Orta-Mascaró, Lozano, and collaborators provide the first analysis of CD6-deficient mice, showing that this molecule modulates T cell receptor signaling and the threshold for thymocyte and peripheral T cell subset selection.
Abstract
The CD6 glycoprotein is a lymphocyte surface receptor putatively involved in T cell development and activation. CD6 facilitates adhesion between T cells and antigen-presenting cells through its interaction with CD166/ALCAM (activated leukocyte cell adhesion molecule), and physically associates with the T cell receptor (TCR) at the center of the immunological synapse. However, its precise role during thymocyte development and peripheral T cell immune responses remains to be defined. Here, we analyze the in vivo consequences of CD6 deficiency. CD6−/− thymi showed a reduction in both CD4+ and CD8+ single-positive subsets, and double-positive thymocytes exhibited increased Ca2+ mobilization to TCR cross-linking in vitro. Bone marrow chimera experiments revealed a T cell–autonomous selective disadvantage of CD6−/− T cells during development. The analysis of TCR-transgenic mice (OT-I and Marilyn) confirmed that abnormal T cell selection events occur in the absence of CD6. CD6−/− mice displayed increased frequencies of antigen-experienced peripheral T cells generated under certain levels of TCR signal strength or co-stimulation, such as effector/memory (CD4+TEM and CD8+TCM) and regulatory (T reg) T cells. The suppressive activity of CD6−/− T reg cells was diminished, and CD6−/− mice presented an exacerbated autoimmune response to collagen. Collectively, these data indicate that CD6 modulates the threshold for thymocyte selection and the generation and/or function of several peripheral T cell subpopulations, including T reg cells.
T cell development is a highly regulated physiological process through which T cells acquire competence for antigen recognition; those recognizing self-antigens with high affinity are physically deleted before migrating to peripheral lymphoid organs (Palmer, 2003). Alterations of this process lead to both defective immune responses to foreign antigens and to autoimmunity. Even though the avidity of the TCR for self-peptide–MHC complexes is a key factor in determining the fate of developing thymocytes and the outcome of peripheral T cell immune responses, concomitant signals provided by other lymphocyte surface receptors are known to influence this process by increasing or reducing the threshold for TCR signaling (Palmer, 2003). One such receptor is CD5 (Soldevila et al., 2011), and this could be also the case for the related molecule CD6, as they are highly homologous receptors encoded by contiguous genes most likely arising from duplication of a common ancestor. Indeed, CD5 associates with the antigen-specific receptor complex and negatively modulates its signaling. In turn, the expression levels of CD5 on T cells reflects the strength of TCR signaling, which reciprocally tunes the threshold of the response.
CD6 is a 105–130-kD surface glycoprotein expressed on all T cells from early stages of their development, but also on some B and NK subsets, BM precursors, and brain areas (Santos et al., 2016). The extracellular domain of CD6 comprises three scavenger receptor cysteine-rich extracellular domains, the most membrane-proximal of which (D3) interacts with the N-terminal immunoglobulin domain of CD166/ALCAM (activated leukocyte cell adhesion molecule), a broadly expressed cell adhesion molecule (Santos et al., 2016). The structure of CD6 and the binding region of ALCAM have recently been resolved (Chappell et al., 2015). In vitro assays revealed that this interaction is critical for optimal T cell activation and proliferative responses (Gimferrer et al., 2004; Hassan et al., 2004; Zimmerman et al., 2006). Indeed, when co-cross-linked with anti-CD3 mAb, CD6 increased proliferation, intracellular Ca2+ levels, and activation of MAPK in human T cells (Santos et al., 2016). The CD6 cytoplasmic region is devoid of intrinsic catalytic activity, but harbors consensus motifs for phosphorylation and association with signal-transducing effectors. Accordingly, CD6 holds two constitutively phosphorylated Ser clusters needed for proper MAPK activation (Bonet et al., 2013), and nine Tyr residues that may be phosphorylated upon TCR activation and serve as docking sites for downstream signaling effectors, such as Syntenin-1 and SLP-76 (Gimferrer et al., 2005; Hassan et al., 2006). The latter has been recently confirmed by quantitative proteomic analysis of primary mouse T cells (Roncagalli et al., 2014), showing that SLP-76 binds to CD6 in a ZAP-70–dependent but LAT-independent manner, and placing CD6 as a signaling molecule that contributes to the diversification of TCR signals. In this regard, CD6 is well positioned to modulate the TCR signaling as it physically associates with the TCR complex and co-localizes with it at the center of the immunological synapse during APC–T cell contacts (Gimferrer et al., 2004; Zimmerman et al., 2006). However, the belief that CD6 behaves as a co-stimulatory molecule has been recently challenged by in vitro data showing that it may also act as a negative modulator of TCR signaling (Hassan et al., 2006; Oliveira et al., 2012).
Besides its role in T cell activation, a single study has also shown CD6 to be involved in thymocyte survival and selection in mice and humans (Singer et al., 2002). This study showed that CD6 surface expression levels increase when double-positive (DP) thymocytes are selected to become single-positive (SP) cells, and that such increases correlate with both expression of the selection marker CD69 and resistance to apoptosis.
All the aforementioned in vitro evidences support a putative involvement for CD6 in the modulation of thymocyte selection and T lymphocyte activation (Santos et al., 2016). However, the lack of proper animal models has hindered further understanding of its in vivo role during relevant physiological processes. This report provides the first analysis on the in vivo consequences of CD6 deficiency on T cell development, homeostasis, and activation. Our results show that CD6 modulates the threshold for negative selection in thymus and influences the generation and/or function of peripheral effector/memory and regulatory (T reg) T cells. The functional defects associated with CD6 deficiency also result in an exacerbated autoimmune response in a model of collagen-induced arthritis (CIA).
RESULTS AND DISCUSSION
CD6 deficiency impacts normal T cell development
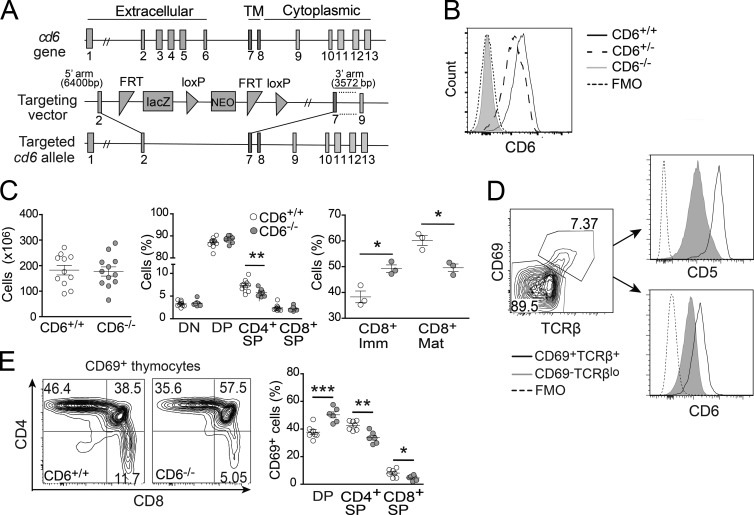
The role of CD6 in T cell development was assessed by generating CD6−/− C57BL/6N mice in which exons coding for the extracellular region of CD6 were deleted by homologous recombination (Fig. 1 A). Flow cytometry analysis of LN CD4+ and CD8+ T cells from CD6−/− mice showed a complete lack of CD6 surface expression as compared with CD6+/+ and CD6+/− littermates (Fig. 1 B). Analysis of thymus from sex- and age-matched CD6−/− and CD6+/+ littermates showed no significant differences in total cell numbers (Fig. 1 C, left). Although no differences were found in the frequency of NKT (CD161+CD3+) and γδ T cells (not depicted), as well as of DP or double-negative (DN) thymocyte subsets, a significant decrease was observed for CD4+SP cells (Fig. 1 C, middle). A detailed analysis of CD8+SP cells showed a reduction in the frequency of mature CD8+SP (TCRβ+CD24−), together with an increase in immature CD8+SP (TCRβ−CD24+; Fig. 1 C, right). These findings suggest the involvement of CD6 in the final steps of thymocyte selection. Consistent with that view, CD6 expression correlated with that of the positive selection marker CD69 in DP thymocytes (Fig. 1 D). A similar situation has been documented for CD5, a negative modulator of TCR signaling (Tarakhovsky et al., 1995) whose expression is developmentally regulated in a TCR signal- and avidity-dependent manner (Azzam et al., 2001). A significant increase in the frequency of DP cells undergoing selection (CD69+DP) and a concomitant reduction in recently selected CD69+CD4+SP and CD69+CD8+SP was found in CD6−/− mice as compared with CD6+/+ mice (Fig. 1 E). These results suggest that, rather than a defect in positive selection per se, CD6 deficiency triggers an increase in the negative selection of CD4+ and CD8+SP cells, the final product of the DP to SP transition.
Figure 1.
Abnormal thymocyte maturation in CD6−/− mice. (A) Schematic representation of the gene targeting strategy used for generating CD6−/− mice. (B) Flow cytometry analysis of CD3+CD19− LN T cells from CD6+/+ (solid line), CD6+/− (dashed line), and CD6−/− (gray shaded line) mice stained for CD6 expression (mAb OX-129) or unstained as control (FMO, dotted line). (C) Total cell numbers in thymi (left) from CD6+/+ (open circles) and CD6−/− (gray filled circles) mice. The percentage (mean ± SEM) of main thymocyte subsets (middle) and of immature (TCRβ−CD24+) and mature (TCRβ+CD24−) CD8+ cells (right) is also shown. (D) Contour plot showing CD69 and TCRβ expression on gated DP cells from a CD6+/+ mouse. Histograms show CD5 (top) and CD6 (bottom) expression on nonselected (CD69−TCRβlo; gray line), and positively selected (CD69+TCRβ+; black line) DP cells from CD6+/+ mice. (E, left) Representative contour plots showing the percentage of DP, CD4+ SP, and CD8+ SP cells among total live CD69+ thymocytes in CD6+/+ and CD6−/− mice. Percentage (mean ± SEM) of CD69+ cells in CD4+SP, CD8+SP, and DP subsets. Each dot represents an individual mouse. *, P < 0.05; **, P < 0.01; ***, P = 0.001 (unpaired Student’s t test).
CD6 deficiency impacts peripheral effector/memory and regulatory T cell compartments
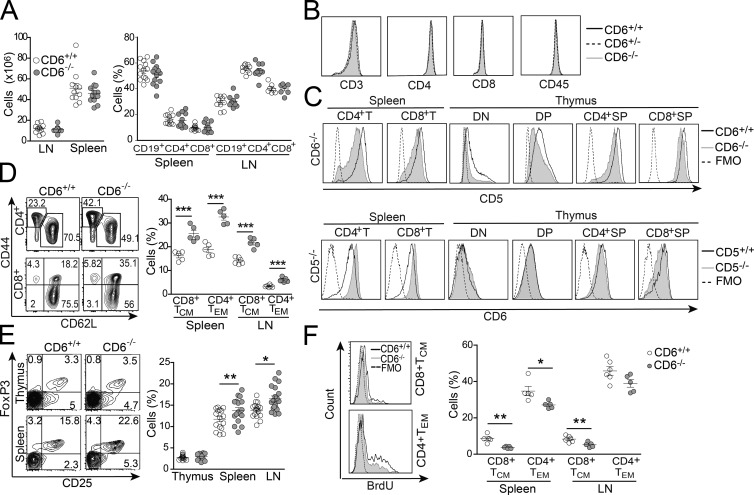
Analysis of spleen and LN from CD6−/− mice revealed total cell numbers (Fig. 2 A, left) and percentage of T (both CD4+ and CD8+), B (CD19+; Fig. 2 A, right), γδT, NK, and NKT cells (not depicted) similar to those found in CD6+/+ littermates. Surface levels of CD3, CD4, CD8, and CD45 from peripheral CD6−/− T cells were also similar to those of CD6+/+ cells (Fig. 2 B). However, a slightly reduced CD5 expression was observed on both CD4+ and CD8+ spleen T cells from CD6−/− mice compared with CD6+/+ littermates (Fig. 2 C). This was also true for thymocytes, from early to late maturational stages (Fig. 2 C). The intriguing fact is that the analysis of CD5−/− mice revealed increased CD6 expression in both CD4+ and CD8+ spleen T cells but not thymocytes. These reciprocal changes observed in both CD6−/− and CD5−/− mice suggest a functional cross-talk in the regulation of CD6 and CD5 surface expression that deserves further exploration.
Figure 2.
Phenotypical changes in peripheral T cell compartments in CD6−/− mice. (A, left) Total cell numbers in LN and spleen from CD6+/+ (open circles) and CD6−/− (gray filled circles) mice. (right) Percentage (mean ± SEM) of CD19+ (B), and CD4+, and CD8+ (T) cells from spleen and LNs. Each dot represents an individual mouse. (B) Flow cytometry analysis of spleen T cell surface marker expression from CD6+/+ (black solid line), CD6+/− (black dashed line), and CD6−/− (gray shaded line) mice. (C) Flow cytometry analysis of CD5 (top) and CD6 (bottom) expression on CD4+ and CD8+ spleen T cells, and DN, DP, CD4+SP, and CD8+SP thymocytes from wild-type (black solid line) and deficient (gray shaded line) mice for CD6 (top) and CD5 (bottom) receptors. FMO (dotted line). (D, left) Representative contour plots showing the percentage of CD4+ TEM (CD44hi CD62Llo) and CD8+ TCM (CD44hi CD62Lhi) cells from spleen of CD6+/+ and CD6−/− mice. (right) Percentage (mean ± SEM) of CD4+ TEM and CD8+ TCM cells in spleen and LN from CD6+/+ (open circles) and CD6−/− (gray filled circles) mice. (E, left) Representative contour plots showing the frequency of CD25+FoxP3+ in gated CD4+ thymus and spleen cells from CD6+/+ (left) and CD6−/− (right) mice. (right) Percentage (mean ± SEM) of T reg cells (CD4+CD25+FoxP3+) in thymus, spleen, and LNs from CD6+/+ (open circles) and CD6−/− (gray filled circles) mice. (F, left) Representative flow cytometry histograms showing in vivo BrdU incorporation into CD8+TCM (top) and CD4+TEM (bottom) spleen cells from CD6+/+ (black solid line) and CD6−/− mice (gray shaded line). (right) Dot density graph showing percentage (mean ± SEM) of BrdU+ CD8+TCM and CD4+TEM cells in spleen and LN from CD6+/+ (open circles) and CD6−/− (gray filled circles) mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired Student’s t test).
Analysis of spleen and LN also revealed that CD6−/− mice present significantly increased percentages of effector/memory CD4+ (CD4+TEM and CD44hiCD62Llo) and central memory CD8+ (CD8+TCM and CD44hiCD62Lhi) T cells (Fig. 2 D). Increased percentages of cells with T reg cell phenotype (CD4+CD25+FoxP3+) were also observed (Fig. 2 E). Interestingly, this increase could not be detected in the thymus, suggesting that CD6 deficiency does not affect the generation of thymic T reg cells. Collectively, the data indicate that CD6 deficiency favors the expansion of antigen-experienced peripheral T cell subsets generated under high TCR signal strength conditions (Lanzavecchia and Sallusto, 2005).
Increased frequency of effector/memory T cells has been attributed to homeostatic proliferation, also known as lymphopenia-induced proliferation (Jameson, 2002). Such an increase could result from the decreased thymic output observed in the absence of CD6 (Fig. 1 C). However, the lower BrdU incorporation observed in spleen and LN CD8+TCM and CD4+TEM cells from CD6−/− mice (Fig. 2 F) argued against the possibility of increased homeostatic proliferation. Instead, the previously reported role of CD6–ALCAM interactions in optimizing T cell activation and proliferative responses (Gimferrer et al., 2004; Hassan et al., 2004; Zimmerman et al., 2006) could feasibly account for the low in vivo proliferation observed.
T cell–intrinsic mechanisms account for changes in the central and peripheral T compartments found in CD6-deficient mice
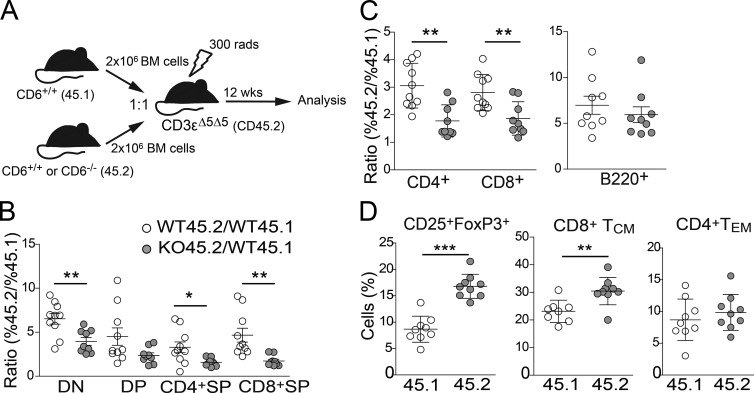
To determine whether the defect in developing and mature lymphocytes observed in CD6−/− mice is T cell–intrinsic, competitive mixed BM chimera experiments were performed. CD3-ε–deficient mice were used as recipients of BM cell mixes (1:1) from CD6+/+ (WT45.1) mice with that of CD6−/− (KO45.2) or CD6+/+ (WT45.2) mice (Fig. 3 A). Control mice injected with only CD6+/+ or CD6−/− BM cells showed no significant differences in total cell numbers from primary and secondary lymphoid organs (unpublished data), indicating that absence of CD6 did not affect BM engraftment and/or thymic seeding. In the mixed chimera control group (WT45.2/WT45.1), a competitive advantage for CD45.2 cells was observed as visible by CD45.2/CD45.1 ratios >1 (Fig. 3 B). However, this advantage was modulated in the experimental group (KO45.2/WT45.1), where the CD45.2/CD45.1 ratio was substantially reduced. In the thymus, this effect was observed for all major thymocyte developmental subsets, though it reached statistical significance only for DN and CD4+SP and CD8+SP. This indicates that CD6−/− T cells present a competitive disadvantage from very early stages of their development in regard to their CD6+/+ counterparts. In the periphery, a significant reduction of CD6−/− SP CD4+ and CD8+ T cells was also apparent in LNs (Fig. 3 C) and spleen (not depicted). Importantly, such a disadvantage was restricted to T cells, as the percentage of CD6−/− CD45.2 B cells in both the LN (Fig. 3 C) and spleen (not depicted) was not affected as compared with the control group. Overall, these results indicate that CD6 deficiency confers a selective disadvantage to developing T cells in a cell autonomous manner and that, in a competitive setting, the presence of peripheral CD6−/− T cells was also reduced. As observed in CD6−/− mice, (Fig. 2, D and E), increased frequencies of CD6−/− T reg and CD8+TCM cells could be detected in peripheral lymphoid organs of KO45.2/WT45.1 mixed-BM chimeras compared with their CD6+/+ counterparts from control WT45.2/WT45.1 mixed chimeras (Fig. 3 D), indicating that such phenotypic alterations are also T cell intrinsic.
Figure 3.
Alterations in central and peripheral T cell compartments in the absence of CD6 are cell intrinsic. (A) Schematic representation of competitive mixed BM chimeras experiment performed. (B) Dot density graph showing the ratio (mean ± SD) between the percentage of CD45.2+ and CD45.1+ cells in DN, DP, and SP subsets from control (WT45.2/WT45.1; open circles) and index (KO45.2/WT45.1; gray filled circles) experimental groups of mixed BM chimera assays. Each dot represents an individual mouse. (C) Dot density graph showing the ratio (mean ± SD) between the percentage of CD45.2+ and CD45.1+ cells in LN CD3+ CD4+ and CD8+ (left) and B220+ (right) cells from the same groups as in B. (D) Dot density graph showing the percent (mean ± SEM) of CD4+CD25+FoxP3+ T cells (left), CD8+TCM cells (middle), and CD4+TEM (right) gated on CD45.1 or CD45.2 cells from the KO45.2/WT45.1 group. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired Student’s t test).
CD6 sets the threshold for negative selection in the thymus
The aforementioned results suggest that CD6 deficiency alters thymocyte selection by lowering the threshold for negative selection and decreasing the number of cells reaching the CD4+SP and CD8+SP stage. Considering that TCR repertoire readjustments may occur and complicate result interpretation in mice with polyclonal T cell populations, we further addressed the role of CD6 in intrathymic T cell section using CD6−/− mice in which all T cells bear a single specific TCR. For that purpose, CD6−/− mice were crossed with either OT-I TCR mice, which produces MHC class I–restricted, OVA-specific CD8+ T cells, or with Marilyn mice, whose CD4+ cells are MHC class–II restricted for the male HY antigen. CD6−/− OT-I mice showed a reduction in the number of thymic CD8+SP cells expressing the TCR transgene, which reached statistical significance in peripheral organs (Fig. 4 A). Interestingly, CD6−/− OT-I mice also showed an increased proportion of peripheral CD8+ TCM cells (CD44hiCD62Lhi) compared with their CD6+/+ counterparts (Fig. 4 B). In Marilyn mice, female CD6−/− showed a reduced number of thymic CD4+SP cells expressing the TCR transgene, which again reached statistical significance in spleen and LNs (Fig. 4 C). Collectively, these data indicate that CD6 regulates TCR signaling on developing and antigen-experienced T cells.
Figure 4.
Effect of CD6 deficiency on SP subsets from TCR transgenic mice. (A, left) Representative contour plots indicating the percentage of CD8+ SP cells from CD6+/+ and CD6−/− OT-I TCR transgenic mice. (right) Total CD8+TCRα-2+ cell numbers (mean ± SEM) in thymus, spleen, and LNs from CD6+/+ (open bars) and CD6−/− (gray filled bars) OT-I mice (n = 4 for each genotype). (B) Representative contour plots showing CD44 and CD62L staining of gated LN CD8+ from CD6+/+ (top) and CD6−/− (bottom) OT-I mice. Percentage of SP and DP cells are indicated. (C, left) Representative contour plots showing the percentage of CD4+ SP cells from male CD6+/+ or CD6−/− Marilyn mice. (right) Total numbers of CD4+TCRβ-6+ cells (mean ± SEM) in thymus, spleen and LN from male CD6+/+ (open bars) or CD6−/− (gray filled bars) Marilyn mice (n = 4 for each genotype). (D) Analysis of Nur-77 expression in CD6hi and CD6lo peripheral T cells. LN cell suspensions from WT C57BL/6 mice were surface stained for CD4, CD8, CD5, and CD6, and then intracellularly for Nur-77. (top) FACS histograms showing CD6 (left) and CD5 (right) expression on pregated CD8+ cells. (bottom) FACS analysis of Nur-77 expression on gated cells representing the 5–6% with the highest (hi, black solid line) and lowest (lo, black dotted line) positivity for CD6 (left) and CD5 (right) expression in CD8+ T cells. FMO (gray shaded line), cells unstained for Nur-77. *, P < 0.05; **, P < 0.01 (unpaired Student’s t test).
This result is reminiscent of the well-acknowledged negative regulatory role of CD5 in T cell development and activation (Tarakhovsky et al., 1995). The CD5 receptor is closely related to CD6 both structurally and genetically (Soldevila et al., 2011) and its expression is developmentally regulated by TCR avidity, contributing to the fine tuning of TCR signaling (Azzam et al., 2001). Consequently, CD5 surface levels on thymocytes and peripheral T cells parallel the avidity of their TCRs, as demonstrated by the higher levels of Nur-77 found in CD5hi peripheral T cells compared with CD5lo (Fulton et al., 2015). Nur-77 levels are considered a sensitive readout of TCR signaling strength and are also found to be elevated in CD6hi peripheral T cells compared with CD6lo counterparts (Fig. 4 D), further supporting the role of CD6 as a putative negative regulator under high-avidity TCR conditions.
Influence of CD6 deficiency on TCR complex-induced responses from developing T cells
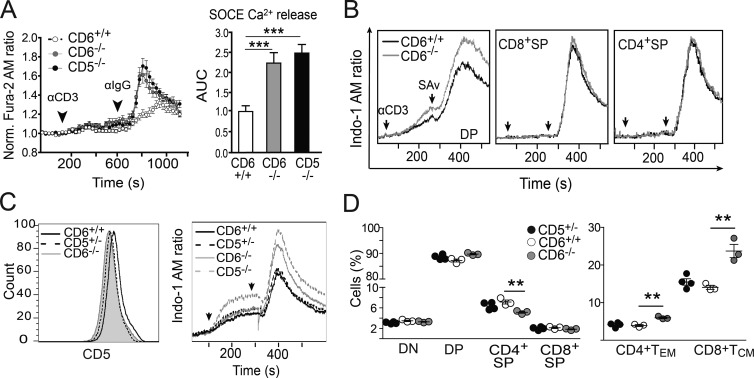
The fate of developing thymocytes is determined by the TCR signal strength resulting from interaction with MHC–self peptide complexes presented by thymic APCs (Palmer, 2003). Such a signal is negatively modulated by CD5, and as a consequence CD5−/− T cells are hyperresponsive to TCR cross-linking (Tarakhovsky et al., 1995). The analysis of TCR-induced Ca2+ flux showed that CD6−/−, similar to CD5−/−, displayed higher intracellular [Ca2+] than CD6+/+ cells (Fig. 5 A). Further analyses showed that CD6−/− DP, but not CD4+SP or CD8+SP, were responsible for the increased Ca2+ responses observed (Fig. 5 B). Consistent with this, no differences in Ca2+ responses were observed for spleen CD4+ or CD8+ T cells from CD6−/− and CD6+/+ mice (unpublished data). The increased Ca2+ responses observed in CD6−/− DP thymocytes could not be attributed to the partial CD5 down-regulation observed in these cells, as Ca2+ fluxes from CD5+/− mice, which display similar CD5 surface levels to those observed in CD6−/− (Fig. 5 C, left), did not differ from those observed with CD6+/+ thymocytes (Fig. 5 C, right). Moreover, CD5+/− mice did not show other alterations found in CD6−/− mice, such as reduced frequency of thymic CD4+SP cells or increase of peripheral CD8+TCM cells (Fig. 5 D). Thus, the phenotype observed for CD6−/− mice, in terms of Ca2+ flux, decrease in thymocyte maturation and increase of memory phenotype in the periphery, is caused by CD6 deficiency and not to the reduced expression of CD5.
Figure 5.
Modulation of TCR-induced responses from CD6−/− developing and mature T cells. (A, left) Fluorescence microscopy assessment of SOCE over time in Fura-2 AM–loaded thymocytes from CD6+/+ (n = 199 cells, open circles), CD6−/− (n = 188 cells, gray filled circles), and CD5−/− (n = 198 cells, black filled circles) mice after exposure to 145.2C11 mAb (αCD3) and Fc-specific antiserum (αIgG; n = 4 mice/genotype). (right) SOCE as represented by mean ± SEM of area under curve (AUC) from the same experiment. (B) FACS analysis of i[Ca2+] over time in Indo-1 AM–loaded DP, CD8+SP, and CD4+SP cells from CD6+/+ (black solid line) and CD6−/− (gray solid line) mice exposed to biotinylated 145.2C11 mAb (αCD3) plus Streptavidin (SAv). Data are representative of three independent experiments. (C, left) FACS analysis of CD5 expression on DP thymocytes from CD6+/+ (black solid line), CD5+/− (black dashed line), and CD6−/− (gray shaded line). (right) FACS analysis of i[Ca2+] over time of Indo-1 AM–loaded DP cells from CD6+/+ (black solid line), CD5+/− (black dashed line), CD6−/− (gray solid line) and CD5−/− (gray dashed line) mice, upon exposure to αCD3 mAb plus SAv, as assessed in B. (D) Dot density graphs showing the percentage (mean ± SEM) of major thymocyte subsets (left) and of LN CD4+ and CD8+ TEM and TCM subsets (right) from CD5+/− (black filled circles), CD6+/+ (open circles), and CD6−/− (gray filled circles) mice. The results shown are from one representative of two experiments performed. **, P < 0.01; ***, P < 0.001 (unpaired Student’s t test).
Increased peripheral CD4+ proliferative responses, low T reg cell suppressive activity, and exacerbated autoimmune response in CD6−/− mice
Overall, Ca2+ measurements indicate that CD6 behaves as a negative regulator of TCR signaling in immature (DP) but not mature (SP) thymocytes nor peripheral T cells. However, in vitro CD3-mediated stimulation of CD6−/− CD4+ and CD8+ peripheral T cells resulted in slightly increased proliferative responses, which reached statistical significance for CD4+ cells (Fig. 6 A) but disappeared under CD28 co-stimulatory conditions. In contrast, CD5−/− T cells, used for comparative purposes, showed significantly higher proliferative responses under both stimulatory conditions (Fig. 6 A). This indicates that, under ligand-independent conditions, CD6 has the capacity of negatively regulating T cell activation. However, it should be mentioned that under ligand-dependent conditions (e.g., MLR) the missing CD6–ALCAM interaction could result in suboptimal cell–cell contacts and net lower proliferative responses of CD6−/− T cells (unpublished data).
Figure 6.
Effect of CD6 deficiency on peripheral T cell proliferative responses, T reg cell activity, and CIA. (A) Proliferative responses of CFSE-labeled CD4+ and CD8+ T cells isolated from spleen and LN of CD6+/+ (open circles; n = 4–7 mice/point), CD6−/− (gray filled circles; n = 4–7 mice/point), and CD5−/− (black filled circles; n = 3 mice/point) cultured for 72 h in the presence of different concentrations of plastic-bound anti-CD3 mAb alone (top) or plus soluble anti-CD28 mAb (bottom; 1 µg/ml). Shown is the percentage (mean ± SEM) of CFSElo in CD4+ and CD8+ cells. **, P < 0.01 (Two-way ANOVA test). Data are cumulative results from four different experiments. (B, top) Gating strategy used to electronically sort regulatory (T reg, CD4+CD25+) and conventional (T conv, CD4+CD25−) spleen T cells from CD6−/− or CD6+/+ mice. (bottom) CFSE-stained T conv cells from CD6+/+ mice were cultured in anti-CD3–coated plates plus soluble anti-CD28 mAb (1 µg/ml) in the absence (w/o) or presence of T reg cells from CD6−/− or CD6+/+ mice at different ratios. The percentage of CFSElo lymphocytes at day 3 of culture from one experiment of four performed is shown. *, P < 0.05 (unpaired Student’s t test). (C, left) Clinical severity (mean ± SEM) at different weeks after CIA induction. (right) Cumulative incidence (percentage of affected mice) at the indicated weeks after CIA. **, P < 0.01; ***, P < 0.001 (two-way ANOVA test, followed by Bonferroni post-tests). (D) Quantitative RT-PCR analysis of joint mRNA levels (mean ± SD fold change of triplicates) of IL-6, TNF, FoxP3, IL-1β, IL-17A, and IFN-γ relative to GAPDH expression, before (NI, n = 3) and 8 wk after CIA of CD6+/+ (n = 8) and CD6−/− (n = 5) mice. *, P < 0.05 (Student’s t test).
The increased proportion of peripheral T reg cells led us to assess their functional status. As shown in Fig. 6 B, CD6−/− spleen CD4+CD25+ T cells exhibited lower suppressive activity in vitro compared with CD6+/+ counterparts, indicating that CD6 expression is required for full T reg cell functionality. These results are in agreement with the fact that T reg cell function has been linked to correct SLP-76–mediated signaling (Schmidt et al., 2015), and that CD6 constitutes a key element of the SLP-76 interactome (Roncagalli et al., 2014). Thus, CD6 deficiency would prevent T reg cells from proper recruitment of SLP-76, an essential signaling effector for their function.
To further explore the possibility that CD6 deficiency may result in dysregulated immune responses to auto-antigens in vivo as a consequence of defective T reg cell function and/or defective modulation of TCR signaling, we evaluated the susceptibility of CD6−/− mice to CIA, a well-established model of T cell–mediated autoimmune disease resembling rheumatoid arthritis. As illustrated by Fig. 6 C, CD6−/− mice showed an earlier CIA onset and a significantly increased CIA incidence and clinical score than their CD6+/+ littermates. This exacerbated CIA form was accompanied by a significantly enhanced joint expression of TNF and IL-6 transcripts and a reduced expression of FoxP3 (Fig. 6 D). Increased expression of IL-17A and IL-1β transcripts were also observed, but these differences did not reach statistical significance (Fig. 6 D). No differences regarding IFN-γ, IL-10, and TGF-β transcript levels were found (unpublished data). These results are compatible with CD6 deficiency potentiating an induced Th17 autoimmune response to collagen in the context of a dysfunctional T reg cell activity. This is particularly relevant in the light of a study correlating polymorphisms in the human CD6 gene with susceptibility to certain autoimmune diseases, such as rheumatoid arthritis or multiple sclerosis (Kofler et al., 2016). Although the functional consequences of CD6 polymorphism associated to rheumatoid arthritis has not been analyzed, carriers of the multiple sclerosis risk allele (rs17828933GG) show altered CD4+ T cell responses and changes in the proportion of full-length and CD6ΔD3 isoforms in both CD4+ and CD8+ cells (Kofler et al., 2016), but their consequences on disease susceptibility merit further study.
Concluding remarks
This study represents the first in vivo characterization of CD6 deficiency. The results show that CD6 acts as a negative modulator of TCR-mediated signaling, setting the threshold for the final steps of T cell selection and maturation, and likely for peripheral T cell activation. The molecular mechanisms by which CD6 restrains TCR signaling are speculative, as CD6 signaling pathway is still relatively unknown. Given the reported association of CD5 and CD6 at the T cell surface (Gimferrer et al., 2003) and the regulatory role of CD6 on CD5 Tyr phosphorylation (Castro et al., 2003), CD5 could mediate such a negative effect. This situation would be analogous to that reported for CD2, which transduces inhibitory signals through its association with CD5 (Teh et al., 1997; Castro et al., 2002). Alternatively, CD6 could directly recruit downstream negative signaling effectors (e.g., protein Tyr phosphatases). The possibility that, apart from decreasing the threshold for negative selection, the absence of CD6 impacts thymocyte selection in other manners cannot be excluded. Indeed, CD6 has been related to resistance to apoptosis in different experimental settings. Accordingly, CD6 is up-regulated both in DP cells undergoing positive selection and thymocytes resistant to in vitro induced-apoptosis (Singer et al., 2002), and CD6 ligation protects chronic lymphocytic leukemia cells from IgM-induced apoptosis (Osorio et al., 1997). Moreover, CD6 expression protects leukemic T cells from galectin-induced apoptosis (Escoda-Ferran et al., 2014). Thus, further investigation into the role of CD6 as a pro-survival protein is required.
The CD6–ALCAM interaction is certainly a relevant driver of the observed effects on T cell development and peripheral T cell homeostasis. However, it has been observed that interactions between developing thymocytes (CD6+) and TECs (ALCAM+) are not sustained by long-term synapses, but rather by transient and repeated contacts with different TECs that maintain NFAT transcription and allow positive selection (Ebert et al., 2008). According to this model, the CD6–ALCAM interaction might be somewhat dispensable in the thymus, where the negative role of CD6 would be more evident.
The generation and maintenance of antigen-experienced T cells, such as CD4+TEM, CD8+TCM, and T reg cells, in CD6−/− mice has been linked to a certain level of TCR signal strength and/or co-stimulation (Lanzavecchia and Sallusto, 2005). This fits with the view of CD6 as a negative regulator of thymic and peripheral TCR-mediated signaling. Indeed, absence of CD6 led to increased negative selection in the thymus and increased activation in response to self- or environmental antigens in the periphery, as shown by expansion of T cell subsets with memory and regulatory phenotypes. These data are reminiscent of those obtained from the analysis of CD5−/− mice, where expansion of both memory and regulatory (Ordoñez-Rueda et al., 2009) T cells are also observed. Thymic generation of T reg cells requires high-avidity interaction of TCR with self-peptide–MHC complexes. Accordingly, increased frequency of T reg cells in thymus and periphery were previously reported for CD5−/− mice, the suppressive functionality of which is debatable (Dasu et al., 2008; Ordoñez-Rueda et al., 2009). However, although increased proportions of T reg cells can be seen in thymus and periphery of CD5−/− mice, they could only be detected in peripheral lymphoid organs of CD6−/− mice. This would indicate that CD6 is particularly relevant for regulating the generation of extrathymic T reg cells during immune responses, rather than during thymic T cell selection. Furthermore, the ex vivo analysis of CD6−/− spleen T reg cells showed a lower suppressive activity, further indicating that CD6 is also relevant to T reg cell functionality. The in vivo biological meaning of our findings is supported by the fact that CD6−/− mice present an exacerbated form of an inducible autoimmune disease model (CIA).
Thus, the CD6-deficient mice described here represent a new and useful tool to fully understand the multifaceted mechanisms by which CD6 exerts its regulatory functions in different physiological and pathological contexts. Importantly, these mice can also provide valuable insights for the CD6-based therapies that are currently under assessment for clinically relevant diseases such as psoriasis, rheumatoid arthritis, or lymphoproliferative disorders (Hernández et al., 2016).
MATERIALS AND METHODS
Mice
CD6-deficient mice were generated in C57BL/6N background as previously described (Skarnes et al., 2011) through the Mouse Biology Program (University of California, Davis, Davis, CA) using targeting vector DPGS00142_B_G12 from the trans-NIH Knock-Out Mouse Project (KOMP), and obtained from the KOMP Repository. The gene-targeting vector was designed to delete exons 3–6 from the cd6 gene coding for the three SRCR extracellular domains and the stalk region. It was successfully used to obtain a recombinant ES cell clone (JM8A1.N3) that gave rise to chimerical mice after injection into pseudo-pregnant female mice. After germ-line transmission of the mutant allele, denoted as C57BL/6N-Atm1Brd, a homozygous (CD6−/−) colony was established. Absence of CD6 protein expression on T cells was confirmed by flow cytometry. The CD6−/− mice were viable and fertile, and their lifespans and behaviors were equivalent to their WT littermates. CD5-deficient mice (Tarakhovsky et al., 1995) on C57BL/6 background were provided by C. Raman (University of Alabama, Birmingham, AL). Mice were maintained at the animal facilities of the Universitat de Barcelona School of Medicine (Barcelona, Spain) under specific pathogen–free conditions.
CD6−/− and CD6+/+ mice were bred with MHC class I–restricted, OVA-specific OT-I (Clarke et al., 2000) and MHC class II–restricted male HY-specific Marilyn (Scott et al., 2000) TCR-transgenic lines at the Centre d’Immunologie Marseille-Luminy (CIML; Marseille, France) and maintained in SPF conditions. CD3ε-deficient (CD3εΔ5/Δ5) mice displaying thymocyte development arrest at DN3 stage (Malissen et al., 1995) used as hosts in BM chimera experiments were also kept at CIML. All the experiments were carried out in accordance with the guidelines of the Comité Étic d’Experimentació Animal from Universitat de Barcelona. Sex- and age-matched (8–12 wk) mice were used in all the experiments.
Calcium-flux measurement
Cytosolic Ca2+ signal was determined by two different approaches. In the case of fluorescence microscopy assessment of TCR-induced store operated calcium release (SOCE) over time, cells were loaded with 4.5 µM Fura-2 AM (30 min) as previously described (Carreras-Sureda et al., 2013). Cytosolic [Ca2+] increases are presented as the ratio of emitted fluorescence (510 nm) after excitation at 340 and 380 nm, relative to the ratio measured before cell stimulation (Fura-2 AM ratio 340/380). All experiments were performed at room temperature, and cells were bathed in a solution containing 140 mM NaCl, 5 mM KCl, 1.2 mM CaCl2, 0.5 mM MgCl2, 5 mM glucose, 10 mM Hepes (300 mosmol/l, pH 7.4). Ca2+ entry was achieved using 1 µg/ml of anti-CD3ε mAb (145.2C11; BD), and cross-linking was performed using goat anti–Armenian hamster IgG (MP Bio). In the case of flow cytometry analysis, thymus and spleen cells were loaded for 20 min at 37°C with Indo-1 AM (Molecular Probes), and then stained with fluorescent-labeled anti-CD4 (RM4-5, PE; BD) and anti-CD8 (53–6.7, APC; BD). Basal fluorescence was recorded with an LSRII-UV (BD) before addition of 2.5 µg biotinylated anti-CD3 (145.2C11; BD). Cross-linking of anti-CD3 mAb was induced by the addition of streptavidin (10 µg/ml; Thermo Fisher scientific). Calcium fluxes were determined on gated CD4+ and CD8+ cells. As loading control, cells were stimulated with Ionomycin (1 µg/ml; Sigma-Aldrich).
FACS analyses
Cell staining was done with fluorescently labeled mAbs to CD5 (53–7.3), CD4 (RM4-5), CD8α (53–6.7), CD24 (M1/69), TCR-Vα2 (B20.1TCR-Vβ6 (RR4-7), TCRγ (GL-3), and TCRβ (H57-597) were purchased from BD; CD62L (MEL-14), CD44 (IM7), CD69 (H1.2F3), CD3 (145.2C11), and FoxP3 (3G3) were obtained from TONBO Bioscience; CD25 (PC61.5), CD19 (6D5), CD161 (PK136), and CD6 (OX129) were purchased from BioLegend; and Nur77 (12.14) was obtained from eBioscience; all were used according to the manufacturer’s instructions. FACS analyses were done on a LSRII 564 or FACS Canto II system (BD) using FlowJo software (TreeStar). Cell viability was evaluated either with SYTOX Blue (Life Technologies) or 633- or 405-nm Viability dyes (Life Technologies). Apoptotic cells were detected by using Annexin V-FITC apoptosis detection kit (Immunostep).
Mixed BM chimeras
Before use, 8-wk-old CD3εΔ5/Δ5 recipient mice received one single dose of irradiation (300 rads) to eliminate partially endogenous immune cells and to allow efficient engraftment. Mice were then injected i.v. with 2 × 106 BM cells from CD6+/+ (CD45.1) and from CD6−/− (CD45.2) mice, to generate mixed chimeras. As control groups, recipient CD3εΔ5/Δ5 mice (CD45.2) were injected with 4 × 106 BM cells from either CD6+/+ or CD6−/− mice. Mouse chimeras were kept on antibiotic-containing water (0.2% Bactrim; Roche) during the whole experiment and euthanized at 12 wk for FACS analyses on gated CD3+ or B220+ lymphocytes.
In vivo BrdU incorporation assay
For the detection of proliferating cells in vivo, CD6−/− and CD6+/+ littermate mice were administered for 1 wk with BrdU (0.8 mg) into drinking water, changing water every 2 d. BrdU+ cells were visualized by FACS with BrdU Cell Proliferation Assay kit (BD) following the manufacturer’s instructions.
In vitro T cell proliferation and T reg cell suppression assay
CD4+ and CD8+ T cells were negatively isolated from spleen and LN (Dynabeads; Life Technologies) and incubated for 15 min with CellTrace CFSE Cell Proliferation kit (Life Technologies), and then cultured in complete RPMI medium (10% FCS, 10 mM Hepes, 1 mM sodium pyruvate, 50 µM β-Mercaptoethanol, and 5 mM Penicillin-Glutamine-Streptomycin; Lonza) in 96-well plates coated with increasing concentrations of purified anti-CD3 mAb (145.2C11; TONBO Bioscience) alone or in combination with 1 µg/ml soluble anti-CD28 mAb (37.51; TONBO Bioscience) for 72 h at 37°C, 5% CO2.
For T reg cell suppression assays, CD4+CD25− (T conv cells) and CD4+CD25+ (T reg cells) T cells were electronically sorted from spleen specimens and, upon CFSE-labeling of the former, co-cultured for 3 d at different ratios in 96-well plates coated with anti-CD3 mAb plus soluble anti-CD28. Proliferative responses were analyzed by determining the percentage of CFSElo cells by FACS.
CIA
Induction and assessment of CIA was done in 8–12-wk-old male mice, as previously described (Postigo et al., 2011). In brief, mice were immunized once at the base of the tail with 150 µg of chicken collagen type II dissolved at a concentration of 2 mg/ml in 0.05 M acetic acid and emulsified with CFA containing 4 mg/ml of Mycobacterium tuberculosis in a final volume of 150 µl. Evaluation of arthritis clinical severity was performed weekly according to a graded scale of 0–3 as follows: 0, no inflammation (normal joint); 1, detectable local swelling and/or erythema; 2, swelling in >1 joint and pronounced inflammation; 3, swelling of the entire paw and/or ankylosis. Each paw was graded, and the scores were summed (with a maximum possible score 12 per mouse). The expression of arthritogenic cytokines and FoxP3 in the joints was explored before and 8 wk after CIA induction by quantitative real time-PCR, as described previously (Postigo et al., 2011). Results (in triplicate) were normalized to GAPDH expression and measured in parallel in each sample.
ACKNOWLEDGMENTS
We are indebted to Elise Bergot and to the Cytomics core facility of the Institut d'Investigacions Biomèdiques August Pi i Sunyer for technical help.
This work is supported by grants from: Worldwide Cancer Research (14-1275) and Fundació La Marató TV3 (201319-30) to F. Lozano; Spanish Ministerio de Economía y Competitividad (Plan Nacional I+D+i) co-financed by European Development Regional Fund “A way to achieve Europe” to F. Lozano (SAF2013-46151-R), J. Merino (SAF2012-34059), R. Merino (SAF2014-55088-R), and R. Vicente (SAF2014-52228-R); IPT2011-1527-010000 (associated to Fibrostatin SL) to J. Merino; and Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, European Research Council (FP7/2007–2013 grant no. 322465), and ANR grant Basilic to B. Malissen and M. Malissen. M. Orta-Mascaró, I. Simões, and E. Carreras are recipients of fellowships from Ministerio de Economía y Competitividad (BES-2011-048415), Fundação para a Ciência e a Tecnologia (SFRH/BD/75738/2011), and European Community Seventh Framework Program (BIOTRACK, FP7/2007/2013;229673), respectively. National Institutes of Health grants to Velocigene at Regeneron Inc. (U01HG004085), and the CSD Consortium (U01HG004080) funded the generation of gene-targeted vectors and ES cells for 8,500 genes in the KOMP Program and archived and distributed by the KOMP Repository at UC Davis and CHORI (U42RR024244).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ALCAM
- activated leukocyte cell adhesion molecule
- CIA
- collagen-induced arthritis
- DN
- double negative
- DP
- double positive
- mAb
- monoclonal antibody
- TCM
- central memory T cell
- T conv
- conventional T cell
- TEC
- thymic epithelial cell
- TEM
- effector/memory T cell
- T reg
- regulatory T cell
References
- Azzam H.S., DeJarnette J.B., Huang K., Emmons R., Park C.S., Sommers C.L., El-Khoury D., Shores E.W., and Love P.E.. 2001. Fine tuning of TCR signaling by CD5. J. Immunol. 166:5464–5472. 10.4049/jimmunol.166.9.5464 [DOI] [PubMed] [Google Scholar]
- Bonet L., Farnós M., Martínez-Florensa M., Martínez V.G., and Lozano F.. 2013. Identification of functionally relevant phoshorylatable serine clusters in the cytoplasmic region of the human CD6 lymphocyte surface receptor. FEBS Lett. 587:2205–2213. 10.1016/j.febslet.2013.05.043 [DOI] [PubMed] [Google Scholar]
- Carreras-Sureda A., Cantero-Recasens G., Rubio-Moscardo F., Kiefer K., Peinelt C., Niemeyer B.A., Valverde M.A., and Vicente R.. 2013. ORMDL3 modulates store-operated calcium entry and lymphocyte activation. Hum. Mol. Genet. 22:519–530. 10.1093/hmg/dds450 [DOI] [PubMed] [Google Scholar]
- Castro M.A.A., Tavares P.A., Almeida M.S., Nunes R.J., Wright M.D., Mason D., Moreira A., and Carmo A.M.. 2002. CD2 physically associates with CD5 in rat T lymphocytes with the involvement of both extracellular and intracellular domains. Eur. J. Immunol. 32:1509–1518. [DOI] [PubMed] [Google Scholar]
- Castro M.A.A., Nunes R.J., Oliveira M.I., Tavares P.A., Simões C., Parnes J.R., Moreira A., and Carmo A.M.. 2003. OX52 is the rat homologue of CD6: evidence for an effector function in the regulation of CD5 phosphorylation. J. Leukoc. Biol. 73:183–190. 10.1189/jlb.0902437 [DOI] [PubMed] [Google Scholar]
- Chappell P.E., Garner L.I., Yan J., Metcalfe C., Hatherley D., Johnson S., Robinson C.V., Lea S.M., and Brown M.H.. 2015. Structures of CD6 and Its Ligand CD166 Give Insight into Their Interaction. Structure. 23:1426–1436. 10.1016/j.str.2015.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S.R., Barnden M., Kurts C., Carbone F.R., Miller J.F., and Heath W.R.. 2000. Characterization of the ovalbumin-specific TCR transgenic line OT-I: MHC elements for positive and negative selection. Immunol. Cell Biol. 78:110–117. 10.1046/j.1440-1711.2000.00889.x [DOI] [PubMed] [Google Scholar]
- Dasu T., Qualls J.E., Tuna H., Raman C., Cohen D.A., and Bondada S.. 2008. CD5 plays an inhibitory role in the suppressive function of murine CD4+ CD25+ T(reg) cells. Immunol. Lett. 119:103–113. 10.1016/j.imlet.2008.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert P.J.R., Ehrlich L.I.R., and Davis M.M.. 2008. Low ligand requirement for deletion and lack of synapses in positive selection enforce the gauntlet of thymic T cell maturation. Immunity. 29:734–745. 10.1016/j.immuni.2008.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoda-Ferran C., Carrasco E., Caballero-Baños M., Miró-Julià C., Martínez-Florensa M., Consuegra-Fernández M., Martínez V.G., Liu F.-T., and Lozano F.. 2014. Modulation of CD6 function through interaction with Galectin-1 and -3. FEBS Lett. 588:2805–2813. 10.1016/j.febslet.2014.05.064 [DOI] [PubMed] [Google Scholar]
- Fulton R.B., Hamilton S.E., Xing Y., Best J.A., Goldrath A.W., Hogquist K.A., and Jameson S.C.. 2015. The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8+ T cells to respond to foreign antigens. Nat. Immunol. 16:107–117. 10.1038/ni.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimferrer I., Farnós M., Calvo M., Mittelbrunn M., Enrich C., Sánchez-Madrid F., Vives J., and Lozano F.. 2003. The accessory molecules CD5 and CD6 associate on the membrane of lymphoid T cells. J. Biol. Chem. 278:8564–8571. 10.1074/jbc.M209591200 [DOI] [PubMed] [Google Scholar]
- Gimferrer I., Calvo M., Mittelbrunn M., Farnós M., Sarrias M.R., Enrich C., Vives J., Sánchez-Madrid F., and Lozano F.. 2004. Relevance of CD6-mediated interactions in T cell activation and proliferation. J. Immunol. 173:2262–2270. 10.4049/jimmunol.173.4.2262 [DOI] [PubMed] [Google Scholar]
- Gimferrer I., Ibáñez A., Farnós M., Sarrias M.-R., Fenutría R., Roselló S., Zimmermann P., David G., Vives J., Serra-Pagès C., and Lozano F.. 2005. The lymphocyte receptor CD6 interacts with syntenin-1, a scaffolding protein containing PDZ domains. J. Immunol. 175:1406–1414. 10.4049/jimmunol.175.3.1406 [DOI] [PubMed] [Google Scholar]
- Hassan N.J., Barclay A.N., and Brown M.H.. 2004. Frontline: Optimal T cell activation requires the engagement of CD6 and CD166. Eur. J. Immunol. 34:930–940. 10.1002/eji.200424856 [DOI] [PubMed] [Google Scholar]
- Hassan N.J., Simmonds S.J., Clarkson N.G., Hanrahan S., Puklavec M.J., Bomb M., Barclay A.N., and Brown M.H.. 2006. CD6 regulates T-cell responses through activation-dependent recruitment of the positive regulator SLP-76. Mol. Cell. Biol. 26:6727–6738. 10.1128/MCB.00688-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández P., Moreno E., Aira L.E., and Rodríguez P.C.. 2016. Therapeutic targeting of CD6 in autoimmune diseases: a review of cuban clinical studies with the antibodies IOR-T1 and itolizumab. Curr. Drug Targets. 17:666–677. 10.2174/1389450117666160201114308 [DOI] [PubMed] [Google Scholar]
- Jameson S.C. 2002. Maintaining the norm: T-cell homeostasis. Nat. Rev. Immunol. 2:547–556. [DOI] [PubMed] [Google Scholar]
- Kofler D.M., Farkas A., von Bergwelt-Baildon M., and Hafler D.A.. 2016. The link between CD6 and autoimmunity: genetic and cellular associations. Curr. Drug Targets. 17:651–665. 10.2174/1389450117666160201105934 [DOI] [PubMed] [Google Scholar]
- Lanzavecchia A., and Sallusto F.. 2005. Understanding the generation and function of memory T cell subsets. Curr. Opin. Immunol. 17:326–332. 10.1016/j.coi.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Malissen M., Gillet A., Ardouin L., Bouvier G., Trucy J., Ferrier P., Vivier E., and Malissen B.. 1995. Altered T cell development in mice with a targeted mutation of the CD3-epsilon gene. EMBO J. 14:4641–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M.I., Gonçalves C.M., Pinto M., Fabre S., Santos A.M., Lee S.F., Castro M.A.A., Nunes R.J., Barbosa R.R., Parnes J.R., et al. 2012. CD6 attenuates early and late signaling events, setting thresholds for T-cell activation. Eur. J. Immunol. 42:195–205. 10.1002/eji.201040528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordoñez-Rueda D., Lozano F., Sarukhan A., Raman C., Garcia-Zepeda E.A., and Soldevila G.. 2009. Increased numbers of thymic and peripheral CD4+ CD25+Foxp3+ cells in the absence of CD5 signaling. Eur. J. Immunol. 39:2233–2247. 10.1002/eji.200839053 [DOI] [PubMed] [Google Scholar]
- Osorio L.M., De Santiago A., Aguilar-Santelises M., Mellstedt H., and Jondal M.. 1997. CD6 ligation modulates the Bcl-2/Bax ratio and protects chronic lymphocytic leukemia B cells from apoptosis induced by anti-IgM. Blood. 89:2833–2841. [PubMed] [Google Scholar]
- Palmer E. 2003. Negative selection--clearing out the bad apples from the T-cell repertoire. Nat. Rev. Immunol. 3:383–391. 10.1038/nri1085 [DOI] [PubMed] [Google Scholar]
- Postigo J., Genre F., Iglesias M., Fernández-Rey M., Buelta L., Carlos Rodríguez-Rey J., Merino J., and Merino R.. 2011. Exacerbation of type II collagen-induced arthritis in apolipoprotein E-deficient mice in association with the expansion of Th1 and Th17 cells. Arthritis Rheum. 63:971–980. 10.1002/art.30220 [DOI] [PubMed] [Google Scholar]
- Roncagalli R., Hauri S., Fiore F., Liang Y., Chen Z., Sansoni A., Kanduri K., Joly R., Malzac A., Lähdesmäki H., et al. 2014. Quantitative proteomics analysis of signalosome dynamics in primary T cells identifies the surface receptor CD6 as a Lat adaptor-independent TCR signaling hub. Nat. Immunol. 15:384–392. 10.1038/ni.2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R.F., Oliveira L., and Carmo A.M.. 2016. Tuning t cell activation: the function of CD6 at the immunological synapse and in T cell responses. Curr. Drug Targets. 17:630–639. 10.2174/1389450116666150531152439 [DOI] [PubMed] [Google Scholar]
- Schmidt A.M., Lu W., Sindhava V.J., Huang Y., Burkhardt J.K., Yang E., Riese M.J., Maltzman J.S., Jordan M.S., and Kambayashi T.. 2015. Regulatory T cells require TCR signaling for their suppressive function. J. Immunol. 194:4362–4370. 10.4049/jimmunol.1402384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D., Addey C., Ellis P., James E., Mitchell M.J., Saut N., Jurcevic S., and Simpson E.. 2000. Dendritic cells permit identification of genes encoding MHC class II-restricted epitopes of transplantation antigens. Immunity. 12:711–720. 10.1016/S1074-7613(00)80221-6 [DOI] [PubMed] [Google Scholar]
- Singer N.G., Fox D.A., Haqqi T.M., Beretta L., Endres J.S., Prohaska S., Parnes J.R., Bromberg J., and Sramkoski R.M.. 2002. CD6: expression during development, apoptosis and selection of human and mouse thymocytes. Int. Immunol. 14:585–597. 10.1093/intimm/dxf025 [DOI] [PubMed] [Google Scholar]
- Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T., et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 474:337–342. 10.1038/nature10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldevila G., Raman C., and Lozano F.. 2011. The immunomodulatory properties of the CD5 lymphocyte receptor in health and disease. Curr. Opin. Immunol. 23:310–318. 10.1016/j.coi.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakhovsky A., Kanner S.B., Hombach J., Ledbetter J.A., Müller W., Killeen N., and Rajewsky K.. 1995. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 269:535–537. 10.1126/science.7542801 [DOI] [PubMed] [Google Scholar]
- Teh S.J., Killeen N., Tarakhovsky A., Littman D.R., and Teh H.S.. 1997. CD2 regulates the positive selection and function of antigen-specific CD4-CD8+ T cells. Blood. 89:1308–1318. [PubMed] [Google Scholar]
- Zimmerman A.W., Joosten B., Torensma R., Parnes J.R., van Leeuwen F.N., and Figdor C.G.. 2006. Long-term engagement of CD6 and ALCAM is essential for T-cell proliferation induced by dendritic cells. Blood. 107:3212–3220. 10.1182/blood-2005-09-3881 [DOI] [PubMed] [Google Scholar]