Abstract
Bcl2 family proteins control mitochondrial apoptosis and its members exert critical cell type and differentiation stage-specific functions, acting as barriers against autoimmunity or transformation. Anti-apoptotic Bcl2a1/Bfl1/A1 is frequently deregulated in different types of blood cancers in humans but its physiological role is poorly understood as quadruplication of the Bcl2a1 gene locus in mice hampers conventional gene targeting strategies. Transgenic overexpression of A1, deletion of the A1-a paralogue or constitutive knockdown in the hematopoietic compartment of mice by RNAi suggested rate-limiting roles in lymphocyte development, granulopoiesis and mast cell activation. Here we report on the consequences of conditional knockdown of A1 protein expression using a reverse transactivator (rtTA)-driven approach that highlights a critical role for this Bcl2 family member in the maintenance of mature B-cell homeostasis. Furthermore, we define the A1/Bim (Bcl-2 interacting mediator of cell death) axis as a target of key kinases mediating B-cell receptor (BCR)-dependent survival signals, such as, spleen tyrosine kinase (Syk) and Brutons tyrosine kinase (Btk). As such, A1 represents a putative target for the treatment of B-cell-related pathologies depending on hyperactivation of BCR-emanating survival signals and loss of A1 expression accounts, in part, for the pro-apoptotic effects of Syk- or Btk inhibitors that rely on the ‘BH3-only' protein Bim for cell killing.
Bcl2 family proteins are gate keepers of mitochondrial integrity that regulate death and survival of developing immune cells by controlling the activity of pro-apoptotic Bax and Bak.1, 2 Although highly redundant upon overexpression in tissue culture or as transgenes in mice,3 loss-of-function studies assigned cell type and differentiation stage-dependent survival roles to the different anti-apoptotic Bcl2 proteins4 that do not always correlate with their established expression patterns. For example, within the hematopoietic system, Bcl2 oscillates during early and late B- and T-cell development5 and Bcl2−/− mice do suffer the loss of mature lymphocytes, but not their early precursors.6, 7 Similarly, although the expression of BclX perfectly complements that of Bcl2 in developing lymphocytes,8 its most prominent role in hematopoiesis seems to be the control of the survival of pre-B cells and enucleated blood cells, such as erythrocytes and platelets.3 In contrast, although CD4+CD8+ double-positive thymocytes express high levels of BclX, they can cope easily without it.9 Of note, although the survival roles of Bcl2 and BclX are rather restricted within the hematopoietic system, Mcl1, which is ubiquitously expressed and often regulated at the posttranslational level, appears essential for the survival of most blood cell types.10
Bcl2a1/Bfl-1/A1, a poorly investigated Bcl2 pro-survival homolog shows rather restricted expression, limited mainly to the hematopoietic system in mice and man.11 Similar to Mcl1, A1 has a very short half-life and in myeloid cells it is responsive to inflammatory cytokines11, 12, 13, 14 as well as to FcɛRI activation.15, 16 A1 is induced at the mRNA level after successful beta selection of the TCR and rapidly increases upon antigenic challenge in mature T- and B-lymphocytes.17, 18, 19, 20 Together, this suggests that A1 is a critical component of a rewired survival network securing the expansion of activated lymphocytes and that of myeloid cells during inflammation (reviewed in Ottina et al.21). Most recently, a critical role of A1 in the survival of monocytic cells with myeloid-derived suppressor activity during chronic inflammation has been reported22 and increased mRNA expression in CML, pre-B ALL and chronic lymphocytic leukemia (CLL) correlates with poor prognosis and drug resistance phenotypes in humans (reviewed in Vogler23). Of note, treatment of CLL with BH3 mimetics targeting BCL2 and BCLX selects for tumors with high A1 expression,24 defining it as a putative drug target in blood cancer.
The physiological role of A1 is less clear as the Bcl2a1 gene locus has undergone gene quadruplication in mice, whereas only one gene is present in rats or humans.25 Out of the four loci in mice (A1-a, b, c, d), three give rise to mRNA species that translate into nearly identical proteins, whereas A1-c encodes a pseudogene.25 Deletion of A1-a in mice supported a contribution to granulocyte and activated mast cell survival. However, no effects were reported in lymphocytes, where A1-b and A1-d are more prominently expressed.17, 26
Hence, RNAi-based strategies were developed to explore the in vivo role of A1. An initial attempt using the RNA-Pol-III-responsive U6 promoter to drive an shRNA targeting all functional A1 paralogues failed to reveal significant phenotypes, probably due to limited knockdown in mature lymphocytes, or counter selection phenomena in cells where A1 may be essential.27 We previously used two alternative RNAi strategies using A1-specific shRNAs expressed in a miR-30 context (hereafter referred to as mi-shRNA) from the hematopoiesis-restricted RNA pol-II-dependent Vav-gene promoter or the Tet-responsive CMVmin promoter allowing constitutive A1 knockdown in the hematopoietic system.28 Although the analysis of these models pointed toward possible roles of A1 in leukocyte development and homeostasis, most prominently in the myeloid compartment, counter selection phenomena became evident.28 Hence, some of the phenotypes noted may have been ameliorated by insufficient knockdown or compensated by increased expression of other Bcl2 family proteins, whereas others may have been caused by tTA transactivator expression.29 In order to overcome these limitations, we studied the consequences of acute doxycycline-induced mi-shRNA-driven A1 knockdown using a reverse Tet-transactivator (rtTA) under control of the ubiquitous CAG promoter.30
Results
Generation of a doxycycline-induced A1 knockdown model
TRE-A1 mice, carrying a Tet-responsive CMVmin promoter controlling expression of a mi-shRNA targeting all functional A1 paralogues, were intercrossed with CAG-rtTA mice that express the rtTA from the ubiquitous CAG-promoter,31 to generate double-transgenic mice (referred to as DTrA1). As a control for RNAi off-target effects or global interference with the miRNA pathway, we also made use of transgenic mice expressing a mi-shRNA targeting Renilla luciferase31 (referred to as DTrRen).
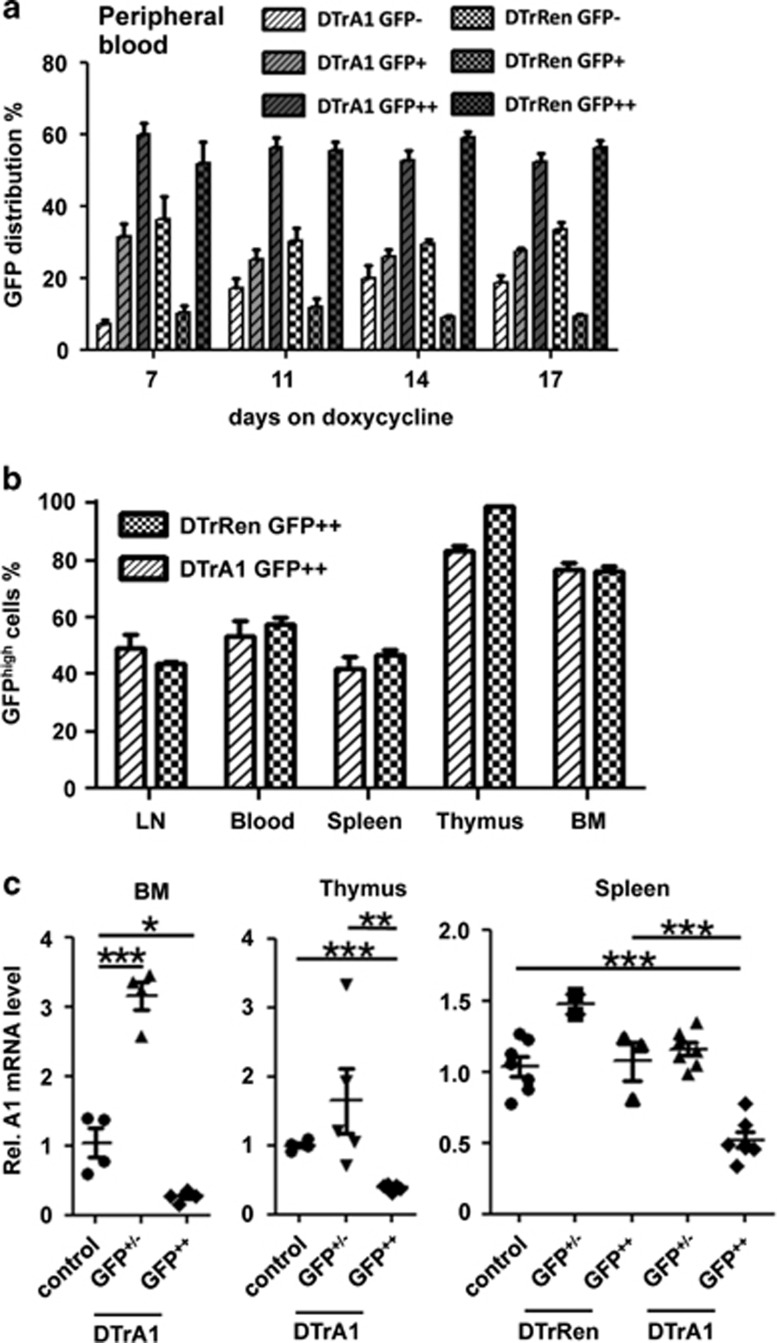
In both models, cells expressing mi-shRNA are GFP-traceable with the minor difference that in steady state, DTrA1 mice already show moderate GFP expression, as the reporter is driven from the UbiP promoter, positioned downstream of the Tet-responsive (TRE) CMVmin cassette controlling mi-shRNA expression. rtTA-binding induces mi-shRNA expression but also potently increases GFP levels. DTrRen mice express GFP plus the mi-shRNA targeting Renilla under control of the Tet-responsive CMVmin promoter.31 Consequently, all cells are initially GFP negative but become GFP positive upon addition of doxycycline (Supplementary Figure S1a). Hence, in both systems, cells that express the mi-shRNAs can be readily detected based on high GFP levels (GFP++).
To assess the flexibility of the system, mice were put on doxycycline-containing food and GFP induction was monitored in the peripheral blood from day 7 up to day 17. We observed strong induction of GFP expression in DTrA1 and DTrRen mice, leveling at around 60% (Figure 1a). Of note, a proportion of cells lost GFP expression (DTrA1) and/or failed to induce GFP (DTrRen), suggesting transgene silencing (Figure 1a, Supplementary Figure S1a). After 17 days of doxycycline treatment, animals were killed and the GFP expression levels were analyzed in all major hematopoietic organs. As in the peripheral blood, the percentage of GFP++ cells was comparable in DTrA1 and DTrRen mice, showing the highest percentages in bone marrow and thymus (Figure 1b,Supplementary Figure S1a).
Figure 1.
GFP-traceable mi-shRNA expression in vivo. (a) Flow cytometric analysis of transgene expression, as monitored by the appearance of GFP++ cells in the peripheral blood of double-transgenic DTrA1 and DTrRen mice kept on doxycycline (DTrA1 n=8, DTrRen n=6). Bars show means±S.E.M. (b) Quantification of the fraction of GFP++ transgene-expressing cells in primary and secondary lymphatic organs derived from DTrA1 (n=14) and DTrRen mice (n=8) killed on day 17 (BM, bone marrow; LN, lymph node). Bars show means±S.E.M. (c) Quantitation of A1 mRNA expression by qRT-PCR in sorted DAPI-negative and GFP++ versus +/- BM cells (wt n=1 or TRE-A1 single transgenic animals n=3, pooled as control; DTrA1 n=4), thymocytes (DTrA1; n=5) or splenocytes (TRE-A1 single transgenic controls, n=6; DTrRen n=3; DTrA1 n=7). Knockdown efficiency was evaluated on cDNA using primers amplifying all A1 paralogues or Actin for normalization. Data are shown as scattergram±S.E.M. with the mean control value set to 1. All experiments were performed in duplicates. ANOVA followed by Bonferroni post-hoc test. *P≤0.05, **P≤0.01, ***P ≤0.001 compared with controls kept on doxycycline
Next, we quantified the efficiency of A1 mRNA knockdown by qRT-PCR in GFP++ versus GFP+/- cells isolated by cell sorting from bone marrow, thymus or spleen of mice kept on doxycycline (Figure 1c, Supplementary Figure S1b). As the expression of A1 mRNA in wt or single transgenic mice (CAG-rtTA, TRE-A1 or TRE-Ren) did not differ significantly, these data were pooled and are generally referred to as ‘controls or CTRL'. Although A1 mRNA levels were reduced by 70–80% in GFP++ cells from bone marrow or thymus of DTrA1 mice (Figure 1c), we observed only about 50% knockdown in spleen, when compared with controls kept on doxycycline (Figure 1c). Of note, mRNA levels appeared to be actually higher in the GFP+/− fraction of bone marrow or thymus of DTrA1 mice when compared with controls, suggesting possible compensatory induction of A1 transcription in these cells. Notably, when analyzing A1 protein in the GFP-positive and GFP-low/negative fractions isolated form bone marrow, thymus or spleen, we were only able to detect basal A1 protein expression in lysates derived from the latter and under these conditions protein levels did not show an appreciable difference (Supplementary Figure S1b).
Together, the data document mosaic transgene expression upon doxycycline treatment that can be traced and define mi-shRNA expression at the target gene mRNA level in different hematopoietic organs. The lack of protein expression in lysates from thymus or bone marrow suggests that only certain subsets of blood cells express A1 in these organs, whereas comparable protein levels in spleen suggest compensatory effects in vivo.
Rapid induction and reversibility of mi-shRNA expression in DTrA1 mice
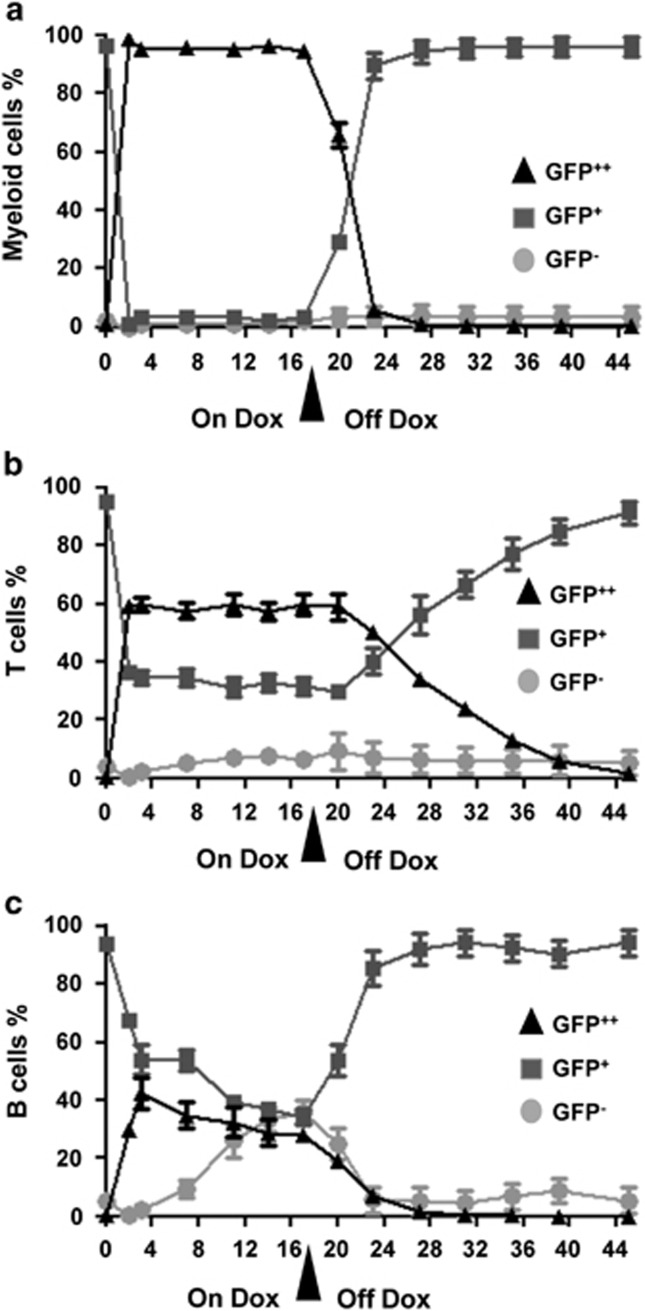
Following the kinetics of GFP induction in peripheral blood, we observed a rapid induction of mi-shRNA in the myeloid compartment, as already after 3 days of dox administration virtually all Mac-1+ myeloid cells qualified as GFP++ (Figure 2a). In contrast, only approximately 60% of blood CD4+ or CD8+ T cells showed such phenotype after 3 days and this fraction did not significantly increase further over time (Figure 2b). Remarkably, only about 40% of CD19+ B cells seemed to induce mi-shRNA expression, as judged by GFP levels, and over the time this population even decreased mildly. This was paralleled by a concomitant increase of GFP-negative B cells (Figure 2c). After 17 days, dox food was removed and mi-shRNA expression was no longer detectable after additional 6 days in myeloid and B cells but persisted for 3 weeks in T cells. These observations are grossly in line with the expected turnover times of the different cell types analyzed, demonstrating that the system is fully reversible.
Figure 2.
mi-shRNA transgene expression is fully reversible in DTrA1 mice. Evaluation of transgene induction, as measured by appearance of GFP++ cells by flow cytometry in double-transgenic DTrA1 mice fed with doxycycline-containing food for 17 days. Peripheral blood was analyzed at the indicated time-points and cells were co-stained for markers specific for (a) myeloid cells (Gr-1+Mac-1+), (b) T (pooled CD4+ and CD8+) or (c) B cells (CD19+). Cells were gated on forward/side scatter, lack of Annexin V binding, lineage markers and GFP. Cells were categorized in three groups: GFP++, GFP+ and GFP-. Symbols represent means±S.E.M. (ON DOX: n=8, OFF DOX: n=5)
CAG-rtTA-driven A1 knockdown does not affect myelopoiesis
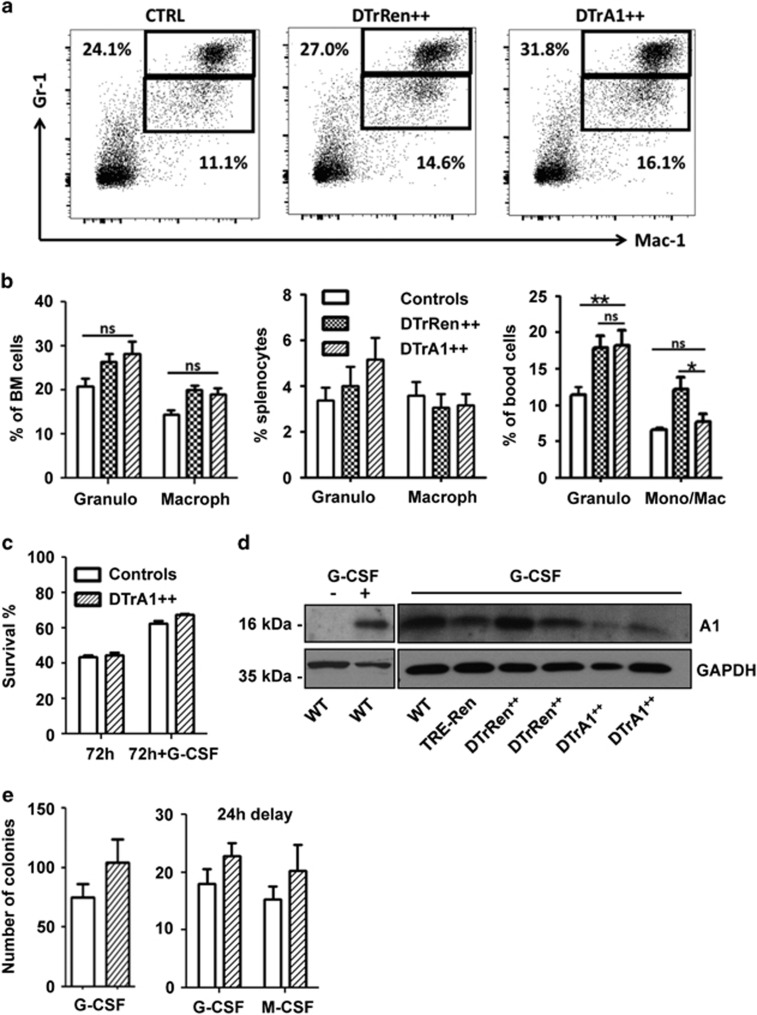
First, we analyzed the distribution of myeloid cells across genotypes. In contrast to our expectation, analysis of Gr-1+Mac-1+ granulocytes and Gr-1lowMac-1+ monocyte/macrophages failed to document an impact of A1 knockdown (Figures 3a and b). Consistently, the in vitro survival of sorted granulocytes from the bone marrow was not affected, when monitored after 24, 48 or 72 h, nor was G-CSF-mediated survival of these cells (Figure 3c, data not shown). Western analysis confirmed strong induction of A1 protein upon G-CSF treatment in wt cells as well as detectable knockdown in GFP++ granulocytes cultured in the presence of G-CSF (Figure 3d). Methylcellulose assays also failed to reveal significant differences in the colony formation potential of DTrA1-derived myeloid progenitors and those derived from control mice (Figure 3e). This suggests that A1 acts redundantly in granulocyte survival or that the knockdown efficacy achieved here versus that achieved in other model systems28 was insufficient to reveal such an effect.
Figure 3.
Induced A1 knockdown does not affect the myeloid compartment. (a) Representative dot plots of single-cell suspensions derived from bone marrow from mice of the indicated genotypes kept on doxycycline for 17 days. Cells were stained with antibodies recognizing Gr-1 or Mac-1 cell surface markers and gated on forward/side scatter profiles and viable (Annexin V-) cells prior analysis. (b) Quantification of data obtained in bone marrow (wt and single transgenic controls n=9; DTrRen n=6 DTrA1 n=8), spleen (wt and single transgenic controls n=7, DTrRen n=6, DTrA1 n=8) or peripheral blood (wt and single transgenic controls n=7, DTrRen n=5, DTrA1 n=7). Bars represent means±S.E.M. (c) Bone marrow-derived GFP++ granulocytes were sorted and cultured for 72 h±G-CSF (50 ng/ml) and doxycycline (1 μg/ml), re-added after 40 h. Viability was assessed by flow cytometry and Annexin V exclusion. Bars represent means±S.E.M. All experiments were performed in technical duplicates (wt and single transgenic controls n=7; DTrA1 n=4). (d) Quantitation of A1 knockdown efficacy at the protein level in sorted Gr-1+ GFP++ bone marrow cells derived from mice of the indicated genotypes kept on doxycycline for 17 days. Cells were cultured±G-CSF and 1 μg/ml doxycycline for 8 h. (e) Bone marrow cells were plated in Methocult medium in the presence of G-CSF or M-CSF plus IL-3 and IL-6. Cultures were maintained in the presence of doxycycline (1 μg/ml), which was re-added on day 3 and day 6. Colonies were scored blinded after 7 days. Data represent means±S.E.M. (Simultaneous addition: wt and single transgenic controls n=4; DTrA1 n=4). Alternatively, cytokines were added with a 24- h delay. Data represent means±S.D. (wt controls n=2; DTrA1 n=2). All experiments were performed in duplicates. ANOVA followed by Bonferroni post-hoc test was performed to evaluate significant differences. *P≤0.05, **P≤0.01 compared with pooled controls kept on doxycycline)
Timed ablation of A1 has minimal impact on T-cell development or activation
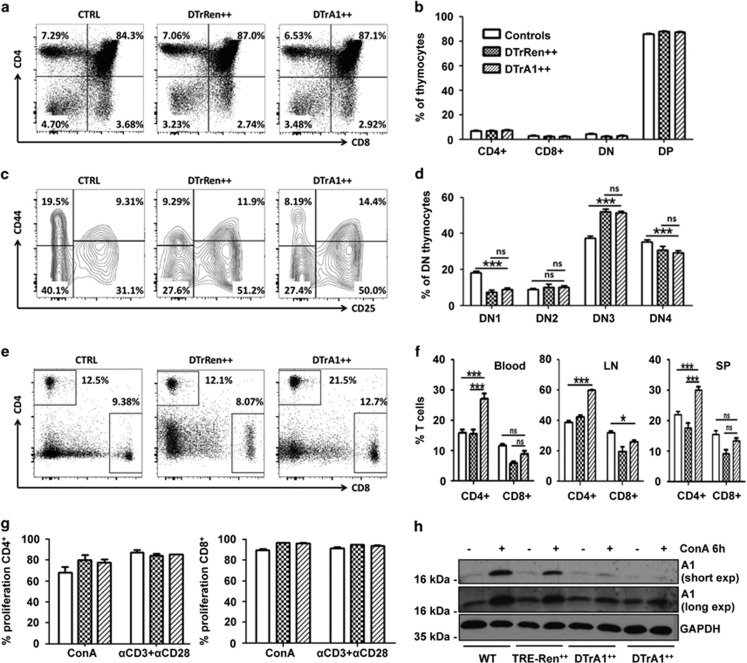
Next, we analyzed the effects of inducible A1 mRNA ablation on T-cell development in the thymus (Figure 1b, Supplementary Figure S1a). Despite a significant reduction of A1 mRNA (Figure 1c), the percentage of CD4+ and CD8+ single- or double-positive thymocytes was not affected in DTrA1 mice when compared with controls (Figures 4a and b). Also, the distribution of early developmental stages, assessed by CD25 and CD44 co-staining of CD4−8− double-negative cells did not differ significantly between DTrA1 and DTrRen mice (Figures 4c and d). Interestingly, both strains showed a cellular distribution that was different from that of single transgenic or wt controls inasmuch that the percentage of DN1 and DN3 cells were found either mildly reduced or increased, respectively (Figures 4c and d). Notably, in blood and secondary lymphatic organs we observed a relative increase of CD4+ T cells, whereas the percentage of CD8+ T cells was similar to that found in controls (Figures 4e and f, Supplementary Figure S2a). Mitogen-driven proliferation of either subset was not affected by A1 knockdown (Figure 4g) that was confirmed at the protein level in activated CD4+ T cells (Figure 4h, Supplementary Figure S6a).
Figure 4.
Inducible A1 ablation shows limited impact on T-cell homeostasis. Flow cytometric analysis of single-cell suspensions stained with antibodies against CD4 or CD8, either alone or in combination with antibodies specific for CD25 or CD44 in GFP++ cells from control, DTrA1 or DTrRen mice kept on dox for 17 days. Cells were gated on forward and side scatter profiles, then gated on alive (Annexin V-) cells and GFP++ levels. Percentages of cells falling within each gate are shown. (a) Representative dot plots of CD4 and CD8 staining profiles of cell suspensions quantified in b. Data shown represent means±S.E.M. (wt and single transgenic controls n=15, DTrRen n=5, DTrA1 n=12). (c) Representative contour plots of flow cytometric analysis defining DN1-4 stages of thymic development, using antibodies recognizing CD25 and CD44 in the CD4-CD8- fraction, quantified in d. Data shown represent means±S.E.M. (wt and single transgenic controls n=9, DTrRen n=4, DTrA1 n=10). (e) Peripheral blood was stained with antibodies recognizing CD4+ or CD8+ and T-cell distribution was assessed in the GFP++ fraction quantified, along with data from lymph node and spleen in f. Data shown represent means±S.E.M. (wt and single transgenic controls n=12, DTrRen n=7, DTrA1 n=12). ANOVA followed by Bonferroni post-hoc test. *P ≤0.05, ***P ≤0.001 compared with controls (CTRL) kept on doxycycline. (g) Proliferation was assessed in GFP++ CD4+ or CD8+ T cells by monitoring CPD-dilution after 72 h of mitogen stimulation. Bars represent means±S.E.M. (wt and single transgenic controls n=5, DTrRen n=3, DTrA1 n=4). All experiments were performed in technical duplicates. (h) Quantitation of A1 knockdown efficacy by western analysis in sorted CD4+ GFP++ splenocytes derived from mice of the indicated genotypes kept on doxycycline for 17 days. Cells were cultured in the presence of IL-2 alone (control) or stimulated with 1 μg/ml ConA in the presence of 1 μg/ml doxycycline for 6 h
Together, this suggests a possible impact of rtTA transgene expression on early thymocyte development that may have compromised prior analyses28 but also a relative increase of CD4+ T cells in response to A1 knockdown in the periphery. The latter turned out to be simply a secondary consequence of mature B-cell loss (see below) but simultaneously implies that CD8+ peripheral T cells may suffer disadvantages upon A1 knockdown.
Acute A1 knockdown triggers loss of mature B cells
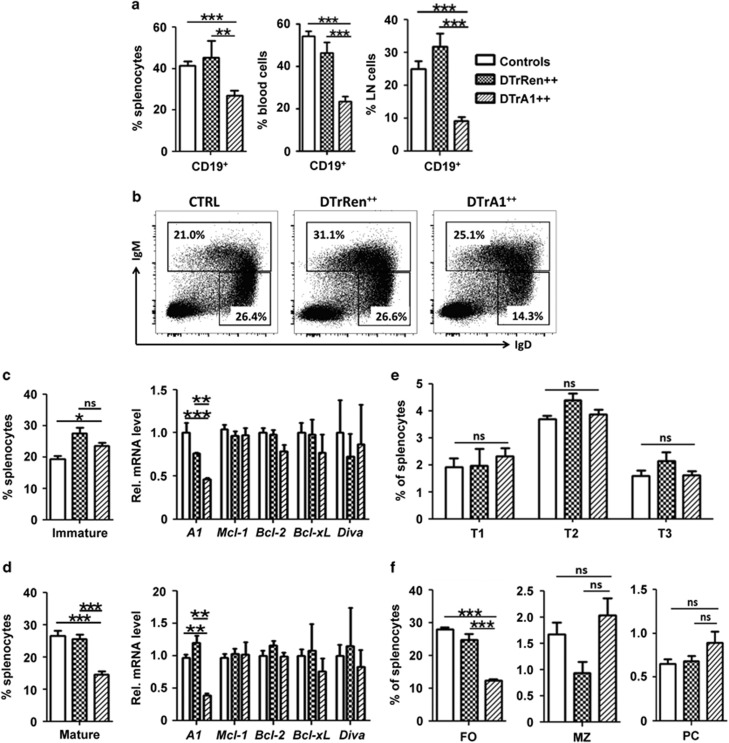
Our analysis of the B-cell compartment revealed that, as noted before,28 early development in the bone marrow was largely unaffected, as the percentage of pro-B and pre-B was comparable between genotypes. Immature B cells were even mildly elevated in the GFP++ fraction of DTrA1 mice (Supplementary Figures S3a–e). Methylcellulose assays also failed to reveal differences in the clonogenic capacity of bone marrow-derived IL-7-dependent pre-B-cell progenitors (Supplementary Figures S3a–e). However, when we analyzed B cells in the periphery we observed a significant reduction of GFP++ CD19+ cells in spleen, peripheral blood and lymph nodes upon A1 knockdown (Figure 5a,Supplementary Figure S2b). Importantly, DTrRen mice did not show such a phenotype, suggesting a specific effect in response to A1 mRNA ablation. A closer examination revealed that mainly mature B cells (defined initially as IgMloIgDhi) were reduced (Figures 5b–d). To test whether the loss of GFP++ mature B cells correlated indeed with reduced levels of A1 mRNA, we sorted immature (IgM+) and mature GFP++ B cells from DTrA1 or control mice kept on doxycycline. qRT-PCR analysis revealed A1 mRNA knockdown to about 35% compared with controls in mature B cells and to about 45% in immature cells (Figures 5c–d), similar to findings made in total spleen (Figure 1c). Compensatory upregulation of other Bcl2 prosurvival family members, however, was not observed at the mRNA level (Figures 5c and d). A more concise analysis of different B-cell subsets in the spleen revealed that mainly follicular (FO) B cells were affected. Neither transitional (T1-3), marginal zone (MZ) B cells nor plasma cells were reduced upon A1 knockdown (Figures 5e and f, Supplementary Figures S4b and c).
Figure 5.
Acute A1 knockdown leads to mature B-cell loss. Single-cell suspensions from mice kept for 17 days on doxycycline were stained with specified antibodies and B-cell distribution was assessed in the GFP++ fraction of cells. (a) Cells were gated on forward and side scatter profiles, then on viable (Annexin V-) cells and GFP++ levels to determine the percentage of CD19+ B cells in spleen, blood and lymph nodes (LN, Blood: wt and single transgenic controls n=12, DTrRen n=7, DTrA1 n=12, spleen: CTRL n=9, DTrRen n=5, DTrA1 n=9). (b) Representative dot plots defining immature (IgM+IgDlo) and mature (IgMloIgD+) B cells, quantified in c and d along with the relative mRNA expression levels of the indicated prosurvival proteins in FACS-sorted cells (c and d). Bars defining percentages represent means±S.E.M. (wt and single transgenic controls n=10, DTrRen=11, DTrA1=12). Bars within the mRNA analysis represent means±S.E.M. (Immature: wt and single transgenic controls n=9, DTrRen n=4, DTrA1 n=5 (for Diva n=3), Mature: wt and single transgenic controls n=7; DTrRen n=4, DTrA1 n=4 (for Diva n=3)). PCR analysis was performed in technical duplicates. Actin levels were quantified and used for normalization. (e) Transitional type 1-3 B cells in the spleen were defined based on the expression of CD19, B220 and AA4.1 before comparing IgM against CD23, whereas (f) FO and MZ B cells were defined as CD19+, B220+, AA4.1- and CD21loCD23+ or CD21+CD23-, respectively (see also Supplementary Figure S4c). Plasma cells were defined as CD138+TACI+. Bars represent means±S.E.M. (wt and single transgenic controls n=9, DTrRen n=5, DTrA1 n=9). ANOVA followed by Bonferroni post-hoc test was used to assess statistical differences *P≤0.05, **P ≤0.01, ***P ≤0.001 compared with pooled controls (wt or single transgenic mice kept on doxycycline)
Together, this suggests that a reduction of A1 mRNA by two-thirds suffices to cause the loss of FO B cells, defining A1 as a key survival factor, whereas other developmental stages are not affected, either because of the comparatively low A1 expression levels (Supplementary Figure S4a) or functional redundancy with other Bcl2 prosurvival proteins.
A1 knockdown impairs survival of activated B cells
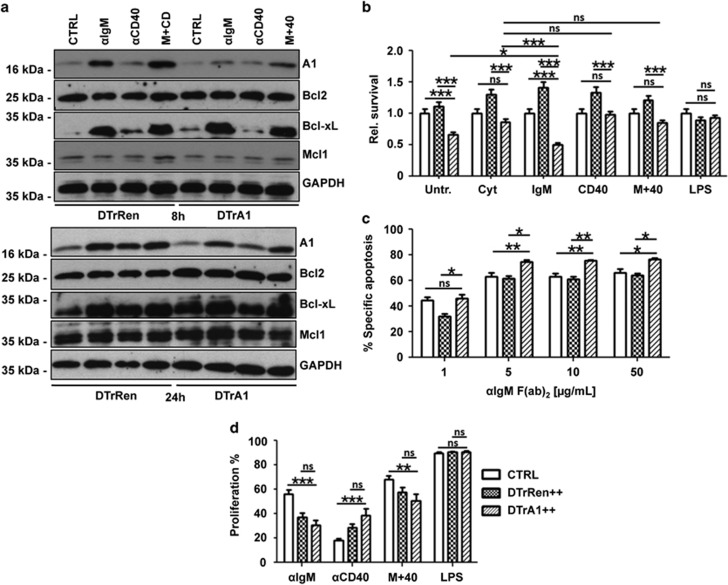
To assess the impact of mi-shRNA expression on A1 protein, B220+ GFP++ cells from DTrRen or DTrA1 spleens were put in culture in the presence of doxycycline and treated for up to 24 h with anti-IgM F(ab)2 fragments and/or anti-CD40 cross-linking antibodies, stimuli reported to induce A1 mRNA.17, 19, 32 Western analysis revealed that A1 was expressed at low levels in freshly isolated B220+ B cells. However, after stimulation with anti-IgM F(ab')2 fragments, we observed a strong induction of A1. CD40 ligation also induced A1 protein and double treatment proved equally potent (Figure 6a). Similar to findings made in T cells, A1 induction was only ameliorated but not abrogated upon mitogen treatment, whereas expression of Bcl2, BclX or Mcl1 was not affected in A1 knockdown B cells, when compared with DTrRen-derived B cells, largely excluding possible compensatory effects.
Figure 6.
A1 knockdown sensitizes mature B cells to apoptosis. (a) Quantitation of A1 knockdown efficacy at the protein level in sorted B220+ GFP++ splenocytes derived from mice of the indicated genotypes kept on doxycycline for 17 days. Cells were cultured in the presence of IL-2, -4, -5 alone (CTRL) or stimulated additionally with 1 μg/ml anti-mouse IgM F(ab')2 fragments (αIgM), 1 μg/ml anti-mouse CD40 mAb (αCD40) or both (M+40) in the presence of 1 μg/ml doxycycline for 8 or 24 h. Membranes were probed with antibodies specific for the indicated Bcl2 family members to assess possible compensatory effects or anti-GAPDH to control for protein loading (one out of two independent experiments yielding similar results is shown). (b) Total spleen cells from mice kept on doxycycline were isolated and incubated in media alone (untreated), cytokines (CYT) or cytokines with the indicated mitogens. Cultures were maintained in the presence of doxycycline (1 μg/ml), which was re-added after 40 h. After 72 h cells were stained with antibodies recognizing IgM or IgD and the percentages of viable (Annexin V-) GFP++ IgM+IgD+ B cells was assessed and compared with the percentage of B cells detected straight after killing. As these percentages differ (see also Figure 5), the bar graphs show the ratio of the percentage of IgM+IgD+ B cells in the culture on day 3, divided by the percentage of the IgM+IgD+ B cells present on day 0. Bars represent means±S.E.M. (wt and single transgenic controls n=11, DTrRen n=7, DTrA1 n=9). (c) GFP++ mature splenic B cells (IgMloIgDhi) from DTrA1 or DTrRen mice were sorted and cultured in the presence of graded concentrations of plate-bound anti-IgM F(ab)2 fragments in the presence of 1 μg/ml doxycycline. Cell viability was assessed after 20 h by Annexin V staining. The extent of apoptosis induced specifically by BCR ligation or SYK inhibition was calculated by the following equation: (induced apoptosis/spontaneous cell death) × 100. All experiments were performed in duplicates. Bars represent means±S.E.M. (wt and single transgenic controls n=5, DTrRen n=3, DTrA1 n≥5). (d) GFP++ B cells were cultured in media containing doxycycline (replenished after 40 h) plus cytokines, or cytokines plus the indicated mitogens. Loss of CPD labeling was assessed after 72 h to calculate the percentage of B-cell proliferation (wt and single transgenic controls n=12, DTrRen n=10, DTrA1 n=9). ANOVA followed by Bonferroni post-hoc test was performed to evaluate results for significant differences. *P≤0.05, **P≤0.01, ***P ≤0.001
We wondered if the reduction of A1 in DTrA1 mice affects survival and/or proliferation of B cells upon activation in vitro, or whether systemic effects imposed by A1 RNAi may contribute to the B-cell phenotype noted in vivo. Therefore, splenocytes from control, DTrA1 and DTrRen mice kept on doxycycline were placed in culture and stimulated with mitogens. After 72 h, we compared the percentages of viable B cells in relation with that detected straight after isolation. In line with a role for A1 in BCR-mediated survival, we observed a strong reduction of GFP++ B cells in DTrA1 splenocyte cultures when compared with DTrRen cultures (Figure 6b,Supplementary Figure S5). This effect was evident in untreated or ‘cytokine-only' controls (tonic signaling impaired), as well as soluble in IgM F(ab)2 treated cultures (simulating antigen-stimulation). In fact, A1 knockdown B cells showed strongly reduced viability upon BCR ligation. B-cell loss in culture after CD40 stimulation was, however, not significantly accelerated over that of untreated controls. In contrast, LPS stimulation did effectively prevent the accelerated loss of A1 knockdown cells, indicating activation of alternative survival pathways (Figure 6b, Supplementary Figure S5). Together, this suggested that lack of BCR signaling leads to a loss of A1 expression thereby facilitating apoptosis.
Excessive BCR stimulation can also lead to apoptosis that depends on the BH3-only protein Bim.33 Hence, we next assessed the sensitivity of mature B cells to BCR cross-linking in more detail. B cells derived from DTrA1 mice showed increased cell death upon activation when compared with cells from DTrRen mice (Figure 6c). Finally, we determined whether A1 knockdown affected B-cell proliferation in culture. To assess this, we used CPD-labeled splenocytes and determined loss of CPD fluorescence in CD19+ B cells after 72 h. Somewhat surprising, we observed that B cells from both, DTrRen and DTrA1 mice, proliferated less efficiently than B cells from wild-type mice upon IgM or IgM+CD40 co-stimulation, suggesting that either mi-shRNA overexpression in general or rtTA expression compromises B-cell proliferation (Figure 6d, Supplementary Figure S6b).
Together, this suggests that A1 levels may co-regulate the susceptibility of B cells to negative selection signals in the periphery and that the degree of A1 knockdown achieved in our model system affects the capacity of B cells to respond to mitogens.
BCR-activated kinases maintain A1 expression to prevent Bim-induced apoptosis
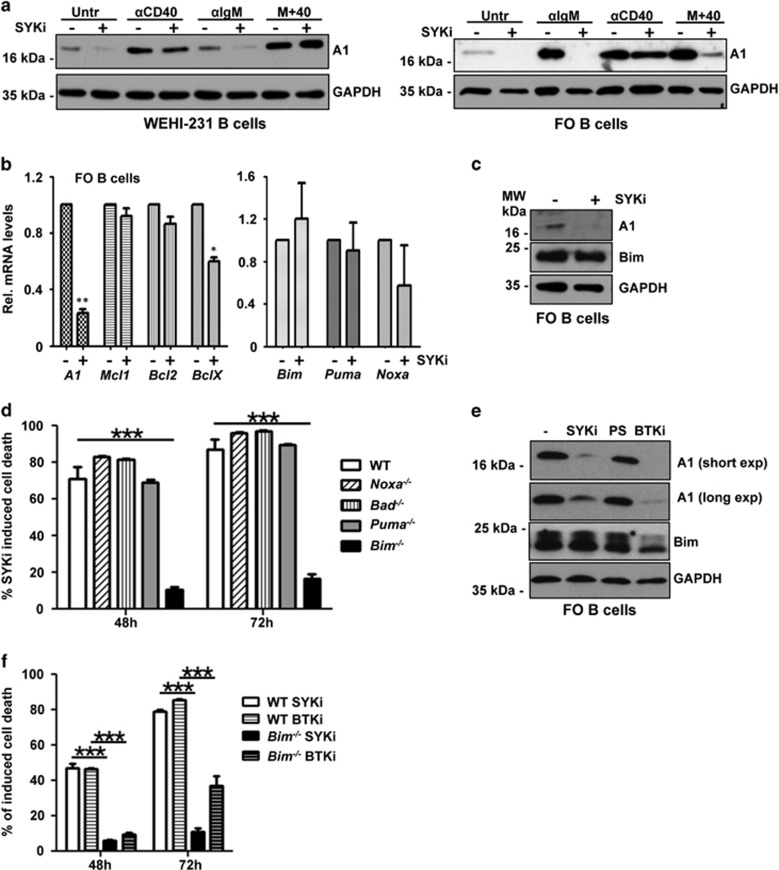
The finding that DTrA1 mice lose mature B cells in vivo suggested that A1 may be a critical target for BCR-emanating survival signaling known to engage spleen tyrosine kinase (Syk). To test the role of Syk in this process, we first analyzed IgM-positive WEHI-231 lymphoma cells that express detectable basal levels of A1. Cells were left untreated, stimulated with anti-IgM, anti-CD40 or the combination of both in the absence or presence of the Syk kinase inhibitor R406. In line with our hypothesis, Syk inhibition per se reduced basal as well as IgM-induced A1 expression but not that induced by CD40 ligation (Figure 7a). In primary sorted FO B cells, ablation of tonic as well as IgM-induced BCR signaling by inhibition of Syk also promoted loss of A1 protein (Figure 7a). Syk inhibition appeared to act at the level of A1 mRNA expression but had limited impact on the mRNA levels of other pro- or anti-apoptotic Bcl2 family proteins (Figure 7b).
Figure 7.
Inhibition of Syk or Btk reduces A1 expression and triggers Bim-dependent apoptosis. (a) WEHI-231 B cells or sorted FO B cells from wild-type mice were left untreated or treated with 2 μM of the Syk inhibitor R406 and stimulated as indicated for 4 h. Cells were lysed and analyzed by immunoblotting using an antibody specific for mouse A1 or GAPDH to control for protein loading (one out of two independent experiments yielding similar results is shown). (b) Sorted follicular B cells were stimulated for 6 h with Syk inhibitor or solvent control. RNA was isolated for qPCR analysis. Bars represent means±S.E.M. (n=3). (c) A1 and Bim protein levels were assessed 4 h after inhibitor treatment (5 μM) by western analysis. (d) FO B cells from mice of the indicated genotypes were sorted and cultured in the absence or presence of 5 μM Syk inhibitor. Survival was monitored by Annexin V staining. Bars represent means±S.E.M. (n=3). (e) Comparison of A1 and Bim protein levels 4 h after 5 μM Syk inhibitor, 10 μM PS-1145 (Ikk) inhibitor or 5 μM Btk inhibitor treatment of sorted splenic B cells pretreated for 24 h with anti-IgM by western analysis. (f) Comparison of FO B-cell survival after inhibition of Syk or Btk (5 μM). *P≤0.05, **P≤0.01, ***P≤0.001
We also explored Bim levels, as a putative effector activated in B cells upon Syk inhibition in WEHI-231 (data not shown) and FO B cells but failed to observe significant changes in protein levels, at least at the time point analyzed (Figure 7c). Hence, in order to define that BH3-only protein family member may be critical for B-cell death noted upon Syk inhibition, we exposed FO B cells derived from cell death gene-deficient mice to Syk inhibitor. This analysis revealed that indeed Bim was the rate-limiting BH3-only protein in this type of cell death (Figure 7d), whereas Noxa, Puma (p53-upregulated modulator of apoptosis) or Bad (B-cell lymphoma 2 (Bcl-2) antagonist of cell death) seemed dispensable. Bim deficiency also protected FO B cells from death induced by Ibrutinib (Figure 7f), a clinically approved inhibitor of Bruton's tyrosine kinase (Btk), a downstream effector of Syk that also strongly reduced A1 expression in WEHI231 cells (data not shown) and FO B cells, whereas Bim expression was largely unaffected (Figures 7c and e). Of note, the IKK inhibitor PS-1145 had only minimal impact on A1 levels under the conditions tested (Figure 7e).
Together, this suggests that Syk inhibition as well as Btk inhibition sensitizes B cells to apoptosis by repressing A1, thereby facilitating Bim-dependent apoptosis and that tonic as well as activation-driven BCR-signaling supports B-cell survival at least in part by controlling A1 expression, most likely needed to restrain Bim.
Discussion
Here we report on the consequences of doxycycline-induced knockdown of A1 expression within the hematopoietic system and provide evidence that A1 is a critical mediator of BCR-dependent B-cell survival. Furthermore, our analysis also documents limitations of in vivo RNAi approaches when used to target prosurvival proteins rather than tumor suppressors or pro-apoptotic genes.34, 35, 36, 37 Despite the successful induction of mi-shRNA expression in different organs and lineages of the hematopoietic system, variegated knockdown efficiencies, possibly due to cell type-specific differences in promoter efficiencies and/or pharmacodynamics of doxycycline as well as counter selection phenomena became evident and not all phenotypes previously attributed to loss of A1 using constitutive knockdown mouse models28 were recapitulated here.
Although analysis of the constitutive knockdown models,28 similar to A1-a deficiency,26 pointed toward a rate-limiting role of A1 in granulocyte survival, doxycycline-driven inducible A1 knockdown did not affect their homeostasis. As granulocytes are short lived and replenished with high efficacy from the bone marrow, we are left to assume that the rtTA-driven knockdown, despite being appreciable in western analysis, is not efficient enough to reveal effects in vitro or is compensated in these cells in vivo.
A1 was also proposed to be critical for TCR-beta selection and subsequent pre-TCR expression in developing thymocytes as well as in antigen-stimulated T- and B cells.19, 20 Our previous analyses of constitutive RNAi-mediated A1 knockdown models provided supportive evidence for a role in TCR-beta selection by demonstrating an accumulation of CD4−CD8− double-negative thymocytes and a relative decrease in CD4+CD8+ double-positive cells in the GFP++ fraction in one mouse strain expressing the A1-selective mi-shRNA from the Vav-gene promoter.28 A second model, using a different mi-shRNA sequence under control of the Vav-tTA transactivator, revealed a similar phenomenon in the thymus.28 As the penetrance of the reported phenotype correlated with A1 mRNA knockdown efficacy, we concluded that A1 is rate limiting for thymocyte survival upon beta selection. However, we were unable to recapitulate these findings here. This may be due to differences in promoter efficacy in individual thymocyte subsets (VavP versus TET-CMVmin).28, 30 Although we noted a mRNA knockdown efficacy of close to 80% in total GFP++ thymocytes derived from DTrA1 mice, weaker knockdown rates in double-negative thymocyte subsets, reported to require A1 for survival,38 may provide a possible explanation. Alternatively, adaptation to chronic versus timed ablation of A1 may account for the different observations made, as may even non-hematopoiesis-driven adaptation processes upon A1 knockdown in other cell types, as the CAG-rtTA activates expression of mi-shRNAs in other tissues.
Notably, none of the model systems used so far revealed a major impact of A1 knockdown on mature T-cell homeostasis.27, 28 This may seem odd in light of the finding that A1 mRNA is strongly induced upon TCR ligation.17, 20, 39 However, knockdown efficiency in all models analyzed appears to be weakest in the spleen were RNAi efficacy seems limited also when using other targets, such as AGO2.27 Noteworthy here, however, remains the observation that only CD4+ T cells showed a relative increase because of B-cell loss, whereas CD8+ T cells remained low. Hence, it is tempting to speculate that CD8+ cells suffer some disadvantage upon A1 knockdown.
The most prominent finding using the different RNAi model systems is the disturbance of mature B-cell homeostasis. Although early B-cell developmental stages were largely unaffected by A1 knockdown,28 we noted a gradual loss of mature B cells in all secondary lymphatic organs. These findings are consistent with the reported gradual increase of A1 mRNA during B-cell development.40 Although the knockdown system is outcompeted upon B-cell activation over time in culture, basal A1 mRNA levels are clearly reduced in steady state and this degree of reduction appears sufficient to cause the loss of resting B cells in vivo.
Interestingly, A1 knockdown mice phenocopy, in part, several knockout strains that show loss of mature B cells. Strikingly, all the respective gene products reportedly control A1. These include mice lacking the TNF family cytokine BAFF41 or its receptor, BAFF-R,42 mice where BCR signaling is blunted or impaired, for example, by loss of Ig-alpha43 or BCAP,44 respectively, or where NF-κB or PI3-kinase signaling is compromised by other means, for example, in c-Rel−/− and PLCγ−/− mutants.19, 45 In most of these cases, B-cell loss can be rescued by transgenic Bcl2 overexpression that may act as an A1 surrogate, supporting a critical role in B-cell survival. Surprisingly, this survival function may actually be shared with Bcl2, as its deficiency also leads to mature B-cell loss (reviewed in Sochalska et al. 3).
Of note, B cells activated by IgM show significantly higher apoptosis than B cells cultured with cytokines alone, suggesting that A1 is a critical survival factor downstream of tonic as well as antigen-driven BCR activation. Consistently, we show that A1 expression is controlled by Syk and Btk, two key regulators of B-cell development and survival downstream of the BCR46 and thereby, most likely, prevent Bim-induced apoptosis. It will also be interesting to see if A1 knockdown affects survival during negative selection in vivo, as apoptosis upon BCR cross-linking was more pronounced when A1 levels were reduced. Analysis of mouse models of negative B-cell selection or B-cell-driven autoimmunity, such as HEL-BCR transgenic47 or BAFF-transgenic mice48 will help to address this issue.
A discrepancy that arose is that B cells from DTrA1 mice showed normal proliferative capacity ex vivo, whereas B cells from Vav-tTA x TRE-A1 mice that constitutively expressed the mi-shRNA targeting A1 showed impaired proliferation upon stimulation when compared with wt or single transgenic controls.28 Next to differences in mRNA knockdown efficacy reported to be higher in the Vav-tTA-driven system in the spleen,28 another possibility may be that transactivator expression may impair B-cell fitness.28 This is supported by the observation here that B cells from DTrRen and DTrA1 mice proliferated less than wt-derived B cells after IgM ligation. Alternatively, we can only speculate that the knockdown achieved in surviving DTrA1-derived B cells is not breaching a critical threshold, allowing their normal proliferation in culture, whereas this threshold may have been exceeded in the other model, causing apoptosis.28
Taken together, our analyses using constitutive and conditional knockdown systems to study A1 physiology (this study and Oberdoerffer et al.27) strongly support a key role for this Bcl2 family member in B-cell survival and identifies, upon extension, A1 as a putative drug target for the treatment of B-cell-associated pathologies in humans. Hence, the A1/BIM axis may constitute a critical target for pharmacological compounds interfering with BCR signaling, such as SYK and BTK inhibitors.
Materials and Methods
Generation of transgenic mice
Animal experiments were performed in accordance with Austrian legislation (codes: 66-011/0058-II/3b/2011 and 66011/0096-WF/V/3b/2015). The generation and genotyping of CAG-rtTA, TRE-A1, TRE-Ren, Bim−/−, Puma−/−, Noxa−/−, Bad−/− mice have been described.28, 30, 31, 49–51 Doxycycline was administered in food pellets (625 mg/kg, Sniff) over a period of up to 17 days.
Cell culture and reagents
Cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. WEHI-231 cells were grown in the DME-medium (Sigma-Aldrich, Dorset, UK; D5671) supplemented with 10% FCS (Sigma-Aldrich, F7524), 250 μM l-glutamine (PAA Laboratories, Pasching, Austria; M11-004), 100 U/ml penicillin and 100 μg/ml streptomycin (PAA Laboratories, P11-010) and 50 μM 2-mercaptoethanol (Applicam, Darmstadt, Germany). Primary hematopoietic cells were cultured in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% FCS, 250 μM l-glutamine, 50 μM 2-mercaptoethanol, 1 mM sodium pyruvate (Gibco, Waltham, MA, USA), 100 μM non-essential amino acids (Gibco) and antibiotics.
Immunoblotting
Cell lysates were prepared in CHAPS-containing lysis buffer and analyzed by immunoblotting as described.28 For detection of proteins by chemoluminescence (Advansta, Menlo Park, CA, USA; K-12049-D50), a rat anti-mA1 mAb, clone 6D6-1-1,52 rat anti-Bim, clone 3C5 (Enzo, Lausen, Switzerland), rabbit anti-BclxL (Cell Signaling, Beverly, MA, USA; cs 2764 S), goat anti-Diva/Bcl2L10 (Santa Cruz, Dallas, TX, USA; sc8739), mouse anti-Bcl2 (BioLegend, London, UK; 633502), rabbit anti-Mcl1 (Rockland, Hamburg, Germany; 600-401-394) or a rabbit anti-GAPDH mAb (Cell Signaling, 2118, 1:5000) were used. Goat anti-rabbit Ig/HRP (Dako, Glostrup, Denmark; P0448), rabbit anti-goat Ig/HRP (Dako, P0449), goat anti-mouse heavy chain IgG1-HRP (Abcam, Cambridge, UK; ab98693) or rabbit anti rat-IgG heavy chain-HRP (Cell Signaling, 7077) were used as secondary reagents.
Immunofluorescence analysis, cell sorting and antibodies
The monoclonal antibodies used were purchased from eBioscience (San Diego, CA, USA), BioLegend or BD (San Diego, CA, USA) and are described in the Supplementary Information Online. Flow cytometric analysis was performed on an LSR-Fortessa or a FACS-Aria-III (both BD).
RNA isolation and quantitative real-time PCR (qRT-PCR) analysis
RNA was isolated using TRIzol (Invitrogen, Waltham, MA, USA). cDNA synthesis was performed using Omniscript-RT-Kit (Qiagen, Germantown, MD, USA) and 1 μg total RNA pretreated with 1 U/μl RQ1 DNase (Promega, Madison, WI, USA). qRT-PCR conditions: 95 °C for 7 min, 40 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s, using DyNAmo Flash SYBR Green (ThermoScientific, Waltham, MA, USA). Reactions were performed in duplicates. Quantification was performed using the ΔΔCt-method.
Survival and proliferation assays
Splenocytes were stimulated with 100 U/ml of mIL-2, 10 ng/ml mIL-4, 10 ng/ml mIL-5 (all PeproTech, Rocky Hill, NJ, USA) and 1 μg/ml goat anti-mouse IgM F(ab')2 fragments (Dianova, Hamburg, Germany), 1 μg/ml hamster anti-mouse CD40 mAb (BioLegend or BD) or 20 μg/ml LPS (Sigma-Aldrich, L2880) in the presence of 1 μg/ml doxycycline (Sigma-Aldrich, D9891). To induce T-cell proliferation, splenocytes were stimulated in the presence of doxycycline with 1 μg/ml Concanavalin A (Sigma-Aldrich) or plate-bound 5 μg/ml hamster anti-mouse CD3 mAb (clone 2C11, BioLegend) plus 1 μg/ml anti-mouse CD28 mAb (clone 37.51 BioLegend). To assess proliferation, cells were labeled with 5 μM CPD eFluor 670 according to the user manual (eBioscience, 65-0840-90). For the induction of cell death, plate-bound goat anti-mouse IgM F(ab')2 fragments (Dianova) was used. BTK inhibitor was applied at 5 μM (Ibrutinib, Selleckchem, Munich, Germany), IKK inhibitor at 10 μM (PS-1145, Sigma-Aldrich) and SYK inhibitor (R406, Selleckchem) at 2 or 5 μM. The percentage of viable cells was determined using DAPI (Sigma-Aldrich) or Annexin-V-Pacific Blue (eBioscience, 88-8006-74).
Colony-forming cell assay
Colony formation of bone marrow-resident progenitor cells was assessed using 3 × 104 or 6 × 104 cells to expand myeloid or pre-B progenitors, respectively, in MethoCult (M03234, M3630, Stem Cells Technologies, Grenoble, France), supplemented with IL-7, G-CSF or M-CSF (10 ng/ml) immediately or with a 24-h delay in the presence of 1 μg/ml doxycycline.
Statistical analysis
Statistical analysis was performed using ANOVA followed by Bonferoni post-hoc test, applying the PRISM Graphpad software (La Jolla, CA, USA). P-values <0.05 were considered to indicate statistically significant differences.
Acknowledgments
We are grateful to K Rossi, C Soratroi and I Gaggl for an excellent technical assistance and Bernd Rieder for animal care as well as to V Labi for help with B-cell analysis and discussion. We thank J Zuber and S Lowe for TRE-Ren mice. This work was supported by grants from the Austrian Science Fund (FWF), grant I1298 (FOR-2036) and the MCBO Doctoral College ‘Molecular Cell Biology and Oncology' (W1101) and the ‘Österreichische Krebshilfe Tirol'. MH is supported by the National Health and Medical Research Council, Australia project grant APP1049720. ST is supported by a Doc-fellowship from the Austrian Academy of Science (ÖAW).
Author contributions
MS performed experiments, analyzed data, contributed to writing and prepared figures; ST and EO performed experiments, analyzed data; SH contributed valuable reagents, analyzed data and edited manuscript; MH provided mice and reagents, edited paper; AV designed research, analyzed data, wrote paper, conceived study.
Glossary
- Btk
Bruton's tyrosine kinase
- Syk
spleen tyrosine kinase
- Bad
Bcl-2 antagonist of cell death
- Bcl-X
Bcl-2-related protein X
- Bcl-2
B-cell lymphoma 2
- BH
Bcl-2 homology domain
- Bim
Bcl-2 interacting mediator of cell death
- PUMA
p53-upregulated modulator of apoptosis
- CLL
chronic lymphocytic leukemia
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by JM Hardwick
Supplementary Material
References
- Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 2014; 15: 49–63. [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondrial regulation of cell death. Cold Spring Harb Perspect Biol 2013; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochalska M, Tuzlak S, Egle A, Villunger A. Lessons from gain- and loss-of-function models of pro-survival Bcl2 family proteins: implications for targeted therapy. FEBS J 2015; 282: 834–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano F, Krajewska M, Ortiz-Rubio P, Krajewski S, Zhai D, Faustin B et al. Nur77 converts phenotype of Bcl-B, an antiapoptotic protein expressed in plasma cells and myeloma. Blood 2007; 109: 3849–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino R, Ding L, Veis DJ, Korsmeyer SJ, Nunez G. Developmental regulation of the Bcl-2 protein and susceptibility to cell death in B lymphocytes. EMBO J 1994; 13: 683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nakayama K, Negishi I, Kuida K, Shinkai Y, Louie MC et al. Disappearance of the lymphoid system in Bcl-2 homozygous mutant chimeric mice. Science 1993; 261: 1584–1588. [DOI] [PubMed] [Google Scholar]
- Veis DJ, Sorenson CM, Shutter JR, Korsmeyer SJ. Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys, and hypopigmented hair. Cell 1993; 75: 229–240. [DOI] [PubMed] [Google Scholar]
- Grillot DA, Merino R, Nunez G. Bcl-XL displays restricted distribution during T cell development and inhibits multiple forms of apoptosis but not clonal deletion in transgenic mice. J Exp Med 1995; 182: 1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhagalov I, Dunkle A, He YW. The anti-apoptotic Bcl-2 family member Mcl-1 promotes T lymphocyte survival at multiple stages. J Immunol 2008; 181: 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT. Life and death during hematopoietic differentiation. Curr Opin Immunol 2007; 19: 497–502. [DOI] [PubMed] [Google Scholar]
- Lin EY, Orlofsky A, Berger MS, Prystowsky MB. Characterization of A1, a novel hemopoietic-specific early-response gene with sequence similarity to bcl-2. J Immunol 1993; 151: 1979–1988. [PubMed] [Google Scholar]
- Herold MJ, Zeitz J, Pelzer C, Kraus C, Peters A, Wohlleben G et al. The stability and anti-apoptotic function of A1 are controlled by its C terminus. J Biol Chem 2006; 281: 13663–13671. [DOI] [PubMed] [Google Scholar]
- Chuang PI, Yee E, Karsan A, Winn RK, Harlan JM. A1 is a constitutive and inducible Bcl-2 homologue in mature human neutrophils. Biochem Biophys Res Commun 1998; 249: 361–365. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Ahmed AA, Moller C, Nakayama K, Hatakeyama S, Nilsson G. Essential role of the prosurvival bcl-2 homologue A1 in mast cell survival after allergic activation. J Exp Med 2001; 194: 1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottina E, Lyberg K, Sochalska M, Villunger A, Nilsson GP. Knockdown of the antiapoptotic bcl-2 family member a1/bfl-1 protects mice from anaphylaxis. J Immunol 2015; 194: 1316–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekoff M, Lyberg K, Krajewska M, Arvidsson M, Rak S, Reed JC et al. Anti-apoptotic BFL-1 is the major effector in activation-induced human mast cell survival. PLoS One 2012; 7: e39117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschelde C, Walzer T, Galia P, Biémont MC, Quemeneur L, Revillard JP et al. A1/Bfl-1 expression is restricted to TCR engagement in T lymphocytes. Cell Death Differ 2003; 10: 1059–1067. [DOI] [PubMed] [Google Scholar]
- Trescol-Biemont MC, Verschelde C, Cottalorda A, Bonnefoy-Berard N. Regulation of A1/Bfl-1 expression in peripheral splenic B-cells. Biochimie 2004; 86: 287–294. [DOI] [PubMed] [Google Scholar]
- Wen R, Chen Y, Xue L, Schuman J, Yang S, Morris SW et al. Phospholipase Cgamma2 provides survival signals via Bcl2 and A1 in different subpopulations of B-cells. J Biol Chem 2003; 278: 43654–43662. [DOI] [PubMed] [Google Scholar]
- Gonzalez J, Orlofsky A, Prystowsky MB. A1 is a growth-permissive antiapoptotic factor mediating postactivation survival in T cells. Blood 2003; 101: 2679–2685. [DOI] [PubMed] [Google Scholar]
- Ottina E, Tischner D, Herold MJ, Villunger A. A1/Bfl-1 in leukocyte development and cell death. Exp Cell Res 2012; 318: 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Echeverz J, Haile LA, Zhao F, Gamrekelashvili J, Ma C, Métais JY et al. IFN-gamma regulates survival and function of tumor-induced CD11b+ Gr-1high myeloid derived suppressor cells by modulating the anti-apoptotic molecule Bcl2a1. Eur J Immunol 2014; 44: 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler M. BCL2A1: the underdog in the BCL2 family. Cell Death Differ 2012; 19: 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler M, Butterworth M, Majid A, Walewska RJ, Sun XM, Dyer MJ et al. Concurrent up-regulation of BCL-XL and BCL2A1 induces approximately 1000-fold resistance to ABT-737 in chronic lymphocytic leukemia. Blood 2009; 113: 4403–4413. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Hamasaki A, Negishi I, Loh DY, Sendo F, Nakayama K et al. Multiple gene duplication and expression of mouse bcl-2-related genes, A1. Int Immunol 1998; 10: 631–637. [DOI] [PubMed] [Google Scholar]
- Hamasaki A, Sendo F, Nakayama K, Ishida N, Negishi I, Nakayama K et al. Accelerated neutrophil apoptosis in mice lacking A1-a, a subtype of the bcl-2-related A1 gene. J Exp Med 1998; 188: 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Kanellopoulou C, Heissmeyer V, Paeper C, Borowski C, Aifantis I et al. Efficiency of RNA interference in the mouse hematopoietic system varies between cell types and developmental stages. Mol Cell Biol 2005; 25: 3896–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottina E, Grespi F, Tischner D, Soratroi C, Geley S, Ploner A et al. Targeting antiapoptotic A1/Bfl-1 by in vivo RNAi reveals multiple roles in leukocyte development in mice. Blood 2012; 119: 6032–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WI, Wiesner SM, Largaespada DA. Vav promoter-tTA conditional transgene expression system for hematopoietic cells drives high level expression in developing B and T cells. Exp Hematol 2007; 35: 1231–1239. [DOI] [PubMed] [Google Scholar]
- Takiguchi M, Dow LE, Prier JE, Carmichael CL, Kile BT, Turner SJ et al. Variability of inducible expression across the hematopoietic system of tetracycline transactivator transgenic mice. PLoS One 2013; 8: e54009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell 2011; 145: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss AW, Knodel M, Berberich-Siebelt F, Lindemann D, Schimpl A, Berberich I. A1 expression is stimulated by CD40 in B-cells and rescues WEHI 231 cells from anti-IgM-induced cell death. Eur J Immunol 1999; 29: 3077–3088. [DOI] [PubMed] [Google Scholar]
- Monroe JG, Bannish G, Fuentes-Panana EM, King LB, Sandel PC, Chung J et al. Positive and negative selection during B lymphocyte development. Immunol Res 2003; 27: 427–442. [DOI] [PubMed] [Google Scholar]
- Dow LE, Nasr Z, Saborowski M, Ebbesen SH, Manchado E, Tasdemir N et al. Conditional reverse tet-transactivator mouse strains for the efficient induction of TRE-regulated transgenes in mice. PLoS One 2014; 9: e95236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GJ, Cimmino L, Jude JG, Hu Y, Witkowski MT, McKenzie MD et al. Pax5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia. Genes Dev 2014; 28: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Rappaport AR, Kitzing T, Schultz N, Zhao Z et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell 2014; 25: 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P, Robati M, Bath M, Strasser A. Polycystic kidney disease prevented by transgenic RNA interference. Cell Death Differ 2005; 12: 831–833. [DOI] [PubMed] [Google Scholar]
- Mandal M, Borowski C, Palomero T, Ferrando AA, Oberdoerffer P, Meng F et al. The BCL2A1 gene as a pre-T cell receptor-induced regulator of thymocyte survival. J Exp Med 2005; 201: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Park SJ, Choi BK, Kim HH, Nam KO, Kwon BS. 4-1BB promotes the survival of CD8+ T lymphocytes by increasing expression of Bcl-xL and Bfl-1. J Immunol 2002; 169: 4882–4888. [DOI] [PubMed] [Google Scholar]
- Tomayko MM, Cancro MP. Long-lived B-cells are distinguished by elevated expression of A1. J Immunol 1998; 160: 107–111. [PubMed] [Google Scholar]
- Schiemann B, Gommerman JL, Vora K, Cachero TG, Shulga-Morskaya S, Dobles M et al. An essential role for BAFF in the normal development of B-cells through a BCMA-independent pathway. Science 2001; 293: 2111–2114. [DOI] [PubMed] [Google Scholar]
- Lentz VM, Cancro MP, Nashold FE, Hayes CE. Bcmd governs recruitment of new B-cells into the stable peripheral B-cell pool in the A/WySnJ mouse. J Immunol 1996; 157: 598–606. [PubMed] [Google Scholar]
- Kraus M, Pao LI, Reichlin A, Hu Y, Canono B, Cambier JC et al. Interference with immunoglobulin (Ig)alpha immunoreceptor tyrosine-based activation motif (ITAM) phosphorylation modulates or blocks B-cell development, depending on the availability of an Igbeta cytoplasmic tail. J Exp Med 2001; 194: 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Kurosaki T. Contribution of BCAP to maintenance of mature B-cells through c-Rel. Nat Immunol 2003; 4: 780–786. [DOI] [PubMed] [Google Scholar]
- Grumont RJ, Rourke IJ, O'Reilly LA, Strasser A, Miyake K, Sha W et al. B lymphocytes differentially use the Rel and nuclear factor kappaB1 (NF-kappaB1) transcription factors to regulate cell cycle progression and apoptosis in quiescent and mitogen-activated cells. J Exp Med 1998; 187: 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol 2009; 9: 195–205. [DOI] [PubMed] [Google Scholar]
- Goodnow CC, Crosbie J, Adelstein S, Lavoie TB, Smith-Gill SJ, Brink RA et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature 1988; 334: 676–682. [DOI] [PubMed] [Google Scholar]
- Mackay F, Woodcock SA, Lawton P, Ambrose C, Baetscher M, Schneider P et al. Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. J Exp Med 1999; 190: 1697–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Köntgen F et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 1999; 286: 1735–1738. [DOI] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Müllauer F, Böck G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036–1038. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Zha J, Harada H, Datta SR, Danial NN, Gilmore AP et al. Bad-deficient mice develop diffuse large B-cell lymphoma. Proc Natl Acad Sci USA 2003; 100: 9324–9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang MJ, Brennan MS, O'Reilly LA, Ottina E, Czabotar PE, Whitlock E et al. Characterisation of a novel A1-specific monoclonal antibody. Cell Death Dis 2014; 5: e1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.