Abstract
The expansion of the GGGGCC hexanucleotide repeat in the non-coding region of the Chromosome 9 open-reading frame 72 (C9orf72) gene is the most common genetic cause of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS). This genetic alteration leads to the accumulation of five types of poly-dipeptides translated from the GGGGCC hexanucleotide repeat. Among these, poly-proline-arginine (poly-PR) and poly-glycine-arginine (poly-GR) peptides are known to be neurotoxic. However, the mechanisms of neurotoxicity associated with these poly-dipeptides are not clear. A proteomics approach identified a number of interacting proteins with poly-PR peptide, including mRNA-binding proteins, ribosomal proteins, translation initiation factors and translation elongation factors. Immunostaining of brain sections from patients with C9orf72 ALS showed that poly-GR was colocalized with a mRNA-binding protein, hnRNPA1. In vitro translation assays showed that poly-PR and poly-GR peptides made insoluble complexes with mRNA, restrained the access of translation factors to mRNA, and blocked protein translation. Our results demonstrate that impaired protein translation mediated by poly-PR and poly-GR peptides plays a role in neurotoxicity and reveal that the pathways altered by the poly-dipeptides-mRNA complexes are potential therapeutic targets for treatment of C9orf72 FTD/ALS.
Introduction
Frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) are adult-onset neurodegenerative disorders. While FTD and ALS are, respectively, characterized by progressive degeneration of frontal and temporal lobes or loss of both upper and lower motor neurons, there is clinical and genetic overlap between them (1). The expansion of the GGGGCC hexanucleotide repeat in the non-coding region of the Chromosome 9 open-reading frame 72 (C9orf72) gene is the most common genetic cause of both FTD and ALS (C9FTD/ALS) (2–5). Several underlying mechanisms have been proposed including the reduced expression levels of C9orf72 (6), toxic RNA foci formation (7,8) and abnormal accumulation of toxic dipeptide-repeat proteins generated by repeat-associated non-ATG (RAN) translation (9). Recent studies have shown that the dipeptide products from C9orf72, which are detected in the cerebrospinal fluid of C9FTD/ALS patients (10), cause cell death in vitro and in a Drosophila model (11,12).
RAN translation from the GGGGCC hexanucleotide can produce five types of dipeptide repeats; glycine-alanine (poly-GA), glycine-proline (poly-GP) and glycine-arginine (poly-GR) from the sense transcripts, and glycine-proline (poly-GP), proline-arginine (poly-PR) and proline-alanine (poly-PA) from the antisense transcripts. It has been shown that poly-PR is toxic to NSC34 cells and mouse motor neurons and poly-PR and poly-GR are toxic in a Drosophila model. However, the underlying mechanisms of toxicity caused by poly-PR and poly-GR are not clear (11–13). Here, we show that poly-PR and poly-GR inhibit protein translation by binding to the translational complex and ribosomal proteins, leading to neurotoxicity.
Results
To understand the mechanisms of neurotoxicity mediated by C9orf72-derived polypeptides, we synthesized C-terminally hemagglutinin (HA)-tagged 20 repeats of Proline-Arginine dipeptide [poly-(PR)20], Glycine-Arginine dipeptide [poly-(GR)20] and Glycine-Alanine dipeptide [poly-(GA)20], and examined the neurotoxicity in NSC34 cells. Poly-(PR)20 caused cell death in motor neuronal NSC34 cells (Fig. 1A). To a lesser extent, poly-(GR)20 showed neurotoxicity, but poly-(GA)20 did not (Fig. 1A).
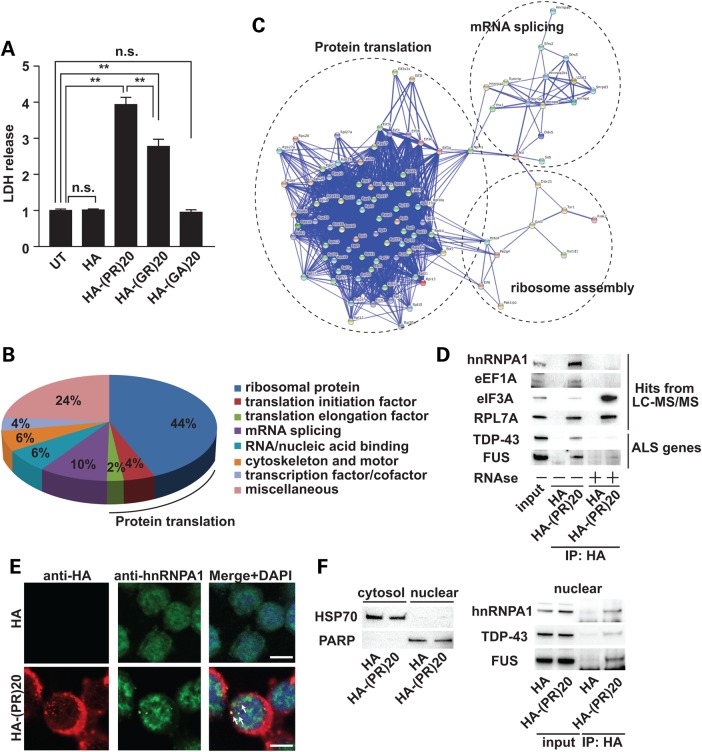
Figure 1.
Poly-PR peptide interacts with RNA-binding proteins. (A) Cytotoxicity of C9orf72-derived dipeptides (10 μm) was monitored by release of LDH. N = 5 biological replicates. Statistical analysis was determined by one-way ANOVA followed by Dunnett's test. **P < 0.01 and *P < 0.05. n.s., not significant. (B) PANTHER classification of poly-PR peptide-interacting proteins identified by LC-MS/MS. (C) Functional protein association network of poly-(PR)20 peptide-binding proteins by STRING classification with high confidence (score = 0.700). (D) Co-immunoprecipitation of hnRNPA1, eEF1A, eIF3A, RPL7A (identified by LC-MS/MS), TDP-43 and FUS (familial ALS-causative RNA-binding proteins) with HA-poly-(PR)20 peptide with or without RNAse treatment. (E) Immunocytochemical analysis of NSC34 cells treated with HA peptide (5 μm) or HA-poly-(PR)20 peptide (5 μm) for overnight. The white bar shows 10 μm. (F) Left panel: Fractionated samples from NSC34 cells treated with HA peptide (5 μm) or HA-poly-(PR)20 peptide (5 μm) for overnight. HSP70 was used as a cytosolic marker and PARP was used as a nuclear marker. Right panel: co-immunoprecipitation of hnRNPA1, TDP-43 and FUS with HA-poly-(PR)20 peptide in the nuclear fraction.
We, next, performed an immunoprecipitation analysis followed by a liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify interacting proteins of poly-(PR)20. NSC34 cells were cultured for 5 days in the presence or absence of poly-(PR)20, and then their lsyates were immunoprecipitated with anti-HA antibody and subjected to LC-MS/MS. We discovered that multiple RNA-binding proteins were bound to the poly-(PR)20. The PANTHER (Protein Annotation Through Evolutionary Relationship) classification system revealed that the most abundant class of interacting proteins was ribosomal proteins (Fig. 1B) (14). Translation initiation factors and translation elongation factors were also identified, suggesting that poly-(PR)20 might interact with proteins involved in protein translation. STRING (Search Tool for the Retrieval of Interacting Genes/proteins) identified three functional clusters involved in: (i) protein translation, (ii) mRNA splicing and (iii) ribosome assembly (Fig. 1C) (15).
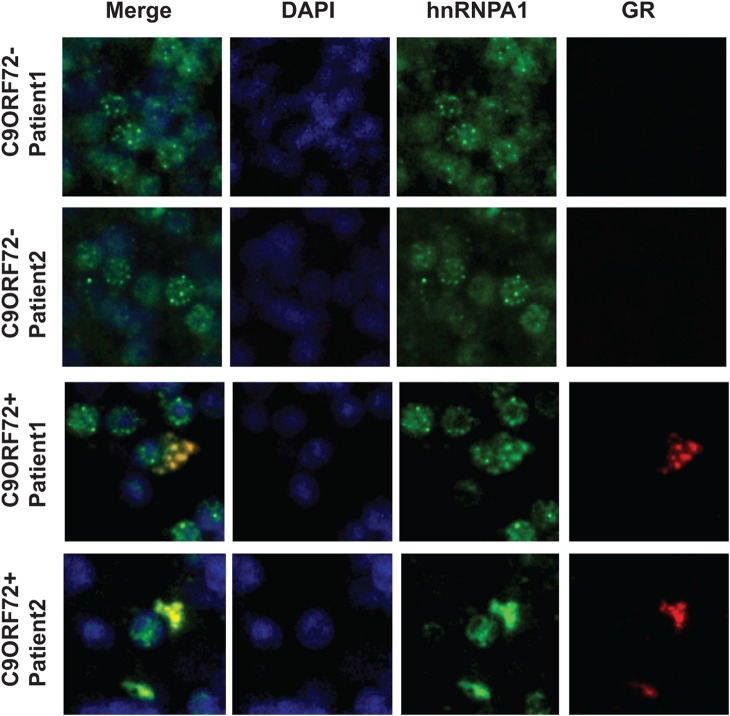
To validate the results of LC-MS/MS, we perfomed co-immunoprecipitation assays and confirmed that poly-(PR)20 interacted with heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), eukaryotic elongation factor 1A (eEF1A), eukaryotic initiation factor 3A (eIF3A) and ribosomal protein L7A (RPL7A) (Fig. 1D). We also discovered that ALS-causative RNA-binding proteins, including TAR DNA-binding protein 43 (TDP-43) and fused in sarcoma (FUS), were co-immunoprecipitated with poly-(PR)20 (Fig. 1D). The interactions between poly-(PR)20 and RNA-binding proteins, such as hnRNPA1, eEF1A, TDP-43 and FUS, were diminished by RNAse treatment, suggesting that these interactions might be mediated by RNA (Fig. 1D). To exclude the possibility that the interaction between poly-(PR)20 and RNA-binding proteins, such as hnRNPA1, TDP-43 and FUS, were artificially formed during cell lysis procedure, we first performed immunocytochemical analysis to show the colocalization of HA-poly-(PR)20 and hnRNPA1. HA-poly-(PR)20 mainly localized to cytosol and partially localized to nucleus, and colocalized with hnRNPA1 (Fig. 1E). Next we examined subcellular fractionation of NSC34 cells treated with HA peptide or HA-poly-(PR)20 followed by co-immunoprecipitation with anti-HA antibody using the nuclear fraction. As Figure 1F shows, HA-poly-(PR)20 interacted with hnRNPA1, TDP-43 and FUS in the nuclear fraction, indicating that the interaction between HA-poly-(PR)20 and RNA-binding proteins including hnRNPA1, TDP-43 and FUS were phyisiologically, but not artificially formed. Consistent with these findings, immunostaining of brain sections from patients with C9orf72 ALS showed that poly-GR was colocalized with hnRNPA1 (Fig. 2).
Figure 2.
GR-aggregates colocalize with hnRNPA1. Immunohistochemistry in the cerebellar granular layer of two C9orf72 ALS cases and two non-C9orf72 cases reveals coaggregation of poly-GR with hnRNPA1.
Poly-(PR)20 peptide and poly-(GR)20 inhibit protein translation
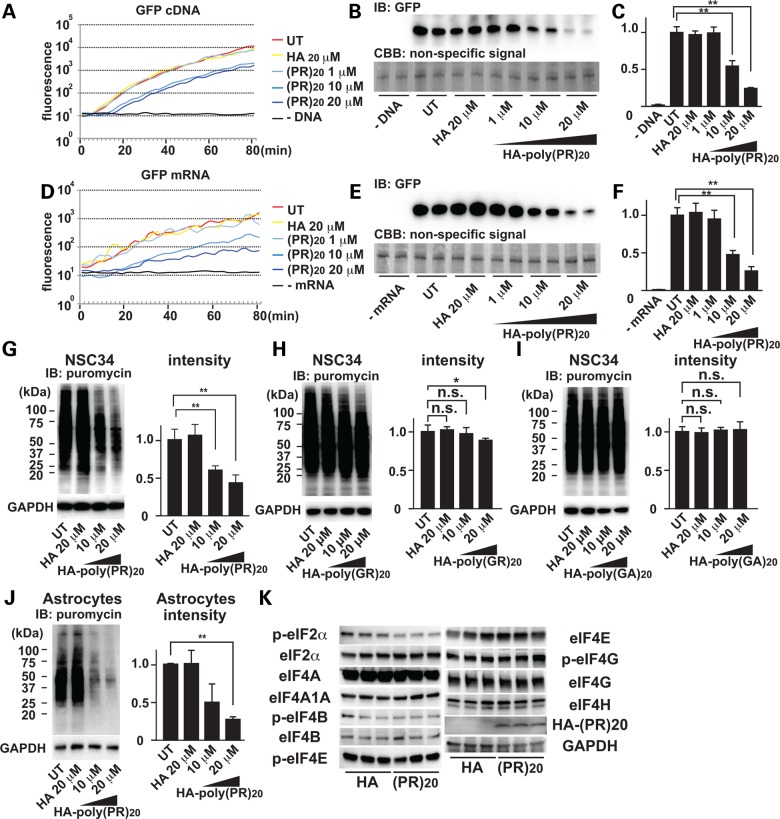
Most of the poly-(PR)20-interacting proteins were involved in protein translation, raising the possibility that poly-(PR)20 may affect protein translation. To test this idea, we perfomed an in vitro translation (IVT) assay using HeLa cell lysates and cDNA encoding green fluorescent protein (GFP) as a reporter. Real-time monitoring of GFP fluorescence and immunoblot analysis showed that poly-(PR)20 inhibited protein translation in vitro in a dose-dependent manner (Fig. 3A–C). To distinguish whether poly-(PR)20 inhibits the expression of GFP protein at the transcriptional level or translational level, we performed IVT using synthesized mRNA of GFP, and poly-(PR)20 inhibited GFP translation from GFP mRNA (Fig. 3D–F), indicating that poly-(PR)20 inhibits protein translation. To confirm the inhibitory effect of C9orf72 dipeptides on translation in vivo, we next treated NSC34 cells with poly-(PR)20, poly-(GR)20 or poly-(GA)20, and then tested the protein translation using a puromycin-based labeling of newly synthesized protein method called SUnSET (16). Poly-(PR)20 and poly-(GR)20, but not poly-(GA)20, inhibited general protein translation in a dose-dependent manner (Fig. 3G–I). We also confirmed that poly-(PR)20 suppressed the translation of proteins in mouse primary astrocytes (Fig. 3J).
Figure 3.
Poly-PR peptide inhibits protein translation. (A) Real-time monitoring of GFP fluorescence translated from GFP cDNA by IVT. (B) Immunoblot analysis of GFP protein. Coomassie stained non-specific signals were used as loading controls. (C) Quantitation of immunoblot of GFP. N = 3 biological replicates. (D) Real-time monitoring of GFP fluorescence translated from GFP mRNA by IVT. (E) Immunoblot analysis of GFP protein. Coomassie stained non-specific bands were used as loading control. (F) Quantitation of immunoblot of GFP. N = 3 biological replicates. (G) Immunoblot analysis of newly translated proteins in NSC34 cells incubated with HA peptide or HA-poly-(PR)20 peptide. Newly translated proteins were labeled with puromycin and visualized by anti-puromycin antibody. (H) Immunoblot analysis of newly translated proteins in NSC34 cells incubated with HA peptide or HA-poly-(GR)20 peptide. Newly translated proteins were labeled with puromycin and visualized by anti-puromycin antibody. N = 3 biological replicates. (I) Immunoblot analysis of newly translated proteins in NSC34 cells incubated with HA peptide or HA-poly-(GA)20 peptide. Newly translated proteins were labeled with puromycin and visualized by anti-puromycin antibody. N = 3 biological replicates. (J) Immunoblot analysis of newly translated proteins in mouse primary astrocytes incubated with HA peptide or HA-poly-(PR)20 peptide. Newly translated proteins were labeled with puromycin and visualized by anti-puromycin antibody. N = 3 biological replicates. (K) Immunoblot analysis of a series of eIF, phosphorylated forms of eIFs, HA and GAPDH in the cell lysates of NSC34 cells treated with 10 μm of HA control peptide or HA-poly-(PR)20 peptide. N = 3 biological replicates. Each experiment was repeated at least three times independently. Representative images were shown. Asterisks indicate a significant difference analyzed by one-way ANOVA followed by Dunnett's test (**P < 0.01 and *P < 0.05).
It has been reported that quantitative changes or phosphorylation levels of eukaryotic initiation factors (eIFs) control protein translation (17–21). Especially phosphorylation status of eIF2α is a major negative regulator of global protein synthesis under various stress conditions (19). Thus, we next examined if poly-(PR)20-mediated translational arrest was caused by quantitative changes or phosphorylation levels of eIFs. Protein expression levels of eIF2α, eIF4A, eIF4A1, eIF4B, eIF4E, eIF4G, eIF4H or phosphorylation levels of eIF4B, eIF4E or eIF4G were not changed by poly-(PR)20. The phosphorylation level of eIF2α was slightly suppressed by poly-(PR)20. These results suggest that poly-(PR)20 might inhibit protein translation by a distinct pathway (Fig. 3K).
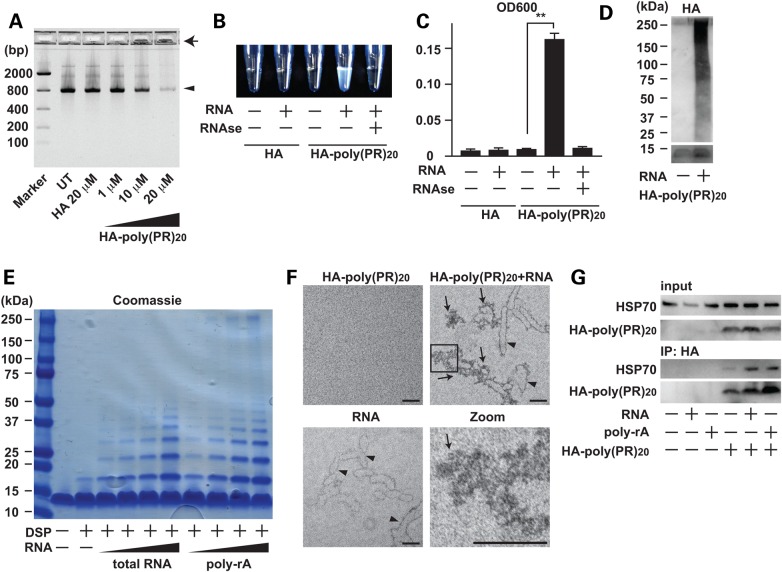
To investigate how poly-(PR)20 inhibits protein translation, we performed agarose gel electrophoresis of the solution containing poly-(PR)20 and GFP mRNA used in the IVT assay shown in Figure 3D and E. The amount of soluble GFP mRNA was decreased by poly-(PR)20 in a dose-dependent manner. Insoluble GFP mRNA accumulated in the loading wells, raising the possibility that poly-(PR)20 and mRNA formed an insoluble complex (Fig. 4A). To test this idea, we mixed yeast total RNA and poly-(PR)20, and then measured the optical density of the mixture at 600 nm. The optical densitiy of yeast RNA mixed with poly-(PR)20 was significantly higher than that of yeast RNA or poly-(PR)20 alone and this was decreased by RNAse treatment, indicating that RNA and poly-(PR)20 formed complexes/aggregates (Fig. 4B and C). To elucidate the nature of these aggregates, we performed immunoblot analysis of poly-(PR)20 mixed with yeast total RNA. In the presence of RNA, poly-(PR)20 formed a high-molecular-weight complex (Fig. 4D). Dithiobis (succinimidyl propionate)-mediated crosslinking assay revealed that RNA enhanced the oligomerlization of poly-(PR)20 in a dose-dependent manner (Fig. 4E).
Figure 4.
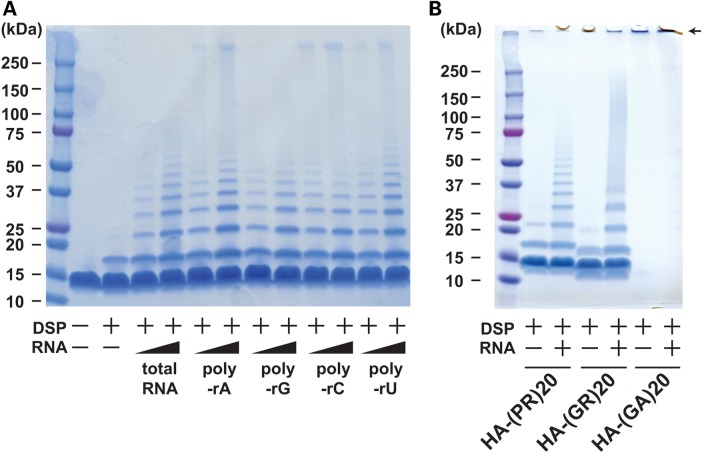
Poly-PR peptide interacts with RNA and forms aggregates. (A) Agarose gel electrophoresis of solutions containing GFP mRNA with HA peptide or HA-poly-(PR)20 peptide at indicated concentrations. An arrowhead shows GFP mRNA and an arrow shows insoluble RNA aggregate. (B and C) The measurement of turbidity using the optical density at 600 nm (OD600) of the solution containing HA peptide or HA-poly-(PR)20 peptides mixed with or without yeast total RNA. The image represents visible formation of poly-PR aggregates that can be reversed by treatment with RNase. N = 3 biological replicates. (D) Immunoblot analysis of HA-poly-(PR)20 peptides mixed with or without yeast total RNA. SDS–PAGE was performed with a 4–20% gradient gel. (E) Coomassie blue staining of poly-PR aggregates induced by yeast total RNA or monopolymeric poly-adenilyc acids (poly-rA). Samples were crosslinked with dithiobis (succinimidyl propionate) (DSP, 2 mm) before applied to SDS–PAGE. (F) Electron microscope images of poly-PR aggregates induced by RNA. The scale bar shows 50 nm. Arrowheads show RNA and arrows show poly-PR aggregate. (G) Co-immunoprecipitation of human recombinant HSP70 with HA-poly-(PR)20 peptides mixed with or without yeast total RNA or poly-rA. Each experiment was repeated at least three times and representative images were shown. Asterisks indicate a significant difference analyzed by one-way ANOVA followed by Dunnett's test (**P < 0.01 and *P < 0.05).
To test if poly-(PR)20 has selective affinity for a specific ribonucleotide, we examined the effects of four homopolymeric ribonucleotides; poly-adenylic acid (poly-rA), poly-guanylic acid (poly-rG), poly citidylic acid (poly-rC) and poly-uridylic acid (poly-rU) on the aggregate formation. All homopolymeric ribonucleotides showed similar effects on the aggregate formation of poly-(PR)20 (Figs 4E and 5A). We also examined the RNA-mediated aggregate formation of poly-(GR)20 and poly-(GA)20. Similar to poly-(PR)20, RNA induced the aggregate formation of poly-(GR)20 (Fig. 5B). Due to the strong hydrophobicity as reported previously, poly-(GA)20 formed high-molecular-weight aggregates without RNA and accumulated in the stacking gel (22). Thus, we could not examine the effect of RNA on the aggregate formation of poly-(GA)20 (Fig. 5B). Electron microscopic images revealed that RNA directely interacted with poly-(PR)20 and enhanced its aggregation (Fig. 4F). These results suggest that RNA may be a possible seed for the poly-PR aggregates seen in patients with c9FTD/ALS.
Figure 5.
Interaction between C9orf72 dipeptides and ribonucleotides in vitro. (A) Coomassie blue staining of poly-PR aggregates induced by yeast total RNA or monopolymeric poly-adenilyc acids (poly-rA, poly-rG, poly-rC or poly-rU). Samples were crosslinked with dithiobis (succinimidyl propionate) (DSP, 2 mm) before applied to SDS–PAGE. (B) Coomassie blue staining of poly-PR, poly-GR or poly-GA aggregates induced by yeast total RNA. Samples were crosslinked with dithiobis (succinimidyl propionate) (DSP, 2 mm) before applied to SDS–PAGE. An arrow shows the aggregate of poly-GA accumulated in the stucking gel.
Proline-Arginine (PR) dipeptides consist of a hydrophobic amino acid (P) and a hydrophilic amino acid (R), and can be dissolved in water due to the poloarity of R (>1 mm). However, the dipeptide products of C9orf72 including poly-PR and poly-GR have been shown to deposit in the cytosol of motor neurons as aggegates (Fig. 2) (9) and poly-(PR)20 formed a water-insoluble complex with RNA shown in Figure 4B. Hydrophobicity of misfolded protein increases its recognition by molecular chaperones, such as heat shock protein 70 (HSP70) and glucose regulated protein 78 (23,24) and the protein aggregates consisting of misfolded proteins and molecular chaperones are frequently observed in the affected tissues of neurodegenerative disorders including ALS (25). These considerations prompted us to test if poly-(PR)20-RNA aggregates are recognized by HSP70 as misfolded proteins. Co-immunoprecipitation analysis indicated that recombinant HSP70 preferentially interacted with poly-(PR)20 in the presense of RNA and poly-rA, suggesting that RNA enhanced hydrophobicity of the poly-(PR)20 peptide and poly-(PR)20-RNA aggregates were recognized as misfolded proteins in vitro (Fig. 4G).
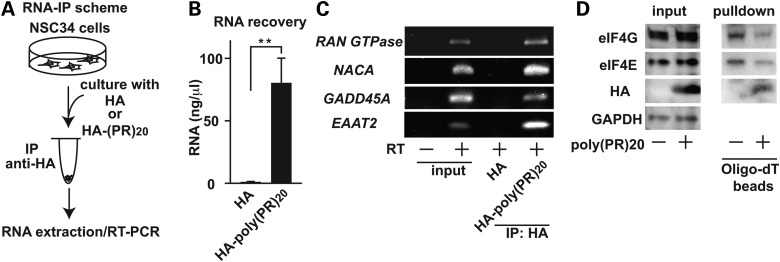
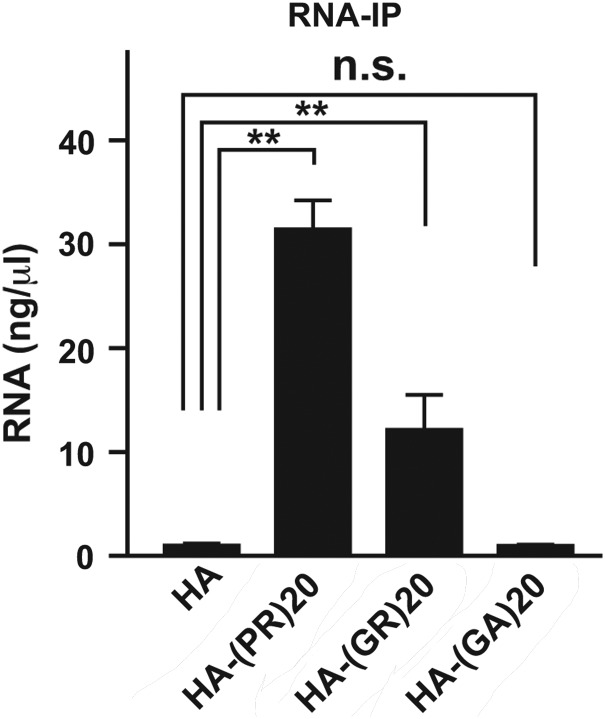
To investgate whether poly-(PR)20 interacts with endogenous RNA in NSC34 cells, we performed an RNA immunoprecipitation assay (RIP). The RIP assay using the lysates from NSC34 cells cultured with HA peptide or poly-(PR)20 revealed that poly-(PR)20 interacted with RNA in NSC34 cells (Fig. 6A and B). Furthermore, poly-(GR)20, but not poly-(GA)20, interacted with RNA, suggesting that the inhibition of protein translation by the C9orf72 dipeptides is mediated by their interaction with RNA (Fig. 7).
Figure 6.
Poly-(PR)20-mRNA complexes inhibits protein translation. (A) A scheme of RNA immunoprecipitation followed by RT-PCR. NSC34 cells cultured with 10 μm of HA peptide or HA-poly-(PR)20 peptide was harvested, followed by RNA immunoprecipitated with anti-HA antibody. After extraction of RNA, RT-PCR was performed. (B) The concentration of RNA extracted from immunoprecipitated samples. N = 3 biological replicates. (C) RT-PCR of Ran GTPase, NACA, GADD45A and EAAT2 from immunoprecipitated samples. (D) mRNA–protein complex in NSC34 cells treated with or without 10 μm of HA-poly-(PR)20 peptide, fixed with formaldeyde, were isolated with oligo-dT beads, followed by immunoblot analysis with anti-eIF4E and anti-eIF4G antibody. Asterisks indicate a significant difference analyzed by Dunnet's test (**P < 0.01). Each experiment and imaging was repeated a least three times independently. Reperesentative images were shown.
Figure 7.
Interaction between C9orf72 dipeptides and ribonucleotides in vivo. The concentration of RNA extracted from immunoprecipitated samples from NSC34 cells cultured with 10 μm of HA peptide, HA-poly-(PR)20, HA-poly-(GR)20 or HA-poly-(GA)20 peptides. N = 3 biological replicates. Asterisks indicate a significant difference analyzed by one-way ANOVA followed by Dunnett's test (**P < 0.01 and *P < 0.05).
It has been reported that aberrant splicing of specific genes, including RAN guanosine triphosphatase (RAN GTPase), nascent polypeptide-associated complex subunit alpha (NACA), growth arrest and DNA damage-inducible 45A (GADD45A) and excitatory amino acid transporter 2 (EAAT2) caused by poly-GR and poly-PR dipeptides from C9orf72 gene contributes to the toxicity in human astrocytes (12). We, therefore, examined whether poly-(PR)20 interacted with endogenous mRNA of these genes and contributed to cell death in NSC34 cells. The RIP assay followed by reverse transcription–polymerase chain reaction (PCR) confirmed that poly-(PR)20 interacted with mRNA of RAN GTPase, NACA, GADD45A and EAAT2 (Fig. 6C). However, we did not observe the alteration of splicing of these mRNA in NSC34 cells (Supplementrary Material, Fig. S1).
Our results shown in Figure 3K suggest that impaired protein translation by poly-(PR)20 may be independent of quantitative changes or phosphorylation levels of translation factors. Poly-(PR)20 peptide showed high affinity to RNA and directly formed hydrophobic complexes in the experiments shown in Figure 4. These results raised the possibility that poly-(PR)20 blocks the access of translation factors to mRNA. To test this idea, mRNA complexes were subjected to pulldown by oligo-dT followed by immunoblot analysis (25). We found that poly-(PR)20 blocked the access of eIF4E and eIF4G to mRNA (Fig. 6D). eIF4E and eIF4G are known to directly interact with mRNA and are essential for the initiation of translation (26). Collectively, our results indicate poly-(PR)20 makes complexes with mRNA and prevents the access of translation factors, leading to impaired protein translation.
Discussion
Since the establishment of disease concept of ALS and FTD, clinicians and geneticists have compiled the genetic information of familial cases, accelerating the research to understand the pathophysiology underlying both familial and sporadic cases. The discovery of C9orf72 is a milestone of those studies, which occupies 40% of familial and 10% of sporadic case of ALS, as well as significant population of FTD. Despite its importance, the mechanism how the expansion of GGGGCC hexanucleotide repeat in the non-coding region causes the disease remains not clear. There are several models explaining the neurotoxicity mediated by the expansion of GGGGCC hexanucleotide repeat in C9orf72 at different levels. At the DNA level, expanded DNA-mediated toxicity caused by DNA-RNA G-quadruplexes formation induces nuclear stress and cell death (27). At the RNA level, it has been shown that RNA-mediated toxicity is mediated by toxic RNA foci formation (7,8) and RNA-G quadruplexes formation (28). At the protein level, repetitive GGGGCC sequence potentially triggers unusual protein translation mechanism called repeat-associated non-ATG (RAN) translation, leading to cell death. In the current study, we focused on the toxicity induced at the protein level because recent studies showed that the dipeptide products from C9orf72, which are detected in C9-FTD/ALS cerebrospinal fluid (10), cause cell death in vitro and in a Drosophila model (11,12). In this study, we showed that RAN-translated poly-dipeptides, especially poly-PR and poly-GR peptide inhibited protein translation by sequestration of RNA. We discovered that mRNA enhanced aggregate formation of poly-PR and poly-GR peptide, a well-known pathological hallmark of C9orf72 dipeptides in patients of FTD/ALS, suggesting that the complex formation between poly-PR/GR peptides and RNA plays an important pathological role in FTD/ALS.
Through the LC-MS/MS, we identified more than one hundred novel interactors with poly-PR peptide including ribosomal proteins and translation initiation factors. These interactions were partially dependent on RNA. When analyzed with bioinformatic tools, translation machinery appeared to be a possible target of poly-PR dipeptide, and both in vitro and in vivo translation assays confirmed that poly-PR as well as poly-GR, which were shown to be neurotoxic, inhibited protein translation at the mRNA level. Furthermore, the degrees of the inhibition of protein translation were consistent with the levels of neurotoxicity, as well as their affinity to RNA, suggesting that the impairement of protein translation through interaction with RNA underly the pathogenesis by C9orf72-derived dipeptides.
Dysregulated protein translation plays a role in the development of neurodegenerative diseases. For example, fragile X mental retardation protein has been reported to directly bind to ribosome and inhibit protein translation (29), and sustained translational repression by eIF2α-P mediates prion neurodegeneration (30). In the case of C9orf72, overexpression of GGGGCC repeats has been shown to inhibit protein translation (31). Here, we demonstrated that poly-PR or poly-GR dipeptides, the products of RAN translation from C9orf72, were sufficient to cause inhibition of protein translation. From the observation of LC/MS-MS and coimmunoprecipitation experiments, we hypothesized that there exists direct interaction between poly-PR/GR peptides and RNA. We discovered that RNA enhanced the aggregate formation mediated by both poly-PR or poly-GR peptides. Because poly-PR had high affinity to mRNA itself and made aggregates with RNA, poly-PR prevented the access of translation initiation factorsm such as eIF4E and eIF4G, to mRNA.
How the poly-PR peptide penerates the plasma membrane and sequesters RNA remains to be elucidated, but natural arginine rich peptides such as human immuno-defficiency virus protein trans-activator of transcription peptide are known to destabilize the plasma membrane and penetrate into cytosol, suggesting that poly-PR peptide might destabilize the plasma membrane in a similar manner (32). We did not observe any significant neurotoxic effect of poly-GA peptide in our experimental system. This might be attributed to the impermeability of poly-GA peptide into the cells. The relationship between cell permeability of dipeptides and cytotoxicity should be further studied in the future.
It has been shown that aberrant splicing of specific genes including RAN GTPase, EAAT2 and GADD45a caused by poly-PR peptide in human astrocytes contributes to the toxicity of poly-PR peptide (12). We demonstrated that poly-PR peptide interacts with mRNA of these genes in mouse motorneuronal NSC34 cells (Fig. 6C). However, we did not observe the alteration of splicing of these genes (Supplementary Material, Fig. S1), suggesting that inhibition of protein translation rather than aberrant splicing of these genes underlies the toxicity seen in this model. It is also possible that poly-PR sequesters mRNA in both cytosol and nucleus, affecting mRNA splicing and protein translation in a cell-type-dependent manner.
Although how sequestration of RNA by poly-PR/GR peptide and other RNA-binding ALS-causative genes functionally interact in the pathogenesis of ALS remains to be determined, a therapeutic strategy targeting the cell-penetrating toxic dipeptide-repeat proteins may potentially rescue the protein translation and protect neurons from the neurotoxicity in patients with C9orf72 FTD/ALS.
Materials and Methods
HA-peptide and poly-dipeptide peptide
HA peptide is purchased from Pierce (Rockford, IL, USA). HA-tagged poly-(PR)20, HA-tagged poly-(GR)20 and HA-tagged poly-(GA)20, peptide were synthesized by Genscript (Piscataway, NJ, USA).
Cell culture
NSC34 cells were maintained in Dulbecco's Modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics.
LDH assay
NSC34 cells were plated on a 96-well plate and incubated with 10 μm of HA peptide, HA-tagged poly-(PR)20, HA-tagged poly-(GR)20 or HA-tagged poly-(GA)20 peptide for 6 h. The release of LDH were assessed by Cytotoxicity LDH detection kit (Clontech).
Antibodies and reagents
Anti-HA antibody was purchased from Genetex (Irvine, CA, USA). Anti-HA antibody-conjugated agarose beads, 1-step human in vitro protein expression kits, anti-turbo GFP antibody and turbo GFP mRNA were purchased from Pierce (Rockford, IL, USA). Anti-hnRNPA1 antibody and anti-FUS antibody were purchased from Abcam (Cambridge, MA, USA). Anti-eEF1A antibody, anti-phosphorylated eIF2A antibody, anti-eIF3A antibody, anti-eIF4A antibody, anti-eIF4A1A antibody, anti-eIF4B antibody, anti-phospho-eIF4B antibody, anti-eIF4E antibody, anti-phospho-eIF4E antibody, anti-eIF4G antibody, anti-phospho-eIF4G antibody, anti-eIF4H antibody, anti-RPL7A antibody, anti-poly (ADP-ribose) polymerase (PARP) antibody, anti-HSP70 antibody and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody were obtained from Cell signaling technology (Danvers, MA, USA). Anti-TDP43 antibody was purchased from Proteintech (Chicago, IL, USA). Yeast total RNA and ribonuclease A were obtained from Sigma (St Louis, MO, USA). Oligo-dT (25) sepharose beads were obtained from New England Biolabs (Ipswich, MA, USA). Anti-poly-GR antibody clone 5A2 were obtained from Millipore (Billerica, MA, USA).
Immunohistochemical staining
Sections of formalin-fixed paraffin embedded brain tissues were dipped with xylene to remove paraffin, then followed by hydration in an ethanol series. Then, heat-induced antigen retrieval was carried out using sodium citrate buffer (pH 6.8) before being incubated with blocking serum. The sections were washed twice with Tris-buffered saline (pH 7.4) containing 0.1% tween-20. The sections were then incubated with 0.5% triton X-100 for 10 min at room temperature in preparation for immunohistochemistry. The sections were incubated with primary antibodies overnight at 4°C. For the double-labeling immunofluorescence staining, fluorescein isothiocyanate -conjugated anti-rabbit IgG and Cy3 conjugated anti-rat IgG secondary antibodies were used. All slides were mounted with 4′,6-diamidino-2-phenylindole containing mounting medium (Vector Laboratories, Burlingame, CA, USA).
liquid chromatography-tandem mass spectrometry/mass spectrometry
NSC34 cells plated on a 15 cm dish were cultured with 5 μm HA peptide (Pierce) or 5 μm of poly-(PR)20 peptide for 5 days. The cells were harvested, washed with phosphate-buffered saline (PBS) and lysed in a immunoprecipitation buffer [150 mm NaCl, 50 mm hydroxyethyl-piperazine ethanesulfonic acid (HEPES) pH 7.4, 0.5% NP-40, 1 mm ethylenediaminetetraacetic acid (EDTA) and protease inhibitor cocktail]. After centrifugation to remove cellular debris, immunoprecipitation was performed with anti-HA antibody-conjugated agarose beads (Pierce). The immunoprecipitates were analyzed with LC–MS/MS by Applied Biomics (Hayward, CA, USA).
Interactome of identified proteins
Prediction of interaction of proteins identified by LC–MS/MS was performed with the Panther software and STRING ver. 9.1 software. For STRING, the confidence score at the level of 0.7 was used to show the best fitted protein-interactive network.
Immunoprecipitation
NSC34 cells were lysed in cell lysis buffer (150 mm NaCl, 0.5% NP40, 50 mm HEPES pH 7.4, 1 mm EDTA, 1 mm dithiothreitol (DTT) and protease inhibitor cocktail) followed by centrifugation at 15 000 rpm to remove cellular debris. The supernatants were incubated with 5 μg/ml HA peptide or poly(PR)20 peptide for 2 h in the presence or absence of RNAse (100 μg/ml). Then, immunorepcipitation was performed with anti-HA antibody-conjugated agarose beads. The beads were washed four times with the lysis buffer and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblot analysis.
RNA immunoprecipitation and RT-PCR
NSC34 cells plated on a six-well plate were treated with 3 μm of HA peptide or HA-tagged poly(PR)20, HA-tagged poly(GR)20 or HA-tagged poly(GA)20 for 6 h. The cells were harvested, washed with PBS and lysed in a RIP immunoprecipitation (RIP) buffer (150 mm KCl, 25 mm Tris–HCl, pH 7.4, 5 mm EDTA, 0.5 mm DTT, 0.5% NP40, 100 U/ml RNAse inhibitor and protease inhibitor cocktail). After centrifugation to remove cellular debris, immunoprecipitation was performed with anti-HA antibody-conjugated agarose beads. RNA were isolated from the immunoprecipitates using RNeasy (Qiagen, Netherland), according to the manufacturer's instructions. The cDNA was synthesized using ImProm-II Reverse Transcription System (Promega, Madison, WI, USA). The primers used in the PCR are following: for mouse EAAT2, ACAATATGCCCAAGCAGGTAGA and CTTTGGCTCATCGGAGCTGA; for mouse Ran GTPase, CCACTTGACGGGCGAGTTT and CCACACAATACAATGGGGATGTT; for mouse NACA, GACAGTGATGAGTCAGTACCAGA and TGCTTGGCTTTACTAACAGGTTC; for mouse Gadd45a, CCGAAAGGATGGACACGGTG and TTATCGGGGTCTACGTTGAGC.
In vitro translation
IVT was performed using 1-Step Human In Vitro Protein Expression Kit (Pierce) according to the manufacturer's instructions. Real-time monitoring of fluorescence of expressed GFP was performed with ViiA7 (Life Technologies, Carlsbad, CA, USA).
Measurement of protein translation in vivo
The rate of protein synthesis was measured by the SUnSET method. Briefly, newly synthesized protein in NSC34 cells pretreated with HA peptide or HA-poly-(PR)20 peptide for 2 h were labeled by incubation with 2 μg/ml puromycin for 30 min. the lysates were immunoblotted with anti-puromycin antibody (Millipore, Billerica, MA, USA).
The measument of turbidity
The measurement of turbidity was performed using NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's instructions.
Electron microscope imaging
HA-tagged poly-(PR)20 peptide (5 μm), RNA (500 ng/μl) and a mixture of HA-tagged poly-(PR)20 peptide and RNA were shaked at 200 rpm at room temperature for 72 h prior to imaging with an electron microscope. The samples were negatively stained with 2% uranyl acetate.
Oligo-dT pulldown
NSC34 cells cultured with 10 μm HA peptide or HA-poly-(PR)20 peptide for 4 h were fixed with 4% paraformaldehyde and then lysed with 1% triton X-100 with syringe and 26-G needle. After centrifugation, the cell lysates were pulldowned with oligo-dT beads (New England Biolab). Briefly, the lysates were incubated with oligo-dT beads for 1 h at 4°C in a pulldown buffer (20 mm Tris–HCl, pH 7.5, 0.5% lithium dodecyl sulfate, 500 mm LiCl), washed twice with washing buffer (20 mm Tris–HCl, pH 7.5, 0.1% lithium dodecyl sulfate, 150 mm LiCl), followed by immunoblot analyses.
HSP70-binding assay
Recombinant HSP70 and HSP40 (Boston Biochem) were incubated with HA-poly-(PR)20 peptide and 1 mm adenosine triphosphate in the presence or absence of 1 mg/ml RNA or poly-rA for 1 h at 37°C. The complex was immunoprecipitated with anti-HA antibody followed by immunoblot analysis.
DSP crosslinking assay
One millimolar of HA-tagged poly-(PR)20, HA-tagged poly-(GR)20, HA-tagged poly-(GA)20, were mixed with 1 mg/ml of RNA or homopolymeric ribonucleotides (poly-rA, poly-rG, poly-rC and poly-rU) and incubated for 5 min at room temperature. Then, 20 mm DSP was added (final 2 mm) and incubated for 30 min at 37°C, followed by Coomassie blue staining.
Reverse transcriptase–polymerase chain reaction
NSC34 cells were treated with 10 μm HA-poly(PR)20, 10 μm HA-poly(GA)20 or untreated for 6 or 24 h. mRNA were purified with RNeasy kit (Promega) and reverse transcribed with M-MLV reverse transcriptase (Promega). Mouse EAAT2 cDNA fragment spanning from Exon 7 to Exon 11 was amplified with primers; 5′-TGCTTGATTTGTGGGAAGATCATCG-3′ and 5′-TGCAGTCAGCTGACTTTCCATTGG-3′. Mouse RAN GTPase cDNA fragment spanning from Exon 1 to Exon 5 was amplified with primers; 5′-AGAGCCGCAGGTCCAGTTCAAG-3′ and 5′-GAGAGCAGTCGTCTGAGCAAC-3′. Mouse GADD45a cDNA fragment spanning from Exon 1 to Exon 4 was amplified with primers; 5′-ATGACTTTGGAGGAATTCTCGGC-3′ and 5′-AAGGCAGGATCCTTCCATTGTGATG-3′.
Immunocytochemistry
NSC34 cells cultured with 5 μm HA peptide or HA-poly-(PR)20 peptide for overnight were fixed with 4% paraformaldehyde for 10 min and then permeabilized with 0.2% triton X-100 for 5 min. After blocking of non-specific binding with bovine serum albumin, cells were incubated with mouse monoclonal anti-hnRNPA1 antibody (Abcam) and rabbit anti-HA antibody (Genetex). After three washes, the cells were incubated with Alexa Fluor 488-conjugated anti-mouse secondary antibody or Alexa Fluor 568-conjugated anti-rabbit secondary antibodies (Thermo Fisher) and examined with a confocal microscope (Eclipse 80i; Nikon, Tokyo, Japan).
Cell fractionation immunoprecipitation
Fractionation of cytosol and nucleus from NSC34 cells treated 5 μm HA peptide or HA-poly-(PR)20 peptide for overnight was done by ProteoExtract Subcellular Proteome Extraction Kit (Millipore) following the manufacturer's protocol. After fractionation, immunoprecipitation using the nuclear fraction was performed with anti-HA antibody-conjugated agarose beads. The beads were washed five times with the immunoprecipitation buffer and analyzed by SDS–PAGE and immunoblot analysis.
Statistical analysis
Statistical analysis of the data was performed by one-way analysis of variance (ANOVA) followed by Dunnet's test or Student's t-test using SPSS 22 (IBM).
Study approval
Studies involving human subjects were approved by the Washington University Human Studies Committee and the Clinical Research Unit Advisory Committee (an Institute of Clinical and Translational Sciences resource unit). Written informed consent was received from all participants prior to inclusion in the study.
Patient samples
Patients samples (C9ORF72-negative patient 1; male, 70 years old, C9ORF72-negative patient 2; male, 74 years old, C9ORF72-positive patient 1; female, 65 years old, C9ORF72-positive patient 2; male, 70 years old) were obtained through appropriate consenting procedures for the collection and use of the human brain tissues. The GGGGCC hexanucleotide expansion in C9ORF72 was confirmed by repeat-primed PCR assay.
Supplementary Material
Authors’ contributions
K.K., T.Y., M.B.H., T.M.M. and F.U. contributed to conception and design of the study. K.K., T.Y., A.J.C., J.M. and M.K. contributed to data collection and analysis. K.K., T.Y., T.M.M. and F.U. wrote and edited the manuscript.
Conflict of Interest statement. None declared.
Funding
This work was partly supported by the grants from the National Institutes of Health/NIDDK (DK067493 and DK020579 to F.U.), the National Institutes of Health/NINDS (NS078398 to T.M.), the Washington University Institute of Clinical and Translational Sciences (grant UL1 TR000448) from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health, and Japan Intractable Diseases Research Foundation to K.K. T.Y. was supported by the fellowship from Japan Society for the Promotion of Science.
Supplementary Material
References
- 1.Ferrari R., Kapogiannis D., Huey E.D., Momeni P. (2011) FTD and ALS: a tale of two diseases. Curr. Alzheimer Res., 8, 273–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L. et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron, 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J. et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron, 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majounie E., Renton A.E., Mok K., Dopper E.G., Waite A., Rollinson S., Chio A., Restagno G., Nicolaou N., Simon-Sanchez J. et al. (2012) Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol., 11, 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters O.M., Ghasemi M., Brown R.H. Jr (2015) Emerging mechanisms of molecular pathology in ALS. J. Clin. Invest., 125, 1767–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciura S., Lattante S., Le Ber I., Latouche M., Tostivint H., Brice A., Kabashi E. (2013) Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann. Neurol., 74, 180–187. [DOI] [PubMed] [Google Scholar]
- 7.Donnelly C.J., Zhang P.W., Pham J.T., Haeusler A.R., Mistry N.A., Vidensky S., Daley E.L., Poth E.M., Hoover B., Fines D.M. et al. (2013) RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron, 80, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sareen D., O'Rourke J.G., Meera P., Muhammad A.K., Grant S., Simpkinson M., Bell S., Carmona S., Ornelas L., Sahabian A. et al. (2013) Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med., 5, 208ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori K., Weng S.M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H.A., Cruts M., Van Broeckhoven C. et al. (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science, 339, 1335–1338. [DOI] [PubMed] [Google Scholar]
- 10.Su Z., Zhang Y., Gendron T.F., Bauer P.O., Chew J., Yang W.Y., Fostvedt E., Jansen-West K., Belzil V.V., Desaro P. et al. (2014) Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron, 83, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizielinska S., Gronke S., Niccoli T., Ridler C.E., Clayton E.L., Devoy A., Moens T., Norona F.E., Woollacott I.O., Pietrzyk J. et al. (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science, 345, 1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon I., Xiang S., Kato M., Wu L., Theodoropoulos P., Wang T., Kim J., Yun J., Xie Y., McKnight S.L. (2014) Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science, 345, 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen X., Tan W., Westergard T., Krishnamurthy K., Markandaiah S.S., Shi Y., Lin S., Shneider N.A., Monaghan J., Pandey U.B. et al. (2014) Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron, 84, 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J. et al. (2005) The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res., 33, D284–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang X., Gold D., Kolaczyk E.D. (2011) Network-based auto-probit modeling for protein function prediction. Biometrics, 67, 958–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt E.K., Clavarino G., Ceppi M., Pierre P. (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods, 6, 275–277. [DOI] [PubMed] [Google Scholar]
- 17.Kuang E., Fu B., Liang Q., Myoung J., Zhu F. (2011) Phosphorylation of eukaryotic translation initiation factor 4B (EIF4B) by open reading frame 45/p90 ribosomal S6 kinase (ORF45/RSK) signaling axis facilitates protein translation during Kaposi sarcoma-associated herpesvirus (KSHV) lytic replication. J. Biol. Chem., 286, 41171–41182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raught B., Peiretti F., Gingras A.C., Livingstone M., Shahbazian D., Mayeur G.L., Polakiewicz R.D., Sonenberg N., Hershey J.W. (2004) Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J., 23, 1761–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baird T.D., Wek R.C. (2012) Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr., 3, 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding H.P., Zhang Y., Ron D. (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature, 397, 271–274. [DOI] [PubMed] [Google Scholar]
- 21.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R. et al. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell, 11, 619–633. [DOI] [PubMed] [Google Scholar]
- 22.Yamakawa M., Ito D., Honda T., Kubo K., Noda M., Nakajima K., Suzuki N. (2015) Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum. Mol. Genet., 24, 1630–1645. [DOI] [PubMed] [Google Scholar]
- 23.Mayer M.P., Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cel. Mol. Life Sci., 62, 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendershot L.M., Valentine V.A., Lee A.S., Morris S.W., Shapiro D.N. (1994) Localization of the gene encoding human BiP/GRP78, the endoplasmic reticulum cognate of the HSP70 family, to chromosome 9q34. Genomics, 20, 281–284. [DOI] [PubMed] [Google Scholar]
- 25.Turturici G., Sconzo G., Geraci F. (2011) Hsp70 and its molecular role in nervous system diseases. Biochem. Res. Int., 2011, 618127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong J., Lasko P. (2012) Translational control in cellular and developmental processes. Nat. Rev. Genet., 13, 383–394. [DOI] [PubMed] [Google Scholar]
- 27.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R. et al. (2014) C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature, 507, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fratta P., Mizielinska S., Nicoll A.J., Zloh M., Fisher E.M., Parkinson G., Isaacs A.M. (2012) C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep., 2, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen E., Sharma M.R., Shi X., Agrawal R.K., Joseph S. (2014) Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell, 54, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno J.A., Radford H., Peretti D., Steinert J.R., Verity N., Martin M.G., Halliday M., Morgan J., Dinsdale D., Ortori C.A. et al. (2012) Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature, 485, 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi S., Serrano A., Gerbino V., Giorgi A., Di Francesco L., Nencini M., Bozzo F., Schinina M.E., Bagni C., Cestra G. et al. (2015) Nuclear accumulation of mRNAs underlies G4C2-repeat-induced translational repression in a cellular model of C9orf72 ALS. J. Cell Sci., 128, 1787–1799. [DOI] [PubMed] [Google Scholar]
- 32.Herce H.D., Garcia A.E., Litt J., Kane R.S., Martin P., Enrique N., Rebolledo A., Milesi V. (2009) Arginine-rich peptides destabilize the plasma membrane, consistent with a pore formation translocation mechanism of cell-penetrating peptides. Biophys. J., 97, 1917–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.