Abstract
Uterine adenosarcomas (UA) are biphasic lesions composed of a malignant mesenchymal (i.e. stromal) component and an epithelial component. UAs are generally low-grade and have a favourable prognosis, but may display sarcomatous overgrowth (SO), which is associated with a worse outcome. We hypothesized that, akin to breast fibroepithelial lesions, UAs are mesenchymal neoplasms where clonal somatic genetic alterations are restricted to the mesenchymal component. To characterize the somatic genetic alterations in UAs and to test this hypothesis, we subjected 20 UAs to a combination of whole-exome (n=6), targeted capture (n=13) massively parallel sequencing (MPS) and/or RNA-sequencing (n=6). Only three genes, FGFR2, KMT2C and DICER1, were recurrently mutated, all in 2/19 cases; however, 26% (5/19) and 21% (4/19) of UAs harboured MDM2/CDK4/HMGA2 and TERT gene amplification, respectively, and two cases harboured fusion genes involving NCOA family members. Using a combination of laser capture microdissection and in situ techniques, we demonstrated that the somatic genetic alterations detected by MPS were restricted to the mesenchymal component. Furthermore, mitochondrial DNA sequencing of microdissected samples revealed that epithelial and mesenchymal components of UAs were clonally unrelated. In conclusion, here we provide evidence that UAs are genetically heterogeneous lesions and mesenchymal neoplasms.
Keywords: Uterine adenosarcoma, massively parallel sequencing, RNA-sequencing, FISH
INTRODUCTION
Uterine adenosarcomas (UAs) comprise a group of mixed epithelial-mesenchymal tumours that account for 5% of uterine sarcomas [1, 2]. UAs typically present as a polypoid mass and are characterized histologically by an admixture of low-grade malignant mesenchymal (i.e. stromal) and neoplastic but benign/atypical epithelial components, with a typical phyllodes tumour-like architecture [2, 3]. The mesenchymal component of UAs generally shows mild-to-moderate nuclear atypia, with variable mitotic rates, while a minority presents with heterologous elements such as immature cartilage or skeletal muscle [1–3]. Sex cord-like differentiation may be present within the mesenchymal component [3]. Most UAs are of low grade and low stage (FIGO stage I), and have low recurrence and mortality rates after surgery (15%–25%)[3, 4]. Approximately 25% of UAs, however, have sarcomatous overgrowth (SO), which is often associated with lymphovascular and/or deep myometrial invasion and high recurrence rates (up to 70%)[2–4].
The genomic features of UAs have yet to be fully characterized. Recently, Howitt et al.[5] employed targeted massively parallel sequencing (MPS) of 275 genes and identified mutations in PI3K/AKT/PTEN pathway members, ATRX and TP53 in 72%, 17% and 11% of UAs analyzed, respectively [5]. Copy number analysis (CNA) revealed recurrent amplifications affecting MDM2/CDK4/HMGA2 (28%) and MYBL1 (22%)[5].
Breast fibroepithelial tumours are biphasic neoplasms with epithelial and mesenchymal components [6], some of which (i.e. phyllodes tumours) bear histological resemblance to UAs. Breast fibroepithelial tumours have been shown to harbour somatic genetic alterations (i.e. MED12 mutations) exclusively in the mesenchymal component, suggesting that these tumours are primarily mesenchymal neoplasms [7, 8]. Furthermore, clonality analyses revealed that the mesenchymal component but not the epithelium is monoclonal in most cases [9].
We hypothesized that, akin to breast fibroepithelial lesions, UAs are mesenchymal neoplasms with clonally unrelated mesenchymal components and epithelium. Here we sought to characterize the landscape of somatic genetic alterations in UAs and to determine whether these alterations are restricted to the mesenchymal component or present in both mesenchymal and epithelial cells of these neoplasms, using a combination of whole-exome or targeted MPS, RNA-sequencing, in situ hybridization and laser capture approaches.
MATERIALS AND METHODS
Case selection
Seven frozen and 13 formalin-fixed paraffin-embedded (FFPE) UAs were retrieved from the pathology files of Memorial Sloan Kettering Cancer Center (MSKCC), New York, NY and the Cleveland Clinic, Cleveland, OH and centrally reviewed by a pathologist with expertise in gynecological pathology (RAS) following the WHO criteria [2](Supplementary Methods). Cases were graded following the criteria outlined in the Supplementary Methods. The study was approved by the Institutional Review Board (IRB) from the respective authors’ institutions. Consent was obtained according to the IRB-approved protocols.
Exome and targeted capture MPS
Tumour and germline DNA from six frozen UAs and from 13 FFPE UAs were subjected to whole-exome sequencing (WES)[10] and targeted capture MPS using the Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) platform [11], respectively (Table 1). WES and targeted MPS were performed at the MSKCC Integrated Genomics Operation (IGO) following validated protocols [10–12](Supplementary Methods).
Table 1.
Histological characteristics of the uterine adenosarcomas included in this study, and sequencing analyses performed
| Case ID | Grade | Heterologous element | Stromal overgrowth | Tissue type | Whole-exome MPS | Targeted capture MPS | RNA-Sequencing |
|---|---|---|---|---|---|---|---|
| AS1 | Low | Absent | Absent | Frozen | Yes | ||
| AS3 | High | Present (rhabdomyoblastic differentiation) | Present | Frozen | Yes | Yes | |
| AS4 | Low | Absent | Absent | Frozen | Yes | Yes | |
| AS5 | Intermediate | Absent | Absent | Frozen | Yes | Yes | |
| AS6 | Intermediate | Present (osseous differentiation) | Present | Frozen | Yes | ||
| AS7 | Low | Absent (sex cord-like features) | Present | Frozen | Yes | Yes | |
| AS8 | Intermediate | Absent (sex cord-like features) | Present | Frozen | Yes | Yes | |
| BAS03 | Low | Absent | Absent | FFPE | Yes | ||
| BAS04 | Low | Absent | Absent | FFPE | Yes | ||
| BAS06 | High | Absent | Present | FFPE | Yes | ||
| BAS07 | Low | Absent | Absent | FFPE | Yes | ||
| BAS09 | Intermediate | Absent | Present | FFPE | Yes | ||
| BAS10 | Low | Absent | Absent | FFPE | Yes | ||
| BAS12 | Low | Absent | Absent | FFPE | Yes | ||
| BAS16 | Low | Present (rhabdomyoblastic differentiation) | Absent | FFPE | Yes | ||
| BAS18 | Low | Absent | Absent | FFPE | Yes | ||
| BAS19 | Low | Absent | Absent | FFPE | Yes | ||
| BAS22 | Low | Absent | Absent | FFPE | Yes | ||
| BAS27 | Low | Absent | Absent | FFPE | Yes | ||
| BAS30 | Low | Absent | Absent | FFPE | Yes |
FFPE, formalin-fixed, paraffin-embedded; MPS, massively parallel sequencing.
Massively parallel RNA-sequencing
Six frozen UAs for which sufficient RNA was available were subjected to RNA-sequencing using a validated protocol employed at the MSKCC IGO [10](Supplementary Methods).
Additional molecular methods
Details of the reverse transcription PCR (RT-PCR)[12], fluorescence in situ hybridization (FISH) with NCOA2 and NCOA3 break-apart probes [13, 14], quantitative PCR (qPCR) for gene copy number analysis of TERT and MDM2, laser capture microdissection and mitochondrial DNA D-loop region (GAMDDL) analysis [15] are available in the Supplementary Methods.
RESULTS AND DISCUSSION
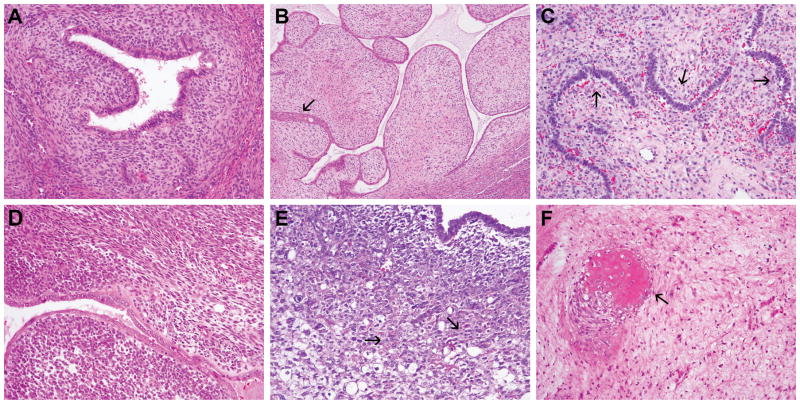
Histopathological review of the 20 UAs revealed that 14 (70%), four (20%) and two (10%) cases were of low-, intermediate- and high-grade, respectively (Table 1). All high-grade (2/2), 75% (3/4) of intermediate-grade and 7% (1/14) of low-grade UAs displayed SO. Three samples had heterologous elements, two displaying rhabdomyoblastic and one osseous differentiation, and two cases had sex cord-like features (Figure 1, Table 1).
Figure 1. Histological features of uterine adenosarcoma.
A, Low-grade uterine adenosarcoma (case BAS10); B, Low-grade uterine adenosarcoma with squamous differentiation (arrow; case BAS7); C, Low-grade uterine adenosarcoma with sex cord-like features (arrows; case AS7); D, Intermediate-grade uterine adenosarcoma with stromal overgrowth (case BAS9); E, High-grade uterine adenosarcoma with stromal overgrowth and heterologous rhabdomyoblastic differentiation (arrows; case AS3); F, Intermediate-grade uterine adenosarcoma with heterologous osseous differentiation (arrow; case AS6). Representative haematoxylin and eosin-stained sections are shown. Magnification A, C-F 100X, B 40X.
UAs are a molecularly heterogeneous group of tumours
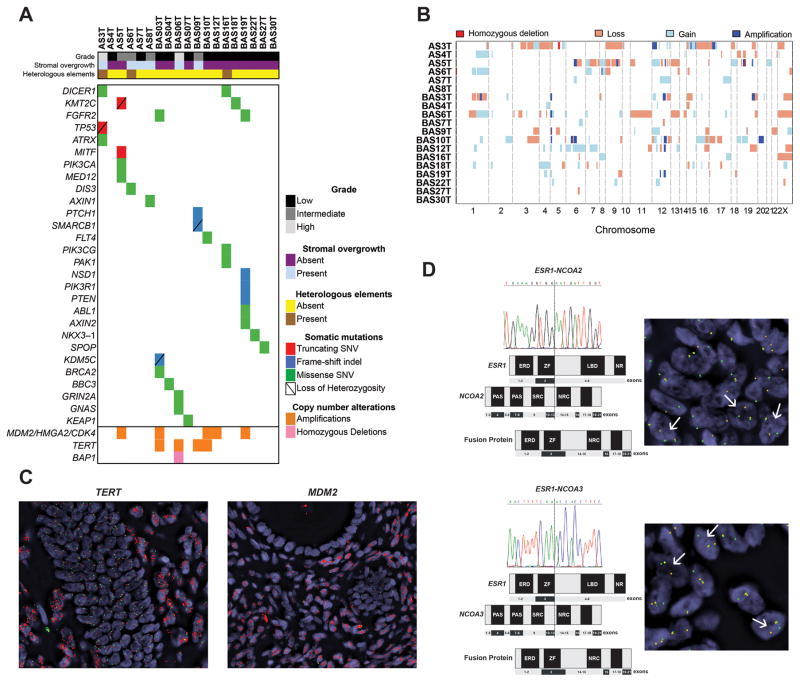
WES and MSK-IMPACT yielded a median depth of coverage of 246x (range 268–402x) and 453x (range 163–1624x), respectively (Supplementary Table S1). Somatic mutation analysis revealed that UAs are heterogeneous, with a median of 32.5 non-synonymous mutations (range 9–75) in the whole-exome, and a median of 1.5 non-synonymous mutations (range 0–4) in MSK-IMPACT (Supplementary Table S2). RNA-sequencing and Sanger sequencing resulted in the validation of 99.9% of the somatic mutations tested (Supplementary Figure S2, Supplementary Table S2). Only three genes, FGFR2, KMT2C and DICER1, were found to be recurrently mutated, all in 10.5% (2/19) of case (Figure 2A; Supplementary Table S2). Interestingly, both cases with DICER1 hotspot mutations displayed rhabdomyoblastic differentiation. Although these cases were re-reviewed by pathologists (RAS, JSR-F) blinded to the results of the sequencing analysis, who favoured a diagnosis of UA with rhabdomyoblastic differentiation, excluding a diagnosis of uterine rhabdomyosarcoma is remarkably challenging. Further studies to investigate whether the mesenchymal heterologous elements may be associated with, or underpinned by, specific somatic genetic alterations are warranted. No associations between SO, present in 6 cases, and specific genetic alterations were found. Likely pathogenic mutations in bona fide cancer genes such as a FGFR2 N549K hotspot mutation, a SMARCB1 mutation coupled with loss of heterozygosity (LOH) of the wild-type allele, and a TP53 truncating mutation coupled with LOH of the wild-type allele (Figure 2A, Supplementary Table S2) were identified. PI3K/AKT/mTOR pathway and ATRX mutations, which were previously reported in >70% and 17% of UAs[5], respectively, were found in only 26% and 5% of the cases studied here.
Figure 2. Somatic mutations, copy number alterations and expressed fusion genes in uterine adenosarcomas.
A, Non-synonymous somatic mutations, amplifications and homozygous deletions identified in the uterine adenosarcomas analyzed. Each column represents one sample; altered genes are reported in rows. Only the 341 genes present on our smallest targeted panel are included. Alteration types are colour-coded according to the legend. SNV, single nucleotide variant; Indel, small insertion/deletion. B, Copy number profiles of uterine adenosarcomas. Samples are represented in rows, chromosomes are represented along the x-axis. Dark red: homozygous deletion; orange: copy number loss; white: copy number neutral; light blue: copy number gain; dark blue: amplification. C, Representative micrographs of amplified TERT (left panel) and MDM2 (right panel) using fluorescent in situ hybridization (FISH). FISH analysis using two-colour probes for MDM2 and hTERT full-length sequence (red) and internal control (green); D, Schematic representation of ESR1-NCOA2 (upper panel) and ESR1-NCOA3 (lower panel) fusion genes in the index cases. Both rearrangements were validated using RT-PCR and FISH using break apart probes. FISH analysis using two-color break-apart probes for NCOA2 and NCOA3, with 5′ NCOA2 and NCOA3 green, 3′ NCOA2 and NCOA3 orange. Split signals are indicated by white arrows. ERD, estrogen receptor domain; LBD, ligand-binding domain; NR, nuclear receptor; NRC, nuclear receptor coactivator; SRC, steroid receptor coactivator; ZF, zinc finger.
CNA analysis revealed amplifications of MDM2/CDK4/HMGA2 (12q14.1-15) and MYBL1 (8q13.1) in 26% (5/19) and 5% (1/19) of UAs, respectively, in agreement with Howitt et al.[5]. In addition, we identified a previously unreported recurrent TERT (5p15.33) gene amplification in 21% (4/19) of UAs (Figures 2A–B). Amplification of TERT and MDM2/CDK4/HMGA2 were validated using qPCR and FISH (Figure 2C, Supplementary Figures S1, S3).
To determine whether a highly recurrent fusion gene would drive UAs, we subjected six UAs to RNA-sequencing. deFuse [16] and ChimeraScan [17] revealed a total of 26 and 46 in-frame fusion transcripts, respectively, including eight unique fusion transcripts identified by both algorithms (Supplementary Table S3). Fusion transcripts with known associated functions and/or with intact functional domains were prioritized for further investigation. Two expressed and RT-PCR-validated in-frame fusion transcripts involved the promoter region and the first three exons of ESR1 and the last ten exons of either NCOA2 or NCOA3. In both fusion genes, the nuclear receptor coactivator domains of NCOA2 and NCOA3 were preserved (Figure 2D). NCOA2 and NCOA3 gene rearrangements, which have been documented in other cancers [18, 19], were confirmed using break-apart FISH probes (Figure 2D, Supplementary Table S4). These fusion genes may provide a functional link between the pathogenesis of UAs and oestrogen signaling; it should be noted, however, that these fusion genes were restricted to their respective index cases.
These findings demonstrate that UAs are molecularly heterogeneous and unlikely to be driven by a highly recurrent expressed in-frame fusion gene. Their repertoire of somatic genetic alterations includes recurrent amplifications of MDM2/CDK4/HMGA2 and TERT and few recurrently mutated genes, which may vary according to the histological features of the sarcomatous component.
The mesenchymal but not the epithelial components of UAs harbour clonal somatic genetic alterations
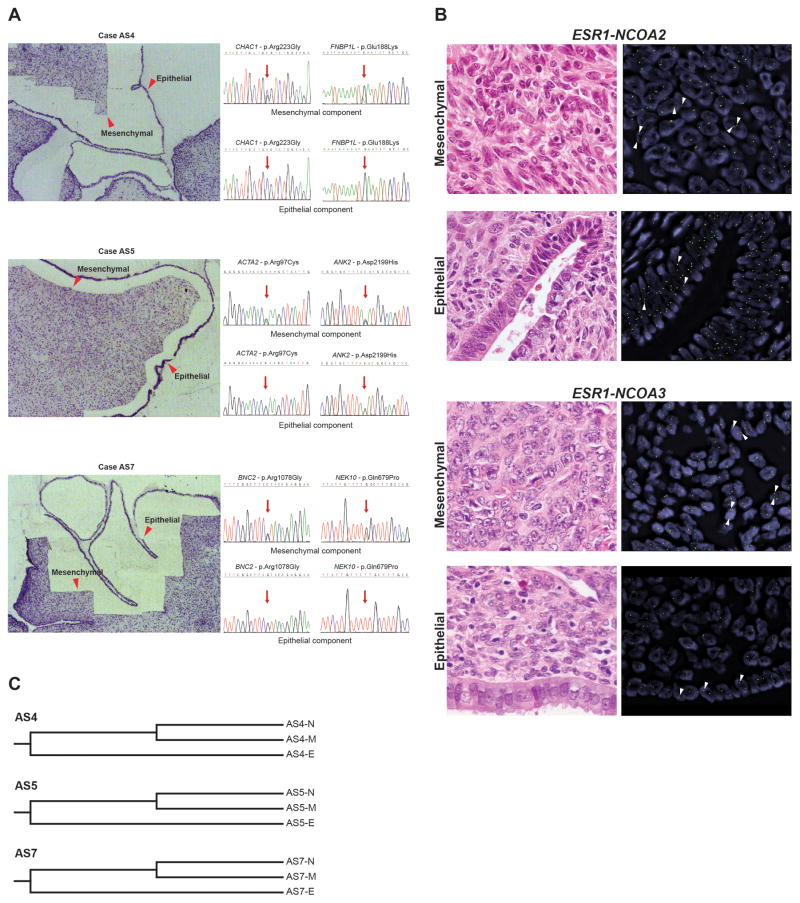
Given that UAs are classified by the WHO as mixed epithelial-mesenchymal tumours [2], we hypothesized that, similar to breast fibroepithelial tumours, the somatic alterations would be restricted to the mesenchymal component and absent in the epithelium. To test this hypothesis, three samples harbouring well-defined mesenchymal and epithelial components were subjected to laser capture microdissection. DNA was extracted from each component separately (Figure 3A). Two truncal somatic mutations identified by MPS in each case were selected and tested in the different components using Sanger sequencing, all of which were found only in the respective mesenchymal component (Figure 3A). Consistent with these findings, FISH analyses demonstrated that TERT and MDM2 amplifications, as well as NCOA2 and NCOA3 rearrangements were found exclusively in the mesenchymal component of cases harbouring these alterations (Figures 2C, 3B). We next sought to define whether the two components were clonally-related by means of GAMDDL sequencing. This analysis revealed that sequences of the mitochondrial-DNA D-loop regions isolated from the epithelial elements consistently differed from the sequences obtained from the mesenchymal component (Figure 3C), supporting the contention that the two components are unlikely to be clonally-related. Our findings provide evidence that UAs are mesenchymal neoplasms and that the epithelium is independent and clonally unrelated to the mesenchymal component. The presence of the non-neoplastic epithelium may be the result of the almost invariable polypoid growth of UAs. One could hypothesize that the neoplastic sarcomatous component arises within the endometrial stromal compartment, and as the stromal component grows it may eventually result in entrapment of non-neoplastic endometrium. With tumour progression, interactions with the mesenchymal neoplastic component may promote the expansion of the non-neoplastic endometrium, and the non-neoplastic endometrium, itself, may also elicit factors that stabilize the relationship between the epithelium and the neoplastic mesenchymal component or even support the growth of the latter.
Figure 3. The presence of somatic genetic alterations in the mesenchymal component but not in the epithelial component of uterine adenosarcomas.
A, Representative micrographs of negative microdissection, and the associated Sanger sequencing traces for selected mutations in the separately microdissected mesenchymal and epithelial components of three uterine adenosarcomas. The regions from which DNA was extracted are indicated by red arrows. Note that the mutations were restricted to the mesenchymal components in each case. B, Representative micrographs (haematoxylin and eosin left, fluorescence in situ hybridization (FISH), right) of the mesenchymal and epithelial components of uterine adenosarcomas AS8 (top) and AS4 (bottom). The presence or absence of the rearrangements using break-apart probes are indicated by white arrows. FISH analysis using two-colour break-apart probes for NCOA2 and NCOA3, with 5′ NCOA2 and NCOA3 green, 3′ NCOA2 and NCOA3 orange. Note that NCOA rearrangements are restricted to the mesenchymal components in both cases. C, Phylogenetic clustering of mitochondrial DNA D-loop regions in mesenchymal and epithelial components, and matched normal tissue of 3 cases (AS4, AS5 and AS7). The relative phylogenetic distances between matched normal tissue (N), mesenchymal component (M) and epithelial component (E) were determined using the neighbour-joining method.
UAs and carcinosarcomas display distinct landscapes of somatic mutations
The mixed epithelial-mesenchymal tumours defined by the WHO include adenofibromas, UAs and carcinosarcomas. A comparison of the repertoire of somatic genetic alterations found in carcinosarcomas [20] and UAs revealed that nearly all recurrently mutated genes in carcinosarcomas were found not to be affected by mutations in this cohort of UAs (Supplementary Table S5). Furthermore, TP53 (73%), PIK3CA (41%) and PIK3R1 (41%), the most frequently mutated genes in carcinosarcomas [20], were significantly less frequently mutated in UAs (5%, 5% and 5%, respectively, p<0.05, Fisher’s exact test). These findings are consistent with the notion that UAs and carcinosarcomas are unlikely to be closely related lesions.
Our study, albeit limited by its relatively small sample size, demonstrates that UAs are a genetically heterogeneous group of lesions, where clonal genetic alterations are found in the sarcomatous but not in the epithelial components, supporting the contention that UAs are mesenchymal neoplasms.
Supplementary Material
A, Copy number profiles of the UAs with TERT and MDM2 amplification. In the genome plots, smoothed log2 ratios (y-axis) were plotted according to their genomic positions (x-axis); B, Copy number quantitative PCR results of the UAs with/without TERT and/or MDM2 amplification.
Sanger sequencing traces of selected mutations identified in uterine adenosarcomas. Mutations are highlighted with a black arrow.
Representative FISH micrographs of A, case BAS03 (TERT); B, case BAS06 (TERT); C, case BAS10 (TERT); D, case BAS03 (MDM2); E, case BAS10 (MDM2); F, case BAS12 (MDM2); and G, case BAS12 (MDM2). FISH analysis was performed using two-colour probes for MDM2 and hTERT full-length sequence (red) and internal control (green).
Whole-exome and targeted capture massively parallel sequencing statistics.
Somatic single nucleotide variants (SNVs) and insertion/deletions (indels) identified by whole-exome and targeted capture massively parallel sequencing in the uterine adenosarcomas studied.
Expressed fusion transcripts identified in uterine adenosarcomas by RNA-sequencing.
Overview of uterine adenosarcomas subjected to fluorescence in situ hybridization (FISH) of NCOA2, NCOA3, TERT and MDM2, to ESR1-NCOA2 and ESR1-NCOA3 RT-PCR, and to TERT and MDM2 copy number alteration quantitative real-time PCR (CNA qPCR) analysis.
Comparisons of the mutational frequencies of the 341 genes included in our targeted capture panel between gynaecological carcinosarcomas reported by Jones et al. and uterine adenosarcomas included in this study.
List of primers used for the validation of mutations by Sanger sequencing, for the validation of selected fusion genes by RT-PCR, and for the mitochondrial DNA D-loop region (GAMDDL) assay.
Acknowledgments
SP is funded by a Susan G Komen Postdoctoral Fellowship Grant (PDF14298348), AMS by a stipend from the German Cancer Aid (Dr. Mildred Scheel Stiftung) and GSM by CAPES (#BEX 5714/14-1). Research reported in this publication was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (Grant No. P30CA008748). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest: The authors have no conflicts of interest to declare.
AUTHORS CONTRIBUTIONS
BW, RAS, BPR and JSR-F conceived and supervised the study. DAL, BPR and RAS provided the samples; RAS performed the histological review. JSR-F, ADP and AMS performed the sample microdissection; KAB, CKYN, MRDF and IdB performed the bioinformatic analysis; SP, GSM, LGM and RAI carried out experiments and analyzed data. SP, CKYN and FCG wrote the first draft that was revised by JSR-F and BW. All authors reviewed and approved the final version of the manuscript.
References
Some references are cited only in Supplementary Information online
- 1.Clement PB, Young RH. Atlas of Gynecologic Surgical Pathology. 3. Saunders/Elsevier; 2014. Mesenchymal and mixed epithelial-mesenchymal tumors of the uterine corpus and cervix; pp. 218–270. [Google Scholar]
- 2.Wells M, Oliva E, Palacios J, et al. Mixed epithelial and mesenchymal tumours. In: Kurman RJ, Carcangiu ML, Herrington CS, Young RH, editors. WHO classification of tumours of female reproductive organs. IARC Press; Lyon: 2014. pp. 148–151. [Google Scholar]
- 3.McCluggage WG. Mullerian adenosarcoma of the female genital tract. Adv Anat Pathol. 2010;17:122–129. doi: 10.1097/PAP.0b013e3181cfe732. [DOI] [PubMed] [Google Scholar]
- 4.Carroll A, Ramirez PT, Westin SN, et al. Uterine adenosarcoma: an analysis on management, outcomes, and risk factors for recurrence. Gynecol Oncol. 2014;135:455–461. doi: 10.1016/j.ygyno.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howitt BE, Sholl LM, Dal Cin P, et al. Targeted genomic analysis of Mullerian adenosarcoma. J Pathol. 2015;235:37–49. doi: 10.1002/path.4442. [DOI] [PubMed] [Google Scholar]
- 6.Tan PH, Tse G, Lee A, et al. Fibroepithelial tumours. In: Lakhani SR, Ellis IO, Schnitt SJ, Tan BH, van de Vijver MJ, editors. WHO Classification of Tumours of the Breast. 4. IARC press; Lyon: 2012. pp. 141–147. [Google Scholar]
- 7.Lim WK, Ong CK, Tan J, et al. Exome sequencing identifies highly recurrent MED12 somatic mutations in breast fibroadenoma. Nat Genet. 2014;46:877–880. doi: 10.1038/ng.3037. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida M, Sekine S, Ogawa R, et al. Frequent MED12 mutations in phyllodes tumours of the breast. Br J Cancer. 2015;112:1703–1708. doi: 10.1038/bjc.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuijper A, Buerger H, Simon R, et al. Analysis of the progression of fibroepithelial tumours of the breast by PCR-based clonality assay. J Pathol. 2002;197:575–581. doi: 10.1002/path.1161. [DOI] [PubMed] [Google Scholar]
- 10.Weinreb I, Piscuoglio S, Martelotto LG, et al. Hotspot activating PRKD1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet. 2014;46:1166–1169. doi: 10.1038/ng.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Natrajan R, Wilkerson PM, Marchio C, et al. Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast. J Pathol. 2014;232:553–565. doi: 10.1002/path.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickson MA, Tap WD, Keohan ML, et al. Phase II trial of the CDK4 inhibitor PD0332991 in patients with advanced CDK4-amplified well-differentiated or dedifferentiated liposarcoma. J Clin Oncol. 2013;31:2024–2028. doi: 10.1200/JCO.2012.46.5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho AS, Kannan K, Roy DM, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;45:791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda S, Kadowaki T, Kumaki N, et al. Analysis of gene alterations of mitochondrial DNA D-loop regions to determine breast cancer clonality. Br J Cancer. 2012;107:2016–2023. doi: 10.1038/bjc.2012.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McPherson A, Hormozdiari F, Zayed A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics (Oxford, England) 2011;27:2903–2904. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosquera JM, Sboner A, Zhang L, et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52:538–550. doi: 10.1002/gcc.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteyries S, Perot C, Adelaide J, et al. NCOA3, a new fusion partner for MOZ/MYST3 in M5 acute myeloid leukemia. Leukemia. 2008;22:663–665. doi: 10.1038/sj.leu.2404930. [DOI] [PubMed] [Google Scholar]
- 20.Jones S, Stransky N, McCord CL, et al. Genomic analyses of gynaecologic carcinosarcomas reveal frequent mutations in chromatin remodelling genes. Nat Commun. 2014;5:5006. doi: 10.1038/ncomms6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martelotto LG, De Filippo MR, Ng CK, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol. 2015;237:179–189. doi: 10.1002/path.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerini-Rocco E, Hodi Z, Piscuoglio S, et al. The repertoire of somatic genetic alterations of acinic cell carcinomas of the breast: an exploratory, hypothesis-generating study. J Pathol. 2015;237:166–178. doi: 10.1002/path.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saunders CT, Wong WS, Swamy S, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 28.De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz JM, Rodelsperger C, Schuelke M, et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 30.Carter H, Chen S, Isik L, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martelotto LG, Ng C, De Filippo MR, et al. Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome Biol. 2014;15:484. doi: 10.1186/s13059-014-0484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronglai S, Seshan V. FACETS: Fraction and allele-specific copy number estimates from tumor sequencing. Memorial Sloan-Kettering Cancer Center, Dept of Epidemiology & Biostatistics Working Paper Series. 2015 [Google Scholar]
- 39.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohsaka S, Shukla N, Ameur N, et al. A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat Genet. 2014;46:595–600. doi: 10.1038/ng.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shugay M, Ortiz de Mendibil I, Vizmanos JL, et al. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013;29:2539–2546. doi: 10.1093/bioinformatics/btt445. [DOI] [PubMed] [Google Scholar]
- 43.Piscuoglio S, Ng CK, Martelotto LG, et al. Integrative genomic and transcriptomic characterization of papillary carcinomas of the breast. Mol Oncol. 2014;8:1588–1602. doi: 10.1016/j.molonc.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Dudley J, Nei M, et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 46.Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 47.Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 2010;4:192–208. doi: 10.1016/j.molonc.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6:718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- 49.Wetterskog D, Lopez-Garcia MA, Lambros MB, et al. Adenoid cystic carcinomas constitute a genomically distinct subgroup of triple-negative and basal-like breast cancers. J Pathol. 2012;226:84–96. doi: 10.1002/path.2974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, Copy number profiles of the UAs with TERT and MDM2 amplification. In the genome plots, smoothed log2 ratios (y-axis) were plotted according to their genomic positions (x-axis); B, Copy number quantitative PCR results of the UAs with/without TERT and/or MDM2 amplification.
Sanger sequencing traces of selected mutations identified in uterine adenosarcomas. Mutations are highlighted with a black arrow.
Representative FISH micrographs of A, case BAS03 (TERT); B, case BAS06 (TERT); C, case BAS10 (TERT); D, case BAS03 (MDM2); E, case BAS10 (MDM2); F, case BAS12 (MDM2); and G, case BAS12 (MDM2). FISH analysis was performed using two-colour probes for MDM2 and hTERT full-length sequence (red) and internal control (green).
Whole-exome and targeted capture massively parallel sequencing statistics.
Somatic single nucleotide variants (SNVs) and insertion/deletions (indels) identified by whole-exome and targeted capture massively parallel sequencing in the uterine adenosarcomas studied.
Expressed fusion transcripts identified in uterine adenosarcomas by RNA-sequencing.
Overview of uterine adenosarcomas subjected to fluorescence in situ hybridization (FISH) of NCOA2, NCOA3, TERT and MDM2, to ESR1-NCOA2 and ESR1-NCOA3 RT-PCR, and to TERT and MDM2 copy number alteration quantitative real-time PCR (CNA qPCR) analysis.
Comparisons of the mutational frequencies of the 341 genes included in our targeted capture panel between gynaecological carcinosarcomas reported by Jones et al. and uterine adenosarcomas included in this study.
List of primers used for the validation of mutations by Sanger sequencing, for the validation of selected fusion genes by RT-PCR, and for the mitochondrial DNA D-loop region (GAMDDL) assay.