrichment in genes related to ribosomal protein-coding genes: ribonucleoprotein complex biogenesis and assembly (P value of 1.46E0.36), rRNA metabolic process (P value of 1.31E−30), and rRNA processing (P value of 5.48E−30). In fact, additional analysis of cluster 4 genes found many highly expressed genes, including PMA1, ADH1, and the ribosomal protein-coding genes.
FIG 2.
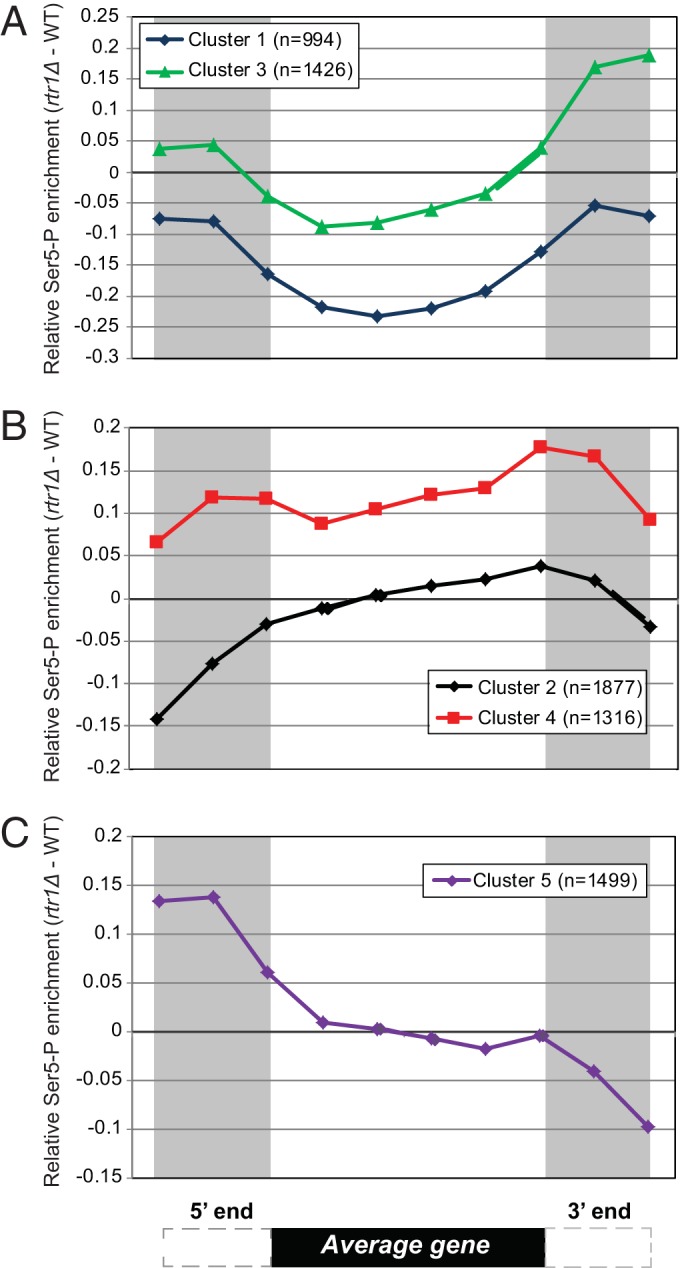
ChIP-chip data plotted as the relative enrichment of Ser5-P (Ser5-P in rtr1Δ − Ser5-P in WT cells). Binned Ser5-P RNAPII ChIP-chip data were subjected to k-means clustering, and the average differential enrichment value [rtr1Δ IP − WT IP)/input] for each cluster was plotted. A schematic representation of an average gene is shown at the bottom of the figure. (A) Data from clusters 1 and 3 showing low relative RNAPII Ser5-P occupancy in the coding region of target genes in rtr1Δ strains. (B) Relative Ser5-P RNAPII enrichment in clusters 2 and 4 shows increased Ser5-P levels across the gene (cluster 4) and/or in the 3′ end of the average gene (clusters 2 and 4). (C) Increased levels of Ser5-P RNAPII were observed in rtr1Δ strains at the 5′ ends of cluster 5 genes.
Analysis of the role of Rtr1 in the regulation of histone H3K36me3 levels in vivo.
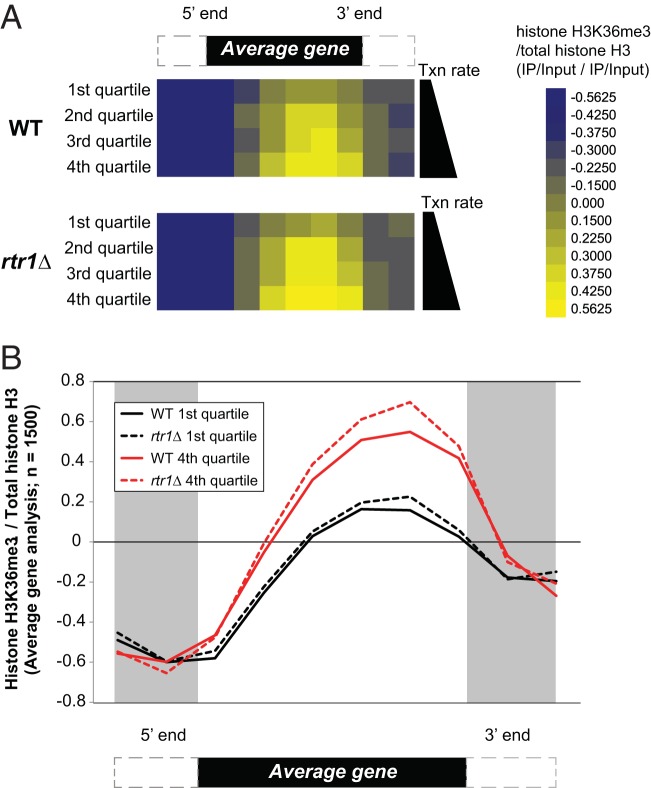
Once RNAPII escapes the promoter, the CTD tail becomes phosphorylated at Ser5 and Ser2 by the action of TFIIH and CTDK-I, respectively. The Ser2-P Ser5-P doubly modified RNAPII CTD has been shown to recruit the histone methyltransferase Set2 (45, 47). In yeast, Set2 catalyzes mono-, di-, and trimethylation of K36 distributed over the coding region of the gene with H3K36 trimethylation enrichment at the 3′ ends of protein-coding genes (59). Set2-facilitated H3K36me3 coordinates the recruitment of the Rpd3S deacetylase complex to the coding regions of genes. Rpd3S is responsible for the deacetylation of coding region histones after the passage of RNAPII to maintain hypoacetylated chromatin architecture, which prevents cryptic transcripts (60–62). Taken together, we hypothesize that changes in Ser5-P distribution caused by loss of Rtr1 function will impact H3K36me3 levels by increasing the number of Set2-CTD-interacting sites during transcription elongation. To test our hypothesis and to better understand the role of Rtr1 in regulating the CTD binding protein Set2, we performed ChIP-chip experiments on H3K36me3 in WT and rtr1Δ strains using high-density Agilent 1x244k (1 × 244,000) arrays with an average probe resolution of 50 nucleotides (Fig. 3). The levels of H3K36me3 throughout RNAPII target genes was normalized by the levels of total histone H3. Average gene enrichment analysis was performed for RNAPII targets and separated into quartiles for transcription rate, since the total levels of histone H3K36me3 have been shown to correlate with RNAPII transcription rate (59, 63). The first quartile contains the most rarely transcribed set of genes, and the fourth quartile contains the most frequently transcribed group of genes. In agreement with previous results, we observed increasing levels of H3K36me3 for both WT and rtr1Δ data sets from the first quartile through the fourth quartile, showing that the total levels of H3K36me3 correlate with transcription rate (Fig. 3A). In addition, we can see increased levels of histone H3K36me3 in rtr1Δ cells relative to WT cells in all four quartiles. Our analysis of total RNAPII levels from Fig. 1B suggests that total RNAPII transcription does not increase in rtr1Δ cells, although we do see dramatic increases in Ser5-P CTD levels. The relative levels of H3K36me3 (adjusted for total histone H3 levels) are shown together for the first and fourth quartile gene sets in Fig. 3B. These data show that the largest differences in H3K36me3 levels occur at the 3′ ends of the RNAPII target genes. Together, these data suggest that Ser5-P CTD dephosphorylation by rtr1Δ regulates H3K36me3 levels by decreasing the number of Ser5-P Ser2-P binding sites available to Set2 during transcription elongation.
FIG 3.
Deletion of RTR1 leads to a genome-wide increase of H3K36me3 at the 3′ ends of coding regions. (A) Average gene analysis from histone H3K36me3 ChIP-chip experiments is shown in WT and rtr1Δ strains following normalization by total histone H3 levels. The heat maps are divided into quartiles based on the transcription rate (Txn rate), with the 1st quartile representing the 25% of yeast genes expressed at low levels and the 4th quartile representing the most highly expressed 25% of yeast genes. The color scale is shown to the right. A schematic representation of an average gene is shown at the top. (B) Average gene analysis from histone H3K36me3 ChIP-chip experiments is shown in WT and rtr1Δ strains following normalization by total histone H3 levels from the 1st quartile and 4th quartile. A schematic of the average gene is shown below the graph.
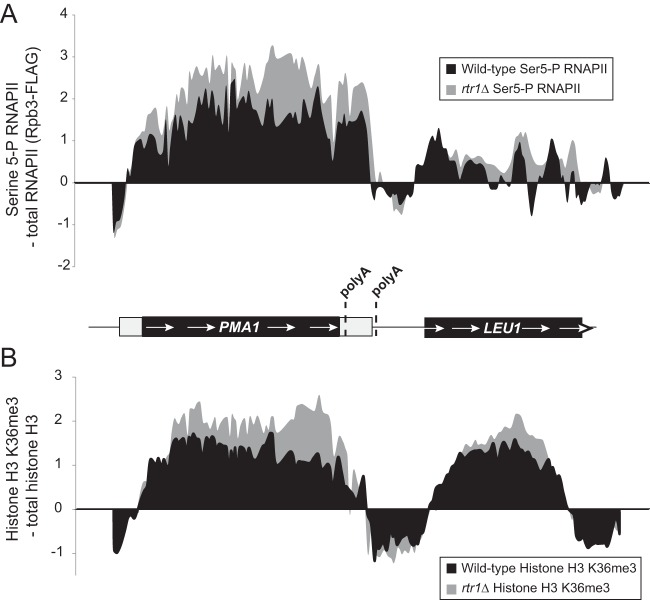
To clearly illustrate the changes we observed in both Ser5-P RNAPII and histone H3K36me3 levels, we have shown the patterns of Ser5-P RNAPII and histone H3K36me3 across a small region of chromosome 7 in Fig. 4. Deletion of RTR1 leads to large increases in total RNAPII Ser5-P (normalized for total RNAPII occupancy) across the coding region of PMA1 (Fig. 4A, left). At the downstream gene LEU1, increased levels of Ser5-P RNAPII are also observed at the 3′ end, although the changes are smaller than those observed at PMA1 (Fig. 4A, right). The pattern of H3K36me3 in WT Ser5-P RNAPII across the PMA1 gene is what would be expected for a typical protein-coding gene, where the H3K36me3 signal peaks in the coding region of the gene and begins to decrease toward the 3′ end just prior to the annotated polyadenylation sites (Fig. 4B). H3K36me3 levels (normalized for total histone H3 occupancy) increase in rtr1Δ cells relative to WT cells at PMA1 (Fig. 4B, left). The largest differences in H3K36me3 levels at PMA1 are observed near the 3′ end similar to the largest differences observed in Ser5-P RNAPII levels (compare the top and bottom representations in Fig. 5). At the downstream gene LEU1, histone H3K36me3 levels are also increased in rtr1Δ cells at the 3′ end of the gene in the same region in which increased levels of Ser5-P RNAPII were also observed (compare the right portions of the top and bottom representations in Fig. 5). We propose that altered histone H3K36me3 enrichment at the 3′ ends of genes occurs as a consequence of RTR1 deletion through increased Set2 recruitment to RNAPII during transcription elongation as a consequence of elevated Ser5-P CTD modification (Fig. 5).
FIG 4.
Changes in Ser5-P RNAPII and histone H3K36me3 levels at PMA1 and LEU1. (A) The relative enrichment of Ser5-P RNAPII occupancy relative to total RNAPII levels (according to Rpb3-FLAG) is shown across a small region of chromosome 7 in WT and rtr1Δ cells. A schematic representation of the PMA1 gene and the downstream LEU1 gene is shown below the graph. The schematic representations of the genes are drawn to scale, and the previously annotated poly(A) sites are indicated by dashed lines. (B) The relative enrichment of H3K6me3 relative to total histone H3 levels is shown across a small region of chromosome 7 in WT and rtr1Δ cells. See the schematic representation of the PMA1 gene and the downstream LEU1 gene for panel A.
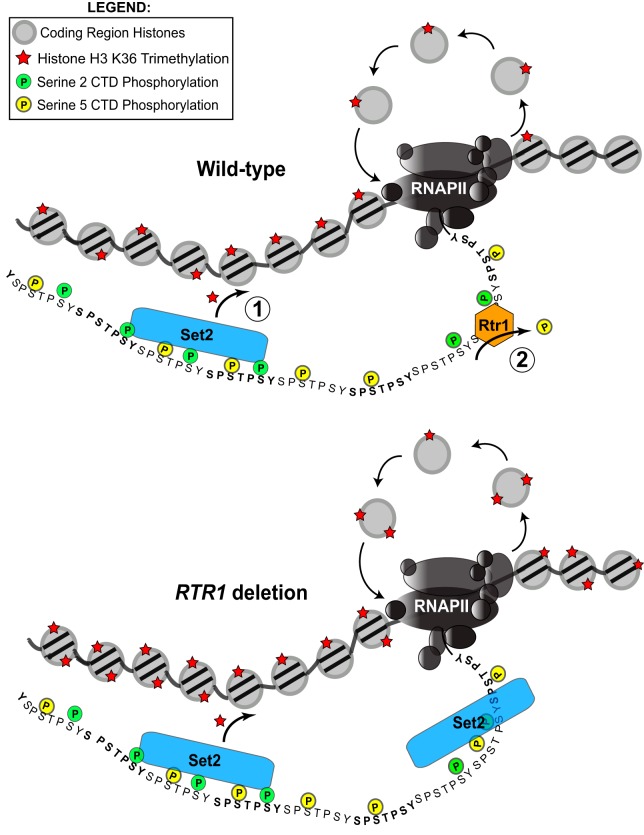
FIG 5.
A model for the role of Rtr1 in regulating cotranscriptional histone modifications. (Top) Ser5-P is at its highest near the 5′ end of the gene. As polymerase travels through the gene body, Ser2-P begins to increase. These modifications recruit Set2 to methylate coding region histones after the passage of RNAPII. Rtr1 dephosphorylates Ser5-P, essentially “turning off” H3K36me3 prior to the transcription termination site. (Bottom) In an rtr1Δ strain, Set2 retains its affinity for the CTD, and H3K36me3 enrichment is elevated in the coding region and the 3′ end of the gene where the histone mark continues into the transcription termination site at specific RNAPII target genes.
DISCUSSION
Multiple reports have shown that deletion of RTR1 in yeast results in elevated Ser5-P at the 3′ ends of a selection of model genes (14, 39, 52). Given that Rtr2 is an orthologue of Rtr1, it was reasonable to assume that Rtr2 may also have phosphatase activity. We tested this hypothesis by ChIP-quantitative PCR (qPCR) on the PMA1 gene and found that RTR2 deletion does not cause significant changes in Ser5-P RNAPII levels (see Fig. S3 in the supplemental material). These data suggest that Rtr2 is not a primary regulator of Ser5-P removal in vivo. We expanded our previous work (39) by addressing the role of Rtr1 on global levels of RNAPII phosphorylation in vivo. We performed ChIP-chip experiments to measure the changes in the Ser5-P levels in RTR1 deletion cells. Using this approach, we clearly demonstrate that Rtr1 is a global regulator of Ser5-P CTD modification, and Rtr1 appears to regulate Ser5-P levels regardless of gene length and independent of total RNAPII occupancy (Fig. 1). These findings are significant because they suggest that even at short genes, Rtr1 works to dephosphorylate RNAPII during the early phases of transcription elongation. Together, these findings indicate that Rtr1 is a Ser5-P CTD phosphatase that regulates RNAPII modification status during transcription elongation.
Using unsupervised k-means clustering, we grouped RNAPII target genes into five clusters based on the pattern of Ser5-P occupancy changes in rtr1Δ cells versus WT cells. We found that most genes, regardless of the cluster, have increased Ser5-P enrichment at the 3′ end (Fig. 2). Moreover, cluster four, in particular, displayed enriched Ser5-P throughout RNAPII target genes. Upon closer inspection of this cluster, we find a high percentage of the genes to be highly expressed yeast genes, including the prototypical yeast model genes, PMA1 and ADH1. We speculate that since highly expressed yeast genes will by definition have higher occupancy of RNAPII, this leads to a larger change in overall Ser5-P levels in the absence of Rtr1 phosphatase activity. Our finding that Rtr1 regulates global Ser5-P RNAPII levels at the 3′ ends of genes bears some similarity to data obtained from global analysis of Ser7-P RNAPII levels in an Ssu72 temperature-sensitive degron strain (Ssu72-td strain) (64). These data are intriguing, since Ssu72 has also been implicated in the removal of Ser5-P following cis isomerization of Pro6 in the RNAPII CTD regulated by Ess1 (65). However, Bataille et al. and Zhang et al. did not observe significant changes in global Ser5-P levels in the Ssu72-td strain at RNAPII target genes by ChIP-chip (18, 64). Recent work by Rosado-Lugo and Hampsey suggests that specific inactivating mutations in Ssu72, such as R129A and C15S, may block Rtr1 recruitment to the CTD during transcription elongation, whereas the loss of all Ssu72 protein in Ssu72-td strains allows Rtr1 to be recruited normally, resulting in near wild-type levels of Ser5-P RNAPII in the Ssu72-td strain (52). Our findings suggest that Rtr1 is a major regulator of Ser5-P RNAPII levels in vivo and that the wild-type Ssu72 in RTR1 deletion strains is not able to compensate for Rtr1. These data suggest that Rtr1 and Ssu72 may target different populations of RNAPII for Ser5-P dephosphorylation in vivo.
By examining the effects of RTR1 deletion on cotranscriptional histone modifications, we were able to test the concept of a CTD code. If precise timing of kinase-mediated CTD phosphorylation and phosphatase-mediated CTD dephosphorylation is needed to maintain CTD-interacting partner recruitment, loss of a CTD phosphatase should result in altered localization or amount of interacting partner function. In Fig. 4 and 5, it is clearly shown that loss of the early CTD phosphatase Rtr1 results in a 3′ spread of histone H3K36me3 by Set2, a well-characterized Ser2-P Ser5-P CTD-interacting protein and the sole H3K36me3 methyltransferase in yeast (66, 67). The changes in H3K36me3 are not caused by changes in overall histone occupancy (Fig. 4 and 5). These data show that Rtr1 has the greatest effect on H3K36me3 at the 3′ end of the gene, and these results directly correlate with the changes we observed in Ser5-P (Fig. 4). Since we observed global increases in H3K36me3 levels rather than spread of the 3′ modification in rtr1Δ cells, we propose that the methyltransferase Set2 is not mislocalized on the gene but likely remains in contact with the CTD for a longer period of time, thus increasing its modification output (Fig. 5). These data suggest that at some genes, Set2 may stay associated with RNAPII further downstream as a result of RTR1 deletion. This could occur as a result of Set2 mislocalization and/or termination defects in RNAPII, resulting in increased occupancy downstream.
On the basis of our observations and previous research from our group and others, we propose a model in which Rtr1 acts upstream of Set2 to regulate the levels of the SRI sites along the RNAPII CTD (Fig. 5). We have previously shown that Rtr1 recruitment to RNAPII occurs in a manner that is dependent upon the Ser2-P kinase Ctk1 (49). In fact, the increased levels of histone H3K36me3 in rtr1Δ cells is additional indirect evidence that could suggest that Rtr1 targets Ser2-P Ser5-P modified RNAPII for dephosphorylation in agreement with studies on RPAP2, the human homologue of Rtr1 (40). We propose the following model: as polymerase enters productive elongation, Ser2-P begins to increase, which leads to the recruitment of Set2 to RNAPII. Rtr1 is recruited to the multiply phosphorylated CTD and starts to remove Ser5-P, which in turn will lower the affinity of Set2 for the CTD, thereby leading to less H3K36me3 by the time RNAPII reaches the polyadenylation site at the 3′ end of the transcribed region (Fig. 5, top). Conversely, in an rtr1Δ strain, Set2 retains its high affinity for the CTD due to a lack of Ser5-P dephosphorylation, thus increasing the output of H3K36me3 toward the 3′ end of the gene, and perhaps in some instances, allowing H3K36me3 to continue past its normal 3′ boundary (Fig. 5, bottom). Maintaining H3K36me3 levels is important for the regulation of the Set2/Rpd3S pathway in yeast. This pathway is vital for the maintenance of chromatin integrity and regulation of cotranscriptional processes. H3K36me3 suppresses histone exchange across the coding region (68), allowing for continuous rounds of transcription and safeguarding transcriptional memory with respect to the transcription status. Future work is needed to determine the effects of RTR1 deletion on these downstream pathways.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Mosley lab for critical reading of the manuscript and technical contributions during the course of the project. We also thank Swaminathan Venkatesh and Jerry Workman for helpful discussions. We thank Michael Washburn and Laurence Florens for their support during the early stages of this project. Finally, we thank Yunlong Liu and Hongyu Gao for average gene analysis of total RNAPII occupancy.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00870-15.
REFERENCES
- 1.Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rino J, Carmo-Fonseca M. 2009. The spliceosome: a self-organized macromolecular machine in the nucleus? Trends Cell Biol 19:375–384. doi: 10.1016/j.tcb.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Shatkin AJ, Manley JL. 2000. The ends of the affair: capping and polyadenylation. Nat Struct Biol 7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 4.Barron-Casella E, Corden JL. 1992. Conservation of the mammalian RNA polymerase II largest-subunit C-terminal domain. J Mol Evol 35:405–410. [DOI] [PubMed] [Google Scholar]
- 5.Nonet M, Sweetser D, Young RA. 1987. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- 6.West ML, Corden JL. 1995. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140:1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zehring WA, Lee JM, Weeks JR, Jokerst RS, Greenleaf AL. 1988. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc Natl Acad Sci U S A 85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartolomei MS, Halden NF, Cullen CR, Corden JL. 1988. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol 8:330–339. doi: 10.1128/MCB.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meininghaus M, Chapman RD, Horndasch M, Eick D. 2000. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J Biol Chem 275:24375–24382. [DOI] [PubMed] [Google Scholar]
- 10.Yang C, Stiller JW. 2014. Evolutionary diversity and taxon-specific modifications of the RNA polymerase II C-terminal domain. Proc Natl Acad Sci U S A 111:5920–5925. doi: 10.1073/pnas.1323616111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman RD, Heidemann M, Hintermair C, Eick D. 2008. Molecular evolution of the RNA polymerase II CTD. Trends Genet 24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Heidemann M, Hintermair C, Voss K, Eick D. 2013. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta 1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 14.Kim M, Suh H, Cho EJ, Buratowski S. 2009. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J Biol Chem 284:26421–26426. doi: 10.1074/jbc.M109.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heidemann M, Eick D. 2012. Tyrosine-1 and threonine-4 phosphorylation marks complete the RNA polymerase II CTD phospho-code. RNA Biol 9:1144–1146. doi: 10.4161/rna.21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris DP, Phatnani HP, Greenleaf AL. 1999. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3′-end formation. J Biol Chem 274:31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 17.Zhang M, Wang XJ, Chen X, Bowman ME, Luo Y, Noel JP, Ellington AD, Etzkorn FA, Zhang Y. 2012. Structural and kinetic analysis of prolyl-isomerization/phosphorylation cross-talk in the CTD code. ACS Chem Biol 7:1462–1470. doi: 10.1021/cb3000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. 2012. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 45:158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, Eick D, Ansari AZ. 2010. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 17:1154–1161. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P. 2012. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336:1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 21.Lu H, Flores O, Weinmann R, Reinberg D. 1991. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci U S A 88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komarnitsky P, Cho EJ, Buratowski S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell 34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. 2010. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn SH, Kim M, Buratowski S. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13:67–76. doi: 10.1016/S1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 26.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9:1101–1111. doi: 10.1016/S1097-2765(02)00518-X. [DOI] [PubMed] [Google Scholar]
- 27.Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL. 2004. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem 279:24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu B, Eick D, Bensaude O. 2013. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res 41:1591–1603. doi: 10.1093/nar/gks1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. 2005. A structural perspective of CTD function. Genes Dev 19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 30.Chambers RS, Dahmus ME. 1994. Purification and characterization of a phosphatase from HeLa cells which dephosphorylates the C-terminal domain of RNA polymerase II. J Biol Chem 269:26243–26248. [PubMed] [Google Scholar]
- 31.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. 2004. Ssu72 is an RNA polymerase II CTD phosphatase. Mol Cell 14:387–394. doi: 10.1016/S1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 32.He X, Khan AU, Cheng H, Pappas DL Jr, Hampsey M, Moore CL. 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev 17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ansari A, Hampsey M. 2005. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev 19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem 278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 35.Steinmetz EJ, Brow DA. 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol Cell Biol 23:6339–6349. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loya TJ, O'Rourke TW, Reines D. 2012. A genetic screen for terminator function in yeast identifies a role for a new functional domain in termination factor Nab3. Nucleic Acids Res 40:7476–7491. doi: 10.1093/nar/gks377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. 2012. Gene loops enhance transcriptional directionality. Science 338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh BN, Hampsey M. 2007. A transcription-independent role for TFIIB in gene looping. Mol Cell 27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 39.Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP. 2009. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell 34:168–178. doi: 10.1016/j.molcel.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni Z, Xu C, Guo X, Hunter GO, Kuznetsova OV, Tempel W, Marcon E, Zhong G, Guo H, Kuo WH, Li J, Young P, Olsen JB, Wan C, Loppnau P, El Bakkouri M, Senisterra GA, He H, Huang H, Sidhu SS, Emili A, Murphy S, Mosley AL, Arrowsmith CH, Min J, Greenblatt JF. 2014. RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat Struct Mol Biol 21:686–695. doi: 10.1038/nsmb.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu PL, Yang F, Smith-Kinnaman W, Yang W, Song JE, Mosley AL, Varani G. 2014. Rtr1 is a dual specificity phosphatase that dephosphorylates Tyr1 and Ser5 on the RNA polymerase II CTD. J Mol Biol 426:2970–2981. doi: 10.1016/j.jmb.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irani S, Yogesha SD, Mayfield J, Zhang M, Zhang Y, Matthews WL, Nie G, Prescott NA, Zhang YJ. 2016. Structure of Saccharomyces cerevisiae Rtr1 reveals an active site for an atypical phosphatase. Sci Signal 9:ra24. doi: 10.1126/scisignal.aad4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. 2007. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Howe L, Anderson S, Yates JR III, Workman JL. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 46.Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. 2005. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci U S A 102:17636–17641. doi: 10.1073/pnas.0506350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuchs SM, Kizer KO, Braberg H, Krogan NJ, Strahl BD. 2012. RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di- and trimethylation at lysine 36. J Biol Chem 287:3249–3256. doi: 10.1074/jbc.M111.273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith-Kinnaman WR, Berna MJ, Hunter GO, True JD, Hsu P, Cabello GI, Fox MJ, Varani G, Mosley AL. 2014. The interactome of the atypical phosphatase Rtr1 in Saccharomyces cerevisiae. Mol Biosyst 10:1730–1741. doi: 10.1039/C4MB00109E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, Moazed D, Gygi SP. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 51.Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S. 2012. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell 45:111–122. doi: 10.1016/j.molcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosado-Lugo JD, Hampsey M. 2014. The Ssu72 phosphatase mediates the RNA polymerase II initiation-elongation transition. J Biol Chem 289:33916–33926. doi: 10.1074/jbc.M114.608695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fox MJ, Gao H, Smith-Kinnaman WR, Liu Y, Mosley AL. 2015. The exosome component Rrp6 is required for RNA polymerase II termination at specific targets of the Nrd1-Nab3 pathway. PLoS Genet 11:e1004999. doi: 10.1371/journal.pgen.1004999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agilent Technologies, Inc. 2007. Agilent yeast ChIP-on-chip analysis protocol, version 9.2, May 2007. Agilent Technologies, Inc., Santa Clara, CA. [Google Scholar]
- 55.Soukas A, Cohen P, Socci ND, Friedman JM. 2000. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev 14:963–980. [PMC free article] [PubMed] [Google Scholar]
- 56.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. 2006. TM4 microarray software suite. Methods Enzymol 411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 57.Beissbarth T, Speed TP. 2004. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol 20:104–112. doi: 10.1128/MCB.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 60.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 61.Lickwar CR, Rao B, Shabalin AA, Nobel AB, Strahl BD, Lieb JD. 2009. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One 4:e4886. doi: 10.1371/journal.pone.0004886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. 2010. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet 6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rando OJ. 2007. Global patterns of histone modifications. Curr Opin Genet Dev 17:94–99. doi: 10.1016/j.gde.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 64.Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ. 2012. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem 287:8541–8551. doi: 10.1074/jbc.M111.335687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. 2011. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem 286:5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hampsey M, Reinberg D. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429–432. doi: 10.1016/S0092-8674(03)00360-X. [DOI] [PubMed] [Google Scholar]
- 67.Shilatifard A. 2004. Transcriptional elongation control by RNA polymerase II: a new frontier. Biochim Biophys Acta 1677:79–86. doi: 10.1016/j.bbaexp.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 68.Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. 2012. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.