Somech and colleagues identify two new mutations in STN1 that causes Coats plus syndrome and telomere abnormalities in human, recapitulated in a zebra fish model.
Abstract
The analysis of individuals with telomere defects may shed light on the delicate interplay of factors controlling genome stability, premature aging, and cancer. We herein describe two Coats plus patients with telomere and genomic defects; both harbor distinct, novel mutations in STN1, a member of the human CTC1–STN1–TEN1 (CST) complex, thus linking this gene for the first time to a human telomeropathy. We characterized the patients’ phenotype, recapitulated it in a zebrafish model and rescued cellular and clinical aspects by the ectopic expression of wild-type STN1 or by thalidomide treatment. Interestingly, a significant lengthy control of the gastrointestinal bleeding in one of our patients was achieved by thalidomide treatment, exemplifying a successful bed-to-bench-and-back approach.
Telomeres are nucleoprotein complexes essential for protecting the chromosome ends from degradation, activation of the DNA damage response, and end-to-end fusion (de Lange, 2009; Jain and Cooper, 2010). Abnormally short or otherwise deprotected telomeres compromise replicative potential and genome stability (Bodnar et al., 1998). Accelerated telomere attrition is associated with several inherited and noninherited human conditions termed telomeropathies (Armanios and Blackburn, 2012; Holohan et al., 2014; Townsley et al., 2014). In addition to the well described shelterin complex (Diotti and Loayza, 2011), the human CST (CTC1–STN1–TEN1) complex plays multiple roles in telomere C-strand synthesis (Chen and Lingner, 2013), as well as genome-wide replication and the recovery from replication stress (Stewart et al., 2012). This conserved trimeric complex binds the telomeric G-strand, promotes the synthesis of the C-strand, and inhibits telomerase-mediated telomere elongation (Miyake et al., 2009; Chen et al., 2012; Stewart et al., 2012; Wang et al., 2012). Disruption of the CST complex in human and mouse cells affects telomere length and telomeric C-strand synthesis, leading to elongated 3′ overhangs (Gu et al., 2012; Stewart et al., 2012; Wang et al., 2012). The importance of the CST complex is evidenced also by the occurrence of Coats plus syndrome in individuals affected by bi-allelic CTC1 mutations (Anderson et al., 2012; Keller et al., 2012; Polvi et al., 2012). Such patients present with cranial calcifications, bone marrow failure, severe anemia, premature hair graying, osteoporosis, exudative retinopathy, and retinal telangiectasia, intestinal ectasia, and liver fibrosis (Anderson et al., 2012; Keller et al., 2012; Polvi et al., 2012; Walne et al., 2013).
To date, germline mutations in either STN1 (also designated OBFC1; Wan et al., 2009) or TEN1 have not been described. Here, we describe two patients with Coats plus syndrome harboring distinct novel mutations in STN1. We characterized their clinical phenotypes, telomeric defects, and genomic replication abnormalities, rescued their cellular phenotypes and recapitulated the disease in a zebrafish model.
RESULTS AND DISCUSSION
Clinical studies
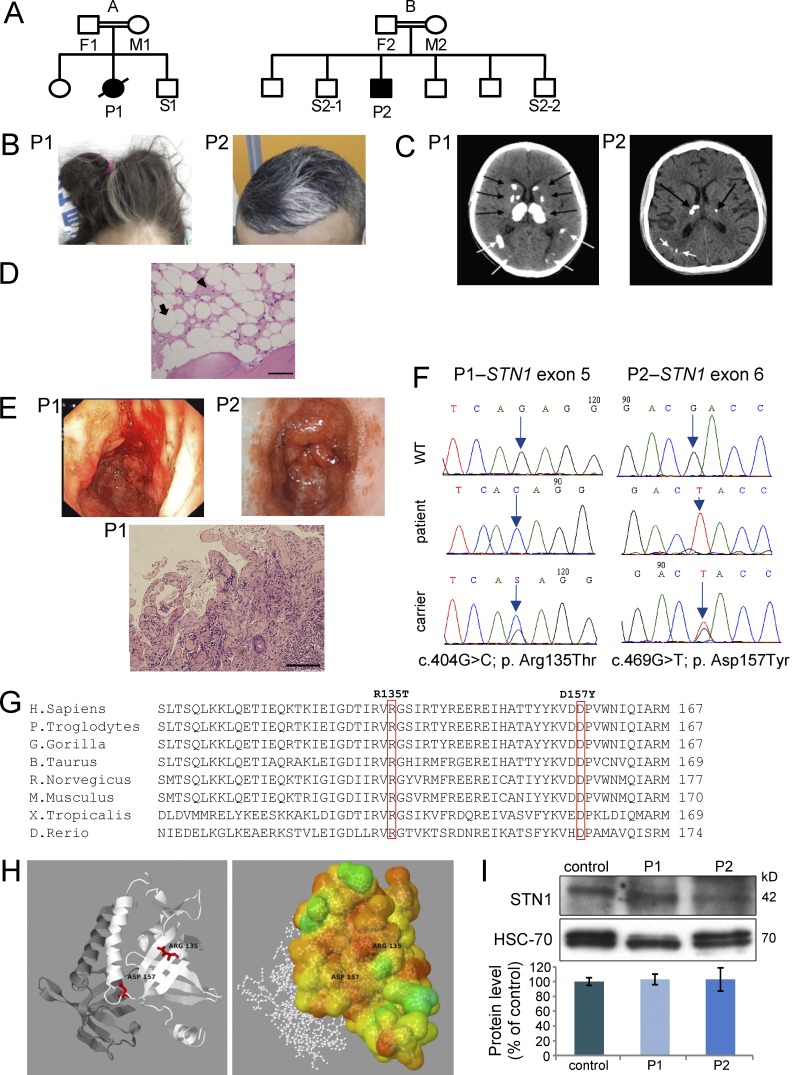
We studied two unrelated patients (Fig. 1 A), born to consanguineous Palestinian parents who presented to our hospital at 12 (P1, female) and 19 (P2, male) years of age (Table S1). Both patients had intrauterine growth retardation and, later in life, presented with premature aging symptoms, including poor growth, graying hair (Fig. 1 B), liver fibrosis, portal hypertension, esophageal varices, brain calcifications, white matter changes (Fig. 1 C), osteopenia, pancytopenia, and hypocellular bone marrow (<5% of normal; Fig. 1 D). Recurrent gastrointestinal hemorrhages caused by gastric antral vascular ectasia and telangiectasia were evident in both patients (Fig. 1 E).
Figure 1.
Genetics and clinical phenotypes of STN1-mutated patients. (A) Family pedigrees of the affected families (A and B). Solid symbols represent the affected subjects P1 and P2, and open symbols represent unaffected relatives. Squares indicate male subjects, and circles female subjects. A slash indicates a deceased subject. (B) Graying hair. (C) Multiple calcifications (arrows) in brain computerized tomography (CT). (D) Bone marrow trephine biopsy of patient P2. The marrow spaces consist primarily of adipose tissue (white areas; example marked with an arrow) and edematous stroma (pink areas; example marked with an arrowhead). Bar, 20 µm. (E) Endoscopy images are noted in both patients (top). Intestine biopsy of patient P1 (bottom). Images are magnified x600. Size bar – Bar, 20 µm. (F) Dideoxy Sanger sequencing was performed to the different STN1 genotypes detected in the studied pedigrees. (G) Alignment analysis using the NCBI HomoloGene tool was performed. The mutated residues R135 (P1) and D157 (P2) are boxed. The aligned vertebrate orthologues from top to bottom are: human, chimpanzee, gorilla, cow, rat, mouse, frog, and zebrafish. (H) Crystal structures of wild-type hSTN1 in complex with TEN1, but without hCTC1 were available in the PDB (4joi; Bryan et al., 2013). (left) A schematic cartoon view of the STN1 (white) and TEN1 (gray) complex with the position of the two mutations highlighted in red. (right) STN1 molecule is colored according to conservation scores of Consurf (Ashkenazy et al., 2010). 2D images were created based on PDBsum (Laskowski, 2001) images. 3D images were created using Jmol. (I) Western blot analysis to STN1 and to heat shock cognate protein 70 (HSC-70) was performed on PBL samples from the patients and healthy control, followed by densitometry using the ImageJ software. STN1 protein level in the control sample was set to 100%. Data represents three repeated analyses.
In addition, P1 had neurological symptoms, which included spasticity, dystonia, and ataxia, and P2 had bilateral retinal telangiectatic changes. P1 succumbed at the age of 16 yr as a result of massive gastrointestinal bleeding, despite multiple treatments. In contrast, gastrointestinal bleeding in P2 was successfully managed with thalidomide treatment (100 mg daily), along with argon plasma coagulation. This patient became free of GI bleeding after initiation of thalidomide treatment. Blood transfusions were required only as a result of his bone marrow failure.
Genetic evaluation
In both patients, consanguinity suggested autosomal recessive inheritance and the clinical constellation was suggestive of Coats plus syndrome (Holohan et al., 2014). Nevertheless, direct dideoxy-sequencing of the CTC1 coding region was normal. Whole-exome sequencing performed for patients 1 and 2, yielded 1,143 and 1,076 homozygous variants that affect protein sequences, respectively. This list of variants was subsequently reduced to 67 and 53 (Table S2) rare variants for patients 1 and 2, respectively, by filtering out variants present in ≥0.01 of our in-house exomes (n = 250) and variants present with a minor allele frequency ≥0.01 in either the 1000 Genomes Project (1KG), dbSNP 135 database, or the National Heart, Lung, and Blood Institute Exome Sequencing Project (ESP). Recessive analysis reduced the variants list of patient 1 to 12 candidate genes (Table S2). The only gene common to both lists was STN1, with a distinct homozygous mutation in each patient. STN1 was pursued because of its telomeric role and interaction with CTC1 within the CST complex (Miyake et al., 2009; Bryan et al., 2013). Dideoxy Sanger sequencing (Fig. 1 F) confirmed the presence of a different missense mutation in each patient (P1: c.404G>C; p. Arg135Thr, AGA to ACA; P2, c.469G>T; p. Asp157Tyr, GAC to TAC) in exons 5 and 6, respectively. Several observations support the pathogenic nature of these STN1 mutations. First, mutation analysis in both families suggested a possible segregation with the disease. Second, they were absent from genome databases, as well as in 160 ethnically matched control alleles (unpublished data). Third, the mutations occurred in evolutionary highly conserved positions in vertebrate species (Fig. 1 G). Fourth, mapping of the mutations on the complete three-dimensional structure of wild-type hSTN1 suggested that they destabilize the protein structure (Fig. 1 H and Table S3). Protein extracts from peripheral blood lymphocytes (PBLs) of both patients demonstrated marginal differences in the level of STN1 protein compared with control (Fig. 1 I).
Abnormal cellular phenotypes of Stn1 mutant patients' cells
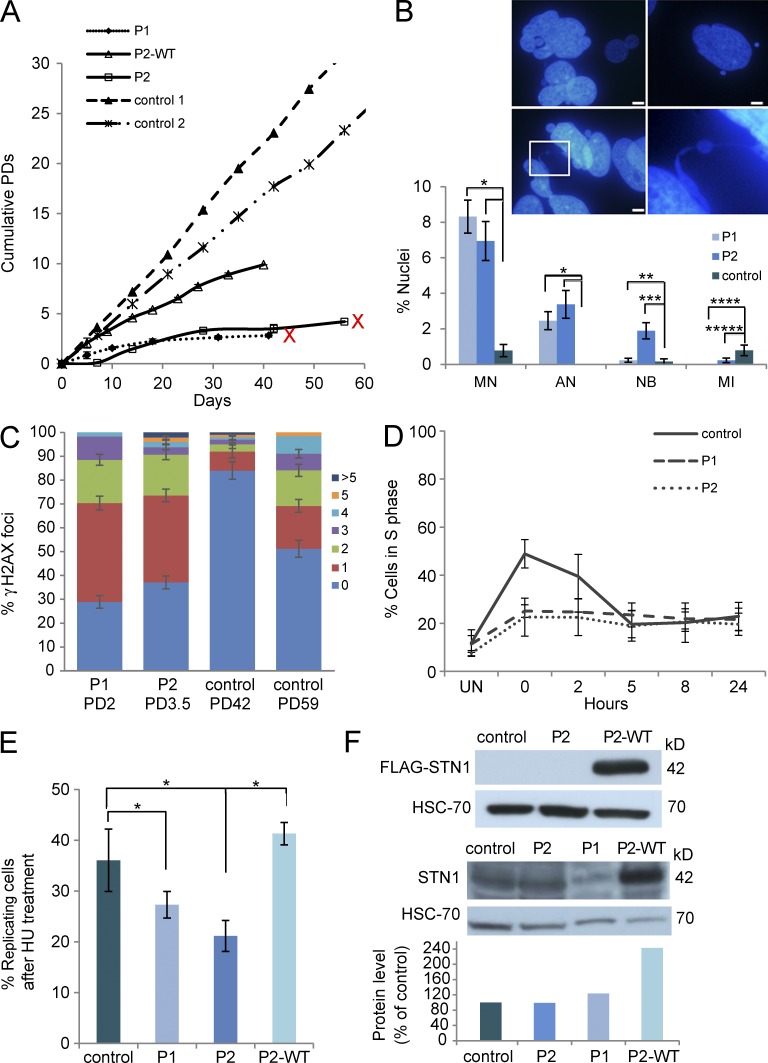
Primary fibroblasts of both patients were abnormally large, contained cytoplasmic vacuoles and extended podia (not depicted), grew poorly in culture, and ceased to proliferate at a very low population doubling (PD; P1 at PD 2.8 and P2 at PD 4.2; Fig. 2 A). Control samples reached senescence at PDs 68.7 and 65.81, after 147 and 126 d in culture, respectively (unpublished data). DAPI staining of both patients' fibroblasts revealed apoptotic nuclei, high rates of micronuclei, intact or torn DNA bridges between nuclei (Fig. 2 B), and γ-H2AX foci at levels comparable to control normal fibroblasts at senescence (Fig. 2 C). These findings support growth arrest caused by premature cellular senescence linked to STN1 loss-of-function, as previously described (Huang et al., 2012; Boccardi et al., 2015). Interestingly, SA-β-Gal staining was not found to be prominent (unpublished data). Apoptosis caused by STN1 depletion has also previously been demonstrated in STN1 knockdown cells (Dai et al., 2010). Together, our findings suggest that both cellular senescence and apoptosis are instigated by STN1 loss-of-function. Cell cycle analysis showed compromised ability to release cells from S-phase after replication stress in both patients compared with control cells (Fig. 2 D and Table S4). Consistent with this observation and similar to previous findings (Stewart et al., 2012), EdU uptake after hydroxyurea (HU) treatment, was significantly lower in both patients compared with control (76% [P1] and 56.7% [P2] of control, Fig. 2 E). These findings support a marked attenuation in the recovery from replication fork stalling induced by HU treatment in STN1 mutant fibroblasts. Both the poor cell growth and the lower EdU uptake were partially or fully rescued, respectively, by WT STN1 overexpression (Fig. 2 F), in P2 fibroblasts (Fig. 2, A and E). Collectively, these results indicate that the patient cells display replication aberrancies caused by the STN1 mutations.
Figure 2.
Mutations in human STN1 result in abnormal cellular phenotypes. (A) Fibroblasts from two healthy controls, the patients P1 and P2, and P2 cells expressing WT STN1 were grown, and cumulative PDs were calculated at each time point. P1 and P2 samples that reached senescence are marked by a red X at the end of the growth curve. P2 fibroblasts expressing WT-STN1 continued to grow without reaching growth arrest as long as they were kept in culture. Data are representative of three independent analyses. (B) Representative nuclei of P1 fibroblasts displaying apoptotisis (top left), micronuclei (top right), and nuclear bridges (bottom). (bottom right) Enlargement of the designated region in the bottom left panel. Bars, 5 µm. The frequencies of micronuclei (MN; *, P < 0.0001), apoptotic nuclei (AN; *, P < 0.0001), nuclear bridges (NB; **, P = 0.7; ***, P = 0.0003), and the mitotic index (MI; ****, P = 0.007; *****, P = 0.08) in P1 and P2 patient fibroblasts and in normal presenescent fibroblasts, appear in the graph below the images. Data represents two to four independent analyses of at least 200 cells per analysis. Statistical analysis was performed by a two-tailed Student’s t test. (C) STN1-mutated fibroblasts P1 (PD 2) and P2 (PD 3.5), and control fibroblasts at two PDs (PD 42 and PD 59) were stained with an antibody for γ-H2AX, and then scored for the number of γ-H2AX foci per cell. The number of foci per cell is depicted by the color code to the right and displayed in the graph as the percentage of total scored cells. At least 100 nuclei were scored for each sample per each independent analysis. Data are representative of two independent repeats for all samples with the exception of the control at PD42, whose analysis was performed once. (D) Patients and control fibroblasts were either untreated (UN) or treated with HU for 48 h, subsequently harvested at 0, 2, 5, 8, and 24 h, and subjected to cell cycle analysis. The percentage of cells in S phase is presented in the graph. Data are representative of three independent experiments. (E) Patients and control fibroblasts were treated with 0.5 mM HU for 48 h, followed by 1-h incubation with 50 µM EdU. EdU incorporation into newly synthesized DNA strands was assayed using the Click-iT EdU Alexa Fluor 488 Imaging kit. The percent of EdU-positive (replicating) cells in control, P1, P2, and P2+WT STN1 (P2-WT) is presented in the graph and represents three independent experiments (*, P < 0.001, two-tailed Student’s t test). (F, top) Western blot analysis of ectopic WT-STN1 expression in P2 fibroblasts, using an anti-FLAG tag antibody. HSC-70 serves as a control for protein loading. The blot represents four independent experiments. (bottom) Western blot analysis of native STN1 expression in control and patients' fibroblasts, and ectopic expression of WT-STN1 in P2 fibroblasts, using anti-STN1 antibodies. HSC-70 serves as a control for protein loading. The protein level in the control sample was set as 100%. The STN1 protein levels in the patient fibroblasts and in P2 fibroblasts ectopically expressing WT-STN1 were compared with the control level. Densitometry was done using the ImageJ software. The presented blot is one of three repetitive independent experiments.
Abnormal telomeric phenotypes of STN1 mutant patients' cells
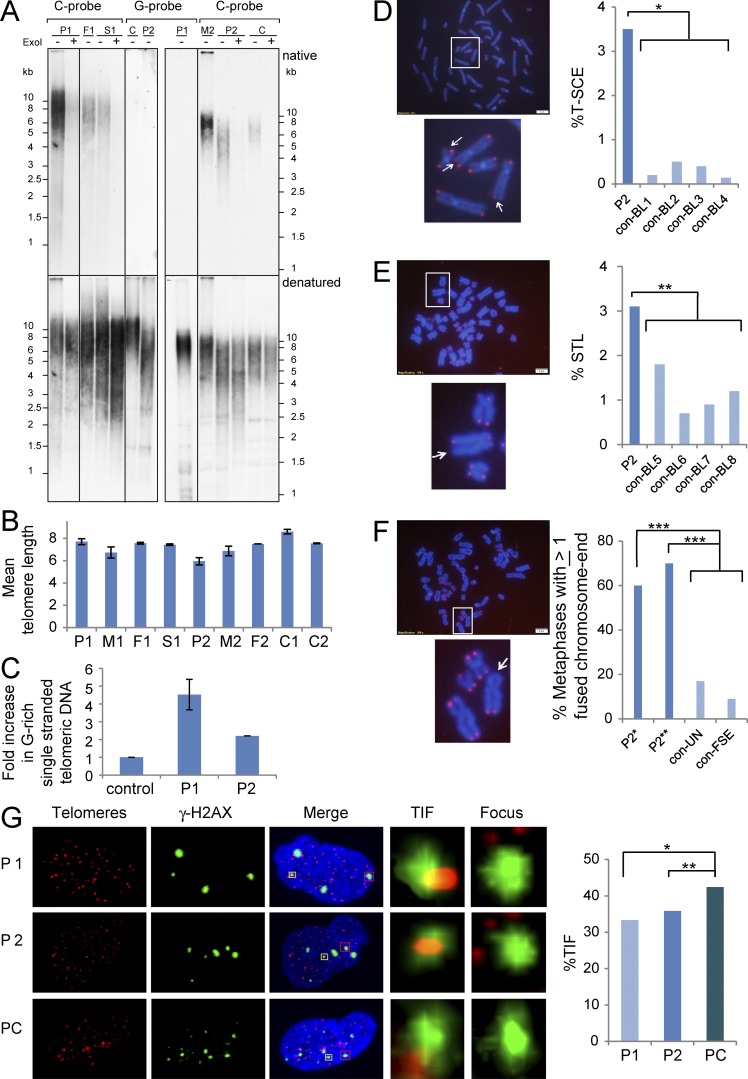
Telomere length of patients’ PBLs was examined by telomere restriction fragment analysis (Fig. 3, A and B), and whereas P1’s telomeres displayed normal length (mean length, 7.7 kb), P2’s telomeres were significantly shorter than expected when compared with control (C2) and his mother (M2), although not severely short (mean length, 5.5 kb; P = 0.031 and 0.018, respectively, by one-tailed Student’s t test). Consistent with the short telomeres detected by Southern analysis, P2 PBLs displayed a significantly higher rate of telomere loss by FISH (Fig. 3 E). CTC1-deficient patients display variable telomere length, thus excessive telomere shortening is not considered a hallmark of Coats plus (Anderson et al., 2012; Keller et al., 2012; Polvi et al., 2012; Walne et al., 2013). STN1 has been shown to interact with polymerase α, and thereby mediate the recruitment/stimulation of polα/primase by CST to synthesize the telomeric C-strand (Gu and Chang, 2013). Because compromised C-strand fill-in synthesis at the telomere ends is expected to result in the elongation of the telomeric G-rich overhang, we examined the G-overhang by in-gel hybridization of a C-rich telomeric probe to native DNA. PBLs from both patients displayed dramatically increased amounts of single-stranded G-rich telomeric DNA when normalized to noncarrier controls (4.5- and 2.2-fold increase for P1 and P2, P values of 0.019 and 0.057 by one-tail Student’s t test, respectively; Fig. 3, A and C).
Figure 3.
Mutations in STN1 result in abnormal telomere phenotypes. (A) DNA samples, prepared from PBLs of patient P1, her heterozygous father (F1), and a noncarrier sibling (S1) and patient P2, his heterozygous mother (M2), and two independent control samples (C), were analyzed by in-gel hybridization. Duplicated lanes were electrophoresed in the same gel, and then separated and hybridized to a G-rich or C-rich telomeric probe, as indicated above the panels. After native hybridization to detect single-stranded telomeric DNA (top), the gels were denatured and rehybridized with the same probes to detect the overall duplex telomeric DNA (bottom). Treatment with exonuclease I is indicated above the lanes. (B) Graphic illustration of the mean telomere length for the patients and their family members, calculated based on the following number of independent measurements of four in-gels and two Southern analyses: P1:6, M1:3, F1:3, S1:3, P2:9, M2:3, F2:1, C1:2, and C2:3. (C) Graphic illustration of the relative native (single strand) per denatured (total) telomeric signal, normalized to the controls. The values represent the mean of four independent measurements for P1 and four for P2. (D, top) A metaphase spread from P2 PBL after CO-FISH. Bar, 5 µm. The area designated by the white frame in enlarged in the bottom panel. Arrowheads point to chromosome ends with hybridization signals on both sister chromatids, indicating that T-SCE occurred at that chromosome end. (right) The frequency of T-SCE in P2 PBLs is compared with PBLs from four age-matched controls (con-BL1-4). For P2 PBLs, 1,150 chromosome ends were analyzed. For control PBLs, between 600 and 930 telomeres were analyzed (*, P < 0.0001; two-tailed Student’s t test). (E, top) A metaphase spread after telomere-FISH. Bar, 5 µm. The area designated by the white frame in enlarged in the bottom panel. Arrow points to a chromosome end missing one signal (STL, sister telomere loss). The frequencies of STLs (percentage of missing signals) are presented in the right panel in comparison to PBLs from four age-matched controls (con-BL5-8). For P2 PBLs, 1,548 chromosome ends were analyzed. For control PBLs, between 1,072 and 1,126 chromosome ends were analyzed (**, P < 0.05; two-tailed Student’s t test). (F, top) A metaphase spread of P2 fibroblasts after telomere-FISH. Bar, 5 µm. The area designated by the white frame in enlarged in the bottom. Arrow points to fused chromatids at a telomeric region with a missing hybridization signal. (right) The frequency of metaphase spreads containing at least one chromosome end with fused chromatids. This parameter was determined in P2 fibroblasts at two-time points of growth (P2*, passage 4+3.2 PDs; 16 metaphases, P2**, passage 4+4.4 PDs; 14 metaphases). Two control fibroblasts were analyzed: con-UN (passage 3 + 4.5 PDS; 12 metaphases) and con-FSE (passage 6 + 14.8 PDs; 11 metaphases; ***, P < 0.007, two-tailed Student’s t test). (G) TIF analysis was performed on STN1-mutated and control presenescent normal fibroblasts (PC). Each row displays a representative nucleus. Telomere signals appear in red and γ-H2AX foci in green. The DNA in the merged nucleus is stained with DAPI. The enlarged regions contain either a TIF (regions enclosed by yellow boxes in the merged image) or a γ-H2AX focus without a telomere signal (regions enclosed by red boxes in the merged image). (right) A graphic representation of TIF levels detected in the analyzed fibroblasts (for P1, *, P = 0.12; for P2, **, P = 0.4; two-tailed Student’s t test).
These hybridization signals were largely eliminated by exonuclease I digestion, indicating that they mostly represented G-rich 3′ overhangs and not internal single-stranded DNA. Single-stranded C-rich telomeric sequences were not detected by the G-rich probe (Fig. 3 B). Dysfunctional telomeres have been associated with enhanced telomeric recombination (Brault and Autexier, 2011), and, indeed, we observed elevated telomere sister chromatid exchange (T-SCE) in P2 PBLs (Fig. 3 D). Collectively, these results suggest that the STN1 mutations identified in the patients are associated with abnormal telomere C-strand synthesis. Southern analysis of telomere length in P2 fibroblasts at two consecutive passages indicated normal telomere length (unpublished data). However, notably, telomere-FISH on these fibroblasts revealed that the majority of the metaphases contain at least one fused chromosome end (Fig. 3 F). Moreover, the short replicative capacity of P1 and P2 fibroblasts (Fig. 2 A) suggested that telomeric defects are present in both patients’ fibroblasts. DNA damage at telomeres constitutes a hallmark of telomere dysfunction in senescent primary fibroblasts, but rarely occurs in early passage fibroblasts (Kaul et al., 2011). Indeed, analysis of telomere dysfunction-induced foci (TIFs) indicated that TIF levels in early passage P1 and P2 fibroblasts are comparable to those detected in late passage, presenescent cells (P1, 33.3%; P2, 35.8%; presenescent control, 42.2%; P > 0.19; two-tailed Student’s t test; Fig. 3 G). Thus, these findings signify that STN1-mutated fibroblasts also display an abnormal telomeric phenotype.
Phenotypic abnormalities of stn1 knockdown zebrafish embryos
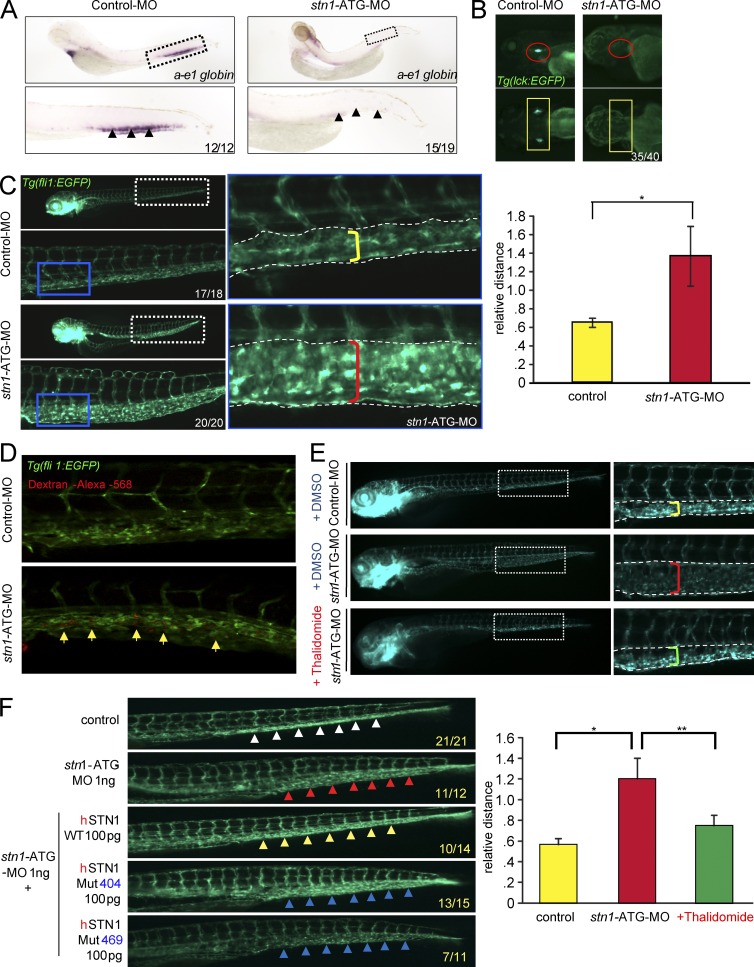
We modeled the STN1 dysfunction disease in zebrafish by injecting embryos with one of two zebrafish stn1-specific morpholino oligonucleotides targeting either the translation initiation site (ATG-MO) or the splice acceptor site of intron 5 (I5E6-MO; unpublished data). Both stn1-morpholino–treated embryos resulted in similar phenotypes. Analogous to the patients, they exhibited a drastic decrease in the number of red blood cells (Fig. 4 A) and an arrest in T cell progenitors (Fig. 4 B). The number of lyc+ myeloid cells and thrombocytes was dramatically elevated in comparison to uninjected controls (unpublished data). We presume that this myeloid expansion results from a myeloid bias among hematopoietic progenitors, which often accompanies dysfunction of hematopoietic stem cells in bone marrow failure syndromes. Interestingly, knockdown of Stn1 also increased vascularity (enlarged vascular bed and expanded, but fragmented, vessels), as indicated by the increased number of fli1:EGFP marked blood vessels (Fig. 4 C). The increased vascularity was further confirmed by angiography in stn1-morpholino–treated embryos (Fig. 4 D) and resembles the telangiectatic changes found in both patients with STN1 mutations. This specific phenotype was improved in stn1-morpholino–treated embryos after thalidomide treatment in a dose-dependent manner (Fig. 4 E). The phenotype correction was similar to the clinical effect observed in P2 after thalidomide treatment. Moreover, the telangiectatic changes were rescued by ectopic expression of STN1, but not by the mutant allele of either patient, thereby directly tying the phenotype to the mutant STN1 alleles (Fig. 4 F).
Figure 4.
A zebrafish model for human STN1 deficiency. (A) To detect anemia in situ hybridization was performed on control and stn1-ATG-morphant embryos at five days after fertilization (dpf) using an anti-sense ɑ-e1 probe. The regions depicted by the boxes in the upper images are enlarged below. Arrows indicate caudal hematopoietic tissue (CHT; lateral view). The number of embryos demonstrating this hybridization pattern out of the total screened embryos appears in the lower right corner. (B) Tg(lck:EGFP) zebrafish embryos were treated with control-MO or stn1-ATG-MO and effects on T cell development were revealed by alterations in lck:EGFP expression (lateral view, red circles; dorsal view, yellow rectangles). The number of embryos demonstrating this phenotype out of the total screened embryos appears in the bottom right corner. (C) Telangiectasis phenotype was determined in stn1 knockdown zebrafish. The yellow and red brackets in the insets on the right indicate the vascular bed widths in control WT and the stn1-knockdown embryos, respectively. The abnormal dilation of vessels and expansion of the vascular bed in the tail region are apparent in a representative 3dpf Tg(fli1:EGFP) embryo. Relative width of the vascular bed was calculated and normalized to control embryos (yellow bracket n = 3, each group), as shown in the graph on the right (*, P = 0.019; two-tailed Student’s t test). (D) Micro-angiography on Tg(fli1:EGFP) zebrafish embryos injected with dextran-alexa-568 at three dpf. Confocal microscopy was performed on fixed embryos. Yellow arrows indicate regions of accumulation of dextran at the CHT in stn1 morphants. (E) Telangiectactic changes in vasculature were detected after thalidomide treatment. Tg(fli1:EGFP) embryos treated with stn1-ATG-MO or Control-MO were incubated with thalidomide (15 µg/ml) from one to three dpf. Relative width of the vascular bed was quantified in control fish and in stn1 morphants with or without thalidomide treatment and expressed graphically, as below (*, P = 0.006; **, P = 0.018; two-tailed Student’s t test). (F) Telangiectactic changes in vasculature were detected after reexpression of wild type human STN1 (hSTN1). Tg(fli1:EGFP) zebrafish embryos were injected with stn1-ATG morphants alone or in conjunction with mRNA encoding WT human STN1 or STN1 bearing either of the patient mutations (Mut404, Mut469). The embryos were photographed at five dpf. White arrows, normal vessel phenotype; Red arrows, telangiectactic phenotype in stn1-ATG morphants; yellow arrows, co-injection with 100 pg WT hSTN1 mRNA. In all panels, the numbers at the bottom right indicate the fraction of embryos exhibiting the depicted phenotype.
In this study, we extend our understanding of the pathophysiology of telomere diseases associated with the CST complex (Anderson et al., 2012; Keller et al., 2012; Gu and Chang, 2013), by showing the first human-inherited defects in STN1. As part of the CST complex, STN1 interacts with and stimulates the activity of DNA polymerase α-primase (Casteel et al., 2009). It also plays a role in genome-wide replication restart after HU-induced replication fork stalling, likely by facilitating dormant origin firing (Stewart et al., 2012; Kasbek et al., 2013). In addition, stable knockdown of STN1 increased the incidence of micronuclei, anaphase bridges, and multitelomeric signals (Stewart et al., 2012). Similarly, we show that cells from STN1 mutant patients display elevated numbers of micronuclei and nuclear bridges, as well as decreased EdU uptake after HU treatment, suggesting defective recovery from induced fork stalling. These findings reinforce the assigned roles of STN1 in nontelomeric genomic regions. Furthermore, our findings of abnormal telomeric phenotypes are in line with previous studies, indicating that STN1 is critical for promoting telomere C-strand synthesis and maintenance (Huang et al., 2012; Stewart et al., 2012). Interestingly, although the overall telomere length of the STN1 mutant PBLs was not strikingly abnormal, with only one of the two patients showing relatively short telomeres, both our patients demonstrated extremely long single-stranded G-rich telomeric sequences, which could abrogate telomere protection and activate DNA damage response and repair by homologous recombination (resulting in telomere sister chromatid exchange) or by nonhomologous end joining (forming telomere–telomere fusions). The high level of TIFs in the STN1-mutated fibroblasts suggests a significant contribution of telomere defects to the severely compromised replication capacity of these cells.
The clinical phenotypes of the patients, including gastrointestinal telangiectatic changes and bleeding can be attributed to the mutant STN1 alleles, as these effects were recapitulated, in our study, in the zebrafish model and rescued by the normal but not the mutated human STN1 alleles. Effective therapeutic strategies for severe GI bleeding caused by telangiectatic changes in patients suffering from telomeropathies are in great need and therapeutic approaches, such as using a combination of oral estrogen and progesterone to control GI bleeding in Coats plus patients, have been proposed (Briggs et al., 2011). Importantly, a significant, lengthy control of the GI bleeding in one of our patients was achieved by thalidomide treatment. Nevertheless, this effect should be viewed in light of the partial response to therapy (the patient still needs blood transfusions) and the limited duration of treatment. The efficacy of thalidomide lies in its potent inhibition of angiogenesis through the regulation of angiogenesis growth factors levels (Folkman, 2001). Strikingly, the stn1-morpholino–treated zebrafish embryos displayed dramatic vascularization regression after thalidomide administration, illustrating a fruitful bed-to-bench-and-back approach.
In conclusion, our findings described here further confirm the role of STN1 in DNA replication at both telomeric and nontelomeric regions and have implications for diagnosis and treatment of Coats plus, as well as other telomeropathies and genome instability diseases. Precisely how the STN1 mutations cause the disease characteristics, and how much of the features can be attributed to telomere or genome-wide replication defects, or to other defects not related to DNA replication, are yet to be explored.
MATERIALS AND METHODS
Patients
Patients were studied at the Edmond and Lily Safra Children's Hospital, Sheba Medical Center, Israel. Patients' parents gave written, informed consent for this study, which was approved by the local institutional review boards at the Sheba Medical Center and Ministry of Health.
Whole exome and Sanger Sequencing
High throughput sequencing was performed by Illumina Hiseq2500 using 2 × 100 bp pair-end. Exome capturing was done using the Illumina Nextera DNA sample preparation kit. Overall, ∼50 M sequence reads were produced for each sample. BWA-mem algorithm (Li and Durbin, 2009) for alignment versus the hg19 version of the human genome was applied. Around 40 M reads were properly aligned. The median coverage was around 40 reads per base. GATK (McKenna et al., 2010) version 2.4.7 with the UnifiedGenotyper algorithm was applied for variant calling including all steps mentioned in the best practice pipeline. KGG-seq (Li et al., 2012) was used for annotation of detected variants and for comparing with allele frequency population databases. In-house scripts were applied for filtering, based on family pedigrees and intersections. STN1 mutations were validated by dideoxy Sanger sequencing in patients and carriers. PCR amplification products were directly sequenced using BigDye 3.1 Terminator chemistry (Applied Biosystems) and separated on an ABI 3500 genetic analyzer (Applied Biosystems). Data were evaluated using Sequencer v5.0 software (Gene Codes Corporation).
STN1 mutation genotyping
80 ethnically matched healthy controls (160 alleles) were examined for the STN1 mutated and wild-type alleles using the TaqMan Pre-Designed SNP Genotyping Assay (Applied Biosystems). The assay contained forward and reverse primers, flanking the mutated nucleotide and two labeled probes, VIC dye-MGB and 6FAM dye-MGB, each bound preferentially to one of the alleles. The reaction was performed using the ABI-7900 Sequence Detection System (Applied Biosystems).
STN1 structural analysis
The coordinates for the STN1–TEN1 complex were taken from the PDB file 4joi (Bryan et al., 2013). Secondary structure diagrams are based on those available in PDBsum (Laskowski, 2001). Jmol was used for visualization and creation of 3D images. A variety of tools, listed in Table S3, were applied for assessing the effect of point mutations on structure and stability.
Cell cultures
Fibroblasts were obtained from forearm skin-biopsy specimens of the affected children and from foreskins of healthy humans (Yalon et al., 2004; Vilboux et al., 2013). Both fibroblasts and peripheral blood lymphocytes (PBLs) were studied. Patient fibroblasts were cultured in DMEM supplemented with 20% FBS, glutamine, and antibiotics. Control fibroblasts were cultured in a similar media, but containing 10% FBS. The population doubling (PD) was determined at each passage, as previously described (Yehezkel et al., 2008). The PDs of patient fibroblasts were calculated after several passages, during which the culture was established. Cells were cultured until the point at which the culture failed to undergo at least one population doubling within 14 d. Peripheral blood cells were grown in Peripheral Blood Karyotyping Medium containing PHA (01–201-1B; Biological Industries) for 72 h before chromosome preparation.
Construction of a STN1-containing vector
The cDNA encoding WT STN1 (251–1355 of NM_024928) was amplified by RT-PCR using total RNA prepared from normal fibroblasts and cloned using the restriction endonucleases XbaI and SalI into a lentivirus vector (pLU-EF1aL-iBLAST) to generate the pLU-EF1aL-STN1-iBLAST plasmid. The primers used for the cloning were: XbaI_kozak_3xFLAG_STN1 5′: 5′-GGACCTCTAGAACCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGATGCAGCCTGGATCCAGCCGGTG-3′ and SalI_STN1_3′: 5′-GCTGGGTCGACGCTCAGAACGCTGTGTAGTAG-3′. The cloned vector was sequenced to verify the entire WT STN1 sequence.
Lentiviral packaging and transduction
Lentiviral particles were produced by transient transfection of 50% confluent 293T cells in 10 cm2 plates with pLU-EF1aL-STN1-iBLAST, together with GAG-POL and pMD2 plasmids, using the jetPEI (Polyplus Transfection) reagent. 30% confluent patient fibroblasts in 10 cm2 plates were infected twice on consecutive days with 2 ml DMEM medium containing the lentiviral particles and 3 µg/ml polybrene (Sigma-Aldrich). The ectopic expression of WT STN1 was verified by Western blot analysis using both, mouse anti-FLAG (Sigma-Aldrich), and rabbit anti-STN1 (Santa Cruz Biotechnology, Inc.) antibodies. Densitometry on scanned immunoblot images was performed using the ImageJ software.
Replication restart after HU treatment
Patient and control fibroblasts were plated onto coverslips in duplicates and grown to 50% confluency. One of the duplicates was incubated with 25 µM EdU for 48 h, to determine the fraction of cycling cells. The other duplicate was incubated with 0.5 mM HU (Sigma-Aldrich) for 48 h, after which the cells were washed with serum-free media to remove the HU, and then incubated with 50 µM EdU for 1 h. EdU incorporation was determined using the Click-iT EdU Alexa Fluor 488 Imaging kit and staining with 5 µg/ml Hoechst 33342 (Invitrogen). Images were produced using fluorescent microscopy (Olympus U-TV0.5XC-3). The percent of cells that incorporated EdU was measured by calculating the number of green nuclei (EDU) out of the total screened nuclei (>1,500; stained blue with Hoechst). The percentage of replicating cells after HU treatment was calculated by the fraction of EdU-positive cells after HU treatment divided by the fraction of EdU-positive cells after the 48-h incubation with EdU.
Cell cycle analysis
Patients and control fibroblasts were grown to 60–70% confluency and treated with 0.5 mM HU for 48 h. Medium was replaced and cells were harvested after 0, 2, 5, 8, and 24 h. Cell cycle analysis was performed using flow cytometry (Epics V; Coulter Electronics) after propidium iodide (PI) staining and analysis was performed with FlowJo software.
In-gel hybridization analyses
Genomic DNA was extracted from peripheral blood samples using the G-DEX IIb genomic DNA extraction kit (iNtRon). The relative amount of G-rich single-stranded telomeric DNA was estimated by in-gel hybridization, as previously described (Lamm et al., 2009). Duplicated samples were electrophoresed in the same gel (0.7% agarose) and, after gel drying, hybridized separately with a C-rich [(TAACCC)3] and G-rich [(AGGGTT)3] probes. The two parts of the gel were washed side-by-side under the same conditions and exposed together to film. Exonuclease I treatment was performed overnight at 37°C with 200 U exonuclease I (Thermo Fisher Scientific) per 5 µg DNA in 500 µl, followed by ethanol precipitation. The hybridization signal intensity for each lane was determined before and after denaturation using Image J (National Institutes of Health). Native hybridization signal was divided by the denatured signal and normalized to the value calculated for the control samples (set as 1). Mean telomere length was calculated using Matelo (Sagie et al., 2014).
Immunofluorescence and microscopy
Analysis for the presence of micronuclei, nuclear bridges, and apoptotic nuclei was performed on fibroblast cells plated on coverslips, cultured for several days, fixed with 4% formaldehyde, and stained with DAPI. Indirect Immunofluorescence (IF) was performed on fibroblast cells, as previously described (Yehezkel et al., 2013) using mouse antiphosphorylated γ-H2AX (05–636; EMD Millipore). Scoring of γ-H2AX foci was done on a BX50 microscope (Olympus).
Telomere FISH and chromosome orientation (CO)-FISH analyses
Metaphase chromosomes were obtained from peripheral blood by standard procedures. Metaphase spreads from fibroblasts were prepared in situ from cells grown for 72 h in chamber slides with BIOAMF-3 media (Biological Industries). During the last 18 h, cells were incubated with 12.5 µg/ml colcemid. Cells were treated in the chamber slides with hypotonic solution (0.28%KCl, 0.25% NaCitrate) for 30 min, and then fixed with Methanol/Acetic acid (3:1). Hybridization was performed with a PNA probe comprised of a (CCCTAA)3 oligo conjugated to Cy3, as previously described (Yehezkel et al., 2008). Chromosome ends (including both chromatids) were scored for the presence of two (double signals), one (sister telomere loss) or absent (signal free end) telomere–hybridization signals and for the presence of telomeric fusions.
CO-FISH analysis was performed as described previously (Yehezkel et al., 2008), after incubation of the cells with 2.5 × 10−4M BrdU for 19 h. Chromosome ends were analyzed for the presence of hybridization signals on both chromatids, indicating T-SCE (Londoño-Vallejo et al., 2004; Yehezkel et al., 2008). Hybridized metaphase spreads were visualized on a BX50 microscope, and images were captured with an Olympus DP73 camera controlled by cellSens Entry Microscopy Imaging software (Olympus).
TIF staining, microscopy, and analysis
Fibroblasts were grown on sterile slides for several days. Cells were then fixed with 4% PFA, permeabilized with 0.5% Triton in PBS, and blocked for 1 h at 37°C in 5%FBS/1xPBS. Following these treatments, slides were incubated overnight at 4°C with the mouse anti phosphorylated γH2AX antibody (#05-636,; EMD Millipore) diluted 1:300 in 3%FCS/0.1% triton/1xPBS. Slides were then washed three times for 5 min in 0.1% Triton/PBS (PBST) at room temperature, and incubated with a secondary antibody, donkey anti-mouse-FITC (715–095-150, Jackson ImmunoResearch Laboratories, INC.) diluted 1:300 in 3% FCS/0.1% Triton X-100/1xPBS for 1 h at 37°C. After three washes in PBST, a second 10-min fixation with PFA was performed, followed by three 5-min washes with PBS and three dehydration steps, each for 5 min in EtOH (70, 90, and 100%), all at room temperature. The telomere-FISH protocol was then continued as previously described (Yehezkel et al., 2008) using a PNA-(CCCTAA)3 oligo conjugated to Cy3. For TIF analysis images were captured on a confocal LSM 510 Meta Inverted Microscope (Zeiss). At least 20 nuclei were analyzed for each cell type by IMARIS software (BITPLANE) on z stack projection images. The γH2AX foci were called as surfaces, and a co-localization channel was built on telomere and foci signals after which the percent of telomeres positive for co-localization was determined.
Zebrafish studies
Wild-type AB strain zebrafish, as well as Tg(cd41:EGFP), Tg(fli1:EGFP)y1, and Tg(lck:EGFP)cz2 transgenic lines, were maintained under standard conditions as previously described (Langenau et al., 2004). After identification of the zebrafish orthologue of human STN1, two different morpholino (MO) oligonucleotides were designed against zebrafish stn1 and purchased from Gene Tools (Gene Tools, LLC). Sequence of stn1-ATG MO: 5′-CGACAGCGAACACTAAAACACAACC-3′), and sequence for targeting the splicing acceptor site of zebrafish stn1 exon6 (stn1-i5e6 MO): 5′-GAACCACCTGCTCTGTAAATGAAAA-3′. All morpholinos were injected at the 1–2 cell stage. The efficacy of translational silencing by the ATG-MO was assessed using an EGFP-biosensor containing the target sequences, as published previously (Nasevicius and Ekker, 2000). The effectiveness of the splicing MO was measured by RT-PCR. Whole mount in situ hybridization for lyc and a-e1 globin was performed as previously described (Thisse and Thisse, 2008). Dextran injection and microangiography in zebrafish was performed as reported previously (Hoeppner et al., 2012). Thalidomide treatment (Sigma-Aldrich; 15 µg/ml) in stn1-morphants was performed as described previously (Mahony et al., 2013). To test the function of the mutant human STN1 alleles, the patient mutations, STN1-Mut-404(c.404G>C) and STN1-Mut-469 (c.469G>T) were recreated using site-directed mutagenesis. After in vitro transcription, capped mRNAs encoding wild-type human STN1 or the aforementioned mutants were co-injected with stn1-ATG-MO into one-cell stage zebrafish embryos to assess their ability to rescue the telangectactic phenotype in the Stn1 morphants. The width of tail vessels in each Tg(fli:EGFP) embryo was quantified using ImageJ software (NIH), to measure the width of the vascular bed relative to that in control embryos. The zebrafish experiments were repeated three times independently. Data were presented as mean values ±SD, and Student’s t test was used to calculate p-values.
Online supplemental materials
Tables S1–S4 show clinical and laboratory data, including brain MRI, whole-exome sequencing analysis, computational prediction for functional, and thermodynamic consequences of the reported mutations and cell cycle analysis. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151618/DC1.
ACKNOWLEDGMENTS
We thank all our patients and their families for their cooperation. Raz Somech is supported by the Jeffrey Model Foundation (JMF). David L. Wiest is supported by NIH grant R21AI111208. Sara Selig is supported by Israel Science Foundation (grant No. 883/12). Yehuda Tzfati is supported by the Israel Science Foundation (grant No. 1729/13), Worldwide Cancer Research (grant No. 15-0338) and a project grant from the Israel Cancer Research Fund. Gideon Rechavi thanks the Kahn Family Foundation for their continuous support. He holds the Djerassi Chair for Oncology at the Tel-Aviv University, Israel and is supported by grants from the Israel Science Foundation (grant no. 1667/12), the Israeli Centers of Excellence (I-CORE) Program (ISF grants No. 41/11 and No. 1796/12), the Ernest and Bonnie Beutler Research Program, the Sagol Neuroscience Network and the Teva National Network of Excellence in Neuroscience. All authors declare no competing financial interests.
Footnotes
Abbreviations:
- CST
- CTC1–STN1–TEN1
- HU
- hydroxyurea
- PBL
- peripheral blood lymphocyte
- PD
- population doubling
- TIF
- telomere dysfunction induced foci
- T-SCE
- telomere sister chromatid exchange
References
- Anderson B.H., Kasher P.R., Mayer J., Szynkiewicz M., Jenkinson E.M., Bhaskar S.S., Urquhart J.E., Daly S.B., Dickerson J.E., O’Sullivan J., et al. 2012. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 44:338–342. 10.1038/ng.1084 [DOI] [PubMed] [Google Scholar]
- Armanios M., and Blackburn E.H.. 2012. The telomere syndromes. Nat. Rev. Genet. 13:693–704. 10.1038/nrg3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H., Erez E., Martz E., Pupko T., and Ben-Tal N.. 2010. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38:W529-W533 10.1093/nar/gkq399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccardi V., Razdan N., Kaplunov J., Mundra J.J., Kimura M., Aviv A., and Herbig U.. 2015. Stn1 is critical for telomere maintenance and long-term viability of somatic human cells. Aging Cell. 14:372–381. 10.1111/acel.12289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., and Wright W.E.. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science. 279:349–352. 10.1126/science.279.5349.349 [DOI] [PubMed] [Google Scholar]
- Brault M.E., and Autexier C.. 2011. Telomeric recombination induced by dysfunctional telomeres. Mol. Biol. Cell. 22:179–188. 10.1091/mbc.E10-02-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs T.A., Hubbard M., Hawkins C., Cole T., Livingston J.H., Crow Y.J., and Pigott A.. 2011. Treatment of Gastrointestinal Bleeding in a Probable Case of Cerebroretinal Microangiopathy with Calcifications and Cysts. Mol. Syndromol. 1:159–162. 10.1159/000321559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan C., Rice C., Harkisheimer M., Schultz D.C., and Skordalakes E.. 2013. Structure of the human telomeric Stn1-Ten1 capping complex. PLoS One. 8:e66756 10.1371/journal.pone.0066756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel D.E., Zhuang S., Zeng Y., Perrino F.W., Boss G.R., Goulian M., and Pilz R.B.. 2009. A DNA polymerase-alphamiddle dotprimase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 284:5807–5818. 10.1074/jbc.M807593200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.Y., and Lingner J.. 2013. CST for the grand finale of telomere replication. Nucleus. 4:277–282. 10.4161/nucl.25701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-Y., Redon S., and Lingner J.. 2012. The human CST complex is a terminator of telomerase activity. Nature. 488:540–544. 10.1038/nature11269 [DOI] [PubMed] [Google Scholar]
- Dai X., Huang C., Bhusari A., Sampathi S., Schubert K., and Chai W.. 2010. Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J. 29:2788–2801. 10.1038/emboj.2010.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. 2009. How telomeres solve the end-protection problem. Science. 326:948–952. 10.1126/science.1170633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diotti R., and Loayza D.. 2011. Shelterin complex and associated factors at human telomeres. Nucleus. 2:119–135. 10.4161/nucl.2.2.15135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkman J. 2001. Angiogenesis-dependent diseases. Semin. Oncol. 28:536–542. 10.1016/S0093-7754(01)90021-1 [DOI] [PubMed] [Google Scholar]
- Gu P., and Chang S.. 2013. Functional characterization of human CTC1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease Coats plus. Aging Cell. 12:1100–1109. 10.1111/acel.12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P., Min J.N., Wang Y., Huang C., Peng T., Chai W., and Chang S.. 2012. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 31:2309–2321. 10.1038/emboj.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner L.H., Phoenix K.N., Clark K.J., Bhattacharya R., Gong X., Sciuto T.E., Vohra P., Suresh S., Bhattacharya S., Dvorak A.M., et al. 2012. Revealing the role of phospholipase Cβ3 in the regulation of VEGF-induced vascular permeability. Blood. 120:2167–2173. 10.1182/blood-2012-03-417824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan B., Wright W.E., and Shay J.W.. 2014. Cell biology of disease: Telomeropathies: an emerging spectrum disorder. J. Cell Biol. 205:289–299. 10.1083/jcb.201401012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Dai X., and Chai W.. 2012. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 22:1681–1695. 10.1038/cr.2012.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D., and Cooper J.P.. 2010. Telomeric Strategies: Means to an End. In Annual Review of Genetics, Vol 44. Vol. 44. A. Campbell, M. Lichten, and G. Schupbach, editors. 243-269. [DOI] [PubMed] [Google Scholar]
- Kasbek C., Wang F., and Price C.M.. 2013. Human TEN1 maintains telomere integrity and functions in genome-wide replication restart. J. Biol. Chem. 288:30139–30150. 10.1074/jbc.M113.493478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul Z., Cesare A.J., Huschtscha L.I., Neumann A.A., and Reddel R.R.. 2011. Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep. 13:52–59. 10.1038/embor.2011.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R.B., Gagne K.E., Usmani G.N., Asdourian G.K., Williams D.A., Hofmann I., and Agarwal S.. 2012. CTC1 Mutations in a patient with dyskeratosis congenita. Pediatr. Blood Cancer. 59:311–314. 10.1002/pbc.24193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm N., Ordan E., Shponkin R., Richler C., Aker M., and Tzfati Y.. 2009. Diminished telomeric 3′ overhangs are associated with telomere dysfunction in Hoyeraal-Hreidarsson syndrome. PLoS One. 4:e5666 10.1371/journal.pone.0005666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau D.M., Ferrando A.A., Traver D., Kutok J.L., Hezel J.P., Kanki J.P., Zon L.I., Look A.T., and Trede N.S.. 2004. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl. Acad. Sci. USA. 101:7369–7374. 10.1073/pnas.0402248101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A. 2001. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 29:221–222. 10.1093/nar/29.1.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.X., Gui H.S., Kwan J.S., Bao S.Y., and Sham P.C.. 2012. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 40:e53 10.1093/nar/gkr1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londoño-Vallejo J.A., Der-Sarkissian H., Cazes L., Bacchetti S., and Reddel R.R.. 2004. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 64:2324–2327. 10.1158/0008-5472.CAN-03-4035 [DOI] [PubMed] [Google Scholar]
- Mahony C., Erskine L., Niven J., Greig N.H., Figg W.D., and Vargesson N.. 2013. Pomalidomide is nonteratogenic in chicken and zebrafish embryos and nonneurotoxic in vitro. Proc. Natl. Acad. Sci. USA. 110:12703–12708. 10.1073/pnas.1307684110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M.A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., and Ishikawa F.. 2009. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell. 36:193–206. 10.1016/j.molcel.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Nasevicius A., and Ekker S.C.. 2000. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26:216–220. 10.1038/79951 [DOI] [PubMed] [Google Scholar]
- Polvi A., Linnankivi T., Kivelä T., Herva R., Keating J.P., Mäkitie O., Pareyson D., Vainionpää L., Lahtinen J., Hovatta I., et al. 2012. Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am. J. Hum. Genet. 90:540–549. 10.1016/j.ajhg.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagie S., Ellran E., Katzir H., Shaked R., Yehezkel S., Laevsky I., Ghanayim A., Geiger D., Tzukerman M., and Selig S.. 2014. Induced pluripotent stem cells as a model for telomeric abnormalities in ICF type I syndrome. Hum. Mol. Genet. 23:3629–3640. 10.1093/hmg/ddu071 [DOI] [PubMed] [Google Scholar]
- Stewart J.A., Wang F., Chaiken M.F., Kasbek C., Chastain P.D. II, Wright W.E., and Price C.M.. 2012. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 31:3537–3549. 10.1038/emboj.2012.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., and Thisse B.. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3:59–69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Townsley D.M., Dumitriu B., and Young N.S.. 2014. Bone marrow failure and the telomeropathies. Blood. 124:2775–2783. 10.1182/blood-2014-05-526285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilboux T., Lev A., Malicdan M.C., Simon A.J., Järvinen P., Racek T., Puchalka J., Sood R., Carrington B., Bishop K., et al. 2013. A congenital neutrophil defect syndrome associated with mutations in VPS45. N. Engl. J. Med. 369:54–65. 10.1056/NEJMoa1301296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne A.J., Bhagat T., Kirwan M., Gitiaux C., Desguerre I., Leonard N., Nogales E., Vulliamy T., and Dokal I.S.. 2013. Mutations in the telomere capping complex in bone marrow failure and related syndromes. Haematologica. 98:334–338. 10.3324/haematol.2012.071068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M., Qin J., Songyang Z., and Liu D.. 2009. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem. 284:26725–26731. 10.1074/jbc.M109.021105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Stewart J.A., Kasbek C., Zhao Y., Wright W.E., and Price C.M.. 2012. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Reports. 2:1096–1103. 10.1016/j.celrep.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalon M., Gal S., Segev Y., Selig S., and Skorecki K.L.. 2004. Sister chromatid separation at human telomeric regions. J. Cell Sci. 117:1961–1970. 10.1242/jcs.01032 [DOI] [PubMed] [Google Scholar]
- Yehezkel S., Segev Y., Viegas-Péquignot E., Skorecki K., and Selig S.. 2008. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 17:2776–2789. 10.1093/hmg/ddn177 [DOI] [PubMed] [Google Scholar]
- Yehezkel S., Shaked R., Sagie S., Berkovitz R., Shachar-Bener H., Segev Y., and Selig S.. 2013. Characterization and rescue of telomeric abnormalities in ICF syndrome type I fibroblasts. Front. Oncol. 3:35 10.3389/fonc.2013.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]