Abstract
The involvement of the nicotinamide adenine dinucleotide (NAD+) salvage pathway in cancer cell survival is poorly understood. Here we show that the NAD+ salvage pathway modulates cancer cell survival through the rarely mutated tumour suppressor p73. Our data show that pharmacological inhibition or knockdown of nicotinamide phosphoribosyltransferase (NAMPT), a rate-limiting enzyme in the NAD+ salvage pathway, enhances autophagy and decreases survival of cancer cells in a p53-independent manner. Such NAMPT inhibition stabilizes p73 independently of p53 through increased acetylation and decreased ubiquitination, resulting in enhanced autophagy and cell death. These effects of NAMPT inhibition can be effectively reversed using nicotinamide mononucleotide (NMN), the enzymatic product of NAMPT. Similarly, knockdown of p73 also decreases NAMPT inhibition-induced autophagy and cell death, whereas overexpression of p73 alone enhances these effects. We show that the breast cancer cell lines (MCF-7, MDA-MB-231 and MDA-MB-468) harbour significantly higher levels of NAMPT and lower levels of p73 than does the normal cell line (MCF-10A), and that NAMPT inhibition is cytotoxic exclusively to the cancer cells. Furthermore, data from 176 breast cancer patients demonstrate that higher levels of NAMPT and lower levels of p73 correlate with poorer patient survival, and that high-grade tumours have significantly higher NAMPT/p73 mRNA ratios. Therefore, the inverse relationship between NAMPT and p73 demonstrable in vitro is also reflected from the clinical data. Taken together, our studies reveal a new NAMPT-p73 nexus that likely has important implications for cancer diagnosis, prognosis and treatment.
Tumour suppressor genes encode various proteins that inhibit tumour formation, and inactivation of this inhibitory function can lead to the development of cancer.1 Of the numerous tumour suppressor genes identified to date, p53 is by far the most extensively studied as it is often found to be absent or mutated in cancer.2, 3 In view of its central role in cellular homeostasis, p53 has been considered a promising cancer therapeutic target. Unfortunately, p53 is often lost, mutated or inactivated in the majority of human cancers, making it unavailable for therapeutic modulation.4, 5 However, recent evidence shows that this problem can be circumvented, at least partially, by targeting another member of the p53 family, the tumour suppressor p73.6, 7 p73 shares some overlapping functions with p53.7 However, unlike p53, mutations in p73 are very rare in the majority of cancer types.8, 9 These characteristics have made p73 an attractive/alternative target for anticancer therapy.
In considering the possibility of exploiting p73's tumour suppressing potential, it is important to understand the various mechanisms involved in its activation. In the case of p53, acetylation is a most important post-translational modification involved in its activation.10, 11 A major regulator of p53 acetylation is a group of Class III histone deacetylases known as sirtuins (SIRTs).11, 12 SIRT-mediated deacetylation causes inactivation of a variety of proteins including tumour suppressor proteins.13 As SIRTs require nicotinamide adenine dinucleotide (NAD+) as a cofactor to function as deacetylases, parameters that affect cellular NAD+ levels should theoretically also impact the functions of those proteins regulated by SIRTs, including p73.
NAD+ has central roles in a variety of biological processes ranging from cellular metabolism and energy production to the regulation of enzymatic activities.14 Accumulating evidence reveals that NAD+ serves as a substrate for non-redox reactions, which are essential for diverse biological functions such as gene expression, Ca2+ mobilization and cell death. Importantly, as mentioned above, NAD+ serves as a key cofactor of SIRTs in removing the acetyl moiety from various proteins, and hence has a crucial role in regulating their activities.15 In view of the multifaceted roles NAD+ has in mediating various important biological functions, it is not surprising that cancer cells generally display aberrant NAD+ metabolism.15, 16 A first consideration in this regard is the Warburg effect, which relates to cancer cells' dependence on cytoplasmic aerobic glycolysis for energy production.17 The high rates of aerobic glycolysis perturb NAD+ metabolism, altering the NADH/NAD+ redox ratio, thereby disrupting the cellular redox homeostasis and further promoting cancer.17 The synthesis of NAD+ is in turn catalysed by two major metabolic pathways: the de novo pathway from tryptophan, and the salvage pathway that involves biosynthesis from other NAD+ precursors.18 The synthesis of NAD+ via the salvage pathway is conducted by precursor enzymes NAMPT (nicotinamide phosphoribosyltransferase) and NMNAT-1, -2 and -3 (nicotinamide mononucleotide adenylyltransferase isoforms 1, 2 and 3).16 Hence, we hypothesize that the SIRTs and NAMPT (the first enzyme involved in NAD+ synthesis) have critical roles in p73 activation, and that their modulation can impact p73-mediated tumour suppression in cancer. Our data show that NAMPT negatively regulates p73 levels independent of p53, thereby modulating autophagy-mediated cell death. The inverse relationship between NAMPT and p73 levels was also evident from breast cancer patient survival analysis, where high levels of NAMPT, and low levels of p73, correlate with poorer patient survival, further underscoring the clinical significance of the NAMPT-p73 nexus in dictating cancer outcome.
Results
NAMPT inhibition or KD reduces viability of cancer cells independent of p53 status
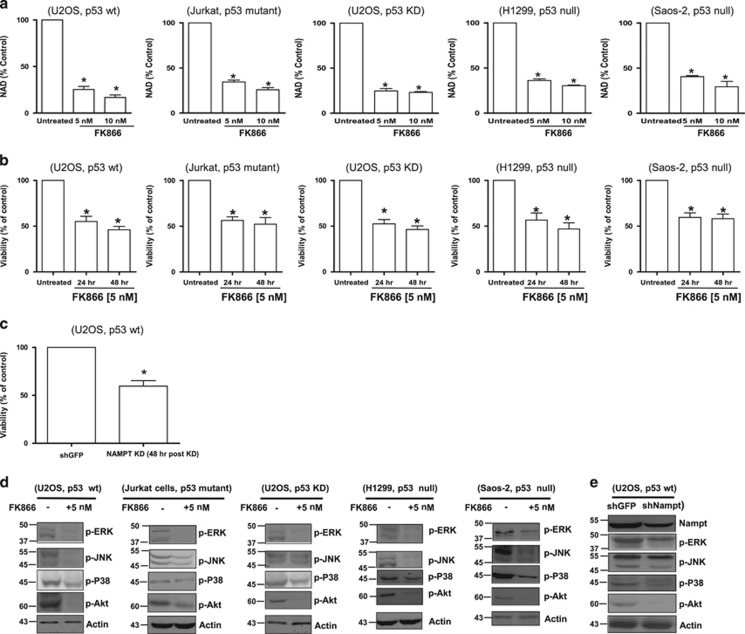
It was previously shown that blockage of the NAD+ salvage pathways using the NAMPT inhibitor FK866 induces autophagic death in neuroblastoma SH-SY5Y cells and multiple myeloma cells.19, 20, 21 Recent data show that FK866 activates AMPK (adenosine monophosphate-activated protein kinase) and downregulates mammalian target of rapamycin (mTOR) signalling in hepatocarcinoma cells.22 In view of the important role NAD+ has in modulating p53 activity (via NAD+-dependent SIRTs) 23, 24 and our recent demonstration that the NAD+ synthesizing enzyme NMNAT2 is a downstream target of p53,16 we wished to first determine if FK866-induced cell death was p53-dependent. To this end, U2OS (p53 wt and p53 knockdown (KD)), Jurkat (p53 mutant), H1299 (p53 null) and Saos2 cells (p53 null) were exposed to FK866 (inhibitor of NAMPT) doses from 0 to 10 nM, and analysed for NAD+/NADH levels and cell viability (by Trypan blue exclusion) at various times post-treatment. The results showed that FK866 effectively inhibited NAD+ production (Figure 1a), and caused significant decrease in cell viability (Figure 1b), independent of the p53 status of the cells. Furthermore, specific KD of NAMPT using shRNA also significantly reduced the viability of U2OS cells (Figure 1c). In all cases, the decrease in cell viability following the inhibition (Figure 1d) or KD (Figure 1e) of NAMPT was accompanied by a marked decrease in the activation of mitogen-activated protein kinases (p-ERK, p-JNK, p-P38) and p-AKT, suggesting a crucial role for NAMPT in promoting cell survival and proliferation. Interestingly, the lack of annexin V staining, the absence of cleaved caspase-3 following FK866 treatment (as opposed to actinomycin D treatment) and the inability of necrostatin (50 μM) to prevent FK866-mediated effect on cell viability (Supplementary Figure S1) suggested that the decrease in cell viability was likely through an alternative cell death or growth arrest pathway rather than apoptosis or necroptosis, as reported previously.19 Collectively, these data demonstrate that inhibition of NAMPT causes reduced viability of cancer cells regardless of their p53 status.
Figure 1.
Pharmacological inhibition of NAMPT (by FK866) or NAMPT KD reduces NAD+ levels and cell viability (cell count), independent of p53 status. (a) U2OS (p53 wild-type (wt) and p53 KD), Jurkat, H1299 and Saos2 cells were treated with 5 or 10 nM of FK866 for 48 h. Intracellular levels of NAD+ were measured using the NAD+/NADH Quantification Kit. (b and c) Cells treated with FK866 (5 nM) for 24 or 48 h were stained with Trypan blue and counted to determine the percentage of viable cells. Results represent three independent experiments. Statistical analysis was performed with one-way analysis of variance (ANOVA) followed by paired ‘t'-test; *P-values ⩽0.05 obtained by comparing the respective data with the untreated control. (d) Western blot analysis of lysates from various cells exposed to FK866 for 48 h or (e) from NAMPT KD cells showing levels of p-ERK, p-JNK, p-P38 and p-Akt
Cell death following NAMPT inhibition or KD is partially reversed by inhibiting the autophagy machinery
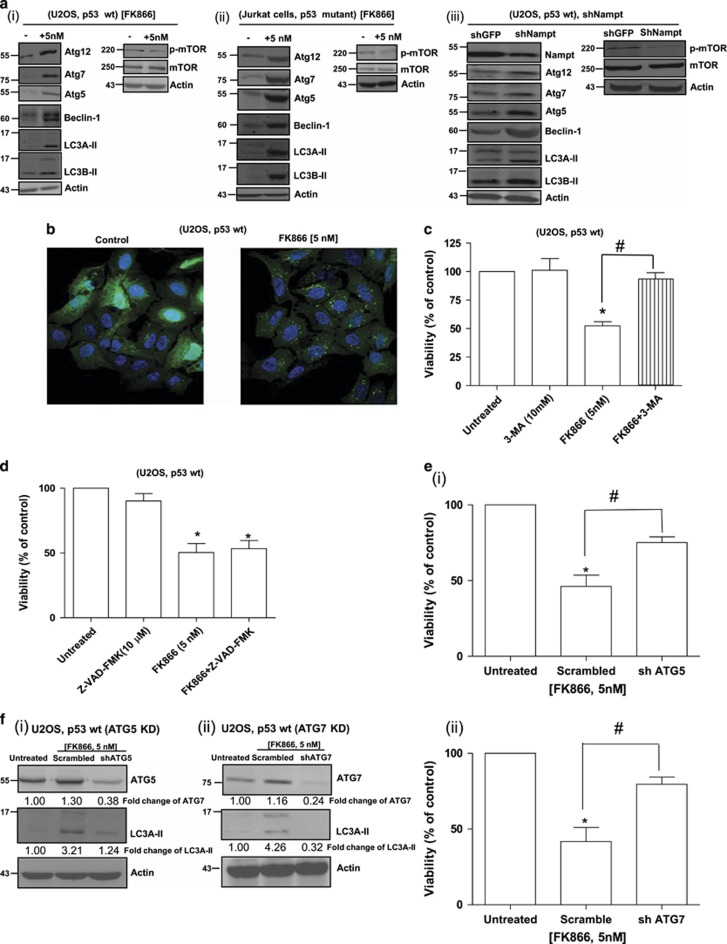
We then proceeded to examine the possible involvement of the autophagy machinery in NAMPT inhibition- or KD-mediated cell death. Our results showed that NAMPT inhibition or KD significantly stimulated autophagy in U2OS (wt p53) and Jurkat cells (mutant p53) as evidenced by increased levels of autophagy-related proteins including Atg5, Atg7, Atg12, Beclin 1, LC3A-II and LC3B-II, and decreased levels of p-mTOR, a major negative regulator of autophagy (Figure 2a). Induction of autophagy by NAMPT inhibition was further confirmed by the presence of LC3B puncta in FK-866-treated cells (Figure 2b). To determine if autophagy induced by NAMPT inhibition, or KD-mediated cell viability under these conditions, cells were treated either with 3-methyladenine (3-MA, 10 mM), which blocks autophagy, or with Z-VAD-FMK (10 μM), which blocks apoptosis. Figure 2c shows that pre-treatment of the cells with 3-MA very effectively reversed the growth-inhibiting effect of FK866, whereas Z-VAD-FMK (Figure 2d) was unable to do so. FK866-induced cell death could also be partially reversed by knocking down Atg5 or Atg7 (Figure 2e) that also reduced the levels of autophagosomal marker LC3A-II (Figure 2f). Taken together, our results suggest that cell death induced by the inhibition of NAMPT is at least partially mediated via autophagy.
Figure 2.
Cell death by NAMPT inhibition or KD is linked to activation of the autophagy machinery. (a) Lysates from FK866-treated or NAMPT KD cells were subjected to western blot analysis using antibodies against Atg12, Atg7, Atg5, Beclin 1, LC3A, LC3B, mTOR and p-mTOR. (b) U2OS cells stably transfected with GFP-LC3 were exposed to FK866 (5 nM) for 48 h and examined by microscopy for the presence of GFP-LC3 puncta. (c and d) Effect of 3-MA (10 mM) and Z-VAD-FMK (10 μM) pre-treatment (2 h) on FK866-induced decrease in viability (based on Trypan blue staining) of U2OS cells. (e and f) U2OS cells (p53 wild-type (wt)) with Atg5 KD or Atg7 KD were exposed to FK866 (5 nM) for 48 h and analysed for (e) viability or (f) LC3A-II levels. Statistical analysis for the cell viability study was performed with one-way analysis of variance (ANOVA) followed by paired ‘t'-test; *, #P-values ⩽0.05 obtained by comparing the respective data with the corresponding controls. Results represent three independent experiments. sh, short hairpin
NAMPT inhibition or KD significantly enhances p73 levels by post-translational modification of p73
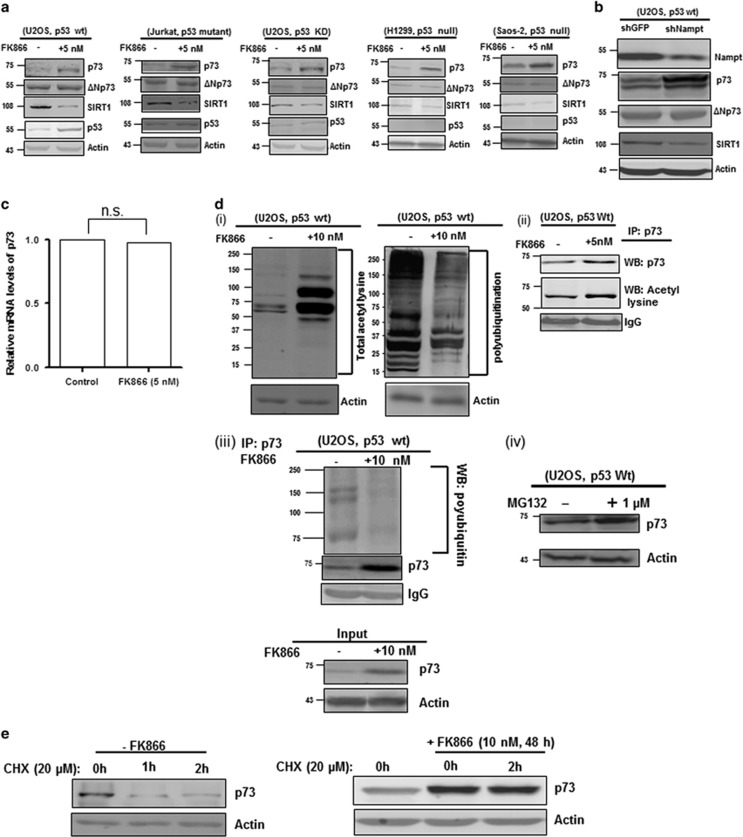
Our observation that cell death caused by NAMPT inhibition or KD was p53-independent led us to consider the possible involvement of other tumour suppressors in this process. A plausible candidate in this regard was p73, a homologue of p53 recently shown to regulate autophagy in hepatocytes.25 Strikingly, NAMPT inhibition or KD caused a marked increase in p73 levels in a p53-independent manner, concomitant with reduced SIRT1 levels (Figures 3a and b). In all cases, the levels of the oncogenic ΔNp73 isoform remained unchanged. As there was no change in p73 mRNA levels following FK866 treatment (Figure 3c), the observed increase in p73 protein levels was likely due to stabilization of p73 by post-translational modification. Indeed, FK866 treatment caused an increase in the overall acetylation and a decrease in the overall ubiquitination of cellular proteins (Figure 3di). Furthermore, immunoprecipitation of p73 followed by western blot using an anti-acetylated lysine antibody showed enhanced p73 acetylation after FK866 treatment (Figure 3dii). This increase in acetylation was coupled with a decrease in ubiquitination of p73 (Figure 3diii), which was expected as acetylation and ubiquitination often target the same lysine residues, resulting in opposite outcomes. The involvement of ubiquitin/proteasome-mediated p73 degradation under normal conditions was further confirmed by the demonstration that the proteasome inhibitor MG132 effectively elevated p73 levels (Figure 3div). To further examine whether p73 accumulation was due to protein stabilization, we measured p73 decay with and without FK866 treatment. U2OS cells were treated with FK866 for 48 h, and then cycloheximide (CHX) was added to inhibit protein synthesis, and cells were collected at the indicated time points and analysed for p73 by western blot (Figure 3e). Remarkably, FK866 treatment resulted in a marked increase in p73 stability, as compared with untreated cells. Collectively, these results show that an inverse relationship exists between NAMPT and p73, and that inhibition of NAMPT increases the stability of p73 through enhanced acetylation and decreased ubiquitination.
Figure 3.
NAMPT inhibition or KD results in increased p73 levels independent of p53. (a and b) Lysates from FK866-treated (a) or NAMPT KD (b) cells were subjected to western blot (WB) analysis for p73, ΔNp73, SIRT1, p53 and actin. (c) Relative expression levels of p73 mRNA from untreated (control) and FK866-treated U2OS cells were quantified by quantitative PCR (Q-PCR). Results represent three independent experiments. (di) Western blot analysis of overall acetylation (left panel) and ubiquitination (right panel) in FK866-treated U2OS cells. (ii and iii) Lysates from FK866-treated cells were subjected to immunoprecipitation (IP) using an anti-p73 antibody followed by western blot analysis for protein acetylation (ii) and ubiquitination (iii). (iv) Western blot showing the effect of MG132 treatment (1 μM for 24 h) on p73 levels in U2OS cells. (e) FK866 enhances p73 stability. U2OS cells were treated with FK866 for 48 h and then CHX (20 μM) was added. Cells were collected at different time points and analysed by immunoblotting to measure p73 protein levels. Wt, wild type; NS, nonsignificant
NAMPT mediates autophagy and cell viability via p73
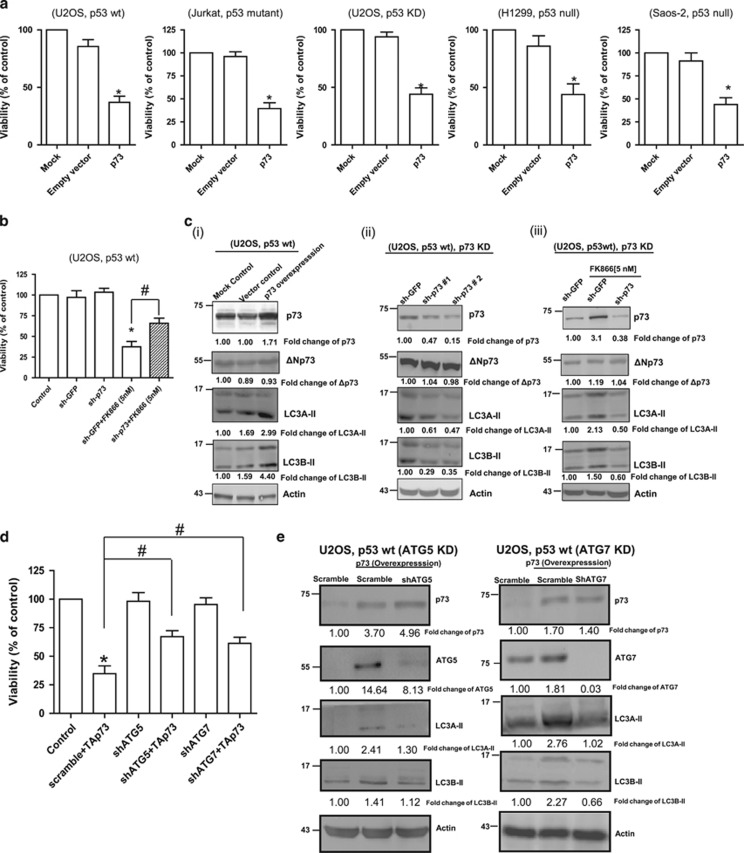
To determine if increased levels of p73 as a result of NAMPT inhibition or KD was accountable, at least partially, for the observed enhanced autophagy and decreased cell viability, we carried out transient overexpression of p73 in U2OS (p53 wt and p53 KD), Jurkat (p53 mutant), H1299 (p53 null) and Saos2 cells (p53 null) cells. In all cases, p73 overexpression alone resulted in significant decrease in cell viability, regardless of the p53 status of the cells (Figure 4a). Conversely, p73 KD significantly reversed the growth-inhibiting effect of FK866 (Figure 4b). The proautophagy nature of p73 was also evident from our demonstration that transient overexpression of p73 in U2OS cells resulted in increased levels of autophagosomal markers LC3A-II and L3CB-II (Figure 4ci), whereas p73 KD in these cells (both untreated and FK866-treated), showed reduced levels of these proteins (Figure 4cii and iii). The role of autophagy in p73-mediated cell death was further confirmed by the demonstration that KD of ATG5 or ATG7 significantly reversed the effect of p73 overexpression on cell viability (Figure 4d), as well as p73-mediated autophagy (Figure 4e). Taken together, these results provide strong evidence that NAMPT regulates cell survival and autophagy, at least partially, via p73, and also highlight the proautophagic nature of p73.
Figure 4.
Role of p73 in NAMPT inhibition- or NAMPT KD-mediated effect on autophagy and cell viability (cell count). (a) The various cell lines were transfected with p73 and cell viability was determined after 48 h using Trypan blue staining. (b) U2OS cells were transfected with either control (sh-GFP) or p73-specific shRNA (sh-p73), cultured in the presence or absence of FK866, and then analysed for cell viability after 48 h. (c) U2OS cells overexpressing p73 (i) or with p73 KD (ii) were analysed for basal levels of LC3A-II and LC3B-II; (iii) U2OS cells with control (sh-GFP) or p73 (sh-p73) KD were exposed to FK866 for 48 h and analysed for LC3A-II and LC3B-II. (d and e) Transient overexpression of p73 (for 48 h) was carried out in U2OS cells with or without ATG5 KD or ATG7 KD. Cell were then analysed for (d) viability, and (e) the levels of LC3A-II and LC3B-II. Statistical analysis for the cell viability study was performed with one-way analysis of variance (ANOVA) followed by paired ‘t'-test; *, #P-values ⩽0.05 obtained by comparing the respective data with the control. Results represent three independent experiments. sh, short hairpin; Wt, wild type
NAMPT inhibition-related effects (decrease in cell viability, induction of autophagy and enhanced stability of p73) are NMN-dependent
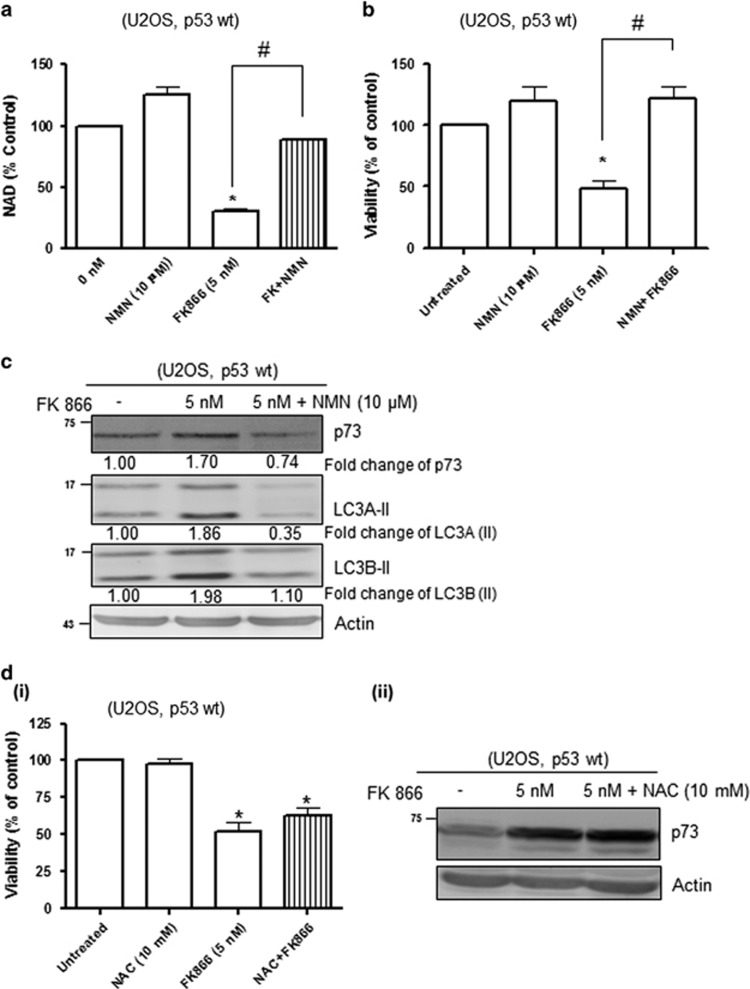
The enzymatic product of NAMPT is nicotinamide mononucleotide (NMN). To determine if the observed effects of NAMPT inhibition (decrease in cell viability, induction of autophagy and increased p73 stability) were because of NMN depletion, NMN was added to the cells exogenously during FK866 treatment, and cell viability, autophagy and p73 levels were measured after 24 h. The results show that NMN effectively reversed the effects of FK866 on NAD+ levels (Figure 5a), cell viability (Figure 5b), autophagy marker (LC3A-II and LC3B-II) protein levels and p73 stability (Figure 5c). These results therefore lend strong support to the notion that the NAD+ salvage pathway regulates cell viability via p73.
Figure 5.
NMN (the enzymatic product of NAMPT) reverses the effect of FK866 on cell viability (cell count) and levels of p73 and autophagy markers LC3A-II and LC3B-II. U2OS cells were cultured in medium containing FK866, or NMN, or FK866+NMN for 48 h, and analysed for (a) intracellular NAD+ levels, (b) cell viability and (c) levels of LC3A-II and LC3B-II. (d) U2OS cells were cultured in medium containing FK866, or NAC, or FK866+NAC for 48 h, and analysed for (i) cell viability (ii) and levels of p73. Results shown in (a–c) represent three independent experiments; *, #P-values ⩽0.05 obtained by comparing the respective data with the untreated and FK866 only control, respectively
To determine if the observed effects of NAMPT inhibition or KD could be because of increased production of reactive oxygen species (ROS), we examined the effects of N-acetylcysteine (NAC, 10 mM), a potent inhibitor of ROS, on FK866-induced cell death and p73 stabilization. The results (Figure 5di and ii) show that NAC slightly reversed the effect of FK866 on cell viability but not p73 levels. It thus appears that FK866 treatment does increase the production of ROS but this amount of ROS is not sufficient to affect cell viability or p73 levels.
The inverse relationship between NAMPT and p73 is also reflected in breast cancer patient survival outcome
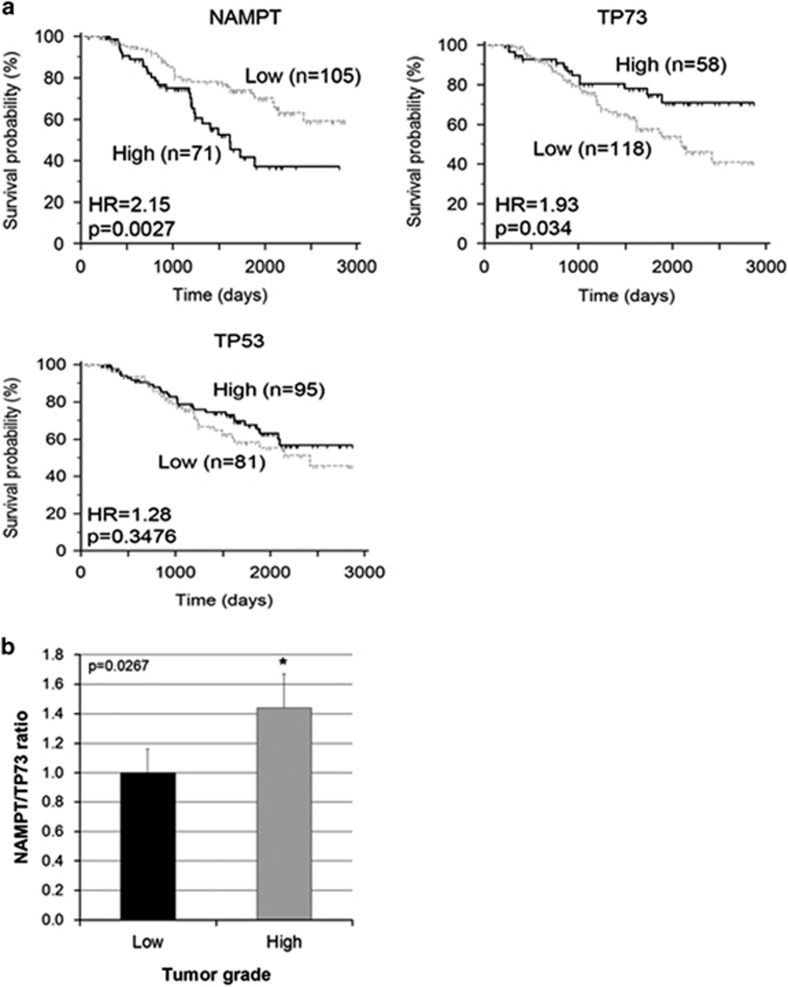
To investigate whether the NAMPT-p73 connection had any clinical relevance, we examined NAMPT and p73 expression levels in breast cancer biopsy tissues from 176 breast cancer patients using gene profiling, and correlated them with patient survival. Strikingly, the data show that high levels of NAMPT are associated with lower patient survival probability, whereas high levels of p73 (but not p53) significantly correlate with better patient prognosis (Figure 6a). The inverse relationship between NAMPT and p73 observed in vitro is therefore mirrored by patient outcome data. Furthermore, the NAMPT/p73 mRNA ratios for individual tumours were calculated and found to correlate with tumour grade, being significantly higher in high-grade tumours compared with low-grade tumours (Figure 6b). These findings highlight the clinical implications of the inverse relationship between NAMPT and p73.
Figure 6.
Inverse relationship between NAMPT and p73 from breast cancer patient outcome analysis. NAMPT and p73 expression levels in breast cancer biopsy tissues from 176 breast cancer patients are correlated with patient survival and tumour grade. (a) High levels NAMPT or low levels of p73, but not p53, correlate with poorer patient survival. (b) NAMPT/p73 ratio was calculated for each tumour sample and a t-test was performed between sub-populations with low and high histological tumour grade. *P-values ⩽0.05 compared with low grade tumors. HR, hazard ratio
Human breast cancer cell lines possess high NAMPT and low p73 levels
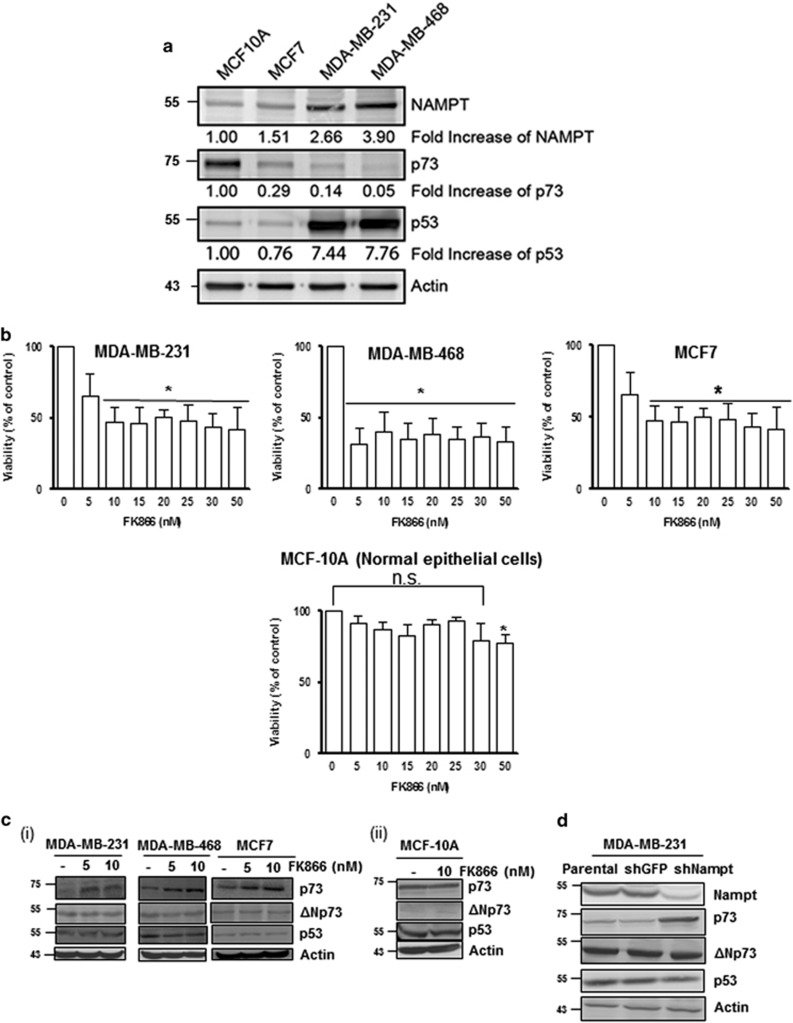
To further support and extend our breast cancer patient outcome data, we compared the basal protein levels of NAMPT and p73 in breast cancer (MDA-MB-231, MDA-MB-468, MCF-7) and normal, non-transformed breast cell lines (MCF-10A). Our results showed that compared with the non-transformed MCF-10A cell line, the three breast cancer cell lines possessed higher NAMPT levels (significantly higher in MDA-MB-231 and MDA-MB-468), but much lower p73 levels (Figure 7a). This again confirms the inverse relationship between NAMPT and p73, and suggests that reduced levels of p73 may be linked to the transformed state of the cancer cell lines.
Figure 7.
Inverse relationship between NAMPT and p73 levels in breast cancer cell lines. (a) Comparison of the basal levels of NAMPT in breast cancer cell lines (MCF-7, MDA-MB-231 and MDA-MB-468) and the normal, non-transformed cell line MCF-10A. (b) The above cell lines were exposed to FK866 for 24 h and analysed for cell count. Results represent three independent experiments; *P-values ⩽0.05 compared with the control. (c) Western blot analysis showing the effect of FK866 treatment (48 h) on p73 and ΔNp73 expression in breast cancer cells (i) and non-transformed cells (ii). (d) Effect of NAMPT KD on p73 levels in MDA-MB-231 cells. NS, nonsignificant
To address the possibility that high levels of NAMPT promotes breast cancer cell survival, we assessed the effect of FK866 on viability of the breast cancer cell lines. The results showed that FK866 significantly decreased the viability of breast cancer cell lines MDA-MB 231, MDA-MB-468 and MCF-7, but had little effect on the viability of normal, non-transformed breast cells (MCF-10A) (Figure 7b). These results show that NAMPT inhibition selectively reduces survival of breast cancer cells while sparing normal, non-transformed cells.
The inverse relationship between NAMPT and p73 observed with the three breast cancer cell lines then led us to investigate whether NAMPT negatively regulates p73 levels. Strikingly, inhibition of NAMPT (by FK866) or NAMPT KD increased p73 levels in all three cancer cell lines, but had no effect on p73 levels of the normal breast cell line MCF-10A (Figures 7c and d). Interestingly, the levels of p53 remained relatively unaffected by FK866 in all the cell lines. These results again demonstrate that NAMPT negatively regulates p73 levels and, coupled with the cell viability data, suggest that high levels of NAMPT in breast cancer cells suppress p73 (but not p53), thereby promoting the survival of breast cancer cells.
Discussion
NAD+ is an essential molecule involved in many important cellular processes ranging from energy transduction to various signalling events that regulate major biological functions including DNA repair, cell cycle progression, transcription control, calcium homeostasis, immune response, metabolism regulation, autophagy and cell death.26, 27, 28, 29, 30 Considering the multifaceted roles NAD+ has in maintaining cellular homeostasis, it is predictable that perturbations that affect NAD+ levels and/or distribution would lead to various diseases, including cancer. Additionally, recent evidence that NAD+ regulates p53 stability and activities via SIRTs while being back regulated by p53 via a feedback loop further attests to the intimate relationship between NAD+ metabolism and cancer control.16 The present study shows that the NAD+ salvage pathway, a crucial pathway for mammalian NAD+ metabolism, modulates cancer cell viability via the tumour suppressor p73.
As a key regulator of NAD+ biosynthesis, NAMPT has received considerable attention in recent years and is deemed to be a potential cancer therapeutic target.31 There is evidence that NAMPT activity promotes cell proliferation and survival following genotoxic stress, suggesting that it can function as an oncoprotein under certain conditions.32 Indeed, overexpression of NAMPT has been observed in many cancers, including colorectal cancer, breast cancer, gastric cancer, prostate cancer, ovarian cancer, oesophageal cancer and lymphoma.32 The progrowth and -survival nature of NAMPT has been attributed primarily to its function as an NAMPT, as FK866, a highly specific non-competitive inhibitor of NAMPT presently undergoing clinical trials, can effectively reduce the intracellular NAD+ pool, resulting in cell death.21, 33
A most important revelation from the present study pertains to the linkage between NAMPT and an important member of the p53 family, the tumour suppressor p73. We show that inhibition of NAMPT by FK866, which effectively induces autophagy and autophagy-mediated cell death, also causes significant increase in p73 levels, and this increase is reversible by the addition of NMN, the enzymatic product of NAMPT. FK866-induced increase in p73 levels is not because of increased p73 mRNA expression but rather because of enhanced stability of p73 as a result of increased p73 acetylation and reduced ubiquitination. Increased p73 acetylation is in turn due to reduced levels of NAD+-dependent deacetylases (SIRTs, particularly SIRT1) as a result of reduced NAD+ levels by FK866 treatment. The demonstration that similar results can be achieved by NAMPT KD confirms that NAMPT is specifically targeted by FK866. That inhibition of NAMPT (and hence the NAD+ salvage pathway) results in autophagy and cell death has recently been reported;20, 21, 22 however, the precise underlying mechanisms are unclear. Our results show that this is at least, in part, due to increased p73 levels as KD of p73 in FK866-treated cells effectively reverses the effects of FK866 on autophagy and cell survival. Furthermore, p73 overexpression alone is sufficient to promote autophagy and cell death. Therefore, p73 is the chief element downstream of the NAD+ salvage pathway that modulates such events, which, interestingly and importantly, can occur in the absence of p53. Our data also show that autophagy-mediated cell death via the NAMPT-p73 nexus is not due to apoptosis. Recently, Liu et al.34 reported the identification of a unique form of autophagy-dependent cell death, termed ‘autosis', which is distinct from apoptosis or necroptosis, and is regulated by Na+, K+-ATPase.34 Whether autophagy-mediated cell death via the NAMPT-p73 nexus is due to autosis remains to be seen.
The clinical relevance of our in vitro findings was also reflected from our breast cancer patient outcome analysis where poor prognosis is associated with higher levels of NAMPT and lower expression of p73 in patient tumour samples. Of note, p53 expression levels in these tumours appear to be irrelevant in the context of patient survival. Importantly, and congruent with the patient survival data, high-grade tumours were found to have a higher NAMPT/p73 ratio than do low-grade tumours. The inverse relationship between NAMPT and p73 levels was also demonstrable in the breast cancer cell lines: basal levels of NAMPT are markedly higher, whereas p73 levels are markedly lower in breast cancer cell lines (MDA-MB-231, MDA-MB-486, and MCF-7) compared with normal, non-transformed breast cells (MCF-10A). Here again, pharmacological inhibition or KD of NAMPT significantly increased p73 levels (irrespective of p53 status) and decreased the viability of breast cancer cell lines while having minimum effects on the non-transformed cell line.
Overall, our observations are consistent with the notion that cancer cells rely heavily on the NAD+ salvage pathway for metabolism, lending further credence to the idea of using inhibitors of this pathway for cancer targeting. It is clear, however, that the use of such drugs should be strictly regimented to limit potential side effects. In view of our present revelation of the inverse relationship between NAMPT and p73, and cancer cells' enhanced sensitivity to p73 levels, it would seem logical to consider coupling controlled targeting of the NAD+ salvage pathway with p73 stabilization to enhance anticancer efficacy. Extending this concept further, if indeed the ultimate effect of FK866 is the promotion of p73-mediated cell death, as demonstrated from the present study, then perhaps direct targeting of p73 alone would be sufficiently efficacious, and maybe with less adverse effects than FK866. Small molecules that selectively target and directly stabilize p73 would be particularly desirable to minimize systemic toxicity. Considering the rarity of p73 mutations in human cancers, such an approach should be highly feasible as the probability of enhancing potentially damaging dominant-negative or oncogenic effects of mutant p73 is not high.
Our study also raises the intriguing aspect of a conceivable NAD+/p73 connection pertaining to neurological diseases including Alzheimer's disease. Accumulating evidence suggests that elements of the NAD+ salvage pathway have important roles in maintaining neuronal health,35, 36 whereas separate and unrelated studies have reported that p73 is a major regulator of neural stem cell maintenance and neurodegeneration during aging and Alzheimer's disease.37, 38 In view of the observations from the present study, it would seem logical to conjecture that these are related events, with p73 as the executioner regulated by the NAD+ salvage pathway. Should this turn out to be the case, the NAMPT-p73 nexus may well represent a major molecular link between cancer development/control and neuronal development and degeneration/regeneration.
Materials and Methods
Reagents
U2OS, Jurkat, H1299, Saos2, MDA-MB-231, MDA-MB-468, MCF-7 and MCF-10A cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). NAMPT and p73 KD were generated using lentiviral plasmids expressing shRNAs (from Open Biosystems (GE Dharmacon, Ottawa, ON, Canada) against NAMPT (accession no.: NM_005746, cat. no. RHS4533-EG10135, clone ID: TRCN0000116177) and p73 (accession nos: NM_001126240, NM_001126242, NM_005427, cat. no. RHS4533-EG7161, clone ID: TRCN0000006510). The 293T cells were transfected with the plasmid of interest together with the packing (psPAX2) and envelope vector (pMD2G) using the calcium phosphate transfection method. For transient overexpression of p73, the pCDNA3 vector containing the p73 gene was used. Actively growing cells were treated either with FK866 at different concentrations or with the vehicle and incubated for the times indicated. Cells were then harvested and centrifuged at 200 × g for 10 min at room temperature; the pellets were resuspended in RIPA buffer (25 mM Tris, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing protease inhibitors (Sigma-Aldrich, Oakville, ON, Canada) and incubated on ice. The cell suspensions were then sonicated and the supernatants were collected after centrifugation at 10 000 × g for 15 min. Protein concentrations were determined using the Micro BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, USA). For western blot analysis, proteins were resolved on 10% SDS-polyacrylamide gels and transferred onto PVDF membranes. Immunoblotting was then performed using specified antibodies.
Antibodies against NAMPT (Bethyl Laboratories, Montgomery, TX, USA), p73 (BD Pharmingen, San Jose, CA, USA), ΔNp73 (Abcam, Toronto, ON, Canada), p53 (Santa Cruz), SIRT1 (Abcam), Atg12, 7, 5, LC3A, LC3B (Cell Signalling, Whitby, ON, Canada), p-ERK, p-JNK, p-P38, p-AKT, p-mTOR (Santa Cruz) were used for immunoblotting. FK866 was purchased from Promega (Madison, WI, USA). NMN, chloroquine and actinomycin D were purchased from Sigma (Oakville, ON, Canada), and MG132 was purchased from Calbiochem (Burlington, ON, Canada). Human primers for TAP73: 5′-GACGGACGCCGATG-3′ and 5′-CTGGTCCATGGTGCTG-3′ were used for RT-PCR. The NAD+/NADH Quantification Kit (Biovision, Burlington, ON, Canada) was used to measure cellular levels of NAD+. To detect LC3B puncta formation, cells were transduced with lentivirus expressing pLJM1-EGFP-LC3B. Briefly, the EGFP cassette of pLJM1-GFP (a gift from David Sabatini, Addgene; no.19319) was replaced with EGFP-LC3 from pDEST-EGFP-LC3B (a gift from Terje Johansen, Tromsø, Norway). The resulting pLJM1-EGFP-LC3B was then engineered to replace the puromycin resistance cassette with a blasticidin resistance cassette. Lentiviruses were prepared and titered. U2OS cells were transduced at a low MOI. Cells were selected in 10 μg/ml blasticidin before experiments (Sigma).
Gene expression microarray analysis of patients
One hundred and seventy-six treatment-naïve primary breast cancer samples were obtained from the CBCF Tumour Bank (Edmonton, AB, Canada) and used for gene expression microarray. Frozen dissected tumour samples had been banked by board-certified breast cancer pathologists who also performed quality assurance reports (histological and cellular composition quantifications of the tumours) of the clinical tissue samples directly adjacent to the banked tissue samples. Patient material and clinical information was collected under Research Ethics Board Protocol ETH-02-86-17. RNA isolation from frozen human breast tumour samples, gene microarray analysis and data processing were as described previously.39 The mRNA levels were estimated based on normalized gene microarray signal intensity. The cutoff values to define ‘high' and ‘low' levels for each gene were determined with receiver operating characteristic (ROC) curve analysis. The NAMPT/p73 ratio was calculated for each tumour sample from a 176 patient cohort and a t-test was performed between sub-populations with low and high histological tumour grade.
Statistical analysis
All values are expressed as means±S.E.M. of three independent experiments. Statistical evaluation was performed with one-way ANOVA test followed by a Tukey's post-hoc test as appropriate using GraphPad Prism software (version 5.03 for Windows; GraphPad Software Inc., La Jolla, CA, USA). P<0.05 was considered as significant.
Acknowledgments
This work was supported by grants from the Canadian Institute of Health Research (CIHR) (to SAG, PWKL, PM and CM), from the Canadian Breast Cancer Foundation – Atlantic (to SAG and PWKL) and from the Canadian Breast Cancer Foundation – Prairies/NWT (to RG). SAG was supported by a CIHR Postdoctoral Fellowship. D-GA, EP, DC and PM are supported by the CIHR-Cancer Research Training Program and the Beatrice Hunter Cancer Research Institute (BHCRI). DC also received support from the Nova Scotia Health Research Foundation, and AN received a Norah Stephen Oncology Award from BHCRI.
Glossary
- NAD+
nicotinamide adenine dinucleotide
- NAMPT
nicotinamide phosphoribosyltransferase
- NMN
nicotinamide mononucleotide
- Ca2+
calcium
- NMNAT
nicotinamide mononucleotide adenylyltransferase
- AMPK
adenosine monophosphate-activated protein kinase
- mTOR
mammalian target of rapamycin
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Guo XE, Ngo B, Modrek AS, Lee WH. Targeting tumor suppressor networks for cancer therapeutics. Curr Drug Targets 2014; 15: 2–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000; 408: 307–310. [DOI] [PubMed] [Google Scholar]
- Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. The p53 network. J Biol Chem 1998; 273: 1–4. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Harris CC. P53 mutation spectrum and load: the generation of hypotheses linking the exposure of endogenous or exogenous carcinogens to human cancer. Mutat Res 1999; 428: 23–32. [DOI] [PubMed] [Google Scholar]
- Sidransky D, Tokino T, Helzlsouer K, Zehnbauer B, Rausch G, Shelton B et al. Inherited p53 gene mutations in breast cancer. Cancer Res 1992; 52: 2984–2986. [PubMed] [Google Scholar]
- Slade N, Horvat A. Targeting p73 – a potential approach in cancer treatment. Curr Pharm Des 2011; 17: 591–602. [DOI] [PubMed] [Google Scholar]
- Vilgelm A, El-Rifai W, Zaika A. Therapeutic prospects for p73 and p63: rising from the shadow of p53. Drug Resist Updat 2008; 11: 152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin AR, Paul RK, Thakur VS, Agarwal ML. A novel role for p73 in the regulation of Akt-Foxo1a-Bim signaling and apoptosis induced by the plant lectin, concanavalin A. Cancer Res 2007; 67: 5617–5621. [DOI] [PubMed] [Google Scholar]
- Amin AR, Thakur VS, Paul RK, Feng GS, Qu CK, Mukhtar H et al. SHP-2 tyrosine phosphatase inhibits p73-dependent apoptosis and expression of a subset of p53 target genes induced by EGCG. Proc Natl Acad Sci USA 2007; 104: 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrylenko S, Baniahmad A. Sirtuin family: a link to metabolic signaling and senescence. Curr Med Chem 2010; 17: 2921–2932. [DOI] [PubMed] [Google Scholar]
- Fan QD, Wu G, Liu ZR. Dynamics of posttranslational modifications of p53. Comput Math Methods Med 2014; 2014: 245610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001; 107: 149–159. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 2008; 135: 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes CM, Schonknecht G. Gene transfers shaped the evolution of de novo NAD+ biosynthesis in eukaryotes. Genome Biol Evol 2014; 6: 2335–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E. The many faces of sirtuins: coupling of NAD metabolism, sirtuins and lifespan. Nat Med 2014; 20: 25–27. [DOI] [PubMed] [Google Scholar]
- Pan LZ, Ahn DG, Sharif T, Clements D, Gujar SA, Lee PW. The NAD+ synthesizing enzyme nicotinamide mononucleotide adenylyltransferase 2 (NMNAT-2) is a p53 downstream target. Cell Cycle 2014; 13: 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo H, Tortorella SM, Ververis K, Karagiannis TC. The Warburg effect: molecular aspects and therapeutic possibilities. Mol Biol Rep 2014; 42: 825–834. [DOI] [PubMed] [Google Scholar]
- Dong WR, Xiang LX, Shao JZ. Novel antibiotic-free plasmid selection system based on complementation of host auxotrophy in the NAD de novo synthesis pathway. Appl Environ Microbiol 2010; 76: 2295–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington RA, Genazzani AA, Travelli C, Condorelli F. NAD depletion by FK866 induces autophagy. Autophagy 2008; 4: 385–387. [DOI] [PubMed] [Google Scholar]
- Cea M, Cagnetta A, Fulciniti M, Tai YT, Hideshima T, Chauhan D et al. Targeting NAD+ salvage pathway induces autophagy in multiple myeloma cells via mTORC1 and extracellular signal-regulated kinase (ERK1/2) inhibition. Blood 2012; 120: 3519–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnetta A, Cea M, Calimeri T, Acharya C, Fulciniti M, Tai YT et al. Intracellular NAD(+) depletion enhances bortezomib-induced anti-myeloma activity. Blood 2013; 122: 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S, Penke M, Gorski T, Gebhardt R, Weiss TS, Kiess W et al. FK866-induced NAMPT inhibition activates AMPK and downregulates mTOR signaling in hepatocarcinoma cells. Biochem Biophys Res Commun 2015; 458: 334–340. [DOI] [PubMed] [Google Scholar]
- Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA 2004; 101: 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Rosenthal DS, Luo RB, Samara R, Jung M, Dritschilo A et al. Poly(ADP-ribosyl)ation of p53 in vitro and in vivo modulates binding to its DNA consensus sequence. Neoplasia 2001; 3: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Liu H, Agostini M, Yousefi S, Perren A, Tschan MP et al. p73 regulates autophagy and hepatocellular lipid metabolism through a transcriptional activation of the ATG5 gene. Cell Death Differ 2013; 20: 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura N, Tanuma S. Involvement of cytosolic NAD+ glycohydrolase in cyclic ADP-ribose metabolism. Biochem Biophys Res Commun 1998; 253: 246–252. [DOI] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA 2008; 105: 3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, He Z. NAD and axon degeneration: from the Wlds gene to neurochemistry. Cell Adhes Migr 2009; 3: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla TA, Denu JM. NAD+ deficiency in age-related mitochondrial dysfunction. Cell Metab 2014; 19: 178–180. [DOI] [PubMed] [Google Scholar]
- Muller-Steffner H, Augustin A, Schuber F. Involvement of bovine spleen NAD+ glycohydrolase in the metabolism of cyclic ADP-ribose-mechanism of the cyclization reaction. Adv Exp Med Biol 1997; 419: 399–409. [DOI] [PubMed] [Google Scholar]
- Bi TQ, Che XM. Nampt/PBEF/visfatin and cancer. Cancer Biol Ther 2010; 10: 119–125. [DOI] [PubMed] [Google Scholar]
- Jieyu H, Chao T, Mengjun L, Shalong W, Xiaomei G, Jianfeng L et al. Nampt/Visfatin/PBEF: a functionally multi-faceted protein with a pivotal role in malignant tumors. Curr Pharm Des 2012; 18: 6123–6132. [DOI] [PubMed] [Google Scholar]
- Gehrke I, Bouchard ED, Beiggi S, Poeppl AG, Johnston JB, Gibson SB et al. On-target effect of FK866, a nicotinamide phosphoribosyl transferase inhibitor, by apoptosis-mediated death in chronic lymphocytic leukemia cells. Clin Cancer Res 2014; 20: 4861–4872. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shoji-Kawata S, Sumpter RM Jr, Wei Y, Ginet V, Zhang L et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci USA 2013; 110: 20364–20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, Coleman MP. Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol 2010; 8: e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Parrish JZ, He R, Zhai RG, Kim MD. Nmnat exerts neuroprotective effects in dendrites and axons. Mol Cell Neurosci 2011; 48: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talos F, Abraham A, Vaseva AV, Holembowski L, Tsirka SE, Scheel A et al. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ 2010; 17: 1816–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel MK, Naska S, Laliberte CL, Rymar VV, Fujitani M, Biernaskie JA et al. p73 regulates neurodegeneration and phospho-tau accumulation during aging and Alzheimer's disease. Neuron 2008; 59: 708–721. [DOI] [PubMed] [Google Scholar]
- Marcato P, Dean CA, Liu RZ, Coyle KM, Bydoun M, Wallace M et al. Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling. Mol Oncol 2015; 9: 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.