Abstract
Meta-analysis strategies have become critical to augment power of genome-wide association studies (GWAS). To reduce genotyping or sequencing cost, many studies today utilize shared controls, and these individuals can inadvertently overlap among multiple studies. If these overlapping individuals are not taken into account in meta-analysis, they can induce spurious associations. In this article, we propose a general framework for adjusting association statistics to account for overlapping subjects within a meta-analysis. The key idea of our method is to transform the covariance structure of the data, so it can be used in downstream analyses. As a result, the strategy is very flexible and allows a wide range of meta-analysis methods, such as the random effects model, to account for overlapping subjects. Using simulations and real datasets, we demonstrate that our method has utility in meta-analyses of GWAS, as well as in a multi-tissue mouse expression quantitative trait loci (eQTL) study where our method increases the number of discovered eQTL by up to 19% compared with existing methods.
Introduction
Meta-analysis of genome-wide association studies (GWAS) has become crucial to scale up genetic discovery (1,2). Using meta-analysis, investigators are able to combine many studies to dramatically increase sample size and thereby to increase statistical power to detect phenotypically associated variants. In recent years, large-scale meta-analyses have become essential to increase the number of known associated loci in many human diseases (3,4). Indeed, almost all GWAS publications today employ meta-analysis methodologies (5).
A range of meta-analysis methods are now commonly used in GWAS (6). The fixed effects (FE) model, which assumes that effect sizes are fixed across studies, is powerful when its assumption holds true (7,8). The random effects (RE) model assumes that the effect sizes can be different between studies, a phenomenon called heterogeneity (9). We recently proposed a RE model that is more powerful than FE if the data are heterogeneous (10). Additional categories of meta-analysis methods include the P-value-based approaches (11,12); the subset approaches, which assume that the effects are present or absent in studies (13,14); and the Bayesian approaches (15–17). The majority of these methods assume that component studies are independent and that the individuals collected in one study are separate and unrelated from those in other studies.
Many studies today utilize shared controls to reduce genotyping or sequencing cost. For example, the 1958 British Birth Cohort (18) is being widely used in recent GWAS (19–23). As a result, there can be inadvertent overlap between individuals across multiple studies. The resulting dependencies can induce spurious associations if the overlapping subjects are not explicitly removed prior to meta-analysis or taken into account during meta-analysis. A simple solution would be to manually split the overlapping subjects into separate studies, but this can be suboptimal in terms of statistical power (24). Moreover, splitting genotype data is often not possible in practice if the primary genotype data are not shared; this is a common situation in current GWAS due to the sensitivities of sharing individual-level data.
Previous studies addressed this challenge, but only focusing on specific meta-analysis models. Lin and Sullivan (24) proposed a FE meta-analysis approach that takes into account overlapping subjects. Zaykin and Kozbur (12) and Bhattacharjee et al. (13) extended this method to the P-value-based approach and subset approach, respectively. None of these approaches are able to account for dependency structure in other meta-analysis frameworks such as the RE models. Application of the RE models to meta-analyses recently became crucial because of its potential to go beyond standard study designs. Investigators are now increasingly using meta-analysis to combine data from multiple related diseases in order to identify shared pathways (25,26), to combine data from multiple tissues to identify shared expression quantitative trait loci (eQTL) (27,28) and to combine neuroimaging data of different psychiatric diseases to identify shared brain features (29). In these new study designs, the use of RE models is ideal because the effect sizes are not necessarily the same between datasets. However, if dependency structure exists in the data, the current RE models cannot be applied due to the independence assumption.
Another practical challenge in using the current solutions to dependency structure is that one cannot use widely used pipelines for GWAS meta-analysis. Many analysis pipelines utilize standard software packages implementing meta-analysis models (2,30). These packages have advantages in solving practical issues in data cleaning, quality control and imputation (2). However, many of these do not support overlapping subjects and therefore cannot be used if subjects are shared between studies.
In this article, we propose a flexible and general framework for meta-analyzing dependent studies. The philosophy of our framework is unique, in that our framework does not compete with existing methods; it enables them to account for overlapping subjects. Given dependent studies, our framework transforms the covariance structure of the data, so it can be used in methods that strictly assume independence between studies. Specifically, our method decouples dependent studies into independent studies, adjusting association statistics to account for uncertainties in dependent studies. As a result, our approach enables general meta-analysis methods, including the FE and RE models, to account for overlapping subjects. Existing pipelines implementing these models can be reused for analyzing dependent studies, as long as our framework is applied as a front end of the analysis procedure.
We applied our approach to the eQTL data of Sul et al. (27), which involves data from multiple mouse tissues. Using our framework to combine information from multiple tissues and to account for tissue heterogeneity with the RE model, we increased the number of discovered eQTL by up to 19% compared with existing methods. We also performed a meta-analysis of three autoimmune diseases with shared controls using the Wellcome Trust Case Control Consortium (WTCCC) data (18) to demonstrate that our framework improved statistical significance of known candidate loci associated to multiple autoimmune diseases.
Results
Overview of the method
We propose a general approach that accounts for the dependency structure between studies in a meta-analysis. As we review in the Materials and Methods section, many traditional as well as recently proposed meta-analysis methods cannot account for overlapping subjects (Supplementary Material, Table S1). Our goal was to develop a framework that could be generally applied to existing approaches, so that methods not yet accounting for dependency could, and so that existing software packages could be reused. To this end, we can transform the covariance structure of the data by decoupling dependent studies into independent studies. Specifically, to account for uncertainties between studies that are dependent, we increase the standard errors in the decoupled studies. In Figure 1, we have three studies, A, B and C, whose statistics are correlated and have unit variances. Our approach decouples these studies into independent studies that have the same information with respect to the summary statistic. The variances of A, B and C increase to 1.39, 2.52 and 1.24, respectively. The variance of Study B increases the most drastically to 2.52 because B was highly correlated to A and C, with correlations of 0.5 and 0.3, respectively. High correlations to other studies imply that this study has the smallest information content as an independent study. We can then use the decoupled studies in the downstream meta-analysis approaches.
Figure 1.
A simple example of our decoupling approach. Ω and are the covariance matrices of the statistics of three studies A, B and C before and after decoupling, respectively. The thickness of the edges denotes the amount of correlation between the studies. After decoupling, the size of the nodes reflects the information that the studies contain in terms of the inverse variance.
False-positive rate
One of the primary goals of decoupling is to prevent spurious associations that can occur if we ignore overlapping subjects in a meta-analysis. Thus, we performed simulations to compare the false-positive rates of different methods: (a) a naive method that does not account for overlapping subjects, (b) a splitting method that splits shared controls into individual studies and (c) a decoupling method that systematically accounts for overlapping subjects. We simulated genotypes assuming a SNP with the minor allele frequency (MAF) P = 0.3. We constructed five studies whose numbers of cases are 1000, 2000, 3000, 4000 and 5000. We assumed that these studies share 10 000 controls. In the splitting method, we equally split controls, allotting 2000 controls per study. Under the null hypothesis of no association of the SNP to the trait, we generated 1 million such study sets. We applied the FE model to all three methods.
Table 1 shows that both decoupling and splitting correctly control the false-positive rate. However, the naive method that ignores overlapping subjects does not control the false-positive rate correctly. We calculated the ratios between the estimated false-positive rates and the significance threshold α. The ratios for decoupling and splitting are consistently close to 1.0 (mean ratio over different thresholds, 0.98 for decoupling and 1.08 for splitting). However, the ratio of the naive method is very large (mean ratio of 6.65). In fact, the ratio becomes larger as the threshold becomes more significant, the maximum being 13.56 at threshold α = 5 × 10−5. This phenomenon emphasizes that ignoring overlapping subjects will incur a large number of false positives at the extreme threshold of 5 × 10−8 that is typically used in GWAS. We also applied the RE model of Han and Eskin (10) to the three methods. Decoupling conservatively controlled false-positive rate (mean ratio of 0.70), and the naive method again had an inflated false-positive rate (mean ratio of 5.40, see Supplementary Material, Table S2). Additionally, to explore a wider range of realistic scenarios, instead of fixing sample sizes of five studies, we randomly sampled sample sizes for simulations. We sampled case sample size of each study from N(3000, 10002), with lower bound 500, and assumed study-specific control samples whose size is sampled from N(3000, 10002). We assumed 10 000 shared controls and similarly repeated 1 million null simulations. Supplementary Material, Table S3 shows that the results are largely unchanged. Decoupling and splitting controlled false-positive rate well. Decoupling RE showed similar conservative tendency (mean ratio of 0.72).
Table 1.
False-positive rates of different methods
| Threshold α | 0.05 | 0.01 | 0.005 | 0.001 | 5 × 10−4 | 1 × 10−4 | 5 × 10−5 |
| Decoupling | 0.0496 (0.99) | 0.00997 (1.00) | 0.00499 (1.00) | 0.000992 (0.99) | 0.0005 (1.00) | 8.7 × 10−5 (0.87) | 4.9 × 10−5 (0.98) |
| Splitting | 0.0499 (1.00) | 0.01 (1.00) | 0.00501 (1.00) | 0.00104 (1.04) | 0.000539 (1.08) | 0.000123 (1.23) | 5.9 × 10−5 (1.18) |
| Naive | 0.1 (2.00) | 0.0308 (3.08) | 0.0186 (3.72) | 0.0059 (5.90) | 0.00354 (7.09) | 0.00112 (11.20) | 0.000678 (13.56) |
Naive refers to a method that ignores overlapping subjects. Splitting refers to a strategy that splits shared controls into individual studies. All three methods used the FE model. The numbers in parentheses are the ratios of estimated false-positive rates and the threshold α. We expect these numbers to be close to 1.0.
Power
Decoupling can allow the effective use of shared controls to increase power, when compared with the power of splitting individuals into individual studies. Using simulations, we compared the power of the decoupling and splitting methods. Among the three methods that we evaluated for false-positive rate, we excluded the naive method that ignored shared controls because the naive method did not control the false-positive rate properly. We assumed the same number of cases and shared controls as in the false-positive rate simulations above (unequal case sample sizes from 1000 to 5000, and shared controls of 10 000 which are equally split in the splitting method). However, this time, we assumed that the SNP had a genetic effect on the trait with relative risk γ. Given population MAF p, the MAF in cases becomes
Based on this frequency, we generated 10 000 simulation sets, each consisting of 5 case/control studies. In each set, we calculate the FE model P-values for both decoupling and splitting. We approximated power as the proportion of the sets in which the P-value exceeded the threshold of 5 × 10−8. We gradually increased γ and evaluated the power of the decoupling and splitting methods. Figure 2 shows that decoupling consistently outperforms splitting at all the relative risks we tested. For example, at relative risk 1.13, the power of decoupling is 78.2%, whereas the power of splitting is 70.4%. This result is in line with Lin and Sullivan's (24) observation that their approach (which is equivalent to decoupling applied to FE) outperforms splitting, unless controls are split into studies proportionally to case sample sizes, in which situation both methods will have similar power.
Figure 2.
Power of different methods. Splitting refers to a strategy that splits shared controls into individual studies.
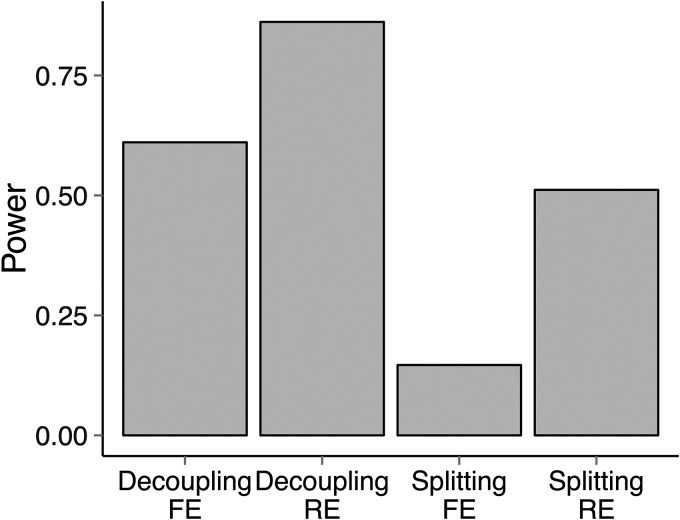
One advantage of decoupling is that it allows a wide range of meta-analysis methods to account for overlapping subjects, including the RE model. Using simulations, we demonstrated that we can gain power by using decoupling with the RE model in situations where effect sizes are heterogeneous between studies. Assuming the same simulation frameworks as in the previous power simulations, we additionally assumed that only two out of the five studies have genetic effects with relative risk 1.2. We assumed that the two studies having the largest case sample sizes (4000 and 5000) have effects. Then, after decoupling, we applied both the FE model and Han and Eskin's (10) RE model. We also used the splitting method and applied FE and RE. Figure 3 shows that in this situation, RE gains power over FE. When applied after decoupling, the power of RE is 86%, whereas the power of FE is 61%. Thus, we gained 25% more power by decoupling, because decoupling allowed the application of RE for dependent studies. Splitting has suboptimal power in this simulation as well. When applied after splitting, the power of RE and FE is only 51 and 14%, respectively. Note that the power difference between decoupling FE and splitting FE is even more prominent in this simulation than in Figure 2, because of our design that assumes effects in the two studies having the largest case sample sizes. Since we equally split controls in our simulation, which is equivalent to down-weighting studies having large case sizes, the power drop of splitting compared with decoupling is dramatic in this design.
Figure 3.
Power of different methods when heterogeneity exists. We assumed that only two out of five studies had an effect size of relative risk 1.2. Splitting refers to a strategy that splits shared controls into individual studies. FE, fixed effects model. RE, random effects model of Han and Eskin (10).
Cross-disease analysis using the WTCCC data
One application of the decoupling approach is cross-disease analysis, in which controls are shared between diseases. Because it is reasonable to assume that effect size varies between different diseases, it is ideal to use the RE model. Thus, in this design, decoupling can help because it allows the application of RE models when studies share controls. As an example of cross-diseases analysis, we applied our decoupling approach to the WTCCC data. The WTCCC has performed GWAS on seven diseases, including three autoimmune diseases (Crohn's disease, rheumatoid arthritis and type 1 diabetes; CD, RA and T1D in short), using a set of shared controls (18). We performed meta-analysis on the three autoimmune diseases to identify shared loci between them.
We compared three methods: the naive method that ignores the fact that the studies share controls, the splitting method that splits controls to each disease and the decoupling method. For all three methods, we applied both the FE model and the Han and Eskin's (10) RE model. In the splitting method, we assumed that each individual is assigned to one of the three diseases with equal probability. We considered 397 450 SNPs that passed quality control for all three diseases in the original publication (18) and whose MAF are greater than 1%.
Supplementary Material, Figure S1 shows that the qq-plot is inflated for the naive method, which ignores overlapping subjects, for both FE and RE. The genomic control factors (31) were and for FE and RE, respectively. In contrast, the splitting and decoupling methods did not have such inflation. The genomic control factors were and for splitting, and and for decoupling.
We then examined the P-values of three non-MHC loci that were originally reported in the WTCCC analyses as candidates for shared loci between autoimmune diseases. Both splitting and decoupling gave significant P-values at those loci (P< 1.2 × 10−7, Bonferroni corrected for 397 450 tests, see Table 2). However, decoupling gave much more significant P-values than splitting did. For example, at PTPN22, the decoupling RE P-value (1.00 × 10−29) is about seven orders of magnitude more significant than the splitting RE P-value (1.88 × 10−22). At these loci, RE gave more significant P-values than FE did for both splitting and decoupling (Table 2 and Supplementary Material, Fig. S2A). This is due to the effect size heterogeneity at these loci between different diseases.
Table 2.
Association results of a meta-analysis combining CD, RA and T1D from the WTCCC data
| Gene | RSID | Chr. | Position | Meta-analysis P-values |
|||
|---|---|---|---|---|---|---|---|
| Splitting FE | Splitting RE | Decoupling FE | Decoupling RE | ||||
| PTPN22 | rs6679677 | 1 | 114 015 850 | 1.45 × 10−13 | 1.88 × 10−22 | 4.48 × 10−23 | 1.00 × 10−29 |
| SH2B3 | rs17696736 | 12 | 110 949 538 | 1.93 × 10−09 | 1.18 × 10−09 | 7.72 × 10−10 | 3.40 × 10−10 |
| PTPN2 | rs2542151 | 18 | 12 769 947 | 1.14 × 10−07 | 9.03 × 10−08 | 3.74 × 10−08 | 5.16 × 10−08 |
The three non-MHC loci reported in the WTCCC combined analysis (18) are presented. The candidate gene for each locus was obtained from the ImmunoBase website (www.immunobase.org).
To test the robustness of methods against noise, we added the other four diseases as noise into the meta-analysis. Supplementary Material, Figure S2B shows that when we meta-analyze all seven diseases, the absolute magnitudes of the significance of PTPN22 are reduced, but it is still significant for both the decoupling FE and the decoupling RE. Moreover, the relative significance gain of decoupling RE compared with decoupling FE is still largely pronounced (PTPN22: and ).
Multi-tissue eQTL analysis
Recently, Sul et al. (27) applied our decoupling approach to multi-tissue eQTL analysis. Their goal was to combine information from multiple tissues to increase power for eQTL identification. They proposed a linear mixed model that accounts for multiple tissues being collected from the same individuals. However, using this approach, the resulting effect size estimates that are to be combined become correlated, which prevents the application of the standard meta-analysis method. Moreover, it is reasonable to assume effect size heterogeneity between tissues. Thus, decoupling with the RE model may assist in analysis. In their study, Sul et al. (27) compared the performance of decoupling FE, decoupling RE and the tissue-by-tissue approach (TBT) in terms of the number of eQTL identified. In TBT, they call an eQTL significant if any of the q-values in the set of 4 or 10 tissues is smaller than the false discovery rate (FDR) threshold. They demonstrated that applying decoupling with FE found a much larger number of eQTL in 4 or 10 mouse tissues than applying TBT, and applying decoupling with RE found even more at the same FDR level of 5%.
We revisited this dataset and examined whether the relative performances between methods remained unchanged if we changed the FDR threshold. See Sul et al. (27) for a detailed description of the dataset. When we varied the FDR threshold down to 0.0001 and up to 0.2, the relative performances of the methods did not change (Fig. 4). For example, at the threshold of FDR = 0.0001, TBT, decoupling FE and decoupling RE discovered 61, 718 and 780 eQTL, respectively, in 10 tissues. At the threshold of FDR = 0.2, TBT, decoupling FE and decoupling RE discovered 3870, 4914 and 5855 eQTL, respectively, in 10 tissues. Thus, decoupling RE improved the number of discovered eQTL by up to 19% compared with the best competing method (decoupling FE). These observations underscore that in multi-tissue eQTL analysis, it is important to take into account both the between-tissue heterogeneity and the between-tissue correlations, and that the decoupling RE approach can handle both of them effectively.
Figure 4.
The number of eQTL identified by different methods in the 4 or 10 mouse tissue dataset of Sul et al. (27). We applied decoupling to account for the correlation of estimates between tissues caused by within-individual correlations between tissues. TBT, tissue-by-tissue approach that examines each tissue separately and takes the minimum q-value.
Simulations of decouplable matrices
We define the covariance matrix of the studies in a meta-analysis as decouplable if the decoupled variances are non-negative. In the Materials and Methods section, we analytically demonstrate that when the number of studies is two and the two studies share controls, the covariance matrix is always decouplable. To demonstrate that the covariance matrix is highly likely to be decouplable for more than two studies sharing controls, we performed extensive simulations. We assumed a situation where 10 studies share controls. We assumed the same MAF of 0.3 and odds ratio of 1.2 for all studies. Then, to simulate differences in the sample sizes between studies, we made the number of cases in five studies large and the number of cases in the other five studies small. Specifically, the number of cases of study i was for five studies and for the other five studies. We then varied from 1000 to 20 000 and δ from 0 to 4500 when μ was fixed at 5000. In this simulation, we observed that for all configurations we tested, the matrices were decouplable. Supplementary Material, Figure S3 shows that for all points, the percentage of studies that have negative variance is consistently zero in this simulation. This empirical result demonstrates that in most practical and common situations where studies share controls, the matrices are highly likely to be decouplable.
Discussion
We proposed a general framework for dealing with dependency structure in a meta-analysis. Our approach is philosophically different from other approaches, in that our method transforms the covariance structure of the data so that it can be used in other approaches that assume independence. Therefore, our approach flexibly allows many meta-analysis methods, such as the RE model, to account for dependency between studies and overlapping subjects. The simulations and applications to the WTCCC data support the utility of our approach. Moreover, our method allows the use of standard software packages designed for meta-analysis of GWAS, even when studies share subjects. Our method can be used beyond the standard meta-analysis designs, such as for multi-tissue eQTL analysis.
In our method, the covariance matrix can be non-decouplable in theory (the decoupled variances can be negative). However, as we analytically and empirically showed, for the common and typical situations where controls are shared, the matrices are likely decouplable. However, a covariance matrix that is not decouplable is certainly possible because of numerical instabilities when estimating the variances of effect size estimates. For such situations where a matrix is not decouplable, we can use a heuristic of dropping the least informative (smallest) studies until the variances become positive. If the test turns out to be moderately significant, we can revisit the data and apply the standard Lin and Sullivan (24) approach.
Our method builds upon the Lin and Sullivan (24) approach. Our method is optimal in the same sense that the Lin and Sullivan (24) approach is optimal for the FE model. Thus, the optimality with respect to power is guaranteed only under the FE model. Our false-positive rate simulations demonstrated that our approach has some conservative tendencies under the RE model. However, we showed that the decoupling RE approach has much higher power than the decoupling FE approach (which is equivalent to the Lin and Sullivan (24) approach) in both simulations and in real data, showing that decoupling can be a flexible and useful tool despite the fact that it might not be optimal for all methods. Optimal solutions to account for dependency structure in each meta-analysis method will be an interesting and important topic for further research.
In this article, we demonstrated the false-positive rate of different methods such as decoupling RE only by simulations. Obviously, the value of simulations is limited to the conditions chosen. Nevertheless, we explored a wide range of situations by using randomly chosen sample sizes as well as fixed sample sizes (Supplementary Material, Table S3) so that our simulations can represent realistic scenarios. In future studies, we expect that analytical characteristics of decoupling can be further investigated, which may establish theoretical connection of decoupling to other meta-analysis methods than FE.
We note that one should be careful in interpreting data based on decoupled studies. For example, the heterogeneity testing such as Cochran's Q test (1) is highly conservative when using decoupled studies and may not detect true heterogeneity. Unfortunately, there is no good alternative to Cochran's Q test and the estimate (32) that can take into account correlated data. Developing such methods will also be an interesting future research area.
Materials and Methods
Review of meta-analysis methods
To better discuss our proposed analysis method, we first briefly describe several existing meta-analysis methods.
Fixed effects model
The FE model assumes that the magnitude of the effect size is fixed across studies (7,8). Two widely used FE methods are the inverse-variance-weighted effect size method (1) and the weighted sum of z-scores method (2). Because these two methods are approximately equivalent (10), we only describe the inverse-variance-weighted effect size method. Let be the effect size estimates in N independent studies (such as the log odds ratios or regression coefficients). Let be the standard error of , and let . Let be the inverse variance. The inverse-variance-weighted summary effect size is
| (1) |
The variance of is
| (2) |
Because the standard error of is , we can construct a summary z-score:
that follows under the null hypothesis of no association. The P-value is calculated as follows:
where Φ is the cumulative density function of the standard normal distribution. The traditional FE model cannot account for overlapping subjects.
Random effects model
The RE model assumes that the effect sizes vary among studies. In this model, the effect size is assumed to follow an underlying probability distribution with mean μ and variance . There are several approaches to estimate , the most common being the moment-based estimator of DerSimonian and Laird (9). Given the estimate , the summary effect size estimate analogous to Equation (1) in FE is
The standard error of is . Then, similarly to FE, one can construct a z-score statistic:
The P-value is computed as
The traditional RE model cannot account for overlapping subjects.
New random effects model of Han and Eskin
Han and Eskin (10) found that the traditional RE is conservative and rarely achieves higher power than the FE model. This is because traditional RE tests by constructing a z-score, and constructing a z-score is equivalent to a likelihood ratio test assuming the same heterogeneity under the null and alternative hypotheses. This is a conservative assumption because the known causes of heterogeneity in GWAS, such as gene-by-gene interactions or linkage disequilibrium, do not cause heterogeneity under the null hypothesis. Han and Eskin (10) therefore proposed a new RE model that assumes no heterogeneity under the null hypothesis. The test statistic is
| (3) |
where and are the maximum likelihood estimators of the mean and variance of the effect size, respectively, which can be found using an iterative procedure. The statistic follows a half and half mixture of and under the null hypothesis (33). The P-value can be calculated using the asymptotic distribution or using a pre-constructed P-value table for a more accurate calculation accounting for the small number of observations. The RE model of Han and Eskin (10) cannot account for overlapping subjects.
P-value-based approaches
There exist meta-analysis approaches that combine P-values instead of effect sizes, the most traditional being Fisher's method (11). Fisher's method combines the P-values by constructing a statistic:
which follows under the null hypothesis. An advantage of the P-value-based approaches is that they can be used even without knowing the directions of effect. Fisher's method strictly assumes independence between studies and cannot account for overlapping subjects. However, a recently proposed P-value-based approach by Zaykin and Kozbur (12) can take into account overlapping subjects.
Subset approaches
Subset approaches are similar to the FE model, but they differ in that they assume the effects can exist in only a subset of the studies. Han and Eskin (10) computed a statistic called the ‘m-value’, a posterior probability that the effect is present in a study. They incorporated m-values into the weighted z-score FE approach to upweight studies with high m-values. The P-value is assessed using importance sampling. This method cannot account for overlapping subjects. Bhattacharjee et al. (13) proposed an approach that computes FE statistics using all possible subsets of studies in a meta-analysis and uses the maximum statistic. The method expedites the enumeration of all possible subsets of N studies by using a novel statistical technique. This method can account for overlapping subjects.
Bayesian approaches
Morris (15) proposed a Bayesian approach optimized for trans-ethnic meta-analysis. This method utilizes the Markov Chain Monte Carlo (MCMC) procedure to navigate through possible disease models. In his MCMC sampling procedure, closely related populations are given higher chance to have similar effect sizes, which increases the statistical power to detect heterogeneity caused by the population spectrum. This method cannot account for overlapping subjects. Wen and Stephen (16) proposed a new method that takes the heterogeneity in the data into account by using a hierarchical model in the Bayesian framework. Wen (17) recently extended this method to a model selection approach in complex linear systems, which can account for overlapping subjects.
Definition of dependency
In this article, we often describe two studies in a meta-analysis as dependent, which needs to be defined. We define two studies as dependent if the statistics from those studies are correlated. The statistics can be correlated in a number of reasons, the most common one being that the control subjects overlap, resulting in positive correlations.
The Lin and Sullivan approach
Lin and Sullivan (24) proposed a systematic approach for dealing with overlapping subjects in the FE model. Although only applicable to the FE model, this approach was an innovative breakthrough that provided a foundation of methods that account for dependency in meta-analysis. Recently, Zaykin and Kozbur (12) and Bhattacharjee et al. (13) extended this approach to the P-value-based approach and the subset approach, respectively. Our proposed approach also builds upon this method.
The first step of their approach is to analytically calculate the correlation structure of the statistics caused by overlapping subjects, where N denotes the number of studies in a meta-analysis. Let x be the vector of effect size estimates, , and let
be the correlation matrix of X where denotes the correlation between and . is analytically approximated with the formula
| (4) |
where , and (or , and ) are the number of cases, the number of controls and the total number of subjects in the ith (or jth) study, respectively; and are the numbers of cases and controls that overlap between the ith and jth studies, respectively. See Bhattacharjee et al. (13) for an extended formula for a more complicated situation where some cases in one study are controls in other studies. Given the correlation matrix C, it is straightforward to calculate the covariance matrix Ω.
The second step of the Lin and Sullivan approach is to optimally take into account Ω in the testing. The optimal FE model meta-analysis statistic is
| (5) |
where e is a vector of ones (e = (1,…,1)). The formal proof for the optimality of this statistic is shown in Wei et al. (34) and Wei and Johnson (35). The variance of the statistic is given,
| (6) |
The z-score is calculated to obtain the P-value. Note that in the special case where are independent (Ω is a diagonal matrix), equals to , and equals to . Thus, the Lin and Sullivan (24) approach becomes equivalent to the standard FE model.
Decoupling approach
Decoupling
We propose the following decoupling transformation, which keeps the same information as the Lin and Sullivan (24) approach, with respect to the summary effect size. Suppose that we are given the effect sizes x, the standard errors s and the correlation matrix C as computed by formula (4). Computing C using formula (4) is feasible in many situations since we typically know the exact numbers of shared cases and controls. If this is not the case, an alternative approach would be empirically approximating C by calculating correlations using genome-wide z-scores of the studies. This empirical approximation requires an assumption that the majority of the SNPs in the genome follow the null hypothesis and do not have an effect, which may not be true if the trait is polygenic (36).
Given x, s and C, we propose the following procedure:
- Keep the original x:
- Compute the covariance matrix of x:
where denotes a diagonal matrix whose diagonals are s. - Compute the decoupled covariance matrix:
- Update the standard errors:
where the brackets [ ] denote the index of an element in a vector or matrix. Use and in the downstream meta-analysis.
In the Supplementary Materials, we present a simple R code that performs this procedure.
Relation to the Lin and Sullivan approach
Our approach has a relationship to the Lin and Sullivan (24) approach; if the decoupled studies are used in the FE model, the results are equivalent. Thus, one can use existing software packages, such as METAL (30) or MANTEL (2), to obtain optimal results after the data transformation. Below is a brief proof of this relationship.
Given an effect size vector x and standard errors s, the standard FE model equations (1) and (2) can be rewritten as
| (7) |
and
| (8) |
where .
Now, we decouple studies and put them into the standard FE model formulae. Given a covariance matrix Ω, our decoupling approach will calculate the updated standard errors by calculating . Note that because is a diagonal matrix, the following relationship holds:
If we plug and into Equation (7), then
where is in Equation (5). Similarly, we can show that equals to in Equation (6) by plugging into Equation (8). Therefore, applying our decoupling approach to the FE model is equivalent to the Lin and Sullivan (24) approach.
Decouplable matrix
We define a decouplable covariance matrix as a covariance matrix whose decoupled variances are non-negative. In theory, not every matrix Ω is decouplable. However, in practice, in typical situations where studies share all or a subset of control samples, the covariance matrix is highly likely to be decouplable (see the Results section).
We can analytically demonstrate that the covariance matrix is always decouplable in a simple situation where two studies share a subset of controls. We assume that the two studies have the same MAFs and the same odds ratios. The correlation formula of equation (4) is
Because only controls are shared, . Thus,
Let and be the standard errors of the two studies. Then, the covariance matrix is
For the matrix to be decouplable, the column sums ( and ) have to be positive. Without loss of generality, assume that (study i is larger than study j). Then, the following inequality is the sufficient and necessary condition for decouplability:
| (9) |
Under the assumption of the same MAF and odds ratio, the variance of the statistic will only depend on the sample size. The effective total sample size can be calculated as
Because the standard error will be approximately inversely proportional to the square root of the total sample size, inequality (9) becomes
which is
This inequality always holds true because
Thus, inequality (9) holds, and therefore, matrix Ω is always decouplable.
Meta-analysis in multi-tissue eQTL analysis
One of the applications of meta-analysis methods is the combination of information from multiple tissues to increase the power of identification of eQTL. Because a gene's expression levels in multiple tissues are correlated within an individual, a study should take into account that correlation. Recently, Sul et al. (27) proposed a linear mixed model approach to account for the fact that eQTL studies collect multiple tissues from the same individuals. Let T be the number of tissues, and let N be the number of individuals for each tissue. To test the statistical significance between single nucleotide polymorphism (SNP) j and the expression of a gene, Sul et al. (27) built a linear mixed model:
| (10) |
where Y is an matrix denoting expression levels of N individuals in T tissues. 1 is an NT × T matrix denoting the intercepts for T tissues. The first column is the intercept for the first tissue; the first N rows are ones, and the next are zeros. Other columns are similarly defined. α is a matrix denoting coefficients of intercepts. is an matrix denoting SNP genotype for T tissues. This is similar to the 1 matrix, and we replace ones in the 1 matrix with SNP information. β is a matrix denoting the coefficients of SNP effects in T tissues. is the random effects due to the repeated measurements of individuals. D is an NT × NT matrix representing how individuals are shared across tissues. is the random noise.
Sul et al. (27) efficiently estimated the two variance components ( and ) using the techniques described in Kang et al. (37). After obtaining the covariance matrix of the model, , we can apply the generalized least squares to obtain the estimates and variances of β:
Our goal is to combine information from multiple tissues in . However, as shown in the variance formula, the estimates in are dependent on each other. Thus, we can apply our decoupling strategy. Note that it is reasonable to assume that the genetic effects on expression level can vary between tissues. Therefore, application of the RE model is appropriate in this situation.
Supplementary Material
Funding
This work was supported by the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea (2015-0222 to B.H.); the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI14C1731 to B.H.); the National Science Foundation (0513612 to D.D. and E.E., 0731455 to D.D. and E.E., 0729049 to D.D. and E.E., 0916676 to D.D. and E.E., 1065276 to D.D. and E.E.); the National Institutes of Health (K25-HL080079 to D.D. and E.E., U01-DA024417 to D.D. and E.E., P01-HL30568 to D.D. and E.E., PO1-HL28481 to D.D. and E.E., NIH-NIAMS 1R01AR062886-01 to B.H., S.R. and P.I.W.d.B.) and the Netherlands Organization for Scientific Research (NWO) (VIDI award to P.I.W.d.B.).
Supplementary Material
Acknowledgements
We thank the WTCCC for data access.
Conflict of Interest statement: None declared.
References
- 1.Fleiss J.L. (1993) The statistical basis of meta-analysis. Stat. Methods Med. Res., 2, 121–145. [DOI] [PubMed] [Google Scholar]
- 2.de Bakker P.I.W., Ferreira M.A.R., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. (2008) Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet., 17, R122–R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eyre S., Bowes J., Diogo D., Lee A., Barton A., Martin P., Zhernakova A., Stahl E., Viatte S., McAllister K. et al. (2012) High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet., 44, 1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jostins L.L., Ripke S.S., Weersma R.K.R., Duerr R.H.R., McGovern D.P.D., Hui K.Y.K., Lee J.C.J., Schumm L.P.L., Sharma Y.Y., Anderson C.A.C. et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature, 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michailidou K., Beesley J., Lindstrom S., Canisius S., Dennis J., Lush M.J., Maranian M.J., Bolla M.K., Wang Q., Shah M. et al. (2015) Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet., 47, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeggini E., Ioannidis J.P.A. (2009) Meta-analysis in genome-wide association studies. Pharmacogenomics, 10, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochran W.G. (1954) The combination of estimates from different experiments. Biometrics, 10, 101–129. [Google Scholar]
- 8.Mantel N., Haenszel W. (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst., 22, 719–748. [PubMed] [Google Scholar]
- 9.DerSimonian R., Laird N. (1986) Meta-analysis in clinical trials. Control. Clin. Trials, 7, 177–188. [DOI] [PubMed] [Google Scholar]
- 10.Han B., Eskin E. (2011) Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet., 88, 586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher R.A. (1967) Statistical Methods for Research Workers. 13th ed Oliver & Boyd, Edinburgh, UK. [Google Scholar]
- 12.Zaykin D.V., Kozbur D.O. (2010) P-value based analysis for shared controls design in genome-wide association studies. Genet. Epidemiol., 34, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharjee S., Rajaraman P., Jacobs K.B., Wheeler W.A., Melin B.S., Hartge P., Yeager M., Chung C.C., Chanock S.J., Chatterjee N. et al. (2012) A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am. J. Hum. Genet., 90, 821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han B., Eskin E. (2012) Interpreting meta-analyses of genome-wide association studies. PLoS Genet., 8, e1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris A.P. (2011) Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol., 35, 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen X., Stephens M. (2014) Bayesian methods for genetic association analysis with heterogeneous subgroups: From meta-analyses to gene-environment interactions. Ann. Appl. Stat, 8, 176–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen X. (2014) Bayesian model selection in complex linear systems, as illustrated in genetic association studies. Biometrics, 70, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burton P.R., Clayton D.G., Cardon L.R., Craddock N., Deloukas P., Duncanson A., Kwiatkowski D.P., McCarthy M.I., Ouwehand W.H., Samani N.J. et al. (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature, 447, 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dehghan A., Yang Q., Peters A., Basu S., Bis J.C., Rudnicka A.R., Kavousi M., Chen M.-H., Baumert J., Lowe G.D.O. et al. (2009) Association of novel genetic loci with circulating fibrinogen levels: a genome-wide association study in 6 population-based cohorts. Circ. Cardiovasc. Genet., 2, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A. et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet., 41, 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hysi P.G., Young T.L., Mackey D.A., Andrew T., Fernández-Medarde A., Solouki A.M., Hewitt A.W., MacGregor S., Vingerling J.R., Li Y.-J. et al. (2010) A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat. Genet., 42, 902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mells G.F., Floyd J.A.B., Morley K.I., Cordell H.J., Franklin C.S., Shin S.-Y., Heneghan M.A., Neuberger J.M., Donaldson P.T., Day D.B. et al. (2011) Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat. Genet., 43, 329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rapley E.A., Turnbull C., Olama Al A.A., Dermitzakis E.T., Linger R., Huddart R.A., Renwick A., Hughes D., Hines S., Seal S. et al. (2009) A genome-wide association study of testicular germ cell tumor. Nat. Genet., 41, 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin D.-Y., Sullivan P.F. (2009) Meta-analysis of genome-wide association studies with overlapping subjects. Am. J. Hum. Genet., 85, 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellinghaus D., Ellinghaus E., Nair R.P., Stuart P.E., Esko T., Metspalu A., Debrus S., Raelson J.V., Tejasvi T., Belouchi M. et al. (2012) Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am. J. Hum. Genet., 90, 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhernakova A., Stahl E.A., Trynka G., Raychaudhuri S., Festen E.A., Franke L., Westra H.-J., Fehrmann R.S.N., Kurreeman F.A.S., Thomson B. et al. (2011) Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet., 7, e1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sul J.-H., Han B., Ye C., Choi T., Eskin E. (2013) Effectively identifying eQTLs from multiple tissues by combining mixed model and meta-analytic approaches. PLoS Genet., 9, e1003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flutre T., Wen X., Pritchard J., Stephens M. (2013) A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet., 9, e1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodkind M., Eickhoff S.B., Oathes D.J., Jiang Y., Chang A., Jones-Hagata L.B., Ortega B.N., Zaiko Y.V., Roach E.L., Korgaonkar M.S. et al. (2015) Identification of a common neurobiological substrate for mental illness. JAMA Psychiatry, 72, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devlin B., Roeder K. (1999) Genomic control for association studies. Biometrics, 55, 997–1004. [DOI] [PubMed] [Google Scholar]
- 32.Higgins J.P.T., Thompson S.G. (2002) Quantifying heterogeneity in a meta-analysis. Statist. Med., 21, 1539–1558. [DOI] [PubMed] [Google Scholar]
- 33.Self S.G., Liang K.Y. (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J. Am. Stat. Assoc., 82, 605–610. [Google Scholar]
- 34.Wei L.J., Lin D.Y., Weissfeld L. (1989) Regression analysis of multivariate incomplete failure time data by modeling marginal distributions. J. Am. Stat. Assoc., 84, 1065–1073. [Google Scholar]
- 35.Wei L.J., Johnson W.E. (1985) Combining dependent tests with incomplete repeated measurements. Biometrika, 72, 359–364. [Google Scholar]
- 36.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W. et al. (2010) Common SNPs explain a large proportion of the heritability for human height. Nat. Genet., 42, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang H.M., Sul J.-H., Service S.K., Zaitlen N.A., Kong S.-Y.Y., Freimer N.B., Sabatti C., Eskin E. (2010) Variance component model to account for sample structure in genome-wide association studies. Nat. Genet., 42, 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.