Abstract
The CD95/Fas/APO-1 death-inducing signaling complex (DISC), comprising CD95, FADD, procaspase-8, procaspase-10, and c-FLIP, has a key role in apoptosis induction. Recently, it was demonstrated that procaspase-8 activation is driven by death effector domain (DED) chains at the DISC. Here, we analyzed the molecular architecture of the chains and the role of the short DED proteins in regulating procaspase-8 activation in the chain model. We demonstrate that the DED chains are largely composed of procaspase-8 cleavage products and, in particular, of its prodomain. The DED chain also comprises c-FLIP and procaspase-10 that are present in 10 times lower amounts compared with procaspase-8. We show that short c-FLIP isoforms can inhibit CD95-induced cell death upon overexpression, likely by forming inactive heterodimers with procaspase-8. Furthermore, we have addressed mechanisms of the termination of chain elongation using experimental and mathematical modeling approaches. We show that neither c-FLIP nor procaspase-8 prodomain terminates the DED chain, but rather the dissociation/association rates of procaspase-8 define the stability of the chain and thereby its length. In addition, we provide evidence that procaspase-8 prodomain generated at the DISC constitutes a negative feedback loop in procaspase-8 activation. Overall, these findings provide new insights into caspase-8 activation in DED chains and apoptosis initiation.
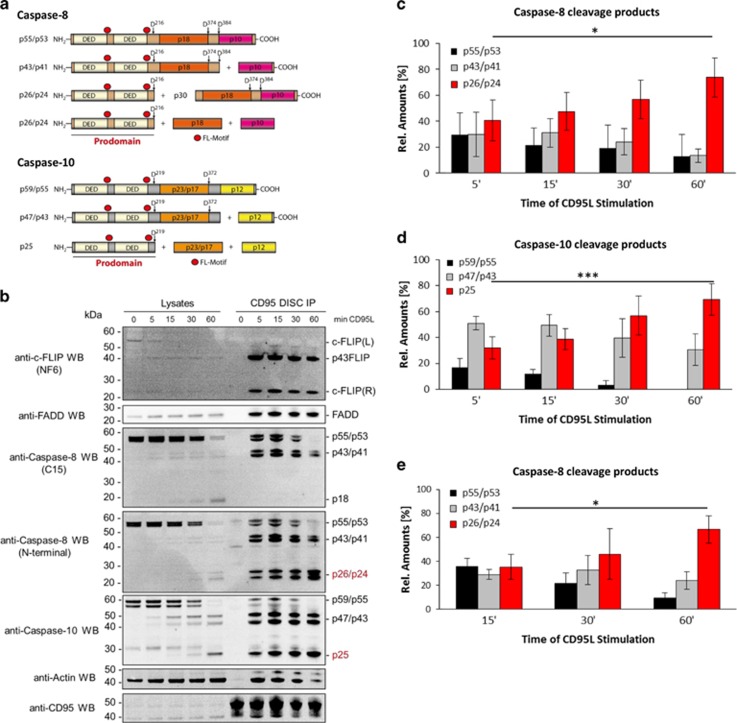
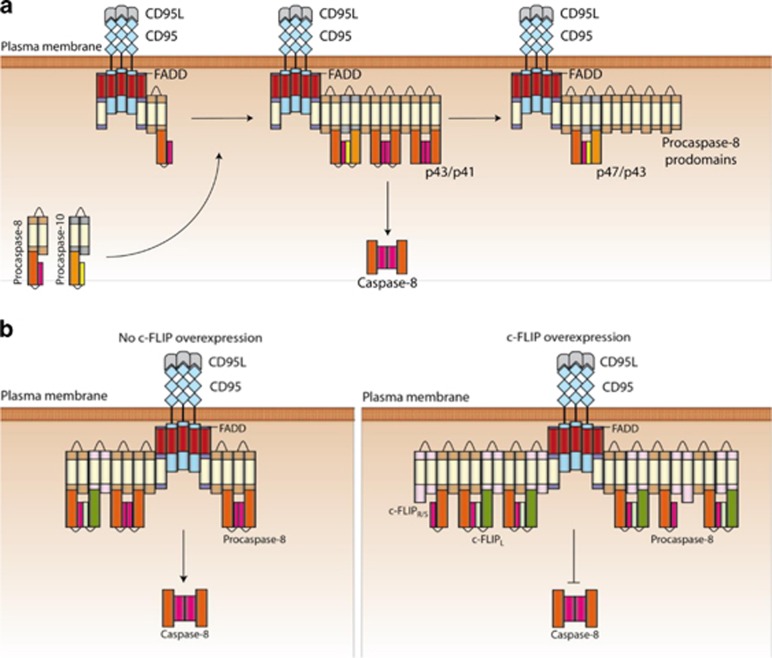
CD95 (Fas/APO-1) is a cell-surface death receptor (DR) that induces apoptosis upon stimulation with its natural ligand CD95L or agonistic antibodies, such as anti-APO-1.1, 2, 3 CD95 engagement also leads to the induction of anti-apoptotic signaling pathways, in particular NF-κB.4, 5, 6 Triggering of CD95 results in the formation of a multiprotein complex, termed death-inducing signaling complex (DISC), consisting of FADD, procaspase-8, procaspase-10, and cellular FLICE inhibitory proteins (c-FLIPs).7, 8, 9, 10 FADD is recruited to CD95 via death domain (DD) interactions whereas procaspase-8, -10, and c-FLIP are recruited to the DISC via death effector domain (DED) interactions.8, 11 At the DISC, procaspase-8 is activated by dimerization followed by internal cleavage, resulting in different cleavage products (Figure 1a).12, 13, 14, 15, 16 Procaspase-8a/b (p55/p53) processing at the DISC results in generation of the N-terminal cleavage products p43/p41, the prodomains p26/p24, as well as the C-terminal cleavage products p30, p18, and p10 (Figure 1a).13, 14, 17, 18 Active caspase-8 heterotetramers p102/p182, generated at the DISC, initiate apoptosis.8
Figure 1.
Kinetics of procaspase-8/-10 cleavage products at the DISC. (a) Schematic representation of procaspase-8 and -10. (b) SKW6.4 cells were stimulated with 1000 ng/ml CD95L for indicated periods of time. CD95 DISC IPs were analyzed using western blot and probed for the indicated proteins. Band intensities of the cleavage products of procaspase-8 and -10 at the DISC were quantified using ImageJ. The amount of each cleavage product for procaspase-8 or -10 was measured in relation to the total amounts of procaspase-8 or -10, respectively. (c) Distribution of procaspase-8 cleavage products measured by quantitative western blot. To determine the distribution of the cleavage products for procaspase-8, the band intensities for the procaspase-8 and its cleavage products at the DISC-IP, i.e., p55/53, p43/p41 and p26/p24, were quantified and divided by the sum of the intensities of all procaspase-8 proteins. The mean±S.D. is shown (n=4). (d) Distribution of procaspase-10 cleavage products measured by quantitative western blot. The mean±S.D. is shown (n=5). (e) Distribution of procaspase-8 cleavage products measured by quantitative mass spectrometry. To determine the distribution of the cleavage products for procaspase-8, the peak areas at the mass spectrometry spectrum for the procaspase-8 and its different cleavage products at the DISC-IP were quantified and divided by the sum of the amounts of all procaspase-8 proteins as in the quantifications using western blot. The mean±S.D. is shown (n=3). Statistical significance: *P<0.05, ***P<0.001
Procaspase-10 is a homolog of procaspase-8 with similar molecular architecture. Like procaspase-8, it contains tandem DEDs followed by the large and small catalytic subunits.10, 19 Full-length procaspase-10a/d (p55/p59) is cleaved between the large and small subunits, generating p47/p43, which is further processed to the prodomain p25 and the large catalytic subunit of caspase-10 (Figure 1a). Procaspase-10 is also suggested to be activated at the DISC via homodimerization.10, 20, 21
C-FLIP is a well-described inhibitor of DR-mediated apoptosis. Three c-FLIP isoforms and two cleavage products have been characterized so far.22, 23, 24, 25 The three c-FLIP isoforms include Long (L), Short (S), and Raji (R), that is, c-FLIPL, c-FLIPS, and c-FLIPR, respectively. All three isoforms possess two DED domains; c-FLIPL also contains catalytically inactive caspase-like domains (p20 and p12). The short c-FLIP isoforms, c-FLIPS and c-FLIPR, block DR-induced apoptosis by inhibiting procaspase-8 activation at the DISC presumably by forming catalytically inactive heterodimers.23, 26 C-FLIPL at the DISC can have both pro- and anti-apoptotic roles. The pro-apoptotic function of c-FLIPL has been reported to be mediated by the formation of catalytically-active procaspase-8/c-FLIPL heterodimers, in which the active loop of procaspase-8 is stabilized by c-FLIPL.27, 28
Recently, we studied the stoichiometry of the CD95 DISC by AQUA peptide-based mass spectrometry.29 In combination with western blot analysis supported by mathematical modeling, we revealed an unexpected stoichiometry in which procaspase-8 is more abundant at the DISC compared with FADD.29 Similar results were obtained for the TRAIL DISC.30 Furthermore, the study by Dickens et al.30 has also shown that these chains are formed via DED interactions based on a hydrophobic patch in DED2 of procaspase-8 (F122/L123), referred to as FL-motif, which is highly conserved for procaspase-8, procaspase-10, and c-FLIP (Supplementary Figure S1). On the basis of these findings, it was proposed that procaspase-8, procaspase-10, and c-FLIP form chains via DED interactions. Our mathematical model used for the analysis of the chain dynamics could well describe caspase-8 activation in silico. However, we had to introduce a restriction of chain lengths into our model in order to re-model our experimental data. This indicates the existence of a yet uncovered mechanism of chain termination. Taken together, the discovery of the DED chains provides a basis for understanding how homodimers of procaspase-8 could be formed in the context of the DISC and subsequently be activated, but poses further numerous questions: How are the DED chains terminated? What is the mechanism of action of c-FLIP and procaspase-10 in the DED chains? How does c-FLIP inhibit procaspase-8 in the DED chains? What are the dynamics of procaspase-8 processing in the chain and how does the composition of the chain change upon procaspase-8 cleavage? To get more insight into these questions, we have further analyzed the dynamics of the DISC assembly.
In this study, we addressed the composition and activity of the DED chains as well as mechanisms of chain termination. We showed that the amounts of the procaspase-8 and -10 cleavage products increase over time and the DED chain comprises mostly procaspase-8 prodomain at later time points. Our data suggest that short tandem DED proteins, that is, c-FLIP-DED-DED or procaspase-8 prodomain, do not block DED chain elongation. We show that short c-FLIP isoforms can inhibit CD95-induced cell death upon overexpression, likely by forming inactive heterodimers with procaspase-8 but not by terminating the chain formation. In addition, mathematical modeling indicated that procaspase-8 binding to the DISC might become less stable with increasing DED chain length leading to chain termination. Thus, we suggest that termination of the chains is not based on the action of one particular DED protein, but they are rather terminated by instability of longer chains. Furthermore, we provide evidence that procaspase-8 prodomain regulates procaspase-8 processing by forming a negative-feedback loop. Taken together, our data demonstrate how procaspase-8 activation can be regulated in the DED chain model by short DED proteins.
Results
The procaspase-8 prodomain is a major part of DED chain
It was reported that procaspase-8 forms DED chains at the DISC.29, 30 To get more insight into the composition of the DED chains at the DISC, we analyzed CD95 DISCs after immunoprecipitation (IP) by quantitative western blot and mass spectrometry. As in our previous study, we used the B lymphoblastoid cell line SKW6.4.29 SKW6.4 cells were stimulated with 1000 ng/ml CD95L and CD95 DISCs were immunoprecipitated using agonistic anti-APO-1 (CD95) antibodies.3 Under these experimental conditions, CD95 could be completely immunoprecipitated.31 For western blot analysis, CD95 DISC IPs were separated by SDS-PAGE and the membranes were probed using antibodies against c-FLIP, FADD, procaspase-8, and procaspase-10 (Figure 1b). To detect both, C-terminal and N-terminal procaspase-8 cleavage products, we used anti-N-terminal and anti-C-terminal (C15) caspase-8 antibodies. The quantification revealed that the amount of p43/p41 cleavage product decreases over time, while the relative amount of the procaspase-8 prodomain p26/p24 at the DISC increases over time reaching from 40% at 5 min to 80% at 60 min of the total procaspase-8 amounts (Figure 1c).
The distribution of procaspase-10 cleavage products was quantified in the same way as for the ones of procaspase-8. Interestingly, the ratio between procaspase-10 cleavage products was different from that of procaspase-8, for example, procaspase-10 appeared to be faster processed to its intermediate cleavage product p47/p43 compared with procaspase-8 processing to its intermediate cleavage product p43/p41 (Figure 1d). Similar to procaspase-8 processing at the later time points, for example, 60 min, we observed substantial amounts of the procaspase-10 prodomain at the DISC.
For the mass spectrometry analysis, the CD95 DISC IPs were separated on a one-dimensional SDS-PAGE, which was subsequently sliced into pieces. The relative abundance of the different cleavage products for procaspase-8 in each slice was quantified by AQUA peptide-based mass spectrometry as described before (Supplementary Figure S2).29, 31 Similar to the western blot quantifications, the relative amounts of the procaspase-8 prodomain p26/p24 at the DISC increased over time (Figure 1e). This quantification further confirmed that the prodomain of procaspase-8a/b p26/p24 is a major part of the DED chains at the CD95 DISC, while the cleavage product p43/p41 is present in lower amounts. This, in turn, suggests that procaspase-8 in the chains is rapidly processed via p43/p41 to the prodomain accompanied by the release of the active caspase-8 subunits. This also fits to our previous studies of caspase-8 prodomain generation at the DISC.13
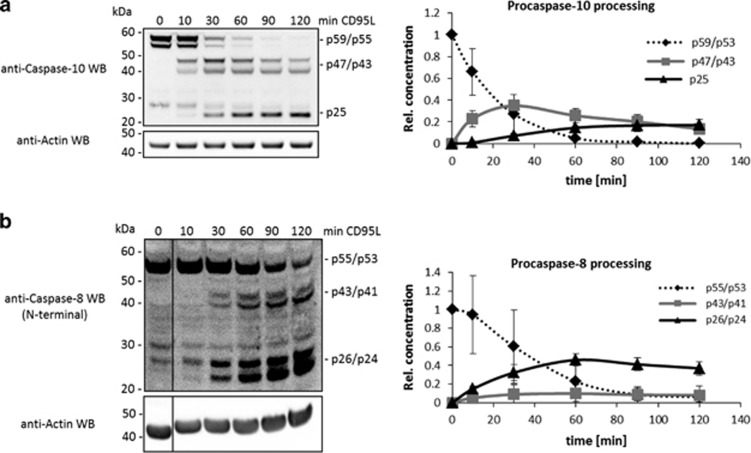
The analysis of procaspase-8 and -10 processing in the lysates at later time points after CD95 stimulation further supported our findings (Figure 2). Indeed, the intermediate procaspase-10 cleavage product increased up to 40% of the total procaspase-10 amounts over the first 30 min following CD95 stimulation (Figure 2a), while the p43/p41 levels of procaspase-8 always remained below 20% (Figure 2b). In addition, the formation of procaspase-10 prodomain occurred later compared with the procaspase-8 prodomain (Figure 2).
Figure 2.
Kinetics of procaspase-8 and -10 in whole cellular lysates. SKW6.4 cells were stimulated with 1000 ng/ml CD95L for different time points and analyzed by western blot using antibodies against procaspase-10 (a), procaspase-8 (b), and actin. Band intensities were quantified using ImageJ. The intensities of the different cleavage products were normalized to actin and the total amounts of the full-length protein. Quantification of procaspase-10 (a) and -8 (b) cleavage products (mean±S.D., n≥3, right panels). The samples in (b) were loaded on the same gel, but the blot was cut (marked by black line) for presentation purposes
The difference in procaspase-8 versus -10 cleavage product distribution indicates different processing regimes for these two procaspases in the chain. Due to the low amounts of procaspase-10 at the DISC,29 probably only very few procaspase-10 molecules can be found in each chain. Indeed, if we consider the chain length of eight DED proteins, as proposed in our previous study, then this would result in one procaspase-10 per chain at most. Thus, only very few procaspase-10 homodimers might be formed in the DED chain. Instead, heterodimers with procaspase-8 would be formed. These procaspase-8/procaspase-10 heterodimers have been reported to be catalytically inactive.32 Consequently, procaspase-10 would be largely processed by neighboring procaspase-8 homodimers, but not by autoprocessing. This difference in the processing mechanisms of procaspase-8 and procaspase-10 in the chain, for example, processing in homodimer versus heterodimer, respectively, might result in the different rates of generation of the intermediate cleavage products and prodomains.
Short c-FLIP proteins and the procaspase-8 prodomain do not block procaspase-8 chains
In previous studies, the inhibition of procaspase-8 by c-FLIP was assigned to the competition of c-FLIP with procaspase-8 for FADD binding. Therefore, in the context of the DED chain model, additional mechanisms have to play a role for procaspase-8 inhibition by c-FLIP. We suggested that short DED proteins might inhibit procaspase-8 activation by chain termination, for example, short c-FLIP isoforms as well as the procaspase-8 prodomain might interrupt chain elongation by causing less stable binding of subsequent DED molecules to the chain. This assumption was motivated by structural considerations. Namely, procaspase-8 homodimers or procaspase-8/c-FLIPL heterodimers form hydrogen bonds between the catalytic subunits,28, 33 which are lacking in the short isoforms of c-FLIP as well as in the procaspase-8 prodomain.
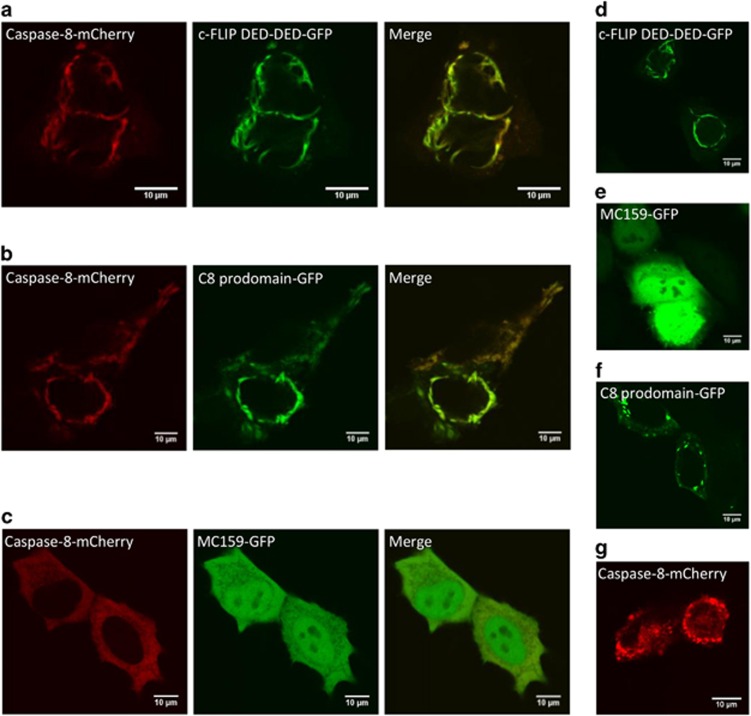
To address the role of short DED proteins in chain formation, we performed immunofluorescence experiments on HeLa cells overexpressing CD95 (HeLa-CD95) that were transiently transfected with c-FLIP DED-DED-GFP (aa 1–197), procaspase-8a prodomain-GFP or MC159-GFP either alone or together with full-length procaspase-8a-mCherry. The c-FLIP construct contained both DEDs and was mimicking short c-FLIP isoforms. MC159 is a viral short FLIP protein, which possesses two DEDs. Interestingly, MC159 has been reported to interact with the DISC via structural motifs different from c-FLIP, acting directly on FADD, and inhibits procaspase-8 activation.34 The co-expression of DED proteins together with procaspase-8-mCherry indicated that neither the N-terminal part of c-FLIP nor the procaspase-8 prodomain blocked filament formation of procaspase-8 (Figures 3a, b, d, f, and g). Here the formation of filament-like structures was observed, associated with the appearance of punctuated staining pattern. MC159 blocked any aggregation of procaspase-8, that is, the formation of filament-like structures was not observed, in accordance with its reported inhibitory role (Figures 3c and e). Taken together, these experiments indicate that the short c-FLIP isoforms and the procaspase-8 prodomain probably do not inhibit procaspase-8 filament formation but rather form filaments or chains with procaspase-8, that is, by incorporating into the DED chains upon overexpression.
Figure 3.
c-FLIP DED-DED and procaspase-8 prodomain do not block procaspase-8 chain formation. Immunofluorescence microscopy was used to analyze procaspase-8 chain formation by (a–c) overexpression of procaspase-8-mCherry together with c-FLIP DED-DED-GFP (a), caspase-8 prodomain-GFP (b), or MC159-GFP (c) in HeLa-CD95 cells. (d–g) Single overexpression of c-FLIP prodomain-GFP (d), MC159-GFP (e), caspase-8 prodomain-GFP (f), or procaspase-8-mCherry (g) in HeLa-CD95 cells. In (a–c) and (g), 50 μM zVAD-fmk was added at the time of transfection
Short DED proteins inhibit CD95-induced cell death
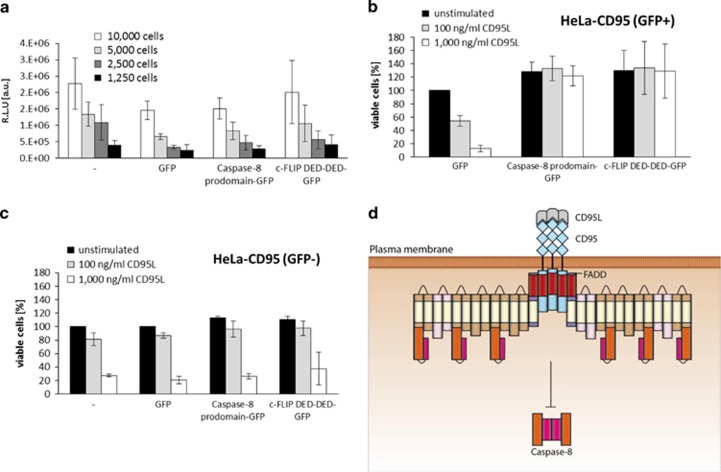
The confocal microscopy analysis of HeLa-CD95 cells overexpressing short DED proteins indicated that they can incorporate into DED chains. To study the impact of the chains on apoptosis induction with high content of short DED proteins, HeLa-CD95 cells were transfected with procaspase-8a prodomain-GFP, c-FLIP DED-DED-GFP (aa 1–197), or GFP. The cell viability was measured by an ATP-based assay, which demonstrated that procaspase-8 prodomain-GFP and c-FLIP DED-DED-GFP transfected cells were equally viable compared with the GFP-transfected cells (Figure 4a). Further, procaspase-8 prodomain-GFP or c-FLIP DED-DED-GFP transfected HeLa-CD95 cells were stimulated with different amounts of CD95L for 4h and cell death was assessed by PI staining (Figures 4b and c, Supplementary Figure S3). GFP-only transfected cells died in a dose-dependent manner (Figure 4b, Supplementary Figure S3A), while procaspase-8 prodomain-GFP and c-FLIP DED-DED-GFP transfected cells were protected from CD95L-induced cell death (Figure 4b,Supplementary Figures S3B and C). The untransfected population of cells also died upon CD95L stimulation in a dose-dependent manner (Figure 4c, Supplementary Figure S3D). Collectively, these data demonstrate that overexpression of the procaspase-8 prodomain and c-FLIP DED-DED-GFP does not reduce cell viability and even protects from CD95-induced cell death. This might be explained by the formation of the DED chains with a high number of heterodimers: procaspase-8/prodomain and procaspase-8/c-FLIP-DED-DED (Figure 4d). In this type of chains the formation of procaspase-8 homodimers, which is a prerequisite for procaspase-8 activation, might be decreased due to the inclusion of short DED proteins into the chain.
Figure 4.
c-FLIP DED-DED and procaspase-8 prodomain inhibit CD95-induced cell death. (a) CellTiter-Glo assay was performed on HeLa-CD95 cells, which were transfected with procaspase-8 prodomain-GFP, c-FLIP DED-DED-GFP, GFP, or left untransfected (−). The experiments were carried out after 16h expression and always performed with different cell numbers (1250, 2500, 5000, and 10 000) in duplicates. The mean±S.D. is shown (n=3). RLU, relative luminescence units. (b) HeLa-CD95 cells overexpressing the indicated constructs were stimulated with different concentrations of CD95L for 4 h and cell death measured by PI staining and flow cytometry. Cell viability was calculated as described in Materials and methods. The results for GFP-positive cells are shown. (c) Same as in (b), but for GFP-negative cells or untransfected cells (−). The data in (b and c) show mean±S.D. (n=3). Representative FACS stainings can be found in Supplementary Figure S3. (d) Upon overexpression of the procaspase-8 prodomain or short c-FLIP protein (depicted in brown), they could be recruited into the DISC alongside full-length procaspase-8 leading possibly to heterodimer formation. Consequently, procaspase-8 cannot efficiently form homodimers and its activation is inhibited
C-FLIP does not block procaspase-8 recruitment into the DISC
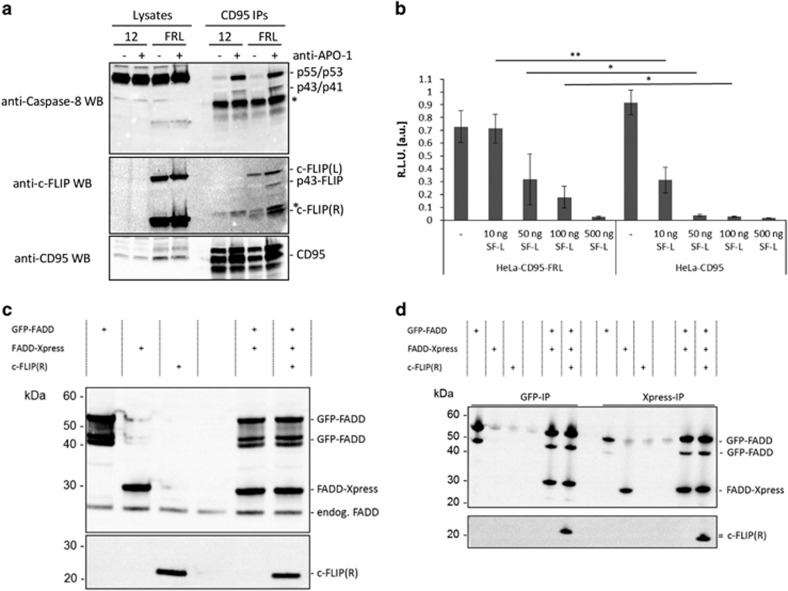
To further address the role of c-FLIP and analyze whether overexpression of c-FLIP prevents procaspase-8 recruitment to the CD95 DISC, we performed DISC IPs from HeLa-CD95 cells, which stably overexpress two c-FLIP isoforms, c-FLIPL and c-FLIPR.35 In accordance with the immunofluorescence data and putative formation of DED chains containing both c-FLIP and procaspase-8, we observed that overexpression of c-FLIPL/R does not prevent recruitment of procaspase-8 into the CD95 DISC (Figure 5a). This is consistent with the analysis of c-FLIP-overexpressing cell lines published by us previously, for example, procaspase-8 was detected at the DISC of HeLa-CD95 cells overexpressing c-FLIPs or c-FLIPL,35 and indicates that c-FLIP probably does not terminate DED chains but rather incorporates into them. Furthermore, stable overexpression of both c-FLIP isoforms in HeLa-CD95 cells protected the cells from CD95-induced cell death upon low stimulation strengths, but did not do so upon high stimulation strengths (Figure 5b). The level of the short c-FLIP isoform in this cell line is very high, while the overexpression of c-FLIPL is 20 times higher than endogenous.35 As we have described before in our studies, this ratio between long and short c-FLIP isoforms likely promotes preferable formation of procaspase-8/c-FLIPL heterodimers, subsequent activation of procaspase-8 and processing to p43/p41, which is followed by apoptosis.35 Interestingly, the high level of short c-FLIP isoforms in these cells does not prevent recruitment of procaspase-8 and c-FLIPL into the chains, which further supports a ‘non-terminating' role of the short c-FLIP isoforms.
Figure 5.
c-FLIP overexpression does not block procaspase-8 recruitment into the DISC. (a) HeLa-CD95 (12) or HeLa-CD95 c-FLIPR/L overexpressing cells (FRL) were stimulated with 1 μg/ml anti-APO-1 and CD95 DISC IPs were prepared. Lysates and DISC IPs were separated by SDS-PAGE and analyzed by western blot. The bands marked by an asterisk denote unspecific bands in the anti-caspase-8 (C15) western blot and IgG light chain in the anti-c-FLIP western blot. (b) HeLa-CD95 cells stably overexpressing c-FLIPR and c-FLIPL (HeLa-CD95-FRL) or HeLa-CD95 cells were stimulated with different concentrations of Super Fas Ligand (SF-L) for 4h and cell viability was measured by ATP assay (n=3). RLU, relative luminescence units. Statistical significance: **P<0.01, *P<0.05. (c and d) c-FLIPR does not interfere with FADD self-association. GFP-FADD, Xpress-FADD, or c-FLIPR was overexpressed in 293T cells either alone or in combination. Cell lysates show equal expression of the two FADD plasmids. (c) Co-expression of both FADD constructs together with c-FLIPR did not block or reduce co-IP of FADD (d)
c-FLIPR does not block FADD self-association
To exclude that short DED proteins inhibit apoptosis by mechanisms independent from their action in the DED chain, we analyzed whether they can block FADD self-association. Indeed, sequence alignments demonstrated that the charge triad motif (E17/R64, D66, L67, L68 in DED1 and D108/R161, D163, L164), which was reported to be important for apoptosis inhibition by the viral analog of c-FLIP, MC159, is highly conserved in c-FLIP (Supplementary Figure S1).34, 36 Due to this sequence similarity, we speculated that c-FLIPR might inhibit procaspase-8 processing by interfering with FADD self-association via the charge triad motif reported for MC159, which would occur upstream of chain formation.34 This was analyzed in 293T cells, using GFP- and Xpress-tagged FADD molecules that were co-expressed with c-FLIPR (Figures 5c and d). However, overexpression of c-FLIPR did not block interactions between differentially tagged FADDs (Figure 5d). In line with this, it has been reported that the charge triad motif of murine c-FLIPR is not required for its recruitment to the DISC and inhibition of procaspase-8 activation.37
Chain termination depends on procaspase-8 dissociation/association rates
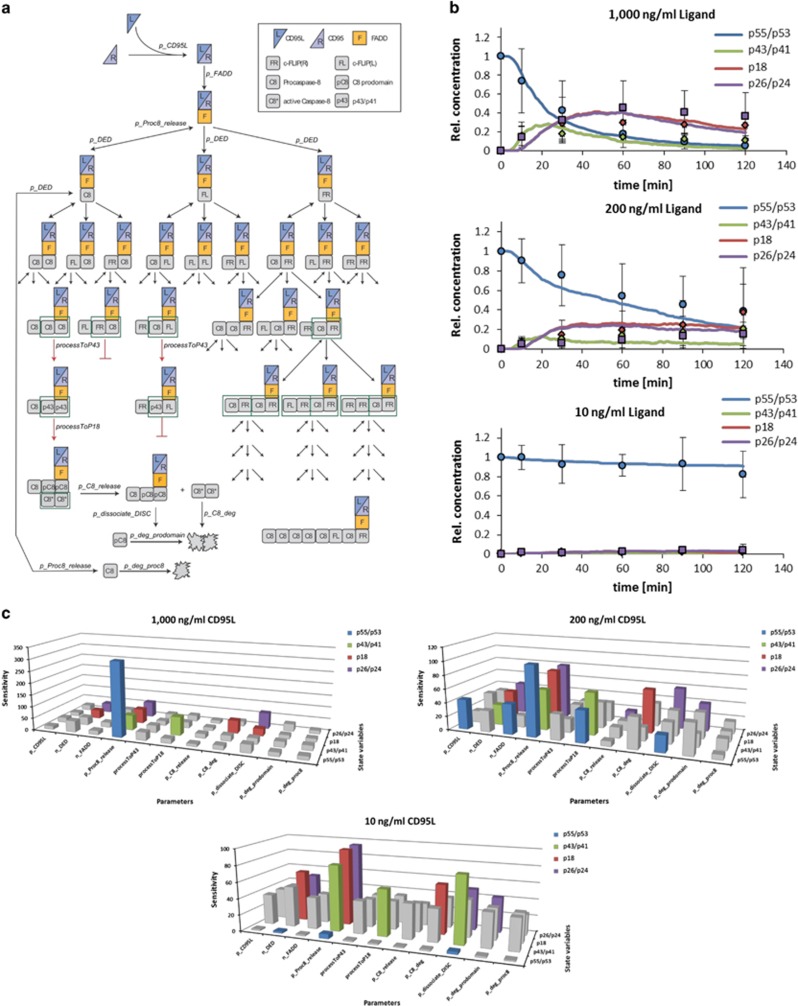
To strive better understanding of chain termination control, we used mathematical modeling. In our previous work of modeling the dynamics of DED chains we developed an agent-based model.29 Our former model was valid only assuming a limitation of chain lengths.29 Thus, in the current study, we modified our previous model in order to search for new parameters controlling chain length and dynamics. Along with interactions of the core DISC proteins: CD95, FADD, c-FLIP, and procaspase-8 leading to DED chain formation, the generation of procaspase-8 prodomain was included into the model followed by processing of p43/p41 to the prodomain and p18 (Figure 6a). Furthermore, procaspase-8 released after DED chain disassembly was assumed to be either degraded or recruited to other receptors (Figure 6a). Cytosolic procaspase-8 prodomain (p26/p24) and p18 were assumed to undergo proteolytic degradation, which was integrated into the model (Figure 6a).
Figure 6.
Mathematical modeling suggests that DED chains can be regulated by ‘increasing instability'. (a) Model Topology. CD95 is stimulated by CD95L followed by recruitment of FADD. Subsequently, DED proteins, i.e., procaspase-8 (C8), c-FLIPL (FL), and c-FLIPR (FR) are recruited resulting in chain formation with different length and composition. Procaspase-8 homodimers are fully processed via p43/p41 (p43) to prodomain (pC8) and p18 (C8*), while procaspase-8/c-FLIPL heterodimers are partially processed to p43/p41 (p43) and procaspase-8/c-FLIPR heterodimers are not processed. Fully processed caspase-8 (C8*) is released into the cytosol, but the prodomain (pC8) remains at the DISC. In addition, complete DED chains can be disassembled at any time point. Full-length procaspase-8 (p55/p53) can be degraded following chain disassembly or recruited to other receptors; p43/p41 can be further processed to p18. Free p18 (C8*) or prodomain (pC8) is degraded. Red arrows indicate procaspase-8 processing steps and black arrows other reactions. (b) Simulations of procaspase-8 processing (solid lines) compared with experimental data (points) (mean±S.D.; n⩾3) for p55/p53 (blue), p43/p41 (green), p18 (red), and p26/p24 (purple). Experimental data are taken from Supplementary Figure S4A and our previous work.29 A full list of parameter values is shown in Supplementary Table S1. (c) Sensitivity analysis performed on the parameters of the model. Each parameter was varied individually from x/5 to x*5, where x was the final value in the simulation (Supplementary Table S1) and the sensitivity to model output was computed. Sensitivities above the average on each state variable upon changing different parameters are indicated using the same color code as in (b). The parameters regulating procaspase-8 association (n_DED) and dissociation (p_Proc8_release) and DISC stability (p_dissociate_DISC) had an above-average effect on multiple state variables at different CD95L levels
Caspase-8 association/dissociation rates were reported previously to have an important role in apoptosis initiation.38 Furthermore, we assumed that the stability of the longer chains might decrease. Following this assumption, the dissociation probability of procaspase-8 was modeled to be depending on the length of the DED chain. Namely, procaspase-8 bound to the end of a long chain had a higher dissociation probability compared with one bound to a shorter chain. The parameters of the model were adjusted based on the experimental data of procaspase-8 processing using 1000 ng/ml CD95L stimulation (Figure 6b, Supplementary Figures S4A and B). There was a good fit between simulations of this model and experimental data of procaspase-8 processing of full-length procaspase-8 p55/p53 to p43/p41 and further to p18 and p26/p24 in the total cellular lysates for intermediate (500, 200 ng/ml) as well as low (50, 10 ng/ml) stimulation strengths (Figure 6b, Supplementary Figures S4A, C and D). The model also well reproduced distribution of procaspase-8 cleavage products at the DISC, especially for the later time points, upon high CD95L stimulation (Supplementary Figure S4B). To test whether the generated model could also describe procaspase-8 activation in another cell line, we used the experimental data, that is, western blots for procaspase-8 processing in total cellular lysates, obtained by us previously in HeLa-CD95 cells.5, 35 Importantly, the model could also verify this experimental data (Supplementary Figure S4E). Thus, the generated model could reasonably well simulate procaspase-8 processing underlining the importance of procaspase-8 association/dissociation rates in regulation of the chain length. This analysis also indicates that the restriction of the chain length could result from decreasing stability of a DED chain upon increase of its length.
There may be, however, other mechanisms controlling chain length such as binding of an inhibitor to a chain or post-translational modifications of procaspase-8 at later stages of processing, for example, TRAF2-mediated ubiquitination resulting in procaspase-8 degradation.39 In addition, the longer chains might cause sterical hindrance within the vicinity of CD95, for example, via possible contacts with adjacent FADDs and receptors. To test this suggestion, we performed analysis of the DED chain in silico using PyMOL.40 The structure of full-length FADD was obtained from the RCSB database (PDB 2GF5). The structure of the procaspase-8 prodomain was modeled using the Phyre2 web service41 from the sequence of procaspase-8 prodomain (aa 1–216, Uniprot ID Q14790) and the structure of the viral FLIP MC159 (PDB 2BBR). The DED chain was assembled using ad-hoc structural considerations based on the reported interactions of the tandem DED with the FADD DED.42 This structural analysis of the DED chain architecture supported our suggestion and indicated that indeed longer chains could potentially contact adjacent receptors (Supplementary Figure S5).
We also investigated the role of c-FLIP proteins in controlling the chain length in our simulations. We took the concentrations as well as association rates of c-FLIP proteins from our previous data sets.29 Importantly, c-FLIP proteins are much less abundant in SKW6.4 cells compared with procaspase-8. In accordance with this, the predictions in silico stated that c-FLIP proteins have only marginal impact in regulation of chain composition. Accordingly, the increase of c-FLIP protein concentrations in silico resulted in the chains initiated by c-FLIP proteins, which led to a reduction of caspase-8 processing upon high concentrations of c-FLIP (data not shown). It has to be noted that the role of c-FLIP proteins in the chain might be different in other cellular models with higher levels of c-FLIP and cannot be generalized based on the results in our cellular model.
To get a systematic overview, we investigated the influence of different parameters on the simulation outcome by applying a sensitivity analysis. Sensitivity analysis was performed by varying each parameter individually and testing the influence on the modeled state variables, that is, p55/p53, p43/p41, p18, and p26/p24, upon different CD95L stimulation strengths. We observed an increased sensitivity for a number of parameters including a high sensitivity on p43/p41 levels upon changing processing to p18 (processToP18). Furthermore, p18 and p26/p24 levels were highly dependent upon their degradation rates p_C8_deg and p_deg_prodomain, respectively. Interestingly, the parameters regulating procaspase-8 association/dissociation to the DISC, namely n_DED and p_Proc8_release, as well as DISC stability (p_dissociate_DISC) had a large impact on multiple state variables at different CD95L concentrations (Figure 6c). This further supports our hypothesis that procaspase-8 dissociation/association rates in combination with procaspase-8 position in the chain have an important role in regulating its processing in the chain model.
Negative feedback on procaspase-8 activation mediated by the procaspase-8 prodomain
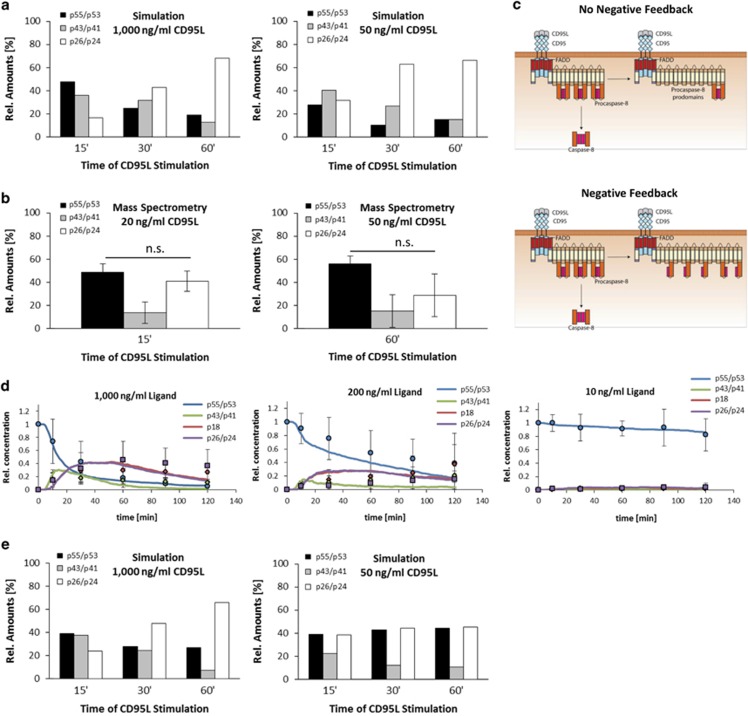
The model could well describe procaspase-8 processing as validated by quantitative western blot of procaspase-8 processing in total cellular lysates (Figure 6b). However, upon evaluation of the quantitative mass spectrometry data of the CD95 DISCs formed upon low CD95L stimulation strength, we found contradictions compared with our simulations (Figures 7a and b). The mass spectrometry analysis indicated that procaspase-8 (p55/p53) is not fully processed to the prodomain (p26/p24) at the 60-min time point (Figures 7a and b). To explain this contradiction, we suggested the existence of a negative feedback loop mediated by the procaspase-8 prodomain. We assumed that upon low stimulation strength there might be more unprocessed procaspase-8 in the DISC leading to the formation of procaspase-8/prodomain heterodimers in the chain (Figure 7c). These procaspase-8/prodomain heterodimers would not be processed to the active caspase-8 heterotetramer and therefore, this would result in inhibition of procaspase-8 activation at the DISC. This would form a negative feedback loop in procaspase-8 activation (Figure 7c). To test this scenario in silico, we added the negative feedback loop to the model. The addition of the negative feedback loop to the model (Figure 7d,Supplementary Figure S6) allowed to bring the simulations (Figure 7e) in good agreement with the experimental data for DISC IPs (Figures 1e and 7b). This suggests that the negative feedback loop in procaspase-8 processing involving the generation of its prodomain might contribute to the regulation of the procaspase-8 processing at the DISC.
Figure 7.
Mathematical modeling unravels the inhibitory mechanism of the procaspase-8 prodomain. (a) Distribution of procaspase-8 cleavage products for the indicated CD95L concentrations over time in simulations. (b) Distribution of procaspase-8 cleavage products at the CD95 DISC IP for 20 ng/ml CD95L after 15 min and 50 ng/ml CD95L after 60 min stimulation measured by quantitative mass spectrometry. The mean±S.D. is shown (n=3). Statistical significance: n.s., not significant (P>0.05). (c) Scheme representing negative feedback in procaspase-8 activation. Negative feedback of the procaspase-8 prodomain was introduced into the model by allowing insertion of procaspase-8 into DED chains thus forming inactive heterodimers (right) compared with allowing only recruitment to the end of the DED chain (left). (d) Simulations of procaspase-8 processing (solid lines) with addition of the negative feedback to the model compared with experimental data (mean±S.D.; n⩾3) for p55/p53 (blue), p43/p41 (green), p18 (red), and p26/p42 (purple). Experimental data were taken from Supplementary Figure S4A and our previous work.29 (e) Distribution of procaspase-8 cleavage products for the indicated CD95L concentrations over time in simulations with negative feedback loop
Discussion
In this study, we have investigated the molecular architecture of the DED chains and the mechanisms of their termination. We considered the role of three DED proteins: procaspase-8, procaspase-10, and c-FLIP in light of the concept of DED chain formation, in particular, focusing on the short DED proteins. This provided new insights into the function of all three DED proteins in apoptosis initiation.
After stimulation, procaspase-8a/b immediately forms the chains at the DISC, which triggers its activation (Figure 8a). Procaspase-8a/b is then processed to a number of cleavage products including p43/p41, p26/p24, p30, p18, and p10 within the chains, which is followed by the generation of the procaspase-8 prodomain platform at CD95 and release of the active caspase-8 heterotetramer p102/p182. Procaspase-10 is randomly incorporated together with procaspase-8 molecules into the DED chain at a ratio of 1–10. Procaspase-8 cleaves procaspase-10 to its intermediate cleavage product p47/p43 and later to the prodomain (Figure 8a). Our data strongly point out that due to its low abundance in the chain procaspase-10 does not have a central role in apoptosis initiation in our cellular model. The different c-FLIP isoforms are also randomly recruited to the chains via the FL-motif, in a ratio to procaspase-8 of 1–10, which is followed by processing of c-FLIPL to p43-FLIP (Figure 8b).34, 37 Remarkably, our data indicate that the previously suggested mechanism of procaspase-8 inhibition by c-FLIP, based on the competition for binding sites at the DISC, is not valid. Our data indicate that likely c-FLIPS/R has an inhibitory potential, if the DED chain is mostly composed of c-FLIPS/R/procaspase-8 heterodimers, which would prevent procaspase-8 homodimer formation. Another modeling study also suggested that dynamic regulation of c-FLIP levels is important for the signaling outcome in DR-induced apoptosis underlining the importance of their amount in the chain.43
Figure 8.
Model of CD95 DISC dynamics. (a) The suggested role of procaspase-10 in the DED chain. Procaspase-10 is only marginally present compared with procaspase-8 resulting in procaspase-8/-10 heterodimers in the chain, which are most likely not catalytically active.32 Therefore, procaspase-10 is mainly processed by procaspase-8 activity in the chain leading to the generation of p47/p43 cleavage product. (b) The suggested role of c-FLIP in the DED chain. c-FLIPL/S/R is recruited to the chain and form heterodimers with procaspase-8. Upon high levels c-FLIPS/R inhibits procaspase-8 activation in the DISC
The data presented here show that the procaspase-8 prodomain inhibits CD95-induced apoptosis upon overexpression. This is in line with other studies reporting an inhibitory function of the procaspase-8 and -10 prodomains as well as short isoforms of both procaspases.21, 44, 45 On the contrary, it has been reported that the procaspase-8 prodomain induces apoptosis upon overexpression.46 These different findings may be cell type specific or dependent on the level of procaspase-8 prodomain overexpression.
Mathematical modeling in combination with quantitative mass spectrometry indicated that the procaspase-8 prodomain may form a negative feedback loop in procaspase-8 activation upon low CD95L stimulation strength. The different stimulation strengths leading to different signaling outcomes, for example, life or death, have been pointed out in recent studies. Thus, this negative feedback might contribute to the induction of ‘pro-life' signaling pathways via proteins associated with procaspase-8 retained at the DISC. An overall challenge of the DED chain model is the order of recruitment of the different DED proteins leading to different homo- or heterodimers upon different stimulation strength. Moreover, each DISC might form its own ‘microenvironment' in which DED chains are constantly disassembled and re-assembled. This might then preferentially lead to inactive procaspase-8/prodomain heterodimers if the amount of prodomain in the DISC is high. Finally, the mechanism of the negative feedback regulation via the formation of inactive heterodimers of procaspase-8 with its prodomain allows making a suggestion upon the unified inhibition mechanism of procaspase-8 inhibition in the chain, which might involve the formation of inactive heterodimers of procaspase-8 with any other DED protein, including short c-FLIP proteins.
After addressing several scenarios of chain termination, we applied mathematical modeling that indicated that the chain length could be regulated by its stability and association/dissociation rates of procaspase-8 to the chain. In line with this, a previous study demonstrated the importance of procaspase-8 dimerization/dissociation in regulating its processing.38 Upon elongation of the DED chain each next molecule is less stably associated with the chain than the previous ones providing clear restrictions on the chain length. A certainly important factor, which was not considered in this study, is the clustering of receptors, which may also differentially regulate procaspase-8 recruitment into the chain and has to be addressed in future work.
In this study, we applied quantitative western blot, mass spectrometry, single-cell analysis, and mathematical modeling to investigate the dynamics of the DED chains and apoptosis initiation. This quantitative analysis gave us new insights into the mechanisms of caspase-8 activation in the DED chains. The understanding of the functions of molecules in their signaling pathway using the knowledge about their concentrations and interactions is a powerful tool, which has to be applied in cellular biology and would undoubtedly provide new insights into a number of the intricate molecular interactions.
Materials and Methods
Cell lines
SKW6.4 cells were maintained in RPMI (Sigma-Aldrich Corporation, St. Louis, MO, USA), with l-glutamine and sodium bicarbonate, supplemented with 10% fetal calf serum in 5% CO2. HeLa-CD95 cells or stably c-FLIPR/L overexpressing HeLa-CD95 cells were maintained in DMEM (Sigma-Aldrich Corporation), with 1000 mg/ml glucose, l-glutamine, and sodium bicarbonate, supplemented with 10% fetal calf serum in 5% CO2.
Antibodies and reagents
Anti–caspase-8 monoclonal antibody C15 (mouse IgG-2b) recognizes the p18 subunit of caspase-8.9 Anti-FLIP monoclonal antibody NF6 (mouse IgG-1) recognizes the N-terminal part of c-FLIP.22 Anti-FADD monoclonal antibody 1C4 (mouse IgG-1) recognizes the C-terminal part of FADD.47 N-terminal anti-caspase-8 antibody was purchased from Millipore (Darmstadt, Germany). Anti-caspase-10 antibody was from MBL (Woburn, MA, USA, clone 4C1). The anti-actin antibody was purchased from Sigma-Aldrich Corporation. Anti-CD95 antibody C-20 for western blot detection was obtained from Santa Cruz Biotechnology (Heidelberg, Germany). Anti-APO-1 (CD95) is an agonistic monoclonal antibody (IgG-3) recognizing an epitope at the extracellular part of CD95.3 Anti-GFP antibody for IP was purchased from eBioscience (Frankfurt am Main, Germany). Anti-Xpress antibody was from Life Technologies (Darmstadt, Germany). Horseradish peroxidase-conjugated goat anti-mouse IgG-1, -2a, and -2b antibodies were from Southern Biotech (Eching, Germany), the goat anti-rabbit antibody was from Santa Cruz Biotechnology. The coding sequence of LZ-CD95L48 was cloned into a pIRESpuro3 plasmid (Takara Bio Inc., Kyoto, Japan). Recombinant LZ-CD95L was produced using 293T cells stably transfected with this vector. All chemicals used were of analytical grade and purchased from Merck (Darmstadt, Germany) or Sigma-Aldrich Corporation.
The Xpress-tagged FADD plasmid was kindly provided by Irmela Jeremias (Helmholtz Center, Munich, Germany). Full-length caspase-8-mCherry, procaspase-8 prodomain-GFP, c-FLIP DED-DED (aa 1–197), and GFP-FADD were kindly provided by Joel Beaudouin (German Cancer Research Center, Heidelberg, Germany). MC159-GFP was cloned from GFP and MC159 plasmids, which were kindly provided by Joel Beaudouin (German Cancer Research Center).
Cell transfection
DNA plasmids were introduced into eukaryotic cell lines using polyethyleneimine (PEI, Sigma-Aldrich Corporation). Cells were seeded in an appropriate cell culture plate or dish to reach 50–70% confluency at the time of transfection. Appropriate amounts of DNA and transfection PEI were diluted in OPTI-MEM in separate 1.5 ml reaction tubes and incubated for 5 min at room temperature. Subsequently, the DNA and transfection reagent solutions were mixed and again incubated for 20 min at room temperature. Finally, the solution of DNA and transfection reagent was added to the cells.
Mass spectrometry
DISC IPs and mass spectrometry were performed as described previously.29, 31 Briefly, 5 × 107 SKW6.4 cells were stimulated with 1 μg/ml or 50 ng/ml CD95L and DISC IPs were prepared and separated by SDS-PAGE. The gel was stained with Coomassie and sliced into pieces. The slices containing the procaspase-8 and its cleavage products were digested and analyzed by LTQ-Orbitrap (Thermo Fisher Scientific, Waltham, MA, USA) using AQUA peptides against procaspase-8 N-terminal part.
Kinetics of procaspase processing in CD95 DISC IPs
For each time point, 107 B lymphoblastoid SKW6.4 cells were harvested by centrifugation for 5 min at 1500 r.p.m., washed once with PBS and resuspended in 1ml RPMI medium in a 6-well plate. Cells were stimulated with CD95L for 5, 15, 30, and 60 min at 37 °C or left unstimulated. Stimulation was stopped by addition of 2 ml cold PBS, cells were centrifuged for 5 min at 1500 r.p.m. and washed once with cold PBS. Cells were lysed in 1 ml lysis buffer (20 mM Tris HCl, pH 7.4, 137 mM NaCl, 2 mM EDTA, 10% glycerine, 1% Triton X-100, 1 mM PMSF, Protease Inhibitor mix (Roche, Penzberg, Germany)) for 30 min on ice and subsequently centrifuged for 15 min at 13 000 r.p.m. In all, 50 μl supernatant was used as a lysate control. The remaining supernatant was immunoprecipitated by mixing with 30 μl protein A-Sepharose and 10 μl anti-APO-1. DISC IPs were rotated for at least 2 h at 4 °C and washed four times with PBS. Samples were subjected to SDS-PAGE (Invitrogen, Darmstadt, Germany) and transferred onto Hybond nitrocellulose membrane using the iBlot system (Invitrogen). Blots were blocked with 5% non-fat dry milk in PBS/Tween (PBS+0.05% Tween-20) for 1 h, washed with PBS/Tween three times for 10 min and incubated with the primary antibody in PBS/Tween for 1 h at room temperature. Blots were developed with the chemiluminescence method according to the manufacturer's protocol (Perkin-Elmer, Rodgau, Germany). Western blots were quantified using ImageJ.49 Special care was taken not to overexpose the membranes. The relative amounts of the different cleavage products, that is, p55/p53, p43/p41, p26/p24 for procaspase-8 and p59/p55, p47/p43, and p25 for procaspase-10 were calculated according to the following formula (representative for procaspase-8).
I(xt) is the measured intensity of one cleavage product (e.g., p55/p53) at time point t. I(p55/p53t), I(p43/p41t), and I(p26/p24t) are the intensities of full-length procaspase-8 (p55/p53), p43/p41 cleavage product, and procaspase-8 prodomain (p26/p24) at time point t, respectively.
Kinetics of procaspase-8 and -10 processing in cell lysates
For each time point, 5 × 106 SKW6.4 cells were plated in 1 ml RPMI in a 6-well plate and stimulated with 1 ml CD95L for 0, 10, 30, 60, 90, and 120 min. CD95 stimulation was stopped by addition of 2 ml cold PBS. Subsequently, the cells were centrifuged at 1500 r.p.m. for 5 min, lysed in 50 μl lysis buffer and western blot was performed as explained for CD95 DISC IPs. The blots were probed using antibodies against caspase-8 (C15), caspase-10, and actin. The bands corresponding to full-length proteins (p55/p53 for procaspase-8, p59/p55 for procaspase-10), their intermediate cleavage products (p43/p41 and p47/p43), and prodomains (procaspase-8 p26/p24 or procaspase-10 p25) or p18 procaspase-8 were quantified using ImageJ.49 Special care was taken not to overexpose the membranes. The band intensity for each individual cleavage product was normalized to actin and the initial amounts of full-length proteins (representative for procaspase-8).
I(xt) is the measured intensity of one procaspase-8 cleavage product (e.g., p55/p53). I(p55/p530) is the intensity of full-length procaspase-8 p55/p53 at time point 0. I(Actint) and I(Actin0) are the intensities of actin at time points t and 0, respectively.
Microscopy
HeLa-CD95 cells were seeded in a 8-well Labtek chambered cover glass at a density of 7000–10000 cells/well and transfected with 100 ng of the DNA corresponding to proteins of interest fused to either GFP or mCherry. Forty-eight hours after transfection, live-cell imaging was performed using a Leica TCS SP5 confocal microscope, equipped with an incubation chamber with temperature and CO2 control. GFP was excited with a wavelength of 488 nm, mCherry was excited at 561 nm. Emission of GFP was recorded in a range of 495–556 nm and mCherry was recorded in a range of 586–677 nm. Fluorescence and transmission light images were acquired using a × 63 1.4 NA oil immersion objective. Experiments were performed at 37 °C and 5% CO2. The images were analyzed in ImageJ.49
Flow cytometry and cell viability assay
HeLa-CD95 cells were seeded in a 6-well plate at a density of 200 000–300 000 cells/well. The next day, the cells were transfected with procaspase-8 prodomain-GFP, c-FLIP DED-DED-GFP (aa 1–197) or GFP or left untransfected. After 6 h, the cells were washed once with PBS and trypsinized. In all, 10000, 5000, 2500, or 1250 cells per well were plated in duplicates in a white 96-well plate. The remaining cells from each well were distributed into five wells in a 24-well plate. The following experiments were performed after approximately 16 h after seeding.
The cells in the 96-well plate were analyzed by CellTiter-Glo luminescent cell viability assay. The medium was removed, leaving 25 μl and then 25 μl CellTiter-Glo solution (1 : 2 dilution, Promega, Mannheim, Germany) added to each well. The plate was incubated on a tumbler in the dark for 15 min. Luminescence was measured using the OrionL Microplate Luminometer (Berthold Detection Systems, Pforzheim, Germany).
The cells in the 24-well plate were washed once with PBS and stimulated with 1 μg/ml or 100 ng/ml CD95L for 4 h or left unstimulated. Subsequently, the medium was transferred to another 24-well plate and propidium iodide added at a final concentration of 10 μg/ml. The cells were washed with PBS and detached using accutase (Sigma-Aldrich Corporation). After cell detachment, the medium was transferred back to the cells and the cells centrifuged at 1500 r.p.m. for 5 min. The supernatant was removed and the cells washed once with PBS and finally resuspended in 500 μl PBS supplemented with 10% FCS. The cells were kept on ice until analysis by flow cytometry using the BD FACSCantoII. Single stained and unstained cells were used to compensate spectral overlap of the GFP and PI signals. The data were analyzed using FlowJo (Treestar Inc., Ashland, OR, USA). Specific cell death was calculated according to the following formula: % specific cell death=(% PI+ in sample−% PI+GFP+ unstimulated) × 100/(100−% PI+GFP+ unstimulated). Specific cell death for the untransfected populations was calculated accordingly, but using the percentage of PI+GFP− cells. Specific cell viability was calculated according to the following formula: (100−% specific cell death).
Cell death induction in HeLa-12 and HeLa-FRL cells was also studied by CellTiter-Glo. In all, 12 000 cells/well were plated in duplicate in a white 96-well plate. After 24 h of attachment, the old medium was removed and the cells were treated with different concentrations of human SuperFasLigand (SFL) (Enzo Life Science, Lörrach, Germany) for 4 h at 37 °C in a cell incubator under an atmosphere containing 5% CO2. The luminescence was detected using Infinite M200 Pro (Tecan). The cell numbers at the time point zero were set to 1 and the cell numbers assessed at the other conditions were normalized to these values.
FADD co-IP
293T cells were seeded in a 10-cm dish 1 day before transfection to reach 50–70% confluency at the day of transfection. The next day each dish was transfected with GFP-FADD, Xpress-FADD, or c-FLIPR, either alone or in combination. From each vector, 3 μg was used for transfection and supplemented with Salmon Sperm DNA to a total amount of 9 μg DNA. Plasmids were expressed for 1 day and then the cells were lysed in 2 ml lysis buffer. Whole cellular lysates were precleared by incubation with 30 μl Protein A-Sepharose beads for 1 h at 4 °C on a tumbler. Lysates were centrifuged at 6000 r.p.m. for 1 min and the cleared supernatant was taken. In all, 40 μl was used as a lysate control. Half of the remaining lysate was used for Xpress IP, the other half for GFP IP using 2 μg of the respective antibodies and 30 μl Protein A-Sepharose beads. IPs were rotated for 2 h at 4 °C and washed four times with PBS. Western blot was performed as explained for CD95 DISC IPs.
Mathematical modeling
In silico analysis of procaspase-8 processing was performed using an improved version of our previously developed agent-based model of DED chain formation.29 The model was refined in the following way. It was assumed that a ligand concentration of 1 μg/ml CD95L would lead to receptor saturation and 1 ng/ml CD95L could stimulate 1% of the receptors.50 The ligand/receptor ratio for 1 ng/ml CD95L was based on theoretical calculations and simulations. Intermediate concentrations were estimated by linear increase. Interaction of molecules and recruitment of the different DISC proteins was modeled using a probabilistic approach as described before (Figure 6a).29 Briefly, a random number in the interval [0, 1] was generated and compared with a given reaction probability, which was also a value between 0 and 1. The reaction could occur if the random number was smaller compared with the reaction probability. Receptor stimulation was modeled in a stepwise manner with CD95L binding probability p_CD95L. For recruitment of DED-only proteins, that is, procaspase-8, c-FLIPL and c-FLIPR, or FADD, a random receptor was drawn from all receptors (stimulated and unstimulated). The recruitment probabilities p_DED and p_FADD were calculated based on the distance of the respective molecule to the chosen receptor.29 Recruitment was repeated for n_DED or n_FADD times, respectively, if recruitment failed, to represent diffusion of the molecules. During each repetition cycle already tested, unstimulated receptors were removed from the selection pool, assuming that molecules would rapidly diffuse to other receptors. p_DED was computed for both, procaspase-8 and c-FLIP, but increased 10-fold for c-FLIP, which was based on previous studies that c-FLIP has a higher affinity to the DISC compared with procaspase-8.50, 51, 52 DISC-bound homodimers of procaspase-8 were processed to p18 and p26/p24 via its intermediate p43/p41 with probabilities processToP18 and processToP43, respectively. Fully processed caspase-8 was released (p_C8_release) and subsequently degraded (p_C8_deg). DED chains could be disassembled with probability p_dissociate_DISC followed by degradation of procaspase-8 prodomain with p_deg_prodomain. Procaspase-8 could be also degraded after dissociation of DED chains with probability p_deg_proc8, p43/p41 could be further processed in the lysate to p18 and p26/p24. Reversible binding of procaspase-8 was added when indicated with a probability p_Proc8_release. This dissociation probability was influenced by the DED chain length by decreasing the random number R depending on the chain length L by R/L. In addition, we tested another implementation of increasing procaspase-8 dissociation depending on the chain length. There, the procaspase-8 dissociation probability was kept constant, but the random number was generated multiple times depending on the chain length, which increases the probability of a random number smaller than the dissociation probability. This second implementation gave very similar simulation results. Upon addition of the negative feedback loop to the model, procaspase-8 dissociation probability was slightly reduced (Supplementary Table S1).
Experimental data for simulations
Experimental data for 10, 200, and 1000 ng/ml CD95L were obtained from quantitative western blot comprising p55/p53, p43/p41, p18, and p26/p24 (Supplementary Figure S4A). Quantifications for p55/p53 and p43/p41 were averaged from both C- and N-Terminal procaspase-8 antibody signals. Data for p55/p53, p43/p41, and p18 were further combined with the corresponding data from our previous work.29 Experimental data for 500 and 50 ng/ml CD95L were obtained from quantitative western blot comprising p55/p53, p43/p41, and p18 (Supplementary Figure S4C). Experimental data for HeLa-CD95 cells were taken from our previous work.35
Parameter estimation
The model parameters were estimated based on experimental data of procaspase-8 processing in whole cellular lysates (Figure 6b,Supplementary Figure S4A) and at the DISC (Figure 1b, Supplementary Figure S4B) at 1000 ng/ml CD95L stimulation. To estimate the modeling parameters, different parameters were varied in a certain range in various combinations. Subsequently, the model error χ2 was calculated for each simulation run according to least squares.
yid(tj) denotes the experimental data of observable i and time point j. yis(tj) denotes the corresponding simulation results and σij2 is the variance of the data. The observables were p55/p53, p43/p41, p18, and p26/p24 in whole cellular lysates at time points 0, 10, 30, 60, 90, and 120 min and p55/p53, p43/p41, and p26/p24 in DISC IPs at time points 15, 30, and 60 min. In total, almost 100 000 different simulations were run with the goal to minimize χ2. The quality of model fit in parameter estimation was assessed by χ2/N, which is the model error divided by the number of data points N. A value of χ2/N<1 indicates a good fit. For lysate and DISC data alone these were 0.5 and 1.16, respectively. Both data taken together gave a χ2/N=0.71, indicating a good fit between simulations and experimental data. Final parameter values are listed in Supplementary Table S1.
Sensitivity analysis
For sensitivity analysis of different parameter values on the simulation results, each parameter j was varied independently from x/5 to x*5, using Np different values, with x being the used parameter value (Supplementary Table S1). The step size for each parameter was computed for 50 variations. Due to rounding of values, the number Np of parameter variations might vary a little. For each parameter value v of parameter j, the relative change in concentration sij for each observable i was computed for each simulation time point t.
The sensitivity Sij for each observable i upon changing parameter j was computed by summing up the concentration changes over the whole simulation time T for all Np parameter values and normalized to the number of parameter values Np to adjust for different number of parameter variations.
Modeling procaspase-8 processing in HeLa-CD95 cells
To model procaspase-8 processing in HeLa-CD95 cells, the estimated kinetic parameters were used (Supplementary Table S1) and molecule numbers for CD95 (170 999), Procaspase-8 (250 000), FADD (130 000), c-FLIPL (230), and c-FLIPR (530) taken from the literature.29, 35 In all, 3 μg/ml CD95L corresponded to 1 500 000 molecules resulting in about 10-fold oversaturation of ligand/receptor.35 To account for ligand oversaturation, the model was adapted to allow CD95 stimulation multiple times with probability p_CD95L depending on the level of oversaturation, for example, in 10-fold oversaturation each receptor might be stimulated by 10 ligands. In addition, a cytosolic volume of HeLa cells of 940 μm3 was used to calculate the grid size of the simulation environment.29, 53
In silico analysis of the DED chain structure
Analysis of the structural features of the DED chain was performed in silico using PyMOL. The structure of full-length FADD was obtained from the RCSB database (PDB 2GF5). The structure of the procaspase-8 prodomain was modeled using the Phyre2 web service41 from the sequence of procaspase-8 prodomain (aa 1–216, Uniprot ID Q14790) and the structure of the viral FLIP MC159 (PDB 2BBR). The DED chain was assembled using ad-hoc structural considerations based on the suggested interaction of the tandem DED with the FADD DED based on structural and mutagenesis studies on MC159, FADD, procaspase-8, and c-FLIP.42 These showed that helix α2/α5 of MC159 DED2, comprising the conserved hydrophobic patch (FL-motif), interacts with α1/α4 of the FADD DED.42 The same structural features make up the DED1/DED2 interface of MC159.42 Helices α1/α4 in DED1 of MC159 are free to interact with another tandem DED.
Statistics
Statistical analysis was carried out in R.54 Statistical significance was analyzed using Student's t-test. Percentage data were transformed before analysis using the arcsine square root transformation according to 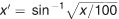 .
.
Acknowledgments
We thank Joel Beaudouin for providing various plasmids and Irmela Jeremias for providing Xpress-tagged FADD. Further, we thank Claudia Arndt for experimental support. We acknowledge the Ministry of Sciences and Economic Affairs of Saxony-Anhalt (Research Centre Dynamic Systems: Biosystems Engineering, MW-21LMS 5), BMBF (eBIO project ‘ImmunoQuant'-TPU-0316170G), (eBIO project ‘AML' 031A304), DFG (LA 2386/6-1), and Russian Science Foundation 14-44-00011 for supporting our work. The support from project 14-44-00011 was used for modeling.
Glossary
- DISC
death-inducing signaling complex
- DED
death effector domain
- DD
death domain
- c-FLIP
cellular-FLICE-like inhibitory protein
- FADD
Fas-associated death domain
- DR
death receptor
- AQUA peptide
Absolute Quantification peptide
- IP
immunoprecipitation
- PDB
protein data bank
- PBS
phosphate-buffered saline
- GFP
green fluorescence protein
- SFL
super Fas-Ligand
- zVAD-fmk
carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]- fluoromethylketone
- RLU
relative luminescence units
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by S Nagata
Supplementary Material
References
- Krammer PH. CD95's deadly mission in the immune system. Nature 2000; 407: 789–795. [DOI] [PubMed] [Google Scholar]
- Lavrik IN, Golks A, Krammer PH. Caspases: pharmacological manipulation of cell death. J Clin Invest 2005; 115: 2665–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauth BC, Klas C, Peters AM, Matzku S, Möller P, Falk W et al. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science 1989; 245: 301–305. [DOI] [PubMed] [Google Scholar]
- O'Reilly LA, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature 2009; 461: 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann L, Pforr C, Beaudouin J, Pappa A, Fricker N, Krammer PH et al. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol Syst Biol 2010; 6: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen SP, Henry CM, Kearney CJ, Logue SE, Feoktistova M, Tynan GA et al. Fas/CD95-induced chemokines can serve as "Find-Me" signals for apoptotic cells. Mol Cell 2013; 49: 1034–1048. [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J 1995; 14: 5579–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer PH, Arnold R, Lavrik IN. Life and death in peripheral T cells. Nat Rev Immunol 2007; 7: 532–542. [DOI] [PubMed] [Google Scholar]
- Scaffidi C, Medema JP, Krammer PH, Peter ME. FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. J Biol Chem 1997; 272: 26953–26958. [DOI] [PubMed] [Google Scholar]
- Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand MA, Walczak H. Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J 2002; 21: 4520–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Lo Y-C, Lin S-C, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol 2007; 25: 561–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J 2004; 384: 201–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golks A, Brenner D, Schmitz I, Watzl C, Krueger A, Krammer PH et al. The role of CAP3 in CD95 signaling: new insights into the mechanism of procaspase-8 activation. Cell Death Differ 2006; 13: 489–498. [DOI] [PubMed] [Google Scholar]
- Hoffmann JC, Pappa A, Krammer PH, Lavrik IN. A new C-terminal cleavage product of procaspase-8, p30, defines an alternative pathway of procaspase-8 activation. Mol Cell Biol 2009; 29: 4431–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrik I, Krueger A, Schmitz I, Baumann S, Weyd H, Krammer PH et al. The active caspase-8 heterotetramer is formed at the CD95 DISC. Cell Death Differ 2003; 10: 144–145. [DOI] [PubMed] [Google Scholar]
- Medema JP, Scaffidi C, Kischkel FC, Shevchenko A, Mann M, Krammer PH et al. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J 1997; 16: 2794–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema JP, Toes RE, Scaffidi C, Zheng TS, Flavell RA, Melief CJ et al. Cleavage of FLICE (caspase-8) by granzyme B during cytotoxic T lymphocyte-induced apoptosis. Eur J Immunol 1997; 27: 3492–3498. [DOI] [PubMed] [Google Scholar]
- Hughes MA, Harper N, Butterworth M, Cain K, Cohen GM, MacFarlane M. Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol Cell 2009; 35: 265–279. [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P et al. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem 2001; 276: 46639–46646. [DOI] [PubMed] [Google Scholar]
- Fischer U, Stroh C, Schulze-Osthoff K. Unique and overlapping substrate specificities of caspase-8 and caspase-10. Oncogene 2006; 25: 152–159. [DOI] [PubMed] [Google Scholar]
- Mühlethaler-Mottet A, Flahaut M, Bourloud KB, Nardou K, Coulon A, Liberman J et al. Individual caspase-10 isoforms play distinct and opposing roles in the initiation of death receptor-mediated tumour cell apoptosis. Cell Death Dis 2011; 2: e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi C, Schmitz I, Krammer PH, Peter ME. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem 1999; 274: 1541–1548. [DOI] [PubMed] [Google Scholar]
- Golks A, Brenner D, Fritsch C, Krammer PH, Lavrik IN. c-FLIPR, a new regulator of death receptor-induced apoptosis. J Biol Chem 2005; 280: 14507–14513. [DOI] [PubMed] [Google Scholar]
- Golks A, Brenner D, Krammer PH, Lavrik IN. The c-FLIP-NH2 terminus (p22-FLIP) induces NF-kB activation. J Exp Med 2006; 203: 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueffing N, Schuster M, Keil E, Schulze-Osthoff K, Schmitz I. Up-regulation of c-FLIP short by NFAT contributes to apoptosis resistance of short-term activated T cells. Blood 2008; 112: 690–698. [DOI] [PubMed] [Google Scholar]
- Krueger A, Schmitz I, Baumann S, Krammer PH, Kirchhoff S. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J Biol Chem 2001; 276: 20633–20640. [DOI] [PubMed] [Google Scholar]
- Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW et al. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J Biol Chem 2002; 277: 45162–45171. [DOI] [PubMed] [Google Scholar]
- Yu JW, Jeffrey PD, Shi Y. Mechanism of procaspase-8 activation by c-FLIPL. Proc Natl Acad Sci USA 2009; 106: 8169–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich K, Warnken U, Fricker N, Oztürk S, Richter P, Kammerer K et al. Stoichiometry of the CD95 death-inducing signaling complex: experimental and modeling evidence for a death effector domain chain model. Mol Cell 2012; 47: 306–319. [DOI] [PubMed] [Google Scholar]
- Dickens LS, Boyd RS, Jukes-Jones R, Hughes MA, Robinson GL, Fairall L et al. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol Cell 2012; 47: 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnken U, Schleich K, Schnölzer M, Lavrik I. Quantification of high-molecular weight protein platforms by AQUA mass spectrometry as exemplified for the CD95 death-inducing signaling complex (DISC). Cells 2013; 2: 476–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Orozco A, Spencer DM, Wang J. Activation of initiator caspases through a stable dimeric intermediate. J Biol Chem 2002; 277: 50761–50767. [DOI] [PubMed] [Google Scholar]
- Keller N, Mares J, Zerbe O, Grütter MG. Structural and biochemical studies on procaspase-8: new insights on initiator caspase activation. Structure 2009; 17: 438–448. [DOI] [PubMed] [Google Scholar]
- Yang JK, Wang L, Zheng L, Wan F, Ahmed M, Lenardo MJ et al. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol Cell 2005; 20: 939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker N, Beaudouin J, Richter P, Eils R, Krammer PH, Lavrik IN. Model-based dissection of CD95 signaling dynamics reveals both a pro- and antiapoptotic role of c-FLIPL. J Cell Biol 2010; 190: 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvey TL, Bertin J, Siegel RM, Wang GH, Lenardo MJ, Cohen JI. Binding of FADD and Caspase-8 to molluscum contagiosum virus MC159 v-FLIP is not sufficient for its antiapoptotic function. J Virol 2002; 76: 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueffing N, Keil E, Freund C, Kühne R, Schulze-Osthoff K, Schmitz I. Mutational analyses of c-FLIPR, the only murine short FLIP isoform, reveal requirements for DISC recruitment. Cell Death Differ 2008; 15: 773–782. [DOI] [PubMed] [Google Scholar]
- Würstle ML, Laussmann MA, Rehm M. The caspase-8 dimerization/dissociation balance is a highly potent regulator of caspase-8, -3, -6 signaling. J Biol Chem 2010; 285: 33209–33218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalvez F, Lawrence D, Yang B, Yee S, Pitti R, Marsters S et al. TRAF2 sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol Cell 2012; 48: 888–899. [DOI] [PubMed] [Google Scholar]
- Schrodinger LC. The PyMOL Molecular Graphics System Version 1.3r1 2010. Available from https://www.pymol.org/citing.
- Kelley LA. Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 2009; 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Yu JW, Shi Y. FLIP and the death effector domain family. Oncogene 2008; 27: 6216–6227. [DOI] [PubMed] [Google Scholar]
- Toivonen HT, Meinander A, Asaoka T, Westerlund M, Pettersson F, Mikhailov A et al. Modeling reveals that dynamic regulation of c-FLIP levels determines cell-to-cell distribution of CD95-mediated apoptosis. J Biol Chem 2011; 286: 18375–18382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama Y, Shen L, Yonetani M, Miyauchi J, Miyashita T, Yamada M. Death effector domain-only polypeptides of caspase-8 and -10 specifically inhibit death receptor-induced cell death. Biochem Biophys Res Commun 2002; 291: 484–493. [DOI] [PubMed] [Google Scholar]
- Himeji D, Horiuchi T, Tsukamoto H, Hayashi K, Watanabe T, Harada M. Characterization of caspase-8L: a novel isoform of caspase-8 that behaves as an inhibitor of the caspase cascade. Blood 2002; 99: 4070–4078. [DOI] [PubMed] [Google Scholar]
- Siegel RM, Martin DA, Zheng L, Ng SY, Bertin J, Cohen J et al. Death-effector filaments: novel cytoplasmic structures that recruit caspases and trigger apoptosis. J Cell Biol 1998; 141: 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi C, Volkland J, Blomberg I, Hoffmann I, Krammer PH, Peter ME. Phosphorylation of FADD/ MORT1 at serine 194 and association with a 70-kDa cell cycle-regulated protein kinase. J Immunol 2000; 164: 1236–1242. [DOI] [PubMed] [Google Scholar]
- Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med 1999; 5: 157–163. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int 2004; 11: 36–42. [Google Scholar]
- Bentele M, Lavrik I, Ulrich M, Stösser S, Heermann DW, Kalthoff H et al. Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J Cell Biol 2004; 166: 839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S et al. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J 2002; 21: 3704–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrik IN, Golks A, Riess D, Bentele M, Eils R, Krammer PH. Analysis of CD95 threshold signaling: triggering of CD95 (FAS/APO-1) at low concentrations primarily results in survival signaling. J Biol Chem 2007; 282: 13664–13671. [DOI] [PubMed] [Google Scholar]
- Milo R, Jorgensen P, Moran U, Weber G, Springer M. BioNumbers-the database of key numbers in molecular and cell biology. Nucleic Acids Res 2010; 38: D750–D753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core TeamR: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.