Abstract
Interleukin-6 (IL-6) has been demonstrated to be involved in Hepatitis B virus (HBV)-associated hepatocarcinogenesis through activation of the STAT3 pathway. The sustained activation of the IL-6/STAT3 pathway is frequently associated with repression of SOCS3, which is both a target gene and a negative regulator of STAT3. However, the silencing mechanism of SOCS3 in hepatocellular carcinoma (HCC) remains to be elucidated. Here, we showed that the repression of SOCS3 and sustained activation of IL-6/STAT3 pathway in HBV-producing HCC cells were caused by HBV-induced mitochondrial ROS accumulation. Mechanistic studies revealed that ROS-mediated DNA methylation resulted in the silencing of SOCS3. Decreased SOCS3 expression significantly promoted the proliferation of HCC cells and growth of tumor xenografts in mice. Further studies revealed that HBV-induced ROS accumulation upregulated the expression of the transcription factor, Snail, which bound to the E-boxes of SOCS3 promoter and mediated the epigenetic silencing of SOCS3 in association with DNMT1 and HDAC1. In addition, we found that the expression of Snail and SOCS3 were inversely correlated in HBV-associated HCC patients, suggesting that SOCS3 and/or Snail could be used as prognostic markers in HCC pathogenesis. Taken together, our data show that HBV-induced mitochondrial ROS production represses SOCS3 expression through Snail-mediated epigenetic silencing, leading to the sustained activation of IL-6/STAT3 pathway and ultimately contributing to hepatocarcinogenesis.
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the second leading cause of cancer death in the world.1 Chronic HBV infection and chronic liver disease that progress from hepatitis to cirrhosis are major risk factors for the development of HCC. Options for the treatment of HCC are quite limited because the molecular, cellular and environmental mechanisms that drive HBV-associated HCC pathogenesis are poorly understood.
Pro-inflammatory cytokines contribute significantly to the pathogenesis of many tumor types.2 Among them, elevated IL-6 is closely related to HCC development and progression.3 Elevated IL-6 was found in patients with viral and alcoholic hepatitis and liver cirrhosis. In these conditions, IL-6 is expressed mainly by myeloid cells/leukocytes.4 Recent studies have confirmed that upregulation of IL-6 in human HCC, where it is expressed by the cancer cells, also has a central role in a gene expression network via an autocrine signaling pathway that drives tumor development and progression.5 However, it remains unknown whether IL-6 autocrine signaling can be stimulated by HBV during hepatocarcinogensis.
Reactive oxygen species (ROS) are continuously generated by cells to regulate cellular responses such as proliferation, differentiation and apoptosis.6 Chronic virus infection may enhance ROS production and cause oxidative stress in host cells.7 Continued oxidative stress can lead to IL-6 overexpression through activation of a variety of redox-sensitive transcription factors, such as Nrf2.8 Interestingly, the sources of ROS are a factor in the specific functions of ROS.9 For example, ROS generated by mitochondrial respiration are important for pro-inflammatory cytokine production, such as IL-6, whereas ROS generated by other sources, such as NADPH oxidases, may not be able to stimulate the overproduction of pro-inflammatory cytokines.10
In the IL-6-mediated network, IL-6/STAT3 signaling has a major impact upon cell growth, survival, differentiation and pathogen resistance.11 Deregulated signaling of IL-6 can cause constitutive activation of STAT3, which promotes the development and progression of HCC.11 Suppressor of cytokine signaling 3 (SOCS3) is a target gene for STAT3 and also a member of the STAT-induced STAT inhibitor family, which functions as a cytokine-inducible negative regulator of cytokine signaling.12 The expression of SOCS3 is induced by various cytokines, including IL-6. SOCS3 can bind to and inhibit the activity of JAK2 kinase, thus blocking the activation of STAT3 and controlling the IL-6/STAT3 signaling pathway by a classic negative feedback loop.12 Recently, reduced expression of SOCS3 has been reported in multiple cancer types, including HCC.13, 14, 15 This is recognized as the major cause of deregulated IL-6/STAT3 signaling. However, the molecular mechanism of SOCS3 silencing and whether it is implicated in HBV-associated HCC development and progression requires further clarification.
In this study, we revealed that HBV-induced mitochondrial ROS overproduction upregulated the expression of IL-6, leading to the activation of the STAT3 pathway and HCC development. Importantly, our data showed that ROS-mediated Snail upregulation repressed SOCS3 expression via hypermethylation of the SOCS3 promoter, contributing to the sustained activation of IL-6/STAT3.
Results
HBV-induced ROS accumulation stimulates the production of IL-6
To determine whether IL-6 was involved in HBV-induced HCC, we examined the expression of IL-6 in HepG2.2.15 cells (which stably express HBV) and HepAD38 cells (which express HBV under tetracycline-off control16). The results showed that both the protein and mRNA expression levels of IL-6 were increased in HBV-producing cells (HepG2.2.15 and non-tetracycline-treated HepAD38) compared with the control cells (HepG2 and tetracycline-treated HepAD38) (Supplementary Figures 1a and b). Similarly, elevated protein and mRNA expressions of IL-6 were observed in Hep3B cells with transient transfection of a plasmid containing a 1.3-fold-overlength HBV genome (HBV1.3) (Supplementary Figures 2a and b). When protein and mRNA expression levels of IL-6 were evaluated in HCC tissues, their levels were higher in liver sections of HBV-positive compared with HBV-negative HCC patients (Supplementary Figures 3a–c). Similar results were observed in peritumor liver tissues of HBV-positive compared with HBV-negative HCC patients (Supplementary Figures 4a–c).
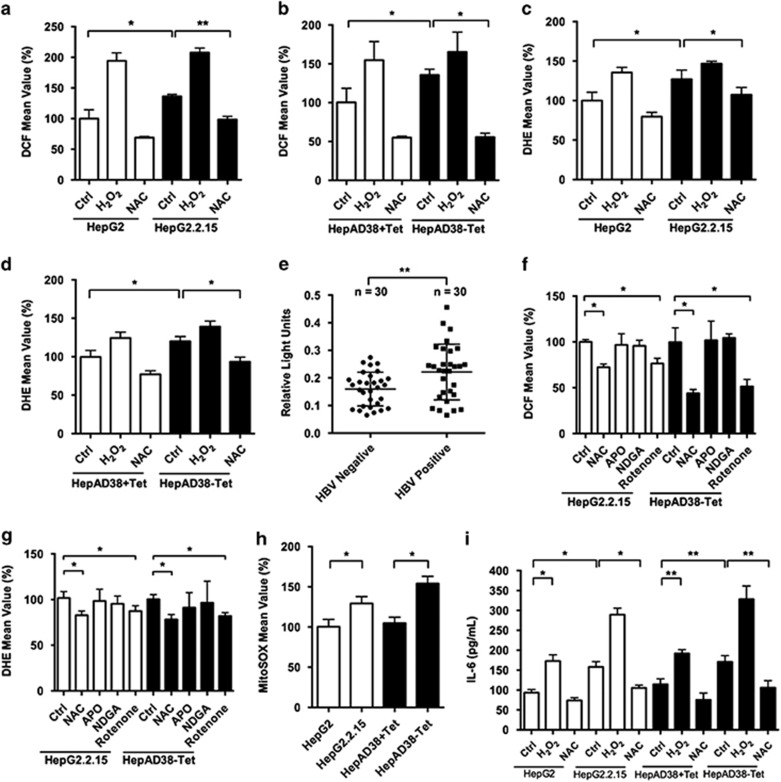
ROS has been demonstrated to induce IL-6 overproduction.9 To determine whether HBV stimulated IL-6 overexpression through induction of ROS levels, the latter was compared between HBV-producing cells and the control cells. The results showed that both H2O2 (as indicated by DCF assay) (Figures 1a and b) and O2− (as indicated by dihydroethidium assay) (Figures 1c and d) levels were increased in HBV-producing cells compared with control cells. To investigate whether HBV could induce ROS levels in vivo, liver lipid peroxidation levels were assessed in HBV-positive and -negative HCC patients. There were increased lipid peroxidation levels in the liver samples of HBV-positive HCC patients compared with HBV-negative HCC patients, suggesting that ROS levels were enhanced in the HBV-infected liver (Figure 1e).
Figure 1.
HBV induces IL-6 expression via stimulation of ROS overproduction. (a–d) ROS levels of HBV-producing HCC cells and control cells were detected by DCF (H2O2) and dihydroethidium (DHE) (O2-) assay. Cells treated with H2O2 (300 μM) or NAC (5 mM) for 2 days served as positive and negative controls, respectively. (e) Lipid peroxidation levels were assessed by thiobarbituric acid-reactive substance (TBARS) assay in liver tissues of HCC patients. HBV-positive patients n=30, HBV-negative patients n=30. (f and g) HBV-producing HCC cells were incubated with the indicated antioxidants for 1 h. ROS levels were detected by DCF and DHE assay. (h) HBV-producing HCC cells and control cells were stained with MitoSOX Red, and ROS levels were determined. (i) HBV-producing HCC cells and control cells were treated as indicated. IL-6 levels in media supernatants of the cells were assessed by ELISA. Average values and S.D.s were calculated from triplicate samples
To investigate the source of HBV-induced ROS levels, HBV-producing cells were treated with selective inhibitors of intracellular sources of ROS. In addition to the general scavenger NAC, apocynin was used to inhibit NAPDH oxidase, nordihydroguaiaretic acid to block 5-lipoxygenase and rotenone to block mitochondrial ROS production. Only NAC and rotenone treatment suppressed intracellular ROS production (Figures 1f and g), suggesting that the source of HBV-associated elevations in ROS is mitochondria. Likewise, experiments using MitoSOX Red dye, a probe of mitochondrial superoxide production in live cells, showed that mitochondrial ROS levels were higher in HBV-producing cells compared with control cells (Figure 1h). To determine whether HBV-induced ROS levels affect IL-6 production in liver cells, IL-6 expression levels were determined in HCC cells following H2O2 and NAC treatment. H2O2 (300 μM) increased the IL-6 expression level of non-HBV control cells to a similar level as that of HBV-producing cells, while NAC treatment suppressed IL-6 production in HBV-producing cells, suggesting that HBV-induced IL-6 overproduction was mediated, at least in part, by H2O2 accumulation (Figure 1i). Taken together, these data indicated that HBV-associated ROS production could enhance IL-6 expression in liver cells both in vitro and in vivo.
Transcriptional factor Nrf2 has been recognized as a sensitive oxidative stress sensor.17 A recent study demonstrated that IL-6 was a target of Nrf2.8 To verify whether HBV-induced ROS levels increased IL-6 production through the activation of Nrf2, we measured the protein expression level of Nrf2 and mRNA levels of IL-6, as well as two other Nrf2 target genes, Gclc and Nqo1, in HepG2 and HepG2.2.15 cells (Supplementary Figures 5a and b). Nrf2 was markedly upregulated by elevated ROS levels and similar results were also observed in mRNA levels of IL-6, Gclc and Nqo1. To confirm that Nrf2 regulated ROS-mediated alterations of IL-6 expression, we blocked the Nrf2 expression in HepG2 and HepG2.2.15 cells with two specific siRNAs. Both protein and mRNA levels of IL-6 were downregulated by underexpression of Nrf2 (Supplementary Figures 5c and d), indicating that Nrf2 had a key role in ROS-mediated IL-6 overproduction.
HBV-induced ROS accumulation activates IL-6/STAT3 pathway through epigenetic silencing of SOCS3
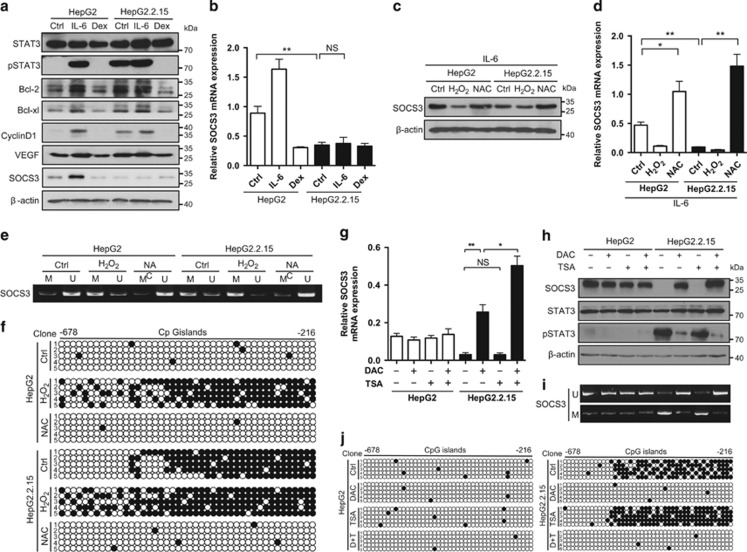
STAT3 signaling is one of the most important pathways in HCC development and progression.11 To determine whether HBV-induced IL-6 overexpression was associated with HCC progression, the activity of STAT3 was determined. As shown in Figure 2a and Supplementary Figure 6, STAT3 was activated by HBV. Phosphorylation of STAT3 and elevated expression of Cyclin D1 were also observed in HBV-positive HCC samples (Supplementary Figures 7a and b). STAT3 target genes, including Bcl-2, Bcl-xl, Cyclin D1 and VEGF, were also elevated at both the mRNA and protein expression levels, while HBV-induced STAT3 activation was blocked by dexamethasone (an inhibitor of IL-6 expression) (Figure 2a and Supplementary Figure 8). However, the protein and mRNA expression levels of SOCS3, which is a target gene and negative regulator of STAT3, were unaltered upon IL-6 treatment in HBV-producing cells, but upregulated in non-HBV control cells (Figures 2a and b and Supplementary Figures 9a and b), suggesting that HBV inhibited the expression of SOCS3. To examine the involvement of HBV-induced ROS accumulation in SOCS3 inhibition, SOCS3 expression was determined in HCC cells treated with H2O2 or NAC. Interestingly, IL-6-induced upregulation of SOCS3 mRNA and protein levels was decreased upon H2O2 treatment in non-HBV control cells, while underexpression of SOCS3 could be overcome by NAC in HBV-producing cells (Figures 2c and d and Supplementary Figures 9c and d). This indicated that ROS repressed the upregulation of SOCS3 mediated by IL-6.
Figure 2.
HBV-induced ROS overproduction leads to sustained STAT3 activation through epigenetic silencing of SOCS3. (a and b) Activation of STAT3 and the protein levels of STAT3 target genes (Bcl-2, Bcl-xl, Cyclin D1, VEGF and SOCS3) were examined in HepG2 and HepG2.2.15 cells by immunoblot (a). Cells treated with IL-6 (10 ng/ml) or dexamethasone (Dex) (10−7 M) for 2 days served as positive and negative controls, respectively. β-Actin serves as a loading control. mRNA level of SOCS3 was determined by RT-PCR (b). (c and d) HepG2 and HepG2.2.15 cells pretreated with IL-6 (10 ng/ml) for 12 h were treated with H2O2 (300 μM) or NAC (5 mM) for 2 days. SOCS3 expression was examined by immunoblot (c). mRNA level of SOCS3 was determined by RT-PCR (d). (e) HepG2 and HepG2.2.15 cells were treated as indicated. Genomic DNA extracted from the cells was treated with bisulfite and then subjected to methylation-specific PCR (MSP) using the methylated DNA (M)- and unmethylated DNA (U)-specific primer sets. (f) PCR products were sequenced for the 44 CpG sites located between nucleotides -678 and -216 of the socs-3 promoter. The horizontal dots represent CpG islands while the vertical dots represent the individual five clones sequenced. Each black dot represents a methylated cytosine residue within the CpG islands. (g–j) HepG2 and HepG2.2.15 cells were treated with either DAC (5 μM) for 48 h or trichostatin A (TSA) (0.5 μM) for 24 h or both (DAC for 24 h, followed by TSA for additional 24 h). Activation of STAT3 and the protein levels of SOCS3 were examined by immunoblot (g). NS, non-significant. mRNA level of SOCS3 was determined by RT-PCR (h). Methylation status of SOCS3 promoter was determined by MSP (i) and BSP (j). Average values and S.D.s were calculated from triplicate samples
SOCS3 is frequently silenced in cancer cells through epigenetic silencing.13 To explore the mechanism of SOCS3 repression, the methylation status of the SOCS3 promoter was determined by methylation-specific PCR. As shown in Figure 2e and Supplementary Figure 9e, increased levels of methylated PCR product were identified in HBV-producing HCC cells, as well as control cells treated with H2O2. In addition, hypermethylation of SOCS3 promoter could be reversed by NAC, suggesting that ROS mediated the methylation process of SOCS3 promoter. Furthermore, a total of 44 CpG sites located between nucleotides -678 and -216 in the SOCS3 promoter were examined by bisulfite sequencing. Dense cytosine methylation was observed in the CpG islands of the SOCS3 promoter of HBV-producing HCC cells (Figure 2f and Supplementary Figure 9f). These data suggested that the SOCS3 promoter was hypermethylated by HBV-induced ROS accumulation in HCC cells. To further determine whether SOCS3 promoter hypermethylation was responsible for SOCS3 gene silencing, the effect of 5-aza-2'-deoxycytidine (DAC), a demethylating agent, was examined. Treatment with DAC significantly increased the mRNA levels of SOCS3 (Figure 2g) and restored the expression of SOCS3 protein (Figure 2h) in HBV-producing HCC cells. On the other hand, addition of a histone deacetylase (HDAC) inhibitor, trichostatin A, had no effect on the expression of SOCS3 (Figures 2g and h). However, trichostatin A was found to augment the effect of DAC, indicating that SOCS3 might be regulated by HDACs in a demethylation-dependent manner. Consistently, similar results were observed in methylation status of SOCS3 promoter upon DAC and trichostatin A treatment (Figures 2I and j), suggesting that the impact of these treatments on SOCS3 expression were mediated by methylation of SOCS3 promoter. Taken together, these observations indicated that SOCS3 repression was due to ROS-mediated SOCS3 promoter methylation in HBV-producing HCC cells.
SOCS3 inhibits the growth of HCC cells both in vitro and in vivo
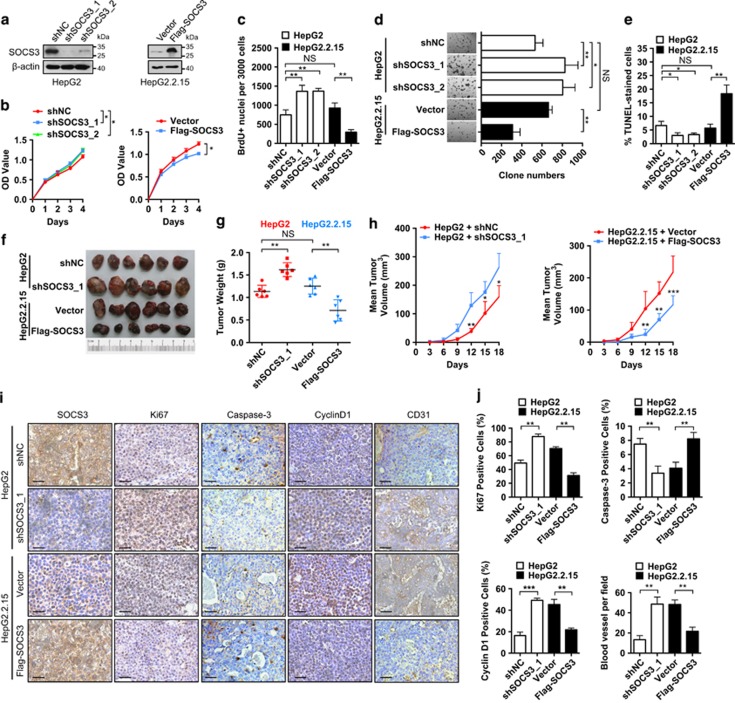
To elucidate the role of SOCS3 in HBV-associated HCC development and progression, SOCS3 was overexpressed in HBV-producing HCC cells that had relatively low endogenous SOCS3 levels. Alternatively, relatively high levels of SOCS3 in control cells were suppressed by the introduction of two specific shRNAs (Figure 3a and Supplementary Figure 10a). As shown in Figure 3b and Supplementary Figure 10b, inhibition of SOCS3 expression enhanced cell proliferation in control cells, whereas SOCS3 overexpression reduced proliferation in HBV-producing HCC cells. The effect of SOCS3 on HCC cell proliferation was further confirmed by bromodeoxyuridine incorporation and colony formation assay (Figures 3c and d and Supplementary Figures 10c and d). Moreover, knockdown of SOCS3 repressed apoptotic cell death in control cells, whereas overexpression of SOCS3 increased cell death rate in HBV-producing HCC cells (Figure 3e and Supplementary Figure 10e). Collectively, these results indicated that SOCS3 inhibited the proliferation and cell survival of HCC cells.
Figure 3.
SOCS3 is a negative regulator of hepatocarcinogenesis. (a and b) HepG2 cells were stably transfected with SOCS3 shRNAs (shSOCS3_1, shSOCS3_2) or negative control vector (shNC). HepG2.2.15 cells were stably transfected with Flag-SOCS3 or Flag-tag vector (Vector). SOCS3 expression was determined by immunoblot (a). β-Actin serves as a loading control. Proliferation of transfected HepG2 and HepG2.2.15 cells were examined over a 4- day period by Cell Counting Kit-8 (b). (c) Cells with bromodeoxyuridine-positive nuclei labeling were counted. (d) Colony formation assays were performed for 14 days. (e) Quantification of cell death in transfected cells by TUNEL assay. (f–h) Transfected HepG2 and HepG2.2.15 cells were subcutaneously injected into nude mice and xenograft tumor growth was determined for 18 days. Tumor mass images (f). Tumor weight (g). Growth curves showing tumor volumes (h). (i) Immunohistochemical staining of SOCS3, Ki67, Caspase-3, Cyclin D1 and CD31 in tumor samples. Scale bar, 50 μm. (j) Ki67-, Caspase-3- and Cyclin D1-positive cells and blood vessel numbers were calculated. Average values and S.D.s were calculated from triplicate samples
To determine whether SOCS3 had the same influence on tumor growth in vivo, HCC cells were injected subcutaneously into nude mice. SOCS3 overexpression repressed the tumor growth over the course of the experiment, as indicated by tumor weight and volume (Figures 3f–h). In addition, immunohistochemistry (IHC) showed less Ki67 staining, more caspase-3 staining and less Cyclin D1 staining in SOCS3-overexpressing HCC cells, indicative of reduced proliferation (Figures 3I and j). Moreover, there were fewer blood vessels in SOCS3-overexpressing tumors, suggesting that SOCS3 could repress the cell growth in vivo through inhibition of angiogenesis (Figures 3i and j). Taken together, these data indicated that SOCS3 inhibits HCC cell growth both in vitro and in vivo.
ROS-induced Snail upregulation is responsible for epigenetic silencing of SOCS3 promoter
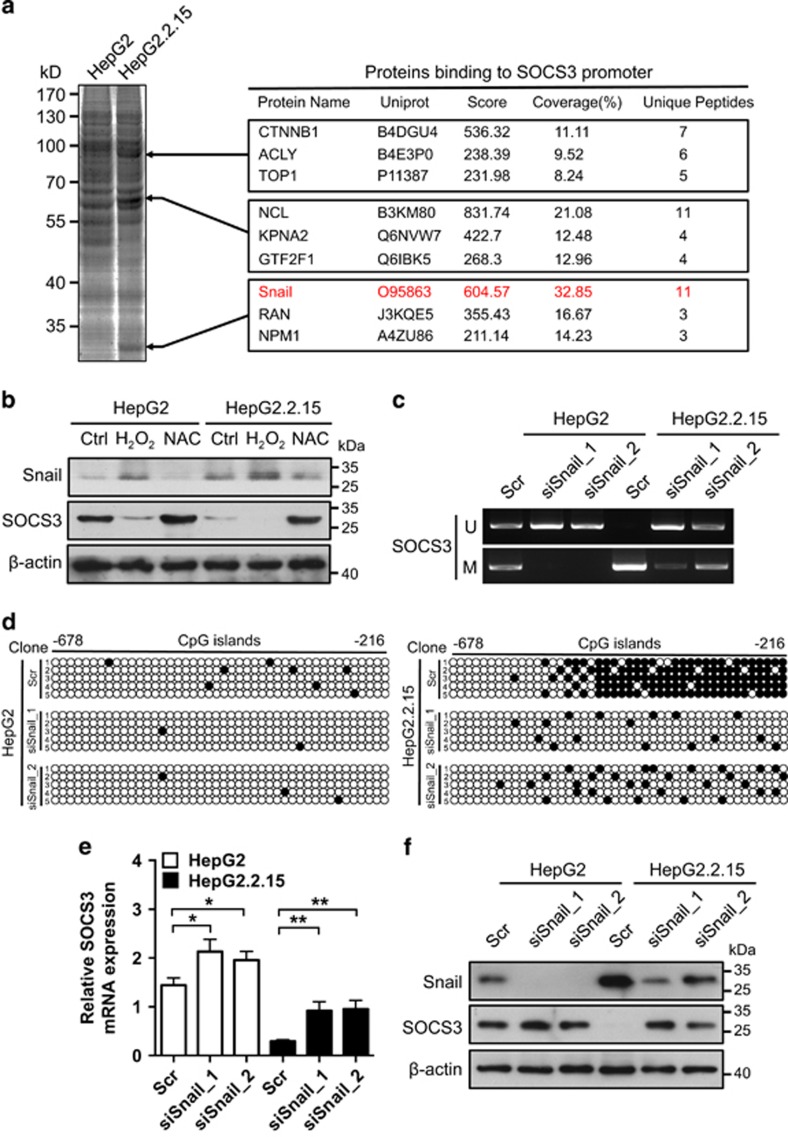
To elucidate the mechanism of SOCS3 promoter hypermethylation, biotin-labeled SOCS3 promoter was used as a probe for proteomic analysis of nuclear proteins that could bind to it in HepG2 and HepG2.2.15 cells. This yielded a series of proteins that showed different binding affinities to the SOCS3 promoter in HepG2 compared with HepG2.2.15 cells (Figure 4a). After bioinformatics analyses of the MS data, Snail was identified as a potential repressor of SOCS3 because Snail mediates the promoter hypermethylation process.18 Moreover, the expression of Snail could be increased by ROS in HCC cells,18 suggesting that HBV-induced ROS levels might lead to SOCS3 promoter hypermethylation through the upregulation of Snail. To verify the hypothesis, Snail expression was analyzed in HBV-producing HCC cells, as well as control cells upon H2O2 or NAC treatment. The results showed that Snail expression was upregulated by ROS and negatively correlated with SOCS3 (Figure 4b and Supplementary Figure 11a). To confirm whether Snail could regulate the expression of SOCS3, Snail was knocked down by two specific siRNAs. This resulted in the demethylation of the SOCS3 promoter (Figures 4c and d and Supplementary Figures 11b and c) and restored the mRNA and protein expression levels of SOCS3 (Figures 4e and f and Supplementary Figures 11d and e), suggesting that Snail had a key role in hypermethyaltion of SOCS3 promoter mediated by HBV-induced ROS accumulation.
Figure 4.
Snail is a repressor of SOCS3 gene expression. (a) Proteins binding preferentially to SOCS3 promoter in HepG2.2.15 cell nuclear lysates. (b) HepG2 and HepG2.2.15 cells were treated as indicated. Protein levels of Snail and SOCS3 were examined by immunoblot. β-Actin serves as a loading control. (c–f) HepG2 and HepG2.2.15 cells were transfected with scrambled control siRNA (Scr) or Snail siRNAs (siSnail_1, siSnail_2). Methylation status of SOCS3 promoter was determined by MSP (c) and BSP (d). mRNA level of SOCS3 was determined by RT-PCR (e). Protein levels of Snail and SOCS3 were determined by immunoblot. β-Actin serves as a loading control (f). Average values and S.D.s were calculated from triplicate samples
To determine whether HBV-induced ROS accumulation promoted hepatocarcinogenesis through Snail-mediated epigenetic silencing of SOCS3, Snail was overexpressed in control cells that had relatively low endogenous Snail levels. Alternatively, relatively high levels of Snail in HBV-producing HCC cells were suppressed by the introduction of specific shRNAs. As shown in Supplementary Figure 12a, Snail overexpression repressed SOCS3 levels in control cells, whereas inhibition of Snail expression increased SOCS3 levels in HBV-producing HCC cells. As indicated by proliferation tests and colony formation assay, Snail-mediated silencing of SOCS3 enhanced cell proliferation in control cells, whereas inhibition of Snail expression negatively regulated cell proliferation in HBV-producing HCC cells (Supplementary Figures 12b–d). In addition, overexpression of Snail repressed apoptotic cell death in control cells, whereas knockdown of Snail increased cell death rate in HBV-producing HCC cells (Supplementary Figure 12e). Collectively, these results indicated that Snail inhibited the proliferation and cell survival of HCC cells through the modulation of SOCS3 expression.
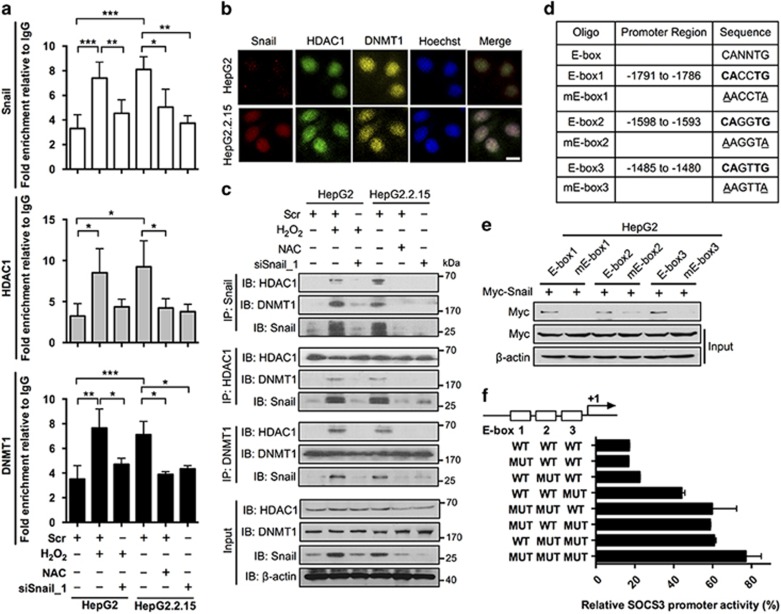
Snail represses SOCS3 expression through binding to the E-boxes of SOCS3 in association with HDAC1 and DNMT1
To investigate the mechanism of Snail-mediated SOCS3 promoter hypermethylation, we analyzed the sequence of SOCS3 promoter and identified three E-boxes, which have been demonstrated to serve as the DNA-binding sites of Snail.19 To determine whether Snail could bind to SOCS3 promoter, the interaction between Snail and SOCS3 promoter was determined by chromatin immunoprecipitation (IP) assay. The results showed that the SOCS3 promoter could bind with Snail and that this binding depended on the Snail expression level (Figure 5a). Snail has been demonstrated to recruit HDAC1 and DNMT1 during the silencing process of its target.18 To determine whether this epigenetically modified complex, Snail/HDAC1/DNMT1, participated in the silencing process of SOCS3, experiments were designed to determine complex localization with immunofluorescence. The results showed nuclear co-localization of Snail, HDAC1 and DNMT1 in HepG2.2.15 cells, suggesting that they had the potential to form a complex (Figure 5b). In addition, IP data showed that Snail, HDAC1 and DNMT1 could bind with each other in HCC cells, reconfirming their interaction (Figure 5c). Moreover, the interaction between HDAC1, DNMT1 and SOCS3 promoter was verified by chromatin IP assay, which revealed that their interactions with SOCS3 promoter also depended on the Snail expression level, suggesting that Snail was crucial for the recruitment of Snail/HDAC1/DNMT1 complex to the promoter of SOCS3 (Figure 5a). To verify whether the E-boxes of SOCS3 were the binding sites of Snail, biotinylated double-stranded oligonucleotides corresponding to the wild type and mutant E-boxes of SOCS3 promoter were used to precipitate Snail from Snail-overexpressing HepG2 cells (Figure 5d). The results showed that mutation in either E-box significantly reduced the binding between Snail and SOCS3 promoter, indicating that all three E-boxes of the SOCS3 promoter were crucial for Snail binding (Figure 5e). Studies were then performed to compare the ability of Snail to repress different reporter constructs carrying combinations of mutated E-boxes (Figure 5f). The results showed that mutation of either E-box impaired the SOCS3 promoter activity and that the furthest downstream E-box (E-box 3) had the strongest repressive activity, suggesting that the three E-boxes cooperated in the Snail-mediated SOCS3 repression.
Figure 5.
Snail binds to the E-boxes of SOCS3 promoter in association with HDAC1 and DNMT1. (a) HepG2 and HepG2.2.15 cells were transfected with scrambled control siRNA (Scr) or Snail siRNA (siSnail_1) and then treated with H2O2 (300 μM) or NAC (5 mM) for 2 days. chromatin IP assay of Snail, HDAC1 and DNMT1 on the promoter of SOCS3 was performed. Quantification of binding was represented as fold enrichment over IgG control. (b) HepG2 and HepG2.2.15 cells were stained with anti-Snail (red), anti-HDAC1 (green), anti-DNMT1 (yellow) and Hoechst (blue). Cellular localization of Snail, HDAC1 and DNMT1 was examined. Scale bar, 10 μm. (c) HepG2 and HepG2.2.15 cells were treated as indicated. CoIP assay was performed using anti-Snail, anti-HDAC1 and anti-DNMT1 antibodies, respectively. β-Actin serves as a loading control. (d) Depiction of WT and mutant E-box sequences of three E-boxes located within -2029 to -763 of SOCS3 promoter. (e) HepG2 cells were transfected with Myc-Snail plasmids. Cells were lysed 48 h post transfection. Lysates were incubated with biotinylated double-stranded oligonucleotides corresponding to the E-boxes of SOCS3 promoter (wild-type sequence or mutants) and with streptavidin-agarose beads. The precipitated complexes were subjected to Myc immunoblot analysis. (f) pGL3-SOCS3(-2029/-763) (WT) or the same plasmid containing different E-box mutations (MUT) was co-transfected into HepG2 cells together with pRL-TK. Data were expressed as Luc activity in the presence of exogenous Snail as a percentage of the activity of the same reporter in transfections supplemented with control vector. Average values and S.D.s were calculated from triplicate samples
SOCS3 and Snail expression are negatively correlated and could serve as prognostic factors in HBV-associated HCC
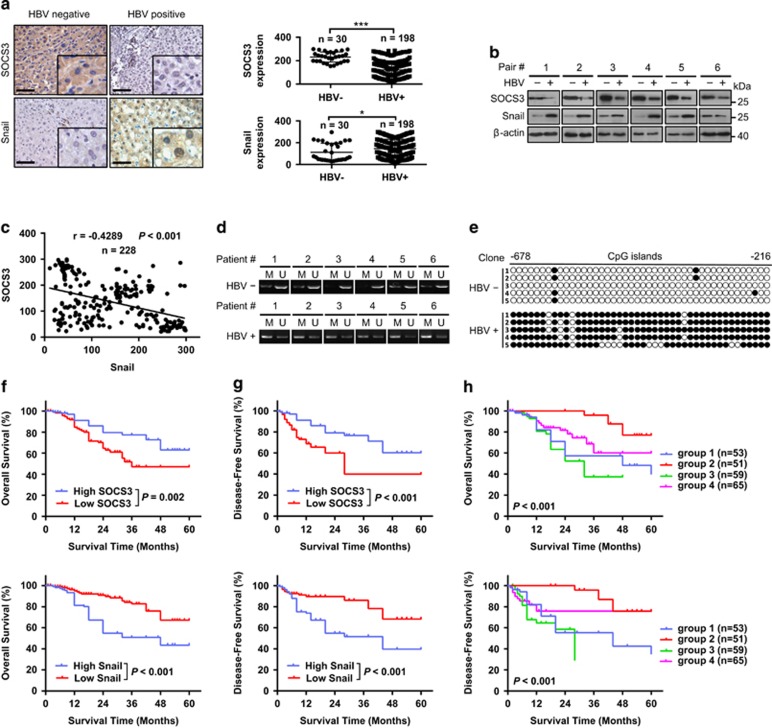
To elucidate the clinical relevance of SOCS3 and Snail, their expression levels were analyzed in clinical liver cancer samples. SOCS3 was shown to be downregulated in HBV-positive HCC tissue samples compared with HBV-negative ones by IHC and immunoblot analysis (Figures 6a and b). In contrast, Snail was overexpressed in HBV-positive HCC tissue samples. Statistical analysis of the IHC quantification data further demonstrated a significant inverse correlation between these markers in HCC samples (Figure 6c). This inverse correlation was also observed in peritumor liver tissues in HBV-positive and HBV negative patients with HCC (Supplementary Figures 13a–d). Methylation-specific PCR (Figure 6d) and bisulfite sequencing (Figure 6e) analysis revealed that the SOCS3 promoter was hyper-methylated in HBV-positive HCC tissue samples, which is consistent with the high expression levels of Snail.
Figure 6.
SOCS3 and Snail can be used as prognostic factors of HBV-associated HCC. (a) Representative images and quantification of SOCS3 (top) and Snail (bottom) expression in HBV-positive and HBV-negative HCC tissue obtained by immunohistochemical analysis. Scale bar, 100 μm. HBV-negative, HBV-; HBV-positive, HBV+. (b) Representative images of SOCS3 and Snail expression obtained by immunoblot analysis in HCC patient liver cancer tissues. (c) Correlation analysis of SOCS3 and Snail expression in HCC patient liver cancer tissues (n=228). (d) Representative images of methylation of SOCS3 promoter obtained by MSP in HBV-positive and HBV-negative HCC tissues. (e) Representative images of BSP analysis in HBV-positive and HBV-negative HCC tissues. (f) Cumulative overall survival curves of patients with high or low SOCS3 (top) and Snail (bottom) expression levels. (g) Cumulative disease-free survival curves of patients with high or low SOCS3 (top) and Snail (bottom) expression levels. (h) Cumulative overall (top) and disease-free (bottom) survival curves of the combination of SOCS3 and Snail. HCC patients were classified into four groups according to SOCS3 and Snail expression: group 1 (n=53): high SOCS3 and Snail expression; group 2 (n=51): high SOCS3 but low Snail expression; group 3 (n=59): low SOCS3 but high Snail expression; and group 4 (n=65): low SOCS3 and Snail expression
The pathological relevance of SOCS3 and Snail expression in HBV-associated HCC was then examined by IHC. On the basis of the IHC quantification data (Figure 6c), we found that low levels of SOCS3 or high levels of Snail expression were correlated with poorly differentiated tumors. In addition, SOCS3 expression correlated with sex and HBsAg expression. By contrast, neither SOCS3 nor Snail expression correlated with age (Supplementary Table 1). HCC patients were then grouped based on the expression levels of SOCS3 and Snail. Log-rank analysis demonstrated that the high SOCS3-expressing group experienced longer overall survival and disease-free survival when compared with the low SOCS3-expressing group, whereas the high Snail-expressing group experienced shorter overall survival and disease-free survival when compared with the low Snail-expressing group (Figures 6f and g). Univariate and multivariate analyses further revealed that the stage of tumor differentiation, TNM stage and Snail expression were independent risk factors for overall survival and disease-free survival (Supplementary Table 2). To determine whether the combination of SOCS3 and Snail could be used as a prognostic factor in predicting the outcome of HBV-associated HCC, patients were classified into four groups according to SOCS3 and Snail expression: group 1 (n=53): high SOCS3 and Snail expression; group 2 (n=51): high SOCS3 but low Snail expression; group 3 (n=59): low SOCS3 but high Snail expression; and group 4 (n=65): low SOCS3 and Snail expression. Differences in both overall survival (P<0.001) and disease-free survival (P<0.001) were significant among the four groups and group 3 indicated the worst prognosis (Figure 6h).
To further elucidate the correlation between SOCS3 promoter methylation and HBV-induced hepatocarcinogenesis, bisulfite sequencing analyses were performed to examine the SOCS3 promoter methylation status of all four groups (n=30 in each group). As shown in Supplementary Figure 14a, the HBV status in the four different groups is as follows: group 1 (HBV-, n=11; HBV+, n=19); group 2 (HBV-, n=19; HBV+, n=11); group 3 (HBV-, n=0; HBV+, n=30); group 4 (HBV-, n=0; HBV+, n=30). The percentage of HBV-positive and -negative patients that show SOCS3 promoter methylation was 86.7% and 10%, respectively. Moreover, SOCS3 promoter methylation was positively correlated with Snail protein levels, whereas negatively correlated with SOCS3 protein levels (Supplementary Figures 14b and c), suggesting that Snail mediated SOCS3 promoter methylation in HBV-associated HCC patients.
Discussion
Over the past 10 years, studies have produced compelling evidence for the functionally important tumor-promoting effects that inflammation can have on neoplastic progression.20 Importantly, inflammation is clearly capable of fostering the development of incipient neoplasias and full-blown cancers.21 In addition, inflammatory cells have been demonstrated to release chemicals, including ROS, that are mutagenic for nearby cancer cells, accelerating the development and progression of malignancy.22 However, the linkage between inflammation and ROS remains unclear. In this study, we found that HBV-induced ROS accumulation could promote inflammation through the upregulation of IL-6 expression in HCC. Moreover, elevated ROS deregulated the IL-6/STAT3 signaling pathway, leading to the constitutive activation of STAT3. Taken together, these results demonstrated that ROS has a key role in promoting the inflammation-mediated hepatocarcinogenesis.
SOCS3 is a target of STAT3, which in turn suppresses the activation of STAT3 through negative feedback.12 Accumulating evidence has revealed that SOCS3 is epigenetically silenced, resulting in the hyper-activation of STAT3 in many cancers, such as cholangiocarcinoma,13 head and neck squamous cell carcinoma23 and HCC.14 However, the epigenetic mechanism underlying SOCS3 repression has not been elucidated. Here, we showed that SOCS3 expression could be downregulated by HBV-induced ROS production through promoter hypermethylation. In addition, demethylation of SOCS3 promoter by DAC restored the SOCS3 expression, which in turn repressed the activation of STAT3, suggesting that the function of SOCS3 as a STAT3 target and negative regulator was controlled by its promoter methylation status. More importantly, we identified Snail as a repressor of SOCS3, which could bind SOCS3 promoter through E-boxes and mediate the methylation process of SOCS3 promoter in association with DNMT1 and HDAC1, similar to the epigenetic silencing process found for E-cadherin.18 These findings indicated that Snail functions as an essential initiating factor which can promote cancer development and progression through epigenetic silencing of certain tumor suppressors, such as SOCS3.
The pathological relevance of the IL-6/STAT3 signaling pathway has been observed in many types of human cancer, and Snail is a validated drug target for treating human cancer. However, IL-6/STAT3 signaling and the Snail-mediated tumor initiation and progression have, to date, been considered as remote pathways.24 The studies herein support the notion that Snail has a regulatory role in the IL-6/STAT3 signaling pathway through hypermethylation of the SOCS3 promoter. As the expression of Snail negatively correlates with HBV-associated HCC patient outcome, while the expression of SOCS3 is positively correlated, it is tempting to speculate that the combination of Snail and SOCS3 may have prognostic value in the prediction of both survival and recurrence. However, it remains to be seen whether the combination of Snail and SOCS3 expression has a better power to predict HBV-associated HCC patient outcome than the prognostic factors that have been used in the clinic, such as alpha-fetoprotein.25
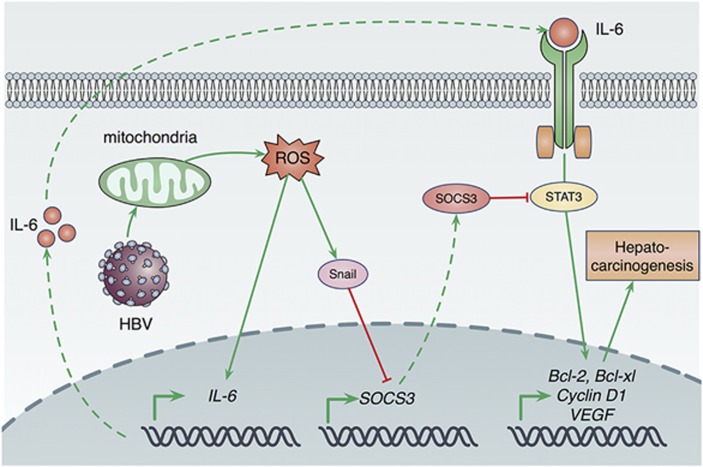
In conclusion, this study has revealed that HBV-induced mitochondrial ROS production stimulates IL-6 expression, which activates STAT3 pathway through autocrine signaling. More importantly, ROS accumulation upregulates Snail expression, which then leads to sustained activation of the IL-6/STAT3 pathway through epigenetic silencing of SOCS3, finally contributing to hepatocarcinogenesis (Figure 7). Further studies will be required to investigate whether the hepatocarcinogenesis process, which is mediated by HBV-induced ROS accumulation, can be observed in HBV transgenic mice that mimic development of human HCC.26
Figure 7.
A mechanism of hepatocarcinogenesis promotion via HBV-induced ROS accumulation. HBV replication induces intracellular ROS levels. Elevated ROS production then increases IL-6 production. IL-6 overexpression activates STAT3 via autocrine signaling, which causes increased expressions of a series of oncogenes, such as Bcl-2, Bcl-xl, Cyclin D1 and VEGF. Commonly, IL-6/STAT3 pathway is negatively regulated by SOCS3, a target gene of STAT3, through a negative feedback loop. However, HBV-induced ROS overproduction can increase the expression of Snail, which silences SOCS3 expression via hypermethylation of SOCS3 promoter and leads to constitutive activation of STAT3, finally contributing to hepatocarcinogenesis of HBV-associated HCC
Materials and Methods
Cell lines
HepG2, HepG2.2.15 and HepAD38 cells were cultivated in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY, USA) containing 10% FBS (Hyclone, Logan, UT, USA), penicillin (107 U/l) and streptomycin (10 mg/l) at 37 °C and 5% CO2. For HepAD38 cells, the medium was further supplemented with 400 μg/ml G418 and 2 μg/ml tetracycline. Confluent HepAD38 cells were washed with prewarmed PBS and then transferred to medium without tetracycline in which the replication of wild-type HBV was induced.16
Assays of reactive oxygen species
The intracellular levels of H2O2 and O2- were measured with 2′, 7′-dichlorofluorescin diacetate and dihydroethidium, respectively. Mitochondrial ROS production was detected by the MitoSOX Red assay (Life Technologies, Grand Island, NY, USA). Fluorescence was detected by Fluorometer (Thermo Fisher, Waltham, MA, USA).
Lipid peroxidation assay
Malondialdehyde is an end product of the lipid peroxidation process and was measured using a colorimetric assay (Calbiochem, San Diego, CA, USA) according to the manufacturer's instructions. Homogenized liver tissue in 62.5 mM Tris-HCl (pH=6.8) supplemented with Complete Mini protease inhibitor (Roche Diagnostics, Indianapolis, IN, USA) at equivalent protein amounts was used in the assay.27
Clinical specimens
All HCCs and corresponding adjacent normal tissues were obtained from de-identified patients who had undergone surgical resections at West China Hospital (Chengdu, China) or Sichuan Provincial People's Hospital (Chengdu, China). All paired samples were immediately frozen in liquid nitrogen. Ethics approval was obtained from the Institutional Ethics Committee of Sichuan University. Tumor differentiation was characterized according to WHO classification, while the pathologic stage was analyzed according to the TNM classification system of the International Union against Cancer.28 Clinico-pathological information for these patients is summarized in Supplementary Table 1.
Methylation-specific PCR
Genomic DNA was extracted and subjected to bisulfite modification as described previously.29 Briefly, 2 μg of genomic DNA was denatured with 0.2 M NaOH at 37 °C for 10 min and treated with 10 mM hydroquinone and 3 M sodium bisulfite (pH 5.0) at 50 °C for 16 h. Modified genomic DNA was analyzed with methylation-specific PCR SOCS3 primers as described.13 The positions and sequences of the primers are shown in Supplementary Figures 15a and b.
Bisulfite sequencing
DNA obtained from surgical samples and cells was treated with bisulfite modification and examined for the methylation status of 44 CpG dinucleotides within the SOCS3 gene promoter region as described previously.13 The SOCS3 gene promoter was amplified from the bisulfite-modified DNA by two rounds of PCR using nested primers specific to the bisulfite-modified sequence of the SOCS3 gene CpG island. The positions and sequences of the primers are shown in Supplementary Figures 15a and b.
Proliferation assays
Cell viability was measured using the Cell Counting Kit-8 (CCK-8; Beyotime, Nanjing, China). Intracellular bromodeoxyuridine incorporation assay was performed according to the manufacturer's instruction (BD Pharmingen, San Diego, CA, USA).
Soft-agar colony formation assay
A total of 5 × 104 cells were suspended in Dulbecco's modified Eagle's medium containing 0.35% agarose and layered on solidified medium containing 0.7% agarose in six-well plates. The medium was replaced every week. Images were taken after 2 weeks and analyzed with ImageJ software.
TUNEL assay
Apoptotic cell death was measured by TUNEL assay according to the manufacturer's instruction (Promega, Madison, WI, USA).
Tumor xenograft model
Male mice (BALB/c, 6 weeks of age) (HFK Bio, Beijing, China) were injected subcutaneously with HCC cells (1 × 107 cells per mouse). Tumor measurements were taken by caliper every 3 days. The tumor volumes were determined by measuring the length (l) and the width (w) and calculating the volume (V=lw2/2). After 18 days, the mice were killed and tumors were excised, pictured and weighed. Use of mice for this study was approved by the Institutional Animal Care and Use Committee (IACUC) at Sichuan University.
Biotinylated oligonucleotide precipitation assay
DNA precipitations were carried out as described previously.30 The biotinylated double-stranded wild-type and mutated SOCS3 oligonucleotide sequences are indicated in Supplementary Table 3. DNA-bound proteins were collected with streptavidin-agarose beads (Pierce, Rockford, IL, USA) and analyzed by western blotting.
Chromatin immunoprecipitation
Chromatin IP assay was carried out as described previously.31 Immunoprecipitated DNA was quantitated by real-time quantitative PCR. Primer sets were chosen to amplify approximately 100 to 150 base pairs on the promoter of SOCS3. The enrichments of Snail, HDAC1 and DNMT1 at the examined regions were quantitated relative to the input amount. The positions and sequences of the primers are shown in Supplementary Figures 15a and b.
Dual-Luciferase reporter assay
The promoter of the human SOCS3 gene (-2029 to -763) was PCR-amplified and then cloned into the pGL3 vector (Promega). HepG2 cells were cotransfected with 1 μg Myc-Snail plasmid, 1 μg of WT or mutant plasmids each and 1 μg of a Renilla luciferase expression construct, pRL-TK (Promega), using Lipofectamine 2000 (Life Technology). Cells were harvested 48 h post transfection and assayed with Dual Luciferase Assay (Promega) according to the manufacturer's instructions.
Statistical analysis
Analysis was performed using SPSS 16.0 for Windows (SPSS Inc, Chicago, IL, USA). All quantitative data were recorded as mean±S.D. Comparisons between two groups were performed by Student's t test. Differences among multiple groups were assessed by one-way ANOVA analysis. Two-way ANOVA was applied to determine significant differences at different time points. Pearson χ2 test or Fisher exact test was used to compare qualitative variables. Kaplan–Meier analysis was used to determine survival. The log-rank test was used to compare patients' survival between sub-groups, and the Cox regression model was used to perform multivariate analysis. The cutoff for the definition of subgroups was the median value. Statistical significance was defined as P<0.05.
Acknowledgments
This work was supported by grants from the National 973 Basic Research Program of China (2011CB910703, 2013CB911300 and 2012CB518900), the Chinese NSFC (81225015, 81430071, 81401951 and 81502441), and Sichuan Science-Technology Innovative Research Team for Young Scientist (2013TD0001).
Glossary
- BSP
bisulfite sequencing
- DCFH-DA
2′,7′-dichlorofluorescein diacetate
- GAPDH
glyceraldehydes-3-phosphate dehydrogenase
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HDI
hydrodynamic tail vein injection
- IHC
immunohistochemistry
- MDA
malondialdehyde
- IP
immunoprecipitation
- MSP
methylation-specific PCR
- NAC
N-acetyl L-cysteine
- ROS
reactive oxygen species
- RT-PCR
reverse-transcription polymerase chain reaction
- SOCS3
suppressor of cytokine signaling 3
- STAT3
signal transducer and activator of transcription 3
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by R Sordella
Supplementary Material
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 2007; 317: 121–124. [DOI] [PubMed] [Google Scholar]
- Soresi M, Giannitrapani L, D'Antona F, Florena AM, La Spada E, Terranova A et al. Interleukin-6 and its soluble receptor in patients with liver cirrhosis and hepatocellular carcinoma. World J Gastroenterol 2006; 12: 2563–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med 2009; 361: 1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol 2003; 15: 247–254. [DOI] [PubMed] [Google Scholar]
- Waris G, Ahsan H. Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog 2006; 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wruck CJ, Streetz K, Pavic G, Götz ME, Tohidnezhad M, Brandenburg LO et al. Nrf2 induces interleukin-6 (IL-6) expression via an antioxidant response element within the IL-6 promoter. J Biol Chem 2011; 286: 4493–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulua AC, Simon A, Maddipati R, Pelletier M, Park H, Kim KY et al. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS). J Exp Med 2011; 208: 519–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik E, Dixit VM. Mitochondrial reactive oxygen species drive proinflammatory cytokine production. J Exp Med 2011; 208: 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 2009; 9: 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D et al. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol 2003; 4: 551–556. [DOI] [PubMed] [Google Scholar]
- Isomoto H, Mott JL, Kobayashi S, Werneburg NW, Bronk SF, Haan S et al. Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing. Gastroenterology 2007; 132: 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Kanda H, Shikauchi Y, Saiura A, Matsubara K, Kitagawa T et al. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene 2005; 24: 6406–6417. [DOI] [PubMed] [Google Scholar]
- Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 2006; 130: 1117–1128. [DOI] [PubMed] [Google Scholar]
- Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 1997; 41: 1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci USA 1996; 93: 14960–14965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SO, Gu JM, Kim MS, Kim HS, Park YN, Park CK et al. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology 2008; 135: 2128–2140. [DOI] [PubMed] [Google Scholar]
- Mauhin V, Lutz Y, Dennefeld C, Alberga A. Definition of the DNA-binding site repertoire for the Drosophila transcription factor SNAIL. Nucleic Acids Res 1993; 21: 3951–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo DG, Andreu P, Coussens LM. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev 2010; 29: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell 2010; 141: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010; 140: 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Hengge UR, Bardenheuer W, Tischoff I, Sommerer F, Markwarth A et al. SOCS-3 is frequently methylated in head and neck squamous cell carcinoma and its precursor lesions and causes growth inhibition. Oncogene 2005; 24: 6699–6708. [DOI] [PubMed] [Google Scholar]
- Tang Y, Kitisin K, Jogunoori W, Li C, Deng CX, Mueller SC et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci USA 2008; 105: 2445–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terris B, Cavard C, Perret C. EpCAM, a new marker for cancer stem cells in hepatocellular carcinoma. J Hepatol 2010; 52: 280–281. [DOI] [PubMed] [Google Scholar]
- Liu S, Li N, Yu X, Xiao X, Cheng K, Hu J et al. Expression of intercellular adhesion molecule 1 by hepatocellular carcinoma stem cells and circulating tumor cells. Gastroenterology 2013; 144: 1031–1041. [DOI] [PubMed] [Google Scholar]
- Yuan K, Huang C, Fox J, Gaid M, Weaver A, Li G et al. Elevated inflammatory response in caveolin-1-deficient mice with Pseudomonas aeruginosa infection is mediated by STAT3 protein and nuclear factor kappaB (NF-kappaB). J Biol Chem 2011; 286: 21814–21825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Union Against Cancer (UICC) Sobin LH, Wittekind C (eds). TNM Classification of Malignant Tumours. 7th edition WILEY-liss: New York, 2009. [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996; 93: 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massaque J. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 2000; 100: 229–240. [DOI] [PubMed] [Google Scholar]
- Yuan K, Xie K, Fox J, Zeng H, Gao H, Huang C et al. Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology 2013; 145: 853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.