Abstract
Human cytochrome P450 oxidoreductase (POR) provides electrons for all microsomal cytochromes P450 (P450s) and plays an indispensable role in drug metabolism catalyzed by this family of enzymes. We evaluated 100 human liver samples and found that POR protein content varied 12.8-fold, from 12.59 to 160.97 pmol/mg, with a median value of 67.99 pmol/mg; POR mRNA expression varied by 26.4-fold. POR activity was less variable with a median value of 56.05 nmol/min per milligram. Cigarette smoking and alcohol consumption clearly influenced POR activity. Liver samples with a 2286822 TT genotype had significantly higher POR mRNA expression than samples with CT genotype. Homozygous carriers of POR2286822C>T, 2286823G>A, and 3823884A>C had significantly lower POR protein levels compared with the corresponding heterozygous carriers. Liver samples from individuals homozygous at 286823G>A, 1135612A>G, and 10954732G>A generally had lower POR activity levels than those from heterozygous or wild-type samples, whereas the common variant POR*28 significantly increased POR activity. There was a strong association between POR and the expression of P450 isoforms at the mRNA and protein level, whereas the relationship at the activity level, as well as the effect of POR protein content on P450 activity, was less pronounced. POR transcription was strongly correlated with both hepatocyte nuclear factor 4 alpha and pregnane X receptor mRNA levels. In conclusion, we have elucidated some potentially important correlations between POR single-nucleotide polymorphisms and POR expression in the Chinese population and have developed a database that correlates POR expression with the expression and activity of 10 P450s important in drug metabolism.
Introduction
Human cytochrome P450 oxidoreductase (POR) is a 78-kDa di-flavin protein anchored to the cytoplasmic side of endoplasmic reticulum membranes (Xia et al., 2011). POR provides electrons to a variety of substrates, including all the microsomal cytochromes P450 (P450s), heme oxygenase, squalene monooxygenase, and cytochrome b5, as well as reducing some therapeutic prodrugs (Miller et al., 2011; Huang et al., 2015).
To date, few studies have characterized the variation in the expression and activity of POR in human liver samples, with only four studies measuring POR content in human samples. A study by Gomes et al. (2009) measured mRNA, protein, and activity of POR in a cohort of liver samples, but the distribution and range of POR mRNA and protein were not reported. A study by Gan et al. (2009) provided both spectral and Western blot analysis of absolute POR content, but there was a 3-fold difference in the mean value as measured by these two methods. Recently, liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS)–based proteomics for absolute quantification of protein has been developed (Picotti et al., 2009). Two studies reported the absolute content of POR using LC-MS/MS, but one assessed only a small number of liver samples (Ohtsuki et al., 2012) and the other determined the POR protein expression in a CYP2C19 genotype-defined pool of human liver microsomal (HLM) samples (Shirasaka et al., 2015). Therefore, determination of the mRNA and protein content and activity of POR across a large number of normal human liver samples is needed to provide reliable physiologic parameters for POR expression in physiologic, pharmacological, and toxicological research.
Although humans have only a single POR gene, it is highly polymorphic. More than 200 mutations and polymorphisms in the POR gene have been reported (Pandey and Flück, 2013). However, very little is known about the effects of POR single-nucleotide polymorphisms (SNPs) on its expression and activity in HLM. Of the 34 POR polymorphisms identified in HLM, only two (L577P and SNP 41301427G>A) were associated with altered POR activity (Hart et al., 2008). Gomes et al. (2009) reported 46 POR SNPs in HLM and found that only one SNP (17148944G>A) directly affected POR mRNA levels and none affected POR protein levels or activity.
P450 is responsible for the metabolism of many endogenous and exogenous substances, including 70–80% of all drugs currently in clinical use (Zhang et al., 2015a), and there are large individual variations in P450-mediated drug metabolism. All microsomal P450s require the input of two electrons for catalysis and POR is the sole donor for the first electron (Bridges et al., 1998). Thus, POR is indispensable in metabolic reactions catalyzed by P450. Several in vitro and in vivo studies have revealed that polymorphisms that affect POR activity can have differing effects on P450 activities, depending on the specific POR mutation, P450 isoform, and the substrate used to assay activity, and hence the activity of a POR variant with one P450 does not predict its activity with other P450s (Yang et al., 2011; Chen et al., 2012). Therefore, the impact of a particular POR mutant needs to be studied individually with each P450. However, the effect of POR protein content on P450 protein content or activities has not been reported to date.
Though POR plays a vital role in drug metabolism, the transcriptional regulation of the POR gene by xenobiotic receptors has not been fully described. Receptor-selective agonists for the pregnane X receptor (PXR) and the constitutive androstane receptor (CAR) induced POR mRNA expression in mouse liver, whereas in human hepatocytes only PXR activators could upregulate POR expression (Maglich et al., 2002). One study has reported that POR expression was associated with the expression level of CAR and hepatocyte nuclear factor 4 alpha (HNF4α) in human livers (Wortham et al., 2007). Thus, it is appropriate to characterize the expression levels of various regulatory factors and determine to what extent they correlate with POR expression.
In this study, 125 liver samples were collected and used to determine the absolute POR protein content by LC-MS/MS, POR mRNA expression, and POR activity. POR SNPs occurring with a frequency >1% in Chinese populations were used to analyze the effect of these SNPs on POR protein content, mRNA levels, and activity. The distribution of POR protein and mRNA was assessed, and relationships between POR expression and the expression of 10 P450s involved in drug metabolism at the protein, mRNA, and activity levels were analyzed. In addition, the regulation of POR expression in human livers was explored.
Materials and Methods
Human Liver Samples and Liver Microsomes
Human liver samples were obtained from 125 Chinese patients undergoing hepatic surgery during 2012 and 2014 at the First Affiliated Hospital of Zhengzhou University, the People’s Hospital of Henan Province, and the Tumors’ Hospital of Henan Province. Detailed information for each patient was obtained, including gender, age, body height, body weight, smoking habits, alcohol consumption, clinical diagnosis, regular drug intake before surgery, previous history, allergic history, pathologic diagnosis, imaging examination, and laboratory test data, as described previously (Zhang et al., 2015b). The study was approved by the ethics committees of Zhengzhou University and written informed consent was obtained from each patient. All experiments were performed in accordance with the Declaration of Helsinki. Samples from normal livers were collected, with liver health confirmed by liver function tests, histopathological analysis, and imaging examination by ultrasonography or CT. All liver samples were frozen immediately after removal and stored in liquid nitrogen until use. HLMs were prepared by differential centrifugation, and total HLM protein concentration was determined by the Bradford method.
Measurement of POR, HNF4α, and PXR mRNA Levels in Human Liver
Primers for POR was designed by Takara Bio Inc. (Otsu, Shiga, Japan) and other primers were from the literature (Table 1) (Wang et al., 2011). mRNA levels were measured as described previously (Zhang et al., 2015a). Briefly, total RNA was isolated from human liver samples using an RNAiso Plus kit (Takara) according to the manufacturer’s instructions. The cDNA for real-time quantitative polymerase chain reaction was synthesized from 1 μg total RNA using a PrimeScript RT reagent kit with gDNA Eraser (Perfect Real Time; Takara). P450 mRNA expression was detected by two-step real-time quantitative polymerase chain reaction using an ABI 7500 Fast Real-Time PCR system (Applied Biosystems). GAPDH was used as a reference gene, and expression of target mRNA was calculated using the 2–△CT method (△CT equals the difference between target gene and GADPH).
TABLE 1.
Primers for quantitative real-time polymerase chain reaction
| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| GAPDH | AACAGGGTGGTGGACCTCAT | GGAGGGGAGATTCAGTGTGG |
| POR | TTTCGCTCATCGTGGGTCT | TCCTCCCCGTTTTCTTCATCT |
| PXR | ACAGCTGGCTAGCATTCCTCA | CTTGCCTCTCTGATGGTCCTG |
| HNF4α | AGCGATCCAGGGAAGATCAAG | AGCAGCAGCAGCTCTCCAA |
Quantification of POR Protein Content in HLM
Preparation of a QconCAT Protein.
Protein quantitation of POR was performed by nano–LC-MS/MS using our previously established quantitative concatemer (QconCAT) strategy combined with stable isotope dilution–multiple reaction monitoring (Wang et al., 2015). Briefly, two signature peptides (GVATNWLR and FAVFGLGNK) were selected to quantify POR on the basis of a genome-wide BLAST search. QconCAT proteins were designed as a concatemer of all the stable isotope-labeled signature peptides. After prokaryotic expression, the QconCAT protein was purified using affinity chromatography and evaluated by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (Beynon et al., 2005).
Protein Digestion.
HLM proteins were denatured, reduced, alkylated, diluted with seven volumes of 50 mM NH4HCO3 solution, and digested with trypsin at a trypsin/substrate ratio of 1:50 at 37°C for 26 hours. The digested QconCAT protein was examined by Fourier Transform–Linear Ion Trap Ion Cyclotron Resonance MS (Thermo Fisher Scientific Inc., Waltham, MA).
Quantification of the QconCAT Protein.
The peptides ASGNLIPQEK and TILDELVQR that composed the QconCAT protein were used to determine the protein using a nano–high-performance LC (HPLC) coupled to multiple-reaction monitoring MS analysis. The limit of quantitation, linear range, and concentration of the QconCAT protein were calculated and reported in our previous work (Wang et al., 2015).
Nano–LC-MS/MS Quantitative Analysis.
The concentration of P450 protein was determined by nano–LC–multiple-reaction monitoring MS using an EASY nano-LC coupled to a TSQ Vantage triple quadrupole mass spectrometer (Thermo Fisher Scientific). Samples were first loaded separately on a trap column (100 μm × 20 mm) packed with SP-300-ODS-AP (5-μm particle diameter; 100-nm pore size in house) Each sample was then eluted into an analytical column (75 μm × 11 mm) and packed with SP-300-ODS-AP, and separated at a flow rate of 300 nl∙min–1 with an elution gradient consisting of mobile phase B (99.9% acetonitrile, 0.1% formic acid) and mobile phase A (99.9% H2O, 0.1% formic acid). Elution gradient solutions were added as follows: B was increased from 2% to 10% in 5 minutes, 10–40% in 60 minutes, and 40–95% in 5 minutes, followed by automatic equilibration of the LC system with mobile phase A for approximately 10 minutes before the next analysis. Fractions were continuously directed into the TSQ Vantage Triple Quadrupole mass spectrometer with a nanoelectrospray ionization source at a capillary temperature of 240°C and spray voltage of 1900 V. Three transitions were selected per peptide for the quantification of each protein with the following MS conditions: Q1 and Q3 resolution, 0.7-Da full-width at half maximum; Q2 pressure, 1.5 mTorr (Ar); cycle time, 1.5 seconds; collision energy, 0.034 × precursor ion m/z + 3.314.
Determination of POR Activity in HLM.
The POR activity assay uses as a basis the rate of cytochrome C reduction by liver microsomes (Guengerich et al., 2009). The reaction was conducted in a 200-μl volume with 0.3 M potassium phosphate buffer (pH 7.7), 0.2 mM horse cytochrome C, and 5 μg microsomal protein. Reactions were initiated by the addition of 20 μl of 10 mM NADPH to a 200-μl assay mixture for a total volume of 220 μl. The rate of cytochrome C reduction was determined from the rate of increase in absorbance at 550 nm produced by the reduced form of cytochrome C using a BioTek (Winooski, VT) Synergy H1MD Multi-Mode microplate reader in the kinetic mode before and after the addition of NADPH (0–5 minutes).
Genotypes of POR.
Genomic DNA was isolated from human liver tissue using a genomic DNA purification kit (Beijing ComWin Biotech Co., Ltd., China). Polymorphisms in POR with frequencies of >1% in the Chinese population were genotyped in this study sample. A total of 18 SNPs in the POR gene were detected. All the POR SNPs were determined by Sequenom method except SNP 3823884A>C and SNP 2302433C>T (by PCR-sequencing).
Determination of P450 Metabolic Activities in HLMs.
Incubation mixtures each contained a single concentration of substrate (400 μM phenacetin for CYP1A2, 20 μM coumarin for CYP2A6, 500 μM bupropion for CYP2B6, 40 μM paclitaxel for CYP2C8, 1500 μM tolbutamide for CYP2C9, 250 μM omeprazole for CYP2C19, 320 μM dextromethorphan for CYP2D6, 500 μM chlorzoxazone for CYP2E1, and 50 μM midazolam for CYP3A4/5); HLMs (0.3 mg protein/ml for CYP1A2, CYP2A6, and CYP2E1; 0.2 mg protein/ml for CYP2D6 and CYP3A; 0.5 mg protein/ml for CYP2B6, CYP2C8, CYP2C9, and CYP2C19); and 1 mM NADPH. The mixture was preincubated for 5 minutes at 37°C. Optimal incubation times for each substrate were as follows: 30 minutes for phenacetin O-deethylation, coumarin 7-hydroxylation, and chlorzoxazone 6-hydroxylation; 60 minutes for bupropion 1-hydroxylation, and tolbutamide 4-hydroxylation; 90 minutes for omeprazole 4-hydroxylation, 120 minutes for paclitaxel 6-hydroxylation; 20 minutes for dextromethorphan O-demethylation; and 5 minutes for midazolam 1′-hydroxylation. Each reaction was terminated after specified incubation period by addition of 20 μl ice-cold acetonitrile or 1 ml ethyl acetate, or perchloric acid and metabolite concentrations were determined by HPLC with ultraviolet detection or HPLC with fluorescence detection. The detailed analysis and results of P450 activities were reported in our previous work (Zhang et al., 2015b).
Relationships between POR and 10 P450s at the mRNA, Protein Content, and Activity Levels.
CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, and CYP3A5 were the most important members of P450 family. The mRNA levels (by qPCR), protein content (by LC-MS/MS), and activities of the 10 P450s were determined in the same set of human liver samples. These data and the analytical methods have been submitted in detail separately (Zhang et al., 2016). In this study we used simple linear regression approaches to evaluate the relationship between POR and the 10 P450s at the mRNA, protein, and activity levels with the goal of identifying possible contributions of POR to P450 phenotypic variation. In addition, we evaluated the effect of POR protein content on the 10 P450 activities.
Statistical Analyses.
The normality of the data distribution was checked using Kolmogorov-Smirnov and Shapiro-Wilk methods. Since most data sets were not normally distributed, nonparametric methods were generally used for statistical analyses. The Mann-Whitney U test was used for pairwise comparison and the Kruskal-Wallis H test was used for multiple pairwise comparisons. Nonparametric Spearman rank correlation analysis was performed to calculate the correlation coefficient (r). A P value <0.05 was considered statistically significant (two-tailed). SPSS Statistics for Windows, Version 17.0. (SPSS Inc. Chicago, IL) was used for data management and statistical analyses. Graphs were generated using GraphPad Prism 5.04 software (La Jolla, CA).
Results
mRNA, Protein, and Activity of POR in Human Livers
POR mRNA.
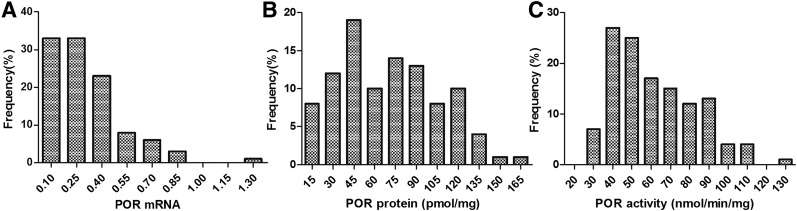
The values for POR mRNA expression in 107 liver samples were not normally distributed (Fig. 1A). The relative POR mRNA expression level varied 26.4-fold (range: 0.05–1.25), with a median value of 0.28. The levels of POR mRNA at the 2.5th and the 97.5th percentiles were 0.05 and 0.83, respectively, exhibiting about a 16-fold variation. As shown in Fig. 1A, there was one liver sample that had an extremely high POR mRNA level (1.25).
Fig. 1.
Frequency distribution of POR mRNA levels (as measured by qPCR, relative to GAPDH, n = 107) (A), POR protein content (as measured by LC-MS/MS, n = 100) (B), and POR activity (as measured by the spectral method, n = 125) (C). The data are presented as the means of three independent experiments.
POR Protein Content.
As with POR mRNA, the protein content of POR in 100 liver samples also was not normally distributed, with a median of 67.99 pmol/mg. The lowest and highest content of POR protein were 12.59 and 160.97 pmol/mg, displaying a 12.8-fold variation (Fig. 1B). The variation in POR protein levels at the 95% prediction interval (PI) was 10.6-fold. Two samples exhibited extreme POR protein content (12.59 and 160.97 pmol/mg, respectively).
Activity of POR.
Compared with mRNA and protein, POR activity was less variable: a 4.7-fold range (26.91–125.77 nmol/min per milligram) was observed across 125 human liver microsomes, with a median value of 56.05 nmol/min per milligram (Fig. 1C). The variation in POR protein content at 95% PI was 3.6-fold.
Correlations in POR mRNA, Protein, and Activity in Human Livers.
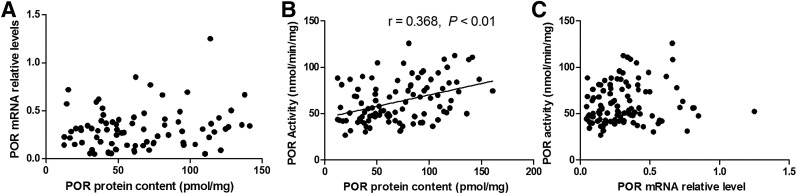
Spearman correlation analysis was used to identify correlations, and the results showed that POR protein content was significantly correlated with POR activity (r = 0.368, P = 0.000). However, we failed to detect significant correlations between POR mRNA and protein level, or POR mRNA and POR activity (Fig. 2).
Fig. 2.
Correlations in POR mRNA, protein, and activity in human livers. (A) Correlation between POR protein content and POR mRNA level; (B) correlation between POR protein content and POR activity; and (C) correlation between POR activity and mRNA level. The data are presented as the means of three independent experiments.
Effect of Demographic Factors on POR mRNA, Protein, and Activity.
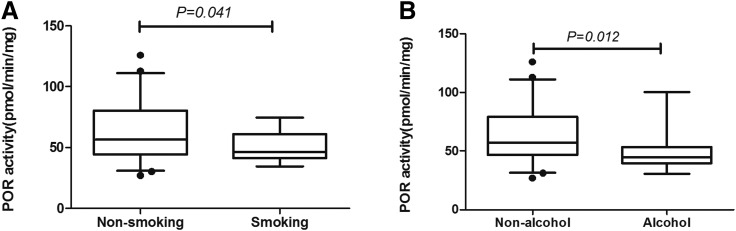
POR mRNA, protein, and activity data were stratified by liver donor age, gender, smoking habit, alcohol consumption, and tissue resource and then analyzed by Mann-Whitney U test or Kruskal-Wallis test. As shown in Fig. 3A, POR activity was significantly higher in nonsmokers than in smokers (P = 0.041). Statistically significant differences were also observed between drinkers and nondrinkers (P = 0.012) (Fig. 3B). The other demographic factors had no noticeable effect on POR mRNA, protein or activity levels (P > 0.05) (Supplemental Table 1).
Fig. 3.
Effect of cigarette smoking (A) and alcohol consumption (B) on POR activity in HLM. The black horizontal line represents the median value with 2.5th to 95th percentile values.
Influence of POR Gene Polymorphisms on POR mRNA, Protein, and Activity in Human Livers
Gene Polymorphisms of POR.
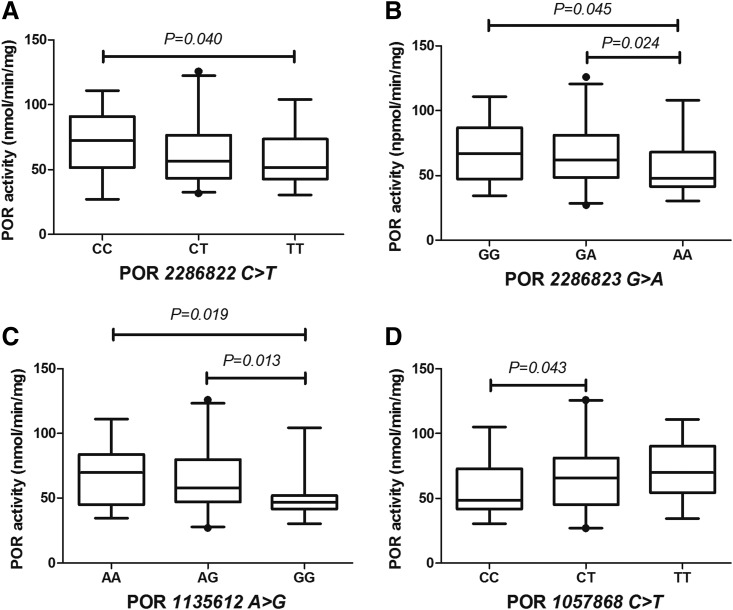
As shown in Table 2, the variant frequencies of 18 SNPs in the POR gene ranged from 0.91% (for 17148944G>A) to 88.2% (for 2302431T>C), in which the frequency of the common mutant 1057868C>T (*28) was 35.5%. This is consistent with previously published results on the Chinese population (Huang et al., 2008). All 18 POR SNPs were evaluated for their effect on POR mRNA, protein, and activity levels with the positive results shown in Figs. 4–6.
TABLE 2.
POR variants detected in this cohort
POR SNP ID, genomic position, coding position, amino acid change, and location were compiled from Hart et al., 2008; Huang et al., 2008; Gomes et al., 2009; and Tomková et al., 2012.
| SNP ID | Genomic Position | Coding Position | Location | Amino Acid Change | Genotype Frequency | Variant Allele Frequency (%) | ||
|---|---|---|---|---|---|---|---|---|
| Genotype | N | Frequency (%) | ||||||
| rs3823884 | 5036A>C | −47A>C | 5′-UTR | AA | 55 | 52.4 | 27.1 | |
| AC | 43 | 41 | ||||||
| CC | 7 | 6.6 | ||||||
| rs17148944 | 62448G>A | 237+88G>A | Intron 2 | GG | 108 | 98.2 | 0.91 | |
| GA | 2 | 1.8 | ||||||
| AA | 0 | 0 | ||||||
| rs10239977 | 69567C>T | 366+89C>T | Intron 3 | CC | 94 | 90.4 | 4.8 | |
| CT | 10 | 9.6 | ||||||
| TT | 0 | 0 | ||||||
| rs1135612 | 70258A>G | 387A>G | Exon 4 | Pro129a | AA | 32 | 28.6 | 46.9 |
| AG | 55 | 49.1 | ||||||
| GG | 25 | 22.3 | ||||||
| rs10954732 | 71730G>A | 931+225G>A | Intron 6 | GG | 27 | 24.1 | 48.2 | |
| GA | 62 | 55.4 | ||||||
| AA | 23 | 20.5 | ||||||
| rs3815455 | 72337C>T | 830+116C>T | Intron 7 | CC | 43 | 41.3 | 34.6 | |
| CT | 50 | 48.1 | ||||||
| TT | 11 | 10.6 | ||||||
| rs41301394 | 73384C>T | 831-35C>T | Intron 7 | CC | 46 | 41.8 | 35.5 | |
| CT | 50 | 45.5 | ||||||
| TT | 14 | 12.7 | ||||||
| rs4732515 | 74610T>C | 1067-66T>C | Intron 9 | TT | 2 | 1.8 | 85.1 | |
| TC | 29 | 26.1 | ||||||
| CC | 80 | 72.1 | ||||||
| rs4732516 | 74663C>G | 1067-13C>G | Intron 9 | CC | 2 | 1.8 | 84.5 | |
| CG | 30 | 27.3 | ||||||
| GG | 78 | 70.9 | ||||||
| rs2286822 | 74869C>T | 1248+12C>T | Intron 10 | CC | 26 | 25 | 51.4 | |
| CT | 49 | 47.1 | ||||||
| TT | 29 | 27.9 | ||||||
| rs2286823 | 74877G>A | 1248+20G | Intron 10 | GG | 23 | 22.3 | 51.5 | |
| GA | 54 | 52.4 | ||||||
| AA | 26 | 25.3 | ||||||
| rs41301427 | 75138G>A | 1398+32G>A | Intron 11 | GG | 100 | 97.1 | 1.46 | |
| GA | 3 | 2.9 | ||||||
| AA | 0 | 0 | ||||||
| rs2302432 | 75445G>T | 1399-33G>T | Intron 11 | GG | 0 | 0 | 87.6 | |
| GT | 25 | 24.8 | ||||||
| TT | 76 | 75.2 | ||||||
| rs2302431 | 75444T>C | 1399-34T>C | Intron 12 | TT | 88.2 | |||
| TC | 24 | 23.5 | ||||||
| CC | 78 | 76.5 | ||||||
| rs1057868 | 75587C>T | 1508C>T | Exon 12 | Ala503Val | CC | 46 | 41.8 | 35.5 |
| CT | 50 | 45.5 | ||||||
| TT | 14 | 12.7 | ||||||
| rs2228104 | 75534T>C | 1455T>C | Exon 12 | Ala485a | TT | 0 | 0 | 88.1 |
| TC | 24 | 23.8 | ||||||
| CC | 77 | 76.2 | ||||||
| rs2302433 | 75781C>T | 1669+33C>T | Intron 12 | CC | 98 | 89.1 | 5.9 | |
| CT | 11 | 10.0 | ||||||
| TT | 1 | 0.9 | ||||||
| rs1057870 | 75868G>A | 1716G>A | Exon 13 | Ser572a | GG | 94 | 91.3 | 4.4 |
| GA | 9 | 8.7 | ||||||
| AA | 0 | 0 | ||||||
5′-UTR, 5′-untranslated region.
SNPs that do not result in amino acid changes.
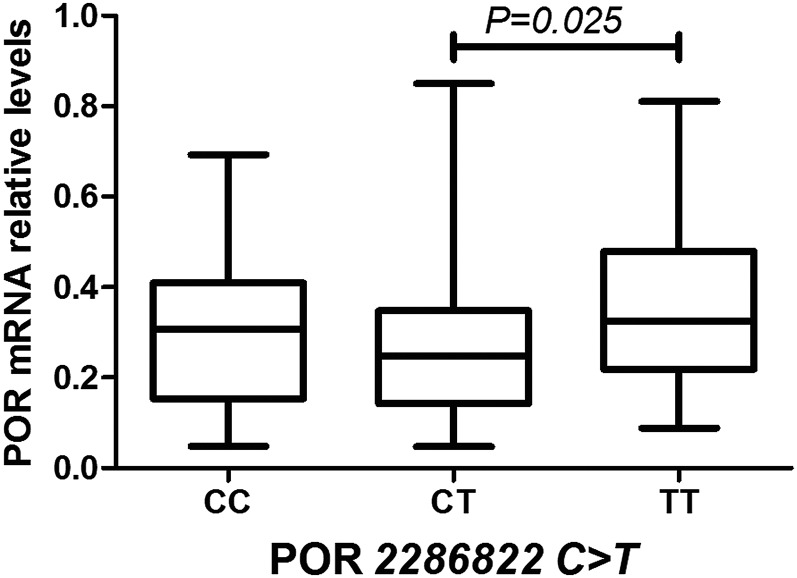
Fig. 4.
Effect of SNP on POR mRNA level in human livers. Median mRNA level of the genotype variant of POR2286822C>T was statistically significant (P < 0.05). Data are shown as box plots representing medians with 2.5th to 97.5th percentile values.
Fig. 6.
Effects of SNPs on POR activity in HLM. Differences in the median activity level of the different genotype variants of POR 2286822C>T (A), 286823G>A (B), 1135612A>G (C), and 1057868C>T (D) were statistically significant (P < 0.05). Data are shown as box plots representing medians with 2.5th to 95th percentile values.
Influence on mRNA Level.
Only SNP 2286822C>T of POR had an impact on mRNA expression. Samples with 2286822 TT genotype had significantly higher POR median mRNA levels than samples with the CT genotype (P = 0.025) (Fig. 4).
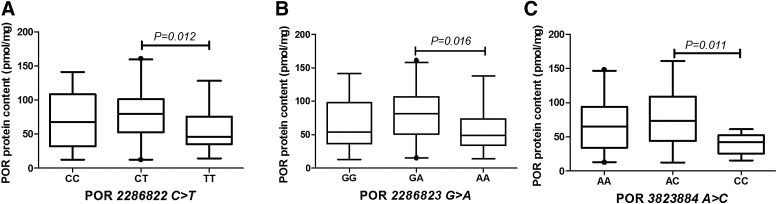
Influence on Protein Content.
Three POR SNPs (2286822C>T, 2286823G>A, and 3823884A>C) had an influence on POR protein content with similar effects. The homozygous carriers of POR 2286822C>T, 2286823G>A, and 3823884A>C had significantly lower protein levels compared with the corresponding heterozygous carriers (Fig. 5).
Fig. 5.
Effect of SNPs on POR protein content in HLM. Differences in the median protein levels of the different genotype variants of POR 2286822C>T (A), 286823G>A (B), and 3823884A>C (C) were statistically significant (P < 0.05). Data are shown as box plots representing medians with 2.5th to 95th percentile values.
Influence on Activity.
As shown in Fig. 6, individuals who exhibited the POR 2286822 TT (C>T) genotype had lower hepatic POR activity compared with 2286822 CC carriers. Individuals genotyped as 286823 AA (G>A) had lower POR activity than those carrying the 286823 GG and GA genotypes. Similarly, 1135612 GG (A>G) carriers also showed significantly decreased POR activity compared with corresponding wild-types as well as heterozygous individuals. However, POR activity in the 1057868 CT (C>T) group was higher than that of wild-type group. Meanwhile, there was a tendency toward increased POR activity in 1057868 TT carriers compared with wild-type and heterozygous carriers, but it did not reach statistical significance.
Correlation between POR and P450 at the mRNA, Protein, and Activity Levels
The mRNA, protein, and activity levels of 10 P450s were simultaneously quantified with POR expression and activity in the same set of 100 HLMs (Zhang et al., 2016). Spearman correlation analysis was used to determine the correlation between POR and the 10 P450s at the mRNA, protein, and activity levels. As shown in Table 3, significant correlations were observed between POR and all 10 P450s at the mRNA level (P < 0.05). There also were significant associations between POR protein content and all P450 isoform content except with CYP2B6. Strong correlations were found between POR protein content and P450 protein content for CYP2C8 and CYP2C9 (r > 0.8, P < 0.001). For CYP2E1 and CYP3A4 the correlation coefficient reached 0.6. However, the association between POR and P450s at the activity level was relatively poor. POR activity was positively associated with CYP2C19 and negatively associated with CYP2C8 activity. In addition, significant associations were found between POR content and the activities of four P450s (CYP2B6, CYP2C8, CYP2C19, and CYP2E1) (P < 0.05).
TABLE 3.
Correlation between POR and P450 at mRNA, protein, and activity levels (n = 100)
| POR mRNA versus P450 mRNA | POR Protein versus P450 Protein | POR Activity versus P450 Activity | POR Protein versus P450 Activity | |
|---|---|---|---|---|
| CYP1A2 | 0.561** | 0.576** | −0.01 | −0.007 |
| CYP2A6 | 0.476** | 0.457** | 0.163 | 0.102 |
| CYP2B6a | 0.787** | 0.172 | 0.067 | −0.247* |
| CYP2C8 | 0.450** | 0.818** | −0.233* | −0.329** |
| CYP2C9 | 0.212* | 0.849** | 0.132 | −0.118 |
| CYP2C19b | 0.602** | 0.474** | 0.507** | 0.438** |
| CYP2D6 | 0.246* | 0.535** | −0.062 | −0.207 |
| CYP2E1 | 0.477** | 0.618** | 0.197 | 0.224* |
| CYP3A4 | 0.494** | 0.661** | 0.158 | 0.017 |
| CYP3A5 | 0.324** | 0.532** | 0.158 | 0.017 |
P<0.01; **P<0.001.
For CYP2B6, n = 91.
For CYP2C19, n = 54.
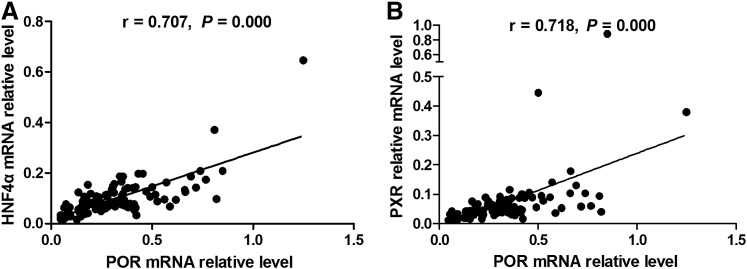
Expression of HNF4α and PXR and Their Relationship with POR in Human Livers
Both HNF4α and PXR mRNA were determined together with POR by qPCR in the same set of 107 human liver samples. Neither HNF4α nor PXR mRNA was normally distributed among the 107 patient samples, with an overall variation of 62.8- and 85.3-fold, respectively. The mRNA levels of both HNF4α and PXR were strongly correlated to the POR transcript levels in all samples (r = 0.707, P = 0.000; r = 0.718, P = 0.000; Fig. 7).
Fig. 7.
Correlation between POR expression and transcriptional factors. (A) Correlation between POR mRNA and HNF4α mRNA levels; (B) correlation between POR mRNA and PXR mRNA levels. The data were determined by qPCR in 107 liver samples and are presented as the means of three independent experiments.
Discussion
This is the first extensive study to quantify the absolute content of POR protein by LC-MS/MS in a large number of normal Chinese liver samples. The non-normally distributed POR values in 100 samples varied from 12.59 to 160.97 pmol/mg with a median of 67.99 pmol/mg. The mRNA expression level and activity of POR was measured in liver tissues and microsomes, respectively, and showed 26.4- and 4.7-fold variations, respectively. Cigarette smoking and alcohol consumption clearly influenced POR activity. Several of the 18 SNPs analyzed had a significant impact on POR expression or activity, and there were significant associations between the POR mRNA level and the 10 P450 mRNA levels, between POR protein content and the protein content of nine P450s, and between POR protein content and the activity of four P450s. In addition, POR transcription was strongly correlated with both HNF4α and PXR mRNA levels in these samples.
Although POR is an enzyme important to physiology, pharmacology, and toxicology, the information concerning POR expression and activity is limited. Gomes et al. (2009) reported a mean POR content of 60.0 ± 29.6 pmol/mg for female and 51.9 ± 29.9 pmol/mg for male Caucasian human liver samples by Western blotting, although there was no statistical difference in the expression between genders. In the Gan et al. (2009) study, POR concentration as measured by immunoblotting was 28 ± 15 pmol/mg, whereas in the same set of HLMs mean POR content was 86 ± 35 pmol/mg as measured by spectrophotometry. The average POR content of 70.67 ± 35.79 pmol/mg from LC-MS/MS analysis of 100 liver tissues in our study was consistent with the result of Ohtsuki et al. (2012) (71.6 ± 17 pmol/mg) by LC-MS/MS. However, owing to the relatively small sample size (n = 17), Ohtsuki et al. observed only a 2.4-fold variation in POR content, whereas the variation in our study was as large as 12.8-fold. The mean POR protein content for pooled HLMs from donors with different CYP2C19 diplotypes by LC-MS/MS was 21.8 ± 11.4 to 31.6 ± 16.4 pmol/mg (Shirasaka et al., 2015), which is much smaller than the results we obtained here. It is possible that the discrepancy is the result of the different ethnicities or the pooled HLMs used in that study.
In two studies on POR mRNA levels (Wortham et al., 2007; Gomes et al., 2009), the largest variation was 18.3-fold, whereas a 26.4-fold difference was observed by our group (Fig. 1). A linear trend analysis displayed no significant correlation between POR mRNA and protein levels (Fig. 2), indicating that POR expression could be regulated post-transcriptionally, consistent with a study showing that miR-214 downregulates POR expression (Dong et al., 2014).
Two earlier studies reported mean POR activities of 139.0 nmol/min per milligram and 177 ± 53 nmol/min per milligram in Caucasian human liver samples (Hart et al., 2008; Gomes et al., 2009). In the present study, we measured POR activity in 125 Chinese human liver samples and found that POR activity was not normally distributed. The median POR activity was 56.05 nmol/min per milligram (Fig. 1C), which is lower than the abovementioned results. Compared with mRNA and protein expression, POR activity was less variable with only a 4.7-fold difference in this study cohort, which is consistent with the result of Gomes et al. (2009).
Although POR is highly polymorphic, the effects of common polymorphisms on POR expression and activity are minimal. Of the 46 SNPs analyzed, only POR 17148944G>A was related to a decrease in POR mRNA (Gomes et al., 2009). We failed to detect such an effect, but we found that POR 2286822C>T mutant was associated with increased POR mRNA expression (Fig. 4). We found that POR mutants correlated with altered POR protein content.
The mutant homozygous carriers of POR 2286822C>T, 2286823G>A, and 3823884A>C had significantly lower protein levels compared with the corresponding heterozygous carriers (Fig. 5). In addition, the effects of SNPs 286823G>A and 1135612A>G, on POR activity were consistent. Homozygous 286823G>A and 1135612A>G, mutants generally had lower POR activity than heterozygous mutants and wild-type samples. However, the common SNP1057868C>T (also known as POR*28 mutant) increased POR activity, although the activity difference between TT and CC groups was not statistically significant (Fig. 6). Both Gomes et al. (2009) and Hart et al. (2008) measured the effect of SNPs 286823G>A, 1135612A>G, and 286823G>A on POR activity in Caucasian populations, but the differences were not statistically significant. However, Hart et al. found that L577P and SNP 41301427G>A were associated with decreased POR activity. No L577P mutant was found in our study or in that of Gomes et al., and we conclude that L577P is a rare mutant.
Of the five SNPs that affected POR expression and activity, both 2286822C>T and 2286823G>A are intronic mutations, whereas 3823884A>C is located in the 5′-untranslated region. Emerging evidence indicates that noncoding genetic variants play an important role in gene regulation by influencing the transcriptional activity or splicing efficiency, or by altering the splicing site of their host genes (Cooper et al., 2010). We also found that a common nonsynonymous variant of POR SNP 1057868C>T increased POR activity, whereas no significant association of this polymorphism with microsomal POR activity was identified in either the Gomes et al. (2009) or Hart et al. (2008) studies. The frequency of POR SNP1057868C>T was 35.5% in this cohort, although relatively lower frequencies (19.1%, 21.9%, 26.4%, and 30.3%) were observed in a Western population (Huang et al., 2008). Therefore the discrepancy may be the result of different frequencies of the POR 1057868C>T variant in different ethnic groups. An in vivo study in healthy Chinese volunteers showed that individuals with the POR 1057868C>T variant were associated with 1.6-fold higher hepatic CYP3A activity, which is in agreement with our result (Yang et al., 2011).
We found that strong correlations existed between POR and nine P450s at the level of protein expression. POR mRNA levels were correlated with all 10 P450 mRNA levels. The strong association between POR and CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, and CYP2D6 at the mRNA level was also observed in an earlier study (Wortham et al., 2007). POR, together with all microsomal P450s, constitutes a complex system responsible for the biosynthesis and degradation of endogenous and exogenous substances. The high correlation between POR and P450s at the protein and mRNA levels provides compelling evidence that the expression of POR and P450s is coregulated. The expression of metabolic enzymes is frequently regulated coordinately by a network of transcription factors such as HNF4α, PXR, and CAR. The strong association between POR and HNF4α and PXR in this study cohort (Fig. 7) suggests that POR expression is controlled, at least in part, by these transcription factors and, importantly, this may partly explain the coregulation of POR and P450s.
Though the expression of POR and P450s was highly correlated at protein and mRNA levels, the correlation at the level of activity was less pronounced. POR activity was significantly associated with the activities of two P450s (CYP2C19 and CYP2C8). This is inconsistent with the result by Hart et al. (2008), in which POR activity was significantly associated with all the analyzed P450 activities, including CYP2C19 and CYP1A2. However, it should be noted that the marker substrates we used for CYP1A2, CYP2A6, CYP2B6, CYP2C9, CYP2C19, and CYP3A4/A5 were different from those used by Hart et al. It may be that the effect of POR activity on microsomal P450 activities is P450- and substrate-dependent. In addition to POR, other factors such as demography, genetic polymorphisms, protein-protein interactions, and epigenetic regulation can also affect the activity of P450s. The weaker correlation between POR and P450 activities hints that POR is not the sole factor that determines P450 activities and the correlation between POR and a specific P450 cannot be extrapolated to other P450s or even the same P450 with a different substrate.
POR content was positively correlated with CYP2C19 and CYP2E1 activities (Table 3). However, a negative correlation was observed between POR content and CYP2B6 and CYP2C8. As only a single POR protein is required to donate electrons to all the microsomal P450s, the protein-protein interaction likely differs between POR and different P450s, and therefore a negative correlation may be reasonable and deserves further investigation.
In conclusion, this is the first report to comprehensively analyze the protein content, mRNA level, enzyme activity of, as well as the effects of gene polymorphisms on POR expression and activity in a Chinese population. There are large individual variations at the mRNA and protein level, whereas POR activity is less variable. Some SNPs have a significant effect on POR expression and activity, including the common variant POR*28. Strong correlations between POR and P450s at the protein and mRNA levels in human livers hint that POR and P450s are coregulated, and this can be explained at least in part by the collinearity of expression of POR and HNF4α and PXR. A poor correlation between POR and the 10 P450s at the level of activity indicates that POR is not the sole determinant of P450 activity. These findings provide an important physiologic and functional database for POR expression and highlight the complex role POR plays in P450-mediated metabolism.
Supplementary Material
Abbreviations
- CAR
constitutive androstane receptor
- HLM
human liver microsomes
- HNF4α
hepatocyte nuclear factor 4 alpha
- HPLC
high-performance liquid chromatography
- LC-MS/MS
liquid chromatography coupled to tandem mass spectrometry
- P450
cytochrome P450
- POR
cytochrome P450 oxidoreductase
- PXR
pregnane X receptor
- qPCR
quantitative real-time polymerase chain reaction
- SNP
single-nucleotide polymorphism
Authorship Contributions
Participated in research design: Qiao.
Conducted the experiments: Zhang, J. Liu, T. Liu, Wang, Fang, N. Gao, Tian.
Contributed new reagents or analytic tools: Zhou, Cui, J. Gao, Wen, Jia.
Performed data analysis: Zhang, Li, J. Liu.
Wrote or contributed to the writing of the manuscript: Zhang, Li, Qiao.
Footnotes
This work was supported by the National Natural Science Foundation of China [No. 81473279], Science and Technology Innovative Scholar Program of Henan Province [No.134200510019], and Scientific and Technical Innovation Team of Zhengzhou City [131PCXTD604].
 This article has supplemental material available
at dmd.aspetjournals.org.
This article has supplemental material available
at dmd.aspetjournals.org.
References
- Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ. (2005) Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat Methods 2:587–589. [DOI] [PubMed] [Google Scholar]
- Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. (1998) Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J Biol Chem 273:17036–17049. [DOI] [PubMed] [Google Scholar]
- Chen X, Pan LQ, Naranmandura H, Zeng S, Chen SQ. (2012) Influence of various polymorphic variants of cytochrome P450 oxidoreductase (POR) on drug metabolic activity of CYP3A4 and CYP2B6. PLoS One 7:e38495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN. (2010) Functional intronic polymorphisms: Buried treasure awaiting discovery within our genes. Hum Genomics 4:284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Liu H, Chen F, Li D, Zhao Y. (2014) MiR-214 promotes the alcohol-induced oxidative stress via down-regulation of glutathione reductase and cytochrome P450 oxidoreductase in liver cells. Alcohol Clin Exp Res 38:68–77. [DOI] [PubMed] [Google Scholar]
- Gan L, von Moltke LL, Trepanier LA, Harmatz JS, Greenblatt DJ, Court MH. (2009) Role of NADPH-cytochrome P450 reductase and cytochrome-b5/NADH-b5 reductase in variability of CYP3A activity in human liver microsomes. Drug Metab Dispos 37:90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AM, Winter S, Klein K, Turpeinen M, Schaeffeler E, Schwab M, Zanger UM. (2009) Pharmacogenomics of human liver cytochrome P450 oxidoreductase: multifactorial analysis and impact on microsomal drug oxidation. Pharmacogenomics 10:579–599. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Martin MV, Sohl CD, Cheng Q. (2009) Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat Protoc 4:1245–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Wang S, Nakamoto K, Wesselman C, Li Y, Zhong XB. (2008) Genetic polymorphisms in cytochrome P450 oxidoreductase influence microsomal P450-catalyzed drug metabolism. Pharmacogenet Genomics 18:11–24. [DOI] [PubMed] [Google Scholar]
- Huang N, Agrawal V, Giacomini KM, Miller WL. (2008) Genetics of P450 oxidoreductase: sequence variation in 842 individuals of four ethnicities and activities of 15 missense mutations. Proc Natl Acad Sci USA 105:1733–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Zhang M, Rwere F, Waskell L, Ramamoorthy A. (2015) Kinetic and structural characterization of the interaction between the FMN binding domain of cytochrome P450 reductase and cytochrome c. J Biol Chem 290:4843–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. (2002) Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol 62:638–646. [DOI] [PubMed] [Google Scholar]
- Miller WL, Agrawal V, Sandee D, Tee MK, Huang N, Choi JH, Morrissey K, Giacomini KM. (2011) Consequences of POR mutations and polymorphisms. Mol Cell Endocrinol 336:174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Schaefer O, Kawakami H, Inoue T, Liehner S, Saito A, Ishiguro N, Kishimoto W, Ludwig-Schwellinger E, Ebner T, et al. (2012) Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab Dispos 40:83–92. [DOI] [PubMed] [Google Scholar]
- Pandey AV, Flück CE. (2013) NADPH P450 oxidoreductase: structure, function, and pathology of diseases. Pharmacol Ther 138:229–254. [DOI] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaka Y, Chaudhry AS, McDonald M, Prasad B, Wong T, Calamia JC, Fohner A, Thornton TA, Isoherranen N, Unadkat JD, et al. (2015) Interindividual variability of CYP2C19-catalyzed drug metabolism due to differences in gene diplotypes and cytochrome P450 oxidoreductase content. Pharmacogenomics J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomková M, Marohnic CC, Gurwitz D, Šeda O, Masters BSS, Martásek P. (2012) Identification of six novel P450 oxidoreductase missense variants in Ashkenazi and Moroccan Jewish populations. Pharmacogenomics 13:543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Jiang Z, Shen Z, Wang H, Wang B, Shou W, Zheng H, Chu X, Shi J, Huang W. (2011) Functional evaluation of genetic and environmental regulators of p450 mRNA levels. PLoS One 6:e24900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang H, Li J, Wei J, Zhai R, Peng B, Qiao H, Zhang Y, Qian X. (2015) A new calibration curve calculation method for absolute quantification of drug metabolizing enzymes in human liver microsomes by stable isotope dilution mass spectrometry. Anal Methods 7:5934–5941. [Google Scholar]
- Wortham M, Czerwinski M, He L, Parkinson A, Wan YJ. (2007) Expression of constitutive androstane receptor, hepatic nuclear factor 4 alpha, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab Dispos 35:1700–1710. [DOI] [PubMed] [Google Scholar]
- Xia C, Hamdane D, Shen AL, Choi V, Kasper CB, Pearl NM, Zhang H, Im SC, Waskell L, Kim JJ. (2011) Conformational changes of NADPH-cytochrome P450 oxidoreductase are essential for catalysis and cofactor binding. J Biol Chem 286:16246–16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Fu Z, Chen X, Yuan H, Yang H, Huang Y, Ouyang D, Tan Z, Tan H, Huang Z, et al. (2011) Effects of the CYP oxidoreductase Ala503Val polymorphism on CYP3A activity in vivo: a randomized, open-label, crossover study in healthy Chinese men. Clin Ther 33:2060–2070. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao N, Liu T, Fang Y, Qi B, Wen Q, Zhou J, Jia L, Qiao H. (2015a) Effect of Cytochrome b5 Content on the Activity of Polymorphic CYP1A2, 2B6, and 2E1 in Human Liver Microsomes. PLoS One 10:e0128547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao N, Tian X, Liu T, Fang Y, Zhou J, Wen Q, Xu B, Qi B, Gao J, et al. (2015b) Content and activity of human liver microsomal protein and prediction of individual hepatic clearance in vivo. Sci Rep 5:17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang H, Gao N, Wei J, Tian X, Zhao Y, Fang Y, Zhou J, Wen Q, Gao J, et al. (2016) Physiological content and intrinsic activities of 10 cytochrome P450 isoforms in human normal liver microsomes. J Pharmacol Exp Ther 358:83–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.