Abstract
Forkhead box O3 (FOXO3) is a multispecific transcription factor that is responsible for multiple and conflicting transcriptional programs such as cell survival and apoptosis. The protein is heavily post-translationally modified and there is considerable evidence that post-transcriptional modifications (PTMs) regulate protein stability and nuclear–cytosolic translocation. Much less is known about how FOXO3 PTMs determine the specificity of its transcriptional program. In this study we demonstrate that exposure of hepatocytes to ethanol or exposure of macrophages to lipopolysaccharide (LPS) induces the c-Jun N-terminal kinase (JNK)-dependent phosphorylation of FOXO3 at serine-574. Chromatin immunoprecipitation (ChIP), mRNA and protein measurements demonstrate that p-574-FOXO3 selectively binds to promoters of pro-apoptotic genes but not to other well-described FOXO3 targets. Both unphosphorylated and p-574-FOXO3 bound to the B-cell lymphoma 2 (Bcl-2) promoter, but the unphosphorylated form was a transcriptional activator, whereas p-574-FOXO3 was a transcriptional repressor. The combination of increased TRAIL (TNF-related apoptosis-inducing ligand) and decreased Bcl-2 was both necessary and sufficient to induce apoptosis. LPS treatment of a human monocyte cell line (THP-1) induced FOXO3 S-574 phosphorylation and apoptosis. LPS-induced apoptosis was prevented by knockdown of FOXO3. It was restored by overexpressing wild-type FOXO3 but not by overexpressing a nonphosphorylatable S-574A FOXO3. Expression of an S-574D phosphomimetic form of FOXO3 induced apoptosis even in the absence of LPS. A similar result was obtained with mouse peritoneal macrophages where LPS treatment increased TRAIL, decreased Bcl-2 and induced apoptosis in wild-type but not FOXO3−/− cells. This work thus demonstrates that S-574 phosphorylation generates a specifically apoptotic form of FOXO3 with decreased transcriptional activity for other well-described FOXO3 functions.
Forkhead box O3 (FOXO3) is a multispecific transcription factor that serves as a longevity factor1, 2 and tumor suppressor and is critically involved in multiple seemingly independent biological process including cell-cycle arrest,3 DNA repair,4 antioxidant and stress responses,5 apoptosis,6 autophagy, glucose metabolism and aging.7 Its many transcriptional programs are frequently in opposition to each other as it induces apoptosis, yet it is also critical for cell survival and longevity. Although there is considerable information regarding the mechanisms that regulate the nuclear/cytosolic distribution and protein stability of FOXO3, the control mechanisms that regulate transcriptional specificity are poorly understood.
FOXO3 function is tightly regulated by multiple post-translational modifications (PTMs) including phosphorylation, acetylation, ubiquitination and arginine and lysine methylation. Specific PTMs regulate the partitioning of FOXO3 between the cytosol and the nucleus8, 9, 10, 11, 12, 13, 14, 15 and its stability and degradation,16 but the links between specific PTMs and FOXO3 transcriptional specificity are less well understood. Brunet et al.17 showed that acetylation favors expression ofpro-apoptotic as opposed to antioxidant genes, but the sites responsible for this are unclear.18 Other studies have suggested that that deacetylation reduces apoptotic transcription primarily by enhancing FOXO3 ubiquitination thus reducing gene expression through FOXO3 degradation.19, 20 Other than acetylation, specific PTMs that control the FOXO3 transcriptional program have not been identified, and effects such as context-dependent alterations in DNA and histone structure21 or binding of transcriptional repressors, such as NFIL3,22 are thought to play the major role in transcriptional specificity tuning.
In our previous work we observed that alcohol treatment of hepatocytes generated specific PTM patterns of FOXO3, particularly those resulting from phosphorylation and arginine demethylation.23 In the present study, we sought to determine whether these modified forms of FOXO3 cause preferential target gene transcription. We observed that ethanol (EtOH) induces a c-Jun N-terminal kinase (JNK)-dependent phosphorylation of FOXO3 at serine-574 (S-574) and this altered the transcriptional specificity of FOXO3, converting it from an antioxidant to an apoptosis-inducing transcription factor. In addition to selectively activating transcription of previously reported pro-apoptotic genes, such as TRAIL (TNF-related apoptosis-inducing ligand), p-574-FOXO3 was a transcriptional repressor of the anti-apoptotic protein B-cell lymphoma 2 (Bcl-2). The combination of an increase in TRAIL and a decrease in Bcl-2 were both sufficient and necessary for apoptosis induction. These results demonstrate that in addition to regulating stability, nuclear translocation and transcriptional efficiency, specific FOXO3 PTMs can alter its transcriptional program, converting it into a potent apoptosis inducer.
Results
EtOH reprograms FOXO3 by selectively enhancing its pro-apoptotic effects
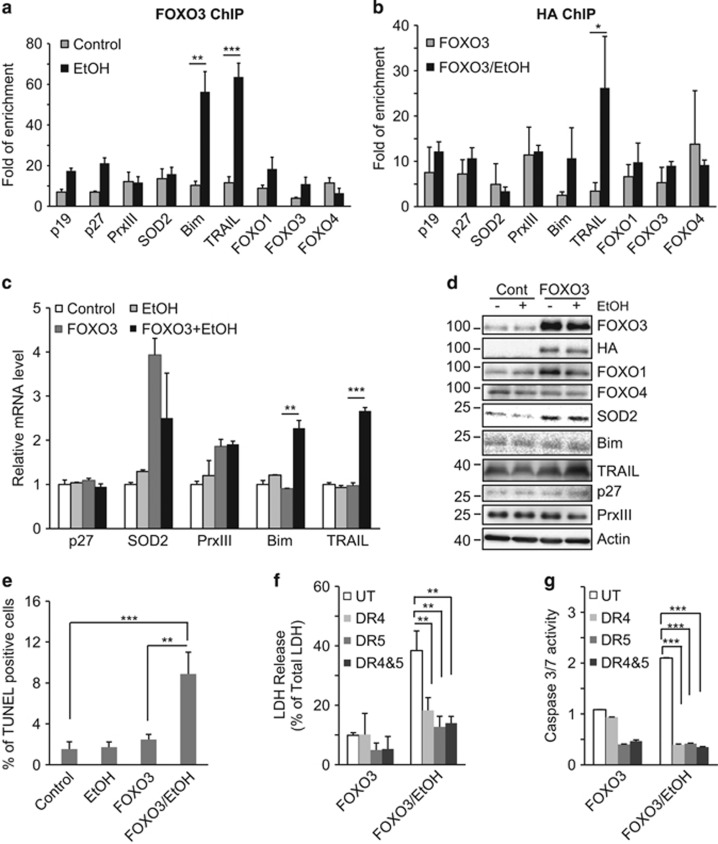
In order to assess the effects of alcohol on FOXO3 we treated Huh7.5 hepatoma cells with or without 50 mM EtOH for 48 h and performed ChIP studies with an endogenous FOXO3 antibody (Figure 1a) or with an anti-HA antibody after overexpression of HA-FOXO3 (Figure 1b). A range of promoter targets were examined to represent the different transcriptional programs of FOXO3. EtOH treatment resulted in increased promoter binding to the pro-apoptotic genes, Bim (B-cell lymphoma 2 interacting mediator of cell death) and TRAIL, but had only minor and nonsignificant effects on other target genes (Figures 1a and b). This effect was present at the mRNA and protein levels as well (Figures 1c and d). HA-FOXO3 overexpression itself increased superoxide dismutase 2 (SOD2) and peroxiredoxin III (PrxIII) mRNA but had no effect on Bim and TRAIL. Subsequent EtOH treatment, however, significantly increased both Bim and TRAIL mRNA (Figure 1c). FOXO3 overexpression increased SOD2 and FOXO1 protein levels and EtOH treatment subsequently increased TRAIL (Figure 1d). The lack of precise agreement between mRNA and protein levels for Bim and PrxIII suggests that these proteins may be controlled by nontranscriptional mechanisms as well.
Figure 1.
EtOH reprograms FOXO3 selectively enhancing pro-apoptotic effects. (a) Huh7.5 cells were cultured with or without 50 mM EtOH for 48 h. ChIP assay was performed with anti-FOXO3 antibody. Promoters assessed were p19, p27, SOD2 (superoxide dismutase 2), PrxIII (peroxiredoxin III), Bim (Bcl-2-interacting mediator of cell death), TRAIL (TNF-related apoptosis-inducing ligand), FOXO3, FOXO1 and FOXO4. (b) Huh7.5 cells were transfected with HA-tagged FOXO3, treated with or without 50 mM EtOH for additional 48 h, and ChIP assay was performed with anti-HA antibody. (c and d) mRNA (c) and protein (d) measurements of FOXO3 target gene expression were evaluated by real-time RT-PCR and western blot, respectively. (e) Cell death was evaluated by TUNEL assay in cells treated with 50 mM EtOH either with or without FOXO3 transfection. (f and g) Effects of TRAIL receptor antagonists on FOXO3/EtOH-induced LDH release and caspase activity. FOXO3-transfected cells were treated with or without EtOH in the presence of TRAIL receptor antagonists DR4 (1 μg/ml) or DR5 (1 μg/ml) or the combination (DR4+5). Values are mean±S.D. of three independent experiments. *P<0.05, **P<0.01, ***P<0.001, Student's t-test
To determine whether these pro-apoptotic gene expression changes cause cell death, FOXO3-overexpressing cells were treated with 50 mM EtOH for 48 h and assessed by TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assay, LDH release and caspase activation. EtOH treatment of untransfected Huh7.5 cells did not cause cell death, but after FOXO3 overexpression, EtOH caused a significant increase in TUNEL positivity, LDH release and caspase 3/7 activity (Figures 1e–g). DR4 and DR5, the TRAIL receptor antagonists,24 significantly blocked FOXO3/EtOH-induced increases of LDH release and caspase 3/7 activity (Figures 1f and g), demonstrating that FOXO3/EtOH-induced cell death was TRAIL dependent. These data indicate that EtOH reprograms FOXO3, enhancing its activation of pro-apoptotic target genes that contribute to TRAIL-dependent apoptosis.
EtOH causes FOXO3 S-574 phosphorylation
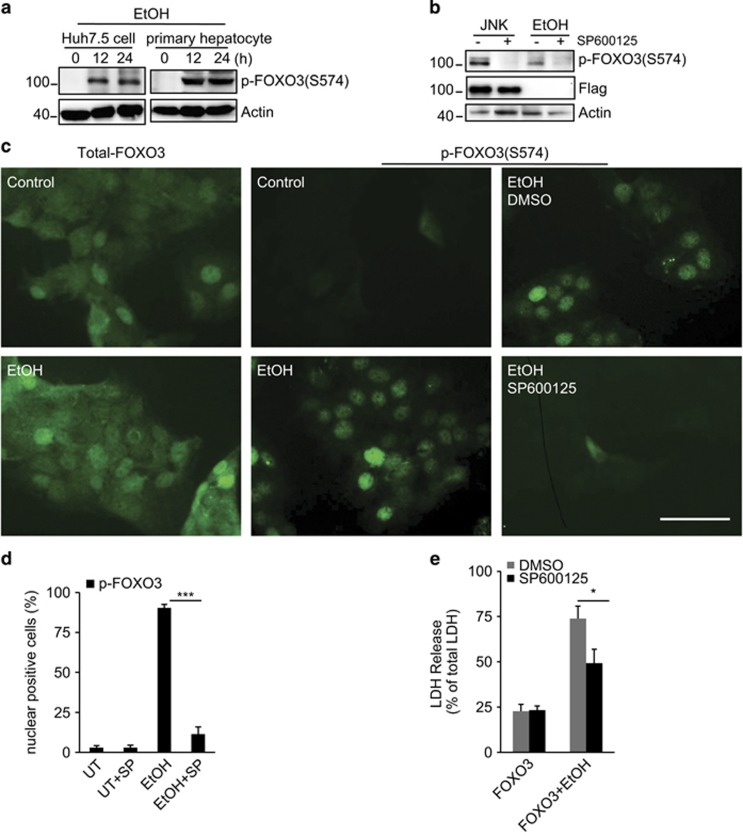
We previously observed that hepatitis C virus (HCV) infection induced JNK-dependent phosphorylation of FOXO3 at S-574.23 The specificity and functional consequences of this phosphorylation are unknown. We generated a specific antibody to phospho-S-574-FOXO3 (p-574-FOXO3) and used it to examine the effects of EtOH on this FOXO3 modification. The antibody showed high specificity for p-574-FOXO3 (Supplementary Figure S1). Huh7.5 cells and primary human hepatocytes showed undetectable levels of p-574-FOXO3 in the absence of EtOH treatment, but exposure to EtOH for 12 h resulted in robust FOXO3 S-574 phosphorylation in both cell types (Figure 2a). S-574 FOXO3 phosphorylation induced by either JNK overexpression or EtOH was JNK dependent as in both cases it was blocked by JNK inhibitor (Figure 2b). Immunofluorescence studies showed that total FOXO3 was present in both nucleus and cytosol and its distribution was not changed by EtOH (Figure 2c). p-574-FOXO3 was only detectable after EtOH exposure, was primarily nuclear and its formation was blocked by JNK inhibitor (Figures 2c and d). The JNK inhibitor significantly reduced LDH release from EtOH-treated FOXO3-overexpressing Huh7.5 cells (Figure 2e).
Figure 2.
EtOH induces FOXO3 S-574 phosphorylation. (a) Primary human hepatocytes or Huh7.5 cells were incubated with 50 mM EtOH. Whole-cell lysates were blotted for p-574-FOXO3 at indicated times. (b) Huh7.5 cells were transfected with FOXO3 and either co-transfected with JNK1 or treated with 50 mM EtOH. Where indicated, cells were treated with JNK inhibitor SP600125 (100 nM). Cells were harvested 24 h after treatment and p-574-FOXO3 was analyzed by western blot. (c) Immunofluorescence for total FOXO3 or p-574-FOXO3 in cells treated with EtOH and JNK inhibitor as indicated. Scale bar, 50 μm. (d) Quantitation of data from imaging studies as in (c). UT, untreated; SP, SP600125. (e) LDH release of FOXO3-transfected cells that were treated with or without 50 mM EtOH in the presence or absence of JNK inhibitor. Values are mean±S.D. of three independent experiments. *P<0.05, ***P<0.001, Student's t-test
Apoptotic effects of p-574-FOXO3
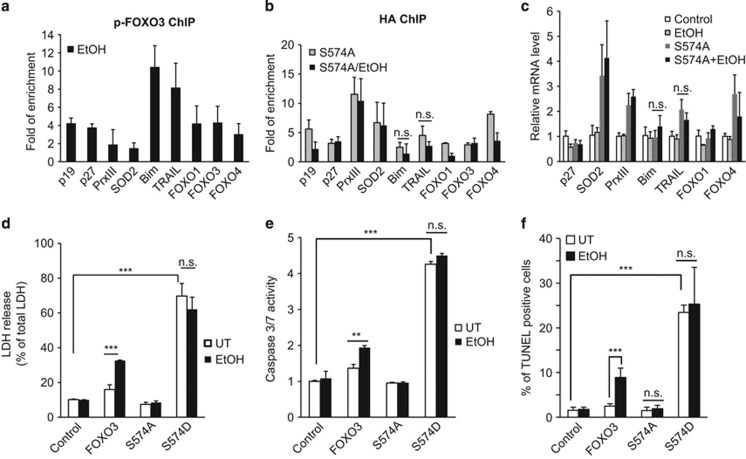
To investigate the functional properties of p-574-FOXO3, we used the p-574-FOXO3 antibody for chromatin immunoprecipitation (ChIP) analysis. The results demonstrated that p-574-FOXO3 bound primarily to apoptotic gene targets Bim and TRAIL, but not to antioxidant gene targets SOD2 and PrxIII (Figure 3a). We next examined the phosphorylation-deficient mutant, FOXO3-574A to determine whether FOXO3/EtOH apoptosis required this modification. Unlike the situation for WT-FOXO3, EtOH treatment of FOXO3 S-574A-expressing cells failed to increase promoter binding to Bim and TRAIL (Figure 3b) and also failed to increase Bim and TRAIL mRNA (Figure 3c). To further confirm the role of S-574 phosphorylation in apoptosis we used a phosphomimetic form, FOXO3 S-574D. The WT-FOXO3 induced apoptosis only after exposure to EtOH, FOXO3 S-574A failed to induce apoptosis with or without EtOH and FOXO3 S-574D induced apoptosis irrespective of whether or not EtOH was present (Figures 3d–f). Alcohol thus induces formation of a novel pro-apoptotic, phosphorylated form of FOXO3 in Huh7.5 cells.
Figure 3.
Apoptotic effects of p-574-FOXO3. (a) Huh7.5 cells were transfected with FOXO3 and treated with 50 mM EtOH for 48 h. ChIP assay was performed as in Figure 1 using p-574-FOXO3 antibody. (b–c) Cells were transfected with HA-FOXO3-S-474A and treated with or without 50 mM EtOH for 48 h. ChIP assay was performed with HA antibody (b) and mRNA expression was determined by RT-PCR (c). (d–f) Cells were transfected with HA-tagged FOXO3, FOXO3-S-547A or FOXO3 S-574D with or without EtOH treatment. Cell death was evaluated by LDH release (d), caspase activity (e) and TUNEL assay (f). Values are mean±S.D. of three independent experiments. **P<0.01, ***P<0.001, Student's t-test
FOXO3/EtOH is a transcriptional repressor of Bcl-2
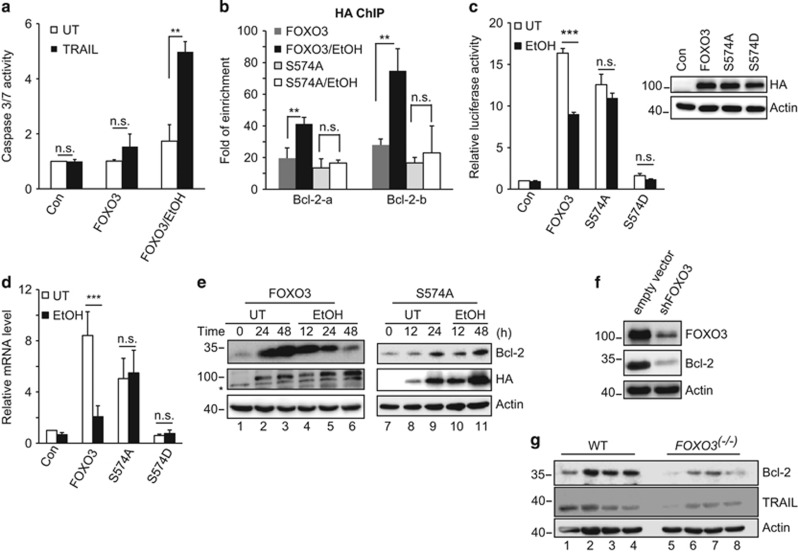
Although p-574-FOXO3 is specifically pro-apoptotic, the observed changes in TRAIL and BIM were modest. Furthermore, the EtOH/FOXO3 combination dramatically increased the sensitivity of cells to exogenous TRAIL (Figure 4a). This suggests that mechanisms other than increased TRAIL account for apoptosis induction. We therefore used an apoptosis-related PCR array and identified nine apoptosis-related gene products that were altered either by FOXO3 or by the combination of FOXO3 and EtOH (Table 1). FOXO3 overexpression induced both pro- and anti-apoptotic genes, but in each case, subsequent addition of EtOH further increased pro-apoptotic genes or decreased anti-apoptotic genes. This effect was particularly striking for Bcl-2 where FOXO3 overexpression induced a 30-fold increase of Bcl-2 mRNA that was then completely abolished by EtOH.
Figure 4.
p-574-FOXO3 is a transcriptional repressor of Bcl-2. (a) FOXO3-transfected Huh7.5 cells were treated with 50 mM EtOH in the absence or presence of exogenous TRAIL (100 ng/ml). Caspase 3/7 activity was determined 48 h after treatment. (b) Cells were transfected with HA-FOXO3 or HA-FOXO3 S-574A, incubated for 16 h and subsequently treated with EtOH (50 mM) for an additional 24 h. ChIP experiments were performed with HA antibody and two sets of Bcl-2 promoter primers. (c) Bcl-2 promoter luciferase assays were performed after expression of HA-tagged WT or mutant FOXO3 proteins followed by incubation with EtOH (50 mM) for an additional 48 h. Relative HA protein expression was demonstrated by western blot. (d) Bcl-2 mRNA levels in response to FOXO3 protein expression and EtOH treatment as in (c). (e) Bcl-2 protein levels in cells that were untransfected (UT) or transfected with HA-FOXO3 (lanes 1–6) or HA-FOXO3 S-574A (lanes 7–11) with subsequent 50 mM EtOH exposure. Time is listed in hours after transfection (lanes 1–3 and 7–9) or subsequent EtOH addition to the same cultures (lanes 4–6 and 10–11). Asterisk indicates nonspecific bands. (f) Bcl-2 expression in FOXO3 knockdown cells. Huh 7.5 cells were infected with vectors coding for FOXO3-specific shRNA or empty vector. Knockdown efficiency and Bcl-2 expression level were verified by western blot. (g) Bcl-2 expression in liver homogenates from WT (lanes 1–4) and FOXO3−/− mice (lanes 5–8). Each lane represents a homogenate from a single mouse. Values are mean±S.D. of three independent experiments. **P<0.01, ***P<0.001, Student's t-test
Table 1. Effects of FOXO3 with and without EtOH on pro- and anti-apoptotic gene mRNA expression.
| Target gene | FOXO3 | FOXO3/EtOH | |
|---|---|---|---|
| Anti-apoptotic | BCL2 | 30.7 | 1.1 |
| BIRC2 | 2.3 | 0.6 | |
| BIRC3 | 9.5 | 3.4 | |
| BNIP3L | 4.0 | 1.3 | |
| TNFRSF1B | 23.1 | 4.0 | |
| Pro-apoptotic | TNF | 11.6 | 14.0 |
| TNFRSF9 | 12.2 | 20.3 | |
| TNFRSF8 | 3.9 | 18.7 | |
| IL10 | 3.4 | 6.3 |
Data are expressed as mRNA fold elevation compared with Huh7.5 cells that were not transfected with FOXO3 or treated with EtOH
The Bcl-2 promoter has three potential FHRE motifs. ChIP studies showed that FOXO3 bound strongly to two of these promoter regions and EtOH treatment increased binding in both cases. In the absence of EtOH, FOXO3 S-574A bound to the Bcl-2 promoter similar to FOXO3, but EtOH had no further effect (Figure 4b). We next performed Bcl-2 promoter luciferase assays to evaluate transcriptional effects. The WT-FOXO3 overexpression induced an ∼15-fold increase of luciferase activity that was reduced by EtOH treatment. FOXO3 S-574A induced a similar increase of luciferase activity but EtOH had no effect. FOXO3 S-574D showed no transcriptional activity before or after EtOH treatment (Figure 4c). FOXO3 further induced Bcl-2 mRNA and this effect was suppressed by EtOH. FOXO3 S-574A also increased Bcl-2 mRNA but this was not suppressed by EtOH. FOXO3 S-574D failed to increase Bcl-2 transcription (Figure 4d).
It has previously been reported that Bcl-2 is minimally expressed in hepatocytes under normal conditions.25, 26 Consistent with these observations, we found low Bcl-2 protein levels in Huh7.5 cells, but FOXO3 overexpression dramatically increased Bcl-2 (Figure 4e, lanes 1–3). Subsequent EtOH exposure decreased Bcl-2 protein back to the basal level (Figure 4e, lanes 4–6). In contrast, FOXO3 S-574A also increased Bcl-2 protein expression (Figure 4e, lanes 7–9), but EtOH did not antagonize this effect (Figure 4e, lanes 10–11). At the steady state, FOXO3 knockdown was sufficient to lower Bcl-2 protein levels as well (Figure 4f). We further examined liver tissue from wild-type (WT) and FOXO3−/− mice. FOXO3−/− mice had decreased hepatic Bcl-2 expression as well as lower basal levels of TRAIL (Figure 4g, lanes 5–8). These data indicate that FOXO3 is important in the basal expression of Bcl-2 and EtOH-induced S-574 phosphorylation suppresses Bcl-2 expression.
Bcl-2 and TRAIL determine apoptosis in FOXO3/EtOH-treated cells
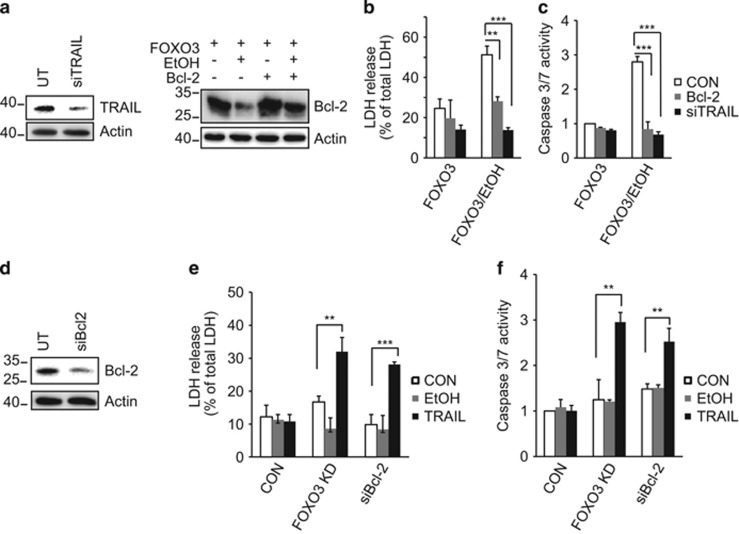
In order to investigate the relative contribution of TRAIL and Bcl-2 in FOXO3/EtOH-induced apoptosis, we examined Huh7.5 cells that were deficient in TRAIL (TRAIL siRNA) or in which Bcl-2 levels were maintained through Bcl-2 overexpression (Figure 5a). Either Bcl-2 overexpression or TRAIL siRNA blocked the ability of FOXO3/EtOH to induce apoptosis (Figures 5b and c). This suggests that both high TRAIL and low Bcl-2 are necessary for FOXO3/EtOH-induced cell death. In addition, either decreased Bcl-2 or increased TRAIL alone did not induce apoptosis but the combination was sufficient to induce apoptosis in the absence of FOXO3/EtOH (Figure 5d). Similarly, decreasing Bcl-2 through FOXO3 siRNA was also sufficient to confer TRAIL-sensitive apoptosis (Figures 5e and f). These results indicate that the observed increase of TRAIL and decrease of Bcl-2 are both necessary and sufficient to explain FOXO3/EtOH-induced apoptosis.
Figure 5.
Bcl-2 and TRAIL determine FOXO3/EtOH-induced apoptosis. (a) Western blots in Huh7.5 cell lysates demonstrating siRNA-induced TRAIL knockdown efficiency and ability to restore EtOH-induced loss of Bcl-2 protein through Bcl-2 overexpression. (b and c) LDH release and caspase 3/7 activity induced by FOXO3 expression and EtOH treatment in cells that are deficient in TRAIL (black bars) or in cells in which Bcl-2 levels were maintained through Bcl-2 overexpression (gray bars). (d) Western blot to assess siRNA knockdown efficiency of Bcl-2. (e and f) LDH release and caspase 3/7 activity induced by exogenous TRAIL (100 ng/ml, black bars) and EtOH (gray bars) in cells that are deficient in FOXO3 or Bcl-2. Values are mean±S.D. of three independent experiments. **P<0.01, ***P<0.001, Student's t-test
FOXO3 mediates macrophage apoptosis
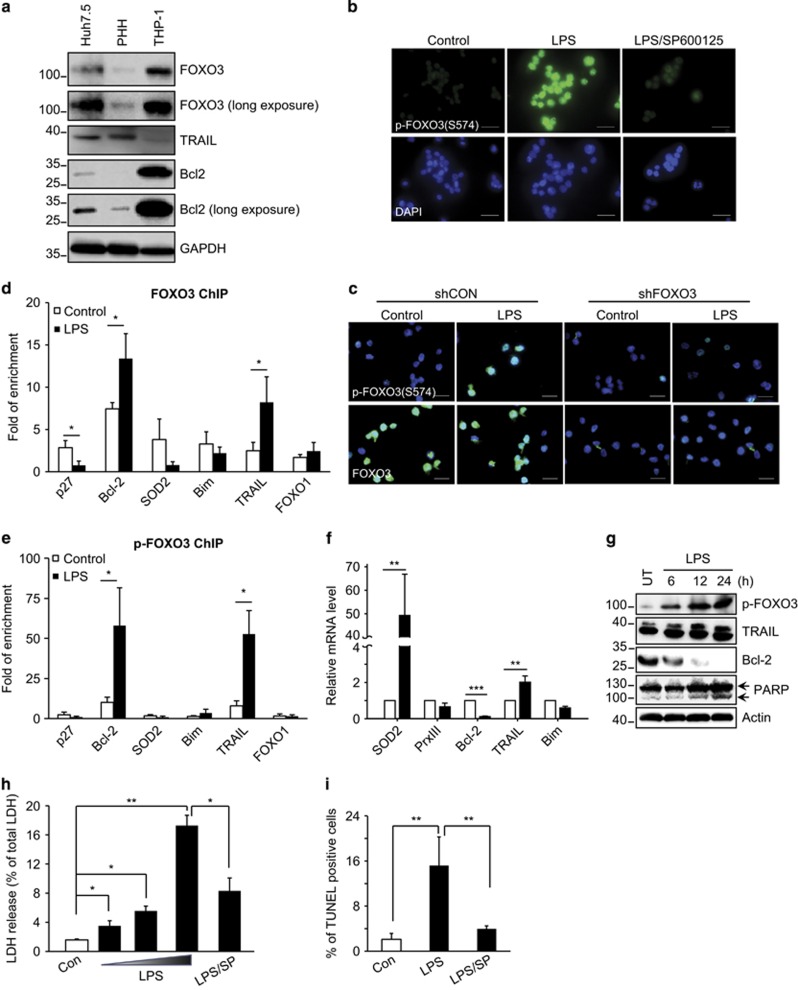
Although the above studies demonstrate the apoptotic function of p-574-FOXO3, the induction of apoptosis in hepatocytes and hepatoma cells required FOXO3 overexpression. As FOXO3-dependent apoptosis results from an increase in TRAIL and a decrease in Bcl-2, we reasoned that differences in basal expression of the proteins involved might influence whether FOXO3 S-574 phosphorylation induces apoptosis. To examine this issue we measured protein levels of FOXO3, Bcl-2 and TRAIL in hepatoma cells, hepatocytes and the human monocyte/macrophage cell line, THP-1 (Figure 6a). THP-1 cells express higher levels of Bcl-2 and FOXO3 and lower levels of TRAIL than hepatocytes. As alcoholic liver disease is known to involve lipopolysaccharide (LPS) release from the gut with subsequent activation of hepatic macrophages,27 we determined the impact of LPS on macrophage FOXO3 and apoptosis.
Figure 6.
Effects of LPS on FOXO3 S-574 phosphorylation, promoter binding and function in monocytes. (a) Western blots of lysates from Huh7.5 hepatoma cells, primary human hepatocytes (PHH), and human monocytic (THP-1) cells demonstrating relative protein levels of FOXO3, Bcl-2 and TRAIL. (b) Immunofluorescence for p-574-FOXO3 in THP-1 cells either without treatment, after treatment with 100 ng/ml LPS or with LPS+JNK inhibitor SP600125 (20 μM). p-574-FOXO3 was determined 24 h after treatment by using p-FOXO3 S-574 antibody. DAPI stain indicates the presence of nuclei. Scale bars, 50 μm. (c) THP-1 cell immunofluorescence images for p-574-FOXO3 or total FOXO3 with or without LPS treatment in cells after infection with lentiviral shRNA constructs for either FOXO3 (shFOXO3) or scrambled control mRNA (shCON). Images are an overlay of immunofluorescence (green) and DAPI (blue). Scale bars, 50 μm. (d and e) THP-1 cells were cultured with or without 100 ng/ml LPS for 12 h. ChIP assays were performed with anti-total FOXO3 or anti p-574-FOXO3 antibodies. (f and g) mRNA and protein expression in untreated (UT) or LPS-treated THP-1 cells. Numbers indicate hours after LPS treatment. (h and i) LDH release and TUNEL assay for THP-1 cells treated with 10, 20 and 100 ng/ml LPS in the absence or presence of SP600125. Values are mean±S.D. of three independent experiments. *P<0.05, **P<0.01, ***P<0.001, Student's t-test
Figure 6b shows immunofluorescence images demonstrating that LPS induces S-574 phosphorylation of FOXO3 in THP-1 cells and this phosphorylation is JNK dependent (Figure 6b). The specificity of this signal for FOXO3 was demonstrated in FOXO3 knockdown cells where LPS no longer induced the immunofluorescence signal (Figure 6c). We next performed ChIP studies using antibodies against total or p-574-FOXO3. Total FOXO3 bound to a range of promoters but LPS treatment increased binding only to the apoptosis-related genes Bcl-2 and TRAIL (Figure 6d). Before LPS stimulation, there was minimal promoter binding of p-574-FOXO3 and we detected significant binding only for Bcl-2 and TRAIL. LPS induced >10-fold increases in the signal for these two genes without altering binding to other FOXO3 targets (Figure 6e). LPS treatment resulted in a 10-fold decrease of Bcl-2, and a 2-fold increase of TRAIL mRNA (Figure 6f) with similar effects on protein levels (Figure 6g). LPS also induced THP-1 cell death as evidenced by LDH release (Figure 6h), TUNEL positivity (Figure 6i) and PARP cleavage (Figure 6g). Both LDH release and TUNEL positivity were suppressed by JNK inhibitor. LPS thus induces p-574-FOXO3 formation and generates the FOXO3-associated apoptosis phenotype characterized by decreased Bcl-2, increased TRAIL and cell death.
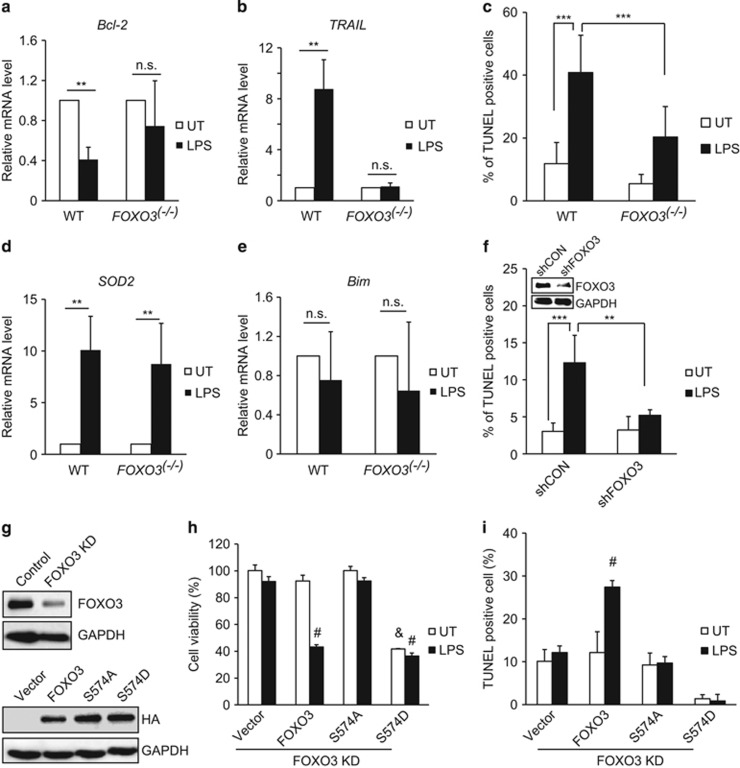
To determine whether LPS-induced macrophage cell death is FOXO3 dependent, we isolated peritoneal macrophages from WT and FOXO3−/− mice. In WT macrophages, LPS treatment decreased Bcl-2 and increased TRAIL mRNA (Figures 7a and b), whereas 40% of the cells became TUNEL positive (Figure 7c). In FOXO3−/− macrophages, LPS treatment had no effect on Bcl-2 or TRAIL and apoptosis was dramatically reduced. LPS also increased SOD2 mRNA but this effect was not dependent on FOXO3 (Figure 7d).Unlike the situation for alcohol treatment of FOXO3-expressing hepatoma cells, Bim mRNA was unaffected (Figure 7e).
Figure 7.
p-574-FOXO3-dependent macrophage apoptosis. (a and b) Real-time RT-PCR analysis of Bcl-2 and TRAIL mRNA levels in peritoneal macrophages isolated from wild-type (WT) or FOXO3−/− mice and subsequently treated for 24 h with 100 ng/ml LPS (WT n=3, FOXO3−/−n=4). (c) TUNEL assays were performed in peritoneal macrophages isolated from WT and FOXO3−/− mice either untreated (con) or treated with 100 ng/ml LPS for 24 h (n=3 each). (d and e) Real-time RT-PCR analysis of SOD2 and Bim mRNA levels in peritoneal macrophages isolated from WT or FOXO3−/− mice after 24 h of treatment with 100 ng/ml LPS (WT n=3, FOXO3−/−n=4). (f) THP-1 cells were infected with vectors coding for FOXO3-specific shRNA or empty lentiviral particles. After 48 h, cells were either left untreated (UT) or treated with 100 ng/ml LPS for 24 h. TUNEL assays were then performed. Values are mean±S.D. **P<0.01, ***P<0.001, Student's t-test. Inset shows western blot demonstrating magnitude of the knockdown achieved. (g, upper panel) Western blot to assess shRNA knockdown efficiency of FOXO3 in THP-1 cells after 5 μg/ml puromycin selection. (g, lower panel) Western blot in these same cells 48 h after electroporation of HA-tagged WT or mutant FOXO3 proteins. (h and i) Cell viability and TUNEL assay for FOXO3 knockdown cells electroporated with empty vector, HA-WT-FOXO3, HA-FOXO3 S-574A or HA-FOXO3 S-574D. Results are shown after 48 h of either no treatment (UT) or 24 h without treatment followed by an additional 24 h of treatment with 100 ng/ml LPS. Values are mean±S.D. #P<0.001 versus vector/UT, &P<0.001 versus vector/LPS
To further explore the role of FOXO3 S-574 phosphorylation in macrophage apoptosis we used lentiviral-mediated FOXO3 shRNA to knock down FOXO3 expression in THP-1 cells. Similar to FOXO3−/− macrophages, these cells were resistant to LPS-induced apoptosis (Figure 7f). We then re-expressed either WT or mutant FOXO3 proteins in these cells (Figure 7g) and challenged them with LPS. WT FOXO3 had no effect on cell viability but its expression restored LPS sensitivity as indicated by decreased viability and increased TUNEL positivity (Figures 7h and i). FOXO3 S-574A was not able to restore LPS sensitivity. The phosphomimetic S-574D mutant reduced cell viability with or without LPS treatment. Because of rapid cell death we were not able to identify TUNEL positivity 72 h after FOXO3 S-574D expression. These results demonstrate that S-574 phosphorylation of FOXO3 is the key event required for LPS-induced apoptosis of macrophages.
Discussion
FOXO3 can be involved in numerous cellular functions, some of which seem contradictory such as promoting both cell survival and apoptosis.28, 29 Although the major mechanisms governing nuclear/cytosolic distribution and stability have been intensively investigated, much less is known about the specific mechanisms that govern transcriptional program specificity. The discovery that p-574-FOXO3 is a specifically pro-apoptotic form of the protein is thus an important advance in our understanding of FOXO3.
S-574 was first reported to be a site of FOXO3 phosphorylation by Brunet et al.17 We previously observed this phosphorylated form after HCV infection23 and now have determined that it occurs after EtOH treatment of hepatocytes or LPS treatment of macrophages as well. We were able to demonstrate a strong connection between JNK activation and the appearance of p-574-FOXO3 as it was induced by activated JNK1 expression, its formation was blocked by JNK inhibitor and the time course of its appearance closely followed that of JNK activation for either LPS-treated macrophages or alcohol-treated hepatoma cells (Supplementary Figure S2). Nonetheless, we do not yet know whether FOXO3 is directly phosphorylated by JNK or by anther kinase that is activated downstream of JNK activation.
This p-574-FOXO3 selectively binds to pro-apoptotic promoters in both hepatocytes and macrophages and not to antioxidant promoters. Targeted substitutions at S-574 showed that alanine substitution was sufficient to abolish this apoptotic activity and aspartate substitution was able to induce the apoptosis program in the absence of EtOH. This indicates that the charge at this site is the critical determinant of the pro-apoptotic function of FOXO3.
In spite of extensive information regarding the large array of FOXO3 PTMs,7 there is relatively little known about how specific PTMs alter the FOXO3 transcriptional program. It has previously been shown that FOXO3 acetylation is related to its relative propensity to induce antioxidant versus apoptotic gene expression.17 In that study, expression or activation of the SIRT family of NAD-dependent deacetylases, particularly SIRT1, favored the antioxidant phenotype, suggesting that acetylated FOXO3 is more apoptotic whereas deacetylated FOXO3 is more antioxidant. However, specific lysine residues responsible for these effects were not determined.17 Our findings with S-574A and S-574D mutants suggest that the final arbiter of whether or not FOXO3 causes apoptosis is related to this phosphorylation event, but it is entirely possible that the ability of this serine to become phosphorylated is regulated by other protein PTMs such as lysine acetylation. This sort of multilevel control of FOXO3 function has been demonstrated for arginine methylation and Akt phosphorylation of FOXO1 where Akt phosphorylation is the final determinant of cytosolic translocation but arginine methylation of FOXO1 controls access of Akt to it substrate serine site.16 It is notable that our studies show that aspartic acid substitution of S-574 alone is sufficient to induce apoptosis, suggesting that once that residue becomes charged, changing acetylation state may not be critical.
The exact identity of the gene expression changes responsible for FOXO3-dependent apoptosis has been a matter of debate. In addition to TRAIL and Bim, there are other well-described FOXO3 targets including the p53 target genes PUMA (p53 upregulated modulator of apoptosis) and Noxa (phorbol-12-myristate-13-acetate-induced protein 1). Although we consistently observed that p-574-FOXO3 increased BIM and TRAIL mRNA, we were not able to observe consistent changes in Bim at the protein level and thus sought out other possible causes of the apoptosis phenotype. Through a PCR array screening we were able to identify Bcl-2 as a novel target that is regulated by FOXO3. Both the unphosphorylated form and p-574-FOXO3 bind to the Bcl-2 promoter, but although the unphosphorylated form is a transcriptional activator, p-574-FOXO3 serves as a transcriptional repressor. In Huh7.5 cells, the combination of the increase in TRAIL and the decrease in Bcl-2 was both necessary and sufficient to produce apoptosis and an increase in Bim was not required. FOXO3/EtOH-induced apoptosis could be prevented either by reducing TRAIL or increasing Bcl-2, and apoptosis could be induced in the absence of EtOH by the combination of Bcl-2 siRNA and exogenous TRAIL exposure. Therefore, although a number of other apoptosis-related genes such as PUMA and Noxa are known to be FOXO3 related,30, 31 the key phenotype of EtOH/FOXO3 apoptosis requires at the minimum, an increase in TRAIL and a decrease in Bcl-2.
We initially used Huh7.5 hepatoma cells as the system in which to investigate FOXO3 effects. These cells are widely used as they are permissive to cell culture infection with HCV and we previously showed that they express alcohol dehydrogenase (ADH) and respond to alcohol.32 They were chosen for this study as previous isoelectric focusing studies showed that HCV infection and alcohol generated specific and different FOXO3 PTMs and these appeared to determine antioxidant responses.23 In this earlier study we observed that HCV caused a specific species of FOXO3 that contained S-574 phosphorylation but this particular species was not identified after EtOH treatment. This implies that the HCV-induced and EtOH-induced p-574 phosphorylated forms of FOXO3 may differ at other sites as they have different pIs.
EtOH induced robust S-574 phosphorylation of endogenous FOXO3 in both hepatocytes and hepatoma cells, but in both cases this was not sufficient to induce apoptosis without first overexpressing FOXO3. In contrast, macrophages undergo endogenous FOXO3 S-574 phosphorylation in response to LPS and this is sufficient to induce apoptosis. Although the reason for the different responses of these two cell types is not certain, our data suggest two possibilities. First, there may be a minimum level of FOXO3 expression below which FOXO3 phosphorylation is not able to induce apoptosis. This possibility is suggested by the observation that endogenous FOXO3 expression is considerably higher in THP-1 cells than hepatocytes or hepatoma cells (Figure 6a) and that an 80% FOXO3 knockdown in THP-1 cells was sufficient to abolish LPS-induced apoptosis (Figure 7). Alternatively, it could result from a difference in how apoptosis is regulated in different cell types. Macrophages have high expression of FOXO3 and express high levels of Bcl-2 and low levels of TRAIL at baseline. Thus, FOXO3 phosphorylation at S-574, which suppresses Bcl-2 and increases TRAIL, is likely to dramatically alter cell death control. Hepatocytes and most hepatoma cells express relatively lower amounts of FOXO3, have relatively high basal TRAIL expression and most importantly express only very low levels of Bcl-2. A further loss of Bcl-2 as a result of FOXO3 phosphorylation is thus likely to be less consequential. After overexpression of FOXO3, however, hepatocellular FOXO3 levels and Bcl-2 levels increase dramatically, thus rendering the cells sensitive to the pro-apoptotic effects of FOXO3 S-574 phosphorylation. We previously showed that FOXO3 is a protective factor that prevents alcohol-induced liver injury.32 The present results suggest that an effect on macrophage apoptosis might be playing a role in this protection. Understanding how p-574-FOXO3-dependent apoptosis is differentially regulated in the different cell types of the liver could provide important insights into the pathogenesis of alcoholic liver disease.
In summary, the present results show that alcohol treatment of hepatocytes or LPS treatment of macrophages induces phosphorylation of FOXO3 at S-574, generating a form of FOXO3 that can promote apoptosis through direct transcriptional activation of TRAIL, and by a previous unrecognized ability of p-574-FOXO3 to act as a transcriptional repressor of Bcl-2. FOXO3-dependent apoptosis occurs in macrophages in response to LPS and may occur in other cell types as well. The finding that a specific PTM is sufficient to induce a FOXO3 apoptosis program is one of the first demonstrations of a transcriptionally selective form of FOXO3 and opens the possibility that modifying this phosphorylation process in both positive and negative ways could be an important therapeutic approach. FOXO3-dependent apoptosis has been postulated to play a role in its ability to serve as a tumor suppressor,33 has been implicated in muscle atrophy and cardiomyopathy34 and appears to play a role in response of cancers to cytotoxic chemotherapy agents.35 Further work will be necessary to understand the factors that regulate the ability of JNK to induce phosphorylation at this site, whether other kinases perform this function as well and whether other factors ultimately determine cell fate in the presence of p-574-FOXO3.
Materials and Methods
Cell culture, plasmids and transfection
Huh7.5 cells (provided by Dr Charles Rice, New York, NY, USA) were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS), 50 U/ml penicillin and 50 mg/ml streptomycin. Cells were treated with 50 mM EtOH, and JNK inhibitor (SP600125, 20 μM) was used 4 h before EtOH treatment. Cells were transfected in the presence of serum-free medium (Opti-MEM, Invitrogen) by using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. pECE-HA-FOXO3, pcDNA3 Bcl-2, LB322 (Bcl-2 from ATG to -3934) and pCDNA3 Flag MKK7B2Jnk1a1 were respectively provided by M Greenberg, S Korsmeyer, L Boxervia and RJ Davis via Addgene (Cambridge, MA, USA). FOXO3 point mutation was generated by site-directed mutagenesis (Quickchange Kit, Stratagene, Santa Clara, CA, USA). For siRNA transfections, 2 × 105 cells were plated and cultured overnight. Thereafter, gene-specific siRNAs were transfected in the presence of Opti-MEM medium (Invitrogen) using X-tremeGENE siRNA Transfection Reagent (Roche, Indianapolis, IN, USA) according to the manufacturer's instructions. siRNA targeting Bcl-2 (s1915) was purchased from Invitrogen and siRNA targeting TRAIL (sc-36719) was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). The lentiviral vectors coding for human FOXO3-specific shRNA (MISSION TRC shRNA TRCN0000040100, TRCN0000040098) and the control vector pLKO.1 were obtained from Sigma-Aldrich (St. Louis, MO, USA). THP-1 cells were purchased from ATCC (Manassas, VA, USA) and maintained in RPMI-1640 Medium (Invitrogen) containing 10% FBS and 0.05 mM 2-mercaptoethanol. For preparation of FOXO3 knockdown cells, THP-1 cells were transduced with lentiviral FOXO3 shRNA supernatants supplemented with polybrene (4 μg/ml). Pools of shRNA-transduced cells were selected by adding 5 μg/ml puromycin (Gibco/Invitrogen, Grand Island, NY, USA) to the culture medium. FOXO3 expression level was determined by western blot 72 h after infection. Expression of FOXO3 or FOXO3 mutants in knockdown cells was performed by electroporation. 4 × 106 FOXO3 stable knockdown THP-1 cells were resuspended in 400 μl serum-free RPMI media and incubated with 10 μg DNA for 5 min on ice before being electroporated at 140 V, 1000 μF. Following electroporation, 600 μl complete RPMI media were added to the cells that were kept on ice for 15 min, and then grown in 5 ml media for 48 h.
Isolation and culture of primary human hepatocytes
Primary human hepatocytes were freshly isolated from liver resections by the Cell Isolation Core of the Department of Pharmacology at the University of Kansas Medical Center (Kansas City, KS, USA). All human tissues were obtained with informed consent from each patient according to ethical and institutional guidelines. The study was approved by the institutional review board at the University of Kansas Medical Center. Cells were isolated using a multi-step collagenase procedure as described in detail previously.36 Media consisted of Williams' Medium E (Life Technologies, Grand Island, NY, USA) supplemented with L-glutamine (2 mM) (Life Technologies), HEPES (10 mM), insulin (10−7 M), dexamethasone (10−7 M), penicillin (100 U/ml), streptomycin (100 μg/ml) and amphotericin B (0.25 μg/ml). The hepatocytes were brought to a concentration of 0.5 × 106 cells/ml in Williams' Medium E, as described above, plus 5% bovine calf serum. The hepatocytes were then seeded on collagen-coated plates and allowed to attach in a humidified 37 °C, 5% CO2 incubator for 12 h and then treated with 50 mM EtOH for indicated times.
Mouse strains
FOXO3−/− mice were provided by Dr Kana Miyamoto (Keio University, Tokyo, Japan) and were generated as previously described.37 Heterozygotes were bred together, obtaining both knockout and WT littermates. All mice were housed in a temperature-controlled, specific pathogen-free environment with 12-h light/dark cycles and fed regular mouse chow and water ad libitum. All animal handling procedures were approved by the institutional animal care and use committees at the University of Kansas Medical Center.
Isolation of mouse peritoneal macrophages
Primary peritoneal macrophages were isolated as described previously.38 C57BL/6 or FOXO3−/− mice at 5 weeks of age were killed by CO2 asphyxiation. Briefly, 10 ml of sterile phosphate buffered saline (PBS) was injected into the caudal half of the peritoneal cavity using a 25-gauge needle (beveled side up), following by shaking the entire body for 10 s. Saline containing resident peritoneal cells was collected and cells were plated on uncoated tissue culture plates and incubated for 60 min at 37 °C. Nonadherent cells were removed by washing five times with warm PBS. Macrophages were maintained in RPMI medium (Invitrogen) containing 10% FBS overnight and then treated with or without LPS (LPS from Escherichia coli 026:B6, Sigma-Aldrich) for additional 24 h before being processed for analysis.
Immunofluorescence
Cells grown on coverslips were washed in PBS three times and fixed for 10 min in 4% paraformaldehyde in PBS at room temperature. For indirect immunofluorescence, fixed cells were permeabilized in 1% Triton X-100 in PBS for 5 min at room temperature. The coverslips were inverted and touched to 40 μl droplets of blocking buffer (4% goat serum and 1% bovine serum albumin in PBS-Tween, 0.05%) on a clean parafilm sheet for 45 min at room temperature. Cells were then incubated in PBS with rabbit anti-FOXO3 antibody (75D8) or rabbit anti-p-FOXO3 S-574 for 1 h at room temperature. After washing with PBS, coverslips were incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG (1 : 5000; Molecular Probes, Waltham, MA, USA) for 1 h in the dark at room temperature. Coverslips were additionally incubated with DAPI for 20 min at room temperature to stain nuclear DNA. Images were acquired by using Nikon Eclipse Ti microscope (Nikon Americas Inc., Melville, NY, USA).
ChIP assay
Huh7.5 cells (1.5 × 107) were transfected with the indicated constructs by Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). At 24 h after transfection, cells were treated with 50 mM EtOH for an additional 48 h. For endogenous FOXO3 ChIP experiments, Huh7.5 cell (1.5 × 107) were treated with 50 mM EtOH for 48 h. DNA and proteins were crosslinked by the addition of formaldehyde to a final concentration of 1% for 10 min before harvesting. The crosslinking reaction was stopped by adding glycine at 0.125 mol/l final concentration for 5 min at room temperature. Flasks were rinsed twice with ice-cold 1 × PBS and cells were scraped off the plates, resuspended in cell lysis buffer (10 mM Tris-HCl (pH 8.0), 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40 and protease inhibitors) and incubated on ice for 15 min. The cell suspension was vortexed briefly every 5 min to aid in release of nuclei. Nuclei were collected by centrifuge, resuspended in nuclei lysis buffer (1% SDS, 5 mmol/l EDTA, 50 mmol/l Tris-HCl (pH 8.0) and protease inhibitor) and sonicated to generate chromatin to an average length of ~200 to 600 bp (10 × 15 s at 55% maximum potency). After centrifugation at 14 000 r.p.m. for 10 min at 4 °C, samples (2–4 mg of protein extracts) were immunoprecipitated overnight at 4 °C with 1 μg anti-HA ChIP-grade antibody (Abcam, Cambridge, MA, USA) or 3 μl anti-FOXO3 (clone 75D8 from Cell Signaling Technology, Danvers, MA, USA). Rabbit IgG was used as a negative control of immunoprecipitation. One percent of supernatant from the immunoprecipitation was saved as total input of chromatin and was processed with the eluted immunoprecipitates beginning at the crosslink reversal step. Next, 10 μl magnetic beads (Dynabeads M-280 Sheep anti Rabbit IgG, Invitrogen) were added into each sample and incubated at 4 °C for 4 h with rotation. Immunoprecipitates were washed with ChIP Low Salt Buffer (0.1% SDS, 1% Triton-100, 2 mM EDTA, 50 mM Hepes, pH 7.5, 150 mM NaCl), ChIP High Salt Buffer (0.1% SDS, 1% Triton-100, 2 mM EDTA, 50 mM Hepes, pH 7.5, 500 mM NaCl), ChIP LiCl Buffer (0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0) and TE buffer (10 mM Tris-HCl, 1 mM EDTA). Immunocomplexes were eluted with 90 μl elution buffer (1% SDS, 50 mmol/l NaHCO3) and 10 μg of RNase A was added to the pooled eluates; crosslinks were reverted by incubation at 65 °C for at least 6 h. Samples were added with 1 μl of 20 μg/μl proteinase K and incubated for 2 h at 45 °C. After incubation at 95 °C for 10 min, samples were purified used Qiagen PCR purify kit (Hilden, Germany). DNA samples were amplified with target promoter-specific primers using SYBR Green quantitative PCR analysis (Applied Biosystems, Waltham, MA, USA). Primers and probe sequences are listed in Supplementary Table S1.
Real-time PCR
RNA was extracted and purified from cultured hepatoma cells using the TRI reagent from Sigma according to the manufacturer's instructions. cDNA was generated with the random primer method using the RNA reverse transcription kit (Applied Biosystems). Quantitative RT-PCR was performed in a CFX96 real-time system (Bio-Rad, Hercules, CA, USA) using specific sense and antisense primers in 25 μl reaction volumes containing 12.5 μl SYBR Green PCR master mix (Applied Biosystems), 10.5 μl of 1 μmol/l primer stock and 2 μl of cDNA (1 : 10 diluted). The amplification consisted of an initial incubation of 95 °C for 5 min (hot start), following by 50 amplification cycles (15 s at 95 °C, 30 s at 55 °C and 30 s at 72 °C) and a final melting curve from 55 °C to 95 °C. Primer sequences are presented in Supplementary Materials.
Apoptosis array analysis
Huh7.5 cell were transfected with HA-FOXO3 by Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). At 24 h after transfection, cells were treated with 50 mM EtOH for an additional 48 h. Cells were washed twice with PBS, total cellular RNA was isolated and cDNA was generated as discussed above. cDNA samples were then assayed using the Apoptosis RT2 Profiler PCR Array (Qiagen cat. no. PAHS-012Z) according to the manufacturer's instructions. Raw CT values were exported from the real-time instrument software, and analyzed using the ΔΔCT method. Data were present as mRNA fold elevation compared with Huh7.5 cells that were not transfected with FOXO3 or treated with EtOH.
Measurement of luciferase activity, LDH release and caspase 3/7 activity
After co-transfection of the two luciferase plasmids, both firefly and Renilla luciferase activities were measured sequentially from a single aliquot of cell lysate using the Dual-Glo Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's instructions. Relative Renilla luciferase light output was normalized to firefly luciferase output. LDH release and total LDH were determined using TOX7 kit (Sigma-Aldrich) according to the manufacturer's instructions. The absorbance was determined at 490 nm. The present data were calculated by dividing the amount released LDH into the media by the total LDH for each experimental group. Caspase 3/7 activity was determined using Caspase-Glo 3/7 Assay Systems (Promega) according to the manufacturer's instructions. Data were presented as mean±S.E. Statistical significance was calculated using Student's t-test.
TUNEL assay
Cells were fixed with 4% paraformaldehyde at room temperature for 10 min at room temperature. After a PBS rinse, cells were stained using the DeadEND Fluorometric TUNEL System (Promega) according to the manufacturer's instructions. Quantification of TUNEL staining was performed by examining five randomly selected fields by an investigator who was blinded to sample identity.
Western blots
Whole-cell lysates were prepared from cells that had been washed and harvested by centrifugation in PBS pH 7.5. Cell pellets were resuspended in RIPA buffer that contained 50 mM Tris, pH 7.5, 150 mM sodium chloride, 1% NP-40, 0.2% SDS, 0.5% sodium deoxycholate, 0.1 mM EDTA and 1% protease and phosphatase inhibitors (Sigma-Aldrich). Lysates were centrifuged at 14 000 r.p.m. for 15 min; supernatants were collected and protein concentration was measured using the Bio-Rad protein assay kit (Bio-Rad). Cell lysates (25 μg) were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon-P membranes; Millipore, Billerica, MA, USA). Membranes were blocked with blocking buffer (5% skim milk, 0.1% Tween-20 in PBS) for 1 h at room temperature. After incubation with appropriate primary antibodies overnight at 4 °C, membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies, detected using the Electrochemiluminescence Plus Western Blotting Detection System (Amersham Biosciences, Piscataway, NJ, USA) with the ODYSSEY Fc, Dual-Mode Imaging system (Li-COR, Lincoln, NE, USA).
Antibodies
A custom p-FOXO3-S-574 rabbit polyclonal antibody was generated by Epitomics using the peptide SAKHQQQS(PO3H2)PVSQSM for immunization. Phospho-specific antibodies were subsequently purified through antigen affinity columns. Anti-HA antibody (ab9110) was purchased from Abcam. Anti-FOXO1 (C29H4), anti-FOXO4 (9472), anti-FOXO3 (75D8), anti-Bcl2 (50D3), anti-TRAIL (C92B9), anti-Bim (C34C5), anti-p27 (2552) and anti-PARP (9542) were purchased from Cell Signaling Technology. Anti-PrxIII was purchased from Upstate (Lake placid, NY, USA). Anti-SOD2 (FL-222) and anti-TRAIL (H-257) were purchased from Santa Cruz Biotechnology. Anti-actin (AC-15) and Anti-Flag (M2) were purchased from Sigma-Aldrich.
Chemicals
SP600125 (Cell Signaling Technology) was solubilized in dimethyl sulfoxide (DMSO) and filter sterilized before use. LPS from E. coli 026:B6 (Sigma-Aldrich), TRAIL-R1 (human):Fc (human) and TRAIL-R2 (human):Fc (human) (Enzo Life Science, Farmingdale, NY, USA) were solubilized in PBS and filter sterilized before use.
Acknowledgments
This study was supported by grant AA012863 from the National Institute on Alcoholism and Alcohol Abuse, grants P20 GM103418 and P20 GM103549 from the National Institute of General Medical Sciences from the National Institutes of Health and a fellowship grant from the Biomedical Research Training Program of the University of Kansas Medical Center to ZL. The human hepatocytes used in this study were derived from samples provided by the University of Kansas Liver Center Tissue Bank. We thank Drs. Jameson Forster, Tim Schmitt and Bashar Abdulkarim for their assistance in obtaining these specimens and Dr. Michele Pritchard for helpful discussions. Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd).
Glossary
- FOXO3
forkhead box O3
- PTM
post-transcriptional modification
- LPS
lipopolysaccharide
- ChIP
chromatin immunoprecipitation
- TRAIL
TNF-related apoptosis-inducing ligand
- Bcl-2
B-cell lymphoma 2
- PrxIII
peroxiredoxin III
- SOD2
superoxide dismutase 2
- Bim
B-cell lymphoma 2 interacting mediator of cell death
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- LDH
lactate dehydrogenase
- JNK
c-Jun N-terminal kinase
- PUMA
p53 upregulated modulator of apoptosis
- Noxa
phorbol-12-myristate-13-acetate-induced protein 1
- ADH
alcohol dehydrogenase
- HCV
hepatitis C virus
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by N Chandel
Supplementary Material
References
- Sedding DG. FoxO transcription factors in oxidative stress response and ageing—a new fork on the way to longevity? Biol Chem 2008; 389: 279–283. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol 2007; 8: 440–450. [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Pals C, Banerji L, Thomas NS, Lam EW et al. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol Cell Biol 2000; 20: 9138–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ Jr, DiStefano PS et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 2002; 296: 530–534. [DOI] [PubMed] [Google Scholar]
- Chung YW, Kim HK, Kim IY, Yim MB, Chock PB. Dual function of protein kinase C (PKC) in 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced manganese superoxide dismutase (MnSOD) expression: activation of CREB and FOXO3a by PKC-alpha phosphorylation and by PKC-mediated inactivation of Akt, respectively. J Biol Chem 2011; 286: 29681–29690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuchner J, Kuznetsov A, Hermann M, Hausott B, Obexer P, Ausserlechner MJ. FOXO3-induced reactive oxygen species are regulated by BCL2L11 (Bim) and SESN3. J Cell Sci 2012; 125: 1191–1203. [DOI] [PubMed] [Google Scholar]
- Calnan DR, Brunet A. The FoxO code. Oncogene 2008; 27: 2276–2288. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999; 96: 857–868. [DOI] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol 2008; 10: 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JL, Cheng X, Yamaguchi H, Chang YW, Hou CF, Lee DF et al. FOXO3a-dependent mechanism of E1A-induced chemosensitization. Cancer Res 2011; 71: 6878–6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Lee YS, Lee S, Kim S, Kim TY. Pachastrissamine from Pachastrissa sp. inhibits melanoma cell growth by dual inhibition of Cdk2 and ERK-mediated FOXO3 downregulation. Phytother Res 2012; 26: 1927–1933. [DOI] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 2006; 8: 1064–1073. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villén J, Becker EB et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 2006; 125: 987–1001. [DOI] [PubMed] [Google Scholar]
- Davila D, Connolly NM, Bonner H, Weisová P, Dussmann H, Concannon CG et al. Two-step activation of FOXO3 by AMPK generates a coherent feed-forward loop determining excitotoxic cell fate. Cell Death Differ 2012; 19: 1677–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J 2004; 23: 4802–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell 2008; 32: 221–231. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004; 303: 2011–2015. [DOI] [PubMed] [Google Scholar]
- Tikhanovich I, Cox J, Weinman SA. Forkhead box class O transcription factors in liver function and disease. J Gastroenterol Hepatol 2013; 28: 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng AH, Wu LH, Shieh SS, Wang DL. SIRT3 interactions with FOXO3 acetylation, phosphorylation and ubiquitinylation mediate endothelial cell responses to hypoxia. Biochem J 2014; 464: 157–168. [DOI] [PubMed] [Google Scholar]
- Wang F, Chan CH, Chen K, Guan X, Lin HK, Tong Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene 2012; 31: 1546–1557. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom A, Mokry M, de Wit E, Smits LM, Polderman PE, van Triest MH et al. Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Mol Syst Biol 2013; 9: 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry M, Mokry M, de Wit E, Smits LM, Polderman PE, van Triest MH et al. Survival factor NFIL3 restricts FOXO-induced gene expression in cancer. Genes Dev 2013; 27: 916–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhanovich I, Kuravi S, Campbell RV, Kharbanda KK, Artigues A, Villar MT et al. Regulation of FOXO3 by phosphorylation and methylation in hepatitis C virus infection and alcohol exposure. Hepatology 2014; 59: 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc HN., Ashkenazi A. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ 2003; 10: 66–75. [DOI] [PubMed] [Google Scholar]
- Feldmann G. Liver apoptosis. J Hepatol 1997; 26(Suppl 2): S1–S11. [DOI] [PubMed] [Google Scholar]
- Skopelitou A, Hadjiyannakis M, Alexopoulou V, Krikoni O, Kamina S, Agnantis N et al. Topographical immunohistochemical expression of bcl-2 protein in human liver lesions. Anticancer Res 1996; 16: 975–978. [PubMed] [Google Scholar]
- Thurman RG II. Alcoholic liver injury involves activation of Kupffer cells by endotoxin. Am J Physiol 1998; 275: G605–G611. [DOI] [PubMed] [Google Scholar]
- Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta 2011; 1813: 1978–1986. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Medema RH. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty. J Leukoc Biol 2003; 73: 689–701. [DOI] [PubMed] [Google Scholar]
- You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med 2006; 203: 1657–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obexer P, Geiger K, Ambros PF, Meister B, Ausserlechner MJ. FKHRL1-mediated expression of Noxa and Bim induces apoptosis via the mitochondria in neuroblastoma cells. Cell Death Differ 2007; 14: 534–547. [DOI] [PubMed] [Google Scholar]
- Tumurbaatar B, Tikhanovich I, Li Z, Ren J, Ralston R, Kuravi S et al. Hepatitis C and alcohol exacerbate liver injury by suppression of FOXO3. Am J Pathol 2013; 183: 1803–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 2005; 24: 7410–7425. [DOI] [PubMed] [Google Scholar]
- Schips TG, Wietelmann A, Höhn K, Schimanski S, Walther P, Braun T et al. FoxO3 induces reversible cardiac atrophy and autophagy in a transgenic mouse model. Cardiovasc Res 2011; 91: 587–597. [DOI] [PubMed] [Google Scholar]
- Wilson MS, Brosens JJ, Schwenen HD, Lam EW. FOXO and FOXM1 in cancer: the FOXO-FOXM1 axis shapes the outcome of cancer chemotherapy. Curr Drug Targets 2011; 12: 1256–1266. [DOI] [PubMed] [Google Scholar]
- Xie Y, Kuravi S, Campbell RV, Kharbanda KK, Artigues A, Villar MT et al. Mechanisms of acetaminophen-induced cell death in primary human hepatocytes. Toxicol Appl Pharmacol 2014; 279: 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 2007; 1: 101–112. [DOI] [PubMed] [Google Scholar]
- Davies JQ, Gordon S. Isolation and culture of murine macrophages. Methods Mol Biol 2005; 290: 91–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.