Abstract
Mitochondria are the energy-producing organelles of our cells and derive from bacterial ancestors that became endosymbionts of microorganisms from a different lineage, together with which they formed eukaryotic cells. For a long time it has remained unclear from which bacteria mitochondria actually evolved, even if these organisms in all likelihood originated from the α lineage of proteobacteria. A recent article (Degli Esposti M, et al. 2014. Evolution of mitochondria reconstructed from the energy metabolism of living bacteria. PLoS One 9:e96566) has presented novel evidence indicating that methylotrophic bacteria could be among the closest living relatives of mitochondrial ancestors. Methylotrophs are ubiquitous bacteria that live on single carbon sources such as methanol and methane; in the latter case they are called methanotrophs. In this review, I examine their possible ancestry to mitochondria within a survey of the common features that can be found in the central and terminal bioenergetic systems of proteobacteria and mitochondria. I also discuss previously overlooked information on methanotrophic bacteria, in particular their intracytoplasmic membranes resembling mitochondrial cristae and their capacity of establishing endosymbiotic relationships with invertebrate animals and archaic plants. This information appears to sustain the new idea that mitochondrial ancestors could be related to extant methanotrophic proteobacteria, a possibility that the genomes of methanotrophic endosymbionts will hopefully clarify.
Keywords: mitochondria, bioenergetics, endosymbiosis, methylotrophs, methanotrophs
Introduction
That mitochondrial organelles derive from ancestral bacteria (Sagan 1967; Margulis 1996) are now widely accepted, but the question of their precise origin remains open (Gray 2012). The endosymbiotic transition that transformed these ancestral bacteria (or proto-mitochondria) into current mitochondria has been fundamental in the evolution of life (Lane and Martin 2010). According to the current consensus, this event occurred only once, at least 1.5 billion years ago, and was followed by divergent evolutionary paths leading to altered forms of the organelles adapted to anaerobic lifestyles (Müller et al. 2012; Mentel et al. 2014). Consistent evidence has indicated that proto-mitochondria emerged from the α lineage of proteobacteria (Andersson et al. 1998, 2003; Williams et al. 2007; Gray 2012; Müller et al. 2012). This α lineage occupies a central position in “phylum” proteobacteria, because it originated after the separation of the predominantly anaerobic δ and ε branches but before the split of the (predominantly) facultatively anaerobic γ and β branches (Battistuzzi et al. 2004).
Proto-mitochondria evolved soon after, when current oxygen levels were established in the atmosphere, emerged land, and the photic zone of Proterozoic oceans, while the rest of the oceans remained fundamentally anoxic (Johnston et al. 2009; Müller et al. 2012). Because at this time all life forms were restricted to the oceans, proto-mitochondria evolved in one of the following ecological environments: (1) photic surface of the ocean; (2) oxic–anoxic interface in oceanic or other aquatic regions; and (3) medium deep ocean, including hydrothermal vents and cold seeps. In environment (1), the ancestors of mitochondria might have been equipped with the protective pigments of purple nonsulfur photosynthetic bacteria such as Rhodospirillum (Esser et al. 2004) or were already adapted to the oligotrophic pelagic lifestyle of the bacteria which are dominant in current seas, for example, Pelagibacter (Thrash et al. 2011). In environment (2), the ancestors of mitochondria should have been facultatively anaerobic, as in the case of extant aquatic bacteria that are free-living (e.g., Magnetococcus; Schübbe et al. 2009) or predatory (e.g., Micavibrio; Davidov et al. 2006). Finally, in environment (3), the ancestors of mitochondria must have had additional metabolic capacities to survive under oligotrophic conditions and eventually exploit, via chemiosynthesis, local areas enriched in chemical nutrients such as H2S, nitrate, reduced metals, and methane. Currently, such metabolic pathways are present in α- and γ-proteobacteria that have become endocellular symbionts of deep-sea invertebrates inhabiting hydrothermal vents, cold seeps, and fallen wood (Dubilier et al. 1999; Blazejak et al. 2006; DeChaine and Cavanaugh 2006; Cavanaugh et al. 2013), or of peat bog mosses (Dedysh 2011; Kip et al. 2011). From a theoretical standpoint, a wide metabolic versatility enabling adaptation to all the above environments would produce maximal probability for being close to the mitochondrial ancestors. At the moment, such a versatility seems to be present in a few families of cultivated α- and γ-proteobacteria and also some uncultivated organisms discovered in metagenomic surveys (Wrighton et al. 2012). However, the requirement for metabolic versatility is not a strict condition for defining the ancestors of mitochondria, which might have been confined to only one of the abovementioned habitats.
Another factor that deserves consideration is bacterial adaptation to endocellular symbiosis. Proto-mitochondria that had evolved in Proterozoic environments must have reduced their metabolic versatility during the transition to symbiotic lifestyle. Although we can only guess the details of this process, we can deduce its contours by comparison with various examples of metabolic adaptation to symbiosis or parasitism that have been documented for extant microorganisms (Vannini et al. 2007; Moran et al. 2009; Manzano-Marín et al. 2010; Sachs et al. 2011; Caffrey et al. 2012). In general, the stability of the host environment reduces evolutionary pressure on organisms that have undergone the transition from free-living to endocellular symbionts, resulting in genome reduction and erosion affecting energy metabolism also (Moran et al. 2009; Caffrey et al. 2012). The original metabolic capacity of mitochondrial ancestors has thus been lost (Huynen et al. 2013; Degli Esposti et al. 2014), contrary to popular reconstructions which depict their bioenergetic capacity as equivalent to that of mammalian mitochondria (Emelyanov 2001; Gabaldón and Huynen 2003). Recently, the progressive gene loss leading to the current bioenergetic capacity of mitochondria has been reconstructed by using a systematic genomic analysis of extant α-proteobacteria and protists’ mitochondria (Degli Esposti et al. 2014). The analysis led to the novel indication that a group of proteobacteria, the methylotrophs, may contain the extant relatives of proto-mitochondria (Degli Esposti et al. 2014). Interestingly, some of these methylotrophs such as Methylocystis possess a metabolic versatility suitable for all environments (1)–(3) mentioned above (Dam et al. 2012). They are also related to the methanotrophic endosymbionts of Spagnum mosses dominating peat bogs and northern wetlands (Raghoebarsing et al. 2005; Dedysh 2011; Kip et al. 2011; Belova et al. 2013).
The work of Degli Esposti et al. (2014) has excluded from mitochondrial ancestry many bacteria that had been previously proposed as mitochondrial relatives or ancestors, for example, Rhodospirillum rubrum (Esser et al. 2004; Abhishek et al. 2011; Thiergart et al. 2012) and Rickettsia (Andersson et al. 1998; Sicheritz-Pontén et al. 1998; Williams et al. 2007; Abhishek et al. 2011; Ferla et al. 2013). Therefore, this work will inevitably generate controversy in the fields of molecular evolution, microbiology, and bioenergetics. Herein, I will examine major issues that may help resolving such controversy within an overview of the common bioenergetic features of proteobacteria and mitochondria.
Common Bioenergetic Features of Proteobacteria and Mitochondria
Mammalian mitochondria constitute the best known model for the energy metabolism in eukaryotic cells (Andersson et al. 2003; Gabaldon and Huyen 2003), but represent only one of the several versions of these organelles that are present in various eukaryotic organisms (Mentel et al. 2014). They all derive from the same unique event of endosymbiosis that occurred in ancestral oceans between proto-mitochondria and other prokaryotes (Müller et al. 2012; Williams et al. 2013). Leaving aside the dehydrogenases that convey electrons from nutrients to ubiquinone (Q) producing its reduced form, ubiquinol, the central and terminal portion of the mitochondrial respiratory chain produces most bioenergy in eukaryotic cells and shows strong similarity with that of aerobic proteobacteria (Sicheritz-Pontén et al. 1998; Emelyanov 2001). It consists of ubiquinol:cytochrome c reductase, commonly called bc1 complex, the soluble electron carrier cytochrome c, and its terminal bioenergy-producing oxidase, usually known as cytochrome c oxidase (COX), but nowadays defined as dioxygen reductase of aa3 type (Sousa et al. 2012). Other mitochondrial enzymes that reoxidize either ubiquinol or cytochrome c without producing bioenergy are not considered in this review.
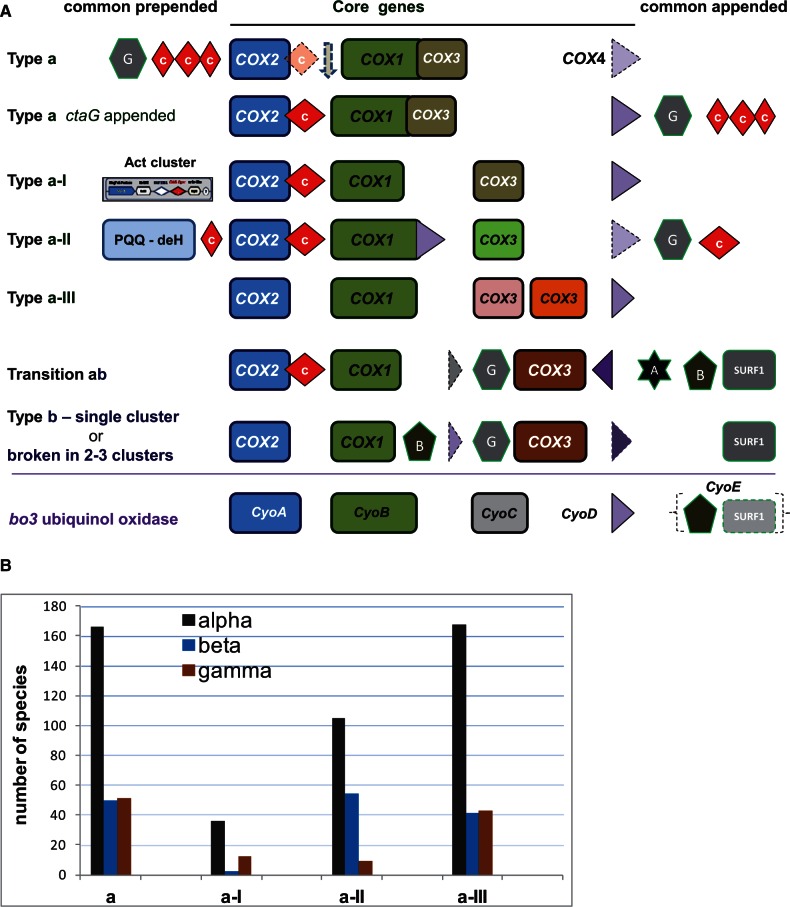
The fundamental difference that distinguishes the bc1 and aa3 enzyme complexes of mitochondria and proteobacteria is the presence of additional, supernumerary subunits that may contribute aspects of fine regulation in eukaryotic cells. In both structure and function, the core catalytic proteins are essentially the same in mitochondrial and proteobacterial complexes (Zhang et al. 1998; Harrenga and Michel 1999; Berry et al. 2004; Radzi Noor and Soulimane, 2012), possessing multiple transmembrane helices and various redox cofactors. Most of these catalytic proteins are retained in mitochondrial DNA (mtDNA), the principal vestige of the bacterial past of the organelles (Sagan 1967). However, the Rieske iron sulfur protein (ISP) and cytochrome c1 subunits of the bc1 complex have migrated into nuclear DNA early after the symbiotic transition of proto-mitochondria, followed by most subunits of the dehydrogenases, the assembly subunits of aa3-type oxidase, and the catalytic subunits of adenosine triphosphate (ATP) synthase (Gray 2012; Burger et al. 2013). Consequently, the core subunits of aa3-type oxidase, COX1, COX2, and COX3, constitute the bioenergetic redox system that has remained most conserved between mitochondria and proteobacteria. Indeed, it appears to show the largest set of common features of potential evolutionary significance, as summarized in table 1. The gene order of these subunits appears to be almost random in the mtDNA of the great majority of eukaryotes, but follows defined sequences in some protists (Burger et al. 2013; Degli Esposti et al. 2014) and most proteobacteria. All such sequences most likely derive from the ancestral operon already present in Firmicutes such as Bacillus subtilis (Radzi Noor and Soulimane 2012; Degli Esposti et al. 2014).
Table 1.
Features of Bioenergetic Systems Specific to Bacteria that Sometimes Are Found in Mitochondria
| System and Feature | Proteobacteria |
Mitochondria (mtDNA) | Origin | ||
|---|---|---|---|---|---|
| Alpha | Beta | Gamma | |||
| COX operon | |||||
| a: fusion COX1 with COX3 | Yes | Yes | Yes | 3 ciliates | Firmicutes |
| a-I: operon linked with Act | Yes | 2 only | Yes | No | Chloroflexi |
| a-II: fusion COX1 with COX4 | Yes | Yes | 9 only | 1 fungus, ciliates? Naegleria? | Proteobacteria |
| a, a-I, a-II, a–b: fusion COX2-cyt c | Yes | Yes | Yes | 1 spongea | Firmicutes |
| a, a-II, a–b, b: ctaG (COX11) in operon | Yes | Yes | Yes | Jakobids | Firmicutes |
| a–b, b: ctaB (COX10) in operon | Yes | Yes | Yes | No | Firmicutes |
| a–b, b: ctaG–ctaE (COX11–COX3) synteny | Yes | No | Yes | Jakobids | γ-proteobacteria? |
| a–b, b: ctaA (COX15) in operon | 2 only | Yes | Yes | 1 Jakobide | Firmicutes |
| N-metabolism | |||||
| NiaD-like nitrate reductase alone | Yes | Yes | Yes | Fungi, Heterokonts, Amoebozoa | Actinomycetes |
| BisC-type nitrate reductase | Yes | Yes | Yes | 1 anellideb | Actinomycetes? |
| bc1 complex | |||||
| Fusion of cyt b with cyt c1/c | Yes | No | 13 only | No | Planctomycetes |
| Long ISP Rieske (ISP2) | 9 only | Yes | Yes | No | Alpha? |
| Whole petABC operon | Yes | Yes | Yes | Daphniac | Cyanophyta |
aCOX2 fused with cytochrome c is found in the genome of the sponge Amphimedon queenslandica (accession: XP_003390963, 391 residues long) and is most closely related to the COX2 protein of COX operon a–b transition of γ-proteobacteria from the genus Thioalkalinivibrio.
bTypical bacterial BisC-type nitrate reductase is embedded in the genome of the anellide Capitella teleta (accession: ELU13386, 644 residues long); it is most closely related to the BisC reductase of γ-proteobacterial endosymbionts of the genus Endozoicomonas.
cThe entire operon of a bacterial bc1 complex is present within the genome of the crustaceon Daphnia (accession for ISP: EFX83890; Colbourne et al. 2011). See text for more information.
The classification of COX gene sequences into defined operon types (fig. 1A, cf. Degli Esposti et al. 2014) has revealed new evolutionary information which had been previously overlooked using standard phylogenetic approaches (Andersson et al. 2003; Brochier-Armanet et al. 2009; Sousa et al. 2012; Burger et al. 2013). In particular, COX operon type b, which is characterized by the insertion of ctaB/COX10 and the conserved synteny ctaG–CtaE (COX11–COX3), is present only in α-proteobacteria; it lies in sister position of the transition operon a–b which is shared by β- and γ-proteobacteria. Recent analysis has shown the insertion of small hydrophobic subunits possibly related to COX4 between COX1 and COX3 in the gene sequences of β-proteobacterial COX operons, thereby providing a new distinguishing feature with respect to the operons of γ-proteobacteria (fig. 1A). The synteny COX11–COX3 previously observed in mtDNA of Jakobide protists (Burger et al. 2013) thus becomes a feature that is common to mitochondria as well as α- and γ-proteobacteria, but no other lineage of the phylum proteobacteria (table 1).
Fig. 1.—
Classification and distribution of COX operons in proteobacteria. (A) COX operons defined by Degli Esposti et al. (2014) are graphically represented with the core genes COX1–4 (corresponding to ctaBCDE in Bacillus) and common other genes including those for assembling the catalytic groups of the oxidase: ctaA (star with A), ctaB (pentagon with B), ctaG (hexagon with G), and SURF-1 (grey square). The sequences falling within COX operon type a can be distinguished from the position of ctaG (supplementary fig. S1, Supplementary Material online). The canonical gene sequence CyoA–E of bo3-type ubiquinol oxidases is shown for comparison. See Matsutani et al. (2014) for a classification of these sequences, which are clearly different from those of COX operons (see also supplementary figs. S1 and S2, Supplementary Material online). (B) Distribution of COX operon type a in the classes of α-, β-, and γ-proteobacteria. Data were extracted from NCBI websites, accessed on September 15, 2014. The total number of species with available genomic data is comparable for α- and γ-proteobacteria (589 and 652, respectively), but approximately one-half (301) for β-proteobacteria. Act, alternative complex three (Refojo et al. 2010); PQQ, deH, methanol dehydrogenase.
Jakobide mtDNA shows the largest set of bacterial bioenergetic proteins among eukaryotes (Gray 2012), including another component of bacterial COX operons, ctaA (COX15; Burger et al. 2013). Intriguingly, the closest bacterial homolog of this protein has been found in the α-proteobacterium Tistrella mobilis, lying within its COX operon type b (Degli Esposti et al. 2014). So far, the presence of ctaA in this type of COX operon has been observed only in another α-proteobacterium, Geminicoccus roseus (accession: NZ_ATYL01000000; Foesel et al. 2007). Hence, it can be considered a novel feature that is shared with the mitochondria of at least one protist, Andalucia (Burger et al. 2013, table 1). Its rare occurrence in both α-proteobacteria and protists contrasts with its frequency in COX operons of β- and γ-proteobacteria (fig. 1 and table 1), suggesting that Tistrella and Geminicoccus might have a common ancestor with the bacteria that then became the mitochondrial organelles of protists. However, phylogenetic analysis of the ctaA/COX15 protein fails to sustain such a possibility, because the sequence of Andalucia COX15 does not form a monophyletic clade with other eukaryotes, while those of α-proteobacteria such as Devosia cluster with eukaryotic sequences rather than with those of other α-proteobacteria (not shown). Presumably, ctaA/COX15 proteins do not carry robust phylogenetic signals that are congruent with the evolution of mitochondria (cf. Hederstedt 2012), a situation that has been found also for other subunits of aa3-type oxidase such as COX2 and ctaB/COX10.
Refining the Evolutionary Path of COX Operons from Bacteria to Mitochondria
In contrast with the abovementioned subunits, COX1 and COX3 possess strong phylogenetic signals that are consistent with the general pattern of mitochondrial evolution, producing monophyletic clades for the mitochondrial homologs within the large group of COX operon type b (Degli Esposti et al. 2014). The molecular variations of COX3 are particularly large, because bacterial homologs of the mitochondrial protein having seven transmembrane helices can be found only in operons b and a–b transition (Degli Esposti et al. 2014). All other bacterial forms of this protein are shorter (generally with five transmembrane helices) and could be considered ancestral homologs of mitochondrial COX3; indeed, they have been categorized in different subfamilies on the basis of their conserved domain features (CDD, Marchler-Bauer et al. 2011). One of these subfamilies is typical of bo3-type ubiquinol oxidase, the operon sequence of which is shown at the bottom of figure 1A, while each of the two consecutive COX3 proteins that are characteristically present in COX operon type a-III belongs to a different recognized subfamily: CD02865 (colored in pink, fig. 1A) and CD02864 (colored in orange, fig. 1A). Of note, the latter is often annotated as “bb3-type cytochrome c oxidase subunit IV” following an old nomenclature of bo3-type oxidases (de Gier et al. 1994)—one of the several confusing annotations for COX genes (see below).
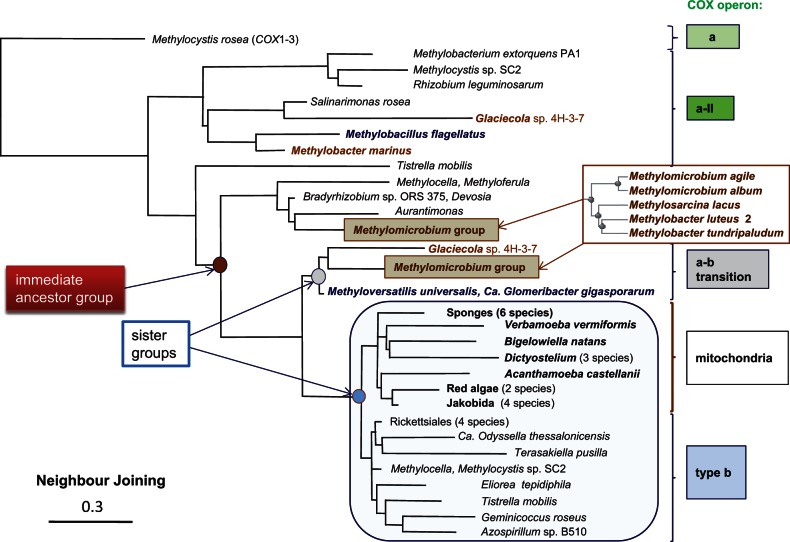
Although not recognized as distinct CDD-based subfamilies, the COX3 subunits of COX operons type a-I and a-II cluster in separate clades of phylogenetic trees and show a different conservation pattern of key protein residues (Degli Esposti et al. 2014). In COX operon type a, instead, COX3 is fused at the C-terminus of COX-1. The resulting fused COX1–3 proteins are distributed in proteobacteria (fig. 1B and supplementary fig. S1, Supplementary Material online), as well as in Thermus, Bacillus (Radzi Noor and Soulimane 2012), and a few mitochondria from ciliates (Degli Esposti et al. 2014, table 1). The constant addition of new proteobacterial genomes has greatly expanded the number of COX1–3 proteins (often, and inexplicably annotated as “cytochrome B561”) from the set previously considered by Degli Esposti et al. (2014). The analysis of these gene clusters has identified two subtypes that are clearly distinguished by the position of the assembly subunit ctaG with respect to the core catalytic subunits, as shown in supplementary figure S1, Supplementary Material online, and figure 1A according to the operon nomenclature of Price et al. (2006). Intriguingly, several proteobacteria have representatives of both subtypes in their genome, for example, Methylocystis and Nitrosococcus (supplementary fig. S1, Supplementary Material online). The increasing complexity and presence of multiple forms within the same organism have hampered a clear definition of the possible evolutionary path of COX operons type a. However, recently I was able to trace this path by extending DELTABLAST (Boratyn et al. 2012) searches to 5,000 bacterial and mitochondrial species using the seven helices form of COX3 from Tistrella as query (fig. 2). The phylogenetic trees thus obtained suggested the following evolutionary sequence: COX operon a, COX operon a-I, bo3-type oxidase, COX operon a-III (with separate clades for the two consecutive COX3 proteins), and a subset of COX operon type a-II, which forms the immediate ancestral precursor of the sister groups of operon b (that surrounds the mitochondrial clade) and a–b transition (including β and γ organisms; fig. 2). This evolutionary sequence of COX operons refines that previously presented in figure 5 of Degli Esposti et al. (2014), which was obtained with searches limited to 1,000 taxa; it is further confirmed by the extended analysis of COX1 and COX4 proteins (not shown).
Fig. 2.—
Phylogenetic tree of COX3 and evolutionary path of COX operons. The sequence of Tistrella COX3 (accession: YP_006372231, 269 residues long) was used as the query of a DELTABLAST search expanded to 5,000 proteins, including β- and γ-proteobacterial taxa for a total selection of 70 species. The Methylomicrobium group of five γ-proteobacterial taxa that is expanded on the right is the same in the branch of COX operon a–b transition and the subbranch of COX-operon a-II (arrow), which appears to be the immediate ancestral group to that containing mitochondria (enclosed in big square—see also supplementary fig. S2, Supplementary Material online). Only Glaceicola contains the same type of protein among other γ-proteobacteria, but in a COX operon that does not show methanol or related dehydrogenases (supplementary fig. S2, Supplementary Material online). The α-proteobacterial taxa in the same group include Devosia sp. DBB001, Bradyrhizobium sp. ORS 375, Hyphomicrobium, and Aurantimonas. Methyloferula and Methylocella have their COX3 proteins of COX operon type a-II that fall in the same group, together with several β-proteobacteria (supplementary fig. S2, Supplementary Material online). The outgroup sequence of Methylocystis rosea corresponds to the fused COX1–3 protein of 850 residues (accession: WP_018409174) belonging to a COX operon type a with prepended ctaG (supplementary fig. S1, Supplementary Material online, and fig. 1A). The α-proteobacterial subbranch close to the mitochondrial clade (species or group outlined in bold characters) contains representative of the order Rickettsiales (Rickettsia prowazekii, Anaplasma, Neorickettisa, and Caedibacter). Sponges include Amphimedon queenslandica and demosponges. Similar trees have been obtained with Geminicoccus COX3 or COX1 as a query.
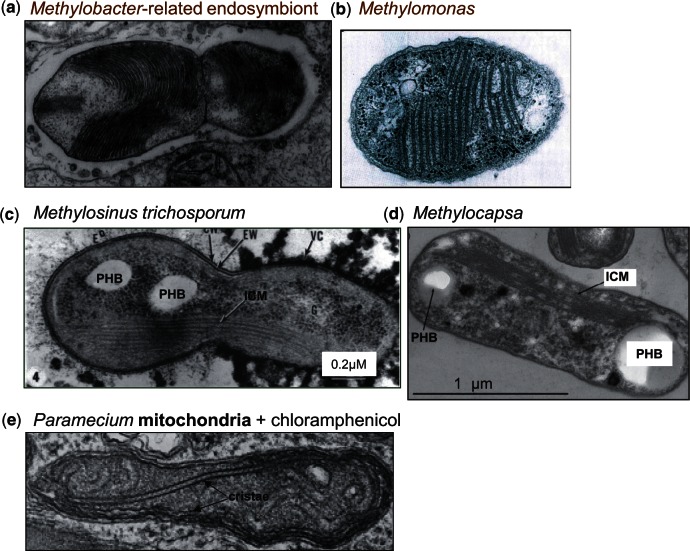
Fig. 5.—
Membrane structures of methanotrophs resembling mitochondrial cristae. Electron microscopy images of methanotrophic bacteria refer to symbionts of clams (A; Fujiwara, Takai, et al. 2000) or plants of bog peats closely related to Methylomonas (B; Kip et al. 2011), or free-living Methylosinus (C, middle; Reed et al. 1980) and Methylocapsa (D; Dunfield et al. 2010). Note the resemblance of their ICMs with mitochondrial cristae, for comparison shown in a micrograph of the protist Paramecium (E; modified from Adoutte et al. 1972). Reserve deposits of poly hydroxy butyrate are labeled as PHB. Note: Images have been modified from the cited references.
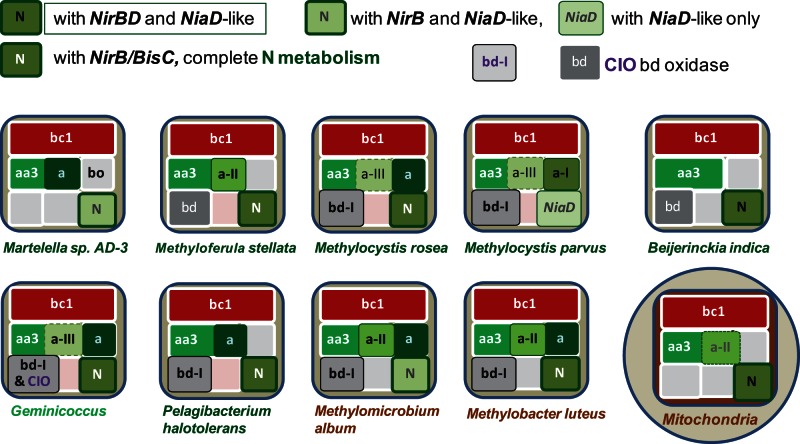
The same trees also revealed the presence of COX operon type a-II in a group of γ methanotrophs including the genera Methylomicrobium, Methylosarcina, and Methylobacter (supplementary fig. S2, Supplementary Material online; cf. pink square in fig. 2, right). These organisms are closely related to the methanotrophic endosymbionts of deep-sea invertebrates and wetland plants (DeChaine and Cavanaugh 2006; Kip et al. 2011; Dedysh 2011; Cavanaugh et al. 2013) and form a separate clade from other methanotrophs of the Methylococcales order (Kip et al. 2011; Cavanaugh et al. 2013), which indeed do not have COX operon type a-II (supplementary fig. S2, Supplementary Material online). They also show a subset of four bioenergetic systems similar to that of α methylotrophs and methanotrophs (fig. 3). The subset is characterized by the presence of at least one type of bd-type ubiquinol oxidase and coincides with that previously suggested to form the most likely path of bioenergetic evolution of bacteria into mitochondria (Degli Esposti et al. 2014). Although previously represented by Beijerinckia indica alone, it is now recognized in additional organisms belonging to either α- or γ-proteobacteria which have methylotrophic metabolism in common, as illustrated in figure 3. Most of such organisms possess COX operons of type a and in some cases have the precursor of eukaryotic assimilatory nitrate reductase, NiaD (fig. 3). Nevertheless, there are multiple combinations of key elements of N-metabolism among the organisms that possess the Beijerinckia subset of bioenergetic systems, which would match the mitochondrial subset of fungi and protists after losing the bd-type oxidase (fig. 3, cf. Degli Esposti et al. 2014).
Fig. 3.—
Bioenergetic systems of bacteria and mitochondria. Proteobacteria possessing only four bioenergetic systems, that is, only one more than those present in the mitochondria of protists and fungi, were identified from the latest NCBI resources (accessed September 15, 2014). Those possessing cbb3-type oxidase and lying in pathway B are not presented here for they are unlikely to be in mitochondrial ancestry (Degli Esposti et al. 2014). COX operons of type a are overlaid on the area of aa3-type oxidase. Different combinations of assimilatory, NAD(P)H-dependent nitrite (NirB) and nitrate reductase (labeled NiaD like when having redox modules that are precursors to those of the fungal NiaD enzyme; Degli Esposti et al. 2014) are color coded as indicated in the legend within the diagram. Specifically, organisms such as Beijerinckia indica that possess both the NirBD nitrite reductase and NiaD-like nitrate reductase are labeled with a different type of symbol for N-metabolism from those having the latter reductase combined with (not fused) NirB or without a NirBD homolog. Organisms having other nitrite or nitrate (e.g., BisC) reductases are differentially labeled too. Note that Beijerinckia indica does not possess COX operons of type a (Tamas et al. 2010) which are instead present in its close relatives such as Methyloferula, which is also shown because it has only four bioenergetic systems. Two major forms of bd-type oxidase that have been previously classified as bd-I and Cyanide-Insensitive-Oxidase (CIO, Degli Esposti et al. 2014) are annotated as shown in the legend within the diagram. Note that Methylomicrobium and Methylobacter are γ-proteobacteria and therefore have ISP proteins with multiple insertions that are not present in several α-proteobacteria and mitochondria (Degli Esposti et al. 2014).
Changes in Bioenergetic Systems of Proteobacteria in the Transition from Free-Living to Symbiont
Because mitochondria derive from ancestral bacteria that became endocellular symbionts, it is of value to analyze the genomic and metabolic changes in closely related bacteria that have undergone equivalent transitions from free-living to host-associated lifestyle (Sachs et al. 2011). There are several examples of such transitions in the literature, but often they are not relevant to the evolution of mitochondria. In some cases, they refer to bacteria that do not possess the respiratory complexes of mitochondria but rather those typical of microaerophilic enterobacteriales, as in insect symbionts of the genus Serratia (Manzano-Marín et al. 2010) and acetic acid bacteria (Chouaia et al. 2014). In other cases, for instance Teredinibacter turnerae versus Saccharophagus degradans, the transition from free-living to endosymbiont is still ongoing and consequently not associated yet with evident bioenergetic changes (Yang et al. 2009). Conversely, the obligate endocellular parasites of the Rickettsiales order still await the recognition of free-living relatives, a major difficulty undermining the proposal that proto-mitochondria originated from these α-proteobacteria (Andersson et al. 1998; Williams et al. 2007; Sassera et al. 2011; Ferla et al. 2013).
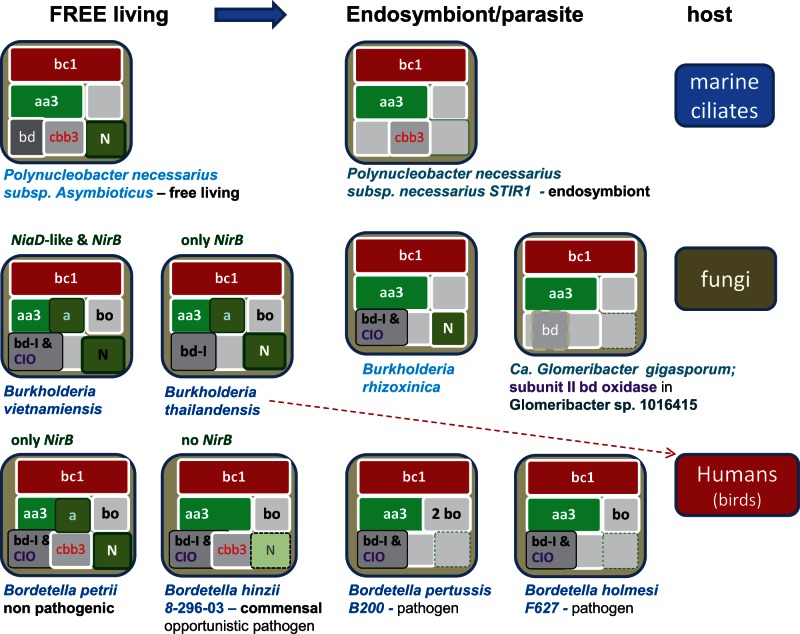
I found three transitions in β-proteobacteria that may be compared with the reconstructed evolution of mitochondria (Degli Esposti et al. 2014) and cover a phylogenetically divergent set of hosts, from unicellular eukaryotes to humans (fig. 4). These organisms belong to the large group of Burkholderiales and show the deletion of one or more terminal oxidases together with N-metabolism in the transition toward endosymbiotic lifestyles. The resulting subsets of bioenergetic systems remaining in symbionts/pathogens have, besides the common bc1 complex (fig. 4) the following: (1) a cbb3-type oxidase together with the aa3-type oxidase, as in the Polynucleatus symbiont of ciliates (Vannini et al. 2007); (2) only the aa3-type oxidase, as in the fungal endosymbiont Glomeribacter (Ghignone et al. 2012); and (3) a bd-type ubiquinol oxidase together with the aa3-type oxidase, as in Bordetella pathogens. Hence, adaptation to a symbiotic lifestyle may follow pathways of serial loss of bioenergetic systems that resemble those previously deduced for proto-mitochondria: One ending with a cbb3-type and another with a bd-type terminal oxidase (Degli Esposti et al. 2014).
Fig. 4.—
Reduction in bioenergetic systems in the transition from free-living to symbionts. Three different lineages of β-proteobacteria from the Burkholderiales order were analyzed on the basis of current genomic data and the reported lifestyle information indicating transitions to symbionts or pathogens (Vannini et al. 2007; Sachs et al. 2011; Ghignone et al. 2012). Bioenergetic systems are labeled as before (Degli Esposti et al. 2014) and variations in their elements of N-metabolism are color coded as indicated within figure 3. The red dash arrow indicates a behavior as human opportunistic pathogen of a plant-associated Burkholderia. See Lackner et al. (2011) and Ghignone et al. (2012) for the related fungal pathogens. Note that subunit II of bd-type oxidase is retained in the indicated Glomeribacter species as a likely relic of this bioenergetic system.
However, proto-mitochondria retained a fully functional system of N-metabolism, which is instead lost in the examples of extant bacteria shown in figure 4. Conversely, N-metabolism is maintained in other bacteria that are phylogenetically close to methanotrophic endosymbionts (fig. 3). For example, Methylocystis parvus is very close to the Methylocystis strains that have been found as moss endosymbionts, which in turn are relatively distant to ecologically diverse organisms of the same genus such as Methylocystis sp. SC2 (Dam et al. 2012; Belova et al. 2013). Notably, M. parvus does not possess the bo3-type ubiquinol oxidase that is found in Methylocystis sp. SC2 and other free-living members of the same group (fig. 3). Clearly, we need to wait until the completion of the genome of Methylocystis bryophila (Belova et al. 2013) or other moss endosymbionts to verify whether the specific loss of this bioenergetic system is a characteristic that methanotrophic α-proteobacteria may share with the presumed adaptation of proto-mitochondria to endosymbiosis (Degli Esposti et al. 2014). The same applies to the methanotrophic symbionts of deep-sea invertebrates (Fujiwara, Kawato, et al. 2010; Cavanaugh et al. 2013), some of which are closely related to the free-living γ-proteobacterium Methylobacter, also shown in figure 3. Intriguingly, these methanotrophic endosymbionts characteristically show stacks of intracytoplasmic membranes (ICMs) that resemble mitochondrial cristae, as documented by the representative microscopic images collected in fig. 5 (Reed et al. 1980; Fujiwara, Takai, et al. 2000; Dunfield et al. 2010; Fujiwara, Kawato, et al. 2010; Dedysh 2011; Cavanaugh et al. 2013). Amazingly, this similarity has not been noted before.
In sum, bd-type ubiquinol oxidase is the bioenergetic system that appears to be most frequently retained in aerobic endosymbionts or pathogens that can be compared with mitochondria. Traces of its progressive loss are present in diverse proteobacterial lineages, such as the α-proteobacterium Rickettsia (Degli Esposti et al. 2014) and the β-proteobacteria Bordetella and Glomeribacter (fig. 4). These lineages include species showing functionally intact bd-oxidases that lie aside species that either retain only one catalytic subunit or exhibit clear degradation of the gene cluster, as well as species that have lost the same gene cluster altogether. Moreover, I found the intact gene for one or both catalytic subunits of bd-type oxidase in the genome of some invertebrates, in particular the fruit fly Ceratitis capitata (accession: XP_004532875 for subunit I and XP_004532875 for subunit II), as well as the anellide Capitella teleta (accession: ELU02357) and the nematode Caenorhabditis remanei (accession: CRE_07978). Because bd-type oxidases are exclusively found in bacteria (Borisov et al. 2011), the presence of their genes in eukaryotic genomes must derive from the sequencing of DNA fragments belonging to associated, most likely symbiotic bacteria. In contrast, catalytic subunits of the cbb3-type oxidase have not been found in currently available genomes of metazoans. All these indications sustain the deduction that loss of bd-type oxidase could be the last step in the bioenergetic evolution of proto-mitochondria (Degli Esposti et al. 2014).
The bc1 Complex of Bacterial Endosymbionts and Evolution of Its Catalytic Subunits
Compared with the bioenergetic systems of terminal oxidases, little is known about the changes in the bc1 complex that may be associated with symbiosis and its transition from free-living lifestyles. So far, I found only two cases that may provide some clues on this topic. The first case regards the presence of the entire petABC operon of the bacterial complex (coding for ISP, cytochrome b, and cytochrome c1 in sequence) within the genome of the small crustacean Daphnia (accession for ISP: EFX83890, DAPPUDRAFT_47869—Colbourne et al. 2011; table 1). The bacterial proteins of the bc1 complex are clearly different from those of Daphnia mitochondria, but very close to those of a uncultivated α-proteobacterium from a groundwater metagenomic survey (accession: EKE09606 for ISP and EKE09607 for cytochrome b fused with cytochrome c1—a protein 653 residues long; Wrighton et al. 2012). Although this metagenomic survey predominantly dealt with anaerobic bacteria, it also reported some potentially aerobic α-, β-, and γ-proteobacteria apparently related to known taxa (supplementary information in Wrighton et al. 2012). Sequence analysis of the ISP protein of the α-proteobacterium above indicates a closeness to Rhodospirillales that is partially confirmed by phylogenetic analysis of the fused cytochrome b protein. The latter shows an immediate ancestral position with respect to the cytochrome b proteins fused with cytochrome c1 that are found in some Rhodospirillales and, characteristically, in most members of the Bradyrhizobiaceae family of α-proteobacteria (Thöny-Meyer 1997; supplementary fig. S3a and b, Supplementary Material online). Interestingly, the previously mentioned T. turnerae—a γ-proteobacterial endosymbiont of wood-boring clams—possesses multiple genes for cytochrome b fused with cytochrome c1, only two of which are associated with an ISP gene thereby producing two separate operons for the bc1 complex (Yang et al. 2009). Although similar proteins are present in closely related free-living bacteria, only Teredinibacter shows multiple genes coding for slightly different versions of ISP and fused cytochrome b proteins, a rare situation in proteobacteria (ten Brink et al. 2013). Among γ-proteobacteria, in fact, only the genus Acidithiobacillus shows the duplication of bc1 operon (ten Brink et al. 2013), without multiple versions of isolated ISP, which are instead found as a result of specific duplication of the ISP gene in Cyanophyta, as well as in some α- and β-proteobacteria (Schneider and Schmidt 2005). Of note, methanotrophs of the Methylocystis genus and some Bradyrhizobiaceae show vestiges of an ancient duplication and diversification of their Rieske ISP, because they retain a long ISP2 form that is now present within an isolated gene cluster resembling some of those present in Teredinibacter (Schneider and Schmidt 2005; Degli Esposti et al. 2014).
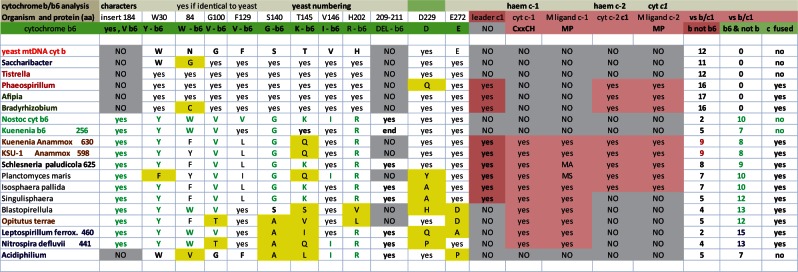
Whether or not duplication and diversification of the bc1 operon can be considered among bioenergetic features associated with the transition from free-living to endosymbiosis, I take the opportunity raised by the above observations to provide an answer to the yet unresolved question: What’s the origin and evolutionary relevance of cytochrome b fusion with cytochrome c1? Previous analyses focused on ancient bacterial lineages, finding diverse forms of cytochrome b6—the “green” paralog of mitochondrial cytochrome b corresponding to its N-terminal domain (Degli Esposti et al. 1993; Lebrun et al. 2006)—fused with c-type cytochromes in Planctomycetes and Nitrospirales (Dibrova et al. 2013; ten Brink et al. 2013). In particular, anammox Planctomycetes such as Kuenenia possess large sets of bioenergetic proteins fused with c-type cytochromes (Dibrova et al. 2013; Kartal et al. 2013). My detailed sequence analysis (fig. 6 and supplementary fig. S3, Supplementary Material online) shows a progressive diversification of proteobacterial cytochrome b proteins fused with cytochrome c1 from some of these Planctomycetes proteins that are fused with two c-type cytochromes, for example, Shlesneria cytochrome b/c subunit (accession: WP_010584211 or ZP11092181—Guo et al. 2012), which has not been considered in recent surveys (Dibrova et al. 2013; ten Brink et al. 2013). This finding supports previous structural deductions suggesting that mitochondrial cytochrome c1 may derive from a collapsed di-heme cytochrome c (Baymann et al. 2004).
Fig. 6.—
Comparison of the molecular characters of cytochrome b from diverse bacterial lineages. The molecular characters which are conserved among all the cytochrome b proteins of the bc1 complex but not in the paralog b6 proteins (in green) are annotated following the numbering of the yeast sequence (Degli Esposti et al. 1993—see supplementary fig. S3a and b, Supplementary Material online for a representative alignment). The two c hemes that are present at the C terminus of some Planctomycetes’ proteins (Kartal et al. 2013) are designated heme c-1 and heme c-2. Fused cytocrome c1 of α- and γ-proteobacteria align only with the heme-binding regions of heme c-2 (supplementary fig. S3b, Supplementary Material online). The amino acid changes that are structurally different from the conserved characters are highlighted in yellow. The total number of characters matching those conserved in either cytochrome b (b not b6) or cytochrome b6 (b6 not b) are listed at the far right of the table—note that the maximal number is 17 for cytochrome b fused with c1 and 12 for normal cytochrome b. Leptospirillum ferrox. refers to the 460 residues protein (accession: BAM07238) from Leptospirillum ferrooxidans C2-3, an organisms related to Nitrospira (Lücker et al. 2010). Note that Acidiphilium cytochrome b is degenerate for it does not have all the conserved histidines that function as heme ligands (annotated in supplementary fig. S3a, Supplementary Material online).
Consequently, the answer to the above question is that cytochrome b fusion with cytochrome c1 in proteobacteria derives from fused proteins present in Planctomycetes such as Shlesneria that have been probably inherited via vertical transmission, given the gradual changes in key amino acids (fig. 6)—but see ten Brink et al. (2013) for a different opinion. In any case, these fused proteins may represent the oldest version of the ancestors of mitochondrial cytochrome b, as in the case of fused COX1–3 for mitochondrial COX1 that have been mentioned before (cf. Degli Esposti et al. 2014). Within proteobacteria and outside the Bradyrhizobiaceae family, fused cytochrome b/c is found only in 13 γ-proteobacteria (table 1), two genera of Rhodospirillaceae, Phaeospirillum and Magnetospirillum, and in Rhodovulum, an organism normally classified among the Rhodobacterales but probably belonging to the Rhizobiales order (Ferla et al. 2013). The ISP proteins from these Rhodospirillaceae organisms and Rhodovulum show at least one conserved insertion that is not present in mitochondrial ISP and are therefore unlikely to be among the immediate ancestors of mitochondrial ISP (Degli Esposti et al. 2014). Conversely, the ISP sequences of Bradyrhizobiaceae appear to be as close to mitochondrial ISP as those of methylotrophs such as Beijerinckia and Methyloferula (cf. Degli Esposti et al. 2014). Such a circumstance underlines the importance of the molecular features of ISP in discriminating among possible candidates for the closest living relative of mitochondrial ancestors.
Conclusion—A Methanotrophic Past for Mitochondria?
The new information presented in this review (figs. 2, 3, and 5 and Supplementary figs. S1–S3, Supplementary Material online) supports and refines recent indications that extant methylotrophs may include the relatives of proto-mitochondria (Degli Esposti et al. 2014). This proposition contrasts with the majority of previous studies that had indicated different bacteria as the possible ancestors of mitochondria. Frequently, such studies have supported the possibility that proto-mitochondria originated from Rickettsia and its relatives (Andersson et al. 1998, 2003; Fitzpatrick et al. 2006; Williams et al. 2007; Sassera et al. 2011; Ferla et al. 2013). However, the possible mitochondrial origin from within the Rickettsiales, which include only obligate endosymbionts, is viewed with increasing scepticism (Esser et al. 2004; Atteia et al. 2009; Abhishek et al. 2011; Gray 2012; Müller et al. 2012), because it suffers from the major problem of long-branch attraction distorting phylogenetic analysis. Moreover, in bioenergetic terms, all Rickettsiales have lost the elements of N-metabolism that are still present in mitochondria from fungi and heterokonts (Takaya 2009), while often retaining the noneukaryotic bd-type ubiquinol oxidase (Degli Esposti et al. 2014). This situation is similar to that encountered in various transitions from free-living to symbiotic lifestyles among extant bacteria (fig. 4), thereby suggesting phenomena of convergent evolution related to endocellular symbiosis. Finally, the proponents of the Rickettsiales origin for proto-mitochondria have as yet to provide a plausible answer to the obvious question: Which organisms were they a parasite of?
Methylotrophs are a general definition for chemiolithotrophic bacteria that utilize C1 compounds as their carbon and energy source and include both methane and ammonia-oxidizing organisms (Chistoserdova et al. 2009). Methane oxidation arose when ambient oxygen levels increased dramatically on the planet, approximately at the time in which β- and γ-proteobacteria separated from the ancestors of current α-proteobacteria (Battistuzzi et al. 2004). Although methylotrophs or methanotrophs had not been considered before as possible ancestors of mitochondria, they now appear to be good candidates for proto-mitochondria, for the following reasons:
they are evolutionary ancient, as just discussed, consistent with the accepted notion that proto-mitochondria evolved early after the separation of the γ- and β-lineages (Williams et al. 2007);
they are related to autotrophic organisms forming permanent symbioses with invertebrate animals living in deep-sea habitats (Dubilier et al. 1999; Fujiwara, Takai, et al. 2000; Cavanaugh et al. 2013), as well as with mosses of northern wetlands (Kip et al. 2011; Dedysh 2011);
they have a wide metabolic versatility which includes the cycles of glyoxylate, citrate, and serine that in eukaryotic lineages are segregated within organellar compartments that may also derive from mitochondria (Mohanty and McBride 2013);
they have a bioenergetically efficient respiratory chain, even under low oxygen concentrations, for the presence of multiple terminal oxidases (Dam et al. 2012; Degli Esposti et al. 2014);
finally, they characteristically contain “ICMs” derived from invaginations of their inner membrane (Hagen et al. 1966; Watson and Mandel 1971; Lynch et al. 1980; Reed et al. 1980; Fujiwara, Takai, et al. 2000; Dunfield et al. 2010; Dedysh 2011; Kip et al. 2011; Cavanaugh et al. 2013). Such tightly packed membranes resemble mitochondrial cristae (fig. 5) and are equally required to expand the surface available to plug in respiratory complexes that provide bioenergy, together with membrane-bound methane mono-oxygenase (Chistoserdova et al. 2009).
It is surprising that such evident structural similarities between methanotrophs and mitochondria have passed unnoticed for almost 50 years (Hagen et al. 1966). However, the overlooked cristae-like structures of methanotrophs may constitute yet another example of how science has become compartmentalized in specialized fields, in this case a niche of microbiology and classical bioenergetics. No doubt, Lynn Margulis has encountered examples of the same kind in her pioneering work that in time established the endosymbiotic theory of eukaryotic cell evolution (Sagan 1967).
Taking together the timeline for bacterial evolution (Battistuzzi et al. 2004) with the restricted distribution of methanotrophic symbionts to α- and γ-proteobacteria (Fujiwara, Takai, et al. 2000; Kip et al. 2011; Dedysh 2011) and the list of rare bioenergetic features shared by proteobacteria and mitochondria, which are more common between α- and γ- than between α- and β-proteobacteria (table 1), it seems possible that proto-mitochondria have evolved from the common ancestors of methanotrophic α- and γ-proteobacteria. The phylogenetic trees of COX proteins (fig. 2) support this possibility. The invention of the metabolic pathways of methane and ammonia oxidation immediately followed the large increase in ambient oxygen that occurred around 2 billion years ago, allowing autotrophic ways of life which are now retained by the abovementioned methanotrophs (Battistuzzi et al. 2004; Chistoserdova et al. 2009; Vlaeminck et al. 2011). These bacteria also possess the largest variety of COX operons and molecular forms of their catalytic subunits (supplementary figs. S1 and S2, Supplementary Material online), as a result of multiple events of operon and gene duplication. Indeed, the reconstruction of the molecular evolution of COX3 proteins and their binding strength for oxygen-modulating phospholipids seems to recapitulate a progressive adaptation to increasing levels of O2, gauged in terms of decreasing oxygen affinity to maintain maximal efficiency of the oxidase reactions (Riistama et al. 1996; Degli Esposti et al. 2014).
These considerations refer to the symbiont independently on the host organism that was involved in the eukaryogenic event leading to the evolution of our cells. Recent evidence points to an archaeal nature for such a host (Williams et al. 2013) without defining ecological and metabolic features that may help indicating the type of symbiosis that it may favor. Certainly, this host was adapted to anaerobic conditions, because the evolution of proto-mitochondria and the earliest eukaryotic cells occurred when oxygen levels were still very low in the oceans (Johnston et al. 2009; Müller et al. 2012). It is therefore plausible that mitochondrial progenitors derive from organisms that had experimented with a wide variety of oxygen-reacting systems and thus retained great plasticity in their adaptation to environments poor in oxygen, a trait that is partially retained in eukaryotes adapted to anaerobic environments (Mentel et al. 2014). Hence, those progenitors were probably related to the ancestors of extant proteobacterial methanotrophs, many of which have the capacity of establishing endosymbiosis with “primitive” animals such as gutless worms, as well as primitive plants such as mosses. A methanotrophic past for mitochondria is thus looming; future research will clarify its validity and evolutionary implications.
Supplementary Material
Supplementary figures S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
I thank Ed Berry (SUNY, Albany, USA), Patricia Lievens, and Roberto Cingolani (IIT, Italy) for support and Paola Bonfante (University of Turin, Italy) plus Claudio Bandi (University Milan, Italy) and Esperanza Martinez-Romero (UNAM, Quernavaca, Mexico) for stimulating discussion.
Literature Cited
- Abhishek A, Bavishi A, Bavishi A, Choudhary M. Bacterial genome chimaerism and the origin of mitochondria. Can J Microbiol. 2011;57:49–61. doi: 10.1139/w10-099. [DOI] [PubMed] [Google Scholar]
- Adoutte A, Balmefrézol M, Beisson J, André J. The effects of erythromycin and chloramphenicol on the ultrastructure of mitochondria in sensitive and resistant strains of Paramecium. J Cell Biol. 1972;54:8–19. doi: 10.1083/jcb.54.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Karlberg O, Canbäck B, Kurland CG. On the origin of mitochondria: a genomics perspective. Philos Trans R Soc Lond B Biol Sci. 2003;358:165–177. doi: 10.1098/rstb.2002.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Zomorodipour A, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Atteia A, et al. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol Biol Evol. 2009;26:1533–1548. doi: 10.1093/molbev/msp068. [DOI] [PubMed] [Google Scholar]
- Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baymann F, Lebrun E, Nitschke W. Mitochondrial cytochrome c1 is a collapsed di-heme cytochrome. Proc Natl Acad Sci U S A. 2004;101:17737–17740. doi: 10.1073/pnas.0407442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belova SE, Kulichevskaya IS, Bodelier PL, Dedysh SN. Methylocystis bryophila sp. nov., a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993. Int J Syst Evol Microbiol. 2013;63:1096–1104. doi: 10.1099/ijs.0.043505-0. [DOI] [PubMed] [Google Scholar]
- Berry EA, et al. X-ray structure of Rhodobacter capsulatus cytochrome bc: comparison with its mitochondrial and chloroplast counterparts. Photosynth Res. 2004;81:251–275. doi: 10.1023/B:PRES.0000036888.18223.0e. [DOI] [PubMed] [Google Scholar]
- Blazejak A, Kuever J, Erséus C, Amann R, Dubilier N. Phylogeny of 16S rRNA, ribulose 1,5-bisphosphate carboxylase/oxygenase, and adenosine 5'-phosphosulfate reductase genes from gamma- and alphaproteobacterial symbionts in gutless marine worms (oligochaeta) from Bermuda and the Bahamas. Appl Environ Microbiol. 2006;72:5527–5536. doi: 10.1128/AEM.02441-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn GM, et al. Domain enhanced lookup time accelerated BLAST. Biol Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta. 2011;1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier-Armanet C, Talla E, Gribaldo S. The multiple evolutionary histories of dioxygen reductases: implications for the origin and evolution of aerobic respiration. Mol Biol Evol. 2009;26:285–297. doi: 10.1093/molbev/msn246. [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 2013;5:418–438. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey BE, et al. Proteome-wide analysis of functional divergence in bacteria: exploring a host of ecological adaptations. PLoS One. 2012;7:e35659. doi: 10.1371/journal.pone.0035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh CM, McKiness ZP, Newton IL, Stewart FJ. Marine chemosynthetic symbioses – Chapter 23. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes. New York: Springer; 2013. pp. 579–607. [Google Scholar]
- Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME. The expanding world of methylotrophic metabolism. Annu Rev Microbiol. 2009;63:477–499. doi: 10.1146/annurev.micro.091208.073600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouaia B, et al. Acetic acid bacteria genomes reveal functional traits for adaptation to life in insect guts. Genome Biol Evol. 2014;6:912–920. doi: 10.1093/gbe/evu062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam B, Dam S, Kube M, Reinhardt R, Liesack W. Complete genome sequence of Methylocystis sp. strain SC2, an aerobic methanotroph with high-affinity methane oxidation potential. J Bacteriol. 2012;194:6008–6009. doi: 10.1128/JB.01446-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidov Y, Huchon D, Koval SF, Jurkevitch E. A new alpha-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory. Environ Microbiol. 2006;8:2179–2188. doi: 10.1111/j.1462-2920.2006.01101.x. [DOI] [PubMed] [Google Scholar]
- DeChaine EG, Cavanaugh CM. Symbioses of methanotrophs and deep-sea mussels (Mytilidae: Bathymodiolinae) Prog Mol Subcell Biol. 2006;41:227–249. doi: 10.1007/3-540-28221-1_11. [DOI] [PubMed] [Google Scholar]
- Dedysh SN. Cultivating uncultured bacteria from northern wetlands: knowledge gained and remaining gaps. Front Microbiol. 2011;2:184. doi: 10.3389/fmicb.2011.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gier JW, et al. The terminal oxidases of Paracoccus denitrificans. Mol Microbiol. 1994;13:183–196. doi: 10.1111/j.1365-2958.1994.tb00414.x. [DOI] [PubMed] [Google Scholar]
- Degli Esposti M, et al. Evolution of mitochondria reconstructed from the energy metabolism of living bacteria. PLoS One. 2014;9:e96566. doi: 10.1371/journal.pone.0096566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degli Esposti M, et al. Mitochondrial cytochrome b: evolution and structure of the protein. Biochim Biophys Acta. 1993;1143:243–271. doi: 10.1016/0005-2728(93)90197-n. [DOI] [PubMed] [Google Scholar]
- Dibrova DV, Cherepanov DA, Galperin MY, Skulachev VP, Mulkidjanian AY. Evolution of cytochrome bc complexes: from membrane-anchored dehydrogenases of ancient bacteria to triggers of apoptosis in vertebrates. Biochim Biophys Acta. 2013;1827:1407–1427. doi: 10.1016/j.bbabio.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubilier N, et al. Phylogenetic diversity of bacterial endosymbionts in the gutless marine oligochaete Olavius loisae. Mar Ecol Prog Ser. 1999;178:271–280. [Google Scholar]
- Dunfield PF, Belova SE, Vorob'ev AV, Cornish SL, Dedysh SN. Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int J Syst Evol Microbiol. 2010;60:2659–2664. doi: 10.1099/ijs.0.020149-0. [DOI] [PubMed] [Google Scholar]
- Emelyanov VV. Evolutionary relationship of Rickettsiae and mitochondria. FEBS Lett. 2001;501:11–18. doi: 10.1016/s0014-5793(01)02618-7. [DOI] [PubMed] [Google Scholar]
- Esser C, et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- Ferla MP, Thrash JC, Giovannoni SJ, Patrick WM. New rRNA gene-based phylogenies of the Alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability. PLoS One. 2013;8:e83383. doi: 10.1371/journal.pone.0083383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, Creevey CJ, McInerney JO. Genome phylogenies indicate a meaningful alpha-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales. Mol Biol Evol. 2006;23:74–85. doi: 10.1093/molbev/msj009. [DOI] [PubMed] [Google Scholar]
- Foesel BU, Gössner AS, Drake HL, Schramm A. Geminicoccus roseus gen. nov., sp. nov., an aerobic phototrophic Alphaproteobacterium isolated from a marine aquaculture biofilter. Syst Appl Microbiol. 2007;30:581–586. doi: 10.1016/j.syapm.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Kawato M, et al. Extracellular and mixotrophic symbiosis in the whale-fall mussel Adipicola pacifica: a trend in evolution from extra- to intracellular symbiosis. PLoS One. 2010;5:e11808. doi: 10.1371/journal.pone.0011808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Takai K, et al. Phylogenetic characterization of endosymbionts in three hydrothermal vent mussels: influence on host distributions. Mar Ecol Prog Ser. 2000;208:147–155. [Google Scholar]
- Gabaldón T, Huynen MA. Reconstruction of the proto-mitochondrial metabolism. Science. 2003;301:609. doi: 10.1126/science.1085463. [DOI] [PubMed] [Google Scholar]
- Ghignone S, et al. The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J. 2012;6:136–145. doi: 10.1038/ismej.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. 2012 Mitochondrial evolution. Cold Spring Harb Perspect Biol. 4:a011403. [Google Scholar]
- Guo M, et al. Genome sequences of three species in the family Planctomycetaceae. J Bacteriol. 2012;194:3740–3741. doi: 10.1128/JB.00639-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen PO, Goldfine H, Williams PJ. Phospholipids of bacteria with extensive intracytoplasmic membranes. Science. 1966;151:1543–1544. doi: 10.1126/science.151.3717.1543. [DOI] [PubMed] [Google Scholar]
- Harrenga A, Michel H. The cytochrome c oxidase from Paracoccus denitrificans does not change the metal center ligation upon reduction. J Biol Chem. 1999;274:33296–33299. doi: 10.1074/jbc.274.47.33296. [DOI] [PubMed] [Google Scholar]
- Hederstedt L. Heme A biosynthesis. Biochim Biophys Acta. 2012;1817:920–927. doi: 10.1016/j.bbabio.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Huynen MA, Duarte I, Szklarczyk R. Loss, replacement and gain of proteins at the origin of the mitochondria. Biochim Biophys Acta. 2013;1827:224–231. doi: 10.1016/j.bbabio.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Johnston DT, Wolfe-Simon F, Pearson A, Knoll AH. Anoxygenic photosynthesis modulated Proterozoic oxygen and sustained Earth’s middle age. Proc Natl Acad Sci U S A. 2009;106:16925–16929. doi: 10.1073/pnas.0909248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartal B, et al. How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev. 2013;37:428–461. doi: 10.1111/1574-6976.12014. [DOI] [PubMed] [Google Scholar]
- Kip N, et al. Detection, isolation, and characterization of acidophilic methanotrophs from Sphagnum mosses. Appl Environ Microbiol. 2011;77:5643–5654. doi: 10.1128/AEM.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner G, Moebius N, Partida-Martinez LP, Boland S, Hertweck C. Evolution of an endofungal lifestyle: deductions from the Burkholderia rhizoxinica genome. BMC Genomics. 2011;12:210. doi: 10.1186/1471-2164-12-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- Lebrun E, et al. The Rieske protein: a case study on the pitfalls of multiple sequence alignments and phylogenetic reconstruction. Mol Biol Evol. 2006;23:1180–1191. doi: 10.1093/molbev/msk010. [DOI] [PubMed] [Google Scholar]
- Lücker S, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci U S A. 2010;107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MJ, Wopat AE, O'connor ML. Characterization of two new facultative methanotrophs. Appl Environ Microbiol. 1980;40:400–407. doi: 10.1128/aem.40.2.400-407.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Lamelas A, Moya A, Latorre A. Comparative genomics of Serratia spp.: two paths towards endosymbiotic life. PLoS One. 2010;7:e47274. doi: 10.1371/journal.pone.0047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. . 2011;39(Database issue):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. Archaeal-eubacterial mergers in the origin of Eukarya: phylogenetic classification of life. Proc Natl Acad Sci U S A. 1996;93:1071–1076. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsutani M, et al. Replacement of a terminal cytochrome c oxidase by ubiquinol oxidase during the evolution of acetic acid bacteria. Biochim Biophys Acta. 2014;1837:1810–1820. doi: 10.1016/j.bbabio.2014.05.355. [DOI] [PubMed] [Google Scholar]
- Mentel M, Röttger M, Leys S, Tielens AG, Martin WF. Of early animals, anaerobic mitochondria, and a modern sponge. Bioessays. 2014;36:924–932. doi: 10.1002/bies.201400060. [DOI] [PubMed] [Google Scholar]
- Mohanty A, McBride HM. Emerging roles of mitochondria in the evolution, biogenesis, and function of peroxisomes. Front Physiol. 2013;4:268. doi: 10.3389/fphys.2013.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- Müller M, et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Arkin AP, Alm EJ. The life-cycle of operons. PLoS Genet. 2006;2:e96. doi: 10.1371/journal.pgen.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzi Noor M, Soulimane T. Bioenergetics at extreme temperature: Thermus thermophilus ba(3)- and caa(3)-type cytochrome c oxidases. Biochim Biophys Acta. 2012;1817:638–649. doi: 10.1016/j.bbabio.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Raghoebarsing AA, et al. Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature. 2005;436:1153–1156. doi: 10.1038/nature03802. [DOI] [PubMed] [Google Scholar]
- Reed WM, Titus JA, Dugan PR, Pfister RM. Structure of Methylosinus trichosporium exospores. J Bacteriol. 1980;141:908–913. doi: 10.1128/jb.141.2.908-913.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refojo PN, Sousa FL, Teixeira M, Pereira MM. The alternative complex III: a different architecture using known building systems. Biochim Biophys Acta. 2010;1797:1869–1876. doi: 10.1016/j.bbabio.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Riistama S, et al. Channelling of dioxygen into the respiratory enzyme. Biochim Biophys Acta. 1996;1275:1–4. doi: 10.1016/0005-2728(96)00040-0. [DOI] [PubMed] [Google Scholar]
- Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci U S A. 2011;108:10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- Sassera D, et al. Phylogenomic evidence for the presence of a flagellum and cbb(3) oxidase in the free-living mitochondrial ancestor. Mol Biol Evol. 2011;28:3285–3296. doi: 10.1093/molbev/msr159. [DOI] [PubMed] [Google Scholar]
- Schneider D, Schmidt CL. Multiple Rieske proteins in prokaryotes: where and why? Biochim Biophys Acta. 2005;1710:1–12. doi: 10.1016/j.bbabio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Schübbe S, et al. Complete genome sequence of the chemolithoautotrophic marine magnetotactic coccus strain MC-1. Appl Environ Microbiol. 2009;75:4835–4852. doi: 10.1128/AEM.02874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicheritz-Pontén T, Kurland CG, Andersson SG. A phylogenetic analysis of the cytochrome b and cytochrome c oxidase I genes supports an origin of mitochondria from within the Rickettsiaceae. Biochim Biophys Acta. 1998;1365:545–551. doi: 10.1016/s0005-2728(98)00099-1. [DOI] [PubMed] [Google Scholar]
- Sousa FL, et al. The superfamily of heme-copper oxygen reductases: types and evolutionary considerations. Biochim Biophys Acta. 2012;1817:629–637. doi: 10.1016/j.bbabio.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Takaya N. Response to hypoxia, reduction of electron acceptors, and subsequent survival by filamentous fungi. Biosci Biotechnol Biochem. 2009;73:1–8. doi: 10.1271/bbb.80487. [DOI] [PubMed] [Google Scholar]
- Tamas I, et al. Complete genome sequence of Beijerinckia indica subsp. indica. J Bacteriol. 2010;192:4532–4533. doi: 10.1128/JB.00656-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Brink F, Schoepp-Cothenet B, van Lis R, Nitschke W, Baymann F. Multiple Rieske/cytb complexes in a single organism. Biochim Biophys Acta. 2013;1827:1392–1406. doi: 10.1016/j.bbabio.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Thiergart T, Landan G, Schenk M, Dagan T, Martin WF. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol. 2012;4:466–485. doi: 10.1093/gbe/evs018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash JC, et al. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep. 2011;1:13. doi: 10.1038/srep00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C, et al. Endosymbiosis in statu nascendi: close phylogenetic relationship between obligately endosymbiotic and obligately free-living Polynucleobacter strains (Betaproteobacteria) Environ Microbiol. 2007;9:347–359. doi: 10.1111/j.1462-2920.2006.01144.x. [DOI] [PubMed] [Google Scholar]
- Vlaeminck SE, Hay AG, Maignien L, Verstraete W. In quest of the nitrogen oxidizing prokaryotes of the early Earth. Environ Microbiol. 2011;13:283–295. doi: 10.1111/j.1462-2920.2010.02345.x. [DOI] [PubMed] [Google Scholar]
- Watson SW, Mandel M. Comparison of the morphology and deoxyribonucleic acid composition of 27 strains of nitrifying bacteria. J Bacteriol. 1971;107:563–569. doi: 10.1128/jb.107.2.563-569.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP, Sobral BW, Dickerman AW. A robust species tree for the alphaproteobacteria. J Bacteriol. 2007;189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- Wrighton KC, et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science. 2012;337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- Yang JC, et al. The complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (shipworms) PLoS One. 2009;4:e6085. doi: 10.1371/journal.pone.0006085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.