Abstract
Thrombotic diseases are among the leading causes of morbidity and mortality in the world. To add insights into the genetic regulation of thrombotic disease, we conducted a genome-wide association study (GWAS) of 6135 self-reported blood clots events and 252 827 controls of European ancestry belonging to the 23andMe cohort of research participants. Eight loci exceeded genome-wide significance. Among the genome-wide significant results, our study replicated previously known venous thromboembolism (VTE) loci near the F5, FGA-FGG, F11, F2, PROCR and ABO genes, and the more recently discovered locus near SLC44A2. In addition, our study reports for the first time a genome-wide significant association between rs114209171, located upstream of the F8 structural gene, and thrombosis risk. Analyses of expression profiles and expression quantitative trait loci across different tissues suggested SLC44A2, ILF3 and AP1M2 as the three most plausible candidate genes for the chromosome 19 locus, our only genome-wide significant thrombosis-related locus that does not harbor likely coagulation-related genes. In addition, we present data showing that this locus also acts as a novel risk factor for stroke and coronary artery disease (CAD). In conclusion, our study reveals novel common genetic risk factors for VTE, stroke and CAD and provides evidence that self-reported data on blood clots used in a GWAS yield results that are comparable with those obtained using clinically diagnosed VTE. This observation opens up the potential for larger meta-analyses, which will enable elucidation of the genetics of thrombotic diseases, and serves as an example for the genetic study of other diseases.
Introduction
Venous thromboembolism (VTE), which includes deep-vein thrombosis (DVT) and pulmonary embolism (PE), is a complex disease determined by well-established environmental and genetic risk factors. VTE is one of the most common cardiovascular diseases with an incidence of 1.5 per 1000 person-years estimated in Europe (1–3). Despite the success of genome-wide association studies (GWAS) in identifying new genetic factors determining other cardiovascular diseases such as coronary artery disease (CAD) (4,5), GWAS results for VTE have, until very recently, yielded little success. One of the reasons for the limited number of loci identified for VTE is most likely lack of power due to the small sample sizes used so far. In an effort to overcome this, a GWAS meta-analysis was recently published, which included 7500 cases in the discovery phase (an almost 4-fold increase over what was previously published) and reported two novel VTE-associated genes (6).
Despite this notable increase in sample size, a fairly small number of cases was included compared with the numbers used for other cardiovascular diseases (for example, 17 900 cases for the International Stroke Genetics Consortium, or 63 750 cases for the CardioGramplusC4D Consortium), and in fact, most GWAS meta-analyses now have sample sizes in the discovery phase exceeding 10 000 individuals (7). As such, concerns about sources of heterogeneity and phenotype definition have been raised, which point to the need for a balance between sample size and specific phenotype definition that avoids diluted effect sizes due to phenotype heterogeneity. While this is true for more complicated phenotypes, certain diseases with less complex phenotype definition could in theory be analyzed just by use of self-reported information about disease from questionnaires. The great advantage of this strategy is that one can take advantage of large cohorts from which genetic information is available and use them to analyze multiple different disease/quantitative/behavioral phenotypes without the need for highly specific clinical confirmation, which is often costly and time-consuming and which frequently represents the limiting factor when gathering sufficiently large disease collections. Here we present, as a proof of concept, the results of a GWAS of VTE based on web-based self-reported data on thrombotic events from the 23andMe research participant cohort, which suggest that strong and reliable association signals can be obtained from questionnaire-defined phenotypes that contribute to the identification and validation of genetic factors affecting this disease. In addition, we present results from expression profile analyses across different tissues to elucidate the possible biological mechanisms explaining the chromosome 19 locus association with disease, our only genome-wide significant thrombosis-related locus that does not harbor likely coagulation-related genes, and we provide evidence for the implications of this novel VTE-associated locus on CAD and stroke.
Results
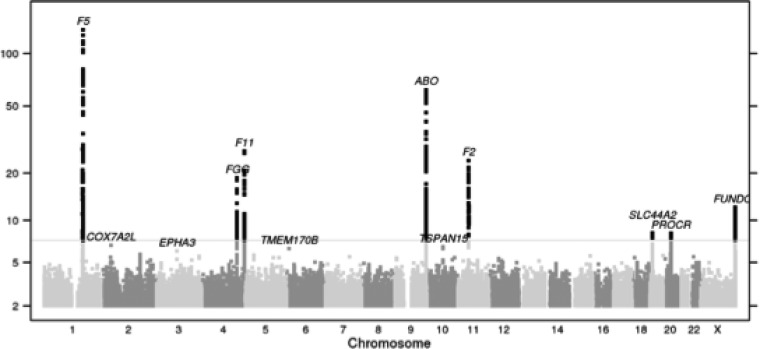
The Manhattan plot for the genome-wide logistic regression analysis of thrombotic events in 6135 cases and 252 827 controls, of which 43.1 and 53.4%, respectively, were male, is shown in Figure 1.
Figure 1.
Manhattan plot showing association between the SNPs tested and blood clots. The Y-axis represents the –log 10 of the P-value for the association for each SNP. SNPs are organized by chromosome and position along the X-axis.
Discovery GWAS
Eight loci exceeded the genome-wide significant threshold of 5 × 10−8: the coagulation factor 5 (F5) locus at chromosome 1q24.2 (P = 3.6 × 10−137), the ABO locus at chromosome 9q34.2 (P = 7.1 × 10−63), the coagulation factor 11 (F11) locus at chromosome 4q35.2 (P = 7.0 × 10−28), the coagulation factor 2 (F2) locus at chromosome 11p11.2 (P = 1.3 × 10−24), the fibrinogen cluster (FGA/FGB/FGG) locus at chromosome 4q31.3 (P = 2.0 × 10−19), the coagulation factor 8 (F8) locus at chromosome Xq28 (P = 7.0 × 10−13), the recently described locus on chromosome 19p13.2 (P = 6.1 × 10−9), which does not contain any coagulation-related candidate gene, and the protein C receptor (PROCR) locus on chromosome 20q11.22 (P = 6.7 × 10−9). Table 1 shows the lead SNPs in each associated region, and the most probable candidate gene (defined as the nearest gene related to coagulation) in the region. Among these, the F8 locus association is reported here for the first time in a GWAS. Results were adjusted for a genomic control inflation factor (λ = 1.033). Regional association plots for the genome-wide significant loci are shown in Supplementary Material, Figure S1.
Table 1.
Association results for the highest associated SNP in each region (cutoff for suggestive association at P < 10−6)
| SNP | Chrom | Position | Alleles | Frequency | P value | OR | 95%CI | Gene context |
|---|---|---|---|---|---|---|---|---|
| rs6025 | 1 | 169519049 | C/T | 0.0253 | 3.6 × 10−137 | 2.927 | [2.715, 3.154] | F5 |
| rs529565 | 9 | 136149500 | C/T | 0.6588 | 7.1 × 10−63 | 0.723 | [0.697, 0.751] | ABO |
| rs4444878 | 4 | 187213883 | A/C | 0.5988 | 7.0 × 10−28 | 0.81 | [0.780, 0.841] | F11 |
| rs1799963 | 11 | 46761055 | A/G | 0.9855 | 1.3 × 10−24 | 0.512 | [0.456, 0.576] | F2 |
| rs7654093 | 4 | 155545072 | A/T | 0.2322 | 2.0 × 10−19 | 1.216 | [1.166, 1.267] | FGG |
| rs114209171 | X | 154278797 | C/T | 0.7756 | 7.0 × 10−13 | 1.153 | [1.108, 1.200] | F8 |
| rs9797861 | 19 | 10743126 | C/T | 0.7601 | 6.1 × 10−9 | 1.145 | [1.093, 1.199] | SLC44A2 |
| rs34234989 | 20 | 33774533 | D/I | 0.7148 | 6.7 × 10−9 | 0.885 | [0.849, 0.922] | PROCR |
| rs72798544 | 2 | 42599605 | G/T | 0.9735 | 1.9 × 10−7 | 0.733 | [0.654, 0.820] | COX7A2L/KCNG3 |
| rs17490626 | 10 | 71218646 | C/G | 0.8782 | 2.9 × 10−7 | 1.165 | [1.098, 1.236] | TSPAN15 |
| rs113092656 | 6 | 11615305 | A/G | 0.9659 | 4.4 × 10−7 | 0.728 | [0.646, 0.820] | TMEM170B/ADTRP |
| rs60942712 | 3 | 89047759 | G/T | 0.101 | 8.0 × 10−7 | 1.211 | [1.124, 1.306] | EPHA3 |
Chorm stands for chromosome number, alleles are expressed as reference allele/effect, OR stands for odds ratio of the effect allele. Frequency refers to the effect allele.
In addition, four other loci were found with suggestive evidence of association with thrombosis (5 × 10−8 < P < 10−6): a locus on chromosome 2p21 (P = 1.9 × 10−7), a locus on chromosome 10q22.1 (P = 2.9 × 10−7), a locus on chromosome 6p24.1 (P = 4.4 × 10−7) and a locus on chromosome 3p11.1 (P = 8.0 × 10−7).
Replication
Since rs114209171, located close to F8 gene, association was reported in our GWAS for the first time, we sought for replication in a subset of the INVENT cohort consortium, including 26 112 individuals from 9 cohorts. Results showed that rs114209171T allele was associated with VTE (OR = 1.08 (1.02–1.14)) at a P-value of 0.011.
Among the significant VTE-associated loci previously reported in genome-wide studies, our study serves as a clear replication for the widely known F5, FGA-FGG, F11, F2, PROCR and ABO loci, and the recently discovered loci in the 19p13.2 and 10q22.1 regions (described as SLC44A2, and TSPAN15 by closest gene name), although the latter was not genome-wide significant in our study. Non genome-wide significant results from other GWAS suggest the implication of glycoprotein 6 (GP6) (8,9), STXBP5 (10), VWF (11) and KNG1 (12) in the etiology of VTE. In agreement with that, our results provided nominally significant replication of these loci (GP6 P = 0.026, STXBP5 P = 0.017, VWF P = 0.0007, KNG1 P = 1.8 × 10−6) (see Supplementary Material, Table S1).
We failed to replicate previously suggested associations within the CNTN6, SV2C, SUSD1, HIVEP1, F13 V34L, SERPINC1, SERPINA10, FX12 46C/T, C4BPB and OTUD7A loci (testing association results for our best proxy, defined as the SNP with highest r2, within 100 kb and with r2 > 0.8).
Locus on chromosome 19
Since the locus on chromosome 19 (19p13.2) was the only genome-wide significant locus in this study for which there did not seem to be an obvious candidate gene, i.e. one known to be involved with hemostasis, we used expression profiles from the EUROBATS and GAIT2 cohorts to identify a causal gene underpinning the genetic signal and to investigate a possible biological mechanism explaining the observed association. First, expression QTL (eQTL) analyses were performed between the top SNP in the region (rs9797861) and expression of all genes located in the 400 Kb flanking region, to identify which genes were affected by the thrombosis-associated SNP. The highest associations with expression levels of adjacent genes for this SNP were with SLC44A2 and ILF3 expression in fat, lymphocytes and whole blood, and with AP1M2 in whole blood (Table 2). Then, correlations between all the genes in the region and expression levels of a list of 54 plasma traits related to thrombosis were calculated in GAIT2 to explore possible pathways that could explain the link between the locus and thromboembolic disease. We found a significant (inverse) genetic correlation (defined as an estimate of the additive genetic effect shared between a pair of traits) between QTRT1 expression and S-adenosylmethionine levels, an intermediary of homocysteine (r = −0.48, P = 1.56 × 10−5), and a significant genetic correlation between TMED1 expression and Factor XII coagulation activity (r = −0.49, P = 4.13 × 10−5). However, when we checked the association between rs9797861 and these two phenotypes in the same individuals from GAIT2, we did not find a significant association. We then looked at the association between expression of all genes in the region and thrombosis outcome in GAIT2 and found no significant associations (Supplementary Material, Table S2).
Table 2.
eQTL data results in blood, fat and LCL
| Source | Transcript | P-value | Beta |
|---|---|---|---|
| EUROBATS (blood) | SLC44A2 | 5.00E−09 | 0.226784 |
| EUROBATS (blood) | ILF3 | 0.002469 | 0.118542 |
| GAIT2 (blood) | AP1M2 | 3.28E−015 | 0.447769 |
| GAIT2 (blood) | SLC44A2 | 2.48E−006 | 0.270386 |
| GAIT2 (blood) | ILF3-AS1 | 0.008084 | 0.145566 |
| EUROBATS (fat) | SLC44A2 | 8.42E−10 | 0.237704 |
| EUROBATS (fat) | ILF3 | 2.24E−07 | 0.201422 |
| EUROBATS (fat) | KRI1 | 0.000473 | 0.136727 |
| EUROBATS (LCL) | ILF3 | 7.63E−10 | 0.238294 |
| EUROBATS (LCL) | SLC44A2 | 1.12E−07 | 0.206283 |
| EUROBATS (LCL) | SMARCA4 | 2.10E−06 | 0.184805 |
| EUROBATS (LCL) | CARM1 | 5.05E−05 | 0.158285 |
| EUROBATS (LCL) | ANGPTL6 | 5.97E−05 | 0.156777 |
| EUROBATS (LCL) | DNMT1 | 0.000204 | 0.145163 |
| EUROBATS (LCL) | PPAN-P2RY11 | 0.000341 | 0.140076 |
| EUROBATS (LCL) | PPAN | 0.001052 | 0.128221 |
| EUROBATS (LCL) | QTRT1 | 0.001846 | 0.121918 |
| EUROBATS (LCL) | ZGLP1 | 0.002041 | 0.120762 |
| EUROBATS (LCL) | DOCK6 | 0.002135 | 0.12024 |
Only results significant after correction for 18 genes in 3 tissues are shown. Beta values refer to the rs9797861T allele.
In an extended search of possible related phenotypes in publicly available GWAS databases, a highly significant association was also found between rs9797861 (using rs6511708 as proxy) and height in 253 288 individuals from the GIANT consortium (P = 6.1 × 10−11) (13), and also between rs9797861 (using rs1560711 as proxy) and total cholesterol (TC) and low density lipoprotein (LDL) levels in the Blood Lipids Consortium (14) (β = 0.049; P = 6.49 × 10−12; n = 88 433). However, a closer look into the region showed that this was not the strongest association in the region for LDL and TC levels (see regional plot in Supplementary Material, Fig. S2), and the most strongly associated SNP for LDL and TC was not the strongest associated SNP with events in 23andMe.
Finally, we investigated the association of rs9797861 with other related cardiovascular diseases. We tested the association with CAD in the CARDIoGRAMplusC4D 1000 genomes-based genome-wide analysis (15) and found that this SNP is also significantly associated with CAD, with the T allele conferring increased risk (β = 0.0449; P = 0.00005); moreover, we tested the association of a proxy for rs9797861 (rs1560711, r2 = 0.90) with ischemic stroke and its subphenotypes in the Metastroke consortium and found that the SNP was significantly associated with ischemic stroke (β (T allele) = 0.055; P = 0.00536), especially with large artery stroke (LAS) (β = 0.1511; P = 0.00048) and to some degree with cardioembolic stroke (CE) (β = 0.0836; P = 0.0361).
Novel suggestive locus on chromosome 6
Since the TMEM170B-ADTRP locus on chromosome 6p24.1 contained a gene that had been previously described to regulate tissue factor pathway inhibitor (TFPI) expression in endothelial cells (16), we tested the association between our top SNP (rs113092656) and expression of TFPI in aortic samples from the ASAP study. The risk allele proved to be significantly associated (P = 0.019) with decreased expression levels of TFPI (Supplementary Material, Fig. S3).
Discussion
In contrast to all prior VTE genetic studies that have relied on clinically diagnosed DVT or PE using compression venous duplex ultrasonography, computed tomography, Doppler ultrasound, impedance plethysmography, magnetic resonance, venography, pulmonary angiography and/or ventilation/perfusion lung scan, the present study used self-reported data on history of blood clots obtained through questionnaires from deeply genotyped individuals in the 23andMe cohort. One recent meta-analysis of GWA studies of clinically defined VTE identified two novel loci associated with venous thrombosis and confirmed multiple other known associated loci (6). Interestingly, the Manhattan plots from the current study and that meta-analysis are almost identical. As the number of people who obtain their SNP profiles through companies such as 23andMe increases rapidly, the database of genotype and phenotype data that can be obtained using these datasets for research is becoming one of the most powerful tools for the discovery of common variants for a wide range of disease-related phenotypes. Several GWAS have already proven the reliability and robustness of self-reported phenotypes obtained by 23andMe (17–20). This study contributes to the knowledge of VTE genetics and shows that collection and analysis of self-reported data is a viable alternative to clinically diagnosed VTE in GWAS of VTE.
Among the previously reported VTE-associated loci, our study provides clear replication for the widely known F5, FGA-FGG, F11, F2, PROCR and ABO loci, and the recently discovered loci near SLC44A2, and TSPAN15 from a previous GWAS meta-analysis, and it strengthens the evidence for other hitherto not genome-wide significant results (GP6, STXBP5, VWF and KNG1). In addition to the previous GWAS findings, our study reports for the first time a significant genome-wide association between rs114209171, located upstream the F8 structural gene, and thrombosis risk. Our results were confirmed in a replication sample of 26 112 VTE cases and controls from the INVENT consortium. High plasma levels of FVIII are a well-established risk factor for VTE (21). There have been reports of association between the coding variant rs1800291 and some coagulation-related phenotypes (FVIII levels, VTE, stroke) all consisting of candidate-gene studies, and an open debate whether this is actually the causal variant is still ongoing. Our lead SNP in this region is in moderate LD with rs1800291 (r2 = 0.28 in 1000 Genomes phase I Europeans) and our results show a substantially weaker association for rs1800291 (P = 9.6 × 10−6), which suggests that rs114209171 represents a novel genetic risk factor for thrombosis. Finally, we have also calculated the LD between rs114209171 and rs2096362, recently found in a gene-centric approach study (22) and is r2 = 0.74 (in March 2012 release of 1000 Genomes), which indicates quite strong LD between these two variants (so, both variants might be tagging the same locus). However, rs114209171 has a strongest association with the phenotype in our cohort, which might indicate that rs114209171 could be a better proxy for the causal SNP. Further studies are needed to elucidate the causal association in this locus.
Locus on chromosome 19
Among our results, all VTE-associated loci are located in genes encoding proteins either known to participate in the coagulation cascade or as inhibitors, except for the recently discovered locus on chromosome 19. Having new candidate genes outside of the canonical coagulation system opens the door for discovery of new pathways for VTE. rs9797861, the highest thrombosis-associated SNP in the region, is located in the SLC44A2 coding region and is in strong linkage disequilibrium (r2 = 0.89) with rs2288904, which encodes an amino acid change that could be responsible for the effect. This gene codes for a human leucocyte alloantigen. However, our results show that expression of other genes in the region is also affected by rs9797861, which indicates that other genes in the region should also be taken into consideration. According to our expression analyses, solute carrier family 44 (choline transporter), member 2 (SLC44A2), Adaptor Protein Complex AP-1 Subunit 2 (AP1M2) and interleukin enhancer binding factor 3 (ILF3), all showed evidence of being differentially expressed by the different alleles on rs9797861.
Further functional analyses will be needed to establish the identity of the causal gene and the mechanism accounting for the association in chromosome 19 clearly. This will be a challenging task, because we and others (6) could not find any association between the most strongly associated SNP in chromosome 19 locus and any intermediate phenotype that is related to coagulation, which limits our ability to identify the mechanistic pathway by which this locus could be associated with disease.
We did observe strong association between a proxy for our most strongly associated SNP and LDL and TC levels in the Blood Lipids Consortium (14), but this was not the strongest association with these lipid phenotypes in the region, and the biological meaning of these seemingly concordant associations needs to be further elucidated. Finally, an association between a proxy for our most strongly associated SNP and height has also been found in the GIANT Consortium (13), which could be interesting given that body height has been considered a risk factor for VTE in men (23,24), and that an inverse association was recently found between genetically determined height and CAD that is partly explained by the association between shorter height and an adverse lipid profile (25). Further work is clearly needed to understand the links between these associations, if any, but exploring this locus further could shed some light on shared biological pathways.
Common pathways between arterial and venous thrombosis
It is also interesting to note that rs9797861 (the strongest associated SNP in the chromosome 19 region) is also associated with CAD and stroke. This is an important novel finding that suggests that still unknown shared pathways exist for venous and arterial thrombosis. Increased risk of CAD among patients previously diagnosed with VTE as well as increased risk of VTE in patients with atherosclerosis and manifest cardiovascular disease have been reported that suggest commonalities between arterial and venous thrombosis (26). Shared disease mechanisms for the two diseases have already been established, including ABO blood group, BMI, activation of blood coagulation, hypofibrinolysis and inflammation. However, no clear common genetic factors have been described, beyond the widely known ABO locus and the recently discovered PROCR gene. This study reveals a novel locus that could represent the link explaining comorbidities between these two diseases. While the main genetic triggers of venous thrombosis are abnormalities in coagulation-related proteins, arterial thrombosis has so far been mainly related to changes in the vessel wall caused by atherosclerosis, promoting plaque rupture. A common genetic determinant linking these diseases opens new views to challenge the understanding of these diseases which have until now been considered to be driven by completely different mechanisms.
Other suggestive loci
Four loci were found with suggestive evidence of association with thrombosis. The locus on chromosome 10q22.1 (near the TSPAN15 gene) has very recently been implicated in VTE by Germain et al. (6). TSPAN15 codes for tetraspanin 15. Although a role in the regulation of hemostasis has been shown for other members of the tetraspanin family, the association of this gene with VTE needs to be further investigated. The new SNP on chromosome 2p21 that exhibits a suggestively associated lies between the COX7A2L and KCNG3 genes, for which no clear role in coagulation/hemostasis has been established. The locus on chromosome 3p11.1 is located upstream of the EPHA3 gene, which codes for a tyrosine kinase receptor involved in contact-dependent bidirectional signaling with neighboring cells, and also lacks a known role in coagulation/hemostasis. The associated SNP on chromosome 6p24.1 is located in the TMEM170B-ADTRP locus. Interestingly, ADTRP codes for the androgen-dependent TFPI-regulating protein. TFPI is a major natural inhibitor of coagulation that inhibits the FVII-TFa complex in the presence of FXa, and it is located mainly on endothelial cells. Reduced levels of TFPI have been associated with ischemic stroke (27), cerebral venous thrombosis (28) and DVT (29–31). ADTRP has been shown to regulate TFPI mRNA expression, cellular distribution and the cell-associated anticoagulant activity of TFPI in endothelial cells, both in native conditions and in response to androgen (16), for which it becomes a novel interesting obvious candidate gene associated with VTE risk. Our results indicate that the strongest VTE-associated SNP in the region is also associated with expression levels of TFPI in the aorta, which supports ADTRP as a novel thrombotic risk factor that regulates TFPI expression in the arterial wall.
Strengths and limitations
A concern of the self-reported phenotype approach used in the present work is the heterogeneity of the phenotype, which includes venous thrombosis but also cases of stroke. While this makes interpretability more challenging, our results show that genetic plots are comparable to clinically diagnosed VTE. Thus, while novel discoveries in a blood clots self-reported phenotype are interesting and suggest implication to disease, further replication on clinically diagnosed specific diseases would provide additional validation.
In conclusion, this study extends the current knowledge of the genetics of VTE by adding for the first time information about associated loci in the sex chromosome, describing a GWAS association within the F8 locus, providing suggestive evidence of a novel gene (ADTRP) that could be implicated in thrombosis by modulating TFPI levels, and describing evidence of a shared genetic factor on chromosome 19, associated with both CAD, stroke and VTE. Expression analyses nail down three plausible causal genes, which may explain the observed comorbidity for these diseases. Moreover, we show that use of self-reported data for VTE yields results comparable with studies based on clinically diagnosed VTE. These latter results open the door to new larger collaborations between groups with more relaxed phenotyping descriptions of VTE, enabling a substantial increase in power and sample size for the next generation of studies.
Methods
Study sample
The study sample consists of 6135 cases self-reporting history of blood clots and 252 827 controls of European ancestry belonging to the 23andMe research participant cohort. All individuals included in the analyses provided informed consent and answered surveys online in accordance with the 23andMe human subjects protocol, which was reviewed and approved by Ethical & Independent Review Services, a private institutional review board (http://www.eandireview.com). A table with demographics of all individuals included in the GWAS is provided in Table 3.
Table 3.
Demographics from individuals included in the GWAS for self-reported blood clots
| Group | N | Male | Female | Age (0, 30] | Age (30, 45] | Age (45, 60] | Age (60, Inf] |
|---|---|---|---|---|---|---|---|
| Case | 6135 | 2644 | 3491 | 161 | 759 | 1643 | 3572 |
| Control | 252 827 | 134 977 | 117 850 | 35 025 | 72 478 | 69 615 | 75 709 |
Phenotype description
Data for the study were collected within a research framework wherein research participants, derived from the customer base of 23andMe, Inc., a direct-to-consumer genetic information company, consented to the use of their data for research and provided with access to their personal genetic information. Most data on blood clots came from the questions ‘Have you ever been diagnosed with any of the following conditions? (A stroke or a blood clot)’ (Yes, No, I'm not sure); if Yes: ‘What types of blood clot or stroke were you diagnosed with? Please check all that apply: (A blood clot in the brain (an ischemic stroke), A blood clot in your arms or legs (deep-vein thrombosis or DVT), A blood clot in your lungs (pulmonary embolism)). Some participants answered only a broadly worded question, either ‘Have you ever been diagnosed by a doctor with any of the following blood conditions? (Blood clots)’ (Yes, No, I'm not sure); or ‘Have you ever been diagnosed with a blood clot?’ (Yes, No, I'm not sure). Cases gave at least one positive response, and controls gave no positive and at least one negative response. Among 4196 individuals who reported a specific diagnosis, 48% reported deep-vein thrombosis (DVT); 14% reported pulmonary embolism (PE); 18% reported ischemic stroke and the remainder reported multiple diagnoses, largely DVT and PE (17%).
DNA sampling, genotyping and imputation
DNA extraction and genotyping were performed on saliva samples by the National Genetics Institute (NGI), a CLIA-licensed clinical laboratory and a subsidiary of Laboratory Corporation of America. Samples were genotyped on one of four genotyping platforms. The V1 and V2 platforms were based on the Illumina HumanHap550+ BeadChip, including about 25 000 custom SNPs selected by 23andMe, with a total of about 560 000 SNPs. The V3 platform was based on the Illumina OmniExpress+ BeadChip, with custom content to improve the overlap with the V2 array, with a total of about 950 000 SNPs. The V4 platform is a fully custom array, including a lower redundancy subset of V2 and V3 SNPs with additional coverage of lower-frequency coding variation, and about 570 000 SNPs. Samples that failed to reach 98.5% call rate were re-analyzed. Individuals whose analyses failed repeatedly were re-contacted by 23andMe customer service to provide additional samples.
We restricted participants to a set of individuals who had >97% European ancestry, as determined through an analysis of local ancestry. The reference population data is derived from public datasets (the Human Genome Diversity Project, HapMap and 1000 Genomes), as well as 23andMe customers who have reported having four grandparents from the same country. Participant genotype data were imputed against the March 2012 ‘v3’ release of 1000 Genomes reference haplotypes (32). Further details about ancestry determinations and imputation are given in Supplementary Material, Section 1.
Association analyses
Genotyped and imputed SNPs were analyzed according to logistic regression assuming an additive model for allelic effects, including covariates for age, gender and the top five principal components to account for residual population structure. Results for the X chromosome are computed similarly, with male genotypes coded as if they were homozygous diploid for the observed allele. Selection of the most associated SNP in each region was done by identifying SNPs with P< 10−5, then grouping these into intervals separated by gaps of at least 250 kb, and choosing the SNP with the smallest P within each interval.
Replication for SNP rs114209171 was sought in a subsample of the INVENT consortium consisting on 26 112 VTE cases and controls. Details of the statistical analysis can be found in Germain et al. (6).
Association with related phenotypes
In order to identify the causal gene within the associated locus on chromosome 19, correlations between expression levels of all genes located within 400 Kb of rs9797861 and 54 phenotypes belonging to the coagulation, anticoagulant and fibrinolytic pathways were tested in GAIT2 (data not shown). GAIT2 includes 35 extended pedigrees with at least 10 living relatives from 3 or more generations (a total of 935 individuals) that were identified based on a proband with idiopathic thrombophilia (at least one spontaneous episode of deep venous thrombosis). Expression levels were measured by RNA-seq in whole blood. Paired reads of 49 bp where sequenced and mapped to the human genome reference using BWA (33). We quantified gene level expression by counting reads that fall in exons as defined in the GENCODE v10 annotation. Quantifications were scaled to the size of the library to make them comparable across samples and then transformed using a rank-normal transformation. Correlations were calculated taking into account the family structure using a bivariate variance components model as implemented in SOLAR (34). Bivariate variance components models allow the partition of the phenotypic correlation in genetic and environmental components (35). We then quantified the genetic correlation between all genes located within 400 Kb of rs9797861 and the thrombosis outcome in the same sample using the same statistical approach.
Since our analyses showed suggestive correlations between two of the genes located in the chromosome 19 locus and FXII coagulant levels as well as levels of an intermediary of homocysteine metabolism (S-adenosylmethionine), we also tested rs9797861 for association with FXII coagulant levels and homocysteine levels in the GAIT2 sample.
Finally, we tested the association of rs9797861 with CAD in the CARDIoGRAMplusC4D GWAS, a meta-analysis of ≈185 000 CAD cases and controls (15) and with 12 389 individuals with ischemic stroke and 62 004 controls, using data from the Metastroke consortium (36).
Expression QTL analyses
The strongest associated SNP on chromosome 19 (rs9797861) was tested for association with the expression levels of all genes located within its 400 Kb flanking region (18 genes) in whole blood, adipose tissue, skin and lymphoblastoid cell lines (LCLs) using available data from EUROBATS (37) and GAIT2 (38). EUROBATS consists of a sample of ∼800 female twins from the TwinsUK cohort. For each individual, there are measures of gene expression obtained from RNAseq in four tissues: adipose tissue, skin, whole blood and LCLs. Methods to quantify gene expression and obtain eQTL have been described (37). The association analysis in the GAIT2 sample was performed with SOLAR Eclipse version 7.6.6 using RNA-seq data from whole blood. Gene expression data were normalized using an inverse normal transformation. We used linear mixed models to allow the decomposition of the phenotypic variance into genetic, environmental and residual terms, adjusting by age and sex, and introducing a pedigree bias correction to correct for family structure. The reported association P-values were determined by likelihood ratio test (LRT). Further information on GAIT2 and RNA extraction methods can be found in Folkersen et al. (38) and in Supplementary Material, Section 2. Association between the strongest associated SNP on chromosome 6 (rs113092656) and expression of TFPI in blood vessels was tested in 132 aortic adventitia samples from the ASAP project (39).
Supplementary Material
Funding
This work was supported by the National Human Genome Research Institute of the National Institutes of Health (grant number R44HG006982-02 to 23andMe). M.S.-L is partially supported by the Swedish Heart-Lung Foundation (20130399) and acknowledges funding from Åke Wiberg, Lars Hiertas Minne, Magnus Bergvalls and Tore Nilssons foundations. Stroke look-ups were partially supported by grants received from the German Federal Ministry of Education and Research (BMBF) in the context of the e:Med program (e:AtheroSysMed) and the FP7 European Union project CVgenes@target (261123).
Supplementary Material
Acknowledgements
We thank the research participants and employees of 23andMe for making this work possible. Data on coronary artery disease/myocardial infarction have been contributed by CARDIoGRAMplusC4D investigators and have been downloaded from www.cardiogramplusc4d.org.
Conflict of Interest statement. D.A.H and J.Y.T are employed by 23andMe, Inc. and disclose funding from NIH. D.Z., A.M. and N.B are employed by Pfizer Worldwide R&D. B.B.W is an Associate Editor for the journal Neurology and discloses grant funding from the NIH.
References
- 1.Nordstrom M., Lindblad B., Bergqvist D., Kjellstrom T. (1992) A prospective study of the incidence of deep-vein thrombosis within a defined urban population. J. Intern. Med., 232, 155–160. [DOI] [PubMed] [Google Scholar]
- 2.Oger E. (2000) Incidence of venous thromboembolism: a community-based study in Western France. EPI-GETBP Study Group. Groupe d'Etude de la Thrombose de Bretagne Occidentale. Thromb. Haemost., 83, 657–660. [PubMed] [Google Scholar]
- 3.Naess I.A., Christiansen S.C., Romundstad P., Cannegieter S.C., Rosendaal F.R., Hammerstrom J. (2007) Incidence and mortality of venous thrombosis: a population-based study. J. Thromb. Haemost., 5, 692–699. [DOI] [PubMed] [Google Scholar]
- 4.Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C. et al. (2011) Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet., 43, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peden J., Hopewell J.C., Saleheen D. (2011) A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet., 43, 339–344. [DOI] [PubMed] [Google Scholar]
- 6.Germain M., Chasman D.I., de Haan H., Tang W., Lindstrom S., Weng L.C., de Andrade M., de Visser M.C., Wiggins K.L., Suchon P. et al. (2015) Meta-analysis of 65,734 individuals identifies TSPAN15 and SLC44A2 as two susceptibility loci for venous thromboembolism. Am. J. Hum. Genet., 96, 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evangelou E., Ioannidis J.P. (2013) Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet., 14, 379–389. [DOI] [PubMed] [Google Scholar]
- 8.Bezemer I.D., Bare L.A., Doggen C.J., Arellano A.R., Tong C., Rowland C.M., Catanese J., Young B.A., Reitsma P.H., Devlin J.J. et al. (2008) Gene variants associated with deep vein thrombosis. JAMA, 299, 1306–1314. [DOI] [PubMed] [Google Scholar]
- 9.Tregouet D.A., Heath S., Saut N., Biron-Andreani C., Schved J.F., Pernod G., Galan P., Drouet L., Zelenika D., Juhan-Vague I. et al. (2009) Common susceptibility alleles are unlikely to contribute as strongly as the FV and ABO loci to VTE risk: results from a GWAS approach. Blood, 113, 5298–5303. [DOI] [PubMed] [Google Scholar]
- 10.Smith N.L., Chen M.H., Dehghan A., Strachan D.P., Basu S., Soranzo N., Hayward C., Rudan I., Sabater-Lleal M., Bis J.C. et al. (2010) Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation, 121, 1382–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith N.L., Rice K.M., Bovill E.G., Cushman M., Bis J.C., McKnight B., Lumley T., Glazer N.L., van Hylckama Vlieg A., Tang W. et al. (2011) Genetic variation associated with plasma von Willebrand factor levels and the risk of incident venous thrombosis. Blood, 117, 6007–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morange P.E., Oudot-Mellakh T., Cohen W., Germain M., Saut N., Antoni G., Alessi M.C., Bertrand M., Dupuy A.M., Letenneur L. et al. (2011) KNG1 Ile581Thr and susceptibility to venous thrombosis. Blood, 117, 3692–3694. [DOI] [PubMed] [Google Scholar]
- 13.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z. et al. (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet., 46, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J. et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CARDIoGRAMplusC4D Consortium (2015) A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet., 47, 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupu C., Zhu H., Popescu N.I., Wren J.D., Lupu F. (2011) Novel protein ADTRP regulates TFPI expression and function in human endothelial cells in normal conditions and in response to androgen. Blood, 118, 4463–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M. et al. (2014) Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet., 46, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira M.A., Matheson M.C., Tang C.S., Granell R., Ang W., Hui J., Kiefer A.K., Duffy D.L., Baltic S., Danoy P. et al. (2014) Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J. Allergy Clin. Immunol., 133, 1564–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinds D.A., McMahon G., Kiefer A.K., Do C.B., Eriksson N., Evans D.M., St Pourcain B., Ring S.M., Mountain J.L., Francke U. et al. (2013) A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat. Genet., 45, 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eriksson N., Macpherson J.M., Tung J.Y., Hon L.S., Naughton B., Saxonov S., Avey L., Wojcicki A., Pe'er I., Mountain J. (2010) Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet., 6, e1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koster T., Blann A.D., Briet E., Vandenbroucke J.P., Rosendaal F.R. (1995) Role of clotting factor VIII in effect of von Willebrand factor on occurrence of deep-vein thrombosis. Lancet, 345, 152–155. [DOI] [PubMed] [Google Scholar]
- 22.The Women's Health Initiative Study Group (1998) Design of the Women's Health Initiative clinical trial and observational study. Control Clin. Trials, 19, 61–109. [DOI] [PubMed] [Google Scholar]
- 23.Braekkan S.K., Borch K.H., Mathiesen E.B., Njolstad I., Wilsgaard T., Hansen J.B. (2010) Body height and risk of venous thromboembolism: the Tromso Study. Am. J. Epidemiol., 171, 1109–1115. [DOI] [PubMed] [Google Scholar]
- 24.Borch K.H., Nyegaard C., Hansen J.B., Mathiesen E.B., Njolstad I., Wilsgaard T., Braekkan S.K. (2011) Joint effects of obesity and body height on the risk of venous thromboembolism: the Tromso Study. Arterioscler. Thromb. Vasc. Biol., 31, 1439–1444. [DOI] [PubMed] [Google Scholar]
- 25.Nelson C.P., Hamby S.E., Saleheen D., Hopewell J.C., Zeng L., Assimes T.L., Kanoni S., Willenborg C., Burgess S., Amouyel P. et al. (2015) Genetically determined height and coronary artery disease. N. Engl. J. Med., 372, 1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prandoni P. (2009) Venous and arterial thrombosis: two aspects of the same disease? Eur. J. Intern. Med., 20, 660–661. [DOI] [PubMed] [Google Scholar]
- 27.Abumiya T., Yamaguchi T., Terasaki T., Kokawa T., Kario K., Kato H. (1995) Decreased plasma tissue factor pathway inhibitor activity in ischemic stroke patients. Thromb. Haemost., 74, 1050–1054. [PubMed] [Google Scholar]
- 28.Javanmard S.H., Shahsavarzadeh T., Saadatnia M. (2015) Low levels of tissue factor pathway inhibitor increase the risk of cerebral venous thrombosis. Adv. Biomed. Res., 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amini-Nekoo A., Futers T.S., Moia M., Mannucci P.M., Grant P.J., Ariens R.A. (2001) Analysis of the tissue factor pathway inhibitor gene and antigen levels in relation to venous thrombosis. Br. J. Haematol., 113, 537–543. [DOI] [PubMed] [Google Scholar]
- 30.Hoke M., Kyrle P.A., Minar E., Bialonzcyk C., Hirschl M., Schneider B., Kollars M., Weltermann A., Eichinger S. (2005) Tissue factor pathway inhibitor and the risk of recurrent venous thromboembolism. Thromb. Haemost., 94, 787–790. [DOI] [PubMed] [Google Scholar]
- 31.Dahm A., Van Hylckama Vlieg A., Bendz B., Rosendaal F., Bertina R.M., Sandset P.M. (2003) Low levels of tissue factor pathway inhibitor (TFPI) increase the risk of venous thrombosis. Blood, 101, 4387–4392. [DOI] [PubMed] [Google Scholar]
- 32.Genomes Project C., Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A. (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Durbin R. (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics, 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almasy L., Blangero J. (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet., 62, 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams J.T., Van Eerdewegh P., Almasy L., Blangero J. (1999) Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am. J. Hum. Genet., 65, 1134–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traylor M., Farrall M., Holliday E.G., Sudlow C., Hopewell J.C., Cheng Y.C., Fornage M., Ikram M.A., Malik R., Bevan S. et al. (2012) Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol., 11, 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buil A., Brown A.A., Lappalainen T., Vinuela A., Davies M.N., Zheng H.F., Richards J.B., Glass D., Small K.S., Durbin R. et al. (2015) Gene–gene and gene–environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet., 47, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camacho M., Martinez-Perez A., Buil A., Siguero L., Alcolea S., Lopez S., Fontcuberta J., Souto J.C., Vila L., Soria J.M. (2012) Genetic determinants of 5-lipoxygenase pathway in a Spanish population and their relationship with cardiovascular risk. Atherosclerosis, 224, 129–135. [DOI] [PubMed] [Google Scholar]
- 39.Folkersen L., van't Hooft F., Chernogubova E., Agardh H.E., Hansson G.K., Hedin U., Liska J., Syvanen A.C., Paulsson-Berne G., Franco-Cereceda A. et al. (2010) Association of genetic risk variants with expression of proximal genes identifies novel susceptibility genes for cardiovascular disease. Circ. Cardiovasc. Genet., 3, 365–373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.