Abstract
Members of Myxozoa, a parasitic metazoan taxon, have considerable detrimental effects on fish hosts and also have been associated with human food-borne illness. Little is known about their biology and metabolism. Analysis of the genome of Thelohanellus kitauei and comparative analysis with genomes of its two free-living cnidarian relatives revealed that T. kitauei has adapted to parasitism, as indicated by the streamlined metabolic repertoire and the tendency toward anabolism rather than catabolism. Thelohanellus kitauei mainly secretes proteases and protease inhibitors for nutrient digestion (parasite invasion), and depends on endocytosis (mainly low-density lipoprotein receptors-mediated type) and secondary carriers for nutrient absorption. Absence of both classic and complementary anaerobic pathways and gluconeogenesis, the lack of de novo synthesis and reduced activity in hydrolysis of fatty acids, amino acids, and nucleotides indicated that T. kitauei in this vertebrate host–parasite system has adapted to inhabit a physiological environment extremely rich in both oxygen and nutrients (especially glucose), which is consistent with its preferred parasitic site, that is, the host gut submucosa. Taking advantage of the genomic and transcriptomic information, 23 potential nutrition-related T. kitauei-specific chemotherapeutic targets were identified. This first genome sequence of a myxozoan will facilitate development of potential therapeutics for efficient control of myxozoan parasites and ultimately prevent myxozoan-induced fish-borne illnesses in humans.
Keywords: Myxozoa, genome and transcriptome, nutrient deprivation, food-borne illness
Introduction
The international market for fish and fish products has grown remarkably, and global fish consumption was estimated to be 140.5 million tons in 2013 (http://www.thefishsite.com/reports/?id=2253&country=WW). Along with the rapid growth in fish consumption, incidence of fish-borne parasitic zoonoses also increased (Dos Santos and Howgate 2011). Of the big family of fish parasites, myxozoans constitute an important group including more than 2,100 species in 58 genera, most of which are coelozoic or histozoic parasites in fish. The life cycles of myxosporeans involve two distinct spore stages, the actinospore in an invertebrate host (usually an annelid), and the myxospore in the vertebrate host (Lom and Dykova 2006). Although most myxozoans are not highly pathogenic for fish hosts, some cause disease and impact upon wild and farmed fish populations (Gomez et al. 2014). The impact of myxozoans has further exacerbated, as indicated by the observed increase in fish-to-fish transmission rate as well as the emergence of fish-to-human transmission (i.e., the recent outbreaks of food poisoning in Japan associated with Kudoa Septempunctata; Kent et al. 2001; Lom and Dykova 2006; Sitja-Bobadilla 2008; Kawai et al. 2012).
Myxozoa has been confirmed as Metazoa rather than Protozoa by the detected metazoan characteristics, multicellularity of some life stages, tight junctions, collagen production (Siddall et al. 1995; Kent et al. 2001; Canning and Okamura 2004), and phylogenetic analyses (Smothers et al. 1994). However, the placement of Myxozoa within Metazoa, either as a sister taxon to Bilateria or within Cnidaria, was still controversial (Jimenez-Guri et al. 2007; Evans et al. 2010; Nesnidal et al. 2013). Recent phylogenomic data supported to place Myxozoa within Cnidaria. However, these phylogenomic analyses either only used expressed sequence tag data which could not reject a bilaterian origin of Myxozoa with the topology test (Jimenez-Guri et al. 2007) or was very weakly supported by a Bayesian inference analysis with the CAT model (Nesnidal et al. 2013). Therefore, the relative phylogenetic position of Myxozoa within Metazoa remains unresolved and is compelling for further study.
In China, more than 600 species of myxozoan parasites have been reported to infest freshwater and marine fish, and some of them cause severe economic losses in the aquaculture industry (Chen and Ma 1998). Particularly, intestinal giant-cystic disease, caused by the myxozoan species Thelohanellus kitauei, has been recognized as the most detrimental disease of farmed carp (Egusa and Nakajima 1981; Chun et al. 1988; Shin et al. 2011), the most commonly farmed fish species with the highest production in China. Around 20% of farmed carp were killed annually due to this disease, leading to an economic loss of approximately 50 million dollars in 2010. Infected carps suffer from profound intestinal swelling and malabsorption (Egusa and Nakajima 1981; Chun et al. 1988; Shin et al. 2011). In addition to an increased erythrocyte sedimentation rate, other hematological parameters such as red blood cells, hemoglobin, hematocrit, albumin, total protein, triglycerides, phosphorus, calcium, and biological mass all decreased significantly (P < 0.05) in T. kitauei-infected carp (Wang et al. 2005). These changes, associated with severe anemia and cacotrophia, lead to secondary enteritis and ultimately death of infected fish (Egusa and Nakajima 1981; Chun et al. 1988; Wang et al. 2005).
In contrast to their evolutionary and economic significance, knowledge of the general biology of myxozoans is very limited, largely due to the inability to culture these parasites in vitro. Genomic studies represent a promising strategy to investigate the biology and metabolism of myxozoans. However, no genome sequence of any myxozoan has been available. Here, we present the genome sequence of T. kitauei. Genomic analysis was conducted to provide insights into the biology of this myxozoan parasite, focusing on the nutrition and metabolism aspect. Phylogenomic analyses using new genomic data of T. kitauei and comparative analyses of minicollagen and mesodermal genes robustly confirmed Myxozoa to be a highly derived Cnidarian taxon, a sister group of Medusozoa (Feng et al. 2014).Taking advantage of this information, we conducted comparative genomic analysis between the parasitic T. kitauei and its two free-living cnidarian relatives, that is, Hydra magnipapillata (Chapman et al. 2010) and Nematostella vectensis (Putnam et al. 2007) to investigate the genomic evolution associated with parasitic lifestyle. Also, aided by transcriptome sequencing of the myxospore stage, we identified a range of potential T. kitauei chemotherapeutic targets, which may facilitate the development of novel therapeutic agents against myxozoan parasites and relieve the potential threat of myxozoan zoonoses to human consumers.
Materials and Methods
Parasite Collection
Infected common carp were collected from a approximately 100-acre aquaculture pond (∼1.5 m deep, 30 °C, pH 7.5–8.0) in Wuqing, Tianjin, China in July 2007. After euthanasia, the intestines of infected fish were obtained by dissection. Cysts found on the surface of the intestines were carefully collected from the intestinal tissue and rinsed with sterile water. The cysts were cut open and spores were washed out with sterile phosphate-buffered saline (pH 7.0). The myxospore suspensions were further filtered with a 300 -µm mesh sieve to remove residual host fish tissue. The filtrates were centrifuged at 6,000 × g for 10 min, and the myxospore pellets were resuspended in 9.5 ml of sterile phosphate-buffered saline (pH 7.0). Host cells were lysed by addition of 0.5 ml of 20% sodium dodecyl sulfate and incubated at 25 °C for 10 min. After centrifugation at 8,000 × g for 10 min, the supernatants were discarded and spores were resuspended in 10 ml of sterile phosphate-buffered saline. The centrifugation and resuspension were repeated once. Finally, the myxospores were microscopically with care examined to ensure that host fish tissue and cells had been removed.
DNA and RNA Extraction
Spore samples (2 g wet weight) were frozen in liquid nitrogen and then ground with a mortar and pestle. DNA was extracted as described previously (Yap and Thompson 1987). The purity and yield of the DNA was assessed spectrophotometrically. The samples were checked by polymerase chain reaction (PCR) with host-specific primers to further ensure that the samples did not contain host fish DNA (Rag1-R: 5′-GACACTATGGAGAAAGGGAGGTGGAGTT-3′; Rag1-F: 5′-GGGAAGCAGAGGTCGCAGTTGGAGG-3′) (Fan et al. 2009).
For RNA extraction, purified spores (1 g wet weight) were frozen in liquid nitrogen and ground with a mortar and pestle. The lysate was transferred to a 1.5-ml RNase-free microcentrifuge tube and homogenized in 350 μl Buffer RLT Plus (Qiagen) with a vortex mixer (MVS-1, Beijing Beide Science Co.). RNA was purified using the RNeasy Plus Mini kit (Qiagen). RNA concentration and purity were assessed with a Nanodrop spectrophotometer (Thermo Scientific).
Species Identification
The species of the spores was determined by morphological examination via light and scanning electron microscopy (supplementary material and fig. S1, Supplementary Material online) and by phylogenetic analysis of 18S rDNA sequences focusing only on Thelohanellus spp. by RAxML version 7.2.6 (Stamatakis 2006) with the best-fitted model GTRGAMMAI.
Genome Sequencing
For Roche 454 library construction and sequencing, two libraries (8- and 20-kb fragments) were prepared using the GS FLX Titanium Library Preparation kit according to the manufacturer’s protocols (Roche Applied Science, Mannheim, Germany). The purified genomic DNA is fragmented by hydrodynamic shearing to generate 8 kb and 20 kb span paired-end libraries. Quantitated DNA fragments from the paired-end library, flanked with proper amplification and sequencing adaptors, were immobilized onto microspheres (beads) and the entire bead-bound library was then emulsified with the amplification reagents (GS FLX Titanium LV emPCR kit). After the emPCR step, the DNA-carrying beads were cleaned and enriched before loaded into the wells of a PicoTiterPlate device (GS FLX Titanium PicoTiterPlate Kit [70 × 75]) at the density of no more than a single DNA bead per well. The loaded PTP device was then inserted into the Genome Sequencer FLX Instrument, and sequencing reagents were sequentially flowed over the plate. The sequencing was conducted using the GS FLX Titanium Sequencing Kit and the Genome Sequencer FLX Instrument strictly following the recommendations of the manufacturer (Roche Applied Science, Mannheim, Germany). The raw output comprising a set of digital images (PIF files) was processed to obtain the sequence of the DNA library fragments/reads and then subjected to downstream analysis. The quality of the data obtained from the sequencing was verified using the Roche 454 Sequencing System Software version 2.6 (Roche 454 Life Sciences, Branford, CT, USA), including normalization, correction, and quality filtering steps, to generate Standard Flowgram Format files containing the base called read sequences and per base quality scores as described in the Software manual.
For short-insert (∼500 bp) DNA libraries, that is, standard DNA libraries, we followed the manufacture’s protocol (Illumina). Briefly, 5 μg of genomic DNA was fragmented by nebulization with compressed nitrogen gas. The DNA ends were polished and an “A” base was added to each end of the fragments. Next, the DNA adaptors (Illumina) with a single “T” overhang at the 3′-end were ligated to the products above. The ligation products were separated on a 2% agarose gel and an approximately 2-mm wide gel slice containing DNA of the desired size was excised (Qiagen Gel Extraction Kit). In this step, DNA fragments in a narrow size range were selected to facilitate the downstream assembly process. For long (∼3 Kb) mate-paired libraries, the manufacture’s mate pair library kit (Illumina) was used. Around 10–30 μg genomic DNA was fragmented by HydroShear and polished with biotin labeled dNTPs. The fragments in the size range of 2–5 Kb were selected by gel excision. The size-selected DNA fragments were circularized by intramolecular ligation of the blunt ends, and the remaining linear DNA fragments from the circularization reaction were digested by DNA exonuclease. Then, the circularized DNA was randomly fragmented to lengths of approximately 400 bp, and the fragments that contain the biotinylated ends of the original size-selected fragment were purified using streptavidin-coated magnetic beads. The fragments bound to the streptavidin beads were end-repaired and A-tailed, followed by attachment of the illumine paired-end oligo adapters by ligation. After the adapter ligation, PCR was carried out to simultaneously enrich and amplify those DNA fragments with adapter molecules on both ends. The product of the PCR amplication is a DNA smear of different fragment sizes. Fragments in the range of 350-650 bp were excised from the gel for cluster formation and sequencing. For quality control of Illumina sequence data, the adaptors were stripped with SeqPrep (http://omictools.com/seqprep-s1982.html), and ConDe Tri_v2.1.pl (Smeds and Kunstner 2011) was used to trim low-quality end sequences of the reads with a Phred quality score lower than 20, as well as non-A/T/G/C bases. Reads with a length lower than 25 bp after trimming were discarded. Paired reads with overlap were merged into single reads by SeqPrep.
Transcriptome Sequencing
mRNA was isolated from total RNA using Oligo-dT beads (Invitrogen) and then fragmented by heating at 94 °C. First-strand complementary DNA (cDNA) was synthesized with random hexamer primers and second-strand cDNA synthesized with DNA polymerase. Double-stranded cDNA was end-repaired/blunt-ended with Klenow polymerase, T4 DNA polymerase, and T4 polynucleotide kinase. A single adenosine moiety was added to the cDNA using Klenow exo- and dATP. All the enzymes used above were included in the kit (mRNA-Seq Sample Pre Kit) from Illumina. Illumina adapters (containing primer sites for sequencing and flowcell surface annealing) were ligated onto the blunt-ended cDNA, and the library of DNA fragments (200–250 bp) was separated from unligated adapters by gel excision. Libraries were amplified by PCR with Phusion polymerase, denatured with 100 mM sodium hydroxide, and diluted in hybridization buffer before they were loaded onto a single lane of an Illumina GA flowcell. Cluster formation, primer hybridization, and paired-end sequencing (101 × 2 cycles) were performed using Illumina reagents. Expression level was determined using the mapped reads per kilobase of exon per million mapped reads (RPKM) in the spore cells.
Genome Assembly
The paired-end reads and mate-paired reads from Solexa sequencing were assembled using SOAPdenovo software, which uses the de Bruijn graph data structure to construct contigs (Li et al. 2010). SOAPdenovo was used to assemble short reads generated by the Illumina Genome Analyzer. Contigs produced by SOAPdenovo were split into 2-kb windows with 1-kb overlap with the EMBOSS splitter program (Rice et al. 2000). The Roche 454 reads and the split Illumina sequences were then processed with the Newbler sequence assembler (version 2.3) to produce super-reads in a hybrid assembly. Finally, gaps between contigs were filled by mapping and local assembly of the Solexa sequencing reads using GapCloser (http://soap.genomics.org.cn/soapdenovo.html) in iterative mode. The GC content of each assembled scaffold was calculated. The mean GC content was 28.8% with a standard deviation of 5.9%. Only sequences with GC content within three standard deviations of the mean were included in the genome assembly, and 70 scaffolds with GC content higher than 47% were discarded (0.11 Mb total).
Repetitive sequences were identified by searching Repbase using RepeatMasker (http://www.repeatmasker.org/) with default parameter settings and by de novo repetitive sequence search using Repeat Modeler (http://www.repeatmasker.org/RepeatModeler.html).
Gene Prediction
A strategy combining RNA-Seq and de novo approaches was used to predict protein-coding genes. First, the RNA-Seq reads were assembled to generate the training set for the de novo algorithms. Generally, a combination of mapping-based assembly (RNA-Seq reads were mapped to reference genome and assembled) and de novo methods were used. For mapping-based assembly, RNA-Seq reads were mapped to the assembled genome using SOAPsplice (Huang et al. 2011), allowing up to two mismatches, and assembled using the Cufflinks software (http://cufflinks.cbcb.umd.edu/). For de novo assembly, transcripts were constructed using Trinity software (Grabherr et al. 2011). The assembled transcripts obtained by the two methods were pooled and validated by aligning them to the assembled genome with PASA (http://pasa.sourceforge.net/) and GMAP (Wu and Watanabe 2005). The PASA-recommended best transcript candidates were used to train the gene prediction programs. Four programs were utilized for de novo gene prediction, that is, GeneMark (Lukashin and Borodovsky 1998), Augustus (Stanke et al. 2008), GlimmerHMM (Majoros et al. 2004), and SNAP (Korf 2004). The final gene set was created by merging all of the gene sets using Evidence Modeler (http://evidencemodeler.sourceforge.net/).
Functional Annotation
Functions were assigned and classified by searching for sequence similarity with BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against the eggNOG database (http://eggnog.embl.de), Kyoto Encyclopedia of Genes and Genomes (KEGG) reference database (http://www.genome.jp/kegg/), and the National Center for Biotechnology Information (NCBI) nonredundant protein sequence database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/). Gene Ontology (http://www.geneontology.org/) was used to classify and functionally annotate genes. Protein domains were predicted using the Pfam database (http://pfam.sanger.ac.uk/) for gene function assignment. The tRNA and rRNA genes were predicted with tRNASCAN-SE (http://lowelab.ucsc.edu/tRNAscan-SE/). Potential secreted proteins were predicted by SignalP v4.1 (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al. 2011) and PrediSi (http://www.predisi.de/) (Hiller et al. 2004). Predicted proteins were classified as proteases by querying the MEROPS database (http://merops.sanger.ac.uk/) (Rawlings et al. 2014) with BLASTp with cut-off E value as 1 e −5. If a predicted protein was found to have more than one functional domain, only the domain with the lowest E value was retained. Putative α/β-hydrolases were classified according to BLASTp results (E value cut-off = 1 e −3) obtained against the Lipase Engineering database (http://www.led.uni-stuttgart.de/). Carbohydrate-active enzymes (carbohydrate esterases, glycoside hydrolases, glycosyltransferases, polysaccharide lyases, and carbohydrate-binding modules) were annotated using the dbCAN Web server (http://csbl.bmb.uga.edu/dbCAN/) (Yin et al. 2012) at an E value cut-off of 1 e −5. Transporters were predicted from the genome sequence by the TransportTP tool (http://bioinfo3.noble.org/transporter/) (Li et al. 2009). Reference organisms for our TransportTP analysis were: Escherichia coli, Saccharomyces cerevisiae, Arabidopsis thaliana, Oryza sativa, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens (E value threshold = 0.1).
Results and Discussion
Species Identification
Cysts of an unknown origin were collected from the intestines of common carp and examined for parasitic infection. Isolated myxosporean-like spores from the cysts showed morphological and morphometric features nearly indistinguishable from those of the previously described T. kitauei (supplementary fig. S1A and B and table S1, Supplementary Material online; Egusa and Nakajima 1981; Zhou et al. 1998; Xie et al. 2000; Liu et al. 2011). BLAST analysis showed that 18S rRNA sequence of the isolated species was 99% identical to that of the T. kitauei-China (HQ115585) and T. kitauei-Korea (HM624024; supplementary table S2, Supplementary Material online). Furthermore, the phylogenetic analysis showed that the parasite from common carp in this study is clustered with T. kitauei-China (HQ115585) and T. kitauei-Korea (HM624024) with 100% bootstrap confidence values (supplementary fig. S1C, Supplementary Material online). These findings, combining with the fact that this parasite showed the same host and tissue specificity as T. kitauei, confirmed our isolate as T. kitauei.
Genome Sequencing and Assembly
To optimize genome assembly, two next-generation sequencing platforms, Roche 454 and Illumina, were used to sequence the genome of T. kitauei. We generated 1.48 Gb (∼8 × genome coverage; 8-kb library with a Q20 percentage of 86.3% and 20-kb library with a Q20 percentage of 90.5%) from the Roche 454 sequencing and 5.5 Gb (∼29 × genome coverage; 3-kb mate-paired library with a Q20 percentage of 75.4%) and 2.5 Gb (∼13 × genome coverage; 500-bp paired-end library with a Q20 percentage of 80.1%) from Illumina sequencing. A hybrid genome assembly of 454 and Illumina sequences was conducted by first assembling the Illumina mate-pair and paired-end data with SOAPdenovo, followed by fragmentation of the SOAPdenovo assembled contigs and assembly with the Roche 454 data using Newbler. This approach improves the performance of Newbler with Roche 454 data by correcting the homopolymer and insertion/deletion errors common to Roche 454 sequencing. Potential contamination of the parasite samples with carp DNA was excluded in four ways: 1) During sample collection, the carp intestinal tissue was carefully removed from the cysts, and the collected T. kitauei spores were repeatedly washed; 2) The purity of the T. kitauei DNA was checked by PCR with carp-specific primers, and no carp DNA was detected; 3) In the bioinformatics analysis, assembled scaffolds showing high similarity (E value ≤ 1 e−20) to fish and bacterial sequences in GenBank were excluded; 4) A mean ± standard deviation (n = 3) approach was used to exclude potential carp scaffolds based on the fact that the mean GC content of T. kitauei (∼0.29; supplementary fig. S2, Supplementary Material online) is lower than that of fish. After the filtering process, 150.7 Mb were assembled into 5,610 scaffolds (N50 = 170 kb), and over 90% of the genome assembly was included in 1,336 scaffolds.
The genome size of T. kitauei was estimated based on 17-mer analysis (Kurtz et al. 2008; Wang et al. 2011; supplementary fig. S3, Supplementary Material online). To further eliminate contaminating fish and bacterial reads, only the reads mapped to the draft assembly (∼1.3 Gb) were used. The peak of 17-mer frequency (M) correlates with sequencing depth (N), read length (L), and k-mer length (K), with the relationship expressed as M = N × (L − K + 1)/L. The genome size of T. kitauei was inferred to be 188.5 Mb, meaning that 80% of the genome sequences were present in our draft assembly of the T. kitauei genome. The loss of the remaining 20% of the genome sequences may be due to the presence of repeat sequences in the genome and/or the limitations of next-generation sequencing technologies.
Repeat Sequences
The assembled T. kitauei genome was found to contain approximately 13.8% interspersed repeat sequences (supplementary table S3, Supplementary Material online), and this number is likely underestimated owing to the limitations of genome sequencing strategies. Among the interspersed repeat sequences, approximately 7% were transposable DNA elements, including hAT-Charlie (2.3%) and TcMar-Tigger (0.2%); and some (0.07%) were long interspersed nuclear elements, while no short interspersed nuclear elements were observed. Moreover, a large number of DNA transposons and retrotransposons were found in the T. kitauei genome (supplementary table S4, Supplementary Material online). These elements can lead to increased expression if inserted in the vicinity of virulence-related genes (Faust and Guillen 2012). Proteins encoded by the transposons and retrotransposons fell into several families, including endonucleases, reverse transcriptases, transposases, and integrases (supplementary table S4, Supplementary Material online). One of these transposons encodes a retroviral aspartyl protease belonging to the expansive retroviral-like protease family, which may be involved in processing of polyproteins in T. kitauei.
Gene Models
With the combination of four gene-scanning programs, 16,638 protein-coding genes were annotated in the 150. 7-Mb draft genome assembly. Transcriptome data were also collected to verify the gene annotations of the T. kitauei genome. About 8,659 predicted genes, accounting for 51.5% of the total predicted genes (16,638 protein-coding genes), were expressed in the myxospores of T. kitauei. The numbers of genes with RPKM values in the range of 10–100, 100–1,000, and over 1,000 were 1,817, 715, and 121, respectively. Thereafter, we then compared the genome features of T. kitauei to those of the two closely related free-living cnidarian species H. magnipapillata and N. vectensis (table 1). Although the estimated genome size of T. kitauei is approximately half of that of N. vectensis and one-fifth of that of H. magnipapillata, the gene numbers among the three species were very similar. The percentages of coding sequences, gene density, and average exon size of T. kitauei were much higher than those of N. vectensis and H. magnipapillata (table 1). Similar to the ciliate Paramecium tetraurelia (Aury et al. 2006), T. kitauei has relatively short introns, with the majority in the range of 20–25 bp (supplementary fig. S4, Supplementary Material online). The short introns may not allow alternative splicing (Aury et al. 2006). However, the benefits, if any, are unclear.
Table 1.
Summary of Genomic Features of Thelohanellus kitauei and Two Free-Living Cnidarians
| Genome features | Parasitic | Free-Living |
|
|---|---|---|---|
| Thelohanellus kitauei | Hydra magnipapillata (Chapman et al. 2010) | Nematostella vectensis (Putnam et al. 2007) | |
| Total genome size (Mb) | 150.7 | 852.2 | 356.6 |
| Total coding size (Mb) | 20 | 21.5 | 30.5 |
| Total coding ratio (%) | 13.3 | 2.5 | 8.6 |
| Gene number | 16,638 | 17,918 | 26,676 |
| Average gene length (kb) | 1,210 | 14,568 | 5,869 |
| Gene density (genes per Mb) | 110 | 21 | 75 |
| Average coding sequence size (kb) | 717 | 1,267 | 1,223 |
| Average exon number | 3 | 6 | 5 |
| Average exon size (bp) | 235 | 218 | 208 |
| Average intron size (bp) | 240 | 2,653 | 1,122 |
| Total GC content (%) | 37.5 | 27.8 | 40.6 |
| GC content in coding region (%) | 39.0 | 33.5 | 46.5 |
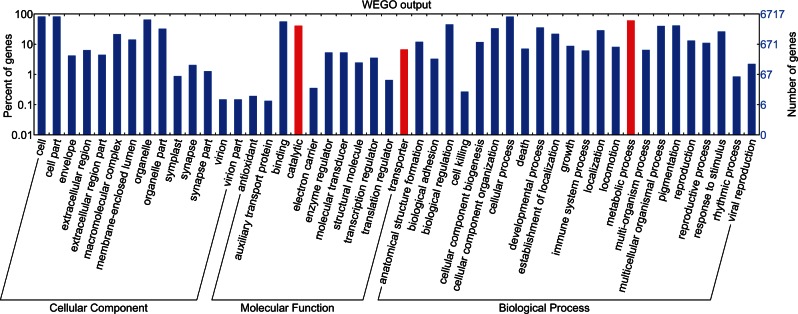
Approximately 55% of the putative genes have homologs in the NCBI database and >3,500 gene products could be assigned to gene ontology terms. As shown in figure 1, a large number of T. kitauei genes appear to be involved in multicellular organismal processes. In addition, KEGG pathway analysis revealed that T. kitauei carries genes encoding components of the Hedgehog, Jak-STAT, Notch, TGF-β, mTOR, and Wnt pathways (supplementary table S5, Supplementary Material online), which are signaling pathways present in multicellular Metazoa but not in fungi or choanoflagellates. Together, these results are coincident with the fact that T. kitauei is a multicellular metazoan.
Fig. 1.—
Overall function catalogs of T. kitauei gene annotation. Function catalogs described in this study are highlighted in red.
Proteases and Protease Inhibitors As Main Enzymes for Nutrient Digestion
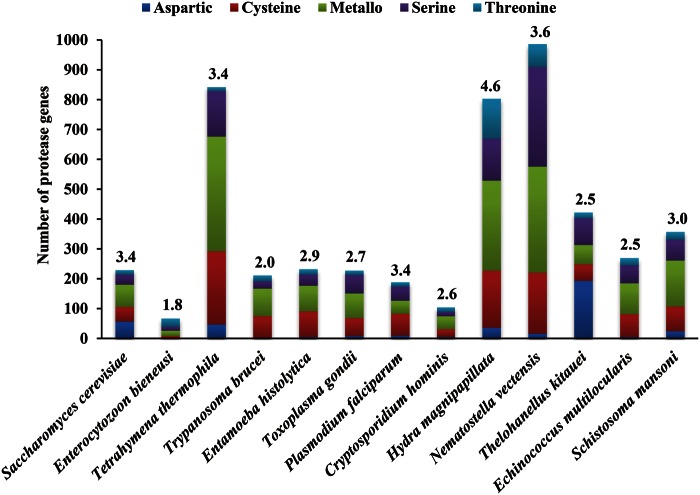
We identified 422 protease (including 34 secreted) homologues in T. kitauei through the MEROPS database, constituting around 2.5% of the proteome (fig. 2 and supplementary tables S6–S8, Supplementary Material online). The number of proteases in T. kitauei is much greater than the number of putative lipid (22 including 1 secreted) and carbohydrate (44 including 4 secreted) digestive enzymes. The number of digestive enzymes in T. kitauei was considerably lower than that in the two free-living relatives, with the number in T. kitauei representing 42.8%/52.6%, 16.3%/37.3%, and 8.5%/19.4% of the numbers in N. vectensis/H. magnipapillata, for proteases, lipid and carbohydrate digestive enzymes, respectively. As noted, the scale of enzyme number reduction in T. kitauei as compared with the free-living relatives was lower for proteases than that of the lipid and carbohydrate digestive enzymes, suggesting that proteases are the main enzymes involved in nutrient digestion (supplementary tables S7–S12, Supplementary Material online).
Fig. 2.—
Comparison of proteases in T. kitauei and other organisms. Values above column are the percentage of the proteases within the total proteome of that species.
Compared with 11 other free-living and parasitic fungi, protozoans, and metazoans (fig. 2 and supplementary table S6, Supplementary Material online), the distribution of proteases in T. kitauei is clearly distinct: T. kitauei has a relatively large proportion of aspartic proteases (46% vs. 0−25%) but small proportion of cysteine proteases (13% vs. 12−38%) and metalloproteases (16% vs. 27−46%; fig. 2). Aspartic proteases subfamily A11X (the transposon endopeptidases) and A33 (skin-specific retroviral-like aspartic protease) are also expanded in T. kitauei relative to H. magnipapillata and N. vectensis (supplementary table S7, Supplementary Material online). These proteins are highly related to hemoglobin catabolism, transposon multiplication, and processing of profilaggrin (Matsui et al. 2011) and therefore likely account for the significant decrease in red blood cells in T. kitauei-infected carp (Wang et al. 2005). The proportion of astacin metalloproteases in T. kitauei (0.2% of total proteases) were much lower than those in two free-living cnidirians (8.5% and 8.9% of total proteases in H. magnipapillata and N. vectensis, respectively; supplementary table S7, Supplementary Material online). As astacin metalloproteases were reported to have the function for degradation of hydra extracellular matrix (ECM; Sarras et al. 2002), which might be beneficial for a myxozoan parasite, the low abundance of astacin metalloproteases may suggest that T. kitauei may have distinct ECM degradation pathway that may involve other metalloproteases or members of serine and cysteine proteinases (Lu et al. 2011).Other abundant protease families in at least one of two free-living cnidarians, such as the ubiquitin-specific proteases (C19 family), lysostaphin-type enzymes (M23B subfamily), and γ-glutamyl transferases (T3 family), were also largely reduced in T. kitauei (supplementary table S7, Supplementary Material online). Furthermore, around 40% of the protease families that were present in the two free-living cnidarians are absent in T. kitauei (supplementary table S7, Supplementary Material online). Taken together, these observations indicated a different life styles of T. kitauei from its free-living relatives.
Relative to the number of proteases, there were more (206) putative protease inhibitors in T. kitauei than in the two free-living cnidarians (94–164), although the diversity (8 vs. 15–18 families) was lower in T. kitauei (supplementary table S13, Supplementary Material online). Moreover, the serpin family (I04), which contains inhibitors of serine and cysteine endopeptidases, was greatly expanded in T. kitauei (182 vs. <5 in H. magnipapillata and N. vectensis).
Thirty-four proteases contain signal peptides (potential secretory pathway-targeted proteins, supplementary table S8, Supplementary Material online). Within them, 5 of the 19 proteases in the C1A subfamily and 2 of the 5 kexin-type peptidases in the S8B subfamily are highly expressed (RPKM values ≥ 100) in the myxospore stage of T. kitauei. Nineteen protease inhibitors were putatively secretory in T. kitauei, with the majority (14) as serine and cysteine protease inhibitors belonging to the serpin, serine carboxypeptidase Y (I51), and Kunitz-type (I02) families, which may regulate the activities of host-derived serine and cysteine proteases. Notably, two of the secretory protease inhibitors from the I25A and I51 subfamily are highly expressed in the myxospore stage. Proteases and their inhibitors have been reported as important factors for invasion and immune evasion of parasites (Toubarro et al. 2010; Doyle et al. 2011; Faria et al. 2011; Alam et al. 2013). The highly expressed (RPKM values ≥ 100) secreted proteases and inhibitors observed in T. kitauei myxospore stage suggest their putative involvement in these functions (supplementary table S8, Supplementary Material online).
Nutrient Absorption Mainly Depends on Endocytosis and Secondary Carriers
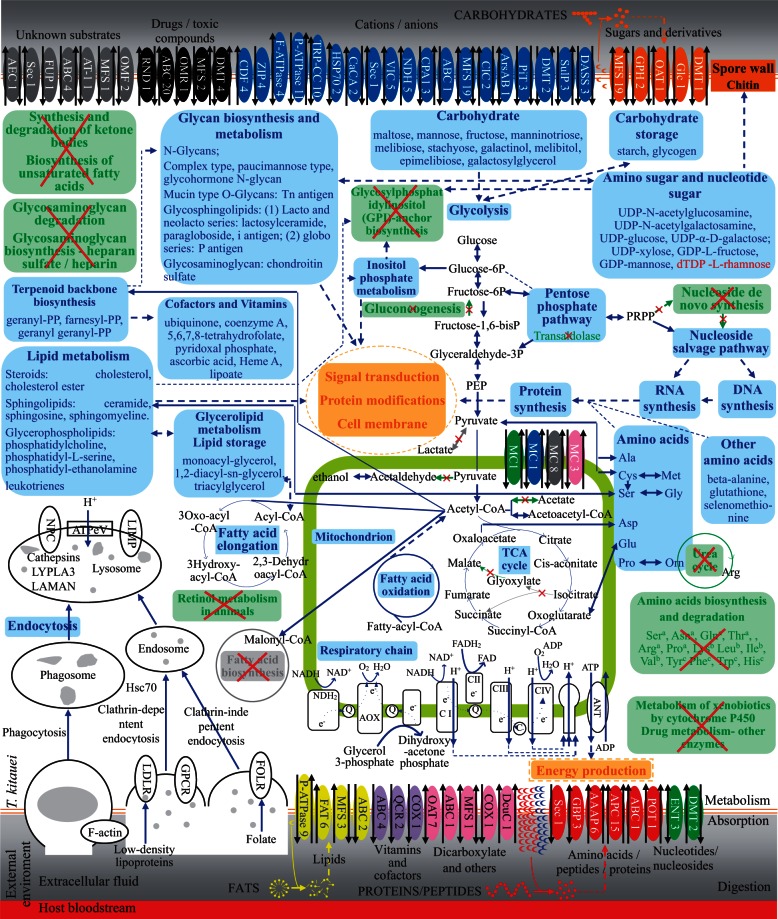
Endocytosis (classified as clathrin-mediated endocytosis, clathrin-independent endocytosis, and phagocytosis) has been confirmed to be central in the pathogenesis of many parasites, including Leishmania spp., Giardia lamblia, trypanosomatids, Entamoeba histolytica, and Toxoplasma gondii (Robibaro et al. 2001; Lopez-Soto et al. 2009; Rivero et al. 2010; Subramanya and Mensa-Wilmot 2010; Andrade-Neto et al. 2011; Rivero et al. 2011). Like the two free-living relatives, T. kitauei has most of endocytosis-related genes (134 including 72 phagosome-related genes; fig. 3 and supplementary fig. S5A, Supplementary Material online). Although failed to map to KEGG endocytosis pathway, the extensive collection of genes in low-density lipoprotein receptor family (122 genes) detected in T. kitauei suggests that low-density lipoprotein receptor-mediated endocytosis (a type of clathrin-dependent pathway) may play an important role in T. kitauei uptake of fish lipids (supplementary fig. S5A1 and table S14, Supplementary Material online). Although caveolin, the central component of caveolae-dependent endocytosis (a type of clathrin-independent pathway), was not detected in both T. kitauei and the two relatives, folate receptors, which are alternatively related to the clathrin-independent pathway, were identified. In addition, actin β/γ 1 is expressed at relatively high level (RPKM = 1,7702.82) in the transcriptome of myxospore-stage T. kitauei, and might be essential for phagocytosis (supplementary fig. S5A2, Supplementary Material online; Doherty and McMahon 2009; Mooren et al. 2012). Moreover, although T. kitauei has fewer lysosomal membrane proteins and acid hydrolases compared with its two free-living relatives (supplementary fig. S5A3, Supplementary Material online), its CD63 antigen (RPKMs of 2 of 3 genes are 2,395.23 and 2,638.33), Niemann-Pick C1 protein 1 (RPKMs of 2 of 7 genes are 355.94 and 589.40), cysteine protease cathepsin L (RPKMs of 5 of 20 genes are ranged from 113.45–4,977.53), and lipase LYPLA3 (RPKM of 1 of 3 genes is 610.83) are highly expressed in the myxospore stage. These results indicate that T. kitauei mainly carries out endocytosis, especially low-density lipoprotein receptor-mediated endocytosis, to acquire main nutrients such as lipids (supplementary fig. S5A, Supplementary Material online).
Fig. 3.—
An overview of nutrient digestion, absorption and metabolism in T. kitauei, as deduced from genome sequence analysis. The color-coding in the secretory digestive enzymes of T. kitauei: red, proteases; blue, proline iminopeptides; orange, carbohydrate-binding module family proteins and glycoside hydrolases; yellow, lipase. Transporters are grouped by substrate specificity: red, amino acids/peptides/proteins; orange, sugars and derivative; yellow, lipids; green, nucleotides/nucleosides; purple, vitamins and cofactors; pink, dicarboxylates and others organic; blue, cations and anions; black, drugs/toxic compounds; gray, unknown substrates; numbers indicated the number of predicted transporters of each type. The color-coding in metabolism of T. kitauei indicates: blue, enzymes or pathways present in all three species, T. kitauei, H. magnipapillata, and N. vectensis; green with a cross, enzymes or pathways present in at least one of the two free-living cnidarians but absent in T. kitauei; gray with a cross, enzymes or pathways absent in all three species; red, enzymes or pathways present only in T. kitauei; orange, putative functions. Amino acids that can be de novo synthesized and catabolized (a) or partially synthesized and catabolized (b) or only catabolized (c) by the two free-living cnidarians. Unbroken arrows indicate direct steps in a pathway, and broken arrows indicate multiple steps in a pathway not shown. Amino acids are indicated in the three letter code. AAAP, amino acid/auxin permease; APC, amino acid-polyamine-organocation family; ArsAB, arsenite-antimonite efflux family; ABC, ATP-binding cassette superfamily; AT-1, autotransporter-1; AEC, auxin efflux carrier; DcuC, C4-dicarboxylate uptake C; CaCA, Ca2+:cation antiporter family; Hsp70, cation channel-forming heat shock protein-70 family; CDF, cation diffusion facilitator; ClC, chloride carrier/channel family; DASS, divalent anion: Na+ symporter; DMT, drug/metabolite transporter; ENT, equilibrative nucleoside transporter; GBP, general bacterial porin; Sec, general secretory pathway family; GPH, glycoside-pentoside-hexuronide: cation symporter; F-ATPase, H+- or Na+-translocating F-type, V-type and A-type ATPase; PiT, inorganic phosphate transporter; MFS, major facilitator superfamily; MC, mitochondrial carrier; CPA1, monovalent cation: proton antiporter-1; OAT, organo anion transporter; OMF, outer membrane factor; FUP, outer membrane fimbrial usher porin family; OMR, outer membrane receptor; FAT, proposed fatty acid transporter; POT, proton-dependent oligopeptide transporter; COX, proton-translocating cytochrome oxidase; NDH, proton-translocating NADH dehydrogenase; QCR, proton-translocating quinol: cytochrome c reductase; Glc, PTS glucose-glucoside; P-ATPase, P-type ATPase; RND, resistance-nodulation-cell division; SulP, sulfate permease; TRP-CC, transient receptor potential Ca2+ channel; VIC, voltage-gated ion channel; ZIP, zinc (Zn2+)-iron (Fe2+) permease; PEP, phosphoenolpyruvic acid; NADH, nicotinamide adenine dinucleotide; Q, quinine; AOX, alternative oxidase; CI to CIV, complex I to complex IV; C, cytochrome c; ANT, adenine nucleotide translocator; FAD, flavin adenine dinucleotide; FADH2, reduced FAD; ADP, adenosine diphosphate; FOLR, folate receptor; GPCR, G protein-coupled receptors; LDLR, low-density lipoprotein receptor; Hsc70, heat shock 70 kDa protein 1/8; ATPev, V-type H+-transporting ATPase; LIMP, lysosomal integral membrane protein; NPC, niemann-pick C protein; LYPLA3, lysophospholipase III; LAMAN, lysosomal alpha-mannosidase; F-actin, actin beta/gamma 1; TCA, tricarboxylic acid.
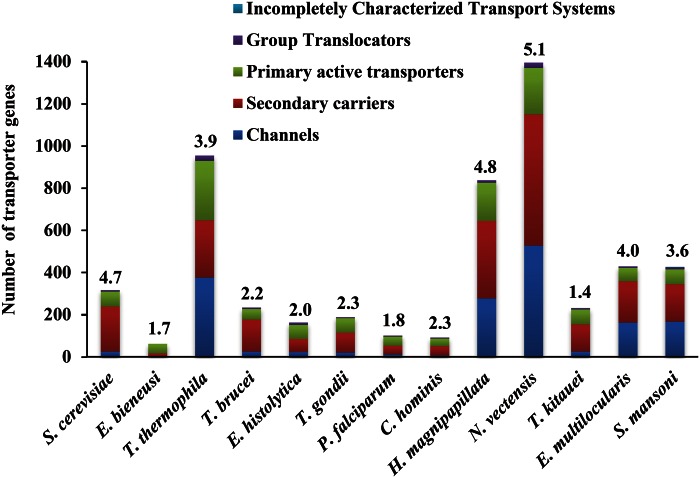
In comparison with the two free-living cnidarians, T. kitauei has a limited repertoire of membrane transporters (231 transport proteins in 38 different transporter superfamilies/families vs. 837−1,395 transporters in 60−73 families; figs. 3 and 4). In addition, there are fewer energy-independent channel proteins in T. kitauei (11% of the total transporters in T. kitauei vs. ∼33% in the two free-living cnidarians). Thelohanellus kitauei seems to rely primarily on secondary carriers (56%) and to a lesser degree on primary active transporters (30%) (fig. 4). The secondary carriers of T. kitauei fall into two main superfamilies, the major facilitator (24% of the total transporters) and the amino acid/polyamine/organocation superfamilies (10%) (supplementary table S15, Supplementary Material online), which together are involved in the utilization of carbohydrates, phosphates, and amino acids (Wong et al. 2012; Yan 2013). On the other side, the primary active transporters of T. kitauei are mainly composed of ATP-binding-cassette transporters (14%), which involve in exporting drug (Leprohon et al. 2011), and P-type ATPases (9%), which transport phospholipids, sodium, potassium, and calcium (Kuhlbrandt 2004). Nearly one-third of the cation transporters are involved in Ca2+ transport, and the majority (∼68%) of the putative anion transporters are involved in phosphate transport (supplementary table S15, Supplementary Material online). Therefore, the low blood phosphorus and calcium levels observed in T. kitauei infected carp might be attributed to the powerful capability of T. kitauei in transporting host calcium and phosphate (Wang et al. 2005).
Fig. 4.—
Comparison of transporters of T. kitauei and other organisms. Values above column are the percentage of the transporters within the total proteome of that species.
Anabolism Rather than Catabolism
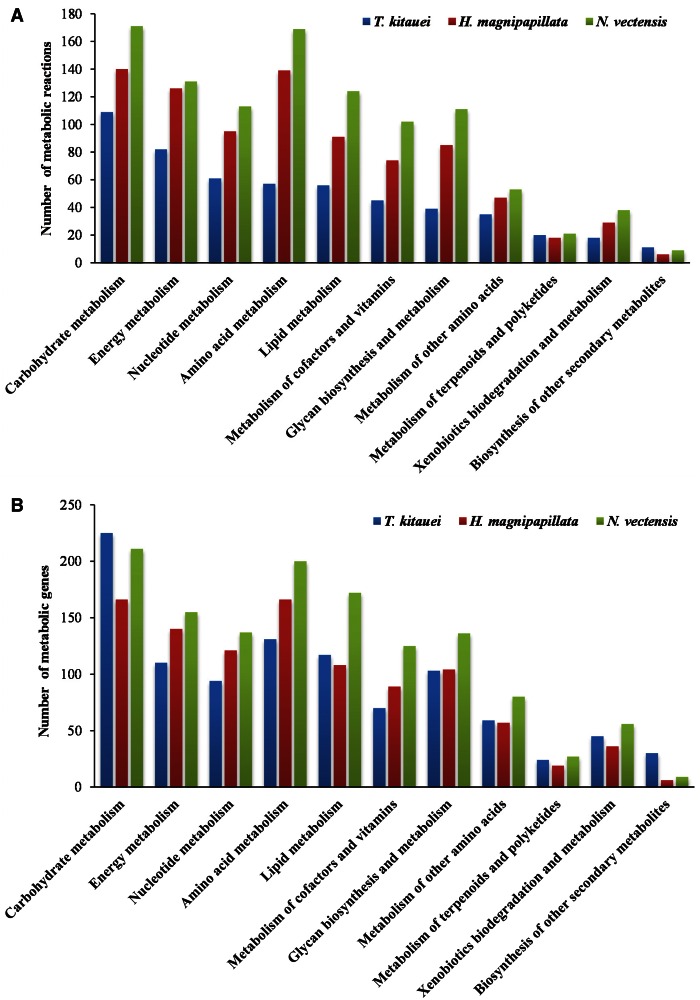
To date, the metabolic pathways of myxozoans have not been well characterized. In this study, comprehensive, genome-wide metabolic pathway reconstruction for a myxozoan was firstly conducted. Compared with the two free-living relatives, T. kitauei genome revealed extensive reductions in overall metabolic capacity, with fewer genes involved in the eight primary metabolic processes, that is, carbohydrate, energy, nucleotide, amino acid, lipid, cofactor/vitamin, glycan, and other amino acid metabolism (figs. 3 and 5 and supplementary fig. S5B–K, Supplementary Material online).
Fig. 5.—
Number of metabolic reactions (A) and number of metabolic genes (B) in comparison between T. kitauei and two free-living cnidarias.
Anaerobic fermentation plays an important role in the energy metabolism of many parasites, such as E. histolytica, Plasmodium falciparum, and tapeworms (Vanacova et al. 2003; Painter et al. 2007; Tsai et al. 2013). Both T. kitauei and its free-living relatives are unlikely to be able to produce lactate, acetaldehyde, and acetate through fermentation, as they lack the requisite lactate dehydrogenase, pyruvate decarboxylase, and acetyl-CoA synthetase (ADP-forming; fig. 3 and supplementary fig. S5B1, Supplementary Material online; Sanchez and Muller 1996; Zhao et al. 2013; Mithran et al. 2014). Moreover, unlike the two free-living relatives, T. kitauei lacks malate dehydrogenase (EC 1.1.1.40) for conversion of malate to pyruvate , as well as the propionyl-CoA cycle which converts succinate to propionate, suggesting a less importance of the anaerobic malate dismutation pathway producing succinate in T. kitauei (supplementary fig. S5B2 and 3, Supplementary Material online; Muller et al. 2012; Mentel et al. 2014). Taken together, these results imply that T. kitauei does not rely much on anaerobic fermentation. On the other hand, some enzymes related to the tricarboxylic acid cycle (RPKMs of 10 genes were higher than 100, and RPKMs of 16 genes were higher than 10, out of 32 genes) and oxidative phosphorylation (RPKMs of 5 genes were higher than 100, and RPKMs of 27 genes were higher than 10, out of 44 genes) were highly expressed in the myxospore stage of T. kitauei (fig. 3 and supplementary fig. S5B4 and C1, Supplementary Material online) further suggesting that T. kitauei may have greater metabolic dependency on respiration than on fermentation.
Compared with the two free-living cnidarians, T. kitauei has a reduced ability to de novo synthesize and hydrolyze amino acids (fig. 3 and supplementary fig. S5D, Supplementary Material online), which is similar to Schistosoma mansoni and tapeworms (Berriman et al. 2009; Tsai et al. 2013). Thelohanellus kitauei seems to only be able to de novo synthesize or hydrolyze a few amino acids including alanine, aspartate, glutamate, glycine, cysteine, and methionine (fig. 3 and supplementary fig. S5D, Supplementary Material online). The polyamines and their derivatives mediate a variety of important functions and are therefore essential to cell growth and proliferation (Birkholtz et al. 2011). Thelohanellus kitauei may use ornithine to synthesize polyamines, as the enzyme (ornithine decarboxylase) that converts ornithine to the polyamine putrescine was highly expressed (RPKM of 1 of 2 genes is 5,246.84) in the myxospore stage of T. kitauei. Other amino acids that can be de novo synthesized (serine, asparagine, glutamine, threonine, arginine, and proline) or partially synthesized (lysine, leucine, isoleucine, and valine) and catabolized by the two free-living cnidarians are unlikely synthesized or catabolized in T. kitauei (supplementary fig. S5D, Supplementary Material online).
Although both T. kitauei and the two free-living relatives lack the pathways for fatty acid biosynthesis, they are capable of fatty acid elongation (fig. 3 and supplementary fig. S5E, Supplementary Material online). Unlike the two free-living relatives, T. kitauei has an incomplete β-oxidation pathway in that it lacks dehydrogenases specific for either short- or medium-chain acyl-CoA (supplementary fig. S5E1, Supplementary Material online), suggesting that the lipid metabolism in T. kitauei inclined to anabolism rather than catabolism.
The reduced ability to hydrolyze fatty acids and amino acids, and the completeness of key carbon metabolism pathways (glycolysis and the tricarboxylic acid cycle) suggest that carbohydrates are the primary energy source for T. kitauei. The two free-living cnidarians have the complete pentose phosphate pathway, whereas T. kitauei appears to have a transaldolase-deficient pentose phosphate pathway, like in P. falciparum (Gardner et al. 2002; supplementary fig. S5B5, Supplementary Material online). Nevertheless, T. kitauei seems to utilize an alternative pathway that contains three enzymes, namely phosphofructokinase, transketolase, and aldolase for hexose-pentose interconversion, which maintains cytosolic NADP+/NADPH and H+ balance. Notably, ribose-phosphate pyrophosphokinase, which converts d-ribose-5′-phosphate to phosphoribosyl pyrophosphate, is highly expressed (RPKMs of 3 genes are 297.03, 592.19, and 968.95, respectively) in the myxospore stage of T. kitauei. Adenosine monophospates (AMP) can be converted to high energy molecule adenosine triphosphate (ATP) and its cyclic structure cyclic AMP plays an important role in intracellular signaling. The product phosphoribosyl pyrophosphate seems to mainly enter the adenine salvage pathway to form AMP, due to the absence of enzymes (amidophosphoribosyl transferase, uridine monophosphate synthetase, and ATP phosphoribosyltransferase) for either de novo nucleotide synthesis or histidine metabolism, and the presence of adenine phosphoribosyltransferase in T. kitauei (supplementary fig. S5B5 and F1, Supplementary Material online). The genomes of the two free-living cnidarians indicate that they can synthesize all glycolytic metabolites by gluconeogenesis. Thelohanellus kitauei, however, lacks fructose-bisphosphatase (supplementary fig. S5B1, Supplementary Material online), implying the absence of gluconeogenesis and the inability to convert fatty acids and amino acids into glucose. Actually, the fructose-bisphosphatase was also absent in other intestinal parasites such as G. lamblia, E. histolytica, Cryptosporidium paryum, and Cryptosporidium hominis (http://www.genome.jp/kegg-bin/get_htext?htext=br08601_map00010.keg&hier=5). Taken together, the absence of gluconeogenesis, high proportion of sugar transporters (10% of total transporters, supplementary table S15, Supplementary Material online), and the high proportion of glycosyltransferases (70.8% of total carbohydrate-active enzymes vs. 27.5–37.8% of those in N. vectensis and H. magnipapillata, supplementary table S11, Supplementary Material online) may underlie the evolution of T. kitauei’s ability to more efficiently harvest carbohydrates (mainly glucose) from the host.
The genomes of the two nonparasitic relatives suggest that they are capable of synthesizing and hydrolyzing chitin. Although failed to map to KEGG metabolic pathways, ten genes encoding cellulose/chitin synthases were annotated in T. kitauei using the dbCAN Web server (supplementary fig. S5B6 and table S11, Supplementary Material online). However, T. kitauei lacks both chitinases and chitosanases (supplementary table S11, Supplementary Material online). Therefore, T. kitauei either degrade chitin through endoglucanases, endo-β-N-acetylglucosaminidases, or other enzymes containing chitin/chitopentaose-binding modules that were identified in T. kitauei genome (supplementary table S11, Supplementary Material online). Alternatively, it is not impossible that T. kitauei does not have the capability for chitin degradation. Further investigations are necessary to uncover the detailed model of chitin metabolism in T. kitauei.
Thelohanellus kitauei seems to have lost the ATP-expensive pathways for de novo biosynthesis of inosine 5′-phosphate and uridine 5′-phosphate. Therefore, it must rely on salvage pathways, which is similar to other parasites such as E. histolytica, G. lamblia, and Trichomonas vaginalis (supplementary fig. S5F, Supplementary Material online; Abrahamsen et al. 2004; Loftus et al. 2005). Genes encoding components of the purine ribonucleotides or pyrimidine ribonucleotides interconversion pathways were identified in the genome of T. kitauei, suggesting that it can generate all necessary nucleotides from individual sources—purines or pyrimidines (supplementary fig. S5F, Supplementary Material online). In contrast to the two free-living cnidarians, T. kitauei lacks the enzymes for degradation of free purine and pyrimidine bases.
The reduced level of metabolic complexity observed in T. kitauei is a convergent trait among endoparasites (Nerima et al. 2010). The lack of de novo synthesis of fatty acids, nucleotides, and many of the amino acids as well as the reduced capacity for hydrolysis of fatty acids, nucleosides, and amino acids suggest that these molecules are mainly directed to anabolic rather than catabolic processes and that carbohydrates are the primary energy source. Also, the lack of gluconeogenesis implies the high reliance of carbohydrates (mainly glucose) from the host. All together, these facts suggest that T. kitauei scavenges nutrients from its host to compensate for its streamlined metabolic capabilities. The high nutrients (especially carbohydrates) requirement and the predominant aerobic respiration are consistent with the parasitic site of T. kitauei, that is, the loose connective tissue of the intestinal submucosa, where a high abundance of capillaries may supply sufficient oxygen and nutrients, especially glucose.
Control Strategy for T. kitauei
Some antiprotozoan drugs have shown efficacy against myxosporean infections, but these compounds either are toxic to fish or have shown limited efficacy or poor palatability (Molnar 1993; Rhee et al. 1993; le Gouvello et al. 1999; Athanassopoulou et al. 2004; Karagouni et al. 2005; Yonar and Yonar 2010; Jones et al. 2012; Manning et al. 2013). Also, many myxozoans elicit little or no host immune response in fish (Sitja-Bobadilla 2008; Gomez et al. 2014), leading to the absence of effective antimyxozoan immunoprophylactic agents till now. Therefore, novel targets and compounds are urgently needed.
The myxospore stage is tied with disease processes of T. kitauei in its carp host. To identify potential drug targets, we first selected the 2,653 most highly expressed genes (RPKMs ≥ 10) out of 8,659 expressed ones in the T. kitauei spore transcriptome. The putative products of these genes were searched against known drug target proteins in the ChEMBL database (https://www.ebi.ac.uk/chembl/), and 513 were found to be homologs of existing drug targets (BLAST E value ≤1 e −10; supplementary table S16, Supplementary Material online).
Nutritional and metabolic pathways have traditionally provided attractive targets for drug development. For example, the widely used quinoline-containing antimalarial drugs inhibit endocytosis (Roberts et al. 2008; Dunmore et al. 2013); vaccines against helminth parasites target digestive proteases (Pearson et al. 2010); several promising chemotherapeutic targets have been identified in the lipid synthetic and salvage pathways of apicomplexan parasites (Coppens 2013); and therapeutic approaches are being developed that exploit auxotrophies and metabolic defects of T. gondii (Coppens 2014). Therefore, targets related to nutritional and metabolic pathways are of particular interest in our study. Among the 513 drug targets we identified, 220 are related to nutrient digestion (27), absorption (68), and metabolism (125). Genes of the 513 drug targets were then compared with the genome of carp to avoid potential drug toxicity arising from chemical cross-reactivity between the orthologous parasite and host targets (Swain et al. 2011; http://zfgenomics.zfscreens.com/sub/carp). Fourty-seven T. kitauei-specific target genes that lack orthologs in the host were selected (BLAST E value >1 e −3; supplementary table S16, Supplementary Material online), with 23 of them related to nutritional and metabolic pathways (table 2). Six T. kitauei-specific targets are related to endocytosis, including two low-density lipoprotein receptor-related proteins, and two sortilins that are involved in the regulation of both apolipoprotein B secretion and low-density lipoprotein catabolism (Strong et al. 2012). Four putative T. kitauei-specific targets are related to lipid metabolism. Other T. kitauei-specific targets related to transport (3) and metabolism of carbohydrates (4), cofactors (3), glycan (1), nucleotides (1), and amino acids (1) (table 2). Homologues of targets for treatment of distantly related protozoan and fungal parasites such as Leishmania spp., Plasmodium spp., and Aspergillus fumigatus were included in the list of T. kitauei-specific targets (table 2), suggesting that some parasitic mechanisms and metabolic pathways are shared by even evolutionarily distant parasites and compound libraries developed against targets in one parasite may contain leads to pursue for chemotherapy of other infections (Renslo and McKerrow 2006). Moreover, the list of T. kitauei-specific targets includes several homologues of targets for cancer chemotherapy, including arachidonate 5-lipoxygenase, 17 beta-estradiol 17-dehydrogenase, diacylglycerol kinase, 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, and ectonucleoside triphosphate diphosphodydrolase (table 2), implying some consistency in the challenges of inhibiting cancer tumors and parasitic infestion (Tsai et al. 2013). Actually, the anthelminthic medicines niclosamine, mebendazole, and albendazole have already been shown to inhibit cancer growth (Tsai et al. 2013).
Table 2.
Promising Thelohanellus kitauei-Specific Drug Targets (23) Relating to Nutrient Process
| Gene ID | Predicted Protein | Function Category | Previous Indications As Drug Target |
|---|---|---|---|
| evm.model.scaffold04826.5 | Peptidyl-prolylcis-trans isomerase FKBP4 | Transport | Antiparasitic targets (Ibrahim and Nishikawa 2012) |
| evm.model.scaffold05481.72 | Vesicular glutamate transporter 3 | ||
| evm.model.scaffold02466.3 | |||
| evm.model.scaffold00373.8 | Integrin beta 2 | Endocytosis | Antiinflammatory target (de Gaetano et al. 2013) |
| evm.model.scaffold04701.1 | Heterogeneous nuclear ribonucleoprotein K | Anticancer target (Benelli et al. 2009) | |
| evm.model.scaffold01071.3 | Sortilin | Target for Alzheimer's disease (Caglayan et al. 2014) | |
| evm.model.scaffold01071.5 | |||
| evm.model.scaffold05633.24 | Low-density lipoprotein-related protein 2 | Target for Alzheimer's disease (Martiskainen et al. 2013) | |
| evm.model.scaffold04423.69 | |||
| evm.model.scaffold01248.6 | Arachidonate 5-lipoxygenase | Lipid metabolism | Anticancer target (Chen et al. 2009) |
| evm.model.scaffold04176.12 | 17 beta-estradiol 17-dehydrogenase | Anticancer target (Szajnik et al. 2012) | |
| evm.model.scaffold00813.1 | Diacylglycerol kinase (ATP dependent) | Targets for cancer (Dominguez et al. 2013) | |
| evm.model.scaffold04610.1 | Phospholipase D3 | Target in disease settings (Huang and Frohman 2007) | |
| evm.model.scaffold05340.25 | Oligosaccharyltransferase complex subunit beta | Glycan biosynthesis | Target for Aspergillus fumigatus (Li et al. 2011) |
| evm.model.scaffold00661.36 | 2,3-bisphosphoglycerate-independent phosphoglyceratemutase | Carbohydrate metabolism | Anticancer (Ren et al. 2010)/antifilarial (Singh et al. 2013) target |
| evm.model.scaffold03068.51 | Inositol-tetrakisphosphate 1-kinase | Antimalarial target (Stritzke et al. 2012) | |
| evm.model.scaffold01538.6 | Inositol-1,4,5-trisphosphate 5-phosphatase | Antimalarial target (Stritzke et al. 2012) | |
| evm.model.scaffold01687.1 | Ribokinase | Antibacterial target (Desroy et al. 2009) | |
| evm.model.scaffold04623.113 | Pyrazinamidase/nicotinamidase PNCA | Cofactor metabolism | Target for Leishmania (Gazanion et al. 2011) |
| evm.model.scaffold02481.37 | |||
| evm.model.scaffold03983.31 | Thiamine pyrophosphokinase | Antimalarial target (Chan et al. 2013) | |
| evm.model.scaffold04174.4 | Ectonucleoside triphosphate diphosphohydrolase 4 | Nucleotide metabolism | Anticancer target (Tada et al. 2011) |
| evm.model.scaffold02448.28 | Glutathione S-transferase | Other amino acid metabolism | Antimesothelioma (De Luca et al. 2013) /antimalarial (Harwaldt et al. 2002) target |
Conclusion
The characterization of the first genome of a myxozoan will provide a platform for exploring basic questions on the biology and evolution of myxozoans. We also expect the T. kitauei genome to provide a useful reference for comparative genomics of eukaryotic fish parasites, particularly to identify the relative importance of shared genes among various evolutionarily distant fish parasites such as Ichthyophthirius multifiliis. Insights gained from this study will promote experimental approaches for myxozoan research and facilitate the development of control strategies for severe fish parasite infections and may ultimately prevent myxozoan infection in humans.
Supplementary Material
Supplementary figures S1–S5, and tables S1–S16 are available at Genome Biology and Evolution online (http://gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Zemao Gu for help in measuring the morphometric parameters of T. kitauei spores. This work was supported by the National Key Technologies R&D Program (grant numbers 2013BAD10B01-2 and 2012BAD25B0203), the Fundamental Research Fund of the Chinese Academy of Agricultural Sciences (grant number 2013ZL043), the Agriculture Science Technology Achievement Transformation Fund (grant number 2013GB23260573), and the Chinese Agriculture Research System (grant number SCGWZJ 20141104-4).
Literature Cited
- Abrahamsen MS, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Alam A, Bhatnagar RK, Relan U, Mukherjee P, Chauhan VS. Proteolytic activity of Plasmodium falciparum subtilisin-like protease 3 on parasite profilin, a multifunctional protein. Mol Biochem Parasitol. 2013;191:58–62. doi: 10.1016/j.molbiopara.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Andrade-Neto VV, et al. The pharmacological inhibition of sterol biosynthesis in Leishmania is counteracted by enhancement of LDL endocytosis. Acta Trop. 2011;119:194–198. doi: 10.1016/j.actatropica.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Athanassopoulou F, et al. Efficacy and toxicity of orally administrated anti-coccidial drugs for innovative treatments of Myxobolus sp. infection in Puntazzo puntazzo. Dis Aquat Organ. 2004;62:217–226. doi: 10.3354/dao062217. [DOI] [PubMed] [Google Scholar]
- Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Benelli R, Monteghirfo S, Balbi C, Barboro P, Ferrari N. Novel antivascular efficacy of metronomic docetaxel therapy in prostate cancer: hnRNP K as a player. Int J Cancer. 2009;124:2989–2996. doi: 10.1002/ijc.24305. [DOI] [PubMed] [Google Scholar]
- Berriman M, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkholtz LM, et al. Polyamine homoeostasis as a drug target in pathogenic protozoa: peculiarities and possibilities. Biochem J. 2011;438:229–244. doi: 10.1042/BJ20110362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglayan S, et al. Lysosomal sorting of amyloid-beta by the SORLA receptor is impaired by a familial Alzheimer's disease mutation. Sci Transl Med. 2014;6:220r–223r. doi: 10.1126/scitranslmed.3007747. [DOI] [PubMed] [Google Scholar]
- Canning EU, Okamura B. Biodiversity and evolution of the Myxozoa. Adv Parasitol. 2004;56:43–131. doi: 10.1016/s0065-308x(03)56002-x. [DOI] [PubMed] [Google Scholar]
- Chan XW, et al. Chemical and genetic validation of thiamine utilization as an antimalarial drug target. Nat Commun. 2013;4:2060. doi: 10.1038/ncomms3060. [DOI] [PubMed] [Google Scholar]
- Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QL, Ma CL. Fauna Sinica: Myxozoa, myxosporea. Beijing, China: Science Press; 1998. [Google Scholar]
- Chen YY, Li DG, Li SG. The Alox5 gene is a novel therapeutic target in cancer stem cells of chronic myeloid leukemia. Cell Cycle. 2009;8:3488–3492. doi: 10.4161/cc.8.21.9852. [DOI] [PubMed] [Google Scholar]
- Chun SK, Choi DL, Park IS. Studies of Thelohanellus infection in carp, Cyprinus carpio. I. Experimental induction of Thelohanellosis. J Fish Pathol. 1988;1:111–116. [Google Scholar]
- Coppens I. Targeting lipid biosynthesis and salvage in apicomplexan parasites for improved chemotherapies. Nat Rev Microbiol. 2013;11:823–835. doi: 10.1038/nrmicro3139. [DOI] [PubMed] [Google Scholar]
- Coppens I. Exploitation of auxotrophies and metabolic defects in Toxoplasma as therapeutic approaches. Int J Parasitol. 2014;44:109–120. doi: 10.1016/j.ijpara.2013.09.003. [DOI] [PubMed] [Google Scholar]
- de Gaetano M, Dempsey E, Marcone S, James WG, Belton O. Conjugated linoleic acid targets beta 2 integrin expression to suppress monocyte adhesion. J Immunol. 2013;191:4326–4336. doi: 10.4049/jimmunol.1300990. [DOI] [PubMed] [Google Scholar]
- De Luca A, et al. Glutathione S-transferase P1-1 as a target for mesothelioma treatment. Cancer Sci. 2013;104:223–230. doi: 10.1111/cas.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desroy N, et al. Towards Gram-negative antivirulence drugs: new inhibitors of HldE kinase. Bioorg Med Chem. 2009;17:1276–1289. doi: 10.1016/j.bmc.2008.12.021. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Dominguez CL, et al. Diacylglycerol kinase alpha is a critical signaling node and novel therapeutic target in glioblastoma and other cancers. Cancer Discov. 2013;3:782–797. doi: 10.1158/2159-8290.CD-12-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos C, Howgate P. Fishborne zoonotic parasites and aquaculture: a review. Aquaculture. 2011;318:253–261. [Google Scholar]
- Doyle PS, et al. The Trypanosoma cruzi protease cruzain mediates immune evasion. PLoS Pathog. 2011;7:e1002139. doi: 10.1371/journal.ppat.1002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunmore BJ, et al. The lysosomal inhibitor, chloroquine, increases cell surface BMPR-II levels and restores BMP9 signalling in endothelial cells harbouring BMPR-II mutations. Hum Mol Genet. 2013;22:3667–3679. doi: 10.1093/hmg/ddt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egusa S, Nakajima K. A new myxozoa Thelohanellus kitauei, the cause of intestinal giant cystic disease of carp. Fish Pathol. 1981;15:213–218. [Google Scholar]
- Evans NM, Holder MT, Barbeitos MS, Okamura B, Cartwright P. The phylogenetic position of Myxozoa: exploring conflicting signals in phylogenomic and ribosomal data sets. Mol Biol Evol. 2010;27:2733–2746. doi: 10.1093/molbev/msq159. [DOI] [PubMed] [Google Scholar]
- Fan SG, Zhang QY, Luo C. Sequence cloning and expression analysis of Rag gene in goldfish. Acta Hydrobiol Sin. 2009;33:603–612. [Google Scholar]
- Faria MS, et al. Leishmania inhibitor of serine peptidase 2 prevents TLR4 activation by neutrophil elastase promoting parasite survival in murine macrophages. J Immunol. 2011;186:411–422. doi: 10.4049/jimmunol.1002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust DM, Guillen N. Virulence and virulence factors in Entamoeba histolytica, the agent of human amoebiasis. Microbes Infect. 2012;14:1428–1441. doi: 10.1016/j.micinf.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Feng JM, et al. New phylogenomic and comparative analyses provide corroborating evidence that Myxozoa is Cnidaria. Mol Phylogenet Evol. 2014;81C:10–18. doi: 10.1016/j.ympev.2014.08.016. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazanion E, et al. The Leishmania nicotinamidase is essential forNAD+ production and parasite proliferation. Mol Microbiol. 2011;82:21–38. doi: 10.1111/j.1365-2958.2011.07799.x. [DOI] [PubMed] [Google Scholar]
- Gomez D, Bartholomew J, Sunyer JO. Biology and mucosal immunity to myxozoans. Dev Comp Immunol. 2014;43:243–256. doi: 10.1016/j.dci.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwaldt P, Rahlfs S, Becker K. Glutathione S-transferase of the malarial parasite Plasmodium falciparum: characterization of a potential drug target. Biol Chem. 2002;383:821–830. doi: 10.1515/BC.2002.086. [DOI] [PubMed] [Google Scholar]
- Hiller K, Grote A, Scheer M, Munch R, Jahn D. PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004;32:W375–W379. doi: 10.1093/nar/gkh378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Frohman MA. The potential for phospholipase D as a new therapeutic target. Expert Opin Ther Targets. 2007;11:707–716. doi: 10.1517/14728222.11.5.707. [DOI] [PubMed] [Google Scholar]
- Huang S, et al. SOAPsplice: Genome-wide ab initio detection of splice junctions from RNA-seq data. Front Genet. 2011;2:46. doi: 10.3389/fgene.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim H, Nishikawa Y. Apicomplexan cyclophilins in host-parasite interaction and their potential as anti-parasitic drug targets. Mol Cell Pharmacol. 2012;3:105–115. [Google Scholar]
- Jimenez-Guri E, Philippe H, Okamura B, Holland PW. Buddenbrockia is a cnidarian worm. Science. 2007;317:116–118. doi: 10.1126/science.1142024. [DOI] [PubMed] [Google Scholar]
- Jones SRM, Forster I, Liao X, Ikonomou MG. Dietary nicarbazin reduces prevalence and severity of Kudoa thyrsites (Myxosporea: Multivalvulida) in Atlantic salmon Salmo salar post-smolts. Aquaculture. 2012 342–343:1-6. [Google Scholar]
- Karagouni E, Athanassopoulou F, Lytra A, Komis C, Dotsika E. Antiparasitic and immunomodulatory effect of innovative treatments against Myxobolus sp. infection in Diplodus puntazzo. Vet Parasitol. 2005;134:215–228. doi: 10.1016/j.vetpar.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Kawai T, et al. Identification of Kudoa septempunctata as the causative agent of novel food poisoning outbreaks in Japan by consumption of Paralichthys olivaceus in raw fish. Clin Infect Dis. 2012;54:1046–1052. doi: 10.1093/cid/cir1040. [DOI] [PubMed] [Google Scholar]
- Kent ML, et al. Recent advances in our knowledge of the Myxozoa. J Eukaryot Microbiol. 2001;48:395–413. doi: 10.1111/j.1550-7408.2001.tb00173.x. [DOI] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Narechania A, Stein JC, Ware D. A new method to compute K-mer frequencies and its application to annotate large repetitive plant genomes. BMC Genomics. 2008;9:517. doi: 10.1186/1471-2164-9-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Gouvello R, Pobel T, Richards RH, Gould C. Field efficacy of a 10-day treatment of fumagillin against proliferative kidney disease in rainbow trout Oncorhynchus mykiss. Aquaculture. 1999;171:27–40. [Google Scholar]
- Leprohon P, Legare D, Ouellette M. ABC transporters involved in drug resistance in human parasites. Essays Biochem. 2011;50:121–144. doi: 10.1042/bse0500121. [DOI] [PubMed] [Google Scholar]
- Li H, Benedito VA, Udvardi MK, Zhao PX. TransportTP: a two-phase classification approach for membrane transporter prediction and characterization. BMC Bioinformatics. 2009;10:418. doi: 10.1186/1471-2105-10-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, et al. Repression of N-glycosylation triggers the unfolded protein response (UPR) and overexpression of cell wall protein and chitin in Aspergillus fumigatus. Microbiology. 2011;157:1968–1979. doi: 10.1099/mic.0.047712-0. [DOI] [PubMed] [Google Scholar]
- Li Y, Hu Y, Bolund L, Wang J. State of the art de novo assembly of human genomes from massively parallel sequencing data. Hum Genomics. 2010;4:271–277. doi: 10.1186/1479-7364-4-4-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. A chitinase from Aeromonas veronii CD3 with the potential to control myxozoan disease. PLoS One. 2011;6:e29091. doi: 10.1371/journal.pone.0029091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus B, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- Lom J, Dykova I. Myxozoan genera: definition and notes on taxonomy, life-cycle terminology and pathogenic species. Folia Parasitol. 2006;53:1–36. [PubMed] [Google Scholar]
- Lopez-Soto F, et al. Use and endocytosis of iron-containing proteins by Entamoeba histolytica trophozoites. Infect Genet Evol. 2009;9:1038–1050. doi: 10.1016/j.meegid.2009.05.018. [DOI] [PubMed] [Google Scholar]
- Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol. 2011;3:pii: a005058. doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- Manning BB, Griffin MJ, Khoo LH, Wise DJ, Greenway T. Palatability of diets for channel catfish that contain Amprolium or Salinomycin using feed conversion ratio as the criterion. N Am J Aquac. 2013;75:99–101. [Google Scholar]
- Martiskainen H, et al. Targeting ApoE4/ApoE receptor LRP1 in Alzheimer's disease. Expert Opin Ther Targets. 2013;17:781–794. doi: 10.1517/14728222.2013.789862. [DOI] [PubMed] [Google Scholar]
- Matsui T, et al. SASPase regulates stratum corneum hydration through profilaggrin-to-filaggrin processing. EMBO Mol Med. 2011;3:320–333. doi: 10.1002/emmm.201100140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentel M, Rottger M, Leys S, Tielens AG, Martin WF. Of early animals, anaerobic mitochondria, and a modern sponge. Bioessays. 2014;36:924–932. doi: 10.1002/bies.201400060. [DOI] [PubMed] [Google Scholar]
- Mithran M, Paparelli E, Novi G, Perata P, Loreti E. Analysis of the role of the pyruvate decarboxylase gene family in Arabidopsis thaliana under low-oxygen conditions. Plant Biol. 2014;16:28–34. doi: 10.1111/plb.12005. [DOI] [PubMed] [Google Scholar]
- Molnar K. Recent achievements in the chemotherapy of myxosporean infections of fish. Acta Vet Hung. 1993;41:51–58. [PubMed] [Google Scholar]
- Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- Muller M, et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerima B, Nilsson D, Maser P. Comparative genomics of metabolic networks of free-living and parasitic eukaryotes. BMC Genomics. 2010;11:217. doi: 10.1186/1471-2164-11-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesnidal MP, Helmkampf M, Bruchhaus I, El-Matbouli M, Hausdorf B. Agent of whirling disease meets orphan worm: phylogenomic analyses firmly place Myxozoa in Cnidaria. PLoS One. 2013;8:e54576. doi: 10.1371/journal.pone.0054576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter HJ, Morrisey JM, Mather MW, Vaidya AB. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007;446:88–91. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- Pearson MS, Ranjit N, Loukas A. Blunting the knife: development of vaccines targeting digestive proteases of blood-feeding helminth parasites. Biol Chem. 2010;391:901–911. doi: 10.1515/BC.2010.074. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, et al. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol Cancer. 2010;9:81. doi: 10.1186/1476-4598-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renslo AR, McKerrow JH. Drug discovery and development for neglected parasitic diseases. Nat Chem Biol. 2006;2:701–710. doi: 10.1038/nchembio837. [DOI] [PubMed] [Google Scholar]
- Rhee JK, Kim HC, Park BK. Efficacy of fumagillin against Thelohanellus kitauei infection of Israel carp, Cyprinus carpio nudus. Korean J Parasitol. 1993;31:57–65. doi: 10.3347/kjp.1993.31.1.57. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rivero MR, et al. Adaptor protein 2 regulates receptor-mediated endocytosis and cyst formation in Giardia lamblia. Biochem J. 2010;428:33–45. doi: 10.1042/BJ20100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero MR, Miras SL, Quiroga R, Ropolo AS, Touz MC. Giardia lamblia low-density lipoprotein receptor-related protein is involved in selective lipoprotein endocytosis and parasite replication. Mol Microbiol. 2011;79:1204–1219. doi: 10.1111/j.1365-2958.2010.07512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L, Egan TJ, Joiner KA, Hoppe HC. Differential effects of quinoline antimalarials on endocytosis in Plasmodium falciparum. Antimicrob Agents Chemother. 2008;52:1840–1842. doi: 10.1128/AAC.01478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robibaro B, et al. Endocytosis in different lifestyles of protozoan parasitism: role in nutrient uptake with special reference to Toxoplasma gondii. Int J Parasitol. 2001;31:1343–1353. doi: 10.1016/s0020-7519(01)00252-1. [DOI] [PubMed] [Google Scholar]
- Sanchez LB, Muller M. Purification and characterization of the acetate forming enzyme, acetyl-CoA synthetase (ADP-forming) fromthe amitochondriate protist, Giardia lamblia. FEBS Lett. 1996;378:240–244. doi: 10.1016/0014-5793(95)01463-2. [DOI] [PubMed] [Google Scholar]
- Sarras MJ, Yan L, Leontovich A, Zhang JS. Structure, expression, and developmental function of early divergent forms of metalloproteinases in hydra. Cell Res. 2002;12:163–176. doi: 10.1038/sj.cr.7290123. [DOI] [PubMed] [Google Scholar]
- Shin SP, et al. Surgical removal of an anal cyst caused by a protozoan parasite (Thelohanellus kitauei) from a koi (Cyprinus carpio) J Am Vet Med Assoc. 2011;238:784–786. doi: 10.2460/javma.238.6.784. [DOI] [PubMed] [Google Scholar]
- Siddall ME, Martin DS, Bridge D, Desser SS, Cone DK. The demise of a phylum of protists: phylogeny of Myxozoa and other parasitic cnidaria. J Parasitol. 1995;81:961–967. [PubMed] [Google Scholar]
- Singh PK, Kushwaha S, Mohd S, Pathak M, Misra-Bhattacharya S. In vitro gene silencing of independent phosphoglycerate mutase (iPGM) in the filarial parasite Brugia malayi. Infect Dis Poverty. 2013;2:5. doi: 10.1186/2049-9957-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitja-Bobadilla A. Fish immune response to Myxozoan parasites. Parasite. 2008;15:420–425. doi: 10.1051/parasite/2008153420. [DOI] [PubMed] [Google Scholar]
- Smeds L, Kunstner A. ConDeTri–a content dependent read trimmer for Illumina data. PLoS One. 2011;6:e26314. doi: 10.1371/journal.pone.0026314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers JF, von Dohlen CD, Smith LJ, Spall RD. Molecular evidence that the myxozoan protists are metazoans. Science. 1994;265:1719–1721. doi: 10.1126/science.8085160. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Stritzke C, Nalaskowski MM, Fanick W, Lin H, Mayr GW. A Plasmodium multi-domain protein possesses multiple inositol phosphate kinase activities. Mol Biochem Parasitol. 2012;186:134–138. doi: 10.1016/j.molbiopara.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Strong A, et al. Hepatic sortilin regulates both apolipoprotein B secretion and LDL catabolism. J Clin Invest. 2012;122:2807–2816. doi: 10.1172/JCI63563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanya S, Mensa-Wilmot K. Diacylglycerol-stimulated endocytosis of transferrin in trypanosomatids is dependent on tyrosine kinase activity. PLoS One. 2010;5:e8538. doi: 10.1371/journal.pone.0008538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain MT, et al. Schistosoma comparative genomics: integrating genome structure, parasite biology and anthelmintic discovery. Trends Parasitol. 2011;27:555–564. doi: 10.1016/j.pt.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajnik M, et al. 17beta Hydroxysteroid dehydrogenase type 12 (HSD17B12) is a marker of poor prognosis in ovarian carcinoma. Gynecol Oncol. 2012;127:587–594. doi: 10.1016/j.ygyno.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, et al. Ectonucleoside triphosphate diphosphohydrolase 6 expression in testis and testicular cancer and its implication in cisplatin resistance. Oncol Rep. 2011;26:161–167. doi: 10.3892/or.2011.1274. [DOI] [PubMed] [Google Scholar]
- Toubarro D, et al. Serine protease-mediated host invasion by the parasitic nematode Steinernema carpocapsae. J Biol Chem. 2010;285:30666–30675. doi: 10.1074/jbc.M110.129346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013;496:57–63. doi: 10.1038/nature12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Liston DR, Tachezy J, Johnson PJ. Molecular biology of the amitochondriate parasites, Giardia intestinalis, Entamoeba histolytica and Trichomonas vaginalis. Int J Parasitol. 2003;33:235–255. doi: 10.1016/s0020-7519(02)00267-9. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. The draft genome of the carcinogenic human liver fluke Clonorchis sinensis. Genome Biol. 2011;12:R107. doi: 10.1186/gb-2011-12-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, et al. Pathogenic mechanism and hematology analysis of the disease of Thelohanellus kitauei Egusa 1981. Tianjin Fisheries. 2005;((4):24–27. [Google Scholar]
- Wong FH, et al. The amino acid-polyamine-organocation superfamily. J Mol Microbiol Biotechnol. 2012;22:105–113. doi: 10.1159/000338542. [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Xie XR, et al. Studies on Thelohanellionsis (Myxosporidiosis) parasitized freshwater fish in China. I. Thelohanelliosis (Myxesporidiosis) Parasitized Carp In Xinyang, Henan Province. J Xinyang Agric Coll. 2000;10:1–5. [Google Scholar]
- Yan N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem Sci. 2013;38:151–159. doi: 10.1016/j.tibs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Yap KW, Thompson RC. CTAB precipitation of cestode DNA. Parasitol Today. 1987;3:220–222. doi: 10.1016/0169-4758(87)90065-2. [DOI] [PubMed] [Google Scholar]
- Yin Y, et al. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonar ME, Yonar SM. Changes in selected immunological parameters and antioxidant status of rainbow trout exposed to malachite green (Oncorhynchus mykiss, Walbaum, 1792) Pestic Biochem Physiol. 2010;97:19–23. [Google Scholar]
- Zhao R, et al. NAD-dependent lactate dehydrogenase catalyses the first step in respiratory utilization of lactate by Lactococcus lactis. FEBS Open Bio. 2013;3:379–386. doi: 10.1016/j.fob.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Gong QL, Yu KK, Meng QX. A preliminary study on the disease of Myxozoa, Thelohanellus Kitauei of Jian carp (Cyprinus Carpio Var. Jian) Period Ocean Univ China. 1998;28:59–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.