Quek and colleagues identify human leukemic stem cells (LSCs) present in CD34− AML. In-depth characterization of the functional and clonal aspects of CD34− LSCs indicates that most are similar to myeloid precursors.
Abstract
Our understanding of the perturbation of normal cellular differentiation hierarchies to create tumor-propagating stem cell populations is incomplete. In human acute myeloid leukemia (AML), current models suggest transformation creates leukemic stem cell (LSC) populations arrested at a progenitor-like stage expressing cell surface CD34. We show that in ∼25% of AML, with a distinct genetic mutation pattern where >98% of cells are CD34−, there are multiple, nonhierarchically arranged CD34+ and CD34− LSC populations. Within CD34− and CD34+ LSC–containing populations, LSC frequencies are similar; there are shared clonal structures and near-identical transcriptional signatures. CD34− LSCs have disordered global transcription profiles, but these profiles are enriched for transcriptional signatures of normal CD34− mature granulocyte–macrophage precursors, downstream of progenitors. But unlike mature precursors, LSCs express multiple normal stem cell transcriptional regulators previously implicated in LSC function. This suggests a new refined model of the relationship between LSCs and normal hemopoiesis in which the nature of genetic/epigenetic changes determines the disordered transcriptional program, resulting in LSC differentiation arrest at stages that are most like either progenitor or precursor stages of hemopoiesis.
Acute myeloid leukemia (AML), the most common human aggressive leukemia, has a poor prognosis. Like many cancers, AML is characterized by differentiation arrest leading to expansion of leukemic stem cell (LSC) populations (also termed leukemia-initiating cells in transplantation experiments in immunodeficient mice). However, there is an incomplete understanding of where arrest occurs in the hemopoietic differentiation hierarchy, which limits development of novel therapeutic approaches in AML aimed at overcoming differentiation arrest.
In most human AML, and in the related preleukemic condition myelodysplastic syndrome, the initiating genetic mutation usually originates in a hemopoietic stem cell (HSC) or very early long-lived multipotent progenitor (MPP) cell (Jan et al., 2012; Corces-Zimmerman et al., 2014; Shlush et al., 2014; Woll et al., 2014). This gives rise to expanded preleukemic stem/progenitor cell populations with clonal advantage but permits differentiation, leaving the hemopoietic hierarchy relatively unperturbed, with subjects often having normal blood counts (Busque et al., 2012; Genovese et al., 2014; Jaiswal et al., 2014). Initiating mutations have been found in epigenetic regulators (e.g., TET2, IDH1, IDH2, DNMT3A, and ASXL1), the cohesin complex (MAU2 and SMC1A), and other AML-associated genes (e.g., NPM1; Busque et al., 2012; Jan et al., 2012; Corces-Zimmerman et al., 2014; Genovese et al., 2014; Shlush et al., 2014). After acquisition of an initiating mutation, it is unclear in which cell population subsequent mutations are acquired and how they perturb the hemopoietic hierarchy. Nevertheless, the net effect of all genetic and epigenetic changes (i.e., leukemic transformation) is characterized by disruption of the hemopoietic hierarchy and differentiation arrest leading to the absence of mature blood cells.
In the fully transformed state, hemopoiesis is dominated by multiple, expanded, immunophenotypically distinct LSC populations that generate bulk non-LSC populations. In AML samples in which the majority of leukemic cells express the normal hemopoietic stem/progenitor cell (HSPC) surface marker CD34, LSCs have been detected in both CD34+CD38− and CD34+CD38+ compartments (Bonnet and Dick, 1997; Ishikawa et al., 2007; Taussig et al., 2010; Goardon et al., 2011) and more rarely in CD34− compartments (Eppert et al., 2011; Sarry et al., 2011). By comparing immunophenotype and global gene expression profiles of CD34+ LSC-containing populations with normal HSPCs, we previously showed that human LSC populations were most closely related to normal progenitors, rather than HSCs, but had co-opted elements of a normal HSC expression signature (Goardon et al., 2011). This was consistent with earlier work in mouse models in which introduction of leukemic oncogenes into progenitors produced transplantable LSCs (Cozzio et al., 2003; So et al., 2003; Krivtsov et al., 2006; Kirstetter et al., 2008).
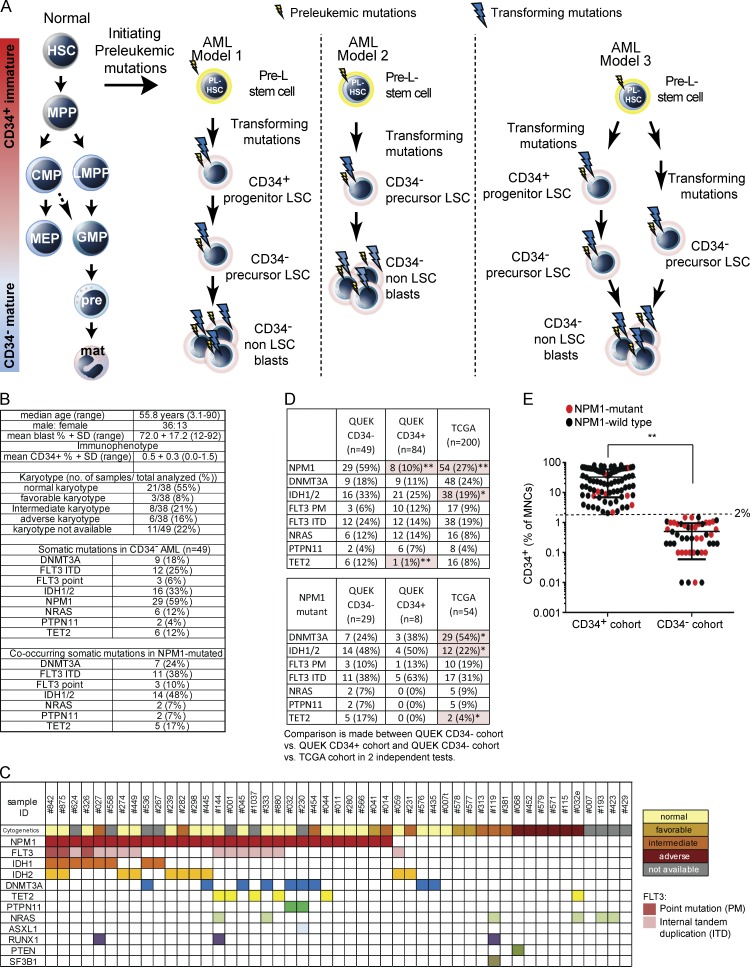
However, in ∼25% of human AML, >90% of leukemic cells do not express CD34. These samples are enriched for the NPM1 mutation (Falini et al., 2005; Martelli et al., 2010; Taussig et al., 2010). Here, LSC activity resides in a small CD34+ fraction and majority CD34− compartment (Martelli et al., 2010; Taussig et al., 2010; Sarry et al., 2011). However, it is unclear whether transformation creating leukemia-propagating cells occurs initially in CD34+ progenitor-like cells or downstream in CD34− cells. It is also unclear whether CD34+ and CD34− LSC populations are hierarchically arranged and what the nature of the clonal relationships between CD34+ and CD34− LSCs is. At least three possible models for leukemic hemopoiesis in CD34− AML exist (Fig. 1 A). In model 1, transformation associated with partial differentiation arrest and expansion of a cell compartment acquiring LSC function occurs at a CD34+ progenitor stage. These CD34+ LSCs partially differentiate into CD34− LSC populations and eventually into CD34− non-LSC bulk blast populations. Here, multiple, distinct, hierarchically arranged LSC populations exist. In model 2, transformation, expansion, and acquisition of the LSC function principally occurs at the CD34− precursor stage (not previously described), with CD34 aberrantly expressed on a small subset of LSCs. Finally, in model 3, there could be a combination of models 1 and 2 in which some clones are transformed at the CD34+ progenitor stage and yet others are transformed at the CD34− precursor stage.
Figure 1.
CD34− AML models of differentiation arrest and genetics of samples. (A, left) Normal hemopoietic hierarchy. Initiating mutations in HSC or very early long-lived progenitors create preleukemic (Pre-L) stem cells with a clonal advantage. Model 1: mutations transform pre-LSCs into CD34+ progenitor-like LSCs that differentiate into CD34− GM-pre–like LSCs, resulting in hierarchically arranged LSC populations. LSC populations then differentiate into CD34− non-LSC blasts. Model 2: mutations transform pre-LSCs into CD34− GM-pre–like LSCs that then differentiate into CD34− non-LSC blasts. Here, CD34 would be aberrantly expressed on a small subset of LSCs. Model 3 combines models 1 and 2. Some clones acquire transforming mutations to create CD34+ progenitor-like LSCs that differentiate into CD34− GM-pre–like LSCs; other clones acquire transforming mutations to create CD34− precursor-like LSCs only. CMP, common myeloid progenitor; mat, mature granulocyte–monocytic effector cells; MEP, megakaryocyte-erythroid progenitor; MPP, MPP/short-term HSC; PL, preleukemic; pre, granulocyte–monocyte precursors. (B) Characteristics of 49 CD34− AML samples: patient demographics, blast percentage, immunophenotype, karyotype, and mutational profile. (C) Karyotype and disease-associated nucleotide variants. The colored boxes denote either karyotype/risk stratification or mutation. No known disease-associated mutations were detected in CEBPA, CBL, ETV6, WT1, and Cohesin genes (see Table S1 B). (D, top) Frequency of the indicated mutations in our CD34− AML (n = 49), our CD34+ AML (n = 84), and our TCGA unselected AML (n = 200) sample cohorts. (Bottom) Mutations within NPM1 mutant samples in all three cohorts. n, number of samples. Statistically significant comparisons assessed by χ2 tests are highlighted and marked with asterisks: *, P < 0.05; **, P < 0.01. (E) Distribution of NPM1 wild-type and mutant AML samples across a continuum of CD34 expression (assessed by flow cytometry as a percentage of MNCs: CD34+ AML, n = 84; CD34− AML, n = 49). The 2% threshold of CD34 expression used to select the CD34− AML cohort in this study is marked. The difference in percentage of NPM1 mutant samples between the two cohorts was statistically significant (**, P < 0.01). PTEN, phosphatase and tensin homologue.
RESULTS
CD34− AML has a distinctive mutational profile
We focused on samples in which ≤2% of mononuclear cells (MNCs) expressed CD34 (hereon referred to as CD34− AML). They accounted for 25% of 270 consecutive de novo AML samples (analyzed between January and July 2010; unpublished data). We analyzed 49 consecutive CD34− AML samples; patient demographics, mean blast percentage, and percentage of MNCs expressing CD34+ (mean, 0.5%; range, 0.0–1.5%) are in Fig. 1 B and Table S1 A. 21/38 (55%) samples, in which karyotyping (segregated into prognostic groups; Grimwade et al., 2010) was available, had a normal karyotype (Fig. 1 B). We screened 24 genes (524 amplicons) by targeted resequencing for mutations commonly found in CD34− AML at a mean read depth of 3,500× (range of 100–13,000×; Fig. 1, B and C; and Table S1 B). Compared with The Cancer Genome Atlas (TCGA) cohort spanning the breadth of adult AML in which 62% of samples were CD34+ (Cancer Genome Atlas Research Network, 2013), our cohort had similar percentages of samples with normal karyotype (21/38 [55%] vs. 92/195 [47%]). However, our cohort of CD34− AML samples was significantly enriched for mutations in NPM1 (29/49 [59%] vs. 54/200 [27%]; χ2 P < 0.0001) and IDH1/IDH2 (16/49 [33%] vs. 38/200 [19%]; χ2 P = 0.038) compared with the TCGA cohort (Fig. 1 D). Within NPM1 mutant samples, our cohort of CD34− AML samples compared with the TCGA cohort was enriched in IDH1/2 (48% vs. 22%; χ2 P = 0.014) and TET2 (17% vs. 4%; χ2 P = 0.034) and depleted for DNMT3A mutations (24% vs. 54%; χ2 P = 0.033). FLT3 mutations were at a similar frequency (48% vs. 50%; Fig. 1 D). In contrast to the TCGA cohort, we did not detect mutations in 14/24 most commonly mutated genes in AML. Finally, we compared the mutation profile in our 49-sample CD34− AML cohort with that of a distinct cohort of 84 sequential AML samples with >2% CD34+ blasts (Fig. 1, D and E). Once again, there was enrichment of NPM1 mutations in CD34− AML (59% vs. 10%; χ2 P < 0.01; Fig. 1 E). Mutations in TET2 were also more frequent in CD34− versus CD34+ AML (12% vs. 1%; P < 0.001). Finally, although there were fewer CD34+ AML samples with IDH1/2 mutations (25%) compared with CD34− AML (33%), this was not statistically significant. Consistent with comparisons with the TCGA cohort, within NPM1 mutant AML, there were fewer TET2 (0% vs. 17%) in CD34+ AML versus CD34− AML. However, as the number of NPM1-mutated AML samples in our 84-sample CD34+ cohort was low (n = 8), these differences did not reach statistical significance. In summary, compared with CD34+ AML, CD34− AML has a distinct, restricted mutational profile within the constraints of the sample size (n = 49) studied.
CD34− AML LSCs exist in CD34+ and CD34− populations and are not hierarchically arranged
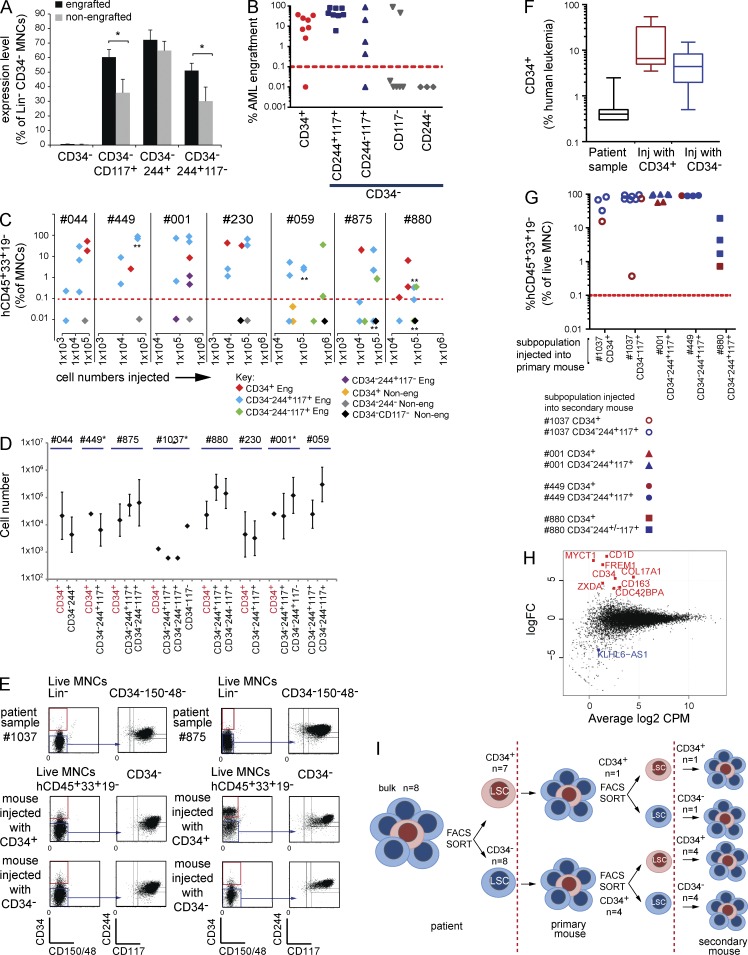
28/49 samples had sufficient cells to test for LSC activity in NSG and/or NRG and/or NSGS mice. 11/28 samples serially engrafted AML with NPM1 and/or FLT3 mutations (Table S2 A), allowing functional LSC identification. Because CD34− AMLs are enriched for more differentiated myelomonocytic/monocytic AML subtypes, we hypothesized that LSCs may express a combination of an immature (C-KIT/CD117, commonly expressed on CD34− blasts; Nomdedeu et al., 2011) and mature (CD244) myelomonocytic marker (Table S1 A). Studying CD244 expression greatly facilitated later comparison of AML LSCs with normal CD34− cell populations (see the next section).
Samples with a higher proportion of CD117+ cells were more likely to engraft (Fig. 2 A). To pinpoint LSC activity, CD34+ and CD34− populations were purified from 8/11 engrafting samples in which sufficient cells were available for subsequent assays (sort purities are in Table S2, B and C). In seven out of eight cases, minor CD34+ populations had LSC activity (Fig. 2 B and Table S2 A). As the mean sort purity in the positive purification of the CD34+ LSC populations used in primary transplantation was 92% (Table S2 B), contaminating CD34− LSCs could make a minor contribution to engraftment. In CD34− fractions, LSC activity principally segregated with CD117 expression, especially when coexpressed with CD244 (eight out of eight samples; Fig. 2 B). Engraftment was also seen from four out of five samples where the CD244−117+ fraction could be purified. CD117− cells engrafted in only two samples (#001 and #1037). Sample #1037 was exceptional, as all sorted cell fractions engrafted aggressively (Fig. 2, B and D). In sample #001, CD244+117− cells engrafted mice ∼80-fold less, with a 3.6-fold–increased cell dose compared with the CD244+117+ fraction (Fig. 2 C).
Figure 2.
CD34+ and CD34− LSCs in CD34− AML are not hierarchically arranged and share near-identical RNA-seq profiles. (A) Mean percentage of cells with immunophenotypes indicated (as a percentage of Lin−CD34− MNCs) in engrafting and nonengrafting populations (total of 28 samples injected). The only populations significantly more abundant in engrafting samples were Lin−CD34−117+ and Lin−CD34−244+117+. *, P < 0.05; Student’s t test. Error bars represent SEM. (B) Primary engraftment (eight patients). Each symbol represents mean engraftment in one to six mice from the indicated population from one patient. Y axis, mean percentage of human (h)CD45+CD33+CD19− cell engraftment/total live MNCs. X axis, injected cell fraction. Red dashed line, engraftment threshold. Leukemic engraftment was confirmed by genetic analysis in each case. (C) Primary engraftment experiments where different numbers of cells were injected from sorted patient subpopulations annotated as engrafting (Eng) or nonengrafting (Non-eng). At least one engrafting subpopulation in each patient sample propagates leukemia at cell numbers lower than nonengrafting subpopulations. Each data point represents one injected mouse. Asterisks indicate cell doses where multiple mice were injected with the same cell numbers. The number of asterisks indicates the number of different mice injected. (D) LSC frequency in sorted subpopulations calculated from limit dilution transplant assays. Error bars indicate the calculated 95% confidence interval. Data points without error bars indicate cell fractions where the threshold nonengrafting cell number was not achieved. Here, lowest injected cell numbers are shown (see Table S2 D). (E) Two points are made here: first, the patient’s CD34+ and CD34− populations when injected into mice produce both CD34+ and CD34− populations; second, the immunophenotype of the patient’s AML is recapitulated in the mouse. Two representative patient samples, #1037 and #875, from experiments with five different patient samples, show the immunophenotype in the patient sample. Blue arrows indicate CD244 and CD117 expression on CD34− AML cells. CD34+ (red boxes) and CD34− (blue boxes) subpopulations are flow sorted with purity for injection into mice. (Middle and bottom) Representative FACS analysis plots of engrafted human cells in mice. Mice injected with CD34+ cells or CD34− cells. In both cases, engrafting leukemic cells contain both CD34+ and CD34− populations and recapitulate the immunophenotype of the patient’s AML (see Table S2 E). (F) Box-whisker plots show percentage of human CD34+ leukemic cells in patient samples or primary recipients injected (Inj) with either CD34+ (samples from five patients injected into five mice) or CD34− populations (samples from five patients injected into 12 mice). The middle line is the median. Box limits are first and third quartile. Bars indicate minimum and maximum values. (G) Serial engraftment shows no hierarchy between CD34+ and CD34− LSCs. X axis, subpopulations from patient samples injected into primary mice; Y axis, mean percentage of human CD45+CD33+CD19− leukemia engraftment of total live MNCs in secondary mice when populations indicated on the left were injected. Each data point represents engraftment in one mouse (see Table S2 E). (H) RNA-seq data showing log2-fold change (logFC) in expression of all expressed genes (5 CD34+ and14 CD34− AML LSC samples on the y axis) plotted against mean log2 cpm (x axis). Statistically significant differentially expressed genes (P < 0.05) are annotated (red, up-regulated in CD34+ LSCs; blue, up-regulated in CD34− LSCs). (I) CD34+ and CD34− LSCs in CD34− AML generate both CD34+ and CD34− populations in primary transplants, each with leukemia-propagating activity in secondary transplants. n, number of patient samples studied per population.
We next tested whether failure to engraft simply reflects low cell numbers injected rather than biological differences. Engrafting populations did so at cell doses equivalent to or lower than nonengrafting (i.e., non-LSC) populations (Fig. 2 C). Limit dilution assays showed similar LSC frequencies between CD34+- and CD34−-engrafting subpopulations within a patient (Fig. 2 D and Table S2 D). CD117 expression in the CD34− compartment also purified LSCs (1.1–232-fold; mean, 27-fold), and the combined CD34−CD244+/−117+ fraction accounted for the majority of LSCs (mean, 72.9%; Table S2 D). CD34 expression also enriched for LSCs (14–263-fold; mean, 133-fold), but CD34+ LSCs only contributed to 3.3% (range, 0.1–9.4%) of LSCs.
As CD34 expression marks normal HSPCs, we asked whether CD34+ and CD34− LSC populations were hierarchically arranged. Injected LSCs recapitulated the immunophenotype of the patient’s AML; CD244 and CD117 expression was maintained in engrafted cell populations in all eight samples (two representative samples in Fig. 2 E and full dataset in Table S2 E). Surprisingly, just as CD34+ LSCs generated both CD34+ and CD34− AML populations in primary recipient mice, CD34− LSCs also produced CD34+ and CD34− progeny (Fig. 2, E and F; and Table S2 E). Both CD34+ and CD34− populations from primary recipients serially engrafted (Fig. 2 G) and generated CD34+ and CD34− cells in secondary recipients. Finally, by RNA sequencing (RNA-seq), only nine protein-coding genes of 15,539 expressed genes, including CD34, were significantly differentially expressed between CD34+ and CD34− LSC populations (P < 0.05; Fig. 2 H). Thus, in CD34− AML, these data are most compatible with functionally equivalent, nonhierarchically arranged CD34+ and CD34− LSC populations with near-identical transcriptomes (Fig. 2 I).
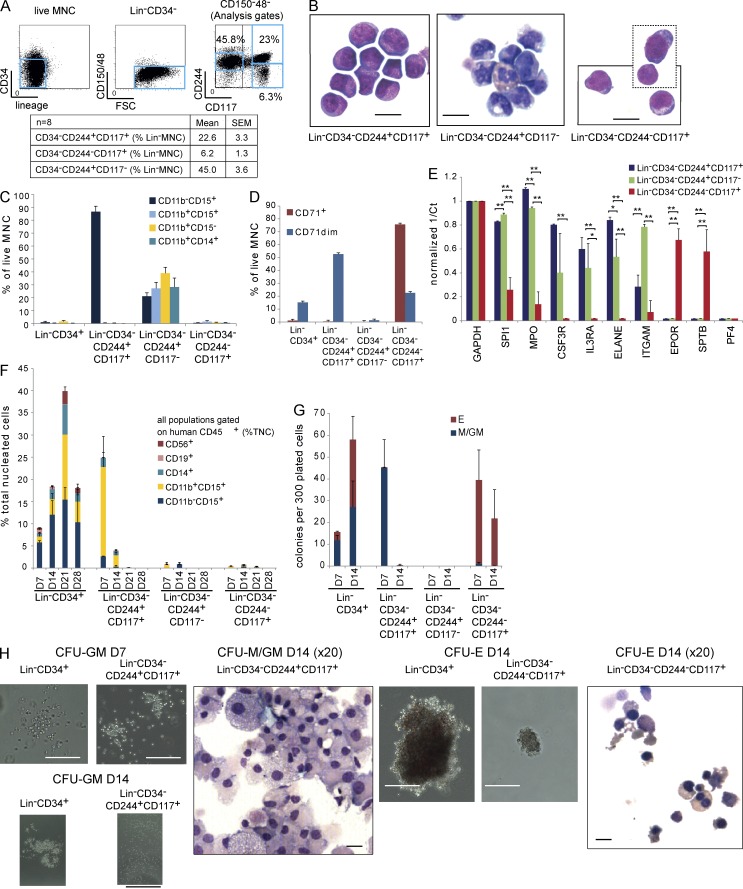
Identification of normal CD34− myeloid and erythroid precursors with limited progenitor potential
Next, we asked which normal BM populations most closely correspond to CD34− AML LSCs. Though myeloid progenitors have been studied, normal precursor/mature myeloid populations downstream of progenitors are poorly characterized, particularly with respect to surface markers used to fractionate AML samples. Distinct normal lineage (Lin)−CD34−244±117± BM populations (Fig. 3 A) were purified, characterized, and compared with normal CD34+ cells (Fig. 3, B–H; and Table S3). Normal CD34−244+117+ cells have promyelocyte morphology (Fig. 3 B), and consistent with this are CD11b−15+ cells (Fig. 3 C; van Lochem et al., 2004). CD34−244+117− cells were a mix of mature granulocytic cells (CD11b−15+, CD11b+15−, and CD11b+15+) and monocytes (CD11b+14+; Fig. 3, B and C). CD34−244−117+ cells are erythroblasts expressing high levels of CD71 (Fig. 3, B and D). CD34−244+117+ and CD34−244+117− cells expressed high levels of granulocyte–macrophage (GM) lineage–affiliated genes SPI1/PU.1, MPO, CSF3R, IL3RA, ELANE, and ITGAM, whereas CD34−244−117+ expressed erythroid-affiliated genes EPOR and SPTB (Fig. 3 E).
Figure 3.
Normal marrow CD34−CD244±CD117+ populations are GM and erythroid precursors. (A, top) FACS plots with analysis gates showing lineage markers and CD34 expression, CD150/CD48 expression and forward scatter (FSC), and CD244 and CD117 expression in normal BM. (Bottom) Mean and SEM of each population as a percentage of MNCs. (B) Photomicrographs of FACS-sorted Lin−CD34−244+117+ (promyelocyte-like), Lin−CD34−244+117− (mature granulocytic series and monocytes), and Lin−CD34−CD244−117+ (erythroblast-like) stained with May-Giemsa-Grunwald. The box with dotted lines shows two amalgamated microscope fields from the same cytospin slide. Images are representative of three separate experiments using BM from three separate donors. Bars, 20 µm. (C and D) FACS-sorted Lin−CD34+ (n = 3 donors) and Lin−CD34− (CD244+117+ and CD244+117−, n = 6; CD244−117+, n = 4) populations were tested for CD11b, CD15, and CD14 (C) and CD71 (D) expression. (E) Normalized gene expression (to GAPDH) of SPI1/PU.1, MPO, CSF3R, IL3RA, ELANE, and ITGAM (expressed in GM lineage cells); EPOR and SPTB (expressed in erythroid cells); and PF4 in Lin−CD34−CD244+117+, Lin−CD34−CD244+117−, and Lin−CD34−CD244−117+ cells (n = 3 donors). *, P < 0.05; **, P < 0.01; two-sided Student’s t test. (F) Liquid culture output (human CD45+ cells and a percentage of total nucleated cells [TNC]) of 200 Lin−CD34+ and Lin−CD34− populations assessed by FACS studied on D7, 14, 21, and 28 (n = 3–4). CD56+, NK cells; CD19+, B cells; CD14+, monocytes; CD11b+15+, granulocytes; and CD11b−15+, promyelocytes.(G) Colonies produced when 300 cells from Lin−CD34+ and Lin−CD34− populations were plated in methylcellulose under myeloid colony formation conditions. Lineage affiliation was assessed by microscopy and confirmed in colony cytospins. E, erythroid; M, macrophage. Colonies were counted at D7 and D14 after plating (n = 5 donors). Error bars represent SEM. (H) Photomicrographs of representative D7 and D14 CFU-GM, D14 CFU-E colonies, and cytospins from the indicated populations (from five separate experiments using BM from five donors). Bars: (CFU-GM D7) 200 µm; (CFU-GM D14) 2,000 µm; (CFU-M/GM D14) 10 µm; (CFU-E D14) 100 µm; (CFU-E D14, right) 10 µm.
We then tested the functional output of normal CD34− fractions in liquid culture (Fig. 3 F). CD34+ cells were tested as a positive control. Whereas CD34+ cells gave rise to multilineage progeny with a peak output at day 21 (D21), CD34−244+117+ cells produced neutrophils (CD11b+15+) and monocytes (CD14+) peaking at D7. CD34+ cells produced 2.4-fold more human cells than the CD34−244+117+ population. Neither CD34−244+117− (mixed mature GM cells) nor CD34−244−117+ (erythroblasts) grew in liquid culture.
We next tested whether CD34− populations had progenitor activity (Fig. 3 G). In contrast to CD34+ cells that gave erythroid and GM colonies peaking at D14, CD34−244+117+ cells gave rise to only short-lived, small macrophage or mixed GM colonies present only at D7 (Fig. 3, G and H). CD34−244−117+ cells gave rise to small, distinct erythroid colonies at D7 that declined by D14. CD34−CD244+117− populations did not give colonies or cell clusters. No CD34− population gave rise to megakaryocyte colonies or expressed megakaryocyte-affiliated genes (PF4; Fig. 3 E; unpublished data). A previous study has shown that CD34−CD38−CD93hi populations have stem cell activity (Anjos-Afonso et al., 2013). The Lin−CD34−CD244±117± populations we purified were CD38−93dim and lacked stem cell activity in NSG mice (105 cells injected; unpublished data). Thus, normal Lin−CD34−117+ populations are GM (GM-pre; CD244+) and erythroid (CD244−) precursors segregated by CD244 expression. Lin−CD34−244+CD117− cells are mature mixed GM cells (GM-mat).
CD34− AML LSCs express a disordered transcriptional profile with both HSPC and mature myeloid gene expression profiles
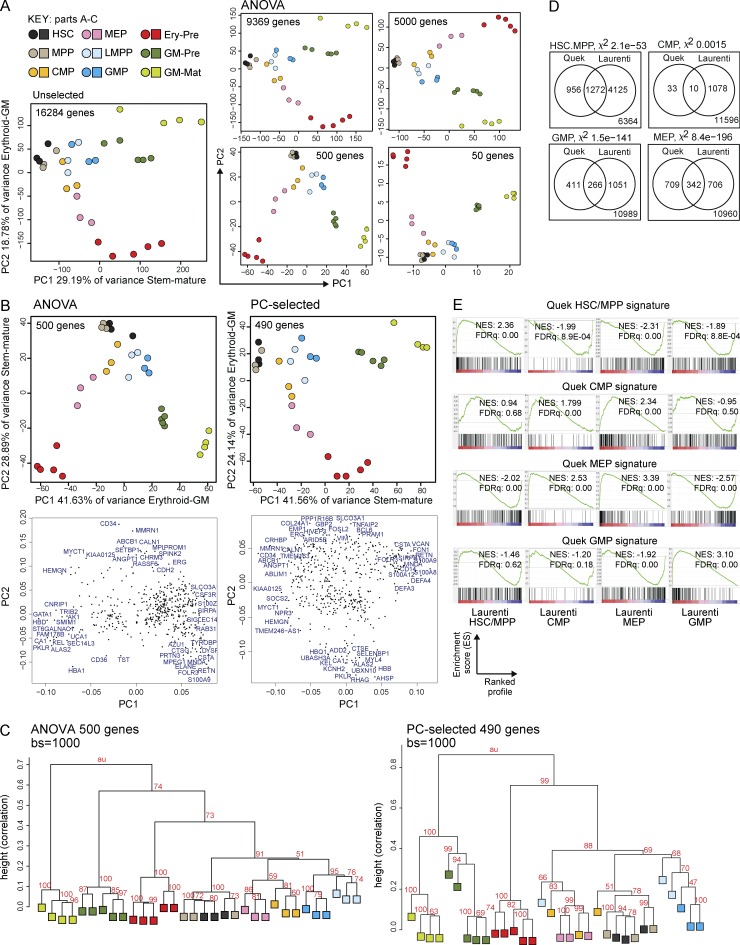
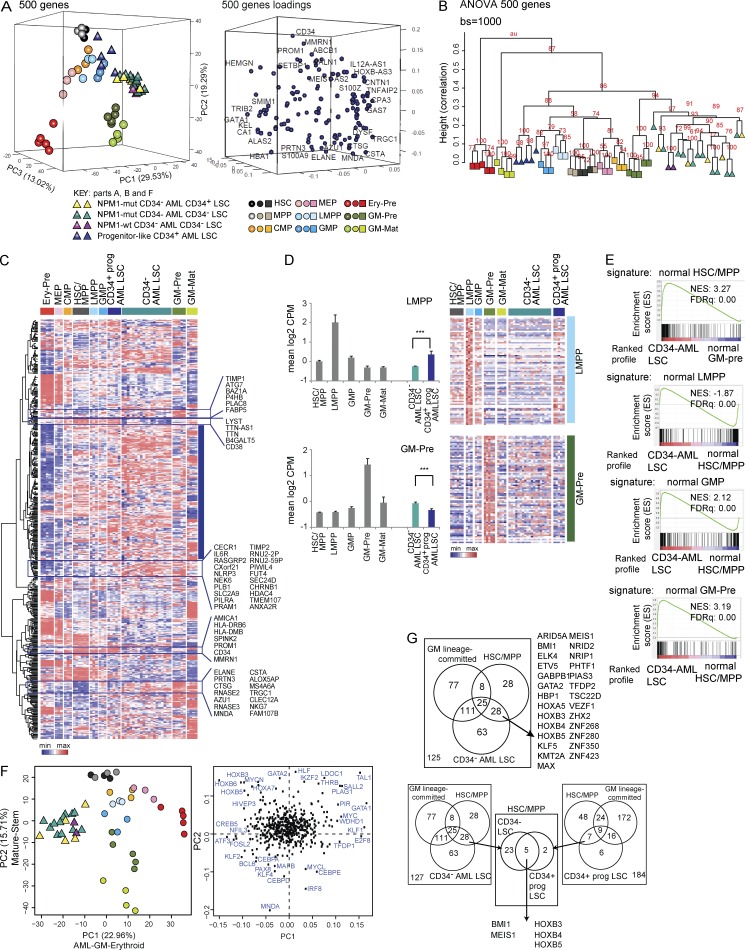
To understand where CD34− AML LSCs are arrested during hemopoietic differentiation, we compared global transcriptional programs of LSCs with purified normal HSPC and precursor/mature populations (Table S4 A) by RNA-seq. We first used all 16,284 expressed genes and performed two-dimensional principal component (PC) analysis (PCA; Fig. 4 A, left). This showed maturation of stem cells (HSCs)/MPPs through to more mature cells along PC1 and differentiation between GM and erythroid cells along PC2. This pattern of differentiation was confirmed by an independent ANOVA approach to identify 9,369 genes (P < 0.05) differentially expressed among all normal populations. PCA showed relationships between normal populations using between 50 and 9,369 differentially expressed genes ranked by p-value (Fig. 4 A, right). As might be expected, there is a greater dispersion of HSPC populations from different individual human BM donors when more genes are used. The 500 most differentially expressed genes (ANOVA 500) gave the best spatial segregation of normal populations and concordance between biological replicates by both PCA (Fig. 4 A and Table S4 B) and hierarchical clustering (unpublished data). To study how comparable the ANOVA 500 gene set is to genes that make the biggest contribution to PC1 and PC2 when using all genes, we selected 600 genes with the most extreme loading scores in PC1 and PC2. After removal of duplicates, 490 of the unique 571 genes had HUGO Gene Nomenclature Committee (HGNC) IDs. We call this the PC-selected gene set. Comparison of PCAs with ANOVA and PC-selected genes showed similar separation of hemopoietic populations with preservation of the stem–mature and erythroid–GM axes (Fig. 4 B). Hierarchical clustering using these two gene sets demonstrated greater donor effect and mixing of cell populations with the PC-selected 490 gene set (Fig. 4 C). As the ANOVA 500 gene set gave a more precise clustering, we used this to define transcriptional profiles of the normal populations for the remainder of the analyses (Fig. 4, D and E; and Fig. 5). To validate our normal expression profiles, we generated gene expression signatures for each normal population, including GM-pre and GM-mat cells, which had not previously been done. Signatures for HSPC populations were validated against published array signatures (Fig. 4, D and E; Laurenti et al., 2013).
Figure 4.
Gene expression of normal HSPC/precursor/mature populations by RNA-seq defines stem–mature and erythroid–GM differentiation axes in unbiased and selected gene sets. (A) PCA plots showing the position of different normal Lin−CD34+ HSPC (n = 3 donors), GM-pre (CD34−117+244+; n = 5 donors), GM-mat (CD34−117−244+; n = 4 donors), and erythroid precursor (Ery-pre; CD34−117+244−; n = 5 donors) populations using different numbers of genes: either all expressed genes (left), ANOVA-selected genes differentially expressed between the normal populations (P < 0.05), or 5,000, 500, or 50 top differentially expressed genes (ranked by ANOVA p-value). The percentage of variance represented by each PC is shown. (B) A comparison of the ANOVA 500 gene set with the PC-selected 490 gene set by two-dimensional PCA. The stem–mature and GM–erythroid axes represented by the PCs are shown together with percentage of variance. Loadings plots are shown directly below the corresponding PCA, showing a selection of genes with the most extreme loadings scores in each PC. (C) Hierarchical clustering of ANOVA 500 and PC-selected 490 gene sets using 1,000 bootstrap (bs) permutation analyses. au = approximate unbiased p-values, which indicate the probability that the cluster is supported by the data. 100 = 100% probability that the cluster is correct. Height values measure dissimilarity expressed as (1 − [correlation coefficient]). Correlation coefficient of 1 = complete correlation. (D) Validation of gene expression signatures from our data for HSCs/MPPs, GMPs, common myeloid progenitors (CMPs), and megakaryocyte-erythroid progenitors (MEPs) with the signatures from a recent publication (Laurenti et al., 2013). In each Venn diagram, the number of genes in our signature (Quek) shared with those from Laurenti et al. (2013) are shown together with the total number of expressed genes in the two gene sets (bottom right corner). The likelihood of having shared genes expressed in both datasets out of total expressed genes is estimated using a χ2 test (χ-value p-value is indicated). (E) GSEA plots showing the enrichment of our HSC/MPP, common myeloid progenitor, megakaryocyte-erythroid progenitor, and GMP signatures against profiles of expressed genes in the indicated populations using published data (Laurenti et al., 2013). Normalized enrichment scores (NES) and FDR q-values (FDRq) are shown.
Figure 5.
GM-pre are the closest normal counterparts to CD34− AML LSCs by gene expression and disorganized transcriptional landscape. (A, left) Three-dimensional PCA separating normal HSPC/precursor/mature and CD34+ AML and CD34− AML LSC populations using the ANOVA 500 gene set (adjusted p-value ≤1.79 × 10−28; Table S4 B). The percentage of variance for each component is indicated. The relationship between NPM1-mutated (mut) CD34+ and CD34− LSCs and NPM1 wild-type (wt) CD34− LSCs (all in CD34− AML) is indicated. (Right) The loadings plot annotated with a selection of most variant genes is shown. (B) Bootstrap hierarchical clustering of normal and LSC populations using the same ANOVA 500 gene set using 1,000 bootstrap (bs) permutations. (C) Heat map showing hierarchical clustering of ANOVA 500 genes and their expression by normal BM HSPC/precursor/mature, CD34+ AML, and CD34− AML LSC populations. Expression values are normalized per gene (by row) and are shown as a minimum–maximum scale (color bar). The blue block indicates genes with a scattered expression profile across normal, multiple normal, and LSC populations. The open rectangles highlight clusters of genes, with their gene names, which show preferential expression in GM-pre, GM-mat, and CD34− AML LSC populations and HSPC and CD34+ progenitor LSC populations. (D, right) Expression of the top 75 differentially expressed genes (rows = genes) in normal LMPPs and GM-pre populations in normal populations, CD34− AML, and CD34+ AML progenitor-like LSCs (indicated by colored horizontal bars; each column represents a different normal or LSC sample). Expression values are normalized per gene (by row) and are shown as a minimum–maximum scale (color bar). (Left) Mean expression of all 75 genes in each normal and LSC population. Triple asterisks indicate highly significant differences in mean expression of 75 genes between CD34− AML and CD34+ progenitor-like AML (P < 0.001, corrected for multiple testing) in a test of 105 permutations of 75 randomly selected genes (of 16,284 expressed genes). Error bars represent SD. (E) GSEA of normal BM HSC/MPP, LMPP, GMP, and GM-pre signatures in preranked profiles (based on mean fold change), comparing CD34− LSCs with normal GM-pre and CD34− LSCs with normal HSCs/MPPs (all others). Normalized enrichment scores (NES) and FDR q-values (FDRq) are shown. All comparisons were statistically significant. (F) PCA using a curated list of 547 TFs adapted from Novershtern et al., 2011, showing the position of normal and CD34− AML LSC populations, with the associated loading plot. (G, top) Venn diagram of the overlap in differentially up-regulated genes encoding TFs in normal HSCs/MPPs, GM lineage–committed populations (GMP, GM-pre, and GM-mat), and CD34− AML LSCs. 28 TFs (of which 27 have HGNC annotation) are up-regulated in both HSCs/MPPs and CD34− AML LSCs and are indicated. (Bottom) Combined analysis of the top Venn diagram with overlap in differentially up-regulated genes encoding TFs in normal HSCs/MPPs, GM lineage–committed populations, and CD34+ progenitor AML LSCs. Seven TFs are coexpressed in CD34+ progenitor AML LSCs and HSCs/MPPs. Of these, five (shown) are also expressed in CD34− AML LSCs. CMP, common myeloid progenitor; Ery-Pre, erythroid precursor; MEP, megakaryocyte-erythroid progenitor; prog, progenitor.
We then used the ANOVA 500 gene in a three-dimensional PCA but included expression data from CD34− AML LSCs (samples selected from those that serially transplant in immunodeficient mice are shown in Fig. 2). The stem–maturation (PC2) and erythroid–GM (PC1) axes were preserved (Fig. 5 A). CD34− AML LSCs were positioned between normal GM-pre and GM progenitors (GMPs). This was independent of CD34 expression and NPM1 mutation status (Fig. 5 A). In bootstrap hierarchical clustering, LSCs clustered closest to normal GM-pre (Fig. 5 B). In contrast, CD34+ progenitor-like LSC populations (samples from Goardon et al., 2011) segregated away from CD34− AML LSCs and were nearest to normal lymphoid-primed MPPs (LMPPs)/GMPs, as previously described (Fig. 5, A and B; Goardon et al., 2011).
Next, we looked at the expression of ANOVA 500 genes by hierarchically clustered heat maps in all normal populations, CD34− AML, and CD34+ progenitor LSC populations (Fig. 5 C). A large proportion of genes showed a scattered expression profile across normal HSCs/MPPs, LMPPs, GMPs, GM-pre, and GM-mat (Fig. 5 C, blue block) and both CD34− AML and CD34+ progenitor LSC populations. Only subsets of genes (Fig. 5 C, highlighted) showed preferential expression in GM-pre and GM-mat, CD34− AML LSC populations, and HSPC and CD34+ progenitor LSC populations.
Given this, we specifically studied the gene expression of signatures of normal LMPP and GM-pre populations and LSC populations (Fig. 5 D). CD34+ progenitor-like AML LSCs compared with CD34− AML LSCs showed higher expression of LMPP/GMP signatures. In contrast, CD34− AML LSCs had higher expression of the normal GM-pre signature. These differences are specific, as there was low expression of the GM-mat signature in LSCs of both AML subtypes (unpublished data).
Despite the close relationship between CD34− AML LSCs with normal GM-pre, we observed that CD34− AML LSCs are clearly distinct from normal populations. We hypothesized that the distinctive transcriptional profile of CD34− AML LSCs is a hybrid, capturing a GM-pre profile (indicative of a stage of differentiation arrest) and elements of an HSC/early progenitor profile (associated with self-renewal). Gene set enrichment analysis (GSEA) showed CD34− AML LSCs are enriched for a normal HSC/MPP signature compared with the normal GM-pre cells. Conversely, unlike normal HSCs/MPPs, CD34− AML LSCs are strongly enriched for genes expressed in normal GM-pre, with gradient enrichment for LMPP and GMP signatures (Fig. 5 E).
As CD34− AML LSCs express components of an HSC/early progenitor transcriptional program, we asked whether LSCs express HSPC transcription factors (TFs). Using 547 previously curated hemopoietic TFs (Novershtern et al., 2011), CD34− AML LSCs occupy a unique position distinct from normal populations, suggesting that disorganization of the normal TF landscape likely results in differentiation arrest at a CD34−117+ GM-pre–like compartment. HOX genes (HOXB5, B3, B6, and A7), oncogenic TF MYCN, and stress- and cytokine-induced TFs CREB5, ATF3, and NFIL3 (Fig. 5 F and Table S4 C) are likely to be key TF candidates mediating this disorganized landscape. In CD34− AML LSCs, 111 TFs are coexpressed with GM lineage populations and 28 TFs are coexpressed exclusively with HSCs/MPPs (Fig. 5 G). The 28 (a) include KMT2A (MLL), (b) select HOXA and HOXB MLL target genes (previously noted in NPM1-mutated human and mouse AML (Alcalay et al., 2005; Vassiliou et al., 2011; Spencer et al., 2015), and (c) include MEIS1, which cooperates with HOX genes in leukemogenesis (Wong et al., 2007), BMI1, and GATA2. All these TFs are associated with self-renewal, stemness, and development of AML. We then compared TF expressed in CD34+ progenitor LSCs, normal HSCs/MPPs, and GM lineage–committed cells (Fig. 5 G). Seven TF genes are coexpressed between normal HSCs/MPPs and CD34+ progenitor LSCs. Of these, five (BMI1, MEIS1, HOXB3, B4, and B5) are also expressed in CD34− AML LSCs. Recent data have pinpointed HOXB5 as a marker of long-term mouse HSCs (Chen et al., 2016). Collectively, CD34− AML LSCs express a hybrid stem-GM TF program. The data are consistent with diverse complements of AML mutations in CD34− AML directing a common aberrant HSC/MPP gene expression signature associated with self-renewal and/or block pathways that promote full differentiation. Furthermore, there is a core set of five TFs expressed in all AML LSCs and normal HSCs/MPPs.
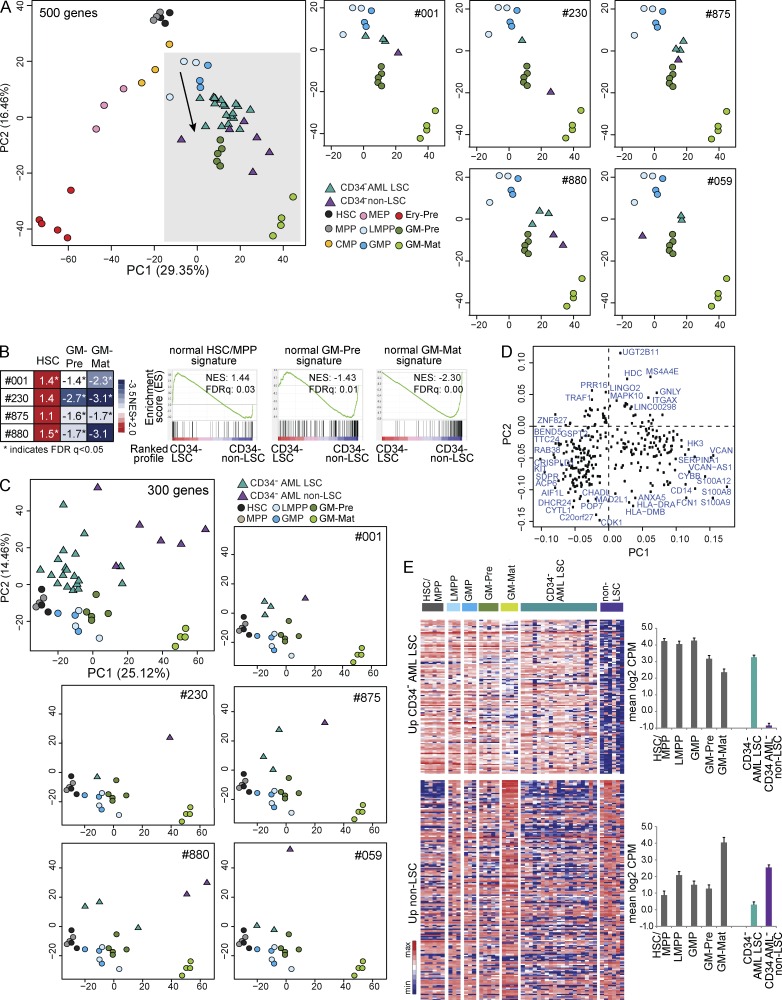
Next, we studied transcriptional differences between AML LSC and non-LSC populations (nonengrafting populations in Fig. 2, B and C). Using the ANOVA 500 gene set from Fig. 4, within any one patient, non-LSCs were positioned further down the axis of GM differentiation than LSCs; i.e., they were more differentiated (Fig. 6 A). Compared with non-LSC populations, CD34− AML LSC RNA profiles are significantly enriched for HSC/MPP signatures and depleted for GM-pre and GM-mat signatures (Fig. 6 B). Rank product analysis identified 125 genes differentially up-regulated in LSC populations and 175 genes more highly expressed in non-LSC populations (predicted false-positive [PFP] rate <0.05; Table S5). Using this 300-gene set in PCA, in PC1, normal HSPCs and a CD34− population of increasing GM maturation were arrayed from left to right (Fig. 6 C) with the loading plot confirming expression of genes in more mature GM cells on the right (Fig. 6 D). Within any individual patient, LSCs lie well to the left of non-LSCs, confirming LSCs are less differentiated. A heat map of the 125 genes more highly expressed in LSC populations shows a gradient of expression (high in HSPCs and low in GM-mat), whereas genes more highly expressed in non-LSCs are most highly expressed in GM-mat (Fig. 6 E). Thus, transcriptome analysis supports a model in which non-LSC populations are more differentiated than LSC populations.
Figure 6.
CD34− non-LSC populations are more differentiated compared with LSC populations. (A, left) PCA of normal populations and CD34− AML LSCs and non-LSC populations using the ANOVA 500 gene set (Fig. 4 A). The arrow shows the axis of normal myelomonocytic differentiation. (Right) PCA comparing LSCs and non-LSCs within five individual patients showing just the area shaded in gray on the main PCA plot on the left. CMP, common myeloid progenitor; Ery-Pre, erythroid precursor; MEP, megakaryocyte-erythroid progenitor. (B) Heat map of the normalized enrichment score (NES) from GSEA of normal HSC/MPP, GM-pre, and GM-mat signatures in profiles of LSC versus non-LSC populations in four patients (#001, #230, #875, and #880). Representative GSEA plots are from patient #001. (C) PCA showing the position of normal HSCs/MPPs, LMPPs, GM-pre, GM-mat, and CD34− AML LSCs and non-LSCs using 300 genes differentially expressed between CD34− LSCs and non-LSCs (from rank product analysis; PFP <0.05). (Top right and bottom) PCA of LSC and non-LSC populations in individual patients (#001, #059, #230, #875, and #880) analyzed with normal HSPC, GM-pre, and GM-mat cell populations. (D) Loadings plot of PCA in C. (E) Heat map of expression pattern of 125 genes with increased expression in CD34− AML LSC versus non-LSC populations (top) and 175 genes with increased expression in non-LSC versus CD34− AML LSC populations (bottom) in normal HSC/MPP, LMPP, GMP, GM-pre, and GM-mat populations. The color bar scale is as in Fig. 5 (B and D). (Right) Mean expression of the two gene sets in each population. Error bars represent SD. CPM, counts per million.
Clonal relationship of CD34+ and CD34− LSCs and expansion of LSC populations in CD34− AML
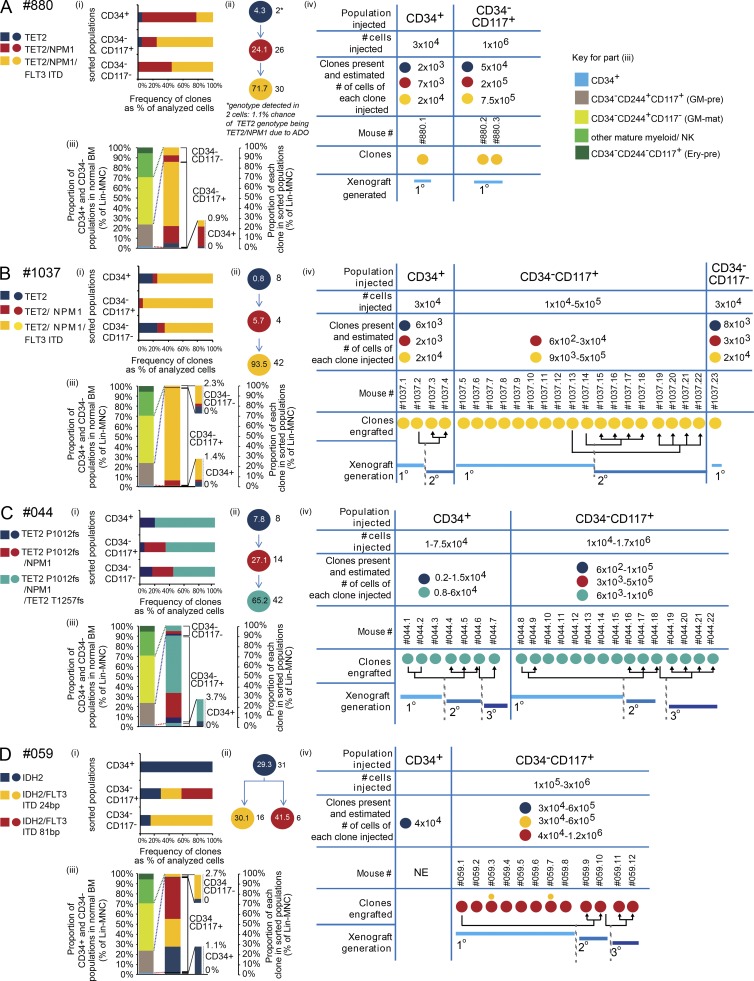
To determine whether clonal structures in CD34+ and CD34− populations varied and the size of each clone within the different leukemic hemopoietic compartments, we first performed exome sequencing (read depth of 30–100×) on all eight patient samples that engrafted. 48 high-confidence recurrent variants per sample were identified. On further filtering, this was reduced to 43 high-confidence AML-associated variants across all samples (Table S6, A–F). We then performed high-depth (100–10,000×) targeted resequencing of these 43 variants on populations and flow sorted single cells (Fig. 7, A–D, i; and Fig. 8, A and B, i). The details of the estimation of allele dropout (ADO) rates and determination of clonal structures (Fig. 7, A–D, ii; and Fig. 8, A and B, ii) are in the Estimation of false-negative homozygous reference calls in single-cell genotyping section of Materials and methods. We studied CD34+ and CD34−117± patient AML cells (Fig. 7, A–D, i–iii; and Fig. 8, A and B, i–iii) and AML cells from engrafted mice in six out of eight samples with sufficient material (Fig. 7, A–D, iv; Fig. 8, A and B, iv).
Figure 7.
Leukemic CD34+ and CD34− populations share clonal structures. (A–D) Partial differentiation arrest leads to expansion of an LSC-containing CD34−CD117+ population. (i) Clonal composition of FACS-purified populations using data from single-cell mutation–specific genotyping. (ii) Clonal hierarchy of AML based on single-cell genotyping. Numbers within the circles indicate the percentage that each clone comprises within a Lin− MNC population. Numbers outside the circles indicate the number of cells called with each genotype. The asterisk indicates genotypes that were detected in less than three cells where they were called with <99% confidence because of ADO. Genotypes detected in more than two cells could be called with 99% confidence. (iii) The proportions of normal BM CD34+, GM-pre (CD34−117+), GM-mat (CD34−117−), other myeloid mature, and NK cell populations and erythroid precursors (Ery-pre; CD34−244+117+) are shown on the left for comparison with that observed in CD34− AML (right), showing expansion or contraction of populations. Within each leukemic population, the contribution of each clone is shown using data from panel i. Data are also in Table S6 G. (iv) Numbers of cells of FACS-purified populations injected into immunodeficient mice, estimated number of cells of each clone based on results in panel iii, and type of leukemic clones engrafted in each mouse are shown. Where more than one clone is detected in a mouse, circle size denotes major and minor clones. Engrafted clones may be serially engrafted in subsequent secondary and tertiary transplants. NE, no engraftment. Data from four patients and the number of single cells analyzed per population are shown. (A) Patient #880: CD34+, n = 18; CD34−CD117+, n = 20; CD34−CD117−, n = 20. (B) Patient #1037: CD34+, n = 16; CD34−CD117+, n = 18; CD34−CD117−, n = 20. (C) Patient #044: CD34+, n = 18; CD34−CD117+, n = 32; CD34−CD117−, n = 18. (D) Patient #059: CD34+, n = 25; CD34−CD117+, n = 14; CD34−CD117−, n = 14.
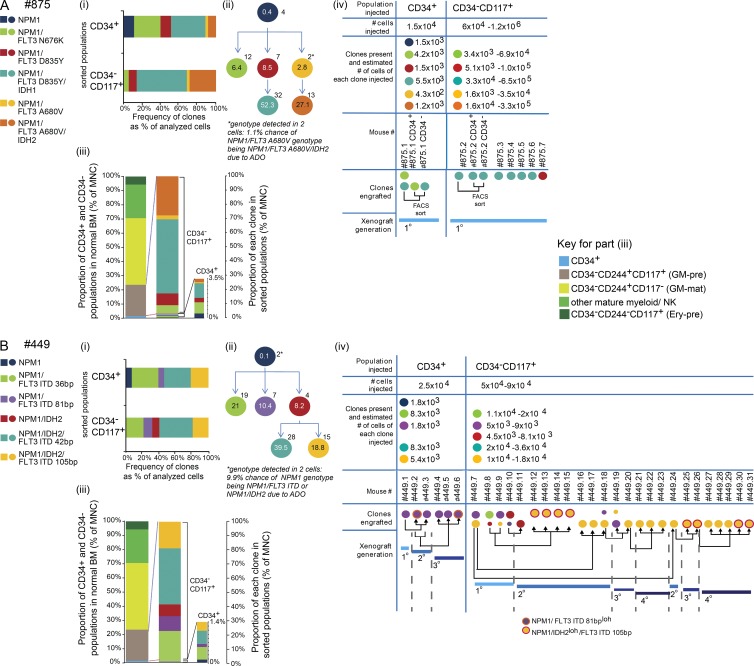
Figure 8.
Complex clonal hierarchies in samples where NPM1 mutation is acquired first also have expanded LSC-containing CD34−CD117+ populations. (A and B) NPM1-mutated clones underwent clonal selection and evolution in xenotransplanted mice. Data from two patients and the number of single cells analyzed per population. (A) Patient #875: CD34+, n = 35; CD34−CD117+, n = 35. (B) Patient #449: CD34+, n = 26; CD34−CD117+, n = 47. Panels i–iv are as in Fig. 7. Ery-pre, erythroid precursor; loh, loss of heterozygosity.
Initiating mutations were present in TET2 (Fig. 7, A–C, #880, #1037, and #044), IDH2 (Fig. 7 D), and NPM1 (Fig. 8, A and B). Mutations in all three genes have been previously shown to give preleukemia (Corces-Zimmerman et al., 2014; Genovese et al., 2014; Jaiswal et al., 2014; Shlush et al., 2014). We could not detect any AML-associated mutations at variant allele frequencies (VAFs) consistent with preleukemia in flow-sorted CD45+CD19+ B cells and CD45+CD3+ T cells available in five out of six samples (#044, #059, #875, #880, and #1037; unpublished data). Downstream of the initiating mutations, subclonal mutations were present in FLT3 (five out of six cases; Fig. 7, A, B, and D; and Fig. 8, A and B), NPM1 (three out of six cases; Fig. 7, A–C), IDH1/2 (two out of six cases; Fig. 8, A and B), and the second allele of TET2 (one out of six cases; Fig. 7 C). In all cases, acquisition of subclonal mutations led to clone expansion.
In five out of six cases, the clones in the CD34+ and CD34− populations were shared. The exception was sample #059, (Fig. 7 D) in which the IDH2 mutant clone accounted for the entire CD34+ compartment. This observation disproves model 3 (Fig. 1 A) as being generally applicable. Interestingly, the CD34+ compartment from #059 was the only CD34+ compartment of any sample that failed to engraft either leukemia or normal hemopoiesis.
By using the abundance of CD34+- and CD34−117±-sorted patient cells in Lin−MNCs (Table S6 G) and the frequency of each clone within a sorted population (Fig. 7, A–D, i; and Fig. 8, A and B, i), we calculated the sizes of all the clones within the entire Lin−MNC compartment (Fig. 7, A–D, ii; and Fig. 8, A and B, ii). In all samples, the terminal clones accounted for the bulk of the Lin−MNCs (58–93%). The genetic basis for different clone sizes was unclear, although in general, clones with more mutations were larger.
By studying the sizes of the CD34+ and CD34−117± populations, we established where leukemia caused differentiation arrest and determined the presence or absence of clones at different stages of leukemic differentiation and the size of clones at each stage of differentiation (Figs. 7 and 8, iii). In normal controls, CD34+ cells comprised 1–2% of Lin−MNCs, and the size of this population was similar (0.9–3.7%) in the leukemic samples. In contrast, the leukemic CD34−117+ population, regardless of CD244 expression, comprised 85–97% of Lin−MNCs (Table S6 G), whereas the normal counterpart population (GM-pre: CD34−244+117+, shown by RNA-seq; Fig. 4) represents on average only 22.6% of Lin−MNCs (Fig. 3 A). The approximately fourfold expansion of the leukemic CD34−117+ population occurred principally at the expense of more mature CD34−117− populations (which were reduced from 45% in normal BM [Fig. 3 A] to ≤14%), consistent with differentiation arrest in a CD34−117+ GM-pre–like compartment that is either complete or partial.
Finally, we analyzed the clonal structures in human AML cells from mice injected with either CD34+ or CD34− populations (Fig. 7, A–D, iv; and Fig. 8, A and B, iv), which defined the clonal composition of experimentally determined LSCs. In four out of six samples, the most abundant clones were most likely to engraft (Fig. 7, samples #880, #1037, #044, and #059). This likely reflects increased cell numbers of the injected terminal clone but does not exclude increased fitness of a terminal clone to engraft. For example, in sample #059 (Fig. 7 D), the majority of engrafted cells came only from one of the two FLT3 internal tandem duplication (ITD; 81 bp) clones. Interestingly, in contrast to the weakly engrafting 24-bp FLT3 ITD clone, the dominant engrafting 81-bp FLT3 ITD clone failed to differentiate into the most mature CD34−117− compartment. Here, complete differentiation arrest of the 81-bp clone may be associated with more potent self-renewal and thus experimental stem cell function. Exome sequencing did not reveal other coding mutations segregating with the 81-bp clone that could account for this behavior.
In two out of six samples (Fig. 8, A and B), experimentally determined LSC clonal architecture was more complex. The CD34+ or the CD34−117+ population from sample #875 (Fig. 8 A) resulted in engraftment of the largest clone (NPM1/FLT3 D835Y/IDH1 mutant). However, single mice engrafted with different minor clones (NPM1/FLT3 N676K and NPM1/FLT3 D835Y). Moreover, the second most abundant clone, NPM1/FLT3 A680V/IDH2, did not engraft. In sample #449 (Fig. 8 B), engrafting clones were either terminal clones (present at 10–40% of MNCs) or an intermediate NPM1/IDH2 clone. In the CD34+ population, only the minor terminal clone (NPM1/FLT3 ITD; 81 bp) engrafted. This clone underwent further evolution on serial transplantation with loss of heterozygosity of the FLT3 wild-type allele. Five clones were present in CD34−CD117+ cells. In primary transplants, four out of five of these engrafted (mice #449.7–449.10). With subsequent transplantation, there was both clonal selection (mice #449.16–449.24) and clonal evolution (mice #449.25–449.26 and #449.30–449.31) with loss of the wild-type IDH2 allele. Comparison of the clonal structure data from patients and engrafting mice consistently demonstrated that the engrafted cells did not recapitulate the entire clonal structure in the patient, providing evidence for different selective pressures for clonal expansion in man and mouse.
DISCUSSION
CD34− AML is genetically distinct and enriched for NPM1 and IDH1/2 mutations. Experimentally defined functional LSCs are present in the major CD34− and minor CD34+ fractions as noted previously (Martelli et al., 2010; Taussig et al., 2010). Our work extends prior work on CD34− AML by showing for the first time that the LSC frequencies are broadly equal in both compartments with the absence of hierarchy and, critically, are highly related transcriptionally. RNA-seq profiles of CD34+ and CD34− LSC populations differ only in the expression of eight genes, one of which is CD34. Thus, in contrast to normal hemopoiesis, CD34 expression is not a fixed immaturity-associated marker. Experimentally defined LSC function in the CD34− fraction is more commonly present in CD117+ cells. The CD34−117+ compartment is greatly expanded with the virtual disappearance of mature myeloid cells. Most functional LSC activity lies within this population. Concordantly, experimentally defined CD34+ and CD34− LSC populations show similar clonal structures, although the preleukemic clone can be enriched or even exclusively present in the CD34+ compartment.
A key problem in cancer biology is defining the stage in normal differentiation where tumor-propagating cells are partially or completely arrested. The key novel finding here is that by using expression signatures of normal HSPC and GM-mat populations, we showed that self-renewing CD34− AML LSCs, regardless of genetic mutation profile, are arrested very late in hemopoiesis, at a GM-pre stage, before terminal maturation. This is in contrast to LSCs in CD34+ progenitor AML (Goardon et al., 2011).
The refined comparison of gene expression profiles between normal HSPCs, GM-pre cells, GM-mat cells, and CD34− AML LSCs shows that CD34− AML LSCs express a small subset of HSC/early progenitor genes, including 28 TFs normally expressed in normal HSCs/MPPs but not in normal GM-pre and GMP cells with oncogenic activity and implicated in stem cell function and myeloid leukemia, including mouse models of Npm1-mutated myeloproliferative disease (Vassiliou et al., 2011) and Mycn-induced AML (Kawagoe et al., 2007). Furthermore, there is a core of only five normal HSC TFs expressed in both CD34+ progenitor LSCs and CD34− GM-pre LSCs.
Collectively, these findings suggest the cellular hierarchy in CD34− AML best fits model 2 in Fig. 1 A, where the sum of genetic and epigenetic changes leads to partial differentiation arrest and marked expansion at a late GM-pre stage of hemopoiesis where cells acquire LSC function, and then LSCs further differentiate into non-LSCs.
LSCs have extensive self-renewal, proliferate, and fail to terminally differentiate. These properties could arise by different means. For example, LSCs could have been most similar to HSCs with extensive self-renewal. However, as HSCs are normally quiescent, oncogenic mechanisms would have to provide proliferative drive and cause differentiation arrest. Alternatively, LSCs could be similar to progenitors/precursors, allowing LSCs to use the proliferative programs intrinsic to these normal populations but requiring oncogenic changes to impart sustained self-renewal and differentiation arrest. Our current and previous observations (Goardon et al., 2011) are compatible with the latter hypothesis. Further work will determine whether aberrant sustained self-renewal is mediated by the core set of five HSC/early progenitor TFs shared between normal HSCs/MPPs, CD34+ progenitor AML (Goardon et al., 2011), and CD34− GM-pre AML identified here, namely, HOXB3, B4 (Sauvageau et al., 1995; Antonchuk et al., 2002; Kyba et al., 2002; Oshima et al., 2011), B5 (Chen et al., 2016), MEIS1 (Ariki et al., 2014), and BMI1 (Lessard and Sauvageau, 2003; Park et al., 2003; Iwama et al., 2004). However, it is also possible that some of these five TFs may also impose differentiation arrest, for example MEIS1 (Wong et al., 2007), possibly working with HOX and PBX (Ficara et al., 2013) partner proteins.
These studies also show that the disease-initiating clone is usually different in a human patient compared with that engrafting an immunodeficient mouse. A disease-initiating clone in a human is the clone with a first or disease-initiating mutation. This creates a preleukemic rather than leukemic state. When primary human AML cells are transplanted into a mouse, the clones that engraft are termed disease initiating in the mouse. These clones are usually fully transformed leukemic cells with multiple mutations.
How do the clonal structures in our six cases of CD34− AML compare with those previously published? Single cell–derived clonal structure data are available in only seven cases of primary human AML (Jan et al., 2012; Corces-Zimmerman et al., 2014; Klco et al., 2014). Additional population-based sequencing of preleukemic and leukemic compartments is available on another 20 samples (Jan et al., 2012; Corces-Zimmerman et al., 2014; Shlush et al., 2014). Population-based data alone do not establish the precise order of mutation acquisition but separate preleukemic from leukemic mutations. Within the limitations of the small datasets, what conclusions can be drawn? In our dataset, as in other AML cases, the initiating (or founder) mutations occur in TET2, NPM1, and IDH2. Combining all 27 cases (7 with single-cell data and 20 samples with preleukemic and leukemic data), NPM1 mutation was more commonly seen in leukemic as opposed to preleukemic cells (19/22 cases). More common preleukemic mutations were in DNMT3A (11/15 cases), IDH1/2 (13/16 cases), and TET2 (2/2 cases). Subclonal acquisition of mutations in NPM1, IDH1/2, and TET2 (the second allele), documented here in CD34− AML, has also been seen in other AML samples (Jan et al., 2012; Corces-Zimmerman et al., 2014; Klco et al., 2014; Shlush et al., 2014). Both in our dataset and in the published dataset, FLT3 mutations are universally acquired as subclonal late events. In our samples, FLT3 ITD is associated with the greatest extent of leukemic expansion, predominantly in the CD34− GM-pre–like compartment. In summary, there are similarities in clonal structure between CD34− AML and other AMLs. The obvious difference is reduced frequency of the DNMT3A mutation in our sample set and its absence in samples in which we performed detailed studies. More generally, although some mutations are more commonly seen in a preleukemic phase and others in a leukemic phase, there is flexibility in the precise order in which mutations in most genes are acquired.
Clonal heterogeneity may also provide insight into how accurately widely used immunodeficient mice model clonal competition and stem cell function in patients. In three out of four samples studied (#059, 449, and 875), our data showed that only a subset of clones with LSC potential in the patient was captured in mice. This mirrors the only other recent publication in which engrafting potential did not match clonal composition in the patient (Klco et al., 2014). Moreover, the more frequent patient subclones are not necessarily the ones that engraft in mice. Finally, clonal competition in mice (sample #449) did not mirror clone fitness in the patient.
A related critical and as yet unanswered question is what are the genetic and epigenetic cell-autonomous determinants that dictate functional differences between clones with respect to achieving clonal dominance in a patient (or experimental in vivo models)? The type of analysis performed here will likely have to be extended with single-cell whole-genome sequencing, clonal epigenetic profiling, and single-cell RNA-seq to establish the full set of genetic and epigenetic differences between clones that could impact stem cell function in a patient and experimentally in vivo. The power of this type of analysis in sequential samples through therapy (and into relapse) will also provide an insight into the selective pressures imposed by therapy and therapy failure. Finally, clonal structure trees coupled with functional interrogation of clonal potential open the door to genome-editing strategies to identify mutations required to sustain common clones and thus help define attractive therapeutic targets. These studies in AML will continue to provide an exemplar for similar studies in other cancers.
MATERIALS AND METHODS
Patient samples
BM or blood samples from normal donors and AML patients were obtained with informed consent and collected by research ethics committee–approved Biobanks (MDSBio Study, MREC 06/Q1606/110; Oxford Musculoskeletal Biobank, MREC 09/H0606/11: South Central Oxford C Research Ethics Committee) and the AML17 clinical trial (MREC 08/MRE09/29: Wales Research Ethics Committee). Karyotype and fluorescent in situ hybridization were carried out by hospital laboratories. MNCs were isolated by ficoll density gradient. In normal BM samples, CD34+ cells were purified using a CD34 Microbead kit and magnetic-activated cell-sorting separation columns (Miltenyi Biotec). Unseparated CD34+- and CD34-deplete fractions were frozen in 90% FCS/10% DMSO, stored in liquid nitrogen, and subsequently thawed on the day of the experiment.
Xenograft assay and LSC frequency assays
10–14-wk-old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ), NRG (NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ), or NSGS (NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg[CMV-IL3,CSF2,KITLG]1Eav/MloySzJ; The Jackson Laboratory) mice were irradiated twice with 100 cGy 4 h apart. Mice were injected intraperitoneally with 10% human immunoglobulin solution (1 mg/g of body weight; Privigen; CSL Behring) immediately after the second irradiation. Cells were administered via intratibial injection within 24 h of the second irradiation. Human CD45+33+19− (myeloid) or human CD45+33−19+ (B lymphoid) engraftment was analyzed by FACS and defined as ≥0.1% of live MNC gate. Experiments were performed in accordance with UK government–approved project license 30/2465. Engraftment was monitored by blood or marrow sampling from 12 wk after transplant, and mice were culled for cell harvesting between 12 and 28 wk. Experiments were performed in accordance with UK government–approved project license 30/2465.
Calculating the frequency of LSCs
Percentages of sorted subpopulations of each sample used in limit dilution assay experiments were expressed as a percentage of live Lin−MNCs. The 95% confidence intervals for stem cell frequency were calculated using L-Calc software (STEMCELL Technologies) based on a Poisson distribution.
FACS protocols
FACS analysis was performed on either a Cyan ADP (Dakocytomation) or LSR Fortessa flow cytometer (BD). Flow sorts were performed on an Aria III SORP (BD). Antibodies used for depletion of cells expressing lineage markers; purification of normal CD34+ HSPCs, CD34− cells, CD34− AML samples, and CD34+ progenitor AML samples; mouse engraftment analysis; and MS5 co-culture experiments (MS5) are included in the the next paragraph.
Antibodies used in the lineage depletion cocktail for purification of (a) CD34− normal and CD34− AML samples were anti-CD2, anti-CD3, anti-CD4, anti-CD8a, anti-CD10, anti-CD19, anti-CD20, and anti-CD235a and of (b) CD34+ normal BM and CD34+ AML samples were anti-CD2, anti-CD3, anti-CD4, anti-CD7, anti-CD8a, anti-CD11b, anti-CD14, anti-CD19, anti-CD20, anti-CD56, and anti-CD235a. Normal CD34+ stem/progenitor populations and CD34+ AML samples were analyzed and sorted using lineage depletion and antibodies to CD34, CD38, CD90, CD123, and CD45RA. Normal CD34− BM and CD34− AML samples were lineage depleted using magnetic bead lineage depletion (anti–mouse IgG microbeads; Miltenyi Biotec) before analysis and sorted using antibodies to CD34, CD150, CD48, CD244, and CD117. Normal subpopulations were analyzed for lineage affiliation using antibodies to CD11b, CD14, CD15, and CD71. Engraftment was assayed using antibodies to human CD45, CD19, and CD33. BM harvested from engrafted mice was analyzed and sorted using antibodies to human CD45, CD33, CD19, CD34, CD150, CD48, CD244, and CD117. See Fig. S1 for FACS gating strategies.
In vitro myeloid/B lymphoid and NK cell differentiation assays
5 × 104 MS5 stromal cells/well were plated onto gelatinized 24-well plates in MEM-α (Thermo Fisher Scientific) with 10% FCS (Thermo Fisher Scientific), 10 ng/ml thrombopoietin, FLT3-ligand, granulocyte CSF, IL-2, IL-15, 20 ng/ml CSF (PeproTech), and 10−7 M DUP697 (Cayman Chemical) 12 h before addition of 200 flow-sorted marrow cells. The medium was half-changed weekly. Harvested cells were FACS analyzed at D7, D14, and D28.
Colony assays
For progenitor colony–forming assays, 300 cells were plated in duplicate in 1.2 ml MethoCult GFH4435 (STEMCELL Technologies). The rest of the procedure was performed as previously described (Goardon et al., 2011).
Gene expression by dynamic arrays
500 cells (>99% purity) were FACS sorted into 96-well plates with 10 µl of reaction buffer (5 µl of 2× reaction mix [CellsDirect; Thermo Fisher Scientific], 1 µl RT/Taq Mix [CellsDirect], 0.4 µl of water, 0.1 µl RNase inhibitor [SUPERase-In; Thermo Fisher Scientific], and 2.5 µl of a mix of 0.2× TaqMan gene expression assays [Thermo Fisher Scientific]). Reverse transcription and specific target preamplification conditions were 15 min at 50°C, 2 min at 95°C, 22 cycles of 15 s at 95°C, and 4 min at 60°C. Preamplified samples were diluted 1:4 and analyzed on a 48.48 dynamic array (Fluidigm). PCR cycling conditions were 10 min at 95°C, 40 cycles of 15 s at 95°C, and 60 s at 60°C. All reactions were performed in three technical replicates. Data were analyzed using the ΔCt (cycle threshold) method; results were normalized to GAPDH expression and expressed as the mean expression level relative to GAPDH. TaqMan assays were obtained from Thermo Fisher Scientific: GAPDH (Hs02758991_g1), ELANE (Hs00975994_g1), EPOR (Hs00959427_m1), MPO (Hs00924296_m1), PF4 (Hs00427220_g1), SPI1 (Hs02786711_m1), SPTB (Hs01024103_m1), CSF3R (Hs00167918_m1), ITGAM (Hs00355885_m1), and IL3RA (Hs00608141_m1).
Nucleic acid manipulation
DNA extraction was performed using a DNeasy Blood and Tissue extraction kit (69506), and RNA extraction was performed using an RNeasy Micro kit (74004; QIAGEN). Whole-genome amplification (WGA) was performed using 3–10 ng of extracted genomic DNA or 3 × 103–104 sorted AML cells using an amplification kit (Illustra GenomiPhiV2; GE Healthcare). Nucleic acids were analyzed and quantified using Qubit assay (Invitrogen) or the appropriate Bioanalyser chip (Agilent Technologies).
Exome sequencing
Libraries for whole-exome sequencing (WES) were prepared using either a SeqCap EZ (v3; Nimblegen) or a SureSelect All Exon Enrichment kit (V5; Agilent Technologies) and sequenced on HiSeq2000 (Illumina) using 100-bp paired-end reads. Raw reads were mapped using a Burrows-Wheeler aligner (Stampy) to the NCBI Human Reference Genome Build hg19 (Lunter and Goodson, 2011). Aligned reads that passed standard quality filters were submitted for variant calling using VarScan2 (Koboldt et al., 2009) for both single nucleotide variants (SNVs) and indels. Variants were annotated with Annovar.
Targeted resequencing
Libraries were created from pooled, indexed amplicons generated using Fluidigm Access Array Primers (see online supplemental material) using Fluidigm Access Array. 500 amplicons were multiplexed per gDNA sample and sequenced on MiSeq (Illumina) using 150-bp or 250-bp paired-end reads. Reads were mapped using Stampy onto hg19. Aligned reads that passed standard quality filters were submitted for variant calling using VarScan2 for both SNVs and indels, and Pindel (Spencer et al., 2013) was used to optimize detection of longer indels, e.g., ITD in FLT3. Variants were annotated with Annovar software.
Mutation detection pipeline
Putative variants were filtered using the following parameters: (a) WES: minimum coverage, 15; minimum variant frequency, 0.01; minimum read depth in variant, 2; and p-value of 0.05; and (b) targeted resequencing: minimum coverage, 100; minimum variant frequency, 0.01; minimum read depth in variant, 5; and p-value of 0.05. As CEBPA had low coverage in next generation sequencing, data mutation analysis for this gene was performed separately (see the CEBPA mutation analysis section).
In both WES and targeted resequencing datasets, additional exclusion criteria for variants were (a) variants predicted to result in a silent amino acid change; (b) known polymorphisms present in human variation databases (Exome Sequencing Project 6500; 1,000 genomes; SNP database) at a population frequency >0.0014 (reflecting the population incidence of myeloid disease) except for when the variant was a known somatic variant in the Catalogue of Somatic Mutations in Cancer (COSMIC); (c) variants present in at least one of the two T cell germline controls with a VAF >10; (d) variants noted to be rare in hematopoietic disease (documented in less than two hematopoietic samples in COSMIC) but which occurred in more than one of our AML cohort samples; and (e) 1-bp indels present adjacent to regions of more than four homopolymer bases. Variants were included on the basis of (a) previous documentation as a somatic mutation in hematopoietic samples in COSMIC or (b) novel variants in ASXL1 or TET2 genes (not previously reported), which have high-impact structural change (nonsense, deleterious missense, and indels) not found in either T cell germline controls or in human variation databases with a frequency >0.0014. Putative variants were further validated by visualization using the Integrated Genome Viewer.
NPM1 quantitative PCR from genomic DNA
To investigate engraftment of bulk and sorted populations from NPM1 mutant AML samples, genomic DNA was extracted from mouse blood or BM (bulk or sorted populations) and subjected to quantitative PCR. Reactions were performed in triplicate and run on the quantitative PCR platform (ABI7900; Applied Biosystems) using standard conditions. Albumin gene (ALB; human and mouse) and NPM1 exon 12 assays (mutant allele and wild-type control) were used to assess the percentage of engraftment, calculated using the ΔCt method, as described previously (Jovanovic et al., 2013). Primers and probe sequences for NPM1 have been previously published (Gorello et al., 2006; Chou et al., 2007): NPM1 common forward, 5′-GTGTTGTGGTTCCTTAACCACAT-3′; NPM1 mutation A reverse, 5′-CTTCCTCCACTGCCAGACAGA-3′; NPM1 mutation D reverse, 5′-CCTCCACTGCCAGGCAGA-3′; NPM1 mutation I reverse, 5′-CTTCCTCCACTGCCTTACAGAGA-3′; NPM1 control reverse, 5′-TCTTAAAGAGACTTCCTCCACTGC-3′; and NPM1 common probe (FAM/minor groove binder), 5′-TTTTCCAGGCTATTCAAGAT-3′.
CEBPA mutation analysis
The entire coding sequence of the CEBPA gene was amplified by PCR in three overlapping fragments, and the products were analyzed by WASP-family verprolin homologous protein (WAVE)–denaturing high-performance liquid chromatography (Transgenomic) as previously described (Green et al., 2010).
Single-cell genotyping
Single cells were flow sorted into 96-well plates containing 4 µl phosphate-buffered saline. WGA was performed using the Single Cell RepliG kit (QIAGEN). In brief, after cell lysis, alkali denaturation, and neutralization, a master mix containing Phi29 polymerase, deoxyribose nucleoside triphosphate, and random oligonucleotide primers was added. WGA was performed at 30°C for 8 h followed by heat inactivation. Diluted (1:20) amplified DNA was used in single-plex PCR with an Access Array system (Fluidigm) using primers relevant to the sample. Barcoding and sequencing oligonucleotides were added by PCR, and sequencing was performed on MiSeq (Illumina). Raw sequences were aligned on BaseSpace using ISAAC (Raczy et al., 2013). Approximately 94% of reads had Phred scores >30. A threshold of 100 reads was set for analysis inclusion, except for NPM1 exon 12, where a threshold of 1,000 reads was used to improve specificity.
VAF estimation of FLT3 ITD and other mutations in bulk cells and single cells
For estimation of VAF of FLT3 ITD sequences in AML samples, we used Pindel (Spencer et al., 2013). We searched fastq files for 20-bp sequences specific for the ITD or unaffected by the ITD. The VAF of the FLT3 ITD was the number of identified ITD sequences/reference sequences. The same approach was used for single-cell data. For VAF estimation of known SNVs in single-cell data, we took a similar approach by creating 20-bp reference and mutation-specific sequences.
Establishing thresholds for detection of variants in single-cell data
Thresholds for detecting a mutation and false-positive rates for each mutation were established by analyzing single cells from samples that were wild type for mutations of interest, for example, sample #449, #1037, and #059, which are all wild type for FLT3 D835Y, A680V, and N676K.
Estimation of false-negative homozygous reference calls in single-cell genotyping
A major issue in single-cell genotyping is ADO, where one allele fails to amplify. This can result in a heterozygous allele being called homozygous. The phenomenon of ADO is well known to occur when performing DNA amplification from low quantities of starting material. Previous studies performing single-cell genotyping have estimated ADO rates of 4–15% (Treff et al., 2011; Hou et al., 2012) using RepliG. To calculate the ADO rate, we performed the following experiments. Heterozygous and homozygous SNVs (synonymous or nonsynonymous) were identified in AML-bulk genomic DNA. These SNVs were then genotyped in single cells. In 80/760 amplicons (i.e., a frequency of 0.105 or 10.5%), heterozygous SNVs were erroneously called homozygous reference alleles in single-cell genotyping. This means that in a population in which two mutations have been called (mutations A and B), there is a 10.5% chance that a single cell in which only a single mutation (e.g., mutation A) is detected may actually have both mutations (mutations A and B). Furthermore, if there are four mutations in the population (mutations A, B, C, and D), the chance that a single cell in which only mutation A is detected but actually has two mutations is 3 × 10.5% = 31.5%.
Where multiple cells are found with only one mutation, the chance that none of the cells are truly of a single mutant clone is (10.5%)n, where n is the number of cells with only a single mutation. For any single cell, the ADO for the full complement of mutations present in that cell depends on the total informative SNVs (and thus the number of amplicons used for genotyping). Thus, the predicted rate at which any single cell would have ADO events in two different loci would be (0.105)2 = 0.011 (or 1.1%). The false-negative rate of 10.5% per amplicon was used to estimate the likelihood of the presence or absence of small clones.
Determination of clonal structures
Each single cell was assessed for detection or nondetection of mutations relevant to that patient sample. To be included for analysis, each cell had ≥100× coverage of all relevant amplicons. We assigned the most likely sequence of acquisition of mutations based on the genotype identified in cells. For example, where mutations A–E were identified in a sample, discrete cells with genotypes A, AB, ABC, ABD, ABCD, and AE may be called. In most cases, the sequence of acquisition, e.g., A→AB, is clear. However, the sequence of acquisition of mutations during the transition, for example, from AB to ABCD may not be clear because of ADO, i.e., the sequence of acquisition may be AB→ABC→ABCD or AB→ABD→ABCD. In such cases, intermediate genotypes represented by the most cells will be more likely to be true. In all cases, models of clonal structures, which require the least number of discrete mutational steps required, will be represented (Potter et al., 2013), although we accept that alternative structures, including ones where the same mutation is acquired twice, are possible.
Analysis of raw RNA-seq data
Duplicates were removed, and reads were filtered for uniquely mapping reads (MAPQ >3) using SAMtools (version 0.1.19; Li et al., 2009). Data analyses were performed using the R software environment for statistical computing (version 3.0.1). Gene-level read summarization was performed using the R package genomic ranges, and gene expression analysis was conducted using edgeR (McCarthy et al., 2012). For read counts and PCAs, read counts were normalized as counts per million (cpm) and log2 transformed. Sequencing data were submitted to ArrayExpress (accession no. E-MTAB-2672).
Analysis of gene expression data
We used a filtering strategy to eliminate nonexpressed or only marginally expressed genes from the 59,689 genes defined in Ensembl. We retained the genes that had a cpm >2 in at least half of the samples of at least one of the experimental conditions considered. Thus, we retained 16,284 genes, of which 13,461 had an HGNC annotation, for further analysis.
We generated gene expression profiles by computing differential gene expression. Our experimental design included comparisons of (a) populations using an ANOVA approach, (b) populations using a selection approach based on genes with the greatest contribution to variance in PCA of total genes (PC-selected), (c) selected populations against other selected populations (e.g., CD34− AML LSCs vs. non-LSCs using rank product analysis), and (d) selected populations against the mean of the remaining populations (e.g., to create expression signatures). Genes present in more than one signature were removed.
Differential gene expression was computed using generalized linear models. Where appropriate, we included the donor as an additive covariate to correct for donor-specific effects. We calculated the log2-fold changes, p-values of differential expression, and the false discovery rate (FDR)–adjusted p-values of differential expression of all genes in all the profiles.
PCA and permutation cluster analysis was performed using Pvclust, with all ANOVA-selected genes (9,369 genes; FDR <0.05) and then the top 5,000, 500, and 50 genes with normal stem, progenitor, precursor, and mature populations. For permutation analysis, we performed analysis with up to 10,000 bootstrap samplings but observed no further change in clustering after 1,000 samplings. We selected the 500-gene set (corresponding to genes with p-values <2.28 E-30) for further analysis of combined data from normal and AML samples. We also performed heat map hierarchical clustering using Hclust.
As set out in Fig. 5 D, the statistical significance of differences in mean expression between CD34− AML LSCs, CD34+ progenitor-like LSCs, and normal populations in terms of normal LMPP and GM-pre signatures was established by permutation testing. Fold-change values of 75 randomly selected genes (from the gene set of 16,284 genes) were assessed over 100,000 samplings and compared with the observed expression difference. The p-values of these comparisons were <0.001 (corrected for multiple testing), indicating that the differences between the overall expression of the 75-gene sets between CD34−AML LSCs and CD34+ progenitor LSCs were highly significant.
GSEA
We used GSEA (Subramanian et al., 2005) to test enrichment of specific population expression signatures in expression profiles. Population-specific signatures were computed from a subset of the gene expression values described in the previous section by selecting genes that are up-regulated with an FDR-adjusted p-value <0.05. The Ensembl gene identifiers of these genes were translated into HGNC symbols, and the 250 genes with the highest fold changes were selected as gene sets for GSEA.
Comparison with published datasets
To estimate the concordance of our population-specific gene signatures (see the previous section at FDR-adjusted p-value <0.05) with published data from equivalent published cell populations (ArrayExpress accession no. E-MTAB-2672; Laurenti et al., 2013), we conducted GSEA with our gene signatures against the Laurenti profiles.
Online supplemental material
Fig. S1 shows FACS analysis and sorting strategies. Table S1 shows data on immunophenotype and karyotypic and genetic analysis of CD34− AML samples. Recurrent nucleotide variants detected by targeted resequencing and/or WES are filtered according to parameters set out in the Mutation detection pipeline section. Table S2 shows FACS data of xenotransplantation assays. Table S3 shows sort purities of normal BM MNC fractions used for in vitro culture assays. Table S4 shows RNA-seq analysis of normal BM and CD34− AML samples. Table S5 lists 300 significantly differentially expressed genes between CD34− LSC and non-LSC populations analyzed by rank product analysis (PFP <0.05). Table S6 contains data from bulk- and single-cell genotyping. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151775/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients and staff in the National Cancer Research Network (NCRN) MDSBio study for samples, the UK NCRN AML Working Party and Genomics facility at the Wellcome Trust Center for Human Genetics, Oxford, and Drs. Simon McGowan, Supat Thongjuea, and Emmanouela Repapi for discussion on data analysis methods.
L. Quek was supported by a Medical Research Council (MRC) Disease Team Award. P. Vyas acknowledges funding from the MRC Disease Team Award (G1000729), the MRC Molecular Haematology Unit (MC_UU_12009/11), the Oxford Partnership Comprehensive Biomedical Research Centre (National Institute for Health Research [NIHR] BRC funding scheme), and Cancer Research UK (C7893/A12796). A. Price acknowledges funding from the Oxford NIHR Musculoskeletal Biomedical Research Unit. D. Grimwade acknowledges the NIHR for its program grants for the Applied Research Program (RP-PG-0108-10093).
The views expressed are those of the authors and not necessarily those of the National Health Service, the NIHR, or the Department of Health.
The authors declare no competing financial interests.
Author contributions: L. Quek designed and performed experiments, analyzed data, and wrote the paper. G.W. Otto performed bioinformatic/statistical analysis. C. Garnett, L. Lhermitte, D. Karamitros, B. Stoilova, I-J. Lau, A. Kennedy, M. Metzner, N. Goardon, A. Ivey, C. Allen, R. Gale, D. Grimwade, and C. Porcher performed experiments and/or analyzed data. B. Usukhbayar and J. Doondeea prepared samples. B. Davies, A. Sternberg, S. Killick, H. Hunter, P. Cahalin, A. Price, A. Carr, M. Griffiths, P. Virgo, S. Mackinnon, S. Freeman, N. Russell, C. Craddock, A. Mead, and A. Peniket provided reagents/samples/clinical data. P. Vyas designed experiments, analyzed data, and wrote the paper. All authors edited the manuscript.
Footnotes
Abbreviations used:
- ADO
- allele dropout
- AML
- acute myeloid leukemia
- cpm
- counts per million
- FDR
- false discovery rate
- GM
- granulocyte–macrophage
- GMP
- GM progenitor
- GSEA
- gene set enrichment analysis
- HSC
- hemopoietic stem cell
- HSPC
- hemopoietic stem/progenitor cell
- ITD
- internal tandem duplication
- LMPP
- lymphoid-primed MPP
- LSC
- leukemic stem cell
- MPP
- multipotent progenitor
- MNC
- mononuclear cell
- PC
- principal component
- PCA
- PC analysis
- PFP
- predicted false positive
- SNV
- single nucleotide variant
- TF
- transcription factor
- VAF
- variant allele frequency
- WES
- whole-exome sequencing
- WGA
- whole-genome amplification
References
- Alcalay M., Tiacci E., Bergomas R., Bigerna B., Venturini E., Minardi S.P., Meani N., Diverio D., Bernard L., Tizzoni L., et al. 2005. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 106:899–902. 10.1182/blood-2005-02-0560 [DOI] [PubMed] [Google Scholar]
- Anjos-Afonso F., Currie E., Palmer H.G., Foster K.E., Taussig D.C., and Bonnet D.. 2013. CD34− cells at the apex of the human hematopoietic stem cell hierarchy have distinctive cellular and molecular signatures. Cell Stem Cell. 13:161–174. 10.1016/j.stem.2013.05.025 [DOI] [PubMed] [Google Scholar]
- Antonchuk J., Sauvageau G., and Humphries R.K.. 2002. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 109:39–45. 10.1016/S0092-8674(02)00697-9 [DOI] [PubMed] [Google Scholar]
- Ariki R., Morikawa S., Mabuchi Y., Suzuki S., Nakatake M., Yoshioka K., Hidano S., Nakauchi H., Matsuzaki Y., Nakamura T., and Goitsuka R.. 2014. Homeodomain transcription factor Meis1 is a critical regulator of adult bone marrow hematopoiesis. PLoS One. 9:e87646 10.1371/journal.pone.0087646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D., and Dick J.E.. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3:730–737. 10.1038/nm0797-730 [DOI] [PubMed] [Google Scholar]
- Busque L., Patel J.P., Figueroa M.E., Vasanthakumar A., Provost S., Hamilou Z., Mollica L., Li J., Viale A., Heguy A., et al. 2012. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 44:1179–1181. 10.1038/ng.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368:2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.Y., Miyanishi M., Wang S.K., Yamazaki S., Sinha R., Kao K.S., Seita J., Sahoo D., Nakauchi H., and Weissman I.L.. 2016. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 530:223–227. 10.1038/nature16943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou W.C., Tang J.L., Wu S.J., Tsay W., Yao M., Huang S.Y., Huang K.C., Chen C.Y., Huang C.F., and Tien H.F.. 2007. Clinical implications of minimal residual disease monitoring by quantitative polymerase chain reaction in acute myeloid leukemia patients bearing nucleophosmin (NPM1) mutations. Leukemia. 21:998–1004. 10.1038/sj.leu.2404637 [DOI] [PubMed] [Google Scholar]
- Corces-Zimmerman M.R., Hong W.J., Weissman I.L., Medeiros B.C., and Majeti R.. 2014. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA. 111:2548–2553. 10.1073/pnas.1324297111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzio A., Passegué E., Ayton P.M., Karsunky H., Cleary M.L., and Weissman I.L.. 2003. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 17:3029–3035. 10.1101/gad.1143403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J., et al. 2011. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17:1086–1093. 10.1038/nm.2415 [DOI] [PubMed] [Google Scholar]
- Falini B., Mecucci C., Tiacci E., Alcalay M., Rosati R., Pasqualucci L., La Starza R., Diverio D., Colombo E., Santucci A., et al. GIMEMA Acute Leukemia Working Party . 2005. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 352:254–266. 10.1056/NEJMoa041974 [DOI] [PubMed] [Google Scholar]
- Ficara F., Crisafulli L., Lin C., Iwasaki M., Smith K.S., Zammataro L., and Cleary M.L.. 2013. Pbx1 restrains myeloid maturation while preserving lymphoid potential in hematopoietic progenitors. J. Cell Sci. 126:3181–3191. 10.1242/jcs.125435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., et al. 2014. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371:2477–2487. 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goardon N., Marchi E., Atzberger A., Quek L., Schuh A., Soneji S., Woll P., Mead A., Alford K.A., Rout R., et al. 2011. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 19:138–152. 10.1016/j.ccr.2010.12.012 [DOI] [PubMed] [Google Scholar]
- Gorello P., Cazzaniga G., Alberti F., Dell’Oro M.G., Gottardi E., Specchia G., Roti G., Rosati R., Martelli M.F., Diverio D., et al. 2006. Quantitative assessment of minimal residual disease in acute myeloid leukemia carrying nucleophosmin (NPM1) gene mutations. Leukemia. 20:1103–1108. 10.1038/sj.leu.2404149 [DOI] [PubMed] [Google Scholar]
- Green C.L., Koo K.K., Hills R.K., Burnett A.K., Linch D.C., and Gale R.E.. 2010. Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J. Clin. Oncol. 28:2739–2747. 10.1200/JCO.2009.26.2501 [DOI] [PubMed] [Google Scholar]
- Grimwade D., Hills R.K., Moorman A.V., Walker H., Chatters S., Goldstone A.H., Wheatley K., Harrison C.J., and Burnett A.K.. National Cancer Research Institute Adult Leukaemia Working Group . 2010. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 116:354–365. 10.1182/blood-2009-11-254441 [DOI] [PubMed] [Google Scholar]
- Hou Y., Song L., Zhu P., Zhang B., Tao Y., Xu X., Li F., Wu K., Liang J., Shao D., et al. 2012. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 148:873–885. 10.1016/j.cell.2012.02.028 [DOI] [PubMed] [Google Scholar]
- Ishikawa F., Yoshida S., Saito Y., Hijikata A., Kitamura H., Tanaka S., Nakamura R., Tanaka T., Tomiyama H., Saito N., et al. 2007. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat. Biotechnol. 25:1315–1321. 10.1038/nbt1350 [DOI] [PubMed] [Google Scholar]
- Iwama A., Oguro H., Negishi M., Kato Y., Morita Y., Tsukui H., Ema H., Kamijo T., Katoh-Fukui Y., Koseki H., et al. 2004. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 21:843–851. 10.1016/j.immuni.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., et al. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371:2488–2498. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M., Snyder T.M., Corces-Zimmerman M.R., Vyas P., Weissman I.L., Quake S.R., and Majeti R.. 2012. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 4:149ra118 10.1126/scitranslmed.3004315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic J.V., Ivey A., Vannucchi A.M., Lippert E., Oppliger Leibundgut E., Cassinat B., Pallisgaard N., Maroc N., Hermouet S., Nickless G., et al. 2013. Establishing optimal quantitative-polymerase chain reaction assays for routine diagnosis and tracking of minimal residual disease in JAK2-V617F-associated myeloproliferative neoplasms: a joint European LeukemiaNet/MPN&MPNr-EuroNet (COST action BM0902) study. Leukemia. 27:2032–2039. 10.1038/leu.2013.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe H., Kandilci A., Kranenburg T.A., and Grosveld G.C.. 2007. Overexpression of N-Myc rapidly causes acute myeloid leukemia in mice. Cancer Res. 67:10677–10685. 10.1158/0008-5472.CAN-07-1118 [DOI] [PubMed] [Google Scholar]
- Kirstetter P., Schuster M.B., Bereshchenko O., Moore S., Dvinge H., Kurz E., Theilgaard-Mönch K., Månsson R., Pedersen T.A., Pabst T., et al. 2008. Modeling of C/EBPα mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 13:299–310. 10.1016/j.ccr.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Klco J.M., Spencer D.H., Miller C.A., Griffith M., Lamprecht T.L., O’Laughlin M., Fronick C., Magrini V., Demeter R.T., Fulton R.S., et al. 2014. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 25:379–392. 10.1016/j.ccr.2014.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D.C., Chen K., Wylie T., Larson D.E., McLellan M.D., Mardis E.R., Weinstock G.M., Wilson R.K., and Ding L.. 2009. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 25:2283–2285. 10.1093/bioinformatics/btp373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 442:818–822. 10.1038/nature04980 [DOI] [PubMed] [Google Scholar]
- Kyba M., Perlingeiro R.C., and Daley G.Q.. 2002. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 109:29–37. 10.1016/S0092-8674(02)00680-3 [DOI] [PubMed] [Google Scholar]
- Laurenti E., Doulatov S., Zandi S., Plumb I., Chen J., April C., Fan J.B., and Dick J.E.. 2013. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat. Immunol. 14:756–763. 10.1038/ni.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J., and Sauvageau G.. 2003. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 423:255–260. 10.1038/nature01572 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G., and Goodson M.. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21:936–939. 10.1101/gr.111120.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli M.P., Pettirossi V., Thiede C., Bonifacio E., Mezzasoma F., Cecchini D., Pacini R., Tabarrini A., Ciurnelli R., Gionfriddo I., et al. 2010. CD34+ cells from AML with mutated NPM1 harbor cytoplasmic mutated nucleophosmin and generate leukemia in immunocompromised mice. Blood. 116:3907–3922. 10.1182/blood-2009-08-238899 [DOI] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., and Smyth G.K.. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40:4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomdedeu J., Bussaglia E., Villamor N., Martinez C., Esteve J., Tormo M., Estivill C., Queipo M.P., Guardia R., Carricondo M., et al. Spanish CETLAM . 2011. Immunophenotype of acute myeloid leukemia with NPM mutations: prognostic impact of the leukemic compartment size. Leuk. Res. 35:163–168. 10.1016/j.leukres.2010.05.015 [DOI] [PubMed] [Google Scholar]
- Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T., et al. 2011. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 144:296–309. 10.1016/j.cell.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima M., Endoh M., Endo T.A., Toyoda T., Nakajima-Takagi Y., Sugiyama F., Koseki H., Kyba M., Iwama A., and Osawa M.. 2011. Genome-wide analysis of target genes regulated by HoxB4 in hematopoietic stem and progenitor cells developing from embryonic stem cells. Blood. 117:e142–e150. 10.1182/blood-2010-12-323212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., and Clarke M.F.. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 423:302–305. 10.1038/nature01587 [DOI] [PubMed] [Google Scholar]
- Potter N.E., Ermini L., Papaemmanuil E., Cazzaniga G., Vijayaraghavan G., Titley I., Ford A., Campbell P., Kearney L., and Greaves M.. 2013. Single-cell mutational profiling and clonal phylogeny in cancer. Genome Res. 23:2115–2125. 10.1101/gr.159913.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczy C., Petrovski R., Saunders C.T., Chorny I., Kruglyak S., Margulies E.H., Chuang H.Y., Källberg M., Kumar S.A., Liao A., et al. 2013. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics. 29:2041–2043. 10.1093/bioinformatics/btt314 [DOI] [PubMed] [Google Scholar]
- Sarry J.E., Murphy K., Perry R., Sanchez P.V., Secreto A., Keefer C., Swider C.R., Strzelecki A.C., Cavelier C., Récher C., et al. 2011. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2Rγc-deficient mice. J. Clin. Invest. 121:384–395. 10.1172/JCI41495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau G., Thorsteinsdottir U., Eaves C.J., Lawrence H.J., Largman C., Lansdorp P.M., and Humphries R.K.. 1995. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 9:1753–1765. 10.1101/gad.9.14.1753 [DOI] [PubMed] [Google Scholar]
- Shlush L.I., Zandi S., Mitchell A., Chen W.C., Brandwein J.M., Gupta V., Kennedy J.A., Schimmer A.D., Schuh A.C., Yee K.W., et al. HALT Pan-Leukemia Gene Panel Consortium . 2014. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 506:328–333. 10.1038/nature13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C.W., Karsunky H., Passegué E., Cozzio A., Weissman I.L., and Cleary M.L.. 2003. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 3:161–171. 10.1016/S1535-6108(03)00019-9 [DOI] [PubMed] [Google Scholar]
- Spencer D.H., Abel H.J., Lockwood C.M., Payton J.E., Szankasi P., Kelley T.W., Kulkarni S., Pfeifer J.D., and Duncavage E.J.. 2013. Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J. Mol. Diagn. 15:81–93. 10.1016/j.jmoldx.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Spencer D.H., Young M.A., Lamprecht T.L., Helton N.M., Fulton R., O’Laughlin M., Fronick C., Magrini V., Demeter R.T., Miller C.A., et al. 2015. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. 29:1279–1289. 10.1038/leu.2015.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussig D.C., Vargaftig J., Miraki-Moud F., Griessinger E., Sharrock K., Luke T., Lillington D., Oakervee H., Cavenagh J., Agrawal S.G., et al. 2010. Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34− fraction. Blood. 115:1976–1984. 10.1182/blood-2009-02-206565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treff N.R., Su J., Tao X., Northrop L.E., and Scott R.T. Jr. 2011. Single-cell whole-genome amplification technique impacts the accuracy of SNP microarray-based genotyping and copy number analyses. Mol. Hum. Reprod. 17:335–343. 10.1093/molehr/gaq103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lochem E.G., van der Velden V.H., Wind H.K., te Marvelde J.G., Westerdaal N.A.C., and van Dongen J.J.. 2004. Immunophenotypic differentiation patterns of normal hematopoiesis in human bone marrow: Reference patterns for age-related changes and disease-induced shifts. Cytometry B Clin. Cytom. 60B:1–13. 10.1002/cyto.b.20008 [DOI] [PubMed] [Google Scholar]
- Vassiliou G.S., Cooper J.L., Rad R., Li J., Rice S., Uren A., Rad L., Ellis P., Andrews R., Banerjee R., et al. 2011. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat. Genet. 43:470–475. 10.1038/ng.796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woll P.S., Kjällquist U., Chowdhury O., Doolittle H., Wedge D.C., Thongjuea S., Erlandsson R., Ngara M., Anderson K., Deng Q., et al. 2014. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 25:794–808. 10.1016/j.ccr.2014.03.036 [DOI] [PubMed] [Google Scholar]
- Wong P., Iwasaki M., Somervaille T.C.P., So C.W.E., and Cleary M.L.. 2007. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 21:2762–2774. 10.1101/gad.1602107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.