Abstract
The objective of this study was to determine the protective effects of the mitochondria-targeted molecules MitoQ and SS31 in striatal neurons that stably express mutant huntingtin (Htt) (STHDhQ111/Q111) in Huntington's disease (HD). We studied mitochondrial and synaptic activities by measuring mRNA and the protein levels of mitochondrial and synaptic genes, mitochondrial function, and ultra-structural changes in MitoQ- and SS31-treated mutant Htt neurons relative to untreated mutant Htt neurons. We used gene expression analysis, biochemical methods, transmission electron microscopy (TEM) and confocal microscopy methods. In the MitoQ- and SS31-treated mutant Htt neurons, fission genes Drp1 and Fis1 were down-regulated, and fusion genes Mfn1, Mfn2 and Opa1 were up-regulated relative to untreated neurons, suggesting that mitochondria-targeted molecules reduce fission activity. Interestingly, the mitochondrial biogenesis genes PGC1α, PGC1β, Nrf1, Nrf2 and TFAM were up-regulated in MitoQ- and SS31-treated mutant Htt neurons. The synaptic genes synaptophysin and PSD95 were up-regulated, and mitochondrial function was normal in the MitoQ- and SS31-treated mutant Htt neurons. Immunoblotting findings of mitochondrial and synaptic proteins agreed with the mRNA findings. TEM studies revealed decreased numbers of structurally intact mitochondria in MitoQ- and SS31-treated mutant Htt neurons. These findings suggest that mitochondria-targeted molecules MitoQ and SS31 are protective against mutant Htt-induced mitochondrial and synaptic damage in HD neurons, and these mitochondria-targeted molecules are potential therapeutic molecules for the treatment of HD neurons.
Introduction
Huntington's disease (HD), a genetic disease with an autosomal dominant inheritance, strikes humans in midlife. HD is characterized by motor dysfunction, involuntary movements, dystonia, cognitive decline, intellectual impairment and emotional disturbances (1,2). HD is a rare disease, afflicting 4–10 per 100 000 persons of mainly Caucasian origin. Histopathological examination of HD postmortem brains has revealed that several regions of the brain are affected, including the caudate and putamen of the striatum, cerebral cortex, hippocampus, hypothalamus and sub-thalamus (1,3,4). Neuronal loss in these regions of the brain has been found in up to 80% of patients with severe HD (1). A progressive loss of body weight is a prominent feature in the progression of HD.
The genetic mutation that causes HD was identified in 1993, via a collaborative effort by an international team of scientists, The Collaborative Research Group (5). They identified expanded polyglutamine repeats (or CAG repeats) as a mutation within exon 1 of the HD gene. In HD patients, the number of polyglutamine repeats ranges from 36 to 120, whereas in healthy individuals, CAG repeats ranges from 6 to 35 (6,7). Onset of HD has been found to be inversely correlated with the number of CAG repeats in the HD gene. Huntingtin (Htt), a product of the HD gene, is a 350 kDa protein, ubiquitously expressed in the brain and peripheral tissues (6,8,9). Both wild-type (WT) and mutant Htt are primarily localized in the cytoplasm of neurons. However, a small portion of mutant Htt has been found to localize abnormally in subcellular organelles, including the nucleus, plasma membrane, mitochondria, lysosomes and endoplasmic reticulum. These abnormal localizations have been found to impair the function of subcellular organelles (7).
Over two decades of intense research using knockout, knockin, and transgenic animal models, cell models, and postmortem of HD have revealed multiple cellular changes that are implicated in HD progression and pathogenesis, including (i) transcriptional dysregulation, (ii) caspase activation, (iii) expanded polyQ repeat protein interactions with other CNS proteins, (iv) NMDAR activation, calcium dyshomeostasis, (v) defective axonal trafficking, and abnormal mitochondrial dynamics (e.g. increased fission and decreased fusion) (7). Among these cellular changes, abnormal mitochondrial dynamics is largely involved in HD pathogenesis.
Several recent studies using postmortem brains of HD patients (10,11); fibroblasts, lymphoblasts and myoblasts from HD patients (12,13); and cell models of HD (13–16) revealed structural and functional abnormalities in mitochondria in cells that express mutant Htt, suggesting that defective mitochondria may be responsible for cell damage in HD.
Using postmortem brains from HD patients and control subjects, Shirendeb et al. measured mRNA and protein levels of mitochondrial dynamics genes and electron transport chain (ETC) genes, and they also assessed mitochondrial function (10). They found increased expression of fission genes (Drp1 and Fis1) and decreased expression of fusion genes (Mfn1, Mfn2 and Opa1) in HD patients relative to the controls. The matrix protein CypD was up-regulated in HD patients. These findings suggest the presence of abnormal mitochondrial dynamics in HD. Further, significantly increased immunoreactivity of 8-hydroxy-guanosine and significantly decreased cytochrome oxidase 1 and cytochrome b were found in HD patients relative to controls, suggesting also that mitochondrial function was defective in HD brains (11).
Using brain tissues from BACHD mice and postmortem brain tissues from HD patients, Shirendeb et al. studied the molecular link between the mitochondrial protein Drp1 and mutant Htt (15). Using primary neurons from the BACHD mice, they also studied the relationship between axonal transport of mitochondria and synaptic degeneration. They found that mutant Htt interacts with Drp1, elevates the enzymatic activity of Drp1 and increases abnormal mitochondrial dynamics, resulting in defective anterograde mitochondrial movement and synaptic deficiencies. These observations suggest that increased mitochondrial fragmentation and defective axonal transport of mitochondria lead to neuronal damage in HD neurons.
In order to understand the mutant Htt effects on mitochondrial structure and function, we recently studied mitochondrial and synaptic activities, mitochondrial function and ultra-structural changes in striatal neurons that stably express mutant Htt neurons (STHDhQ111/Q111) relative to WT Htt neurons (STHDhQ7/Q7). We also studied these parameters in mitochondrial division inhibitor 1 (Mdivi1) treated and untreated WT and mutant Htt neurons to determine the Mdivi1's beneficial effects (17). In both groups of studies, we found increased levels of mRNA and of fission gene proteins, and decreased levels of fusion genes and synaptic and dendritic genes, in the mutant Htt neurons relative to the WT neurons. In addition, we found significantly increased numbers of functional mitochondria and of dysfunctional mitochondria in the mutant Htt neurons. In contrast, in the Mdivil-treated mutant Htt neurons, we found the opposite: we found significantly decreased numbers of functional mitochondria and of dysfunctional mitochondria in the Mdivil-treated mutant Htt neurons. These results indicate that Mdivi1 beneficially affects healthy neurons. Taken together, these findings suggest that Mdivi1 is protective against mutant Htt-induced mitochondrial damage in HD neurons.
Mitochondrial dysfunction and defective bioenergetics have been implicated in HD (7–18). If mitochondrial dysfunction proves to be significant in HD, it may be possible to treat HD by developing antioxidant molecules that target mitochondria for treatment. The use of antioxidants to treat mitochondria in patients with neurodegenerative diseases, including HD, has received great attention. Recent epidemiological studies have suggested that an increased intake of natural antioxidants, such as vitamin E, vitamin C, and β-carotene, might reduce the risk of developing HD, Alzheimer's disease (AD) and Parkinson's disease (PD). However, not all such epidemiological studies agree with this approach to treat neurodegenerative diseases (19,20). Although antioxidant approaches to treating neurodegenerative diseases have been promising, particular currently available antioxidant approaches may not be effective in treating neurodegenerative diseases because naturally occurring antioxidants, such as vitamins E, vitamin C and β-carotene, might not cross the blood–brain barrier and so could not reach the relevant sites of mitochondrial dysfunction (21). To deliver antioxidant molecules to the dysfunctional mitochondria for treatment, antioxidant molecules are needed, in addition to a system that is capable of targeting and treating the dysfunctional mitochondria.
Recently, considerable progress has been made in developing molecules that are capable of targeting to mitochondria; such molecules include (i) MitoQ, (ii) MitoVitE MitoαLipoic acid and (iii) MitoPBN (22–24). These molecules were developed by conjugating the antioxidant molecule to a positively charged lipophilic phosphonium cation. The resulting molecule, i.e. the moiety, was capable of being dragged to the cell plasma membrane because of the charge difference between the positively charged moiety (lipophilic phosphonium cation + antioxidant) and negative charged cell (22,24,25). Further, the moiety could be dragged to mitochondrial matrix several hundred times due to the charge difference. Once being dragged, the moiety exhibited reduced mitochondrial dysfunction by scavenging free radicals rapidly—(i) in the cytosol and (2) in the mitochondrial matrix—and was able to maintain its neuronal viability.
MitoQ, the molecular formula and weight of which are C37H45O4PB4 and 665.65, respectively, has been studied using cell culture and mouse models of neurodegeneration to determine its beneficial effects (26–30) and results were positive. Since mitochondrial abnormalities and oxidative damage are known to be involved in HD pathophysiology, there has been strong interest in determining whether mitochondria-targeted antioxidants are capable of decreasing oxidative damage in mutant Htt neurons.
SS31 is a tetra-peptide, cell-permeable, mitochondria-targeted molecule. The SS31 is one of the four molecules (SS31, SS02, SS19, SS20) developed by Szeto and Schiller (31) that is capable of targeting and permeating to mitochondria. The chemical structure of SS31 is H-D-Arg-Dmt-Lys-Phe-NH2, and its structural motif centers on alternating aromatic residues and basic amino acids. SS31 has a sequence motif that allows it to target mitochondria, scavenge free radicals, including H2O2 and ONOO−, and inhibit lipid peroxidation. Its capability of protecting to be attributed to tyrosine, or dimethyltyrosine, a residue that scavenges oxyradicals and forms relatively unreactive tyrosyl radicals. The unreactive tyrosyl radicals trigger the coupling of tyrosyl radicals to each other, giving rise to dityrosine, which reacts with a superoxide to form tyrosine hydroperoxide. Recently, the efficacy of SS31 was studied in rodent models of ischemic brain injury (32), diabetes (33), myocardial infarction (34) and amyotrophic lateral sclerosis (ALS) (35). These researchers found that SS31 protects cells from mitochondrial toxicity in all of these disease states. However, the efficacy of SS31 has yet to be studied in the HD neurons.
In this study, we sought to investigate the beneficial effects of MitoQ and SS31. We determined (i) mitochondrial structure by measuring mRNA and the protein levels of mitochondrial fission and fusion genes, mitochondrial biogenesis, ETC, and synaptic genes; (ii) mitochondrial function by measuring free radical production, lipid peroxidation, GTPase Drp1 enzymatic activity, mitochondrial ATP, and cell viability; and (iii) ultra-structural changes of subcellular organelles in the cell, including mitochondria in mutant Htt neurons treated with MitoQ and SS31.
Results
Gene expression differences between MitoQ-treated and untreated mutant Htt neurons
To determine the effects of MitoQ on mitochondrial structure and on synaptic genes, we treated mutant Htt neurons with MitoQ, and we used real-time RT–PCR to measure mRNA levels before and after treatment.
mRNA expression levels of Drp1 and Fis1 were significantly decreased in neurons treated with MitoQ relative to untreated mutant Htt neurons (Table 1). In contrast, the levels of mRNA expression of the mitochondrial fusion genes Mfn1, Mfn2, and Opa1 increased after treatment with MitoQ. These findings indicate that MitoQ enhances fusion activity in mutant Htt neurons. Significantly reduced mRNA levels were found in CypD, in mutant Htt neurons treated with MitoQ. Significantly increased mRNA expression levels were found in ETC genes, ND1, Cyt-B, and COX1-3 in mutant Htt cells treated with MitoQ. The levels of mRNA were unchanged for some ETC genes, including ND3, ND6, and ATPase 6 in mutant Htt neurons treated with MitoQ (Table 1).
Table 1.
mRNA fold changes of mitochondrial dynamics, mitochondrial biogenesis, electron transport chain, and synaptic genes in mutant Htt neurons (HDhQ111/Q111) treated with MitoQ and SS31
| Genes | MitoQ | SS31 |
|---|---|---|
| Fold changes | Fold changes | |
| Mitochondrial dynamics genes | ||
| DRP1 | −1.5* | −2.1* |
| FIS1 | −1.4* | −1.9* |
| Mfn1 | 1.8* | 1.2 |
| Mfn2 | 1.7* | 1.4* |
| Cyclophilin D | −1.2 | −2.0* |
| OPA1 | 1.2 | 1.4* |
| VDAC1 | −1.4* | −1.1 |
| Mitochondrial biogenesis genes | ||
| PGC1α | 1.9** | 1.3* |
| PGC3β | 1.5* | 2.0** |
| Nrf1 | 1.5* | 1.1 |
| Nrf2 | 1.4* | 1.2 |
| TFAM | 1.4* | 1.3* |
| Mitochondrial-encoded electron transport chain genes | ||
| ND1 - Complex I | 1.3* | 1.4* |
| ND3 - Complex I | 1.2 | 1.1 |
| ND6 - Complex I | 1.1 | 1.1 |
| Cyt B - Complex III | 1.3* | 1.2 |
| COX1 - Complex IV | 1.3* | 1.4* |
| COX2 - Complex IV | 1.5* | 1.6* |
| COX3 - Complex IV | 1.6* | 1.5* |
| ATP6 - Complex V | 1.2 | 1.4* |
| Synaptic genes | ||
| Synaptophysin | 1.1 | 1.4* |
| PSD-95 | 1.9* | 1.8* |
| Synapsin 1 | 1.5* | 1.6* |
| Synapsin 2 | 1.3* | 1.1 |
| Synaptobrevin 1 | 1.5* | 1.7* |
| Synaptobrevin 2 | 1.2 | 1.1 |
| Neurogranin | 1.5* | 1.2 |
| GAP43 | 1.7** | 1.4* |
| Synaptopodin | 1.2 | 1.2 |
As shown in Table 1, mRNA levels of mitochondrial biogenesis genes PGC1α, PGC1β, Nrf1, Nrf2 and TFAM increased in MitoQ-treated mutant Htt neurons relative to untreated neurons, indicating that MitoQ enhances biogenesis activity in HD neurons.
mRNA expression levels were increased in synaptic genes, including synaptophysin, PSD95, synapsin 1, synapsin 2, synaptobrevin 1, synaptobrevin 2, neurogranin, GAP43, and synaptopodin in mutant Htt neurons treated with MitoQ relative to the untreated Htt neurons (Table 1). These findings suggest that MitoQ enhances synaptic gene expression in mutant Htt neurons.
Gene expression differences between SS31-treated and untreated mutant Htt neurons
As shown in Table 1, in SS31-treated mutant Htt neurons, mRNA expression level of the fission genes Drp1 and Fis1 decreased relative to the untreated Htt neurons. In contrast, the levels of mRNA expression of the mitochondrial fusion genes Mfn1, Mfn2, and Opa1 increased after treatment with SS31. These findings indicate that SS31 enhances fusion activity in mutant Htt neurons. Significantly reduced mRNA levels were found in the matrix gene CypD, in SS31-treated mutant Htt neurons. mRNA expression levels were unchanged for the ETC genes ND3, ND6 and Cy-B in mutant Htt cells treated with SS31, and mRNA expression levels were increased for ND1, COX1-3 and ATP6.
In SS31-treated mutant Htt neurons, increased mRNA levels were found in the mitochondrial biogenesis genes PGC1α, PGC1β, Nrf1, Nrf2 and TFAM relative to the untreated Htt neurons (Table 1), suggesting that SS31 enhances biogenesis activity in mutant Htt neurons.
In mutant Htt neurons treated with SS31 relative to untreated Htt neurons, mRNA expression levels were increased in synaptic genes, including synaptophysin, PSD95, synapsin 1, synapsin 2, synaptobrevin 1, synaptobrevin 2, neurogranin, GAP43 and synaptopodin (Table 1). These findings suggest that SS31 enhances synaptic gene expressions in mutant Htt neurons.
Immunoblotting analysis
Protein levels in MitoQ-treated and untreated mutant Htt neurons
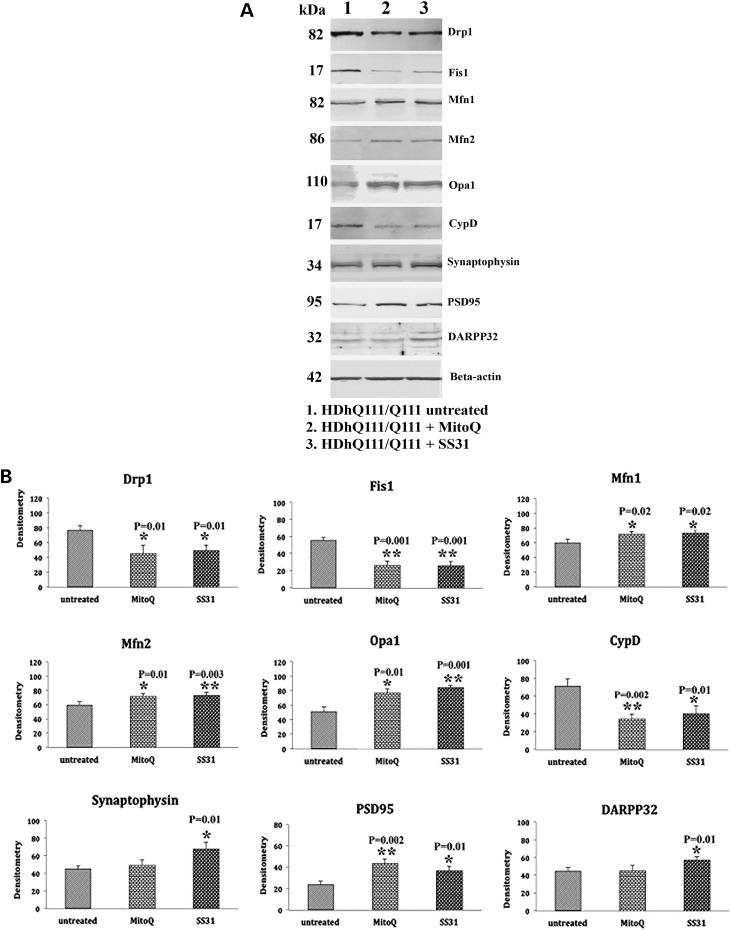
To understand the effects of mutant Htt on mitochondrial and synaptic proteins, we performed immunoblotting analysis, using protein lysates of MitoQ-treated mutant Htt (HDhQ111/Q111) neurons, and we assessed protein levels using densitometry analysis. Our quantitative densitometry revealed significantly decreased levels of the fission proteins Drp1 (P = 0.01) and Fis1 (P = 0.001) proteins in the MitoQ-treated mutant Htt neurons relative to the untreated mutant Htt neurons (Fig. 1A and B). In contrast, the fusion proteins Mfn1 (P = 0.02), Mfn2 (P = 0.01) and Opa1 (P = 0.01) were found to be increased in MitoQ-treated mutant Htt neurons, relative to the untreated Htt neurons (Fig. 1A and B). Levels of the matrix protein CypD (P = 0.002) were significantly decreased in MitoQ-treated mutant Htt neurons. Expression levels of the synaptic protein PSD95 (P = 0.002) were significantly increased in the MitoQ-treated mutant Htt neurons relative to the untreated Htt neurons (Fig. 1A and B).
Figure 1.
Immunoblotting analysis of proteins in mutant Htt neurons treated with MitoQ and SS31 and in untreated mutant Htt neurons. (A) Representative immunoblotting analysis of mutant Htt neurons treated with MitoQ and SS31 and untreated Htt neurons. (B) Quantitative densitometry analysis of mitochondrial dynamics and the matrix proteins Drp1, Fis1, Mfn1, Mfn2, Opa1 and CypD in the mutant Htt neurons treated with MitoQ and SS31 and in the untreated Htt neurons. The fission proteins Drp1 and Fis1 were significantly decreased; and the fusion proteins Mfn1, Mfn2, and Opa1, and the synaptic, synaptophysin, PSD95, and DARPP32 proteins were significantly increased in the MitoQ- and SS31-treated Htt neurons, indicating that MitoQ and SS31 reduce fission activity and enhance fusion and synaptic activity.
Protein levels in SS31-treated and untreated mutant Htt neurons
As shown in Figure 1A and B, in SS31-treated mutant Htt neurons compared with untreated mutant Htt neurons, significantly decreased levels of fission proteins Drp1 (P = 0.01) and Fis1 (P = 0.001) were found (Fig. 1A and B). In contrast, increased fusion protein levels of Mfn1 (P = 0.02), Mfn2 (P = 0.003) and Opa1 (P = 0.001) were found in SS31-treated mutant Htt neurons compared with the untreated Htt neurons (Fig. 1A and B). Levels of the matrix protein CypD were significantly decreased (P = 0.01) in the SS31-treated mutant Htt neurons. Significantly increased levels of synaptophysin (P = 0.01) and PSD95 (P = 0.01) were present in the SS31-treated mutant Htt neurons relative to untreated neurons (Fig. 1A and B). Expression levels of the medium-spiny neuronal protein marker DARRP32 (P = 0.01) were also significantly increased in the SS31-treated mutant Htt neurons relative to untreated mutant Htt neurons (Fig. 1A and B).
Immunofluorescence analysis
Immunofluorescence levels between MitoQ-treated and untreated mutant Htt neurons
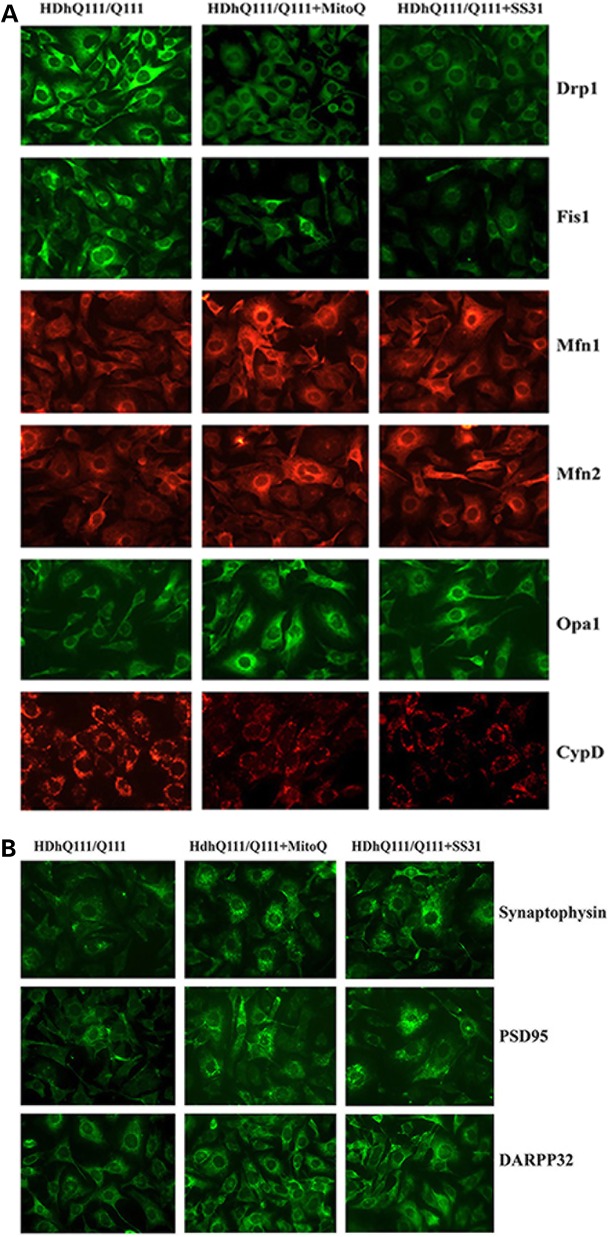
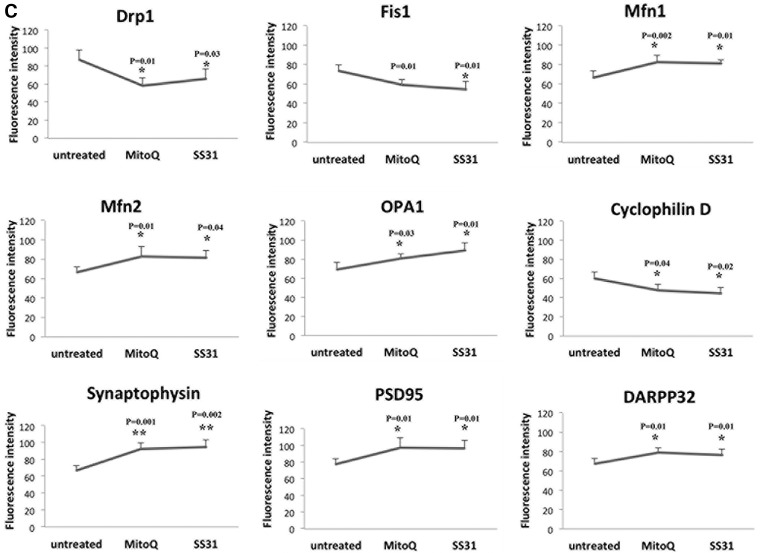
To determine the effect of mutant Htt on protein levels and localizations, we performed immunofluorescence analysis of MitoQ-treated and untreated mutant Htt neurons on mitochondrial fission proteins (Drp1 and Fis1), fusion proteins (Mfn1, Mfn2, and Opa1), the matrix protein CypD and synaptic proteins (synaptophysin and PSD95). As shown in Figure 2A and C, we found significantly decreased levels of Drp1 (P = 0.01) and Fis1 (P = 0.01), and of CypD (P = 0.04); and significantly increased levels of Mfn1 (P = 0.02), Mfn2 (P = 0.01) and Opa1 (P = 0.03) in the MitoQ-treated mutant Htt neurons relative to untreated Htt neurons, indicating that MitoQ enhances fusion activity and reduces fission activity in mutant Htt neurons. The synaptic proteins synaptophysin (P = 0.001) and PSD95 (P = 0.01) were significantly increased in the MitoQ-treated mutant Htt neurons. DARRP32 (P = 0.01) was also significantly increased in the MitoQ-treated mutant Htt neurons relative to the untreated neurons (Fig. 2B and C).
Figure 2.
Immunofluorescence analysis of proteins in MitoQ- and SS31-treated mutant Htt neurons and in untreated mutant Htt neurons. (A) Representative images of MitoQ- and SS31-treated mutant Htt neurons and untreated Htt neurons from matrix proteins. (B) Representative images of Mdivi1-treated and untreated mutant Htt neurons from synaptic proteins. (C) Quantitative immunofluorescence analysis of mitochondrial dynamics, and matrix and synaptic proteins Drp1, Fis1, Mfn1, Mfn2, Opa1, CypD, synaptophysin and PSD95 treated with MitoQ and SS31 and untreated. The fission and matrix proteins were significantly decreased, and the fusion proteins were significantly increased upon treatment with MitoQ and SS31, indicating that MitoQ and SS31 reduce mitochondrial fission activity and enhance fusion activity. Synaptic and DARPP32 proteins increased in the mutant Htt neurons that were treated with MitoQ and SS31 relative to the untreated mutant Htt neurons, indicating that MitoQ and SS31 enhance synaptic activity.
Immunofluorescence levels between SS31-treated and untreated mutant Htt neurons
Significantly decreased levels of Drp1 (P = 0.03), Fis1 (P = 0.01) and CypD (P = 0.02) were found in SS31-treated mutant Htt neurons relative to untreated neurons (Fig. 2A and C). The fusion proteins Mfn1 (P = 0.01), Mfn2 (P = 0.04) and Opa1 (P = 0.01) were increased in the SS31-treated mutant Htt neurons. These findings suggest that SS31 reduces fission activity and increases fusion activity in mutant Htt neurons. Our immunofluorescence findings agreed with these immunoblotting results.
Immunofluorescence analysis showed significantly increased levels of synaptophysin (P = 0.002) and PSD95 (P = 0.01) in the SS31-treated mutant Htt neurons relative to the untreated neurons, indicating that SS31 increases synaptic activity. Immunofluorescence analysis showed significantly increased levels of synaptophysin (P = 0.002) and PSD95 (P = 0.01) in the SS31-treated mutant Htt neurons relative to the untreated neurons (Fig. 2B and C), indicating that SS31 increases synaptic activity.
Transmission electron microscopy
The number of mitochondria in MitoQ-treated and untreated mutant Htt neurons
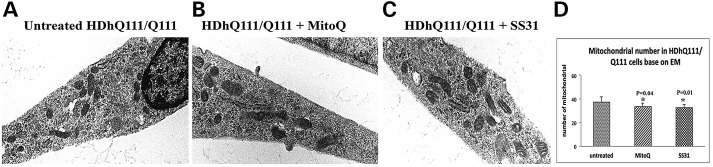
To determine the effects of MitoQ on the number of mitochondria, mutant Htt neurons were treated with MitoQ. The number of mitochondria was assessed, using transmission electron microscopy (TEM). The number of mitochondria significantly decreased following MitoQ treatment (P = 0.04) in the mutant Htt neurons relative to the untreated mutant Htt neurons (Fig. 3B and D).
Figure 3.
TEM of MitoQ- and SS31-treated mutant Htt neurons and untreated Htt neurons. (A) Fragmented and structurally damaged mitochondria in the mutant Htt neurons. (B) Intact mitochondria in the MitoQ-treated mutant Htt neurons. (C) Intact mitochondria in the SS31-treated mutant Htt neurons. (D) Results from the quantitative analysis of mitochondria. Significantly decreased numbers of mitochondria were found in the mutant neurons treated with MitoQ (P = 0.04) and SS31 (P = 0.01) compared with the mitochondria in the untreated mutant Htt neurons.
The number of mitochondria in SS31-treated and untreated mutant Htt neurons
To determine the effects of SS31 on the number of mitochondria, we also treated mutant Htt neurons with SS31 and determined the number of mitochondria using TEM. As shown in Figure 3C and D, we found that the number of mitochondria significantly decreased in mutant Htt neurons treated with SS31 (P = 0.01) relative to the number of mitochondria in the untreated SS31 mutant Htt neurons, indicating that SS31 reduces mitochondrial fission.
Mitochondrial function
Mitochondrial function in MitoQ-treated and untreated mutant Htt neurons
To determine differences in mitochondrial function in MitoQ-treated and untreated mutant Htt neurons, we measured and compared H2O2 production, lipid peroxidation, ATP production, cell viability and GTPase Drp1 enzymatic activity.
H2O2 production: As shown in Figure 4A, significantly decreased levels of H2O2 were found in the mitochondria from mutant Htt neurons treated with MitoQ (P = 0.03) relative to the mitochondria from untreated mutant Htt neurons, indicating that MitoQ decreases H2O2 levels.
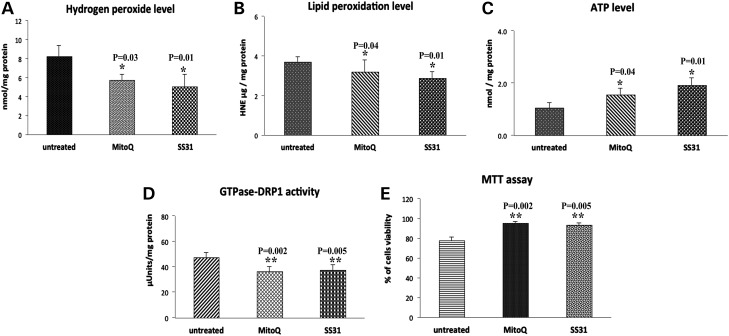
Figure 4.
Mitochondrial functional parameters in MitoQ- and SS31-treated mutant Htt neurons and in untreated Htt neurons. Mitochondrial function was assessed by measuring: (A) H2O2 production, (B) lipid peroxidation, (C) ATP levels, (D) GTPase Drp1 activity and (E) cell viability. Significantly decreased levels were found in the following parameters, in the mutant Htt neurons upon MitoQ and SS31 treatment: H2O2 with MitoQ (P = 0.03) and SS31 (P = 0.01); and 4-hydroxy-2-nonenol with MitoQ (P = 0.04) and SS31 (P = 0.01) and GTPase Drp1 activity for MitoQ (P = 0.002) and SS31 (P = 0.005). In contrast, significantly increased levels were found in the following parameters upon MitoQ and SS31 treatment: ATP production with MitoQ (P = 0.04) and SS31 (P = 0.01); and cell viability for MitoQ (P = 0.002) and SS31 (P = 0.005) concentrations.
Lipid peroxidation: Significantly decreased levels of 4-hydroxy-2-nonenol (HNE) lipid were found in mutant Htt neurons treated with MitoQ (P = 0.04) relative to the untreated mutant neurons, indicating that MitoQ reduces lipid peroxidation in MitoQ-treated mutant Htt neurons (Fig. 4B).
ATP production: Significantly increased levels of ATP were found in the MitoQ-treated mutant Htt neurons (P = 0.04) relative to the untreated Htt neurons, indicating that MitoQ enhances ATP levels in mutant Htt neurons (Fig. 4C).
GTPase Drp1 activity: Significantly decreased levels of GTPase Drp1 activity were found in the mutant Htt neurons treated with MitoQ (P = 0.002) relative to the untreated mutant Htt neurons, indicating that MitoQ decreases GTPase Drp1 activity in mutant Htt neurons. These findings suggest that MitoQ reduces fission-linked GTPase activity (Fig. 4D).
Cell viability: Cell viability was significantly increased in the MitoQ-treated mutant Htt neurons (P = 0.001) relative to the untreated mutant Htt neurons, indicating that MitoQ enhances cell viability (Fig. 4E).
Mitochondrial function in SS31-treated and untreated mutant Htt neurons
To determine the effect of SS31 on mitochondrial function in mutant Htt neurons, we measured mitochondrial function in SS31-treated and untreated mutant Htt neurons.
H2O2 production: Significantly decreased levels of H2O2 were found in the mitochondria from SS31-treated mutant neurons (P = 0.01) relative to untreated mutant Htt neurons (Fig. 4A).
Lipid peroxidation: As shown in Figure 4B, significantly decreased levels of lipid peroxidation were found in the SS31-treated mutant Htt neurons (P = 0.02) relative to the untreated mutant Htt neurons, indicating that SS31 reduces lipid peroxidation in mutant Htt neurons.
ATP production: Significantly increased levels of ATP were found in the SS31-treated Htt neurons (P = 0.01) relative to the untreated mutant Htt neurons (Fig. 4C).
Cell viability: Cell viability was significantly increased in the SS31-treated mutant Htt neurons (P = 0.002) relative to the untreated mutant Htt neurons (Fig. 4E).
GTPase Drp1 activity: Significantly decreased levels of GTPase activity were found in the SS31-treated mutant Htt neurons (P = 0.005) relative to the untreated mutant Htt neurons (Fig. 4D), indicating that SS31decreases GTPase Drp1 activity in the treated mutant Htt neurons.
Discussion
In this study, the effects of mitochondria-targeted molecules MitoQ and SS31 were studied using gene expression, biochemical methods, and TEM methods and mutant Htt (STHDhQ111/Q111) neurons. In the mutant Htt neurons treated with MitoQ and SS31, fission genes were down-regulated and fusion genes were up-regulated, suggesting that mitochondria-targeted molecules reduce fission activity, increase fusion activity and maintain mitochondrial dynamics in mutant Htt neurons. Interestingly, mitochondrial biogenesis was up-regulated in MitoQ- and SS31-treated mutant Htt, indicating that the targeting of mitochondria by molecules activate mitochondrial biogenesis in diseased neurons. Synaptic genes were up-regulated, and mitochondrial function was enhanced in MitoQ- and SS31-treated mutant Htt neurons, suggesting that the mitochondria-targeted molecules also enhance mitochondrial function and synaptic activities. The TEM studies revealed that reduced numbers of mitochondria were present in the MitoQ- and SS31-treated mutant Htt neurons compared with the untreated Htt neurons. These findings suggest that MitoQ and SS31 are protective against mutant Htt-induced mitochondrial and synaptic damage in HD neurons.
mRNA and protein levels
Recent studies using HD postmortem brains, HD cell cultures and HD mouse models revealed impaired mitochondrial dynamics (excessive fragmentation and reduced fusion), defective mitochondrial function, impaired mitochondrial axonal transport, and synaptic damage in HD pathogenesis (10,11,13,14,16,37). Earlier studies also reported that mutant Htt interacts with Drp1, induces the production of free radicals and causes excessive fragmentation of mitochondria, ultimately leading to mitochondrial dysfunction and synaptic damage in HD neurons (15,16). Multiple therapeutic strategies have been suggested and are currently being tested, using cell and mouse models of HD, to reduce (i) excessive mitochondrial fragmentation, using Mdivi1, which is known to inhibit excessive mitochondrial division (17) and (ii) mitochondrially generated free radicals, using MitoQ and SS31, which are known to reduce mitochondrial dysfunction and enhance neuronal activity (30,36).
To determine the effects of Mdivi1, we recently examined the beneficial effects of the mitochondria-targeted inhibitor, Mdivi1 on mutant Htt neurons (17). We found that Mdivil reduced mitochondrial fission in striatal neurons that express expanded polyQ (HDhQ111/111) repeats. We found increased mRNA levels of fission genes and decreased levels of fusion genes in the Mdivil-treated mutant Htt neurons, indicating that Mdivi1 reduces fission activity and enhances fusion machinery and synaptic activities in mutant Htt neurons (17). In this study, to determine the effects of mitochondria-targeted molecules MitoQ and SS31 against mutant Htt-induced mitochondrial damage and synaptic dysfunction in mutant Htt neurons, we measured mRNA levels of mitochondrial fission and fusion genes, ETC genes, and mitochondrial biogenesis and synaptic genes, using MitoQ and SS31 in mutant Htt neurons. In the MitoQ- and SS31-treated mutant Htt neurons, fission genes were down-regulated, and fusion genes were up-regulated, suggesting that mitochondria-targeted molecules reduce fission activity and enhance fusion activity, and maintain mitochondrial dynamics in HD neurons.
The levels of mRNA of mitochondrial biogenesis genes PGC1α, PGC1β, Nrf1, Nrf2 and TFAM were increased in the MitoQ- and SS31-treated mutant Htt neurons, indicating that mitochondrial biogenesis machinery is enhanced in mutant Htt neurons. Our TEM results showed reduced fragmentation of mitochondria and increased elongated and healthy mitochondria in MitoQ- and SS31-treated mutant Htt neurons relative to untreated mutant Htt neurons. These observations suggest that ‘defective mitochondrial biogenesis’ is reduced and ‘healthy mitochondrial biogenesis’ is elevated in MitoQ- and SS31-treated mutant Htt neurons. It is possible that mRNA levels of biogenesis genes were increased per mitochondrion in MitoQ- and SS31-treated mutant Htt neurons compared with single mitochondrion in MitoQ- and SS31-untreated mutant Htt neurons. Further research is needed to determine the mRNA levels of biogenesis genes in each of fragmented and elongated mitochondrion.
It is interesting to observe mRNA levels of all ETC genes were increased in MitoQ- and SS31-treated mutant Htt neurons relative to untreated mutant Htt neurons. However, statistical significance was found only for ND1, COX1-3 and ATP6 genes. The precise reasons for these altered mRNA levels are currently unknown. As discussed later, MitoQ- and SS31-treated mutant Htt neurons showed reduced mitochondrial dysfunction, particularly increased levels of mitochondrial ATP and cytochrome oxidase activity. Further, MitoQ and SS31 scavenged free radicals and maintained/boosted mitochondrial function in mutant Htt neurons. It is possible that MitoQ and SS31 increased mitochondrial ATP by activating mRNA levels of ETC genes. However, further research is needed to determine the underlying protective mechanism(s) of MitoQ and SS31 in mutant Htt neurons.
In addition, synaptic activity was found to be enhanced in MitoQ- and SS31-treated mutant Htt neurons by elevating synaptic mRNA levels of synaptophysin, PSD95, synapsin 1–2, synaptobrevins 1–2, neurogranin, GAP43 and synaptopodin, indicating that MitoQ and SS31 have a positive effect on synapses in mutant Htt neurons.
Mitochondrial dynamics, mitochondrial biogenesis and synaptic activity are critical components in maintaining mitochondrial function, particularly at synapses, and are defective in aging and neurodegenerative diseases, such as AD, HD, PD and ALS. Findings from this study strongly suggest that MitoQ and SS31 reduce impaired mitochondrial dynamics, enhance mitochondrial biogenesis, increase synaptic activity, and maintain neuronal function in HD neurons. Findings from this study may have implications to other neurodegenerative diseases such as AD (36–40), HD (11,15–17), PD (41), multiple sclerosis (42), ALS (43) and other diseases with expanded polyQ repeats, such as spinocerebellar ataxias 1, 2, 3, 6, 7, 17, spino-bulbar muscular atrophy and dentatorubral-pallidolusian atrophy (7).
In this study, we quantified protein levels of mitochondrial dynamics proteins, synaptic proteins and DARPP32, using immunoblotting and immunofluorescence analysis. Our protein data agreed with our mRNA data, indicating that MitoQ and SS31 affect not only mRNA but also protein levels in mutant Htt neurons. These observations confirm that MitoQ and SS31 affect phenotypic behavior of HD neurons by enhancing synaptic activities with elevated, or at least normal, mitochondrial ATP, suggesting that MitoQ and SS31 are promising drugs that may be able to treat HD neurons with increased numbers of polyQ repeats.
Transmission electron microscopy
It is well-established that structurally damaged mitochondria are present in HD neurons and peripheral cells from HD patients and in the primary neurons from HD mice (13,15–17,44,45). These structurally damaged mitochondria are mainly due to the interactions between Drp1-mutant Htt, and due to the subsequent elevation of GTPase Drp1 activity in HD neurons. In using TEM to better understand the ultra-structural changes induced by MitoQ and SS31, we found that Drp1 and Fis1 were reduced in both the MitoQ- and the SS31-treated mutant Htt neurons. We also found a reduction in the number of mitochondria in the MitoQ- and SS31-treated neurons relative to untreated mutant Htt neurons. These results indicate that mitochondria-targeted molecules reduce fission activity, enhance fusion machinery and synaptic activity and, most importantly, reduce defective mitochondrial function in mutant Htt neurons. These results suggest that mitochondria-targeted molecules are promising candidates for treating HD patients.
Mitochondrial function
Mitochondrial function was defective in neurons from HD patients and HD mice; in peripheral cells and primary cultures from HD mice; and in striatal neurons from HD knockin mice that express 111 CAG repeats (11,15–17,46–49). To determine the effects of MitoQ and SS31 on mitochondrial function, in this study we measured mitochondrial function in mutant Htt neurons following MitoQ and SS31 treatment. As expected, we found that the MitoQ- and SS31-treated mutant Htt neurons exhibited reduced free radicals and lipid peroxidation, and increased ATP production and neuronal viability. Further, the levels of GTPase Drp1 enzymatic activity were significantly decreased in the mutant Htt neurons treated with MitoQ and SS31. These findings strongly suggest that mitochondria-targeted molecules enhance or maintain mitochondrial function and neuronal viability by simply scavenging free radicals in the HD neurons.
In summary, findings from our study strongly indicate that the mitochondria-targeted molecules MitoQ and SS31 are promising candidates to treat HD neurons.
Materials and Methods
Cell cultures and Mdivi1 treatments
Immortalized striatal progenitor neurons expressing the homozygous mutant Htt (STHdh Q111/Q111) were used in this study. Cell lines were prepared from homozygous HdhQ111/Q111 knockin mice. Originally, mutant knockin HD mice were generated with 111 CAG repeats using homologous recombination approach—meaning exon 1 of mouse HD gene is replaced with human exon 1 with 111 CAG repeats (50,51). The cells were grown at 37°C in Dulbecco's modified Eagle's medium (Gibco, Carlsbad, CA, USA) and supplemented with 10% fetal bovine serum, 1% nonessential amino acids, 2 mm l-glutamine and 400 μg/ml G418 (Geneticin, Invitrogen, Carlsbad, CA, USA). The cells were treated with MitoQ (25 ηM) and SS31 (2.5 ηM) for 24 h and then were harvested. Pellets were prepared for RNA and protein studies.
Quantitative real-time RT–PCR
Using the reagent TriZol (Invitrogen), total RNA was isolated from three independent MitoQ and SS31 treatments of mutant Htt neurons (n = 3) from control, untreated mutant Htt (n = 3) and mutant Htt (n = 3) neurons. Using primer express software (Applied Biosystems, Carlsbad, CA, USA), we designed the oligonucleotide primers for the housekeeping genes β-actin, GAPDH, mitochondrial structural genes, fission genes (Drp1 and Fis1), fusion genes (MFN1, MFN2, Opa1 and VDAC1), the mitochondrial matrix protein CypD, mitochondrial biogenesis genes (PGC1α, PGC1β, Nrf1, Nrf2 and TFAM), mitochondrial-encoded ETC genes (Complex I: subunits 1, 3 and 6; Complex III: CytB; Complex IV: Cox1–3; and Complex V: ATPase), synaptic genes, synaptophysin, PSD95, synapsins 1–2, synaptobrevins 1–2, neurogranin, GPA43 and synaptopodin. The primer sequences and amplicon sizes are listed in Table 2. Using SYBR-Green, chemistry-based quantitative real-time RT–PCR, we measured mRNA expression of the genes identified earlier, as described by Manczak and Reddy (17) and Reddy et al. (52).
Table 2.
Summary of quantitative RT–PCR oligonucleotide primers used in measuring mRNA expression in mitochondrial dynamics and biogenesis genes, electron transport chain genes, and synaptic genes in mouse Htt-mutant cell lines treated with MitoQ and SS31
| Gene | DNA Sequence (5′-3′) | PCR product size |
|---|---|---|
| Mitochondrial structural genes | ||
| Drp1 | Forward primer ATGCCAGCAAGTCCACAGAA | 86 |
| Reverse primer TGTTCTCGGGCAGACAGTTT | ||
| Fis1 | Forward primer CAAAGAGGAACAGCGGGACT | 95 |
| Reverse primer ACAGCCCTCGCACATACTTT | ||
| MFN1 | Forward primer GCAGACAGCACATGGAGAGA | 83 |
| Reverse primer GATCCGATTCCGAGCTTCCG | ||
| MFN2 | Forward primer TGCACCGCCATATAGAGGAAG | 78 |
| Reverse primer TCTGCAGTGAACTGGCAATG | ||
| Cyclophilin D | Forward primer AGATGTCAAATTGGCAGGGGG | 91 |
| Reverse primer TGCGCTTTTCGGTATAGTGCT | ||
| Opa1 | Forward primer ACCTTGCCAGTTTAGCTCCC | 82 |
| Reverse primer TTGGGACCTGCAGTGAAGAA | ||
| VDCA1 | Forward primer CTCCCACATACGCCGATCTT | 58 |
| Reverse primer GCCGTAGCCCTTGGTGAAG | ||
| Mitochondrial biogenesis genes | ||
| PGC1α | Forward primer GCAGTCGCAACATGCTCAAG | 83 |
| Reverse primer GGGAACCCTTGGGGTCATTT | ||
| PGC1β | Forward primer CAGTGACATGAGCTCTCGGG | 73 |
| Reverse primer CAGCACCTGGCACTCTACAA | ||
| Nrf1 | Forward primer AGAAACGGAAACGGCCTCAT | 96 |
| Reverse primer CATCCAACGTGGCTCTGAGT | ||
| Nrf2 | Forward primer ATGGAGCAAGTTTGGCAGGA | 96 |
| Reverse primer GCTGGGAACAGCGGTAGTAT | ||
| TFAM | Forward primer TCCACAGAACAGCTACCCAA | 84 |
| Reverse primer CCACAGGGCTGCAATTTTCC | ||
| Mitochondrial-encoded electron transport chain genes | ||
| ND1 - CI | Forward primer CGGGCCCCCTTCGAC | 72 |
| Reverse primer GGCCGGCTGCGTATTCT | ||
| ND3 - CI | Forward primer TGTACTCAGAAAAAGCAAATCCATATG | 73 |
| Reverse primer AATAATAGAAATGTAATTGCTACCAAGAAAAA | ||
| ND6 - CI | Forward primer CCGCAAACAAAGATCACCCAG | 79 |
| Reverse primer GAAGGAGGGATTGGGGTAGC | ||
| CYT B - CIII | Forward primer TTATCGCGGCCCTAGCAA | 75 |
| Reverse primer TAATCCTGTTGGGTTGTTTGATCC | ||
| COX1 - CIV | Forward primer ATCACTACCAGTGCTAGCCG | 84 |
| Reverse primer CCTCCAGCGGGATCAAAGAA | ||
| COX2 - CIV | Forward primer CATCCCAGGCCGACTAAATC | 74 |
| Reverse primer TTTCAGAGCATTGGCCATAGAA | ||
| COX3 - CIV | Forward primer CAGGATTCTTCTGAGCGTTCTATCA | 72 |
| Reverse primer AATTCCTGTTGGAGGTCAGCA | ||
| ATP6 - CV | Forward primer TCCCAATCGTTGTAGCCATCA | 76 |
| Reverse primer AGACGGTTGTTGATTAGGCGT | ||
| Synaptic genes | ||
| Synaptophysin | Forward primer CTGCGTTAAAGGGGGCACTA | 81 |
| Reverse primer ACAGCCACGGTGACAAAGAA | ||
| PSD95 | Forward primer CTTCATCCTTGCTGGGGGTC | 90 |
| Reverse primer TTGCGGAGGTCAACACCATT | ||
| Synapsin 1 | Forward primer TGAGGACATCAGTGTCGGGTAA | 64 |
| Reverse primer GGCAATCTGCTCAAGCATAGC | ||
| Synapsin 2 | Forward primer TCCCACTCATTGAGCAGACATACT | 63 |
| Reverse primer GGGAACGTAGGAAGCGTAAGC | ||
| Synaptobrevin 1 | Forward primer TGCTGCCAAGCTAAAAAGGAA | 68 |
| Reverse primer CAGATAGCTCCCAGCATGATCA | ||
| Synaptobrevin 2 | Forward primer GGGACCAGAAGTTGTCGGAG | 89 |
| Reverse primer CTTGAGCTTGGCTGCACTTG | ||
| Neurogranin | Forward primer CTCCAAGCCAGACGACGATA | 83 |
| Reverse primer AACTCGCCTGGATTTTGGCT | ||
| GAP43 | Forward primer GCTGCGACCAAAATTCAGGC | 83 |
| Reverse primer GCTGGTGCATCACCCTTCT | ||
| Synaptopodin | Forward primer TCCTGCGCCCTGAACCTA | 70 |
| Reverse primer GACGGGCGACAGAGCATAGA | ||
| Housekeeping genes | ||
| β-Actin | Forward primer AGAAGCTGTGCTATGTTGCTCTA | 91 |
| Reverse primer TCAGGCAGCTCATAGCTCTTC | ||
| GAPDH | Forward primer TTCCCGTTCAGCTCTGGG | 59 |
| Reverse primer CCCTGCATCCACTGGTGC | ||
The mRNA transcript level was normalized against β-actin and GAPDH at each dilution. The standard curve was the normalized mRNA transcript level, plotted against the log-value of the input cDNA concentration at each dilution. To compare β-actin, GAPDH and neuroprotective markers, relative quantification was performed according to the CT method (Applied Biosystems). Briefly, the comparative CT method involved averaging triplicate samples, which were taken as the CT values for β-actin, GAPDH and neuroprotective markers. β-Actin normalization was used in the present study because the β-actin CT values were similar for the Htt neurons treated with MitoQ and SS31 and the untreated Htt neurons, for the mitochondrial ETC genes, the mitochondrial structural genes and the synaptic genes. The ΔCT-value was obtained by subtracting the average β-actin CT value from the average CT value of the synaptic mitochondrial ETC genes and the mitochondrial structural genes. The ΔCT of untreated Htt neurons was used as the calibrator. Fold change was calculated according to the formula 2^ − (ΔΔCT), where ΔΔCT is the difference between ΔCT and the ΔCT calibrator value. To determine the statistical significance of mRNA expression, the differences in CT value between the untreated WT and mutant Htt neurons, and the Mdivi1-treated WT and mutant Htt neurons were used in relation to β-actin normalization. Statistical significance was calculated using one-way ANOVA.
Immunoblotting analysis
To determine whether MitoQ or SS31 alters the protein levels of mitochondrial structural, ETC and synaptic genes that showed altered mRNA expression, we performed immunoblotting analyses of protein lysates from MitoQ- and SS31-treated and untreated mutant Htt neurons, as described in Manczak and Reddy (17). Twenty μg Protein lysates (20 μg) from the MitoQ- and SS31-treated and untreated mutant Htt neurons were resolved on a 4–12% Nu-PAGE gel (Invitrogen). The resolved proteins were transferred to nylon membranes (Novax Inc., San Diego, CA, USA) and were then incubated for 1 h at room temperature with a blocking buffer [5% dry milk dissolved in a tris-buffer saline tween 20 (TBST) buffer]. The nylon membranes were incubated overnight with the primary antibodies shown in Table 3. The membranes were washed with a TBST buffer three times at 10-min intervals and were then incubated for 2 h with appropriate secondary antibodies, followed by three additional washes at 10-min intervals. Mitochondrial and synaptic proteins were detected with chemiluminescence reagents (Pierce Biotechnology, Rockford, IL, USA), and the bands from immunoblots were quantified on a Kodak Scanner (ID Image Analysis Software, Kodak Digital Science, Kennesaw, GA, USA). Briefly, image analysis was used to analyze gel images captured with a Kodak Digital Science CD camera. The lanes were marked to define the positions and specific regions of the bands. An ID fine-band command was used to locate and to scan the bands in each lane and to record the readings.
Table 3.
Summary of antibody dilutions and conditions used in the immunoblotting analysis of mitochondrial dynamics, and synaptic and DARPP32 proteins, in the HDhQ111/Q111 cell lines treated with MitoQ and SS31
| Marker | Primary antibody, species and dilution | Purchased from company, city and state | Secondary antibody, dilution | Purchased from company, city and state |
|---|---|---|---|---|
| Drp1 | Rabbit Polyclonal 1:500 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Fis1 | Rabbit Polyclonal 1:500 | Protein Tech Group, Inc., Chicago, IL | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Mfn1 | Rabbit Polyclonal 1:400 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Mfn2 | Rabbit Polyclonal 1:400 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Opa1 | Mouse Monoclonal 1:500 | BD Biosciences, San Jose, CA | Sheep anti-mouse HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| CypD | Mouse Monoclonal 1:500 | EMD, Calobiochem Chemicals Inc., Gibbstown, NJ | Sheep anti-mouse HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| SYN | Rabbit Polyclonal 1:400 | Thermo Fisher Scientific Inc., Rockford, IL | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| PSD95 | Rabbit Monoclonal 1:300 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| DARPP-32 | Rabbit Polyclonal 1:200 | Santa Cruz Biotechnology, Inc., Dallas, TX | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| β-Actin | Mouse Monoclonal 1:500 | Sigma-Aldrich, St Luis, MO | Sheep anti-mouse HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
Immunofluorescence analysis and quantification
Immunofluorescence analysis was performed with the MitoQ- and SS31-treated and untreated mutant Htt neurons, as described in Manczak and Reddy (17). The Htt neurons were washed with warm phosphate buffered saline (PBS), fixed in freshly prepared 4% paraformaldehyde in PBS for 10 min, and then washed with PBS and permeabilized with 0.1% Triton-X100 in PBS. They were blocked with a 1% blocking solution (Invitrogen) for 1 h at room temperature. All neurons were incubated overnight with primary antibodies (see Table 4). After incubation, the neurons were washed three times with PBS, for 10 min each. The neurons were incubated with a secondary antibody conjugated with Fluors 488 and 599 (Invitrogen) for 1 h at room temperature. The neurons were washed three times with PBS and then mounted on slides. Photographs were taken with a multiphoton laser scanning microscope system (ZeissMeta LSM510). To quantify the immunoreactivity of mitochondrial and synaptic antibodies for each treatment, 10–15 photographs were taken at ×40 magnification. Signal intensity, indicating the immunoreactivity of the cell body, and neurite length were quantified for several randomly selected images, and statistical significance was determined using one-way ANOVA for mitochondrial and synaptic proteins.
Table 4.
Summary of antibody dilutions and conditions used in the immunohistochemistry/immunofluorescence analysis of mitochondrial dynamics, synaptic, and DARPP32 proteins in the HDhQ111/Q111 cell lines treated with MitoQ and SS31
| Marker | Primary antibody, species and dilution | Purchased from company, city and state | Secondary antibody, dilution, Alexa fluor dye | Purchased from company, city and state |
|---|---|---|---|---|
| Drp1 | Rabbit Polyclonal 1:300 | Novus Biological, Littleton, CO | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
| Fis1 | Rabbit Polyclonal 1:300 | Protein Tech Group, Inc., Chicago, IL | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
| Mfn1 | Rabbit Polyclonal 1:300 | Abcam, Cambridge, MA | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa594 | KPL, Gaithersburg, MD VECTOR Laboratories Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
| Mfn2 | Rabbit Polyclonal 1:200 | Novus Biological, Littleton, CO | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa594 | KPL, Gaithersburg, MD VECTOR Laboratories Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
| OPA1 | Mouse Monoclonal 1:500 | BD Biosciences, San Jose, CA | Goat anti-mouse Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
| CypD | Mouse Monoclonal 1:500 | EMD, Calobiochem Chemicals Inc., Gibbstown, NJ | Goat anti - mouse Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa594 | KPL, Gaithersburg, MD VECTOR Laboratories Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
| SYN | Rabbit Polyclonal 1:400 | Thermo Fisher Scientific Inc., Rockford, IL | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
| PSD95 | Rabbit Monoclonal 1:300 | Abcam, Cambridge, MA | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
| DARPP32 | Rabbit Polyclonal 1:200 | Santa Cruz Biotechnology, Inc., Dallas, TX | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
Transmission electron microscopy
To determine the effects of MitoQ and SS31 on the numbers of mitochondria in the mutant Htt neurons, we used TEM on mutant Htt neurons that underwent (n = 4) and that did not undergo (n = 4) MitoQ and SS31 treatment as described in Manczak et al. (30).
MitoQ- and SS31-treated and untreated Htt neurons were fixed in 100 mm sodium cacodylate (pH 7.2), 2.5% glutaraldehyde, 1.6% paraformaldehyde, 0.064% picric acid and 0.1% ruthenium red. They were gently washed and post-fixed for 1 h in 1% osmium tetroxide plus 08% potassium ferricyanide, in 100 mm sodium cacodylate, pH 7.2. After a thorough rinsing in water, the Htt neurons were dehydrated overnight in 1:1 acetone/Epon 812, for 1 h with 100% Epon 812 resin. The Htt neurons were then embedded in the resin. After polymerization, 60–80 nm thin sections were cut on a Reichert ultramicrotome and stained for 5 min in lead citrate. They were rinsed and post-stained for 30 min in uranyl acetate, and then were rinsed again and dried. Electron microscopy was performed at 60 kV on a Philips Morgagne TEM equipped with a CCD, and images were collected at magnifications of 1000–37 000 x. The numbers of mitochondria were counted in the MitoQ- and SS31-treated mutant Htt neurons, and statistical significance was determined using one-way ANOVA.
Mitochondrial functional assays
H2O2 production
Using an Amplex® Red H2O2 Assay Kit (Molecular Probes, Eugene, OR, USA), the production of H2O2 was measured in independent experiments (n = 4) of Mdivil-treated and untreated mutant and mutant Htt neurons, as described in Manczak and Reddy (17). Briefly, H2O2 production was measured in mitochondria isolated from the MitoQ-treated and untreated mutant Htt neurons. A BCA Protein Assay Kit (Pierce Biotechnology) was used to estimate protein concentration. The reaction mixture contained mitochondrial proteins (μg/μl), Amplex Red reagents (50 μM), horseradish peroxidase (0.1 U/ml) and a reaction buffer (1X). The mixture was incubated at room temperature for 30 min, followed by spectrophotometer readings of fluorescence (570 nm). Finally, H2O2 production was determined, using a standard curve equation expressed in nmol/μg mitochondrial protein.
Lipid peroxidation assay
Lipid peroxidates are unstable indicators of oxidative stress in neurons (53). The final product of lipid peroxidation is HNE, which was measured in the cell lysates from MitoQ- and SS31-treated (n = 4) and untreated mutant Htt neurons (n = 4), using an HNE-His ELISA Kit (Cell BioLabs, Inc., San Diego, CA, USA). Briefly, freshly prepared protein as added to a 96-well protein binding plate and was incubated overnight at 4°C. It was then washed three times with a buffer. After the last wash, the anti-HNE-His antibody was added to the protein in the wells, which was then incubated for 2 h at room temperature and was washed again three times. Next, the samples were incubated for 2 h at room temperature with a secondary antibody conjugated with peroxidase, followed by incubation with an enzyme substrate. Optical density was measured (at 450 nm) to quantify the level of HNE.
ATP levels
ATP levels were measured in mitochondria isolated from MitoQ- (n = 4) and SS31-treated (n = 4) and untreated (n = 4) mutant Htt neurons, using an ATP determination kit (Molecular Probes). The bioluminescence assay is based on the reaction of ATP with recombinant firefly luciferase and its substrate luciferin. Luciferase catalyzes the formation of light from ATP and luciferin. It is the emitted light that is linearly related to the concentration of ATP, which is measured with a luminometer. ATP from mitochondrial pellets was measured using a standard curve method.
Cell viability test (MTT assay)
Mitochondrial respiration, an indicator of cell viability, was assessed in the MitoQ- (n = 4) and SS31-treated (n = 4) and untreated (n = 4) mutant Htt neurons, using the mitochondrial-dependent reduction of 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) to formazan. Briefly, in this reduction, the Htt neurons were seeded in 12-well plates at a density of ∼105 neurons per well. After treatment, MTT (5 mg/ml in PBS) was added to the plates, and the neurons (control and experimental) were incubated for 3 h. The medium was then replaced with an sodium dodecyl sulfate/dimethyl solfoxide lysis buffer, and MTT absorption was measured at 570 nm. Results were expressed as the percentage of MTT reduction, assuming the absorbance of untreated Htt neurons was 100%.
GTPase Drp1 enzymatic activity
Using a calorimetric kit (Novus Biologicals, Littleton, CO, USA), GTPase enzymatic activity was measured in MitoQ- (n = 4) and SS31-treated (n = 4) and untreated (n = 4) mutant Htt neurons, following GTPase assay methods, described in Shirendeb et al. (15). The enzymatic activity was based on GTP hydrolyzing to GDP and to inorganic Pi. GTPase activity was measured on the basis of the amount of Pi that the GTP produces. By adding the ColorLock Gold (orange) substrate to the Pi generated from GTP, we assessed GTP activity, based on the inorganic complex solution (green). Colorimetric measurements (green) were read in the wavelength range of 650 nm. GTPase activity in the MitoQ- and SS31-treated and untreated mutant Htt neurons was compared.
Funding
This research was supported by National Institutes of Health grants (AG042178 and AG047812) and the Garrison Family Foundation.
Acknowledgements
We thank Coriell cell repositories of the Coriell Institute for Medical Research for the HDhQ111/Q111 cell lines.
Conflict of Interest statement: None declared.
References
- 1.Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P. Jr. (1985) Neuropathological classification of Huntington's disease. J. Neuropathol. Exp. Neurol., 44, 559–577. [DOI] [PubMed] [Google Scholar]
- 2.Montoya A., Price B.H., Menear M., Lepage M. (2006) Brain imaging and cognitive dysfunctions in Huntington's disease. J. Psychiatry. Neurosci., 31, 21–29. [PMC free article] [PubMed] [Google Scholar]
- 3.Byers R.K., Gilles F.H., Fung C. (1973) Huntington's disease in children. Neuropathologic study of four cases. Neurology, 23, 561–569. [DOI] [PubMed] [Google Scholar]
- 4.Spargo E., Everall I.P., Lantos P.L. (1993) Neuronal loss in the hippocampus in Huntington's disease: a comparison with HIV infection. J. Neurol. Neurosurg. Psychiatry, 56, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Huntington's Disease Collaborative Research Group. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 6.Reddy P.H., Williams M., Tagle D.A. (1999) Recent advances in understanding the pathogenesis of Huntington's disease. Trends Neurosci., 22, 248–255. [DOI] [PubMed] [Google Scholar]
- 7.Reddy P.H., Mao P., Manczak M. (2009) Mitochondrial structural and functional dynamics in Huntington's disease. Brain Res. Rev., 61, 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S., Li X.J. (2006) Multiple pathways contribute to the pathogenesis of Huntington disease. Mol Neurodegener., 1, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bates G.P. (2005) History of genetic disease: the molecular genetics of Huntington disease - a history. Nat. Rev. Genet., 6, 766–773. [DOI] [PubMed] [Google Scholar]
- 10.Shirendeb U., Reddy A.P., Manczak M., Calkins M.J., Mao P., Tagle D.A., Reddy P.H. (2011) Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington's disease: implications for selective neuronal damage. Hum. Mol. Genet., 20, 1438–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J., Moody J.P., Edgerly C.K., Bordiuk O.L., Cormier K., Smith K., Beal M.F., Ferrante R.J. (2010) Mitochondrial loss, dysfunction and altered dynamics in Huntington's disease. Hum. Mol. Genet., 19, 3919–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Squitieri F., Falleni A., Cannella M., Orobello S., Fulceri F., Lenzi P., Fornai F. (2010) Abnormal morphology of peripheral cell tissues from patients with Huntington disease. J. Neural. Transm. (Vienna), 117, 77–83. [DOI] [PubMed] [Google Scholar]
- 13.Costa V., Giacomello M., Hudec R., Lopreiato R., Ermak G., Lim D., Malorni W., Davies K.J., Carafoli E., Scorrano L. (2010) Mitochondrial fission and cristae disruption increase the response of cell models of Huntington's disease to apoptotic stimuli. EMBO Mol. Med., 2, 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Lim P.J., Karbowski M., Monteiro M.J. (2009) Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum. Mol. Genet., 18, 737–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirendeb U., Calkins M., Manczak M., Dufour B., McBride J., Mao P., Reddy P.H. (2012) Mutant huntingtin's association with mitochondria protein Drp1, and impaired axonal transport of mitochondria in Huntington's disease neurons. Hum. Mol. Genet., 21, 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., Poquiz P., Tjong J., Pouladi M.A., Hayden M.R. et al. (2011) Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med., 17, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manczak M., Reddy P.H. (2015) Mitochondrial division inhibitor 1 protects against mutant huntingtin-induced abnormal mitochondrial dynamics and neuronal damage in Huntington's disease. Hum. Mol. Genet., 24, 7308–7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy P.H., Shirendeb U.P. (2012) Mutant huntingtin, abnormal mitochondrial dynamics, defective axonal transport of mitochondria, and selective synaptic degeneration in Huntington's disease. Biochim Biophys Acta., 1822, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy P.H. (2008) Mitochondrial medicine for aging and neurodegenerative diseases. Neuromolecular Med., 10, 291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy P.H., Tripathi R., Troung Q., Tirumala K., Reddy T.P., Anekonda V., Shirendeb U.P., Calkins M.J., Reddy A.P., Mao P., Manczak M. (2012) Abnormal mitochondrial dynamics andsynaptic degeneration as early events in Alzheimer's disease: implications to mitochondria-targeted antioxidant therapeutics. Biochim. Biophys. Acta, 1822, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szeto H.H. (2008) Development of mitochondria-targeted aromatic-cationic peptides for neurodegenerative diseases. Ann. N. Y. Acad. Sci., 1147, 112–1121. [DOI] [PubMed] [Google Scholar]
- 22.Reddy P.H. (2006) Mitochondrial oxidative damage in aging and Alzheimer's disease:implications for mitochondrially targeted antioxidant therapeutics. J. Biomed. Biotechnol. 2006, 31372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheu S.S., Nauduri D., Anders M.W. (2006) Targeting antioxidants to mitochondria: a new therapeutic direction. Biochim Biophys Acta., 1762, 256–265. [DOI] [PubMed] [Google Scholar]
- 24.Murphy M.P., Smith R.A. (2007) Targeting antioxidants to mitochondria by conjugation to lipophilic cations. Annu. Rev. Pharmacol. Toxicol., 47, 629–656. [DOI] [PubMed] [Google Scholar]
- 25.Smith R.A., Murphy M.P. (2010) Animal and human studies with the mitochondria-targeted antioxidant MitoQ. Ann. N Y Acad. Sci., 1201, 96–103. [DOI] [PubMed] [Google Scholar]
- 26.Hwang P.M., Bunz F., Yu J., Rago C., Chan T.A., Murphy M.P., Kelso G.F., Smith R.A., Kinzler K.W., Vogelstein B. (2001) Ferredoxin reductase affects p53–dependent, 5 fluorouracil-induced apoptosis in colorectal cancer cells. Nat. Med., 7, 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jauslin M.L., Meier T., Smith R.A., Murphy M.P. (2003) Mitochondria-targeted antioxidants protect Friedreich Ataxia fibroblasts from endogenous oxidative stress more effectively than untargeted antioxidants. FASEB J., 17, 1972–1974. [DOI] [PubMed] [Google Scholar]
- 28.Bedogni B., Pani G., Colavitti R., Riccio A., Borrello S., Murphy M., Smith R., Eboli M.L., Galeotti T. (2003) Redox regulation of cAMP-responsive element-binding protein and induction of manganous superoxide dismutase in nerve growth factor-dependent cell survival. J. Biol. Chem., 278, 16510–16519. [DOI] [PubMed] [Google Scholar]
- 29.Dhanasekaran A., Kotamraju S., Kalivendi S.V., Matsunaga T., Shang T., Keszler A., Joseph J., Kalyanaraman B. (2004) Supplementation of endothelial cells with mitochondria-targeted antioxidants inhibit peroxide-induced mitochondrial iron uptake, oxidative damage, and apoptosis. J. Biol. Chem., 279, 37575–37587. [DOI] [PubMed] [Google Scholar]
- 30.Manczak M., Mao P., Calkins M.J., Cornea A., Reddy A.P., Murphy M.P., Szeto H.H., Park B., Reddy P.H. (2010) Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer's disease neurons. J. Alzheimers Dis., 20(Suppl. 2), S609–S631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szeto H.H. (2006) Cell-permeable, mitochondrial-targeted, peptide antioxidants. AAPSJ. 8, E277–EW283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho J., Wo K., Wu D., Soong Y., Liu S., Szeto H.H., Hong M.K. (2007) Potent mitochondria targeted peptides reduce myocardial infarction in rats. Coron. Artery. Dis., 18, 215–220. [DOI] [PubMed] [Google Scholar]
- 33.Thomas D.A., Stauffer C., Zhao K., Yang H., Sharma V.K., Szeto H.H., Suthanthiran M. (2007) Mitochondrial targeting with antioxidant peptide SS-31 prevents mitochondrial depolarization, reduces islet cell apoptosis, increases islet cell yield, and improves post transplantation function. J. Am. Soc. Nephrol., 18, 213–222. [DOI] [PubMed] [Google Scholar]
- 34.Cho S., Szeto H.H., Kim E., Kim H., Tolhurst A.T., Pinto J.T. (2007) A novel cell-permeable antioxidant peptide, SS31, attenuates ischemic brain injury by down-regulating CD36. J. Biol. Chem., 282, 4634–4642. [DOI] [PubMed] [Google Scholar]
- 35.Petri S., Kiaei M., Damiano M., Hiller A., Wille E., Manfredi G., Calingasan N.Y., Szeto H.H., Beal M.F. (2006) Cell-permeable peptide antioxidants as a novel therapeutic approach in a mouse model of amyotrophic lateral sclerosis. J. Neurochem., 98, 1141–1148. [DOI] [PubMed] [Google Scholar]
- 36.Calkins M.J., Manczak M., Mao P., Shirendeb U., Reddy P.H. (2011) Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer's disease. Hum. Mol. Genet., 20, 4515–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manczak M., Calkins M.J., Reddy P.H. (2011) Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Hum. Mol. Genet., 20, 2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Su B., Siedlak S.L., Moreira P.I., Fujioka H., Wang Y., Casadesus G., Zhu X. (2008) Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc. Natl. Acad. Sci. USA., 105, 19318–19323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Su B., Lee H.G., Li X., Perry G., Smith M.A., Zhu X. (2009) Impaired balance of mitochondrial fission and fusion in Alzheimer's disease. J. Neurosci., 29, 9090–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva D.F., Selfridge J.E., Lu J., Lezi E.L., Roy N., Hutfles L., Burns J.M., Michaelis E.K., Yan S., Cardoso S.M., Swerdlow R.H. (2013) Bioenergetic flux, mitochondrial mass and mitochondrial morphology dynamics in AD and MCI cybrid cell lines. Hum. Mol. Genet., 22, 3931–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X., Yan M.H., Fujioka H., Liu J., Wilson-Delfosse A., Chen S.G., Perry G., Casadesus G., Zhu X. (2012) LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum. Mol. Genet., 21, 1931–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao P., Manczak M., Shirendeb U.P., Reddy P.H. (2013) MitoQ, a mitochondria-targeted antioxidant, delays disease progression and alleviates pathogenesis in an experimental autoimmune encephalomyelitis mouse model of multiple sclerosis. Biochim. Biophys. Acta, 1832, 2322–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magrané J., Cortez C., Gan W.B., Manfredi G. (2014) Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum. Mol. Genet., 23, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trushina E., Dyer R.B., Badger J.D. II, Ure D., Eide L., Tran D.D., Vrieze B.T., Legendre Guillemin V., McPherson P.S., Mandavilli B.S. et al. (2004) Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell Biol., 24, 8195–8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo X., Disatnik M.H., Monbureau M., Shamloo M., Mochly-Rosen D., Qi X. (2013) Inhibition of mitochondrial fragmentation diminishes Huntington's disease-associated neurodegeneration. J. Clin. Invest., 123, 5371–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borrell-Pagès M., Zala D., HμMbert S., Saudou F. (2006) Huntington's disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol. Life Sci., 63, 2642–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Browne S.E., Bowling A.C., MacGarvey U., Baik M.J., Berger S.C., Muqit M.M., Bird E.D., Beal M.F. (1997) Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann. Neurol., 41, 646–653. [DOI] [PubMed] [Google Scholar]
- 48.Tabrizin S.J., Cleeter M.W., Xuereb J., Taanman J.W., Cooper J.M., Schapira A.H. (1999) Biochemical abnormalities and excitotoxicity in Huntington's disease brain. Ann. Neurol., 45, 25–32. [DOI] [PubMed] [Google Scholar]
- 49.Pandey M., Varghese M., Sindhu K.M., Sreetama S., Navneet A.K., MohanakμMar K.P., Usha R. (2008) Mitochondrial NAD+-linked State 3 respiration and complex-I activity are compromised in the cerebral cortex of 3– nitropropionic acid-induced rat model of Huntington's disease. J. Neurochem., 104, 420–434. [DOI] [PubMed] [Google Scholar]
- 50.Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V.C., Sharp A.H., Persichetti F., Cattaneo E., MacDonald M.E. (2000) Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet., 9, 2799–2809. [DOI] [PubMed] [Google Scholar]
- 51.Wheeler V.C., White J.K., Gutekunst C.A., Vrbanac V., Weaver M., Li X.J., Li S.H., Yi H., Vonsattel J.P., Gusella J.F. et al. (2000) Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum. Mol. Genet., 9, 503–513. [DOI] [PubMed] [Google Scholar]
- 52.Reddy T.P., Manczak M., Calkins M.J., Mao P., Reddy A.P., Shirendeb U., Park B., Reddy P.H. (2011) Toxicity of neurons treated with herbicides and neuroprotection by mitochondria targeted antioxidant SS31. Int. J. Environ. Res. Public Health, 8, 203–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hall E.D., Bosken J.M. (2009) Measurement of oxygen radicals and lipid peroxidation in neural tissues. Curr. Protoc. Neurosci., 7, 1–51. [DOI] [PubMed] [Google Scholar]