Abstract
Background
Nonsyndromic orofacial cleft is a common birth defect with a complex etiology, including multiple genetic and environmental risk factors. Recent whole genome analyses suggested associations between nonsyndromic orofacial cleft and up to 18 genetic risk loci (ABCA4, BMP4, CRISPLD2, GSTT1, FGF8, FGFR2, FOXE1, IRF6, MAFB, MSX1, MTHFR, MYH9, PDGFC, PVRL1, SUMO1, TGFA, TGFB3, and VAX1), each of which confers a different relative risk in different populations. We evaluate the nonsynonymous variants in these 18 genetic risk loci in nonsyndromic orofacial clefts and normal controls to clarify the specific variants in Taiwanese population.
Methods
We evaluated these 18 genetic risk loci in 103 cases of nonsyndromic orofacial clefts and 100 normal controls using a next-generation sequencing (NGS) customized panel and manipulated a whole-exon targeted-sequencing study based on the NGS system of an Ion Torrent Personal Genome Machine (IT-PGM). IT-PGM data processing, including alignment with the human genome build 19 reference genome (hg19), base calling, trimming of barcoded adapter sequences, and filtering of poor signal reads, was performed using the IT platform-specific pipeline software Torrent Suite, version 4.2, with the plug-in “variant caller” program. Further advanced annotation was facilitated by uploading the exported VCF file from Variant Caller to the commercial software package Ion Reporter; the free online annotation software Vanno and Mutation Taster. Benign or tolerated amino acid changes were excluded after analysis using sorting intolerant from tolerant and polymorphism phenotyping. Sanger sequencing was used to validate the significant variants identified by NGS. Furthermore, each variant was confirmed in asymptomatic controls using the Sequenom MassARRAY (San Diego, CA, USA).
Results
We identified totally 22 types of nonsynonymous variants specific in nonsyndromic orofacial clefts, including 19 single nucleotide variants, 2 deletions, and 1 duplication in 10 studied genes(ABCA4, MYH9, MTHFR, CRISPLD2, FGF8, PVRL1, FOXE1, VAX1, FGFR2, and IRF6). Nonsynonymous variants in MYH9 and ABCA4, which were detected in 6 and 5 individuals, respectively, were identified to be the most frequent risk loci in nonsyndromic orofacial clefts in the Taiwanese population.
Conclusions
Nonsynonymous variants in MYH9 and ABCA4 were identified to be the most frequent risk loci in nonsyndromic orofacial clefts in the Taiwanese population. These findings in our study have provided additional information regarding specific variants associated with nonsyndromic orofacial clefts in different population and demonstrate the power of our customized NGS panel, which is clinically useful for the simultaneous detection of multiple genes associated with nonsyndromic orofacial clefts.
Electronic supplementary material
The online version of this article (doi:10.1186/s12881-016-0322-2) contains supplementary material, which is available to authorized users.
Keywords: Nonsyndromic orofacial clefts, Next-generation sequencing, MYH9, ABCA4
Background
Nonsyndromic orofacial clefts, which include cleft lip with or without cleft palate (CL/P) and isolated cleft palate (CPI), are among the most common congenital malformations worldwide. The prevalence of this major birth defect widely ranges from 1/700 to 1/1000, with fluctuations attributed to different areas and ethnicities [1, 2]. Epidemiological data reveal that the prevalence of nonsyndromic CL/P is lowest among African populations (0.4/1000), intermediate among European populations (1/1000), and highest among Asian populations (2/1000) [3]. Nonsyndromic orofacial cleft is not only associated with increased infant morbidity and mortality but also has enormous effects on speech, hearing, appearance, and mental disability, thereby increasing long-term medical costs and placing substantive burdens on families and societies [4, 5].
Nonsyndromic orofacial cleft is an etiologically heterogeneous disease with multiple genetic and environmental risk factors [6]. Maternal smoking, alcohol consumption, and folate and vitamin deficiencies, particularly during the first trimester of pregnancy, have been suggested to increase the occurrence of nonsyndromic CL/P [7, 8]. Previous gene identification studies of nonsyndromic CL/P were generally based on genome-wide association studies (GWASs) [9–12], genome-wide linkage studies [13], and GWAS meta-analyses [14, 15]. These studies identified genetic risk loci associated with nonsyndromic CL/P on chromosomes 1p22, 1p36, 2p21, 3p11.1, 8q21.3, 8q24, 9q22, 10q25, 15q22, 17p13, 17q22, and 20q12.
Although GWAS studies have identified multiple nonsyndromic CL/P-associated genetic loci, further progress in the identification of casual variants has been limited because these approaches focus on common variants and neglect low-frequency variants [16]. To identify novel and low-frequency variants, next-generation sequencing (NGS)-based techniques facilitate the simultaneous detection of causal variants in large genomic regions. We selected 18 nonsyndromic orofacial cleft-related candidate genes, including ABCA4, BMP4, CRISPLD2, GSTT1, FGF8, FGFR2, FOXE1, IRF6, MAFB, MSX1, MTHFR, MYH9, PDGFC, PVRL1, SUMO1, TGFA, TGFB3, and VAX1, based on previous associated studies [7, 17] to conduct a customized NGS panel and subsequently manipulated a whole-exon targeted-sequencing study based on the NGS system of an Ion Torrent Personal Genome Machine (IT-PGM). A total of 103 patients with nonsyndromic orofacial clefts and 100 independent asymptomatic normal controls were enrolled to investigate potential variants associated with nonsyndromic orofacial clefts and identify specific nonsynonymous variants in Taiwanese population.
Methods
Case enrollment and ethics statement
The study population included 103 Taiwanese patients with isolated, nonsyndromic orofacial clefts recruited from the Linkou Chang Gung Memorial Hospital from 1995 to 2014; patients’ diagnoses had been confirmed via neonatal photographs or chart descriptions written by plastic surgeons or clinical geneticists. Individuals with other systemic abnormalities, developmental or mental delays, and confirmed chromosomal abnormalities were excluded from this study. For the control group, we recruited 100 asymptomatic Taiwanese volunteers who had no family history of orofacial clefts from among the employees of Linkou Chang Gung Memorial Hospital.
This study was approved by the institutional review board (IRB 101-4637A3) at Linkou Chang Gung memorial hospital, and written informed consent was obtained from all adult participants and the parents or guardians of pediatric participants.
Sample collection and genomic DNA preparation
Genomic DNA was extracted from 10-ml peripheral blood samples using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) as follows: 20 μL of QIAGEN Protease (or protease K) was mixed with 200 μL of buffy coat via a 15-s vortex step. The mixture was then incubated at 56 °C for 10 min after adding 200 μL of Buffer AL. Next, 200 μL of 96–100 % ethanol was added, followed by a 15-s vortex and transfer to a QIAamp Mini spin column. The silica membrane was washed via centrifugation with Buffers AW1 and AW2. Genomic DNA was eluted with Buffer AE, and the quantity and quality were determined using a Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA) and Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA).
Design of a customized NGS panel for nonsyndromic orofacial cleft
We analyzed 18 genetic loci associated with the risk of nonsyndromic orofacial cleft, including IRF6, VAX1, ABCA4, BMP4, FGFR2, FOXE1, MAFB, MSX1, MYH9, CRISPLD2, FGF8, GSTT1, MTHFR, PDGFC, PVRL1, SUMO1, TGFA, and TGFB3 (Table 1). We used the Ion AmpliSeq™ Designer v2.2.1 cloud-based software program, which was supplied free of charge by Life Technologies, to design our customized panel. Moreover, we used the National Center for Biotechnology Information (NCBI) ClinVar database to identify pathogenic variants in these 18 genes and set up a hotspot database.
Table 1.
List of 18 selected genes studied in patients with nonsyndromic orofacial clefts and normal controls
| Gene | Gene size (bp) |
Map location | Protein |
|---|---|---|---|
| ABCA4 | 128,313 | 1p21-p22.1 | ATP-binding cassette, sub-family A, member 4 |
| BMP4 | 9026 | 6p12 | Bone morphogenetic protein 4 |
| CRISPLD2 | 100,788 | 16q24.1 | Cysteine-rich secretory protein LCCL domain containing 2 |
| GSTT1 | 8548 | 22q11.23 | Glutathione S-transferase theta 1 |
| FGF8 | 10,240 | 10q24.32 | Fibroblast growth factor 8 |
| FGFR2 | 120,129 | 10q26.13 | Fibroblast growth factor receptor 2 |
| FOXE1 | 3462 | 9q22 | Forkhead box E1 |
| IRF6 | 20,553 | 1q32.2 | Interferon regulatory factor 6 |
| MAFB | 3393 | 20q12 | V-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| MSX1 | 4272 | 4p16.2 | Msh homeobox 1 |
| MTHFR | 21,198 | 1p36.22 | Methylenetetrahydrofolate reductase |
| MYH9 | 106,741 | 22q12.3 | Myosin, heavy chain 9 |
| PDGFC | 210,941 | 4q32 | Platelet derived growth factor C |
| PVRL1 | 105,675 | 11q23. | Poliovirus receptor-related 1 |
| SUMO1 | 32,429 | 17p13.1 | Small ubiquitin-like modifier 1 |
| TGFA | 106,914 | 6p21.3 | Transforming growth factor, alpha |
| TGFB3 | 24,893 | 14q24.3 | Transforming growth factor, beta 3 |
| VAX1 | 9781 | 10q25.3 | Ventral anterior homeobox 1 |
IT-PGM AmpliSeq library preparation and IT-PGM sequencing
AmpliSeq multiplexed libraries were constructed using the Ion AmpliSeq Library Kit 2.0 according to the manufacturer’s protocol (Life Technologies, Part #4475345), with some modifications. The preparation was started with 30 ng of genomic DNA in a volume of ≤6 μL; the 501 amplicons were amplified by PCR and divided into two primer pools. We increased the annealing and extension steps of PCR program from 4 to 8 min to improve the efficiency of longer amplicons. Primer sequences were partially digested with FuPa reagent, and barcoded adapters were ligated with DNA ligase. Following purification and size selection using AMPure beads (Beckman Coulter, Brea, CA, USA), the prepared library was quantified using a Qubit 2.0 Fluorometer (Life Technologies) and Bioanalyzer high-sensitivity DNA chip (Agilent Technologies Inc., Santa Clara, CA, USA). Quantified libraries were pooled and diluted further to generate a 10-pmol/L working stock. To clonally amplify library DNA onto IonSpheres (ISPs), we used emulsion PCR, emulsion breaking, and template enrichment using the Ion OneTouch™ 200 system and Template Kit v2.0 (Life Technologies) according to the manufacturer’s protocols. Enriched ISPs were prepared for sequencing using the Ion PGM 200 Sequencing Kit v2.0 and loaded on an Ion 316 chip v2 or Ion 318 chip v2, depending on whether 7 or 14 samples were to be sequenced, respectively. To sequence an authentic variant, an ideal average coverage for each amplicon of 500× and variant frequency of at least 5 % in the wild-type background were used in this study.
Bioinformatics analysis
IT-PGM data processing, including alignment with the human genome build 19 reference genome (hg19), base calling, trimming of barcoded adapter sequences, and filtering of poor signal reads, was performed using the IT platform-specific pipeline software Torrent Suite, version 4.2, with the plug-in “variant caller” program (Life Technologies). Further advanced annotation was facilitated by uploading the exported VCF file from Variant Caller to the commercial software package Ion Reporter (Life Technologies); the free online annotation software Vanno [18] and MutationTaster. Benign or tolerated amino acid changes were excluded after analysis using sorting intolerant from tolerant (SIFT) and polymorphism phenotyping (PolyPhen). In addition, we used the Integrative Genomics Viewer to visualize the status of each read alignment and the presence of variants from the reference genome to clarify possible strand biases or sequencing errors.
Experimental validation
Validation by alternative sequencing methods was required for NGS-identified variants that passed the in-house filtering steps. Sanger sequencing was used to validate the significant variants identified by NGS. Furthermore, each variant was confirmed in asymptomatic controls using the Sequenom MassARRAY (San Diego, CA, USA).
Results
Cases
The clinical features of the 103 patients with nonsyndromic orofacial clefts and 100 normal controls are listed in Table 2.
Table 2.
Clinical characteristics of patients with nonsyndromic orofacial clefts and normal controls
| Characteristics | Nonsyndromic orofacial clefts | Normal controls | |
|---|---|---|---|
| Number (%) | Number (%) | ||
| Gender | Male | 48 (46.6 %) | 45 (45.0 %) |
| Female | 55 (53.4 %) | 55 (55.0 %) | |
| Age | Range | 1–41 | 21–58 |
| Cleft type | Cleft lip only | 12 (11.6 %) | absent |
| Cleft palate only | 32 (31.0 %) | absent | |
| Cleft lip and palate | 56 (54.4 %) | absent | |
| Unclassified | 3 (3.0 %) | absent | |
| Cleft site | Unilateral | 53 (51.4 %) | absent |
| Bilateral | 46 (44.7 %) | absent | |
| Unclassified | 4 (3.9 %) | absent | |
Customized NGS panel for nonsyndromic orofacial cleft
Detailed information about this customized panel is listed in Additional file 1: Table S1. This panel comprises 501 amplicons divided into two primer pools: 254 amplicons in primer pool 1 and 247 amplicons in primer pool 2. The amplicon sizes are 125–275 bp. Details regarding the numbers of exons and amplicons in the 18 selected genes are listed in Additional file 1: Table S2. The average target region coverage rate was 94.09 %.
Performance of the customized NGS panel
Our quality control standard for defining a true variant is a gene locus coverage depth >50×. In other words, the average coverage depth of each amplicon should receive more than 50× reads to reduce the risk of misjudgment in subsequent PGM sequencing. Figure 1 shows the average coverage depths of the 501 amplicons in the 203 evaluated samples (103 nonsyndromic orofacial cleft and 100 normal control samples); 95 % of amplicons had a gene locus coverage depth >50 × .
Fig. 1.
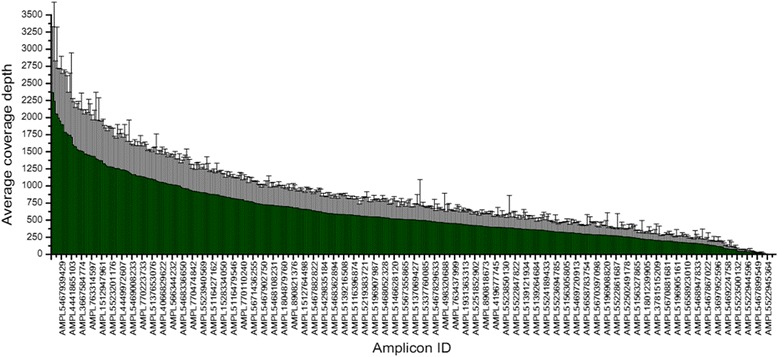
Nonsyndromic orofacial clefts customized next-generation sequencing panel performance with an average coverage depth of 501 amplicons
All nonsynonymous variants in patients with nonsyndromic orofacial clefts
The distributions of all nonsynonymous variants of the 18 analyzed genes among patients with nonsyndromic orofacial clefts are shown in Additional file 1: Table S3. We identified 29 types of nonsynonymous variants, including 23 single nucleotide variants, 4 deletions, 1 duplication, and 1 insertion.
All nonsynonymous variants in normal controls
The distributions of all nonsynonymous variants of the 18 analyzed genes among normal controls are shown in Additional file 1: Table S4. We identified 15 types of nonsynonymous variants, including 12 single nucleotide variants, 2 deletions, and 1 insertion.
Specific nonsynonymous variants in patients with nonsyndromic orofacial cleft
A comparison of nonsynonymous variants between patients with nonsyndromic orofacial clefts and normal controls identified 22 types of specific variants within 10 studied genes in the former group (Table 3). Among these, 19 were single-nucleotide variants (in ABCA4, MYH9, MTHFR, CRISPLD2, FGF8, PVRL1, FOXE1, and FGFR2), 2 were deletions (in CRISPLD2 and IRF6), and 1 was a duplication (VAX1). The nonsynonymous variants in MYH9 and ABCA4, which affected 6 and 5 individuals with nonsynonymous variants, respectively, were the most frequent risk loci among this Taiwanese population. The distributions of nonsynonymous variants in MYH9 and ABCA4 are shown in Fig. 2.
Table 3.
Specific variants found to be associated with nonsyndromic orofacial cleft
| Gene | Coding | Amino acid change | PolyPhen | SIFT | Mutation Taster | Phenotype | Number of cases |
|---|---|---|---|---|---|---|---|
| MTHFR | c.1816C > T | p.R606C | Possibly damaging | Damaging | Disease causing | BL CLP | 1/103 |
| MTHFR | c.62G > A | p.S21N | Benign | Damaging | Disease causing | BL CP | 1/103 |
| MYH9 | c.5722G > A | p.D1908N | Possibly damaging | Damaging | Disease causing | L CLP | 1/103 |
| MYH9 | c.3676C > T | p.R1226W | Benign | Damaging | Disease causing | L CLP | 1/103 |
| MYH9 | c.3320G > A | p.R1107Q | Possibly damaging | Damaging | Disease causing | BL CP | 1/103 |
| MYH9 | c.3262G > A | p.A1088T | Possibly damaging | Damaging | Disease causing | R CL | 1/103 |
| MYH9 | c.2606C > T | p.T869M | Benign | Damaging | Polymorphism | BL CP | 1/103 |
| MYH9 | c.452A > G | p.Y151C | Possibly damaging | Damaging | Disease causing | L CLP | 1/103 |
| CRISPLD2 | c.119_121del | p.40_41del | — | — | Polymorphism | L CLP | 1/103 |
| CRISPLD2 | c.1337C > G | p.A446G | Possibly damaging | Damaging | Disease causing | L CLP | 1/103 |
| ABCA4 | c.6498C > G | p.I2166M | Possibly damaging | Tolerated | Disease causing | R CLP | 1/103 |
| ABCA4 | c.4610C > T | p.T1537M | Possibly damaging | Damaging | Disease causing | L CL; unknown CP | 2/103 |
| ABCA4 | c.4297G > A | p.V1433I | Possibly damaging | Tolerated | Polymorphism | BL CLP | 1/103 |
| ABCA4 | c.763C > T | p.R255C | Possibly damaging | Tolerated | Disease causing | L CL | 1/103 |
| FOXE1 | c.1090G > A | p.G364S | Possibly damaging | Tolerated | Disease causing | BL CLP | 2/103 |
| FGF8 | c.251C > T | p.P84L | Possibly damaging | Damaging | Disease causing | L CL | 1/103 |
| FGF8 | c.250C > T | p.P84S | Possibly damaging | Damaging | Disease causing | L CL | 1/103 |
| VAX1 | c.363dupT | p.C122fs | — | — | Disease causing | R CLP | 1/103 |
| PVRL1 | c.334 T > A | p.S112T | Benign | — | Disease causing | Unknown CLP | 1/103 |
| PVRL1 | c.52C > T | p.L18F | Benign | — | Polymorphism | R CLP; L CLP | 2/103 |
| FGFR2 | c.293C > T | p.T98M | Possibly damaging | Damaging | Disease causing | L CL | 1/103 |
| IRF6 | c.421_423del | p.141_141del | — | — | Disease causing | R CLP | 1/103 |
BL bilateral, CL cleft lip, CLP cleft lip with palate, CP cleft palate, L left, R right
Fig. 2.
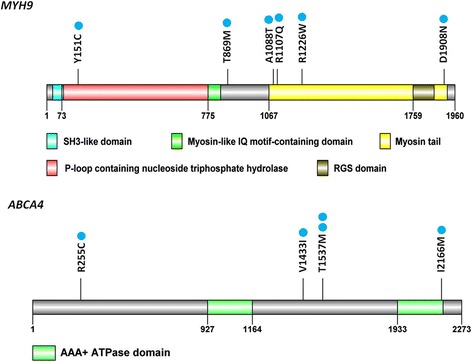
Distribution of nonsynonymous variants specific to orofacial clefts in MYH9 and ABCA4 (blue dots indicate the numbers of affected individuals)
It is worth noticing that in our population, no nonsynonymous variants were found in eight genes (BMP4, GSTT1, MAFB, MSX1, PDGFC, SUMO1, TGFA, and TGFB3). These eight genes appear to have a weaker association with nonsyndromic orofacial cleft in the Taiwanese population. As most hotspot variants in the NCBI database were previously collected from foreign populations, our findings may reflect specific variants associated with nonsyndromic orofacial cleft in the Taiwanese population.
Discussion
Orofacial cleft is among the most common human congenital malformations, affecting 135,000 neonates worldwide each year [2]. Orofacial clefts have been associated with both environmental and genetic risk factors and the prevalence of this major birth defect widely ranges among different areas and ethnicities.
In the last decade, major advances in the identification of the causative genetic mutations underlying syndromic forms of CL/P have revealed more than 60 syndromic cleft-associated genes [19]. By contrast, the genetic cause of nonsyndromic forms of CL/P remains mostly unclear. The lack of progress in our understanding of the genetic etiology of nonsyndromic CL/P is obviously associated with the lack of a recognizable mode of inheritance and reduced penetrance of these patients, as well as the low rate of positive family history among affected persons [20]. With the advances in the genomic era, the recent development of powerful and cost-effective genomic tools has opened new routes for phenotyping nonsyndromic orofacial clefts. Recent GWAS [9–12], genome-wide linkage studies [13], and GWAS meta-analyses [14, 15] have suggested that nonsyndromic orofacial clefts might be associated with up to 18 genetic loci, each with a different relative risk in different populations. These candidate loci include IRF6 (1q32.3-q41), VAX1 (10q26.1), ABCA4 (1p22.1-p21), BMP4 (14q22-q23), FGFR2 (10q26), FOXE1 (9q22), MAFB (20q11,2-q13.1), MSX1 (4p16.3-p16.1), MYH9 (22q13.1), CRISPLD2 (16q24.1), FGF8 (10q24), GSTT1 (22q11.23), MTHFR (1p36.3), PDGFC (4q32), PVRL1 (11q23.3), SUMO1 (2q33), TGFA (2p13), and TGFB3 (14q24) [19, 21–28]. These genetic risk loci carry a different relative risk in different populations. Our study is the first to analyze these 18 genetic loci associated with nonsyndromic orofacial cleft in the Taiwanese population.
As nonsyndromic orofacial cleft is a complex condition affected by multiple genes, genetic testing must be robust and cover a wide spectrum of potential mutations. Unlike traditional sequencing, which screens one gene at a time, exon by exon, NGS techniques allow massive parallel sequencing of as many genes as desired, thereby leveling the economic and technological barriers to detecting mutations on a genome-wide scale. Although this technology is suitable for the detection of any mutation within a rational target, targeted NGS is considered to be particularly useful for detecting mutations in disorders with a highly heterogeneous genetic background. With this understanding, we used a customized NGS panel to rapidly detect possible variants in these 18 statistically validated candidate genes among patients with nonsyndromic orofacial clefts.
In our study, we evaluated 18 genetic risk loci in 103 cases of nonsyndromic orofacial clefts and 100 normal controls from the Taiwanese population using customized NGS, which revealed 22 types of specific variants within 10 studied genes in individuals with nonsyndromic orofacial clefts. Among these, 19 were single nucleotide variants (in ABCA4, MYH9, MTHFR, CRISPLD2, FGF8, PVRL1, FOXE1, and FGFR2), 2 were deletions (in CRISPLD2 and IRF6 gene), and 1 was a duplication (VAX1). The nonsynonymous variants in MYH9 and ABCA4, which were detected in 6 and 5 individuals, respectively, were the most frequent risk loci in our Taiwanese population.
MYH9, or myosin heavy chain 9, has been shown to associate with nonsyndromic CL/P in several populations [29]. Abundant, specific expression of MYH9 was observed in the epithelial cells of palatal shelves prior to fusion. The expression level of MYH9 was shown to decrease and be restricted to epithelial triangles before disappearing upon the completion of fusion [30]. In our study, MYH9 is the most frequent risk loci in the Taiwanese population, providing further evidence for the involvement of MYH9 in the etiology of nonsyndromic CL/P.
A recent GWAS of several populations revealed markers in/near the gene encoding ABCA4, indicating a novel susceptibility locus for CL/P [9]. In Honduran and Colombian populations, ABCA4 is a candidate gene associated with nonsyndromic orofacial clefting [31]. In the Brazilian population, ABCA4 rs540426 associated strongly with CL/P, unilateral and right CL/P, and bilateral CL/P, whereas the SNP rs481931 exhibited borderline associations with CL/P and bilateral CL/P [32]. However, in a Chinese Han population, ABCA4 was not found to associated with nonsyndromic orofacial clefts [33]. In our study, several nonsynonymous variants in ABCA4 were specifically found in individuals with nonsyndromic orofacial clefts from a Taiwanese population.
Conclusions
Nonsynonymous variants in MYH9 and ABCA4 were identified to be the most frequent risk loci in nonsyndromic orofacial clefts in the Taiwanese population. Our findings provide us more information about specific variants associated with nonsyndromic orofacial clefts in different population, as well as demonstrate the power of our customized NGS panel, which is clinically useful for the simultaneous detection of multiple genes associated with nonsyndromic orofacial clefts. Furthermore, recent NGS studies have shown that fetal DNA from a few milliliters of maternal plasma is sufficient for fetal whole genome sequencing. Importantly, using parental genomes as guides, fetal genome sequences could be scanned for mutations prenatally and noninvasively [34, 35]. In the near future, it will be possible to predict whether a fetus will be affected by a nonsyndromic orofacial cleft based on a targeted NGS-based investigation of genetic risk loci.
Acknowledgements
This study was supported by the following grants: CMRPG3C1411 (PJ Cheng), CMRPG3C1412 (PJ Cheng), CMRPG3C1413 (PJ Cheng) from Linkou Chang Gung memorial hospital, Taiwan.
Authors’ contributions
HHP participated in the design of the study and drafted the manuscript. NCC participated in the NGS experiments and sequence alignment. KTC participated in the design of the study and case enrollment. JJL participated in the design of the study. PYC participated in the design of the study. SCC participated in the sequence alignment. YHWC participated in data analysis. YTC participated in data analysis. WP participated in case enrollment. PJC conceived of the study, participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The author(s) declare that they have no competing interests.
Additional file
Customized NGS panel information for nonsyndromic orofacial clefts. Table S2. Detailed panel information about the 18 selected genes studied in nonsyndromic orofacial clefts. Table S3. All nonsynonymous variants found in individuals with nonsyndromic orofacial clefts. Table S4. All nonsynonymous variants found in normal controls. (DOC 164 kb)
Contributor Information
Hsiu-Huei Peng, Email: hhpeng@cgmh.org.tw.
Nai-Chung Chang, Email: frank90237@cgmh.org.tw.
Kuo-Ting Chen, Email: philip@cgmh.org.tw.
Jang-Jih Lu, Email: jjlpcp@cgmh.org.tw.
Pi-Yueh Chang, Email: changpy@cgmh.org.tw.
Shih-Cheng Chang, Email: changsc137@cgmh.org.tw.
Yah-Huei Wu-Chou, Email: yhwc8876@cgmh.org.tw.
Yi-Ting Chou, Email: yp740212@cgmh.org.tw.
Wanni Phang, Email: wanni7233@gmail.com.
Po-Jen Cheng, Email: pjcheng@cgmh.org.tw.
References
- 1.Yuan Q, Blanton SH, Hecht JT. Genetic causes of nonsyndromic cleft lip with or without cleft palate. Adv Otorhinolaryngol. 2011;70:107–113. doi: 10.1159/000322486. [DOI] [PubMed] [Google Scholar]
- 2.Mossey PA, Modell B. Epidemiology of oral clefts 2012: an international perspective. Front Oral Biol. 2012;16:1–18. doi: 10.1159/000337464. [DOI] [PubMed] [Google Scholar]
- 3.Croen LA, Shaw GM, Wasserman CR, et al. Racial and ethnic variations in the prevalence of orofacial clefts in California, 1983–1992. Am J Med Genet. 1998;79:42–47. doi: 10.1002/(SICI)1096-8628(19980827)79:1<42::AID-AJMG11>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 4.Christensen K, Juel K, Herskind AM, et al. Long term follow up study of survival associated with cleft lip and palate at birth. BMJ. 2004;328:1405. doi: 10.1136/bmj.38106.559120.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai L, Zhu J, Mao M, et al. Time trends in oral clefts in Chinese newborns: data from the Chinese National Birth Defects Monitoring Network. Birth Defects Res A Clin Mol Teratol. 2010;88:41–47. doi: 10.1002/bdra.20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahimov F, Jugessur A, Murray JC. Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac J. 2012;49:73–91. doi: 10.1597/10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon MJ, Marazita ML, Beaty TH, et al. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bezerra JF, Oliveira GHM, Soares CD, et al. Genetic and non-genetic factors that increase the risk of non-syndromic cleft lip and/or palate development. Oral Dis. 2015;21:393–399. doi: 10.1111/odi.12292. [DOI] [PubMed] [Google Scholar]
- 9.Beaty TH, Murray JC, Marazita ML, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birnbaum S, Ludwig KU, Reutter H, et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 11.Mangold E, Ludwig KU, Birnbaum S, et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- 12.Grant SFA, Wang K, Zhang H, et al. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr. 2009;155:909–913. doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Marazita ML, Lidral AC, Murray JC, et al. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype-specific differences in linkage and association results. Hum Hered. 2009;68:151. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig KU, Mangold E, Herms S, et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet. 2012;44:968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y-Y, Qin C-Q, Deng M-H, et al. Association between BMP4 rs17563 Polymorphism and NSCL/P Risk: A Meta-Analysis. Dis Markers. 2015;2015:763090. doi: 10.1155/2015/763090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leslie Elizabeth J, Taub Margaret A, Liu H, et al. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS Loci. Am J Hum Genet. 2015;96:397–411. doi: 10.1016/j.ajhg.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brito LA, Meira JGC, Kobayashi GS, et al. Genetics and management of the patient with orofacial cleft. Plast Surg Int. 2012;2012:782821. doi: 10.1155/2012/782821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang PJ, Lee CC, Tan BCM, et al. Vanno: a visualization-aided variant annotation tool. Hum Mutat. 2015;36:167–174. doi: 10.1002/humu.22684. [DOI] [PubMed] [Google Scholar]
- 19.Kohli SS, Kohli VS. A comprehensive review of the genetic basis of cleft lip and palate. J Oral Maxillofac Pathol. 2012;16:64–72. doi: 10.4103/0973-029X.92976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray J. Cleft lip and palate. From origin to treatment. J Med Genet. 2004;41:80. doi: 10.1136/jmg.2003.013359. [DOI] [Google Scholar]
- 21.Alkuraya FS, Saadi I, Lund JJ, et al. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- 22.Marazita ML, Murray JC, Lidral AC, et al. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32-35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neiswanger K, Deleyiannis FW, Avila JR, et al. Candidate genes for oral-facial clefts in Guatemalan families. Ann Plast Surg. 2006;56:518–521. doi: 10.1097/01.sap.0000210261.65455.9d. [DOI] [PubMed] [Google Scholar]
- 24.Prescott NJ, Lees MM, Winter RM, et al. Identification of susceptibility loci for nonsyndromic cleft lip with or without cleft palate in a two stage genome scan of affected sib-pairs. Hum Genet. 2000;106:345–350. doi: 10.1007/s004390051048. [DOI] [PubMed] [Google Scholar]
- 25.Riley BM, Schultz RE, Cooper ME, et al. A genome-wide linkage scan for cleft lip and cleft palate identifies a novel locus on 8p11-23. Am J Med Genet A. 2007;143A:846–852. doi: 10.1002/ajmg.a.31673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyszynski DF, Albacha‐Hejazi H, Aldirani M, et al. A genome‐wide scan for loci predisposing to non‐syndromic cleft lip with or without cleft palate in two large Syrian families. Am J Med Genet A. 2003;123:140–147. doi: 10.1002/ajmg.a.20283. [DOI] [PubMed] [Google Scholar]
- 27.Park JW, McIntosh I, Hetmanski JB, et al. Association between IRF6 and nonsyndromic cleft lip with or without cleft palate in four populations. Genet Med. 2007;9:219–227. doi: 10.1097/GIM.0b013e3180423cca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanton SH, Cortez A, Stal S, et al. Variation in IRF6 contributes to nonsyndromic cleft lip and palate. Am J Med Genet A. 2005;137:259–262. doi: 10.1002/ajmg.a.30887. [DOI] [PubMed] [Google Scholar]
- 29.Jia Z-L, Li Y, Chen C-H, et al. Association among polymorphisms at MYH9, environmental factors, and nonsyndromic orofacial clefts in western China. DNA Cell Biol. 2010;29:25–32. doi: 10.1089/dna.2009.0935. [DOI] [PubMed] [Google Scholar]
- 30.Martinelli M, Di Stazio M, Scapoli L, et al. Cleft lip with or without cleft palate: implication of the heavy chain of non-muscle myosin IIA. J Med Genet. 2007;44:387–392. doi: 10.1136/jmg.2006.047837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lennon CJ, Birkeland AC, Nunez JA, et al. Association of candidate genes with nonsyndromic clefts in Honduran and Colombian populations. Laryngoscope. 2012;122:2082–2087. doi: 10.1002/lary.23394. [DOI] [PubMed] [Google Scholar]
- 32.Fontoura C, Silva RM, Granjeiro JM, et al. Further evidence of association of the ABCA4 gene with cleft lip/palate. Eur J Oral Sci. 2012;120:553–557. doi: 10.1111/eos.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan Y, Zhang W, Du Y, et al. Different roles of two novel susceptibility loci for nonsyndromic orofacial clefts in a Chinese Han population. Am J Med Genet A. 2011;9:10. doi: 10.1002/ajmg.a.34170. [DOI] [PubMed] [Google Scholar]
- 34.Saito H, Sekizawa A, Morimoto T, et al. Prenatal DNA diagnosis of a single-gene disorder from maternal plasma. Lancet. 2000;356:1170. doi: 10.1016/S0140-6736(00)02767-7. [DOI] [PubMed] [Google Scholar]
- 35.Chiu RW, Lo YM. Non-invasive prenatal diagnosis by fetal nucleic acid analysis in maternal plasma: the coming of age. Semin Fetal Neonatal Med 2011;16:88–93. [DOI] [PubMed]


