Abstract
Almost all genetic risk factors for autism spectrum disorders (ASDs) can be found in the general population, but the effects of that risk are unclear in people not ascertained for neuropsychiatric symptoms. Using several large ASD consortia and population based resources (total n>38,000), we find genomewide genetic links between ASDs and typical variation in social behavior and adaptive functioning. This finding is evidenced through both LD score correlation and de novo variant analysis, indicating that multiple types of genetic risk for ASDs influence a continuum of behavioral and developmental traits, the severe tail of which can result in an ASD or other neuropsychiatric disorder diagnosis. A continuum model should inform the design and interpretation of studies of neuropsychiatric disease biology.
Autism spectrum disorders (ASDs) are a group of neuropsychiatric conditions defined through deficits in social communication, as well as restricted and repetitive interests. Consistent with traditional approaches to psychiatric phenotypes, most genetic studies of ASDs compare cases to controls in order to identify risk-associated variation. This approach has been highly productive—recent studies have linked common polygenic as well as de novo and inherited rare variation to ASD risk 1, 2. Common genotyped SNPs are estimated to account for at least 20% of ASD liability 1, 3, 4. Contributing de novo variants are found in 10–20% of cases but de novo mutations collectively explain less than 5% of overall ASD liability 1, 5, 6.
Almost all genetic risk factors for ASDs can be found in unaffected individuals. For example, most people who carry a 16p11.2 deletion, the most common large mutational risk factor for ASDs, do not meet criteria for an ASD diagnosis 7. Across healthy populations there is also significant variability in capacity for social interaction and social communication 8. While such phenotypic variation is well established, the genetic relationship between neuropsychiatric disorders and typical social and behavioral variation remains unclear. From the first published descriptions of ASDs, clinical and epidemiologic reports have commonly noted subthreshold traits of autism in the family members of many diagnosed individuals 9, 10. Twin and family studies have suggested that those similarities are at least in part inherited and also suggest that traits and the diagnosis are correlated genetically 11–13, but the correlation has yet to be estimated using measured genetic data.
We examined the association between genetic risk for ASDs and social and behavioral variation in the general population, and examined the model through which genetic risk for ASDs is conferred. Traditional categorical psychiatric diagnoses (e.g., yes/no ASD) ignore the possibility of intermediate outcomes, long known to be relevant to phenotypes like intellectual disability and IQ that are more easily quantified. Several studies have now associated neuropsychiatric disease associated CNVs with cognitive or educational differences in the general population 14, 15. De novo deletions at 16p11.2 were recently reported to confer a quantitative effect on intelligence (an average two standard deviation reduction from mean IQ of the parents), rather than creating risk for categorical yes/no intellectual disability 16. The extent to which such patterns extend to social and behavioral traits is unknown and could substantially influence: 1) the design and interpretation of biological studies of ASDs and other severe mental illnesses, as well as 2) the designation of therapeutic treatment thresholds. We aimed to resolve this question using multiple categories of genetic variation that create risk for ASDs—common, inherited alleles as well as rare, de novo variants.
Like nearly all common diseases, common variant risk for ASDs is distributed across the genome, with many thousands of contributing loci of small effect 1, 3. The cumulative contribution of common SNPs to ASD risk (SNP heritability17) has been estimated using several methods, most recently and efficiently via LD score regression. LD score regression makes use of GWAS summary statistics to estimate SNP heritability 4. The method can also be used to estimate the correlation between common variant influences on two phenotypes (genetic correlation) 18. As LD score correlation requires only GWAS summary statistics, genetic correlations can be estimated between distinct data sets and cohorts.
We used three data sets to examine the common variant association between ASDs and social and communication difficulties in the general population (Table S1). First, traits of social and communication impairment were measured using the Social and Communication Disorders Checklist (SCDC) in the Avon Longitudinal Study of Parents and Children (ALSPAC), a general population cohort born in 1991–1992 in Bristol, United Kingdom 19, 20. The SCDC is a parent-rated quantitative measure of social communication impairment that is continuously distributed and has been extensively studied 21–23. There is substantial trait overlap between the SCDC and canonical ASD symptomology (e.g. “Not aware of other people’s feelings”) and children with ASDs have very high average scores (indicating many difficulties) on the SCDC 21. The measure does not include items on restricted and repetitive interests. For the purposes of this project, we used summary statistics from a published GWAS of the SCDC, administered when the children were 8 years old (n=5,628) 23. The SNP heritability of the SCDC was 0.17 (s.e.=0.09) using LD score regression, similar to the estimate derived from residual maximum likelihood using the software package GCTA (h2g=0.24, s.e.=0.07; n=5,204) 23.
We correlated the genetic influences on the SCDC with those on diagnosed ASDs using ASD data from two large consortium efforts. The Psychiatric Genomics Consortium autism group (PGC-ASD) has completed a GWAS of 5,305 ASD cases and 5,305 pseudocontrols constructed from untransmitted parental chromosomes (see Online Methods). Summary statistics from that GWAS are publicly available through the PGC website (URLs: PGC). As a replication set, we recently completed an independent ASD case-control GWAS with 7,783 ASD cases and 11,359 controls from the Danish iPSYCH project (iPSYCH-ASD; Online Methods). Using LD score regression, we estimated that the liability-scale SNP heritability of PGC-ASD was 0.23 (s.e.=0.03; assumed population prevalence 1%), suggesting that approximately one quarter of ASD liability reflects common genotyped variation. The estimated liability-scale SNP heritability of iPSYCH-ASDs was 0.14 (se=0.03; assumed population prevalence 1%). The genetic correlation between PGC-ASD and iPSYCH-ASD was 0.74(se=0.07; p<1e-20), indicating similar common, polygenic architecture between ASDs diagnosed primarily in the United States and Denmark.
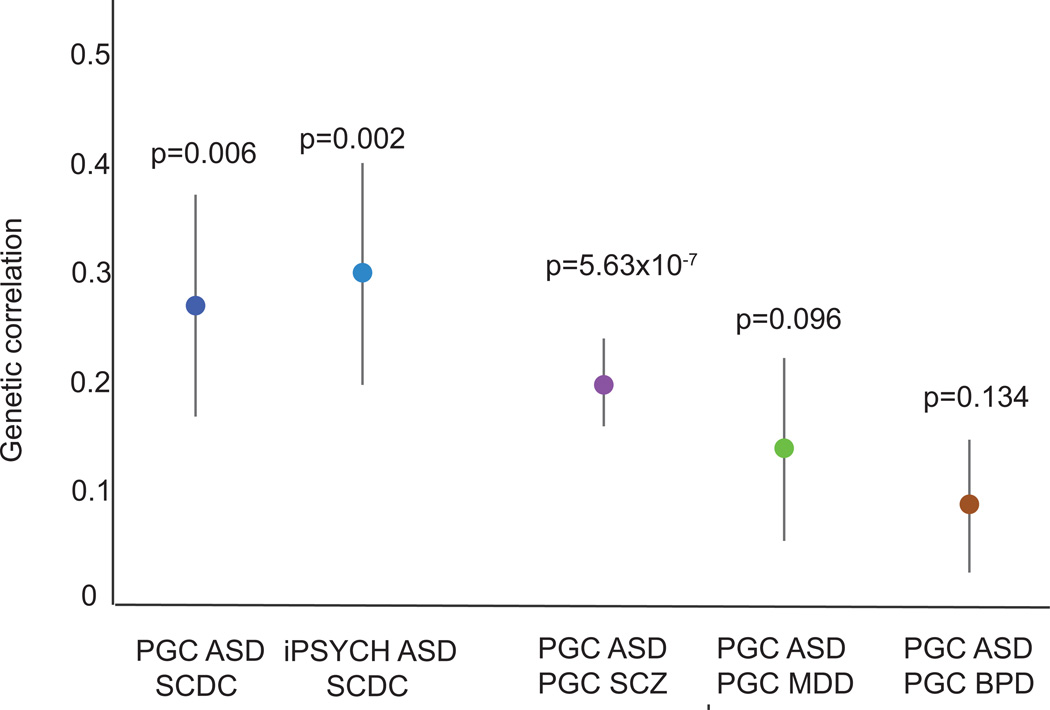
The estimated genetic correlations between the SCDC in ALSPAC and the two ASD GWAS data sets are shown in Figure 1. The estimated genetic correlation between PGC-ASD and the SCDC was 0.27 (s.e.= 0.13, p=0.006), suggesting that approximately one quarter of the genetic influences on ASDs share that influence with the SCDC. The estimated genetic correlation between iPSYCH-ASD and the SCDC was similar (rg=0.30; s.e.=0.10; p=0.002), evidencing substantial and replicable etiologic overlap between ASDs and typical variation in social and communication ability in childhood. The estimated genetic correlations between ASDs and the SCDC meet or exceed those previously estimated between PGC ASD and each of PGC schizophrenia, PGC bipolar disorder, and PGC major depressive disorder as shown in Figure 1. This suggests that ASDs are at least as strongly associated with social and communication trait variation in the population as they are several other categorically diagnosed psychiatric disorders. The observed genetic correlation between ASDs and the SCDC is similar to that estimated between type 2 diabetes and obesity, as well as other phenotypes that are strongly associated epidemiologically (Bulik-Sullivan and Finucane, 2015). Many behavioral and cognitive features are captured in an ASD diagnosis: social communication impairment, restricted and repetitive interests, functional impairments, and often co-occurring phenotypes that increase probability of diagnosis (e.g. intellectual disability, hyperactivity). The SCDC captures only traits of social and communication impairment, and the genetic association between ASDs and ASD traits may be stronger if more dimensions of the ASD phenotype were included in the traits measure. Supplemental analyses suggest that the estimated ASD-SCDC correlations are not driven by a negative association between ASDs and IQ (Table S2).
Figure 1.
The genetic correlation between ASDs and pediatric social and communication difficulties in the general population
Note: Genetic correlations are shown +/− one standard error; p-values indicate probability with which true genetic correlation is zero; PGC= Psychiatric Genomics Consortium; ASD= Autism Spectrum Disorders; iPSYCH= iPSYCH-SSI-Broad Autism project; SCZ=schizophrenia; MDD=major depressive disorder; BPD= bipolar disorder. Genetic correlations were estimated using constrained intercept LD score correlation. The correlations between PGC-ASD and each of PGC-SCZ, PGC-MDD, and PGC-BPD are modified from Bulik-Sullivan and Finucane et al. (2015).
The continuum of behavioral and developmental outcomes associated with de novo variation
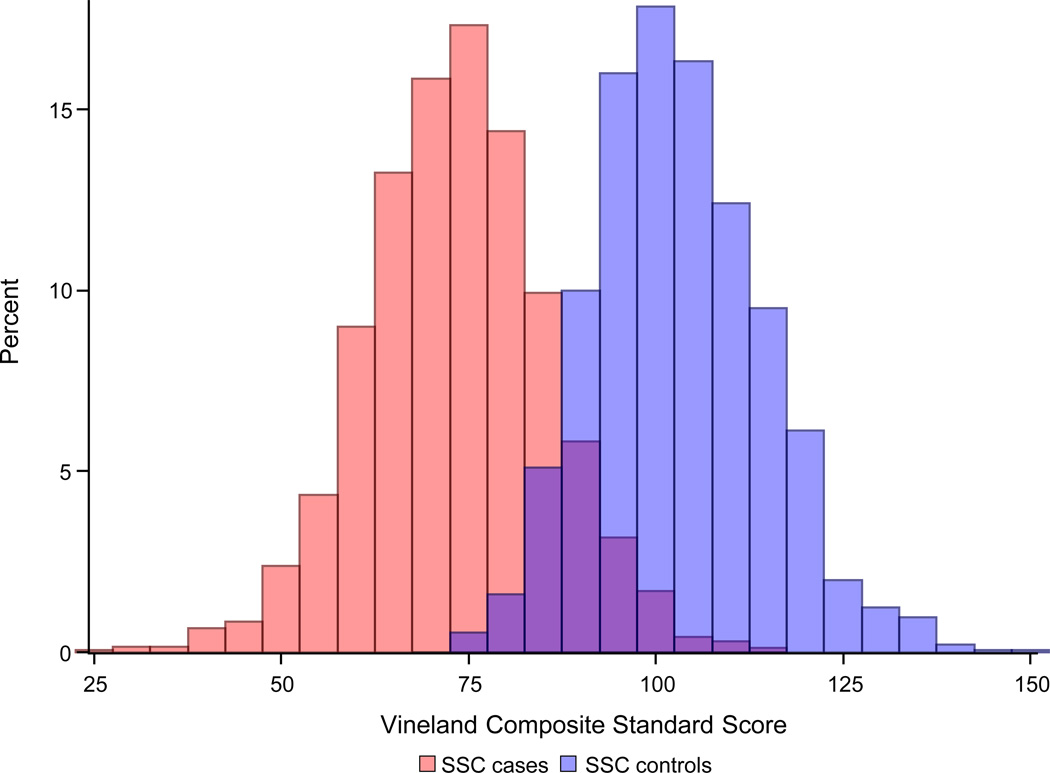
We next aimed to examine whether de novo variant data similarly gives evidence of a genetic relationship between ASDs and a continuum of behavioral outcomes in the population. The Simons Simplex Collection (SSC) is a sample of over 2,800 individuals with ASDs and their nuclear family members, with extensive data on unaffected siblings 24. To our knowledge, the SSC unaffected siblings are currently the only deeply phenotyped control sample with de novo variant information available. The Vineland Scales of Adaptive Behavior (Vineland) was used in the sequenced SSC cases (n=2,497) and sibling controls (n=1,861). The Vineland captures parent-rated variation in social, communication, and daily living skills, and normalizes those abilities to a mean of 100 and standard deviation of 15 in the population, corrected for age 25. On average, individuals with ASDs have mean Vineland scores approximately two standard deviations below the mean (SSC case mean = 73.3; s.d. = 12.17), consistent with the social and communication impairments definitional to the diagnosis. The distribution of the Vineland overlaps between cases and controls, as shown in Figure 2a.
Figure 2.
a. The distribution of Vineland scores overlaps between SSC cases and controls
Note: SSC= Simons Simplex Collection. The Vineland Composite Standard Score is normed, across ages, at mean 100, standard deviation (SD) 15 in the general population. The SSC case mean (73.3, SD=12.2) is significantly lower than the SSC control mean (103.0, SD=11.3, p<1e-20).
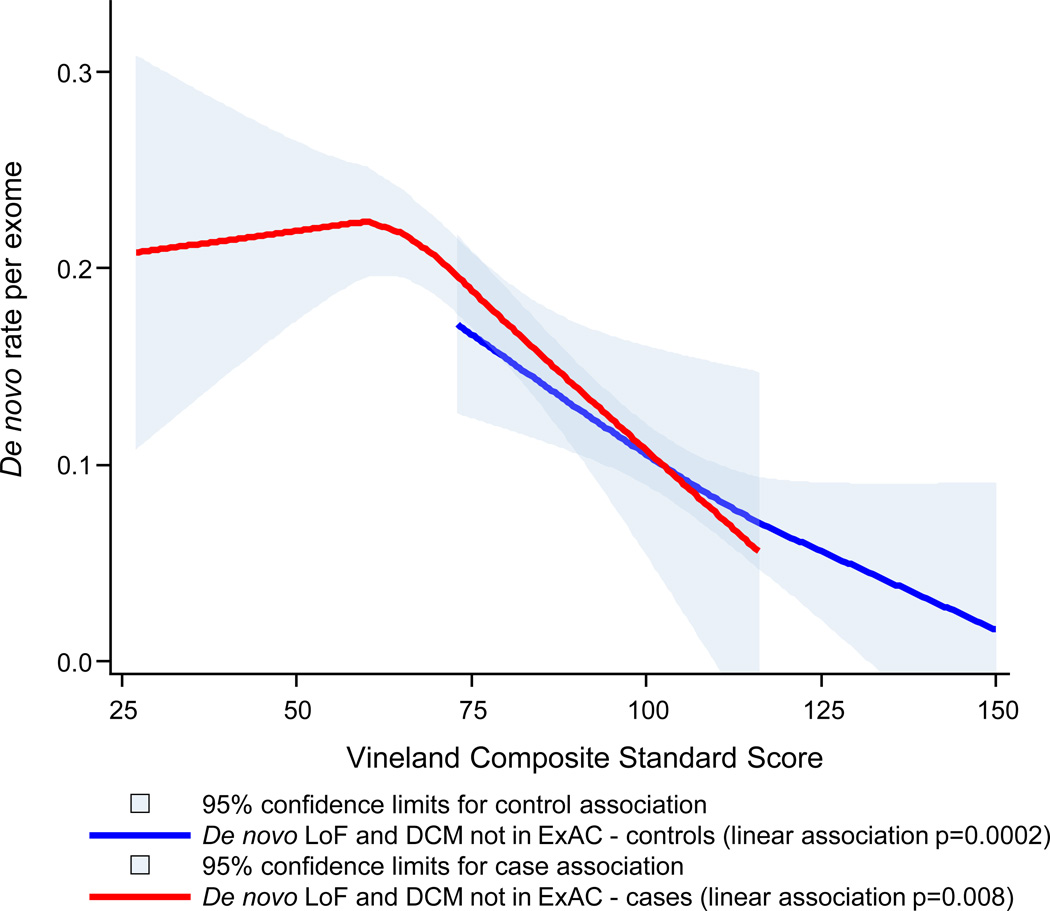
b. De novo variation influences a continuum of functional outcomes in ASD cases and controls
Note: Natural (loess) association shown for both cases and controls; p-values derived from poisson regression; LoF= Loss of Function; DCM = probably damaging constrained missense; ExAC= Exome Aggregation Consortium.
We examined the association between Vineland scores and de novo variant burden in individuals with and without ASDs. Our previous work demonstrated genotype to phenotype relationships using variant classes that are strongly associated with ASD risk 26; subsequently these analyses focused on: 1) de novo loss of function (LoF) variants and 2) de novo missense variants predicted to be damaging by the variant annotation program PolyPhen2 and occurring in a gene known to be intolerant of heterozygous missense variation (DCM) 27, 28. We found that variants in one of these two categories can be found in 22.1% of SSC cases and 13.2% of SSC unaffected siblings (LoF+DCM RR=1.68; p=1.8e-11; online methods). To enhance signal, we further filtered this list to remove de novo variants that were also seen in adult individuals in the Exome Aggregation Consortium (ExAC) resource (URLs: ExAC) 29. The ExAC database includes 60,706 exomes. Variants absent from that reference panel, which is a proxy for standing variation in the human population, are more likely to be deleterious. For example, 18.3% of the de novo LoF and DCM variants in the SSC are found in ExAC and, once those are removed, the relative de novo LoF+DCM burden in cases increases (LoF+DCM not in ExAC RR=1.91; p=7.6e-15).
The natural association between a) de novo LoF+DCM variants not seen in ExAC and b) Vineland score in SSC cases and controls is shown in Figure 2b. The LoF+DCM rate is predicted linearly by functional impairment in both cases (p=0.008) and controls (p=0.0002) using poisson regression, controlling for sex. Cases and controls with equivalent quantitative levels of functional impairment, a key component of all psychiatric diagnoses, are highly similar with regard to de novo variant burden, suggesting that the current categorical clinical threshold is largely arbitrary with regard to the social and communication impairments captured by the Vineland. The strength of the association between LoF+DCM burden and case status (at p=7.6e-15 without controlling for Vineland scores), is only nominally significant after Vineland score is controlled for (p=0.05). The associations are weaker but similar without the ExAC filter (Figure S1).
These data strongly suggest that genetic influences on ASD risk – both inherited and de novo— influence typical variation in the population in social and communication ability. They also link clinically significant problems to impairments that are less likely to be ascertained. The results have significant implications for genetic models of neuropsychiatric disorder risk. It is likely that inherited liability to ASDs is reflected in the behavioral traits of some affected individuals’ family members. This links genetic and phenotypic burden in an intuitively consistent fashion with complex, and continuously distributed, polygenic disease risk. For traits such as height, it is simple to conceptualize a model in which tall parents, (e.g. two standard deviations above the mean) are more likely to have a child that is very tall (e.g. three standard deviations above the mean). Historically this has been more complicated in neuropsychiatric disorders. Despite extensive evidence, some have even questioned the role of inheritance given that the parents of individuals with ASDs or schizophrenia rarely carry a diagnosis themselves. These results suggest that familiality should be studied in a manner beyond a count of categorically affected family members, and that trait variation in controls can provide insight into the underlying etiology of severe neurodevelopmental and psychiatric disorders. The behavioral influence of de novo and inherited genetic risk for ASDs can be quantified, and studies assessing continuous trait variation are likely better equipped to examine the phenotypic correlates of neuropsychiatric disease risk.
Methods
The datasets used in these analyses are summarized in Supplementary Table 1.
LD Score genetic correlation analyses
Genetic correlation quantifies the extent to which two phenotypes share genetic etiology. A genetic correlation of one suggests all influences are shared; a genetic correlation of zero suggests the phenotypes are genetically independent. SNP-based genetic correlations have traditionally been estimated using data sets with individual-level genotype and phenotype data. A new method, LD Score correlation, allows one to estimate genetic relationships using SNP data when the contributing data sets are siloed 18. Requiring only GWAS summary statistics, the method uses linkage disequilibrium (LD) patterns to estimate genetic correlations. The resulting correlations are highly similar to those derived from residual maximum likelihood (e.g., as implemented in GCTA or BOLT-REML 17, 30) and Haseman-Elston regression (URLs: Haseman-Elston). LD Score regression is implemented in the free and open source software package LDSC (URLs: LDSC).
We estimated the genetic association between diagnosed ASDs and traits of social and communication impairment in the general population using LD score correlation. Traits of social and communication impairment were measured using the Social and Communication Disorders Checklist (SCDC) in the Avon Longitudinal Study of Parents and Children (ALSPAC). ALSPAC is a population-based, longitudinal cohort study that initially recruited 14,541 pregnancies in Bristol, United Kingdom with expected dates of delivery between April 1, 1991 and December 31, 1992. All women in the study area with expected delivery dates in that time frame were recruited for participation 19, 20. Ethical approval was obtained from the ALSPAC Law-and-Ethics Committee (IRB00003312) and the Local Research Ethics Committees, and written informed consent was provided by all parents. The study website contains details of all available data (URLS: ALSPAC). The GWAS of the SCDC in ALSPAC, from which we obtained the SCDC summary statistics, has already been published 23. Exome sequencing data from trios is not available in ALSPAC.
The SCDC is a 12-item parent-rated scale that counts traits of social and communication impairment quantitatively. Each question has a 0, 1, or 2 response option; the subsequent range of total scores is 0–24. Individuals with diagnosed ASDs, on average, have very high scores on the SCDC, consistent with the disorders’ definitional social and communication impairment 21. As described in the SCDC GWAS, the SCDC was used at multiple time points in ALSPAC 23. The SCDC is stable over time 22 and individual scores over time are genetically correlated 23. To reduce multiple testing, we used the age 8 SCDC data as childhood autistic traits are well studied, and have been linked to diagnosed ASDs through twin studies 11, 12. We also focused on the age 8 data as it maximizes SCDC sample size (n= 5,628), and accordingly maximizes the power of the correlation tests.
The ASD case-control GWAS summary statistics come from the PGC Autism (PGC-ASD) group and the iPSYCH-SSI-Broad Autism group (iPSYCH-ASD). The PGC-ASD summary statistics were generated using publically available data from a meta-analysis of 5,305 ASD diagnosed cases of ASD and 5,305 pseudocontrols of European descent (URLS: PGC). The pseudocontrol methodology used by the PGC Autism group, and the PGC GWAS analytic pipeline, have been reported on extensively 3, 31. Pseudocontrols are built using the untransmitted alleles from each parent at each locus. The subsequent GWAS is immune to population stratification as the pseudocontrols are ancestrally matched to the cases. LD score correlations have already been estimated using these data, as described in Bulik-Sullivan and Finucane18. In Figure 1, we highlight a subset of the correlations from that manuscript, specifically those associating PGC-ASD with publically available summary statistics from PGC schizophrenia, PGC bipolar disorder, and PGC major depressive disorder (URLS: PGC).
The iPSYCH-ASD data are from a new population based case-control ASD sample derived from the Danish Neonatal Screening Biobank hosted by Statens Serum Institut, comprising dried bloodspots (Guthrie cards) from all individuals born in Denmark since 1981. The samples can be linked to the Danish register system, including the Danish Psychiatric Central Register. DNA extracted from the bloodspots can be successfully amplified and employed in GWAS 32, 33. The iPSYCH-ASD project aims to genotype all Danish individuals with an available bloodspot and an ASD diagnosis in their medical record (ICD codes F84.0, F84.1, F84.5, F84.8, and F84.9). This study has been approved by the Danish research ethical committee system. This analysis employs the iPSYCH-ASD data generated to date, specifically the first 10 genotyping waves of that collection which contain 7,783 ASD cases and 11,359 controls. All individuals in the sample were born between 1981 and 2005. Genotyping was performed at the Broad Institute. The data were cleaned and analyzed using the same analysis pipeline described in Ripke et al. 2014 and other previous PGC GWAS publications.
The GWAS summary statistics used for the secondary genetic correlation analyses are published and publicly available. Summary statistics from a GWAS of child full scale IQ have been made public by Benyamin et al. 2014, and were used in Bulik-Sullivan and Finucane et al. 18 to estimate a genetic association between PGC-ASD and IQ in the general population. More than forty percent of contributing individuals in the Benyamin GWAS were from the ALSPAC cohort (mean age 9 years). We used LD score correlation to similarly estimate a genetic association between iPSYCH-ASD and general population full scale IQ in childhood.
Each of the genetic correlation estimates were obtained using constrained intercept cross-trait LD Score regression, which yields much lower standard errors (by ~30%) than unconstrained intercept LD Score regression 18. Unlike unconstrained intercept LD Score regression, constrained intercept LD Score regression can give biased estimates of genetic correlation if the two studies have either (a) hidden sample overlap or (b) shared population stratification. We can rule out both of these concerns in the present study, because the PGC-ASD sample is family-based (case-pseudocontrol), and does not have case or control overlap with any other PGC analyses. The iPSYCH-ASD and ALSPAC samples do not contain any cases or pseudocontrols that were used in PGC-ASD. In addition, case-pseudocontrol analyses are immune to confounding from population stratification, so it is not possible for genetic correlation estimates to be biased by population stratification when at least one of the studies used a case-pseudocontrol design. As a robustness check, we verified that the point estimates of genetic correlation obtained with unconstrained intercept LD Score regression are similar to the constrained intercept results presented in Figure 1 (although the standard errors are higher, due to the lower statistical efficiency of unconstrained intercept LD Score regression). These results are presented in Supplementary Table 3.
De novo variant analyses
The Simons Simplex Collection (SSC) resource is unique in its combination of detailed phenotypic and genotypic characterizations. The SSC ascertained over 2800 individuals with ASDs and their nuclear family members, restricting recruitment to families in which no other cases of ASD have been diagnosed out to the level of first cousins. Families were also excluded in the event of intellectual disability in a sibling or a history of parental schizophrenia 24. To our knowledge, the SSC siblings currently constitute the most deeply phenotyped control sample with available de novo variant information. We used those data to examine the relationship between de novo variant burden and phenotypic variation in the SSC, in both siblings (n=1,861) and probands (n=2,497).
We limited the analysis to de novo variant classes that are strongly associated with ASD risk, specifically loss of function mutations (LoF) and probably damaging missense mutations found in constrained genes (DCM). LoF mutations include frameshift, splice site, and nonsense mutations. DCM mutations include variants that are a) predicted to be damaging by PolyPhen2 and b) occurring in a gene known to be intolerant of heterozygous missense variation 27, 28. In the SSC, LoF mutations are found in 8.9% of controls and 15.0% of cases; DCM mutations are found in 4.3% of controls and 7.1% of cases. Restricting the classes of de novo variants analyzed increases the probability of association to case status as well as to phenotypic variation within cases 26. Synonymous variants, for example, are associated with neither case status (p=0.23) nor phenotypic variation in IQ (p=0.87) or Vineland scores (p=0.91) in SSC cases. To increase signal, we further filtered observed de novo variants on their presence/absence in the publically available Exome Aggregation Consortium database (ExAC; URLS: ExAC). The ExAC database contains jointly called exomes from 60,706 adult individuals, recruited for an array of exome sequencing studies. These individual exomes form a reference panel of consistently processed human exonic variation, with particular utility towards the consideration of rare variants. Just as selection reduces the probability with which LoF variants will be seen in genes with low tolerance for functional disruption 27, reference genomes will be less likely to contain variants that create strong risk for reproductively deleterious phenotypes like ASDs. In other words, one expects that de novo variants not seen in 60,706 reference exomes will be, on average, more deleterious. We considered an SSC de novo variant recurrent if a variant with the same chromosome, position, reference allele, and alternate allele was seen in ExAC. After filtering out the variants seen in ExaC, the LoF+DCM rate was 19.0% in cases and 9.9% in controls.
We have previously associated de novo variant burden with variation in case IQ and other measures of case severity, including the Vineland Scales of Adaptive Behavior (Vineland), in the SSC 26. IQ was not measured in SSC siblings, however the Vineland was used in a manner consistent with its use in cases. The Vineland assesses overall adaptive functioning, with subscales assessing 1) social and 2) communication ability, as well as 3) daily living skills. The Vineland is commonly used as one measure of case severity in ASD, though its social and communication subscales are not designed to capture canonical autism-like symptomology. In this analysis, we used Poisson regression, in both cases and controls, to associate Vineland scores with de novo variant burden. Sex was controlled for in all de novo variant analyses.
Supplementary Material
Acknowledgments
We thank Steven Hyman, Thomas Lehner, and Nancy Kanwisher for comments on the manuscript. We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. Autism Speaks (7132) provided support for the analysis of autistic-trait related data in ALSPAC to BSTP. This work was also supported by the Medical Research Council Integrative Epidemiology Unit (MC_UU_12013/1-9). This publication is the work of the authors and Elise Robinson and Mark Daly will serve as guarantors for the contents of this paper. The ALSPAC GWAS data was generated by Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute and LabCorp (Laboratory Corportation of America) using support from 23 and Me. E.B.R. was funded by National Institutes of Mental Health Grant 1K01MH099286-01A1 and NARSAD Young Investigator grant 22379. We thank the families who took part in the Simons Simplex Collection study and the clinicians who collected data at each of the study sites. The iPSYCH project is funded by The Lundbeck Foundation and the universities and university hospitals of Aarhus and Copenhagen. Genotyping of iPSYCH and PGC samples was supported by grants from the Stanley Foundation, the Simons Foundation (SFARI 311789 to MJD), and NIMH (5U01MH094432-02 to MJD). The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found on the ExAC website (URLs: ExAC). This work was also supported by a grant from the Simons Foundation (SFARI #307705, S.J.S.).
Collaborators
iPSYCH-SSI-Broad Collaborators
Thomas D. Als1–3, Marie Baekvad-Hansen4, Richard Belliveau5, Ditte Demontis1–3, Ashley Dumont5, Jacqueline Goldstein5–7, Jonas Grauholm4, Christine S. Hansen4, Thomas F. Hansen1, 8, Daniel Howrigan5–7, Francesco Lescai1–3, Manuel Mattheisen1–3, Jennifer Moran5, Ole Mors1, 9, Merete Nordentoft1, 10, Bent Norgaard-Pedersen4, Timothy Poterba5–7, Jesper Poulsen4, Christine Stevens5, and Raymond Walters5–7
The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH, Denmark
Centre for Integrative Sequencing, iSEQ, Aarhus University, Aarhus, Denmark
Department of Biomedicine - Human Genetics, Aarhus University, Aarhus, Denmark
Center for Neonatal Screening, Department for Congenital Disorders, Statens Serum Institut, Copenhagen, Denmark
Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge MA 02142, USA
Analytic and Translational Genetics Unit, Department of Medicine, Massachusetts General Hospital and Harvard Medical School, Boston MA 02114, USA
Medical and Population Genetics Program, Broad Institute of MIT and Harvard, Cambridge MA 02142, USA
Institute of Biological Psychiatry, MHC Sct. Hans, Mental Health Services Copenhagen, Roskilde, Denmark
Research Department P, Aarhus University Hospital, Risskov, Denmark
Mental Health Services in the Capital Region of Denmark, Mental Health Center Copenhagen, University of Copenhagen, Copenhagen, Denmark
Footnotes
URLs
LDSC: http://www.github.com/bulik/ldsc
Haseman-Elston: http://www.biorxiv.org/content/early/2015/04/20/018283).
ALSPAC: http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary
Authorship contributions
EBR, BSTP, VA, JK, BBS JG, KES, SS, DME, SR, MVH, TW, DMH, PBM, and ADB generated data and/or conducted analyses. EBR, BSTP, BBS, BMN, JM, DS, and MJD designed the experiment and tools. PBM, ADB, AR, GDS, and MJD supervised the research. EBR, BSTP and MJD wrote the paper.
COI
We have no conflicts of interest to report.
Contributor Information
Elise B. Robinson, Email: erobinson@atgu.mgh.harvard.edu.
Mark J. Daly, Email: mjdaly@atgu.mgh.harvard.edu.
References
- 1.Gaugler T, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krumm N, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee SH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson E, et al. The cognitive and behavioral phenotype of the 16p11.2 deletion in a clinically ascertained population. Biol Psychiatry. 2015;77:785–793. doi: 10.1016/j.biopsych.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009;10:872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 9.Kanner L. Autistic disturbances of affective contact. Nervous Child. 1943;2:217–250. [PubMed] [Google Scholar]
- 10.Constantino JN, Zhang Y, Frazier T, Abbacchi AM, Law P. Sibling recurrence and the genetic epidemiology of autism. Am J Psychiatry. 2010;167:1349–1356. doi: 10.1176/appi.ajp.2010.09101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson EB, et al. Evidence that autistic traits show the same etiology in the general population and at the quantitative extremes (5%, 2.5%, and 1%) Arch Gen Psychiatry. 2011;68:1113–1121. doi: 10.1001/archgenpsychiatry.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundstrom S, et al. Autism spectrum disorders and autisticlike traits: similar etiology in the extreme end and the normal variation. Arch Gen Psychiatry. 2012;69:46–52. doi: 10.1001/archgenpsychiatry.2011.144. [DOI] [PubMed] [Google Scholar]
- 13.Ronald A, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45:691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 14.Stefansson H, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 15.Mannik K, et al. Copy number variations and cognitive phenotypes in unselected populations. JAMA. 2015;313:2044–2054. doi: 10.1001/jama.2015.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno-De-Luca A, et al. The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatry. 2015;72:119–126. doi: 10.1001/jamapsychiatry.2014.2147. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyd A, et al. Cohort Profile: The 'Children of the 90s'--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2012 doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser A, et al. Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skuse DH, Mandy WP, Scourfield J. Measuring autistic traits: heritability, reliability and validity of the Social and Communication Disorders Checklist. Br J Psychiatry. 2005;187:568–572. doi: 10.1192/bjp.187.6.568. [DOI] [PubMed] [Google Scholar]
- 22.Robinson EB, et al. Stability of autistic traits in the general population: further evidence for a continuum of impairment. J Am Acad Child Adolesc Psychiatry. 2011;50:376–384. doi: 10.1016/j.jaac.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.St Pourcain B, et al. Variability in the common genetic architecture of social-communication spectrum phenotypes during childhood and adolescence. Mol Autism. 2014;5:18. doi: 10.1186/2040-2392-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Sparrow SS, Cicchetti DV, Balla DA. Vineland Adaptive Behavior Scales. Pearson; 2005. [DOI] [PubMed] [Google Scholar]
- 26.Robinson EB, et al. Autism spectrum disorder severity reflects the average contribution of de novo and familial influences. Proc Natl Acad Sci U S A. 2014;111:15161–15165. doi: 10.1073/pnas.1409204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ExAC. EAC. Cambridge, MA: [Google Scholar]
- 30.Loh PR, et al. Contrasting regional architectures of schizophrenia and other complex diseases using fast variance components analysis. 2015 doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borglum AD, et al. Genome-wide study of association and interaction with maternal cytomegalovirus infection suggests new schizophrenia loci. Mol Psychiatry. 2014;19:325–333. doi: 10.1038/mp.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollegaard MV, et al. Robustness of genome-wide scanning using archived dried blood spot samples as a DNA source. BMC Genet. 2011;12:58. doi: 10.1186/1471-2156-12-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Online Methods References
- 1.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loh PR, et al. Contrasting regional architectures of schizophrenia and other complex diseases using fast variance components analysis. 2015 doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd A, et al. Cohort Profile: The 'Children of the 90s'--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2012 doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser A, et al. Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.St Pourcain B, et al. Variability in the common genetic architecture of social-communication spectrum phenotypes during childhood and adolescence. Mol Autism. 2014;5:18. doi: 10.1186/2040-2392-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skuse DH, Mandy WP, Scourfield J. Measuring autistic traits: heritability, reliability and validity of the Social and Communication Disorders Checklist. Br J Psychiatry. 2005;187:568–572. doi: 10.1192/bjp.187.6.568. [DOI] [PubMed] [Google Scholar]
- 8.Robinson EB, et al. Stability of autistic traits in the general population: further evidence for a continuum of impairment. J Am Acad Child Adolesc Psychiatry. 2011;50:376–384. doi: 10.1016/j.jaac.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson EB, et al. Evidence that autistic traits show the same etiology in the general population and at the quantitative extremes (5%, 2.5%, and 1%) Arch Gen Psychiatry. 2011;68:1113–1121. doi: 10.1001/archgenpsychiatry.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lundstrom S, et al. Autism spectrum disorders and autisticlike traits: similar etiology in the extreme end and the normal variation. Arch Gen Psychiatry. 2012;69:46–52. doi: 10.1001/archgenpsychiatry.2011.144. [DOI] [PubMed] [Google Scholar]
- 11.Lee SH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borglum AD, et al. Genome-wide study of association and interaction with maternal cytomegalovirus infection suggests new schizophrenia loci. Mol Psychiatry. 2014;19:325–333. doi: 10.1038/mp.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollegaard MV, et al. Robustness of genome-wide scanning using archived dried blood spot samples as a DNA source. BMC Genet. 2011;12:58. doi: 10.1186/1471-2156-12-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson EB, et al. Autism spectrum disorder severity reflects the average contribution of de novo and familial influences. Proc Natl Acad Sci U S A. 2014;111:15161–15165. doi: 10.1073/pnas.1409204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.