mTEC progenitors differentiate to mature Aire+ mTECs through a pathway that initiates with RANK and LtβR signaling via the nonclassical NF-κB, followed by TRAF-6–driven maturation.
Abstract
Medullary thymic epithelial cells (mTECs) expressing autoimmune regulator (Aire) are critical for preventing the onset of autoimmunity. However, the differentiation program of Aire-expressing mTECs (Aire+ mTECs) is unclear. Here, we describe novel embryonic precursors of Aire+ mTECs. We found the candidate precursors of Aire+ mTECs (pMECs) by monitoring the expression of receptor activator of nuclear factor-κB (RANK), which is required for Aire+ mTEC differentiation. pMECs unexpectedly expressed cortical TEC molecules in addition to the mTEC markers UEA-1 ligand and RANK and differentiated into mTECs in reaggregation thymic organ culture. Introduction of pMECs in the embryonic thymus permitted long-term maintenance of Aire+ mTECs and efficiently suppressed the onset of autoimmunity induced by Aire+ mTEC deficiency. Mechanistically, pMECs differentiated into Aire+ mTECs by tumor necrosis factor receptor-associated factor 6-dependent RANK signaling. Moreover, nonclassical nuclear factor-κB activation triggered by RANK and lymphotoxin-β receptor signaling promoted pMEC induction from progenitors exhibiting lower RANK expression and higher CD24 expression. Thus, our findings identified two novel stages in the differentiation program of Aire+ mTECs.
Medullary thymic epithelial cells (mTECs) are essential for induction of self-tolerance (Kyewski and Klein, 2006). mTECs uniquely express various self-antigens, including proteins whose expression is normally restricted in specific peripheral tissues (tissue-specific antigens [TSAs]; Kyewski and Klein, 2006; Klein et al., 2009). These diverse self-antigens are presented to developing T cells in the thymic medulla directly by mTECs expressing major histocompatibility complex class II (MHCII) and co-stimulatory molecules or indirectly by thymic dendritic cells (Klein et al., 2009). Consequently, T cells interacting with the TSA peptide–MHC complex with high avidity undergo apoptosis or conversion into immune-suppressive regulatory T cells (Kyewski and Klein, 2006; Klein et al., 2009; Hsieh et al., 2012). Nuclear protein autoimmune regulator (Aire) enhances the expression of some TSAs, controlling the induction of mTEC-dependent self-tolerance (Mathis and Benoist, 2009).
The peri- and neonatal functions of Aire-expressing mTECs (Aire+ mTECs) are critical for induction of long-lasting self-tolerance (Guerau-de-Arellano et al., 2009; Yang et al., 2015). Moreover, the frequency of autoimmunity onset is minimized when Aire is eliminated after weaning (Guerau-de-Arellano et al., 2009), and thymic regulatory T cells generated in an Aire-dependent manner during the neonatal period have distinctive properties from those generated in the adult thymus and are essential for lifelong self-tolerance (Yang et al., 2015). Hence, elucidation of the molecular and cellular mechanisms underlying embryonic and neonatal differentiation of Aire+ mTECs is critical.
Several studies using mutant mice have shown the dependence of mTEC development on TNF receptor family, receptor activator of nuclear factor-κB (RANK), CD40, and lymphotoxin β-receptor (LtβR; Boehm et al., 2003; Rossi et al., 2007; Akiyama et al., 2008, 2012; Hikosaka et al., 2008; Irla et al., 2008). These receptor signaling pathways activate the transcription factor NF-κB via two distinct intracellular signaling pathways, i.e., the classical NF-κB pathway and the nonclassical NF-κB pathway (Akiyama et al., 2012). TNF receptor-associated factor 6 (TRAF6) mediates RANK and CD40 signaling and activates the classical NF-κB pathway. RANK, CD40, and LtβR signaling pathways activate the NF-κB complex containing RelB via nonclassical NF-κB signaling mediated by NF-κB–inducing kinase (NIK). Dysfunction of TRAF6, NIK, or RelB abolishes the development of Aire+ mTECs (Weih et al., 1995; Weih and Caamaño, 2003; Kajiura et al., 2004; Akiyama et al., 2005; Shinzawa et al., 2011). Thus, these NF-κB pathways have nonredundant functions in mTEC development. However, the specific differentiation stages of mTECs regulated by these cytokines and signaling pathways remain to be determined.
mTECs and cortical TECs differentiate from common progenitors during embryonic and postnatal thymic development (Bennett et al., 2002; Gill et al., 2002; Bleul et al., 2006; Rossi et al., 2006; Ucar et al., 2014; Wong et al., 2014). mTECs are derived from TEC progenitors transiently expressing mature cTEC markers (Baik et al., 2013; Ohigashi et al., 2013; Ribeiro et al., 2013). Moreover, the TEC fraction expressing claudin 3/4 and the stem cell marker SSEA-1 exhibits stem cell–like features and gives rise to mTECs (Sekai et al., 2014). Additionally, the CD80– fraction in cTEC marker Ly51– embryonic TECs and the Ly51– adult TEC fraction expressing low levels of MHCII molecules contain immature mTECs convert into relatively short-lived Aire+ mTECs in reaggregation thymus organ culture (RTOC) in vitro (Gäbler et al., 2007; Gray et al., 2007). Podoplanin-expressing TECs that localize to the cortico-medulla junction have also been shown to develop into mTECs in the adult thymus (Onder et al., 2015). Thus, Aire+ mTECs may differentiate from common progenitors through several stages. However, the mTEC differentiation program involved in this process remains unclear.
The aim of this study is to elucidate the differentiation program of Aire+ mTECs and the mechanisms regulating this program. In this study, we describe novel embryonic precursors of Aire+ mTECs and their differentiation mechanisms. We identified candidate precursors of Aire+ mTECs (pMECs) expressing RANK and cTEC molecules. pMECs were found to differentiate into Aire+ mTECs in RTOC in vitro and in vivo. Introduction of pMECs into the thymus permitted long-term maintenance of Aire+ mTECs and was sufficient for inhibiting the onset of autoimmunity provoked by Aire+ mTEC deficiency. Nonclassical NF-κB activation by RANK and LtβR signaling induced the differentiation of pMECs from their progenitors expressing high levels of CD24. TRAF6-dependent RANK signaling in turn promoted the differentiation of pMECs into Aire+ mTECs. Thus, our results revealed two novel stages in the differentiation program of Aire+ mTECs.
RESULTS
RANK was expressed in UEA-1+ TECs and up-regulated with thymic development in embryos
RANK expression may identify the mTEC lineage because of its critical role in mTEC differentiation. To elucidate the mTEC differentiation program, we used mutant mice in which an internal ribosome entry site (IRES) linking the coding sequence for an enhanced GFP (EGFP)-Cre recombinase fusion protein was inserted into the final exon of the RANK gene (creating RANK-EGFP mice; Maeda et al., 2012). In these mice, mTEC development was intact, and EGFP expression was detected in adult mTECs in which RANK mRNA expression was confirmed by quantitative PCR (qPCR) analysis (unpublished data).
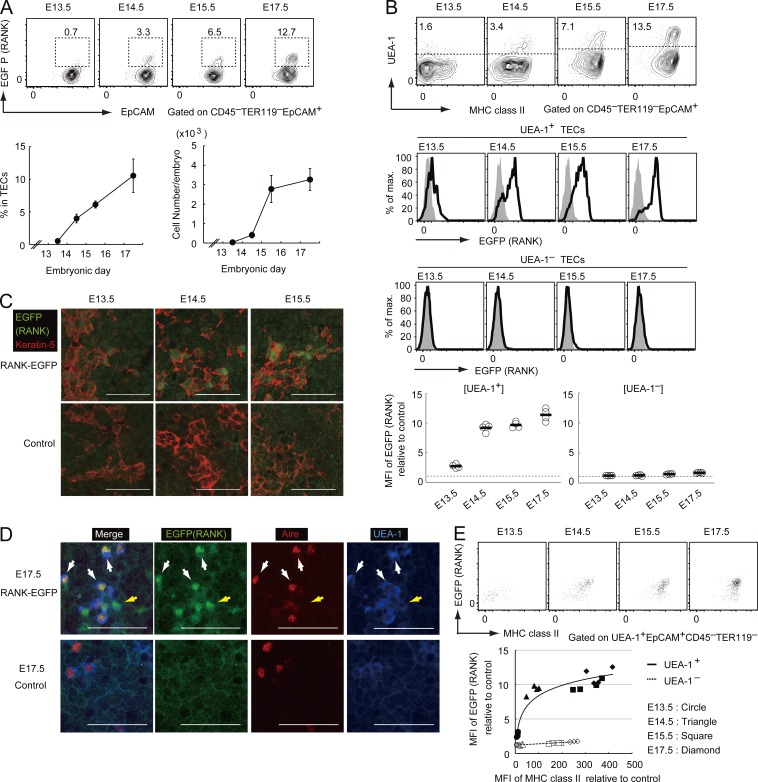
EGFP-expressing cells in the thymic stromal fraction (CD45–TER119–) were identified on embryonic days E13.5, E14.5, E15.5, and E17.5 by flow cytometry. The frequency of EGFP-expressing cells gradually increased in the TEC marker EpCAM+ fraction during embryonic development (Fig. 1 A). EpCAM+ cells were further divided according to the expression of an mTEC marker, UEA-1 ligand. EGFP expression was detected in UEA-1+ TECs, but not in UEA-1– TECs (Fig. 1 B). Immunohistochemical analysis also revealed EGFP expression in cells expressing the mTEC marker keratin-5 in the thymus at E13.5, E14.5, and E15.5 (Fig. 1 C). In the thymus at E17.5, the majority of EGFP+ cells expressed Aire and UEA-1 ligand (Fig. 1 D, white arrows), and a few EGFP+ UEA-1+ cells were negative for Aire expression (Fig. 1 D, yellow arrow). Overall, these data supported the selective expression of RANK in UEA-1+ TECs. Interestingly, the expression level of EGFP in UEA-1+ TECs was gradually increased during embryonic development (Fig. 1 B) and was positively correlated with the expression level of MHCII, a maturation marker of TECs (Fig. 1 E). Thus, the expression level of RANK may be correlated with mTEC differentiation.
Figure 1.
RANK expression in UEA-1+ TECs was increased with embryonic thymic development. (A) Flow cytometric analysis of EGFP expression in TECs from embryonic RANK-EGFP mice. Percentages of EGFP+EpCAM+ cells in TECs and cell numbers are shown on the bottom. Error bars represent the SD. n = 4 for each age group. (B) Flow cytometric analysis of EGFP expression in UEA-1+ TECs from RANK-EGFP embryonic mice. (top) UEA-1 and MHCII. (bottom and bottom middle) RANK expression (EGFP) in UEA-1+ TECs and UEA-1– TECs, respectively. Bold lines represent profiles of RANK-EGFP mice. Shaded regions indicate backgrounds of age-matched control mice. (bottom) median fluorescence intensity (MFI) values relative to the control. Black bars, mean values. n = 4 for each age group. Dotted lines, relative MFI of 1. (C) Immunohistochemical analysis of thymic sections from RANK-EGFP and control mice. EGFP (green) and keratin-5 (red) are shown. Bars, 50 µm. Data are representative of three independent experiments. (D) Immunohistochemical analysis of thymic sections from E17.5 RANK-EGFP mice (top) and wild-type mice (bottom) stained with anti-EGFP (green), anti-Aire (red), and anti–UEA-1 (blue). White arrows, cells expressing EGFP, Aire, and UEA-1 ligand; yellow arrows: cells expressing EGFP and UEA-1-ligand. Bars, 50 µm. Data are representative of three independent experiments. (E) Correlations between EGFP (RANK) and MHCII expression. (top) Profiles of EGFP and MHCII expression. (bottom) MFIs of EGFP expression in TECs of RANK-EGFP mice relative to that in control EpCAM– cells are plotted against the MFI of MHC class II in TECs relative to that in control EpCAM– cells. Closed and open notations indicate UEA-1+ TECs and UEA-1– TECs, respectively. Each data point is derived from one embryo. The solid line shows the fitting curve between relative MFIs of RANK and MHCII expression. n = 4 for each age group.
pMECs expressed low levels of mTEC maturation markers and high levels of cTEC molecules
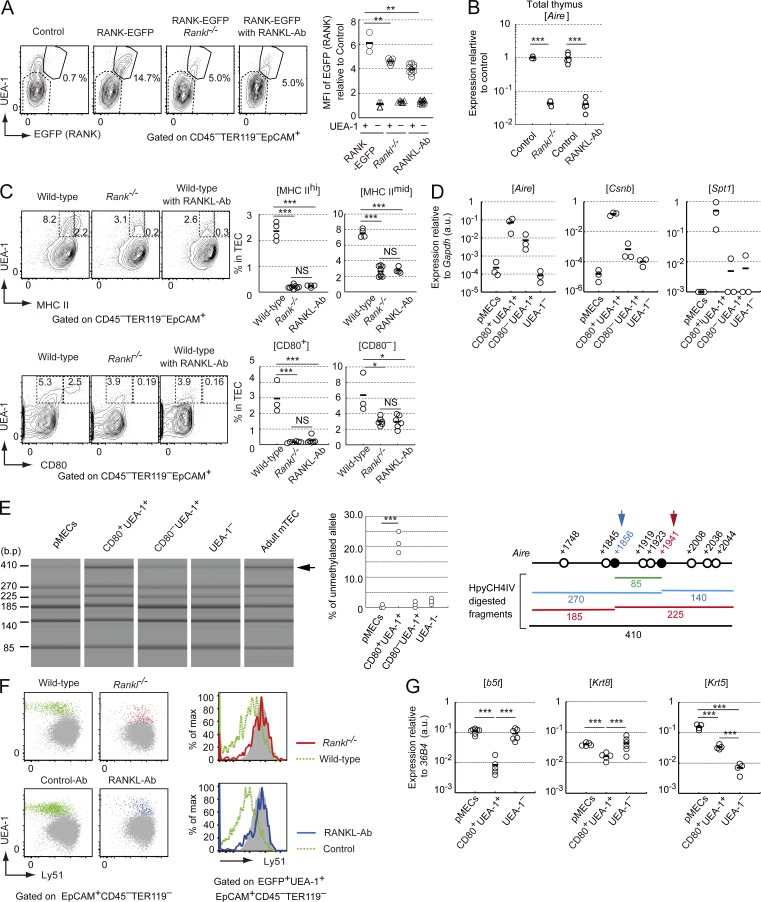
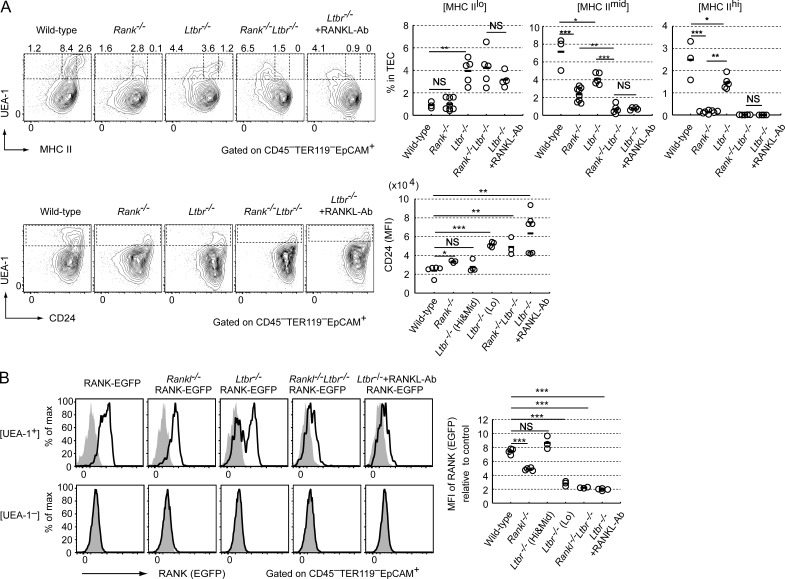
We next aimed to identify precursor cells of Aire+ mTECs that received RANKL signaling. Inhibition of RANKL signaling may block mTEC differentiation in RANK-expressing precursor cells of Aire+ mTECs. Indeed, UEA-1+ TECs expressing EGFP were found in the thymus of Rankl−/− RANK-EGFP embryos and RANK-EGFP embryos treated with an anti-RANKL antibody (RANKL-Ab; Fig. 2 A), which abolished the RANKL–RANK interaction (Furuya et al., 2011; Sugiyama et al., 2012; Khan et al., 2014). qPCR analysis of Aire expression confirmed that the RANKL-Ab treatment efficiently blocked mTEC differentiation (Fig. 2 B). The expression level of EGFP in UEA-1+ TECs was slightly lower in these mice than that in control RANK-EGFP mice (Fig. 2 A). Moreover, UEA-1+ TECs in embryonic mice deficient in RANKL signaling showed a moderate level of MHCII expression (MHCIImid) and a low level of CD80 expression (CD80–; Fig. 2 C). Consequently, we speculated that RANK+UEA-1+MHCIImidCD80– TECs in embryonic mice deficient in RANKL signaling could be candidate precursor cells of Aire+ mTECs (hereafter referred to as pMECs).
Figure 2.
pMECs expressed low levels of mTEC maturation markers and high levels of cTEC molecules. (A) Flow cytometric analysis of EGFP+UEA-1+ TECs from RANK-EGFP mice, Rankl−/− RANK EGFP mice, and RANK-EGFP mice treated with anti–RANKL-Ab at E17.5. The left panel (control) shows the profile of wild-type mice. Percentages of EGFP+UEA+ in TECs are shown. MFIs of EGFP relative to each control in UEA-1+ TECs (circles) and UEA-1− TECs (triangles) are summarized in the graph. n = 3 for RANK-EGFP. n = 6 for Rankl−/− RANK EGFP mice and n = 10 for RANK-EGFP mice treated with anti-RANKL-Ab. Black bars: mean values. **, P < 0.01, two-tailed Student’s t test. (B) Aire mRNA expression in the embryonic thymi of RANKL-Ab–treated mice and Rankl−/− mice. Aire expression in the total thymus of E17.5 Rankl−/− mice, E17.5 mice treated with RANKL-Ab, and corresponding control treated mice was analyzed by qPCR. Expression levels were calculated as arbitrary units normalized to 36B4 mRNA expression. Expression levels relative to the mean of the control were plotted in the graph for comparisons between two sets. Black bars indicate mean values. n = 3 for Rankl−/− and control mice. n = 5 for RANKL-Ab–treated mice and control mice. ***, P < 0.001, two-tailed Student’s t test. (C) Flow cytometric analysis of mTECs from embryonic Rankl−/− mice, Rank−/− mice, wild-type mice treated with RANKL-Ab, and control wild-type mice at E17.5. (left) typical profiles of TECs. (top and bottom) Percentages of UEA-1+MHCIIhi and UEA-1+MHCIImid in total TECs and UEA-1+CD80+ and UEA-1+CD80–, respectively. n = 4 for wild-type embryos and wild-type embryos treated with RANKL-Ab. n = 8 for Rank−/− embryos in upper graphs. n = 3 for wild-type embryos. n = 6 for Rankl−/− embryos and wild-type embryos treated with RANKL-Ab in lower graphs. Black bars indicate mean values. *, P < 0.05; ***, P < 0.001; and NS, not significant (two-tailed Student’s t test). (D) Aire, Csnb, and Spt1 mRNA expression in pMECs, CD80+ TECs, CD80– TECs, and UEA-1– TECs, as determined by qPCR. CD80+ TECs, CD80– TECs, and UEA-1– TECs were sorted from E17.5 wild-type thymi (Fig. S1). n = 3 for each samples. The values are arbitrary units normalized to Gapdh mRNA expression. Black bars indicate mean values. (E) CpG methylation analysis of the Aire gene in pMECs, CD80+ TECs, CD80– TECs, and UEA-1– TECs by combined bisulfite restriction analysis (COBRA). CD80+ TECs, CD80– TECs, and UEA-1– TECs were sorted from E17.5 wild-type thymi (Fig. S1). A typical gel electropherogram of COBRA is shown. The arrow indicates the band that was not digested with the methylation-sensitive restriction enzyme HpyCH13IV. Percentages of unmethylated CpGs are summarized in the right graph. n = 3 for each sample. ***, P < 0.001, two-tailed Student’s t test. (right) Aire gene structure and predicted length of the HpyCH13IV digestion fragment. The red and blue arrows in the lower panels indicate the T-DMR of the Aire gene. (F) Flow cytometric analysis of Ly51 expression in pMECs at E17.5. Typical profiles of UEA-1 and Ly51 staining (left) in TECs and Ly51 expression in RANK+UEA-1+ TECs (right) are shown. Profiles of RANK+UEA-1+ TECs are shown as color dots and lines. Green dots and lines, RANK-EGFP mice (wild-type, n = 3) or control-Ab-treated RANK-EGFP mice (control, n = 3); red dots and lines, Rankl−/− RANK-EGFP mice (n = 3); and blue dots and lines, RANK-EGFP mice treated with RANKL-Ab (blue line, n = 3) at E17.5. Shaded dots and regions indicate UEA-1– TECs. Data are representative of each sample. (G) b5t, Krt8, and Krt5 mRNA expression in pMECs, CD80+ TECs, and UEA-1– TECs, as determined by qPCR. pMECs and other TECs were sorted from RANKL-Ab–treated E17.5 RANK-EGFP mice and untreated E17.5 wild-type mice (see also Fig. S1). Values are arbitrary units normalized to 36B4 mRNA expression. n = 8 for pMECs, n = 5 for CD80+ TECs, and n = 8 for UEA-1– TECs for b5t and Krt8 expression. n = 4 for Krt5 expression. Black bars, mean values. ***, P < 0.001, two-tailed Student’s t test.
pMECs were sorted from the thymus of E17.5 RANK-EGFP mice treated with RANKL-Ab (Fig. S1). qPCR analysis indicated that expression of Aire and TSAs was practically absent in pMECs (Fig. 2 D). Analysis of the CpG methylation status using combined bisulfite restriction analysis (COBRA) indicated that tissue-specific differentially methylated regions (T-DMRs) in the Aire gene (Yagi et al., 2008; Wu et al., 2012) were significantly hypermethylated in pMECs as compared with that in the CD80+ mature mTEC fraction containing Aire-expressing cells (Fig. 2 E). Thus, Aire gene expression may be silenced at the epigenetic level in pMECs.
Surprisingly, the expression of the cTEC marker Ly51 was high in pMECs (Fig. 2 F). Moreover, qPCR analysis indicated that pMECs expressed high levels of the cTEC markers β5t and keratin-8 in addition to keratin-5 (Fig. 2 G). Hence, pMECs were different from the previously reported Ly51–CD80– immature mTEC fraction (Gäbler et al., 2007; Gray et al., 2007). Overall, pMECs expressed low levels of mature mTEC markers and high levels of cTEC molecules.
pMECs differentiated into Aire-expressing mTECs
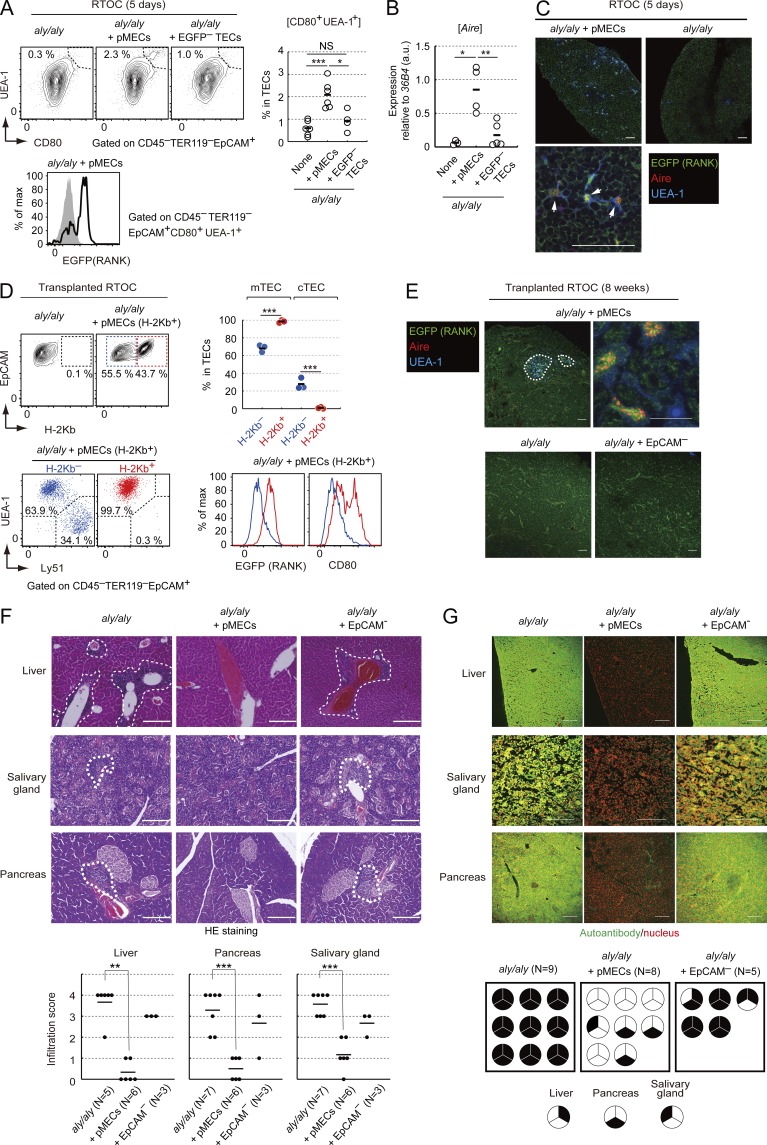
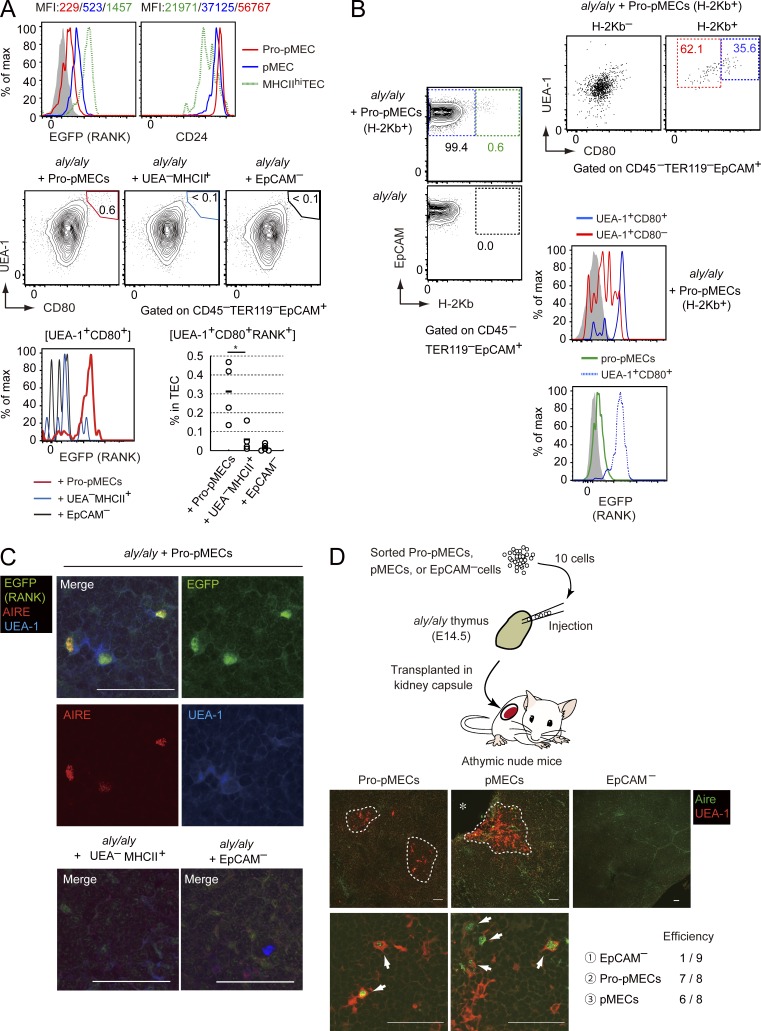
We next examined whether pMECs were capable of differentiating into Aire-expressing mature mTECs by using RTOC. We used an RTOC derived from an aly/aly fetus that lacked Aire+ mTECs because of a dysfunctional mutation in NIK (Kajiura et al., 2004). pMECs were reaggregated with thymic cells of E15.5 aly/aly mice. When RTOC was analyzed after 5 d of culture, UEA-1+CD80+ mature mTECs developed in the RTOC were mixed with pMECs (Fig. 3 A). These UEA-1+CD80+ mature mTECs expressed EGFP (Fig. 3 A), confirming their development from the introduced pMECs. qPCR analysis revealed induction of Aire mRNA in RTOC mixed with pMECs (Fig. 3 B). Moreover, immunohistochemical staining indicated generation of Aire+ mTECs in the RTOC mixed with pMECs (Fig. 3 C). These data strongly suggested that pMECs were capable of differentiating into Aire-expressing mature mTECs.
Figure 3.
pMECs differentiated into Aire-expressing mTECs inducing self-tolerance. (A) Flow cytometric analysis of reaggregation thymic organ culture (RTOC) derived from aly/aly embryonic thymic cells and pMECs. (left) typical flow cytometric profiles. Data are summarized in a right figure. (bottom) EGFP expression profiles in CD80+UEA-1+ TECs (solid line) and UEA-1– TECs (shaded region). n = 6 for aly/aly RTOC with pMECs and aly/aly RTOC. n = 4 for aly/aly RTOC with UEA-1–EGFP– TECs. pMECs and UEA-1–EGFP– TECs were sorted from RANKL-Ab–treated E17.5 RANK-EGFP mice. Black bars, mean values. *, P < 0.05; ***, P < 0.001, two-tailed Student’s t test. (B) qPCR analysis of Aire mRNA in aly/aly RTOC, aly/aly RTOC with pMECs, and aly/aly RTOC with UEA-1–EGFP– TECs. Values are arbitrary units normalized to 36B4 mRNA expression. n = 3 for aly/aly RTOC, n = 4 for aly/aly RTOC with pMECs, and n = 5 for aly/aly RTOC with UEA-1–EGFP– TECs. Black bars, mean values. *, P < 0.05; ***, P < 0.001, two-tailed Student’s t test. (C) Immunohistochemical analysis of RTOCs consisting of the aly/aly embryonic thymus and pMECs. EGFP (green), Aire (red), and UEA-1 (blue) are shown. Arrows: cells expressing Aire, EGFP (RANK), and UEA-1 ligand. Bars, 50 µm. Data are representative of three independent experiments. (D) Flow cytometric analysis of the thymus generated from RTOC by kidney transplantation. Typical profiles of EpCAM and H-2Kb staining (top left) and UEA-1 and Ly51 staining (bottom left) are shown. Percentages of mTECs (UEA-1+Ly51–) and cTECs (UEA-1–Ly51+) in H-2Kb– and H-2kb+ (derived from pMECs) TEC fractions are shown in the top right upper graph. n = 3. ***, P < 0.001, two-tailed Student’s t test. Typical expression profiles of EGFP and CD80 in H-2Kb+ (red) and H-2kb– (blue) TECs are shown in the lower right panel. (E) Immunohistochemical analysis of the thymus generated from RTOC by kidney transplantation. Aire (red), EGFP (green), and UEA-1 (blue) are shown. White dotted lines: medulla regions. A higher magnification image of aly/aly RTOC with pMECs is shown in an upper right panel. Bars, 50 µm for lower magnification images and 10 µm for the higher magnification image. n = 8 for aly/aly RTOC and aly/aly RTOC with pMECs, and n = 5 for aly/aly RTOC with EpCAM– cells. Data are representatives of each sample. (F) Inflammatory cell infiltration in nude mice receiving RTOC. Hematoxylin and eosin staining of organs is shown. White lines: areas of inflammatory cell infiltration in liver (n = 5 for aly/aly only, n = 6 for pMECs, and n = 3 for EpCAM–), pancreas (n = 7 for aly/aly only, n = 6 for pMECs, and n = 3 for EpCAM–) and salivary gland (n = 7 for aly/aly only, n = 6 for pMECs, and n = 3 for EpCAM–). Data are representatives of each sample. Bars, 200 µm. **, P < 0.01; ***, P < 0.001, Mann-Whitney U test. (G) Detection of autoantibodies in serum of nude mice transplanted with RTOC. The liver, salivary glands, and pancreas of RAG2-deficient mice were stained with sera (green) and propidium iodide (red). Data are representative of each sample. Bars, 100 µm. (bottom) A summary of autoantibody. Each circle represents an individual mouse. Autoantibody generation is represented as filled regions. n = 9 for aly/aly RTOC, n = 8 for aly/aly RTOC with pMECs, and n = 5 for aly/aly RTOC with EpCAM– cells.
We further investigated the differentiation potential of pMECs in vivo. pMECs derived from C57BL/6 mice (H-2Kb+) were reaggregated with thymic cells of E15.5 aly/aly mice on the BALB/cA background (H-2kb–; Shinzawa et al., 2011). Aly/aly RTOCs mixed with pMECs then were transplanted on kidney capsules of athymic mice and analyzed 4 wk after grafting. Flow cytometric analysis indicated that H-2Kb+ TECs expressed UEA-1-ligand, RANK, and CD80 (Fig. 3 D), suggesting that pMECs differentiated into mature mTECs. In contrast, Ly51+UEA-1– cTECs were nearly absent in the H-2Kb+ TEC fraction (Fig. 3 D). These data strongly suggested that pMECs could differentiate into mTECs and would lack differentiation potential into cTECs.
Introduction of pMECs in the thymus permitted long-term maintenance of Aire+ mTECs and inhibited autoimmunity onset by mTEC dysfunction
Next, we tested whether pMECs could differentiate into self-tolerance–inducing mTECs by RTOC transplantation in kidneys of athymic nude mice. Aly/aly mice exhibited autoimmunity owing to developmental defects in mature mTECs. We therefore examined whether introducing pMECs suppressed the onset of autoimmunity by the aly/aly mutation. In this experiment, aly/aly mice in the BALB/cA background (Shinzawa et al., 2011), RANK-EGFP mice in the BALB/cA background, and BALB/cA nude mice were used for MHC-matched congenic transplantation. Aly/aly RTOC mixed with pMECs was grafted on kidney capsules of athymic mice and analyzed 8 wk after grafting. Immunohistochemical staining of the thymus generated from the transplanted RTOC supported the development of medullar regions containing Aire+ mTECs when pMECs were mixed with the aly/aly RTOC (Fig. 3 E). This suggested that the introduction of pMECs in the thymus allowed for long-term maintenance of Aire+ mTECs. The number of generated medullas in each grafted thymus may be lower than that in wild-type grafted thymus (unpublished data), likely owing to the requirement of ligand-expressing niches (Rossi et al., 2007; Roberts et al., 2012) for the proliferation of pMECs. We further analyzed autoimmune responses in nude mice receiving aly/aly RTOCs. As expected, nude mice receiving aly/aly RTOCs exhibited substantial inflammatory cell infiltration in the liver, salivary glands, and pancreas of recipient nude mice. Strikingly, introduction of pMECs in the aly/aly RTOC inhibited cell infiltration (Fig. 3 F). Moreover, whereas sera from nude mice receiving aly/aly RTOC and aly/aly RTOC with EpCAM– cells contained autoantibodies against these organs, such autoantibody production was suppressed in nude mice receiving aly/aly RTOC with pMECs (Fig. 3 G). These data strongly suggested that introduction of pMECs in the thymus was sufficient for inhibiting autoimmunity provoked by mTEC dysfunction. Hence, we concluded that pMECs could differentiate into self-tolerance–inducing mTECs. Collectively, our data supported that pMECs were, indeed, precursor cells of Aire+ mTECs.
TRAF6 and RelB promoted the progression of distinct stages of mTEC differentiation
Because RANK signaling initiates pMEC differentiation by activating intracellular signaling pathways, we next aimed to examine the signaling pathways downstream of RANK that drive pMEC differentiation. We then examined whether differentiation of pMECs into Aire+ mTECs required TRAF6 and RelB.
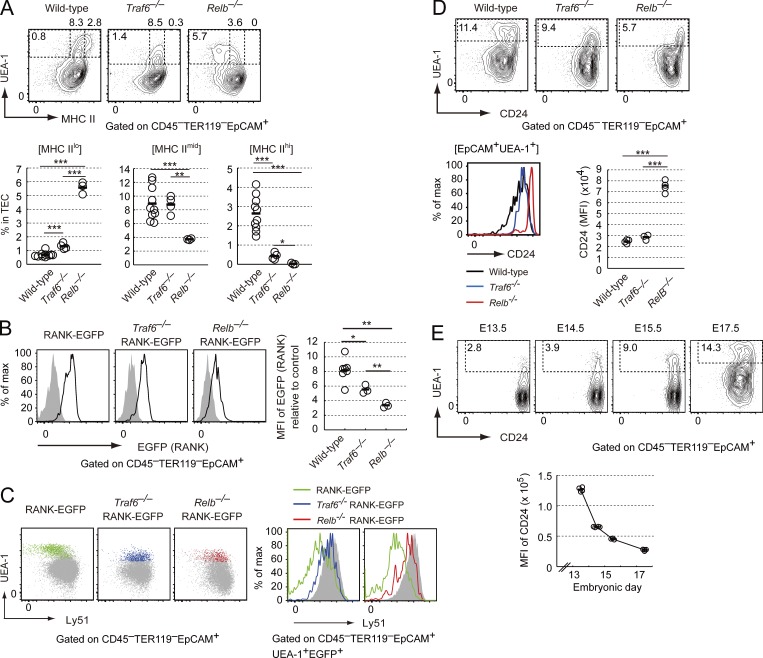
Flow cytometric analysis revealed that there was a dramatic reduction in MHCIIhiUEA-1+ TECs of the Traf6−/− thymus at E17.5 (Fig. 4 A). The expression level of UEA-1 ligand in TECs was also reduced in the Traf6−/− thymus, as previously reported (Akiyama et al., 2005), implying that TRAF6-dependent NF-κB activation may control UEA-1 ligand expression in TECs. Moreover, analysis of Traf6−/− RANK-EGFP mice suggested that MHCIImidUEA-1+ TECs accumulated in the Traf6−/− embryonic thymus expressing RANK and high levels of Ly51 (Fig. 4 B and 4C). These data suggested that the absence of TRAF6 arrested mTEC differentiation at pMECs. Thus, differentiation of pMECs into Aire+ mTECs induced by RANK signaling was likely to be dependent on TRAF6.
Figure 4.
Distinct roles of TRAF6 and RelB in embryonic mTEC differentiation. (A) Flow cytometric analysis of TECs in thymi of Traf6−/−, Relb−/−, and wild-type embryos (E17.5). (top) Typical profiles of UEA-1 and MHCII in TECs. Percentages of specific cell types are shown. n = 10 for wild-type, n = 4 for Traf6−/−, and n = 3 for Relb−/− embryos. Black bars, mean values. *, P < 0.05; **, P < 0.01; ***, P < 0.001, two-tailed Student’s t test. (B) Flow cytometric analysis of EGFP expression in TECs of Traf6−/− RANK-EGFP, Relb−/− RANK-EGFP, and RANK-EGFP embryos (E17.5). Solid lines indicate EGFP expression in UEA-1+ TECs of each type of RANK-EGFP mice. Shaded region shows background determined in each EGFP-negative wild-type or mutant embryos. Relative MFIs of EGFP to control is shown in the right graph. n = 7 for RANK-EGFP mice; n = 3 for Traf6−/− RANK-EGFP mice and Relb−/− RANK-EGFP mice. Black bars, mean values. *, P < 0.05; **, P < 0.01, two-tailed Student’s t test. (C) Flow cytometric analysis of Ly51 expression in EGFP+UEA-1+ TECs at E17.5. Typical profiles of UEA-1 and Ly51 staining (left) in TECs and Ly51 expression in RANK+UEA-1+ TECs (right) are shown. Profiles of RANK+UEA-1+ TECs are shown as colored dots and lines. Green dots and lines, RANK-EGFP mice (wild-type); blue dots and lines, Traf6−/−RANK-EGFP mice; red dots and lines, Relb−/− RANK-EGFP mice. Shaded dots and regions indicate UEA-1– TECs. Data are representative of each experiment. n = 3 for RANK-EGFP mice, n = 2 for Traf6−/− RANK-EGFP mice, and n = 4 for Relb−/− RANK-EGFP mice. (D) Flow cytometric analysis of CD24 expression in UEA-1+ TECs of E17.5 Traf6−/− RANK-EGFP, Relb−/− RANK-EGFP, and RANK-EGFP embryos (E17.5). (top) Typical flow cytometric profiles of UEA-1 and CD24 expression. (bottom left) CD24 expression in EpCAM+UEA-1+ cells. (bottom right) MFIs of CD24. n = 4 for wild-type embryos, n = 3 for Traf6−/− embryos, and n = 4 for Relb−/− embryos. Black bars, mean values. *, P < 0.05; ***, P < 0.001, two-tailed Student’s t test. (E) Flow cytometric analysis of CD24 expression in TECs at E13.5, E14.5, E15.5, and E17.5. (left) typical flow cytometric profiles of UEA-1 and CD24. (right) MFIs of CD24. n = 4 for each age group. Black bars, mean values.
Unexpectedly, Relb−/− and Relb−/− RANK-EGFP mice showed dramatically different profiles of UEA-1+ TEC differentiation at E17.5. The expression of UEA-1 ligand in Relb−/− TECs was reduced to a similar extent as that in the Traf6−/− thymus, which also suggested the contribution of NF-κB activation to UEA-1 ligand expression in TECs. Importantly UEA-1+ TECs in the embryonic Relb−/− thymus expressed significantly lower levels of MHCII than that in the Traf6−/− thymus (Fig. 4 A). Moreover, RANK expression levels were significantly lower in UEA-1+ TECs in the embryonic Relb−/− thymus than that in the Traf6−/− thymus (Fig. 4 B). Interestingly, UEA-1+ TECs in the Relb−/− thymus expressed significantly higher levels of CD24, which has been reported to function as a stem cell marker in other tissues (Rietze et al., 2001; Shackleton et al., 2006), as compared with that in the wild-type and Traf6−/− thymus (Fig. 4 D). Given that mTEC differentiation was arrested at pMECs in the Traf6−/− thymus, the absence of RelB arrested mTEC differentiation at different precursor cells from pMECs. Despite such differences in MHCII and RANK expression levels, Ly51 expression in UEA-1+ TECs in the Relb−/− thymus was as high as that in pMECs, implying that UEA-1+MHCIIloCD24hi TECs in the Relb−/− thymus and pMECs may have the same origin (Fig. 4 C). During early differentiation, UEA-1+ TECs expressed low levels of RANK and MHCII (Fig. 1, B and D). Moreover, CD24 expression in UEA-1+ TECs was down-regulated during embryonic thymus development (Fig. 4 E). Accordingly, UEA-1+RANKloMHCIIloCD24hi TECs accumulated in the Relb−/− embryonic thymus were likely to be in an earlier stage of differentiation than pMECs. We therefore hypothesized that UEA-1+RANKloMHCIIloCD24hi TECs may be candidate progenitors of pMECs (hereafter referred to as pro-pMECs).
RANK and LtβR signaling cooperatively promoted the differentiation of pro-pMECs
Given that RelB activation promotes the differentiation of pro-pMECs into pMECs, we next examined the upstream receptors promoting the differentiation of pro-pMECs through RelB activation. Because LtβR is known to activate the nonclassical NF-κB pathway, causing nuclear translocation of RelB, we first investigated embryonic TEC development in LtβR-deficient mice (Ltbr−/−). Interestingly, all MHCIIhi, MHCIImid, and MHCIIlo UEA-1+ TEC fractions were detected in the LtβR-deficient thymus (Fig. 5 A). Thus, LtβR signaling may be upstream of RelB in the differentiation of pro-pMECs, and other signaling pathways could cooperate with LtβR signaling. Because RANK expression was detected in MHCIIloUEA-1+ TECs in Relb−/− embryos (Fig. 4 B), we hypothesized that RANK signaling may be involved in the differentiation of pro-pMECs. Indeed, double deficiency of RANK and LtβR signaling (Rank−/−Ltbr−/− and Ltbr−/− treated with RANKL-Ab) caused accumulation of the UEA-1+MHCIIlo TEC fraction in E17.5 embryos (Fig. 5 A), similar to that observed in Relb−/− embryos (Fig. 4 A). UEA-1+ TECs in the thymus of Rank−/−Ltbr−/− embryos and Ltbr−/− embryos treated with RANKL-Ab exhibited high CD24 expression at E17.5 (Fig. 5 A). Moreover, analysis of Rankl−/−Ltbr−/− RANK-EGFP embryos and Ltbr−/− RANK-EGFP embryos treated with RANKL-Ab indicated that RANK expression was low in UEA-1+ TECs (Fig. 5 B). These data suggested that UEA-1+ TECs in embryos deficient in RANK and LtβR signaling were identical to pro-pMECs detected in the Relb−/− embryonic thymus. Thus, we hypothesized that LtβR and RANK signaling may cooperatively promote the differentiation of pro-pMECs into pMECs.
Figure 5.
RANK and LtβR signaling induced pMECs from their progenitors. (A) Flow cytometric analysis of TECs in Rank−/−, Ltbr−/−, Rank−/− Ltbr−/−, anti-RANKL-Ab-treated Ltbr−/−, and control wild-type embryos (E17.5). (top left) Typical flow cytometric profiles of UEA-1 and MHCII. Numbers in figures indicate percentages of different TEC subtypes. n = 3 for wild-type, n = 8 for Rank−/−, n = 5 for Ltbr−/−, n = 5 for Rank−/− Ltbr−/−, and n = 4 for Ltbr−/− treated with RANKL-Ab. Black bars, mean values. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant; two-tailed Student’s t test. (bottom left) Typical flow cytometric profiles of UEA-1 and CD24 in TECs. MFIs of CD24 in UEA-1+ TECs are summarized in the bottom right graph. Ltbr−/−(Hi&Mid) and Ltbr−/− (Lo) indicates MHCIIhi and MHCIImidUEA-1+TECs and MHCIIloUEA+TECs in Ltbr−/−. n = 5 for wild-type, n = 3 for Rank−/−, n = 4 for Ltbr−/−, n = 3 for Rank−/− Ltbr−/−, and n = 7 for Ltbr−/− treated with RANKL-Ab. Black bars, mean values. *, P < 0.05; ***, P < 0.001; NS, not significant; two-tailed Student’s t test. (B) Flow cytometric analysis of EGFP expression in TECs of E17.5 Rankl−/− RANK-EGFP, Ltbr−/− RANK-EGFP, Rankl−/− Ltbr−/− RANK-EGFP, anti–RANKL-Ab–treated Ltbr−/− RANK-EGFP, and control wild-type mice. Solid lines of top and bottom left panels show EGFP expression in UEA-1+ TECs and UEA-1– TECs, respectively. Shaded regions show background intensities determined in control embryos. n = 4 for RANK-EGFP mice, n = 4 for Rankl−/− RANK-EGFP mice, n = 3 for Ltbr−/− RANK-EGFP mice, n = 3 for Rankl−/− Ltbr−/− RANK-EGFP mice, n = 4 for Ltbr−/− RANK-EGFP mice treated with RANKL-Ab. Black bars, mean values. ***, P < 0.001; NS, not significant; two-tailed Student’s t test.
Pro-pMECs were capable of differentiating into Aire+ mTECs
We next investigated whether pro-pMECs were capable of differentiating into Aire+ mTECs. Pro-pMECs were sorted from the thymus of Ltbr−/− RANK-EGFP mice at E15.5 treated with RANKL-Ab (Fig. S2). Sorted pro-pMECs showed higher levels of CD24 expression and lower levels of EGFP expression as compared with pMECs (Fig. 6 A) and were reaggregated with the aly/aly embryonic thymus. Flow cytometric analysis of the RTOC after 5 d of culture indicated the generation of EGFP+UEA-1+CD80+ mTECs in RTOCs mixed with pro-pMECs (Fig. 6 A). Further RTOC experiments suggested that pro-pMECs (H2-Kb+) were converted into RANK+UEA-1+TECs, whereas fewer pro-pMECs were converted into UEA-1– TECs (Fig. 6 B). In addition to CD80+RANK+UEA-1+ TECs, the H2-Kb+ fraction contained CD80–UEA-1+ TECs that expressed relatively high levels of EGFP as compared with that in pro-pMECs (Fig. 6 B). This implied that pro-pMECs may be converted into pMECs in addition to mature mTECs. Moreover, immunohistochemical staining suggested the development of EGFP+Aire+UEA-1+ mTECs in RTOCs with pro-pMECs (Fig. 6 C). Overall, these data supported the differentiation of pro-pMECs into Aire+ mTECs in RTOC. Conversion of pro-pMECs into Aire+ mTECs was less efficient than that of pMECs, which may be explained by the absence of LtβR in addition to the limited niches expressing RANKL.
Figure 6.
Pro-pMECs differentiated into Aire+ mTECs. (A) Flow cytometric analysis of RTOCs prepared from aly/aly embryonic thymi and pro-pMECs, UEA-1– TECs, and EpCAM– stroma cells. Pro-pMECs, UEA-1–MHCII+ TECs, and EpCAM– stroma cells were sorted from Ltbr−/− RANK-EGFP mice treated with anti-RANKL-Ab (Fig. S2). (top) EGFP expression and CD24 expression of pro-pMECs (red), pMECs (blue), and MHCIIhi TECs (green dots). Shade regions (top left) show UEA-1-TECs. RTOCs were analyzed after 5 d of culture (middle). Numbers indicate percentages of UEA-1+CD80+ cells in TECs. (bottom left) EGFP expression in UEA-1+CD80+ cells in RTOCs. (bottom right) Percentages of UEA-1+EGFP+CD80+ cells in total TECs. n = 4 for RTOC with pro-pMECs, n = 4 for RTOC with UEA-1–MHCII+ TECs, and n = 5 for RTOC with EpCAM– stroma cells. *, P < 0.05, two-tailed Student’s t-test. (B) Flow cytometric analysis of RTOCs prepared from pro-pMECs after 5 d of culture. Expression of CD80, UEA-1 (top right), and EGFP (middle right) in pro-pMEC–derived cells (H-2Kb+; left) is shown. Data are representative of three independent experiments. EGFP expression profiles of pro-pMECs and CD80+UEA-1+ TECs are shown (bottom right). (C) Immunohistochemical analysis of RTOC sections after 5 d of culture. EGFP (green), Aire (red), and UEA-1 (blue) are shown. Only composite images (merge) are shown for RTOCs with UEA-1–MHCII+ cells and EpCAM– stroma cells. Data are representative of three independent experiments. (D) Injection of limited numbers of pMECs and pro-pMECs (10 cells) into aly/aly embryonic thymi. Pro-pMECs (UEA-1+CD24hiMHCIIlo TECs) and pMECs (EGFP+UEA-1+MHCIImid TECs) were sorted from E14.5 RANK-EGFP mice treated with anti-RANKL-Ab. The sorting strategy is shown in Fig. S3. Immunohistochemical staining for Aire (green) and UEA-1 (red) is shown. White dotted lines show areas containing UEA-1+ cells. Asterisk: a kidney. Efficiency in the bottom panels indicates the ratio of the number of thymi possessing thymic medulla-like areas containing UEA-1+Aire+ cells to the number of trials. n = 8 for pMECs and pro-pMECs, and n = 9 for EpCAM– cells.
To further confirm our hypothesis, we directly injected limited numbers of pro-pMECs and pMECs (10 cells) into the aly/aly thymus (Fig. 6 D), which would minimize the possibility of contamination of other cell types. Pro-pMECs and pMECs were then sorted from RANK-EGFP mice treated with RANKL-Ab at E14.5 (Fig. S3). Immunohistochemical staining indicated that areas consisting of UEA-1+ cells containing Aire+ mTECs were detected in the grafted thymus injected with pro-pMECs or pMECs (Fig. 6 D). On the other hand, UEA-1+ areas were not found in thymi injected with EpCAM– cells, with the exception of one sample. These data further supported the idea that pro-pMECs and pMECs were capable of differentiating into Aire+ mTECs.
Nonclassical NF-κB activation by RANK and LtβR signaling induced pMECs
In vivo data indicated the requirements of RANK/LtβR and RelB for pMEC induction and RANK and TRAF6 for pMEC differentiation into Aire+ mTECs, respectively. Finally, we confirmed that these receptors and downstream molecules were indeed linked.
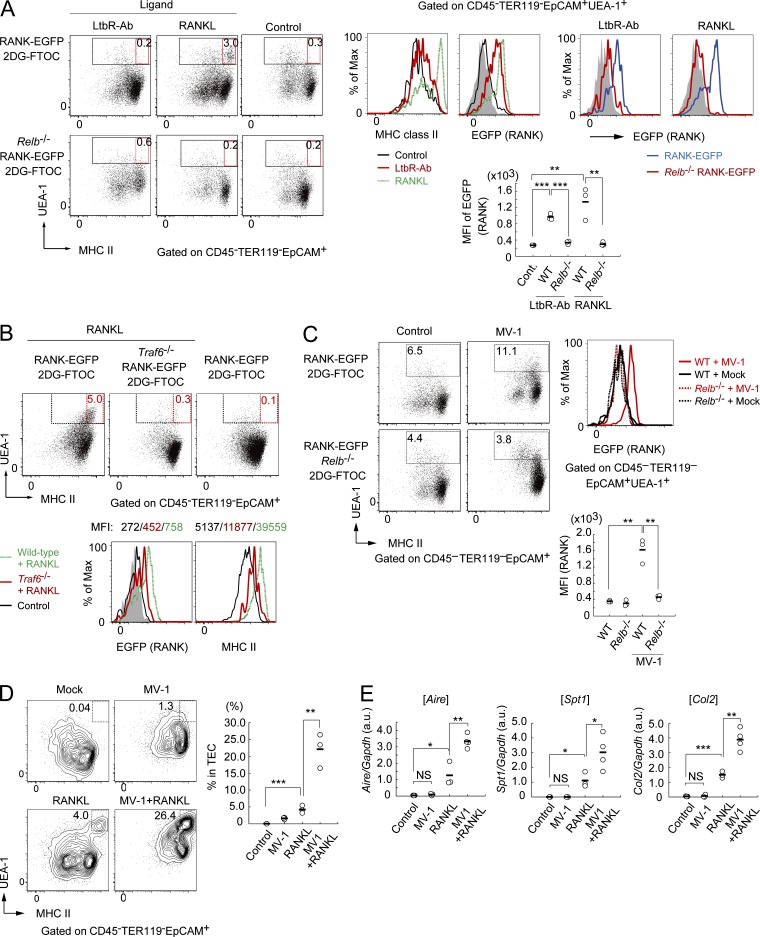
Stimulation of fetal thymic stroma organ culture (2DG-FTOC) with recombinant RANKL induces Aire+ mTECs (Rossi et al., 2007; Akiyama et al., 2008, 2014). Furthermore, stimulation with agonistic LtβR-Ab enhances RANK mRNA expression in 2DG-FTOC without inducing Aire+ mTECs (Mouri et al., 2011). However, cells in which RANK expression is up-regulated by LtβR signaling have not been examined. Stimulation of 2DG-FTOC from RANK-EGFP embryos treated with an agonistic LtβR antibody induced EGFP+UEA-1+MHCIImid TECs (Fig. 7 A). Such induction was completely abolished in Relb−/− 2DG-FTOC. These data suggested that LtβR signaling induced pMECs in a RelB-dependent manner, consistent with in vivo genetic data.
Figure 7.
Nonclassical NF-κB activation by RANK and LtβR signaling induced pMECs. (A) Flow cytometric analysis of fetal thymic stroma stimulated with RANK and LtβR signaling. (left) typical flow cytometric profiles of UEA-1 and MHCII. The ligands used for stimulation are shown at the top. Numbers in red rectangles show percentages of UEA-1+MHCIIhi TECs of the total TECs. Black rectangles represent gating of UEA-1+TECs. (middle) Expression profiles of EGFP and MHCII in UEA-1+ TECs of 2DG-FTOCs stimulated with LtβR-Ab or RANKL. Red lines show profiles in anti–LtβR-Ab–treated 2DG-FTOC. Green dotted lines show profiles in RANKL-treated 2DG-FTOC. (right) EGFP profiles in 2DG-FTOC from Relb−/− RANK-EGFP mice. Blue and red lines show EGFP expression in 2DG-FTOC from RANK-EGFP and Relb−/− RANK-EGFP mice, respectively. Ligands are shown at the top. Shaded regions: EGFP-negative UEA-1+TECs from wild-type 2DG-FTOC. Data are representatives of three independent experiments. (bottom) MFIs of EGFP. n = 3. **, P < 0.01; ***, P < 0.001 (two-tailed Student’s t test). (B) Flow cytometric analysis of Traf6−/− 2DG-FTOC stimulated with RANKL. (top) typical profiles of UEA-1 and MHCII expression in TECs of RANKL-treated 2DG-FTOCs from Traf6−/− RANK-EGFP and RANK-EGFP mice. (bottom) EGFP and MHCII expression in UEA-1+ TECs in RANKL-treated 2DG-FTOC. Green dotted line: profile of RANK-EGFP 2DG-FTOC. Red line shows profile of Traf6−/−RANK-EGFP 2DG-FTOC. Black line shows profile of untreated RANK-EGFP 2DG-FTOC. Shaded region shows UEA-1– TECs from RANK-EGFP 2DG-FTOC. Data are representative of three independent experiments. (C) Flow cytometric analysis of 2DG-FTOC stimulated with the cIAP inhibitor MV-1. Numbers in rectangles show percentages of UEA-1+ cells of total TECs. (right) EGFP expression in UEA-1+ cells. Data are representative of three independent experiments. (bottom) MFIs of EGFP. n = 3. **, P < 0.01, two-tailed Student’s t test. (D) Flow cytometric analysis of 2D-FTOC stimulated with MV-1, RANKL, or MV1 plus RANKL. Numbers in rectangles: percentages of UEA-1+MHCIIhi cells of total TECs. n = 3. **, P < 0.01; ***, P < 0.001, two-tailed Student’s t test. (E) qPCR analysis of 2DG-FTOC stimulated with the cIAP inhibitor MV-1, RANKL, or MV1 plus RANKL. Values are arbitrary units normalized to 36B4 mRNA expression. n = 4 for MV1 and RANKL stimulation and n = 3 for others. *, P < 0.05; **, P < 0.01; ***, P < 0.001, two-tailed Student’s t test.
In contrast to LtβR signaling, stimulation with RANK ligand induced both EGFP+MHCIIhi mTECs and EGFP+UEA-1+MHCIImid TECs in 2DG-FTOC (Fig. 7 A). The absence of RelB completely inhibited RANKL-dependent induction of these cell types (Fig. 7 A). Thus, RANKL induced mature mTECs and pMECs in a RelB-dependent manner. RelB expression should be required in RANKL-dependent mature mTECs because mature mTECs are derived from pMECs induced by RANK signaling. In contrast to Relb−/− 2DG-FTOC, EGFP+UEA-1+MHCIImid TECs were induced by RANKL stimulation in the absence of TRAF6 (Fig. 7 B). RANKL-dependent induction of MHCIIhi mTECs did not occur in Traf6−/− 2DG-FTOC (Fig. 7 B). Thus, although RANK signaling required TRAF6 for differentiation of pMECs into mature mTECs, TRAF6 was dispensable for RANK signaling-dependent pMEC induction in the fetal thymic stroma. Consequently, these data strongly suggested that RANK signaling promoted two different stages in mTEC differentiation through two distinct intracellular signaling pathways (i.e., TRAF6 and RelB dependent) for each differentiation stage.
The requirement for RelB in RANK and LtβR signaling-dependent pMEC induction suggested that activation of the nonclassical NF-κB pathway may promote pMEC induction. To further verify this mechanism by gain-of-function experiments, we used MV-1, a synthetic inhibitor of cellular inhibitors of apoptosis (cIAPs) 1 and 2 (Varfolomeev et al., 2007). Because cIAPs function as negative regulators of the nonclassical NF-κB pathway by inducing degradation of NIK in resting cells (Akiyama et al., 2012), inhibition of cIAPs by MV-1 activates the nonclassical NF-κB pathway through accumulation and activation of NIK without ligand stimulation (Varfolomeev et al., 2007). MV-1 treatment efficiently induced RANK+UEA-1+MHCIImid TECs (Fig. 7 C), despite unexpected increases in UEA-1 ligand expression in the cTEC fraction (Fig. 7 C). Thus, MV-1 treatment was sufficient for inducing pMECs in 2DG-FTOC. As expected, the MV-1–dependent induction of pMECs was abolished by the absence of RelB (Fig. 7 C), confirming that pMEC induction by MV-1 was mediated by the nonclassical NF-κB pathway, leading to RelB activation. Furthermore, treatment of 2DG-FTOC with a combination of MV-1 and RANKL demonstrated that MV-1 significantly enhanced the RANKL-dependent induction of MHCIIhi mature mTECs (Fig. 7 D). Moreover, qPCR analysis indicated that MV-1 enhanced the RANKL-dependent up-regulation of Aire and TSA, whereas treatment with MV-1 alone did not (Fig. 7 E). These data suggested that MV-1 induced large numbers of pMECs by efficient activation of the nonclassical NF-κB pathway; in turn, RANKL induced an increase in the number of mature Aire+ mTECs.
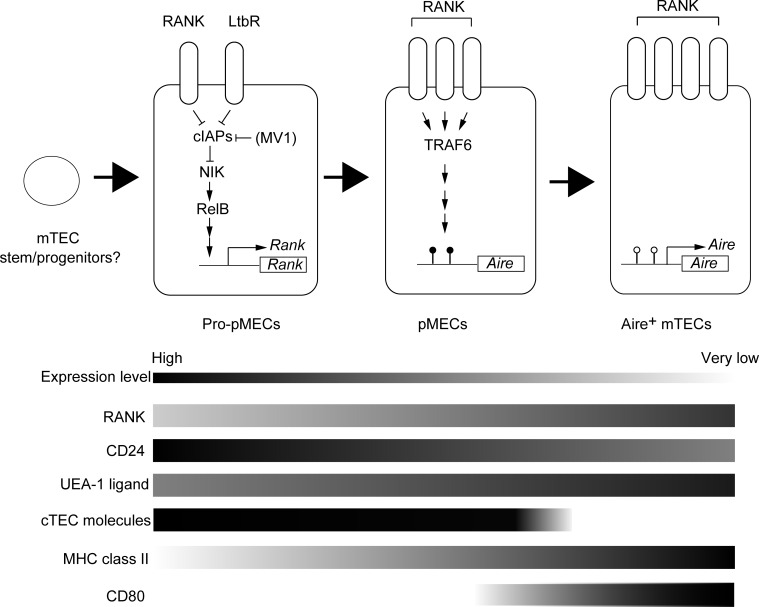
From these results, we propose a mechanism of embryonic Aire+ mTEC differentiation as follows. First, RANK and LtβR signaling activate RelB through the nonclassical NF-κB pathway in pro-pMECs expressing low levels of RANK, thereby initiating differentiation of pro-pMECs into pMECs expressing high levels of RANK. Subsequently, TRAF6-dependent RANK signaling drives the differentiation of pMECs into Aire+ mTECs (Fig. 8).
Figure 8.
A proposed mechanism for embryonic mTEC differentiation. Pro-pMECs derived from earlier progenitor or stem cells receive RANK or LtβR signaling. Nonclassical NF-κB activation results in differentiation of pro-pMECs into pMECs expressing higher levels of RANK. When pMECs receive RANK-TRAF6 signaling, pMECs differentiate into Aire-expressing mTECs.
DISCUSSION
Ly51– mTECs expressing low levels of MHCII and CD80 are thought to be immature mTECs that differentiate into Aire+ mTECs (Gäbler et al., 2007; Gray et al., 2007). In contrast, pMECs and pro-pMECs show Ly51 expression, which is down-regulated when pMECs differentiate into mature mTECs. Thus, pMECs should represent an earlier differentiation stage than Ly51– immature mTECs. Immature Ly51– mTECs may represent a transitional stage between pMECs and Aire+ mTECs and are converted in Aire+ mTECs without additional signaling.
Recent studies have proposed that common precursor cells for mTECs and cTECs express the cTEC molecules β5t and CD205 (Baik et al., 2013; Ohigashi et al., 2013, 2015; Ribeiro et al., 2013; Mayer et al., 2016). Our data suggested that pMECs express high levels of the cTEC molecules β5t, Ly51, and keratin-8. Thus, the cTEC gene expression profile of common precursor cells may be retained in pro-pMECs and pMECs during their differentiation. A recent study suggested that claudin3/4 (Clds)+ SSEA-1+ TECs show properties of mTEC stem cells (Sekai et al., 2014). Importantly, this cell fraction does not express UEA-1 ligand and RANK (Sekai et al., 2014; Baik et al., 2016). Furthermore, a recent study indicated that differentiation of Clds+SSEA-1+ TECs did not require RelB (Baik et al., 2016). Therefore, pro-pMECs and pMECs may be differentiated from Clds+SSEA-1+ TECs. The mechanisms linking pro-pMECs and pMECs with SSEA-1+Clds+ TECs should be clarified in the future.
In vitro RTOC experiments suggested that pMECs could be converted into Aire+ mTECs after 5 d of culture. Intriguingly, Aire+ mTECs were also observed in the thymus derived from RTOCs mixed with pMECs at 8 wk after kidney transplantation. Because Aire+ mTECs turn over rapidly (Gray et al., 2007), these data suggested that pMECs may give rise to Aire+ mTECs from 5 d to 8 wk after transplantation. Moreover, inoculation of the transplanted fetal thymus with 10 pMECs resulted in a medullar region containing Aire+ mTECs at 4 wk after transplantation, supporting the proliferative nature of pMECs or their progeny. Further studies are needed to elucidate the proliferative and stem cell-like properties of pMECs.
Recent studies have suggested the high heterogeneity of mTECs in the adult thymus (Lkhagvasuren et al., 2013; Danzl et al., 2014). LtβR signaling preferentially promotes the development of a subset of mTECs and expression of some chemokines (Seach et al., 2008; White et al., 2010; Lkhagvasuren et al., 2013). Our data suggested that both RANK and LtβR signaling induced pMECs. However, pMECs induced by RANK signaling may be inherently different from pMECs induced by LtβR signaling. Such differences in pMEC stages could indirectly influence the properties of mature mTECs, even if RANK signaling promotes the differentiation of both types of pMECs into mature mTECs. Additionally, our findings may clarify the role of lymphotoxin signaling in Aire+ mTECs. A previous study showed that excess lymphotoxin signaling increases Aire+ mTECs in vivo (Chin et al., 2003); however, other studies have reported that lymphotoxin is not directly associated with differentiation of Aire+ mTECs (Venanzi et al., 2007) and is not essential for Aire+ mTEC differentiation (Boehm et al., 2003). Our model explains the sufficiency of excess lymphotoxin signaling in Aire+ mTEC induction, even though this pathway may not be essential for Aire+ mTEC differentiation.
Our finding suggested distinct roles of the TRAF6-dependent pathway and nonclassical NF-κB pathway induced by RANK signaling. To the best of our knowledge, this is the first example of the physiological function of RANK signaling independent of TRAF6. Interestingly, TRAF6-independent RANK signaling up-regulated RANK expression. High surface expression of RANK may be a prerequisite for RANK signaling controlled by a higher-order TRAF6-oligomerization complex (Yin et al., 2009). The involvement of this auto-amplification mechanism for RANK expression should be verified in other RANK signaling-mediated cell differentiation events.
In summary, our data suggested that introduction of pMECs into the thymus efficiently suppressed autoimmunity induced by mTEC dysfunction. Given that autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy and/or other autoimmune diseases are caused by mTEC dysfunction as a result of Aire mutations (Mathis and Benoist, 2009; Akiyama et al., 2013), pMECs may be used to prevent the onset of such autoimmune diseases. Further characterization of pMECs is needed to develop such cell-based therapies.
MATERIALS AND METHODS
Animals
C57BL/6, BALB/cA, BALB/cA nu/nu, and aly/aly mice were obtained from CLEA. Aly/aly mice on the BALB/cA background (backcrossed 10 times) were described previously (Shinzawa et al., 2011). RANK-EGFP mice on the C57BL/6 background were also described previously (Maeda et al., 2012). RANK-EGFP mice on a BALB/cA background were prepared by backcrossing with BALB/cA mice for 10 generations. Traf6−/−, Rank−/−, Ltbr−/−, and Rankl−/− mice were described previously (Akiyama et al., 2005, 2008; Mouri et al., 2011). Relb−/− mice were purchased from the Jackson Laboratory. Littermate mice were used as controls. All mice were maintained under specific pathogen–free conditions and were handled in accordance with the Guidelines for Animal Experiments of the Institute of Medical Science (The University of Tokyo, Tokyo, Japan). The morning of finding a vaginal plug was designated as E0.5.
Antibodies and reagents
The following fluorescent anti–mouse antibodies were used for flow cytometry: anti-CD16/32, APCCy7-anti-CD45 (clone 30 F-11), APCCy7- or PECy7-anti-TER119 (clone TER-119), PE- or PECy7-anti-EpCAM (CD326, clone G8.8), FITC- or PECy7-anti-MHCII (I-A/I-E, clone M5/114.15.2), APC-anti-Ly51 (clone 6C3; BioLegend), PE-anti-CD80 (clone 16-10A1), PE-anti-CD24 (clone M1/69; eBioscience), and biotinylated UEA-1 (Vector Laboratories) antibodies. An agonistic anti-LtbR antibody was purchased from Alexis Biochemicals. Recombinant RANKL was a generous gift from Oriental Yeast Co. Anti-RANKL antibodies were prepared from a subclone of hybridoma obtained by fusing mouse myeloma cells with B cells.
Flow cytometric analysis and cell sorting
Embryonic thymi were digested in RPMI-1640 medium containing Collagenase/Dispase (Roche) at 0.125% wt/vol plus DNase I (Sigma-Aldrich) at 0.01% wt/vol by incubation at 37°C for 20 min (twice), with pipetting after each incubation. Reactions were stopped by addition of 10 mM EDTA and washing with FACS buffer. Single-cell suspensions were stained with anti–mouse antibodies as described above. Dead cells were excluded with 7-aminoactinomycin D staining. Cells were sorted using a cell sorter (FACSAria; BD). The sorting efficiency was routinely >98%. To compare MFI values in different experiments, external positive controls and/or internal positive and negative controls were used to adjust staining and measurement conditions. Otherwise, MFI values were compared in same day experiments.
RTOC and implantation
For isolation of pMECs, 100 µg anti-RANKL-Ab was intraperitoneally injected into RANK-EGFP mice at 13 d after vaginal plug detection. pMECs (3 × 104) from E17.5 RANK-EGFP mice treated with anti-RANKL-Ab were reaggregated together with trypsin-digested thymic cells (5 × 105) from E15.5 or E16.5 aly/aly mice on the BALB/cA background. Anti-RANKL-Ab (100 µg) was subcutaneously injected into LtbR−/− RANK-EGFP mice at 9 d after vaginal plug detection when pro-pMECs were sorted. Pro-pMECs (3 × 104) from E15.5 LtbR−/− RANK-EGFP mice treated with RANKL-Ab were reaggregated with digested thymic cells (5 × 105) from E15.5 or E16.5 aly/aly mice. RTOCs were cultured on Nucleopore filters (Whatman) placed in R10 medium containing RPMI-1640 (Invitrogen) supplemented with 10% FBS, 2 mM l-glutamine (Wako), 1× nonessential amino acids (NEAAs; Sigma-Aldrich), 100 U/ml penicillin (Ban-yu), 100 µg/ml streptomycin (Meiji), and 50 µM 2-mercaptoethanol (Wako) for 5 d. RTOCs were analyzed by FACS, RT-PCR, and immunohistochemical staining analysis. For implantation experiments, RTOCs were grafted under the kidney capsule of 6-wk-old female nude mice.
FTOC
Thymic lobes were isolated from E15.5 mice and cultured for 4 d on Nucleopore filters (Whatman) placed on R10 medium containing 1.35 mM 2′-deoxyguanosine (2DG; Sigma-Aldrich). Fetal thymic stroma (2DG-FTOC) was subsequently cultured in R10 with recombinant RANKL (1 µg/ml; Oriental Yeast Co.), agonistic anti-LtbR monoclonal antibodies (1 µg/ml), or MV1 (5 µM). 5 d after ligation, 2DG-FTOC was used for the analysis.
Immunohistochemistry
For immunohistochemical staining, samples were embedded in OCT compound frozen on dry ice and sectioned into 5-µm slices. Cryosections were fixed for 20 min in 4% paraformaldehyde in PBS on ice and washed with PBS containing 10 mM glycine. After treatment with 0.1% Triton X in PBS for 5 min on ice, sections were blocked with 10% goat serum in PBS. RANK expression in RANK-EGFP mice was detected using a polyclonal chicken anti-GFP antibody (1:300; Abcam) followed by a secondary goat anti–chicken Alexa Fluor 488–conjugated antibody (1:100; Invitrogen). Keratin-5 was detected using a polyclonal rabbit anti–mouse K5 antibody (1:300; Covance), followed by a secondary goat anti–rabbit Alexa Fluor 546–conjugated antibody (1:100; Invitrogen). Aire was detected using a monoclonal rat anti-Aire antibody (1:300; clone 5H12; eBioscience) and a secondary goat anti–rat Alexa Fluor 546–conjugated antibody (1:100; Invitrogen). Biotin UEA-1 (1:50; Vector Laboratories) and streptavidin Alexa Fluor 647 (1:100; Invitrogen) were used to detect UEA-1–binding protein.
qRT-PCR
Total RNA was extracted using an RNeasy Micro kit (QIAGEN). cDNA was obtained by random-primed RT using a PrimescriptII First Strand cDNA Synthesis kit (Takara Bio, Inc.). qPCR was performed using an ABI PRISM 7300 Sequence Detection System and SYBR Green Master Mix (Toyobo). Expression values for each sample were normalized to GAPDH or 36B4.
Histological examination and detection of autoantibodies
Organs from nude mice were stained with hematoxylin and eosin at 8 wk after implantation. The degrees of inflammatory cell infiltration were scored as follows: 0, no detectable infiltration; 1, a focus of perivascular infiltration; 2, several foci of perivascular infiltration; 3, cellular infiltration in >50% of the vasculature; 4, severe infiltration throughout the interstitial region. To detect autoantibodies in serum, tissue samples from Rag2−/− mice were embedded in OCT compound, frozen on dry ice, and sectioned into 5-µm-thick slices. Cryosections were fixed with ice-cold acetone for 5 min and washed with PBS. Samples were blocked with 10% goat serum in PBS and incubated with serum (100× dilution) for 1 h at room temperature. The slides were subsequently incubated with secondary antibody (anti–mouse IgG-Alexa Fluor 488) and 1 µg/ml propidium iodide for 40 min at room temperature. Confocal color images were obtained using an FV1000D (Olympus).
Intrathymic injection
100 µg anti–RANKL-Ab was injected subcutaneously into RANK-EGFP mice (B6) 9 d after vaginal plug detection. Thymic lobes were dissected from E14.5 embryos, and thymic stromal cells were prepared as previously described (Akiyama et al., 2014). Cells were sorted using a cell sorter (BD Aria; BD). Sorted cells were injected into E14.5 thymic lobes of aly/aly mice (10 cells/lobe). After culture for 1 d, thymic lobes were transplanted under the kidney capsule of 6-wk-old female nude mice. At 4 wk after transplantation, grafted thymi were subjected to immunohistochemical analysis.
COBRA
DNA methylation analysis was performed as described previously (Wu et al., 2012). In brief, cells isolated from the embryonic thymus were subjected to bisulfite treatment using an EZ DNA Methylation-Direct kit (Zymo Research Cooperation). PCR of Aire was performed using bisulfite-converted genomic DNA with the following forward and reverse primers: 5′-TTTGATTTAAATATTTGTTGGATAAGGA-3′ and 5′-CCCATTTTAATACTCCCAATCTTCT-3′, respectively. COBRA was performed by evenly dividing the PCR products to incubate with HpyCH4IV for 5 h at 37°C, and samples were then analyzed with a microchip electrophoresis system (MultiNA; Shimadzu Biotech). Unmethylated DNA levels were calculated as the percentage of undigested fragment compared with the sum of the digested and undigested fragments.
Synthesis of Boc-MV-1 and MV1
N- and C-terminal protected Boc-MV-1 was synthesized using conventional solution-phase methods, in which 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDC) hydrochloride and 1-hydroxybenzotriazole (HOBt) hydrate were used as coupling reagents. The coupling conditions were as follows: N-terminal free amino acid or peptide (1 Eq.), Boc-protected amino acid (1.2 Eq.), EDC (1.5 Eq.), HOBt (1.5 Eq.), and N,N-diisopropylethylamine (DIPEA, 2 Eq.) in dichloromethane (CH2Cl2) at room temperature for 5 h. The deprotection conditions were as follows: Boc-protected peptide (1 Eq.), trifluoroacetic acid (TFA, 10 Eq.) in CH2Cl2 at room temperature for 30 min. Boc-MV-1: 1H NMR (400 MHz, CDCl3): δ 7.19–7.33 (m, 11H), 6.70 (br s, 1H), 5.37 (t, 1H, J = 8.4 Hz), 4.47 (m, 1H), 4.38 (d, 1H, J = 8.4 Hz), 3.62 (m, 1H), 3.49 (s, 3H), 3.38 (m, 1H), 2.79 (s, 3H), 2.07 (m, 1H), 1.61–1.82 (m, 10H), 1.49 (s, 9H), 1.29 (d, 3H, J = 10.8 Hz), 0.96–1.24 (m, 6H); [HR-ESI(+)]: m/z calculated for C38H53N4O7 [M+H]+: 677.3914, found: 677.3920. A solution of Boc-MV-1 (1.35 g, 2.0 mmol) in 4 M HCl in 1,4-dioxane (6 ml) was prepared at 0°C, and the mixture was stirred at room temperature for 4 h. Removal of the solvent afforded MV-1 hydrochloride salt (1.22 g, >99%), as a colorless crystal. MV-1: [HR-ESI(+)]: m/z calc’d for C33H45N4O5 [M+H]+: 577.3390, found: 577.3368.
Statistical analysis
P-values were calculated using Student’s t tests with a two-tailed distribution and two-sample equivalent variance parameters or Mann-Whitney U test.
Online supplemental material
Figs. S1–S3 include gating strategies for isolation of pMECs, pro-pMECs, and other TECs. Online supplemental materials are available at http://www.jem.org/cgi/content/full/jem.20151780/DC1.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Challenging Exploratory Research from Japanese Society for the Promotion of Science (26670234; T. Akiyama); a Grant-in-Aid for Scientific Research on Innovative Areas from Ministry of Education, Culture, Sports, Science, and Technology in Japan (15H01150 and 16H01631; T. Akiyama); Princess Takamatsu Cancer Research Fund (T. Akiyama); and a grant from the Specially Promoted Project of the Toyota Physical and Chemical Research Institute (T. Akiyama).
H. Yasuda belongs to Oriental Yeast Co. Ltd., which provided the recombinant RANKL and anti–RANKL-Ab used in this study. The authors declare no additional competing financial interests.
Footnotes
Abbreviations used:
- 2DG-FTOC
- fetal thymic stroma organ culture
- cIAP
- cellular inhibitor of apoptosis
- COBRA
- combined bisulfite restriction analysis
- LtβR
- lymphotoxin β-receptor
- mTEC
- medullary thymic epithelial cell
- RANK
- receptor activator of nuclear factor-κB
- RTOC
- reaggregation thymus organ culture
- TRAF
- TNF receptor-associated factor
- TSA
- tissue-specific antigen
References
- Akiyama N., Shinzawa M., Miyauchi M., Yanai H., Tateishi R., Shimo Y., Ohshima D., Matsuo K., Sasaki I., Hoshino K., et al. 2014. Limitation of immune tolerance-inducing thymic epithelial cell development by Spi-B-mediated negative feedback regulation. J. Exp. Med. 211:2425–2438. 10.1084/jem.20141207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T., Maeda S., Yamane S., Ogino K., Kasai M., Kajiura F., Matsumoto M., and Inoue J.. 2005. Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science. 308:248–251. 10.1126/science.1105677 [DOI] [PubMed] [Google Scholar]
- Akiyama T., Shimo Y., Yanai H., Qin J., Ohshima D., Maruyama Y., Asaumi Y., Kitazawa J., Takayanagi H., Penninger J.M., et al. 2008. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 29:423–437. 10.1016/j.immuni.2008.06.015 [DOI] [PubMed] [Google Scholar]
- Akiyama T., Shinzawa M., and Akiyama N.. 2012. TNF receptor family signaling in the development and functions of medullary thymic epithelial cells. Front. Immunol. 3:278 10.3389/fimmu.2012.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T., Shinzawa M., Qin J., and Akiyama N.. 2013. Regulations of gene expression in medullary thymic epithelial cells required for preventing the onset of autoimmune diseases. Front. Immunol. 4:249 10.3389/fimmu.2013.00249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik S., Jenkinson E.J., Lane P.J., Anderson G., and Jenkinson W.E.. 2013. Generation of both cortical and Aire(+) medullary thymic epithelial compartments from CD205+ progenitors. Eur. J. Immunol. 43:589–594. 10.1002/eji.201243209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik S., Sekai M., Hamazaki Y., Jenkinson W.E., and Anderson G.. 2016. Relb acts downstream of medullary thymic epithelial stem cells and is essential for the emergence of RANK+ medullary epithelial progenitors. Eur. J. Immunol. 46:857–862. 10.1002/eji.201546253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett A.R., Farley A., Blair N.F., Gordon J., Sharp L., and Blackburn C.C.. 2002. Identification and characterization of thymic epithelial progenitor cells. Immunity. 16:803–814. 10.1016/S1074-7613(02)00321-7 [DOI] [PubMed] [Google Scholar]
- Bleul C.C., Corbeaux T., Reuter A., Fisch P., Mönting J.S., and Boehm T.. 2006. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature. 441:992–996. 10.1038/nature04850 [DOI] [PubMed] [Google Scholar]
- Boehm T., Scheu S., Pfeffer K., and Bleul C.C.. 2003. Thymic medullary epithelial cell differentiation, thymocyte emigration, and the control of autoimmunity require lympho-epithelial cross talk via LTβR. J. Exp. Med. 198:757–769. 10.1084/jem.20030794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin R.K., Lo J.C., Kim O., Blink S.E., Christiansen P.A., Peterson P., Wang Y., Ware C., and Fu Y.X.. 2003. Lymphotoxin pathway directs thymic Aire expression. Nat. Immunol. 4:1121–1127. 10.1038/ni982 [DOI] [PubMed] [Google Scholar]
- Danzl N.M., Jeong S., Choi Y., and Alexandropoulos K.. 2014. Identification of novel thymic epithelial cell subsets whose differentiation is regulated by RANKL and Traf6. PLoS One. 9:e86129 10.1371/journal.pone.0086129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya Y., Mori K., Ninomiya T., Tomimori Y., Tanaka S., Takahashi N., Udagawa N., Uchida K., and Yasuda H.. 2011. Increased bone mass in mice after single injection of anti-receptor activator of nuclear factor-κB ligand-neutralizing antibody: evidence for bone anabolic effect of parathyroid hormone in mice with few osteoclasts. J. Biol. Chem. 286:37023–37031. 10.1074/jbc.M111.246280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gäbler J., Arnold J., and Kyewski B.. 2007. Promiscuous gene expression and the developmental dynamics of medullary thymic epithelial cells. Eur. J. Immunol. 37:3363–3372. 10.1002/eji.200737131 [DOI] [PubMed] [Google Scholar]
- Gill J., Malin M., Holländer G.A., and Boyd R.. 2002. Generation of a complete thymic microenvironment by MTS24+ thymic epithelial cells. Nat. Immunol. 3:635–642. 10.1038/ni812 [DOI] [PubMed] [Google Scholar]
- Gray D., Abramson J., Benoist C., and Mathis D.. 2007. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J. Exp. Med. 204:2521–2528. 10.1084/jem.20070795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerau-de-Arellano M., Martinic M., Benoist C., and Mathis D.. 2009. Neonatal tolerance revisited: a perinatal window for Aire control of autoimmunity. J. Exp. Med. 206:1245–1252. 10.1084/jem.20090300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka Y., Nitta T., Ohigashi I., Yano K., Ishimaru N., Hayashi Y., Matsumoto M., Matsuo K., Penninger J.M., Takayanagi H., et al. 2008. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 29:438–450. 10.1016/j.immuni.2008.06.018 [DOI] [PubMed] [Google Scholar]
- Hsieh C.S., Lee H.M., and Lio C.W.. 2012. Selection of regulatory T cells in the thymus. Nat. Rev. Immunol. 12:157–167. 10.1038/nri3155 [DOI] [PubMed] [Google Scholar]
- Irla M., Hugues S., Gill J., Nitta T., Hikosaka Y., Williams I.R., Hubert F.X., Scott H.S., Takahama Y., Holländer G.A., and Reith W.. 2008. Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity. 29:451–463. 10.1016/j.immuni.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Kajiura F., Sun S., Nomura T., Izumi K., Ueno T., Bando Y., Kuroda N., Han H., Li Y., Matsushima A., et al. 2004. NF-κ B-inducing kinase establishes self-tolerance in a thymic stroma-dependent manner. J. Immunol. 172:2067–2075. 10.4049/jimmunol.172.4.2067 [DOI] [PubMed] [Google Scholar]
- Khan I.S., Mouchess M.L., Zhu M.L., Conley B., Fasano K.J., Hou Y., Fong L., Su M.A., and Anderson M.S.. 2014. Enhancement of an anti-tumor immune response by transient blockade of central T cell tolerance. J. Exp. Med. 211:761–768. 10.1084/jem.20131889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L., Hinterberger M., Wirnsberger G., and Kyewski B.. 2009. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 9:833–844. 10.1038/nri2669 [DOI] [PubMed] [Google Scholar]
- Kyewski B., and Klein L.. 2006. A central role for central tolerance. Annu. Rev. Immunol. 24:571–606. 10.1146/annurev.immunol.23.021704.115601 [DOI] [PubMed] [Google Scholar]
- Lkhagvasuren E., Sakata M., Ohigashi I., and Takahama Y.. 2013. Lymphotoxin β receptor regulates the development of CCL21-expressing subset of postnatal medullary thymic epithelial cells. J. Immunol. 190:5110–5117. 10.4049/jimmunol.1203203 [DOI] [PubMed] [Google Scholar]
- Maeda K., Kobayashi Y., Udagawa N., Uehara S., Ishihara A., Mizoguchi T., Kikuchi Y., Takada I., Kato S., Kani S., et al. 2012. Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat. Med. 18:405–412. 10.1038/nm.2653 [DOI] [PubMed] [Google Scholar]
- Mathis D., and Benoist C.. 2009. Aire. Annu. Rev. Immunol. 27:287–312. 10.1146/annurev.immunol.25.022106.141532 [DOI] [PubMed] [Google Scholar]
- Mayer C.E., Žuklys S., Zhanybekova S., Ohigashi I., Teh H.Y., Sansom S.N., Shikama-Dorn N., Hafen K., Macaulay I.C., Deadman M.E., et al. 2016. Dynamic spatio-temporal contribution of single β5t+ cortical epithelial precursors to the thymus medulla. Eur. J. Immunol. 46:846–856. 10.1002/eji.201545995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouri Y., Yano M., Shinzawa M., Shimo Y., Hirota F., Nishikawa Y., Nii T., Kiyonari H., Abe T., Uehara H., et al. 2011. Lymphotoxin signal promotes thymic organogenesis by eliciting RANK expression in the embryonic thymic stroma. J. Immunol. 186:5047–5057. 10.4049/jimmunol.1003533 [DOI] [PubMed] [Google Scholar]
- Ohigashi I., Zuklys S., Sakata M., Mayer C.E., Zhanybekova S., Murata S., Tanaka K., Holländer G.A., and Takahama Y.. 2013. Aire-expressing thymic medullary epithelial cells originate from β5t-expressing progenitor cells. Proc. Natl. Acad. Sci. USA. 110:9885–9890. 10.1073/pnas.1301799110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohigashi I., Zuklys S., Sakata M., Mayer C.E., Hamazaki Y., Minato N., Hollander G.A., and Takahama Y.. 2015. Adult thymic medullary epithelium is maintained and regenerated by lineage-restricted cells rather than bipotent progenitors. Cell Reports. 13:1432–1443. 10.1016/j.celrep.2015.10.012 [DOI] [PubMed] [Google Scholar]
- Onder L., Nindl V., Scandella E., Chai Q., Cheng H.W., Caviezel-Firner S., Novkovic M., Bomze D., Maier R., Mair F., et al. 2015. Alternative NF-κB signaling regulates mTEC differentiation from podoplanin-expressing presursors in the cortico-medullary junction. Eur. J. Immunol. 45:2218–2231. 10.1002/eji.201545677 [DOI] [PubMed] [Google Scholar]
- Ribeiro A.R., Rodrigues P.M., Meireles C., Di Santo J.P., and Alves N.L.. 2013. Thymocyte selection regulates the homeostasis of IL-7-expressing thymic cortical epithelial cells in vivo. J. Immunol. 191:1200–1209. 10.4049/jimmunol.1203042 [DOI] [PubMed] [Google Scholar]
- Rietze R.L., Valcanis H., Brooker G.F., Thomas T., Voss A.K., and Bartlett P.F.. 2001. Purification of a pluripotent neural stem cell from the adult mouse brain. Nature. 412:736–739. 10.1038/35089085 [DOI] [PubMed] [Google Scholar]
- Roberts N.A., White A.J., Jenkinson W.E., Turchinovich G., Nakamura K., Withers D.R., McConnell F.M., Desanti G.E., Benezech C., Parnell S.M., et al. 2012. Rank signaling links the development of invariant γδ T cell progenitors and Aire(+) medullary epithelium. Immunity. 36:427–437. 10.1016/j.immuni.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S.W., Jenkinson W.E., Anderson G., and Jenkinson E.J.. 2006. Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature. 441:988–991. 10.1038/nature04813 [DOI] [PubMed] [Google Scholar]
- Rossi S.W., Kim M.Y., Leibbrandt A., Parnell S.M., Jenkinson W.E., Glanville S.H., McConnell F.M., Scott H.S., Penninger J.M., Jenkinson E.J., et al. 2007. RANK signals from CD4+3− inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J. Exp. Med. 204:1267–1272. 10.1084/jem.20062497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seach N., Ueno T., Fletcher A.L., Lowen T., Mattesich M., Engwerda C.R., Scott H.S., Ware C.F., Chidgey A.P., Gray D.H., and Boyd R.L.. 2008. The lymphotoxin pathway regulates Aire-independent expression of ectopic genes and chemokines in thymic stromal cells. J. Immunol. 180:5384–5392. 10.4049/jimmunol.180.8.5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekai M., Hamazaki Y., and Minato N.. 2014. Medullary thymic epithelial stem cells maintain a functional thymus to ensure lifelong central T cell tolerance. Immunity. 41:753–761. 10.1016/j.immuni.2014.10.011 [DOI] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., and Visvader J.E.. 2006. Generation of a functional mammary gland from a single stem cell. Nature. 439:84–88. 10.1038/nature04372 [DOI] [PubMed] [Google Scholar]
- Shinzawa M., Maruyama Y., Qin J., Akiyama N., Miyauchi M., Yanai H., Takami M., Inoue J., and Akiyama T.. 2011. Splenic extramedullary hemopoiesis caused by a dysfunctional mutation in the NF-κB-inducing kinase gene. Biochem. Biophys. Res. Commun. 414:773–778. 10.1016/j.bbrc.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Nakato G., Jinnohara T., Akiba H., Okumura K., Ohno H., and Yoshida H.. 2012. Expression pattern changes and function of RANKL during mouse lymph node microarchitecture development. Int. Immunol. 24:369–378. 10.1093/intimm/dxs002 [DOI] [PubMed] [Google Scholar]
- Ucar A., Ucar O., Klug P., Matt S., Brunk F., Hofmann T.G., and Kyewski B.. 2014. Adult thymus contains FoxN1(-) epithelial stem cells that are bipotent for medullary and cortical thymic epithelial lineages. Immunity. 41:257–269. 10.1016/j.immuni.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev E., Blankenship J.W., Wayson S.M., Fedorova A.V., Kayagaki N., Garg P., Zobel K., Dynek J.N., Elliott L.O., Wallweber H.J., et al. 2007. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell. 131:669–681. 10.1016/j.cell.2007.10.030 [DOI] [PubMed] [Google Scholar]
- Venanzi E.S., Gray D.H., Benoist C., and Mathis D.. 2007. Lymphotoxin pathway and Aire influences on thymic medullary epithelial cells are unconnected. J. Immunol. 179:5693–5700. 10.4049/jimmunol.179.9.5693 [DOI] [PubMed] [Google Scholar]
- Weih F., and Caamaño J.. 2003. Regulation of secondary lymphoid organ development by the nuclear factor-κB signal transduction pathway. Immunol. Rev. 195:91–105. 10.1034/j.1600-065X.2003.00064.x [DOI] [PubMed] [Google Scholar]
- Weih F., Carrasco D., Durham S.K., Barton D.S., Rizzo C.A., Ryseck R.P., Lira S.A., and Bravo R.. 1995. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-κB/Rel family. Cell. 80:331–340. 10.1016/0092-8674(95)90416-6 [DOI] [PubMed] [Google Scholar]
- White A.J., Nakamura K., Jenkinson W.E., Saini M., Sinclair C., Seddon B., Narendran P., Pfeffer K., Nitta T., Takahama Y., et al. 2010. Lymphotoxin signals from positively selected thymocytes regulate the terminal differentiation of medullary thymic epithelial cells. J. Immunol. 185:4769–4776. 10.4049/jimmunol.1002151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K., Lister N.L., Barsanti M., Lim J.M., Hammett M.V., Khong D.M., Siatskas C., Gray D.H., Boyd R.L., and Chidgey A.P.. 2014. Multilineage potential and self-renewal define an epithelial progenitor cell population in the adult thymus. Cell Reports. 8:1198–1209. 10.1016/j.celrep.2014.07.029 [DOI] [PubMed] [Google Scholar]
- Wu G., Hirabayashi K., Sato S., Akiyama N., Akiyama T., Shiota K., and Yagi S.. 2012. DNA methylation profile of Aire-deficient mouse medullary thymic epithelial cells. BMC Immunol. 13:58 10.1186/1471-2172-13-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi S., Hirabayashi K., Sato S., Li W., Takahashi Y., Hirakawa T., Wu G., Hattori N., Hattori N., Ohgane J., et al. 2008. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 18:1969–1978. 10.1101/gr.074070.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Fujikado N., Kolodin D., Benoist C., and Mathis D.. 2015. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 348:589–594. 10.1126/science.aaa7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q., Lin S.C., Lamothe B., Lu M., Lo Y.C., Hura G., Zheng L., Rich R.L., Campos A.D., Myszka D.G., et al. 2009. E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 16:658–666. 10.1038/nsmb.1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.