Abstract
Objective Semantic role labeling (SRL), which extracts a shallow semantic relation representation from different surface textual forms of free text sentences, is important for understanding natural language. Few studies in SRL have been conducted in the medical domain, primarily due to lack of annotated clinical SRL corpora, which are time-consuming and costly to build. The goal of this study is to investigate domain adaptation techniques for clinical SRL leveraging resources built from newswire and biomedical literature to improve performance and save annotation costs.
Materials and Methods Multisource Integrated Platform for Answering Clinical Questions (MiPACQ), a manually annotated SRL clinical corpus, was used as the target domain dataset. PropBank and NomBank from newswire and BioProp from biomedical literature were used as source domain datasets. Three state-of-the-art domain adaptation algorithms were employed: instance pruning, transfer self-training, and feature augmentation. The SRL performance using different domain adaptation algorithms was evaluated by using 10-fold cross-validation on the MiPACQ corpus. Learning curves for the different methods were generated to assess the effect of sample size.
Results and Conclusion When all three source domain corpora were used, the feature augmentation algorithm achieved statistically significant higher F-measure (83.18%), compared to the baseline with MiPACQ dataset alone (F-measure, 81.53%), indicating that domain adaptation algorithms may improve SRL performance on clinical text. To achieve a comparable performance to the baseline method that used 90% of MiPACQ training samples, the feature augmentation algorithm required <50% of training samples in MiPACQ, demonstrating that annotation costs of clinical SRL can be reduced significantly by leveraging existing SRL resources from other domains.
Keywords: semantic role labeling, shallow semantic parsing, clinical natural language processing, domain adaptation, transfer learning
INTRODUCTION
Natural language processing (NLP) technologies are important for unlocking information embedded in narrative reports in electronic health record systems. Although various NLP systems have been developed to support a wide range of computerized medical applications, such as biosurveillance and clinical decision support, extracting semantically meaningful information from clinical text remains a challenge. Semantic role labeling (SRL)1 (also known as shallow semantic parsing),2 which extracts semantic relations between predicates and their arguments from different surface textual forms, is an important method for the extraction of semantic information. State-of-the-art SRL systems have been developed and applied to information extraction in open domains and various biomedical subdomains.3–12 However, very few SRL studies have been conducted in the clinical domain,13,14 probably due to the lack of large-scale annotated corpora. The creation of such clinical SRL corpora would be both time-consuming and expensive.13
In this study, we approach SRL on clinical narratives as a domain adaptation problem. The goal is to adapt existing the SRL corpora of newswire text15,16 and biomedical literature17 to the clinical domain. By transferring knowledge from existing corpora in other domains to the clinical domain, we aim to improve the performance of clinical SRL and reduce the cost of developing one de novo. We used three existing SRL corpora outside the clinical domain and evaluated three state-of-the-art domain adaptation algorithms on the task of SRL for clinical text. Our results showed that domain adaptation strategies were effective for improving the performance or reducing the annotation cost of SRL on clinical text. To the best of our knowledge, this is the first work that has introduced domain adaptation algorithms for clinical SRL.
BACKGROUND
The task of SRL is to label semantic relations in a sentence as predicate argument structures (PASs) to represent propositions.18 The definition of PAS originated from the predicate logic for proposition representation in semantics theory.2 There is a large body of work on extracting semantic relations in biomedical text.4–12,19–23 Many are based on the sublanguage theory by Harris,24 which describes the properties of language in closed domains. Typically, in a closed domain such as medicine, there are a limited number of primary semantic types and a set of constraints that can determine how different semantic types of the arguments can be linked to form semantic predications.25 Linguistic String Project (LSP)21 and Medical Language Extraction and Encoding System (MedLEE),22 which use sublanguage grammar, are two early NLP systems for the extraction of semantic relations in the medical domain. SemRep is another biomedical semantic relation extraction system, which extracts semantic predications defined in the Unified Medical Language System Semantic Network from biomedical literature.19,20 Recently, Cohen et al.26 examined the syntactic alternations in the argument structure of domain-specific verbs and associated nominalizations in the PennBioIE corpus, and found that even in a semantically restricted domain, syntactic variations are common and diverse. Currently, many sublanguage-based clinical NLP systems often recognize semantic relations24 by manually extracted patterns using rule-based methods.22,27 SRL, however, focuses on unifying variations in the surface syntactic forms of semantic relations based on annotated corpora. It is inspired by previous research into semantic frames28,29 and the link between semantic roles and syntactic realization.30 Although current SRL approaches are primarily developed in open domains (thus, types of semantic roles or arguments may not be sufficient or appropriate for the medical domain), they can be refined and extended to the medical domain,13,14 thus providing alternative or complementary approaches for clinical semantic relation extraction.
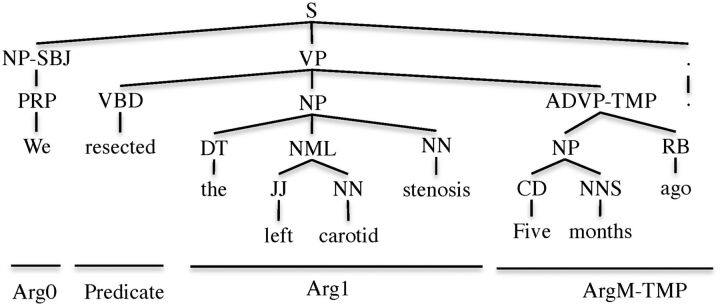
In SRL, a predicate usually refers to a word indicating a relation or an attribute, and arguments refer to syntactic constituents acting as different semantic roles to the predicate. The core arguments represent the essential arguments of a predicate, whereas adjunctive arguments express general properties of a predicate such as time and location. For example, in Figure 1, in the sentence “We resected the left carotid stenosis 5 months ago,” “resected” is the predicate, “We” comprises the role of agent (initiator/executor in the relation), indicating the surgeon performing the action “resect”; “the left carotid stenosis” comprises the role of patient (receptor in the relation), indicating the entity removed by the action “resect,” and “5 months ago” is the time. The roles of agent and patient are the core arguments and time is the adjunctive argument of the predicate.
Figure 1.
Syntactic parse tree annotated with semantic roles.
Automatic SRL was first introduced by Gildea and Jurafsky in 2002.31 Since then, SRL has attracted attention owing to its usefulness for multiple NLP tasks and applications, such as information extraction and question answering.3,32,33 With public availability of large-scale annotated corpora such as PropBank,18 and promotion by CoNLL shared tasks,34,35 many data-driven approaches have been developed for SRL in open domains such as newswire. This approach is standardized and divides SRL into several successive tasks. Argument identification (AI) finds all syntactic constituents with semantic roles, that is, arguments in the sentence. Argument classification (AC) determines the specific semantic role of an argument. Global inference refines the output of preceding tasks using global constraints. These tasks can be conquered individually36,37 or as a combined task.12,31 Other approaches include resolving syntactic parsing and SRL jointly by integrating them into a single model,37 or by using Markov Logic Networks as the learning framework.38
In the last few years, efforts have focused on building SRL systems for biomedical literature. Wattarujeekrit et al.7 developed PASBio, which analyzes and annotates the PASs of over 30 verbs for molecular events. Kogan et al.8 annotated PAS in medical case reports. The LSAT system, developed by Shah and Bork,9 used SRL to identify information about gene transcripts. Bethard et al.10 extracted information about protein movement by using an SRL system, in which 34 verbs and four semantic roles focusing specifically on protein movement were defined and annotated in their corpus. Barnickel et al.11 presented a neural network–based SRL system for relation extraction with emphasis on improving processing speed. Paek et al.12 semantically analyzed abstracts from randomized trials with SRL; however, the predicate set only contained five verbs. The BIOSMILE system by Tsai et al.17 was built on the BioProp corpus, in which PASs of 30 verbs were annotated following the PropBank guideline. Interestingly, their results showed that the SRL system trained on PropBank alone did not perform well on BioProp.
All the previously described SRL systems were built on annotated corpora of biomedical literature, facilitating literature-based information extraction applications.23 However, very few studies have been conducted to investigate SRL techniques for clinical text from electronic health records.39 For example, Wang et al.14,40 analyzed and annotated PASs of 30 predicates in operative notes following the PropBank style, but they did not build an automatic SRL system. The first clinical SRL system was reported by Albright et al.,13 who created an annotated corpus, Multisource Integrated Platform for Answering Clinical Questions (MiPACQ), containing multiple syntactic and semantic layers including SRL information. The SRL dataset in MiPACQ contains 1137 predicates and the SRL performance on that dataset was 79.91% by adopting an existing SRL method developed for newswire.13 The primary limitation for clinical SRL research is apparently the lack of annotated SRL corpora in the medical domain. It is time-consuming and expensive to create large annotated clinical corpora, because this often requires manual review by domain experts such as physicians. Moreover, medicine consists of different subdomains (e.g., internal medicine, pathology, and radiology) and the languages of different subdomains can be distinct,25 which makes it challenging to transfer machine learning–based models from one subdomain to another. For example, the MiPACQ dataset was a collection of Mayo Clinic’s clinical notes (CNs) and pathology notes regarding colon cancer.13 When the SRL model built from MiPACQ was tested on two other CN datasets of different genres and note styles, namely radiology notes from the Strategic Health IT Advanced Research Projects and colon cancer clinical and pathology notes from Temporal Histories of Your Medical Events, the performance dropped significantly.13 Therefore, to construct high-performance SRL systems for each subdomain, we may have to create annotated corpora for every specific subdomain, which would require substantial effort and resources.
To address this limitation in clinical SRL development, we propose an investigation of domain adaptation techniques for SRL. The task of domain adaption is to adapt a classifier that is trained on a source domain to a new target domain. This improves performance and reduces dependency of the classifier on large annotated datasets in the target domain.41,42 Transfer learning algorithms are often employed for domain adaptation.43 Given a source domain, Ds, and its learning task, Ts, a target domain, DT, and its learning task, TT, transfer learning aims to improve the learning of the target predictive model in DT by using the knowledge from Ds and Ts.39 Most commonly used transfer learning algorithms can be categorized into instance-level and feature-level approaches. Let LS denote the labeled dataset of DS, LT denote the labeled dataset of DT, and UT denote the unlabeled dataset of DT; here, instance-level transfer learning algorithms aim to select or weight instances in LS for use in the target domain.43 This does not depend on the machine learning algorithm used for building classifiers, but requires a small gap between the source and target domains. Feature-level transfer learning algorithms aim to find a new feature representation that reduces the difference between features in DS and DT and highlights the similarity between them. This has a moderate dependency on machine learning algorithms, but is more tolerant of domain gap. Transfer learning algorithms have been effective in solving the problem of data scarcity in DT for several key bioinformatics areas, such as sequence classification and gene expression data analysis.44 Dahlmeier and Ng45 addressed SRL on BioProp by using domain adaptation algorithms for the first time, with PropBank as the source domain dataset. Their results demonstrated that the cost of developing an SRL system for interpreting molecular events could be significantly reduced. Ferraro et al.46 also showed improved performance of POS tagging on clinical narratives using the feature-level transfer learning algorithm Easy Adapt. More recently, Laippala et al.47 investigated the use of “source only,” “target only,” and “source + target” in statistical parsing of clinical Finnish; however, no domain adaptation algorithm was employed in their work.
In this study, we explored both instance-level and feature-level domain adaptation algorithms for SRL on clinical narratives. We used PropBank, NomBank, and BioProp as source domain datasets and MiPACQ as the target domain dataset. Dahlmeier and Ng45 previously conducted a domain adaptation study on biomedical literature using BioProp as the target dataset and PropBank as the source dataset, and they obtained promising results on molecular event interpretation. Our study design is similar to the work of Dahlmeier and Ng.45 However, we focus on clinical text instead of biomedical literature on molecular events. Previous studies have shown that clinical reports and biomedical literature are two very different sublanguages in terms of semantic relation types and complexity.25 Their differences were also demonstrated in various other NLP tasks, such as word sense disambiguation and medical term identification.48,49 Furthermore, BioProp only contains semantic roles of 30 verb predicates with 1962 PASs; MiPACQ has 722 verb predicates and 415 nominal predicates with 12 575 PASs. Therefore, it is important to assess domain adaptation methods on the larger clinical SRL corpus, in addition to biomedical literature. Moreover, we investigated the effect of additional external corpora (i.e., BioProp and NomBank) and their combinations on clinical SRL, which was not reported in Dahlmeier and Ng’s work. To the best of our knowledge, this is the first work that has introduced domain adaptation algorithms for clinical SRL. Our evaluation showed that domain adaptation algorithms can improve performance or reduce annotation costs for clinical SRL.
METHODS
Datasets
We used four annotated SRL datasets in our study, three as source domain datasets and one as the target dataset, as described in the following.
Source domain datasets
The PropBank corpus18 is the most widely used corpus for developing SRL systems.45 The corpus is built from news articles of the Wall Street Journal and is available through the Linguistic Data Consortium (http://www.ldc.upenn.edu). Semantic roles of verb predicates are annotated in this corpus. The PropBank corpus has 25 sections, denoted as sections 00–24. We used the standard training set of sections 2–21 as the source domain dataset.
The NomBank corpus16 contains annotated semantic roles of nominal predicates. Similar to PropBank, it is built from news articles of the Wall Street Journal, based on the Penn TreeBank. It is available online at http://nlp.cs.nyu.edu/meyers/NomBank.html. Following PropBank, the standard training set of sections 2–21 was used as the source domain dataset.50
The BioProp corpus17 is annotated based on the GENIA Treebank.51 The GENIA Treebank facilitates information extraction from biomedical literatures about proteins. It is available for downloading from the GENIA project web site (http://www.nactem.ac.uk/genia/genia-corpus/treebank). Specifically, BioProp was created from 500 MEDLINE abstracts. The articles were selected based on the keywords “human,” “blood cells,” and “transcription factor.” Semantic roles of verb predicates are annotated in BioProp.
Target domain dataset
MiPACQ is built from randomly selected CNs and pathology notes of Mayo Clinic related to colon cancer.13 Annotations of layered linguistic information including part of speech tagging, PAS for SRL, named entities, and semantic information from Unified Medical Language Systems are available for building NLP components. The predicate–argument semantic annotations follow PropBank guidelines. Both verb and nominal predicates are annotated in MiPACQ.
Table 1 displays the statistics of the four corpora. MiPACQ contains 722 verb predicates with 9780 PAS and 415 nominal predicates with 2795 PASs. PropBank and BioProp (PB) have only verb predicates; NomBank has only nominal predicates. Among the 722 verb predicates in MiPACQ, 644 are common with PropBank and 15 are common with BioProp. Among the 415 nominal predicates, 265 are common with NomBank. As displayed in Table 1, among the three source domain datasets, the sizes of PropBank/NomBank are significantly larger than BioProp (∼50 times more PASs). Moreover, the size of BioProp is much smaller than MiPACQ, with a ratio of 1:38 for predicates.
Table 1.
Corpus statistics for MiPACQ, PropBank, NomBank, and BioProp
| MiPACQ | PropBank | NomBank | BioProp | |
|---|---|---|---|---|
| Sentences | 6145 | 36 090 | 41 964 | 1635 |
| Unique Predicate | 1137 | 3257 | 4706 | 30 |
| PAS | 12 575 | 112 917 | 114 574 | 1962 |
| ARG0 | 5633 | 66 329 | 49 823 | 1464 |
| ARG1 | 10 343 | 92 958 | 80 102 | 2124 |
| ARG2 | 3080 | 20 547 | 34 850 | 325 |
| ARG3 | 162 | 3491 | 7611 | 8 |
| ARG4 | 134 | 2739 | 494 | 5 |
| ARG5 | 2 | 69 | 23 | 0 |
| ALL_ARGM | 8793 | 60 962 | 25 166 | 1762 |
Domain Adaptation Algorithms
The three transfer-learning algorithms employed in this study are described in detail in the following.
Instance pruning
Instance pruning (InstancePrune) trains a classifier on LT and uses this classifier to predict class labels for LS.52 The top p instances that are predicted wrongly, ranked by prediction confidence, are removed from LS. The intuition here is that instances that are very different from the target domain will affect the prediction ability of the classifier model. The remaining instances, L′S, are added to LT as training data.
Transfer self-training
Transfer self-training (TransferSelf) borrows the idea of self-training from the framework of semisupervised learning into transfer learning.53 It iteratively trains the classifier by transferring a subset of L′S with high similarity to instances in DT from LS to enrich LT as the training data.54 First, a classifier trained using LT is applied to LS. For each category in LS, the top n correctly classified instances ranked by prediction confidence are selected and added into LT as the training data. The classifier is then retrained on the enriched training data and applied to LS–L′S again to select more instances. The iteration terminates when the prediction confidence of instance in LS–L′S is less than a specified threshold or the maximum allowed iterations is exceeded. The final classifier is obtained by training on the combination of LT and L′S.
Both InstancePrune and TransferSelf are instance-level transfer learning algorithms. The differences between them include: (1) InstancePrune attempts to remove wrongly predicted source instances of high confidence; TransferSelf selects correctly predicted source instances of high confidence into the training set; and (2) the source instance selection in InstancePrune is conducted only once; TransferSelf adds source instances iteratively into the training set, thus leveraging selected source instances in previous iterations. In Dahlmeier and Ng,45 another instance level transfer learning algorithm named instance weighting52 was employed for domain adaptation. However, the experimental results were not promising.45 In our pilot study, we tried another instance weighting algorithm called TrAdaBoost55 and it did not perform well either. Hence, we did not employ instance weighting in our study.
Feature augmentation
For feature augmentation (FeatureAug), Daumé III56 proposed a domain adaptation algorithm that maps feature vectors into a higher dimension. This algorithm is also called Easy Adapt, because it can be implemented simply with a few lines of Perl script.56 Denote Xs and XT as the original feature vectors for source and target domain, respectively, then mapping is conducted as follows:
where 0 is a zero vector of length |X|. By this transformation, the feature spaces of both DS and DT are augmented. Three versions of features are generated from each original feature vector, namely, “general,” “source-specific,” and “target-specific” versions. The intuition of this algorithm is to leverage the aggregation of the three feature space versions to learn an efficient feature representation for DT. Common features between DS and DT are assigned with higher weights in instances of both domains; whereas features unique to DS or DT are assigned with higher weights only in instances of DS or DT. A standard machine-learning algorithm will assign weights differently to features in each version. Effective features for DT will be emphasized from the general and target-specific versions.
Experiments
System description
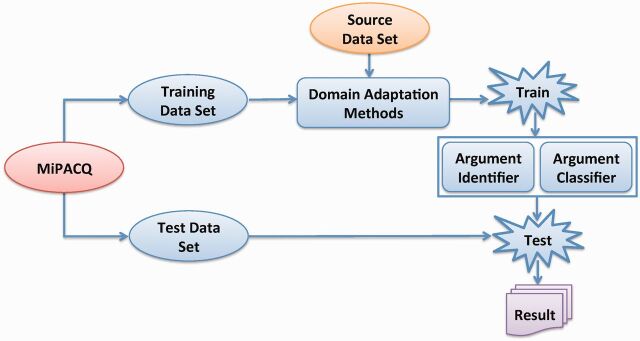
Figure 2 shows the study design of the domain adaptation–based SRL. The SRL system can be viewed as consisting of the training stage and the testing stage. In the training stage, SRL is split into two subtasks: the AI and AC subtasks. First, a binary nonargument versus argument classifier is built as the argument identifier on the entire dataset for all predicates, instead of building one model per predicate. For AC, a multiclass classifier is built to assign semantic roles to arguments of all predicates. In the testing stage, for each predicate, the argument candidates first pass through the argument identifier. If one candidate is identified as an argument, it will go through the argument classifier that assigns the semantic role.
Figure 2:
Overview of the study design of domain adaptation based semantic role labeling. Experimental processes are indicated in blue.
Features and machine-learning algorithm
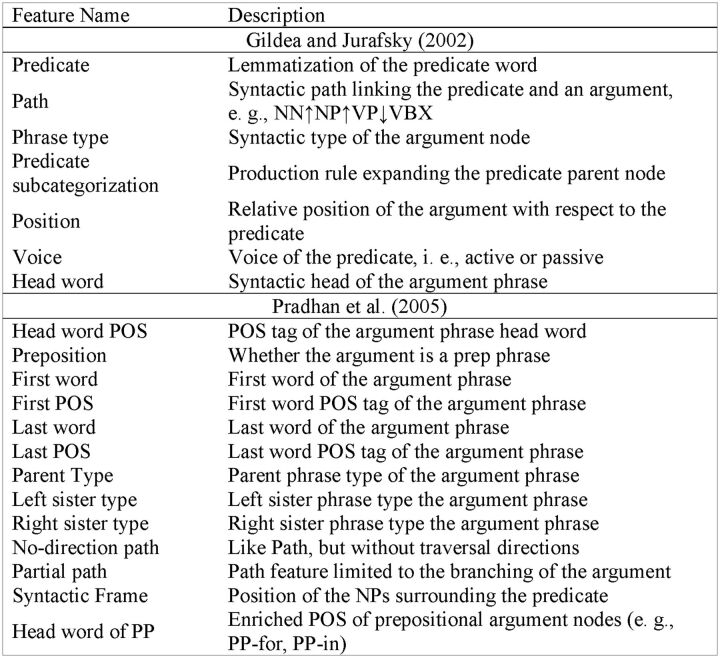
Similar to the work of Tsai et al.17 and Dahlmeier and Ng,17,45 we adopted common features used in current state-of-the-art SRL systems. These features include seven baseline features from the original work of Gildea and Jurafsky31 and additional features taken from Pradhan et al.57 All were extracted from the syntactic parse tree, and are shown in Figure 3. The “Voice” feature is not used for nominal predicates.
Figure 3:
Feature list for semantic role labeling.
We used the open source toolkit Liblinear58 for implementations of machine-learning algorithms. The logistic regression algorithm was applied to select confidence-based source domain training instances for InstancePrune and TransferSelf, because it outputs predictions with their probabilities, which can serve as the prediction confidence.52 The linear support vector machine algorithm was used to build SRL models owing to its high generalization ability for new data.59
Experimental setup
Following the work of Dahlmeier and Ng,45 we used the gold-standard parsing annotations of PropBank, NomBank, BioProp, and MiPACQ in our SRL experiments. In addition to separately using each source domain dataset, the combination of PropBank and NomBank (PN), PB, and all three source datasets (PNB) are used as Ds in our experiments to examine the influence of multiple sources on domain adaptation. Only the PASs with at least one argument were used. For each implemented method, all parameters were tuned for optimal performance.
Experiments and systematic analysis were conducted as discussed in the following.
Algorithms for domain adaptation: InstancePrune, TransferSelf, and FeatureAug were employed in this study, as described in the “Methods” section. To examine the effectiveness of these algorithms, three baseline methods were also developed for comparison: the “Source Only” method uses only Ds to train a classifier; the “Target Only” method uses only DT to train a classifier; the “Source & Target” method directly combines both DS and DT to train a classifier.
Influence of sample size on domain adaptation: To determine the effect of sample size on SRL performance, classifiers were also trained using varying sample sizes of DS and DT. We examined the performance of FeatureAug as the representative of the three domain adaptation algorithms, and used combinations of three sources as the DS, because FeatureAug with combined sources showed optimal SRL performance in our study.
Domain adaptation for different predicate types: As described in the “Datasets” section, MiPACQ contains both verb and nominal predicates. PB have verb predicates only and NomBank has nominal predicates only. Among the 722 verb predicates in MiPACQ, 644 are common with PropBank and 15 are common with BioProp. Among the 415 nominal predicates, 265 are common with NomBank. The effects of domain adaptation on the performance of the common/uncommon predicates between DS and DT as well as the performance of the verb/nominal predicates were examined.
Evaluation
Precision (P), recall (R), and F1 measure were used as evaluation metrics for AI and combined SRL tasks. Precision measures the percentage of correct predictions of positive labels made by a classifier. Recall measures the percentage of positive labels in the gold standard that were correctly predicted by the classifier. F1 measure is the harmonic mean of precision and recall. During the process of AC, the boundaries of candidate arguments are already identified by the AI step. Therefore, the accuracy of the classifier was used for evaluation, which is defined as the percentage of correct predictions with reference to the total number of candidate arguments correctly recognized in the AI step.
Ten-fold cross-validation was employed for performance evaluation. Nine folds of MiPACQ were merged with DS as the training set and one fold was used for testing. In experiments evaluating the influence of source domain sample size, nine folds of MiPACQ were merged with an increasing percentage of PropBank to generate the training set. In experiments evaluating the effect of target domain sample size, an increasing percentage of the nine-fold MiPACQ was added to the entire PropBank as the training set.
RESULTS
Table 2 lists the results of the implemented methods using both individual and combined source domain corpora as DS. Training on MiPACQ alone, that is, the “Target Only” baseline (via 10-fold cross validation) yielded a combined F1 measure of 81.53%. When only source domain corpora were used for training (i.e., the “Source Only” baseline), the performance of SRL systems on MiPACQ was poor (58.87% for PropBank, 11.75% for NomBank, and 20.67% for BioProp). Simply merging PropBank with MiPACQ, i.e., the “Source&Target” baseline, dropped the combined F1 to 79.79%. However, the use of domain adaptation algorithms increased the performance compared to the Target Only baseline. Among the three, FeatureAug with PropBank achieved the highest combined F1 value of 82.83%, an increase of 1.3% over the Target Only baseline, which was statistically significant, as determined by the Wilcoxon signed-rank test60 (p < 0.05). Using NomBank as DS, the best performance was achieved by FeatureAug, with F1 of 81.86%. The performances of InstancePrune with NomBank were worse than the Target Only baseline (80.52% versus 81.53%). The “Source & Target” baseline using BioProp as DS obtained a combined F1 of 81.68%. InstancePrune outperformed the other two domain adaptation algorithms, with a combined F1 value of 81.95%. However, the performance of FeatureAug with BioProp dropped slightly from the Source & Target baseline (81.63% versus 81.68%), making domain adaptation ineffective. When multiple sources were combined as DS, FeatureAug consistently performed the best among the three algorithms; it was significantly better than the Target Only baseline, with a p < 0.05 (83.14% for PN, 82.73% for PB, 83.18% for PNB). The highest combined F1 measure was 83.18% when FeatureAug algorithm was applied to the Ds consisting of all three sources.
Table 2:
Performance with and without domain adaptation using PropBank, NomBank, BioProp, and their combinations (%)
| Data | Methods | AI | AC | AI + AC |
||||
|---|---|---|---|---|---|---|---|---|
| P | R | F 1 | Accuracy | P | R | F 1 | ||
| MiPACQ | Target Only | 93.24 | 92.81 | 93.02 | 87.65 | 81.72 | 81.34 | 81.53 |
| PropBank + MiPACQ | Source Only | 86.54 | 76.36 | 81.13 | 72.57 | 62.80 | 55.41 | 58.87 |
| Source & Target | 93.99 | 90.36 | 92.14 | 86.60 | 81.39 | 78.25 | 79.79 | |
| InstancePrune | 94.22 | 92.70 | 93.45 | 86.95 | 81.93 | 80.60 | 81.26 | |
| TransferSelf | 93.23 | 92.77 | 93.00 | 87.68 | 81.74 | 81.33 | 81.54 | |
| FeatureAug | 94.08 | 93.82 | 93.95 | 88.17 | 82.95 | 82.71 | 82.83 * | |
| NomBank + MiPACQ | Source Only | 84.20 | 11.03 | 19.50 | 60.18 | 50.68 | 6.65 | 11.75 |
| Source & Target | 95.59 | 85.46 | 90.24 | 86.53 | 82.71 | 73.95 | 78.08 | |
| InstancePrune | 94.53 | 90.28 | 92.35 | 87.18 | 82.41 | 78.71 | 80.52 | |
| TransferSelf | 93.21 | 92.49 | 92.85 | 87.94 | 81.98 | 81.34 | 81.66 | |
| FeatureAug | 93.31 | 92.70 | 93.00 | 88.01 | 82.13 | 81.59 | 81.86 | |
| BioProp + MiPACQ | Source Only | 53.48 | 30.62 | 38.95 | 53.06 | 28.38 | 16.25 | 20.67 |
| Source & Target | 93.43 | 92.07 | 92.74 | 88.07 | 82.28 | 81.08 | 81.68 | |
| InstancePrune | 93.43 | 92.68 | 93.05 | 88.07 | 82.28 | 81.62 | 81.95 | |
| TransferSelf | 93.41 | 92.77 | 93.09 | 87.82 | 82.04 | 81.48 | 81.75 | |
| FeatureAug | 93.13 | 92.83 | 92.98 | 87.79 | 81.76 | 81.50 | 81.63 | |
| PN + MiPACQ | Source Only | 91.04 | 78.42 | 84.26 | 75.56 | 68.79 | 59.26 | 63.66 |
| Source & Target | 95.37 | 90.39 | 92.82 | 86.28 | 82.28 | 77.99 | 80.08 | |
| InstancePrune | 95.13 | 92.68 | 93.89 | 87.16 | 82.59 | 80.47 | 81.51 | |
| TransferSelf | 93.15 | 92.42 | 92.78 | 87.96 | 81.93 | 81.29 | 81.61 | |
| FeatureAug | 94.50 | 93.89 | 94.19 | 88.27 | 83.41 | 82.87 | 83.14 * | |
| PB + MiPACQ | Source Only | 90.33 | 75.68 | 82.36 | 72.96 | 65.91 | 55.22 | 60.09 |
| Source & Target | 94.38 | 89.44 | 91.84 | 86.95 | 82.07 | 77.77 | 79.86 | |
| InstancePrune | 93.75 | 91.71 | 92.72 | 87.38 | 81.92 | 80.14 | 81.02 | |
| TransferSelf | 92.37 | 91.23 | 91.80 | 87.70 | 81.01 | 80.01 | 80.50 | |
| FeatureAug | 94.06 | 93.54 | 93.80 | 88.20 | 82.96 | 82.50 | 82.73 * | |
| PNB + MiPACQ | Source Only | 91.30 | 78.25 | 84.27 | 75.65 | 69.07 | 59.20 | 63.75 |
| Source & Target | 95.37 | 90.27 | 92.75 | 86.41 | 82.40 | 78.00 | 80.14 | |
| InstancePrune | 94.70 | 92.57 | 93.62 | 87.30 | 82.67 | 80.81 | 81.73 | |
| TransferSelf | 93.20 | 92.42 | 92.81 | 87.87 | 81.90 | 81.21 | 81.55 | |
| FeatureAug | 94.43 | 93.85 | 94.14 | 88.35 | 83.43 | 82.92 | 83.18 * | |
*Statistically significant with p-value<0.05 by the Wilcoxon signed-rank test.
Table 3 lists the performance of core arguments and adjunctive arguments with the highest frequencies. The remaining adjunctive arguments are listed in “ARGM-Others.” The scores listed in the parenthesis stand for the F1 value improvement by domain adaptation, which measures the extent of increase over using the target domain dataset only. Denoting the F1 values of using MiPACQ only and after domain adaptation as and , respectively, the F1 value improvement by domain adaptation is calculated by . For example, FeatureAug with PropBank increases the F1 of ARG0 by 1.20% over using MiPACQ only. As shown in the table, FeatureAug with PropBank increased the performance of each argument. For NomBank, FeatureAug decreased the performance of ARG0, ARG3-5, ARGM-MOD, and ARGM-NEG, but increased the performance of the other arguments. For BioProp, InstancePrune increased performance on most arguments, but decreased the performance of ARG0, ARG3-5, ARGM-MNR, and ARGM-MOD.
Table 3:
Combined SRL Performance for each Argument using MiPACQ only, FeatureAug with PropBank, FeatureAug with NomBank, and InstancePrune with BioProp (%)
| Argument | MiPACQ |
PropBank_FeatureAug |
NomBank_FeatureAug |
BioProp_InstancePrune |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F 1 | P | R | F 1 | P | R | F 1 | P | R | F 1 | |
| ARG0 | 87.55 | 87.32 | 87.44 | 88.25 | 88.72 | 88.49 (1.20) | 87.30 | 87.28 | 87.29 (−0.16) | 87.19 | 86.93 | 87.06 (−0.43) |
| ARG1 | 84.06 | 87.48 | 85.74 | 84.82 | 88.39 | 86.57 (0.97) | 84.44 | 87.81 | 86.09 (0.41) | 83.97 | 88.10 | 85.98 (0.29) |
| ARG2 | 79.28 | 75.80 | 77.50 | 80.42 | 76.58 | 78.46 (1.23) | 81.71 | 76.75 | 79.15 (2.12) | 80.78 | 76.42 | 78.54 (1.34) |
| ARG3-5 | 77.07 | 53.02 | 62.82 | 80.00 | 57.72 | 67.06 (6.74) | 77.11 | 52.01 | 62.12 (−1.11) | 75.98 | 52.01 | 61.75 (−1.70) |
| ARGM-ADJ | 55.19 | 57.32 | 56.23 | 55.77 | 58.94 | 57.31 (1.92) | 55.54 | 62.20 | 58.68 (4.35) | 56.32 | 59.76 | 57.99 (3.12) |
| ARGM-ADV | 66.15 | 69.03 | 67.56 | 68.69 | 70.79 | 69.72 (3.20) | 65.85 | 69.87 | 67.80 (0.35) | 66.14 | 70.48 | 68.24 (1.01) |
| ARGM-LOC | 77.22 | 78.49 | 77.85 | 78.89 | 81.61 | 80.23 (3.05) | 79.24 | 78.33 | 78.78 (1.19) | 79.22 | 80.13 | 79.67 (2.34) |
| ARGM-MNR | 74.51 | 69.30 | 71.81 | 75.95 | 69.86 | 72.78 (1.35) | 77.31 | 68.85 | 72.84 (1.42) | 74.60 | 68.28 | 71.30 (−0.71) |
| ARGM-MOD | 98.87 | 86.72 | 92.40 | 99.27 | 89.57 | 94.17 (1.92) | 96.84 | 87.38 | 91.86 (−0.58) | 99.36 | 85.29 | 91.79 (−0.66) |
| ARGM-NEG | 91.07 | 89.47 | 90.27 | 95.51 | 89.47 | 92.39 (2.36) | 88.81 | 90.53 | 89.66 (−0.67) | 91.55 | 89.30 | 90.41 (0.16) |
| ARGM-TMP | 88.52 | 87.63 | 88.07 | 90.22 | 88.82 | 89.52 (1.64) | 89.73 | 87.47 | 88.58 (0.59) | 89.41 | 87.78 | 88.59 (0.59) |
| ARGM-Others | 63.83 | 55.64 | 59.45 | 66.20 | 56.83 | 61.16 (2.87) | 66.24 | 54.59 | 59.85 (0.67) | 66.93 | 54.31 | 59.96 (0.85) |
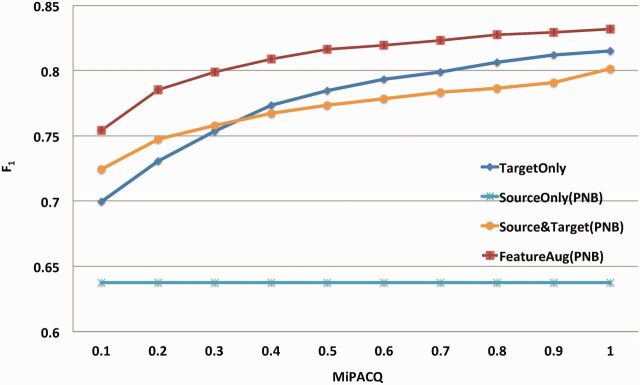
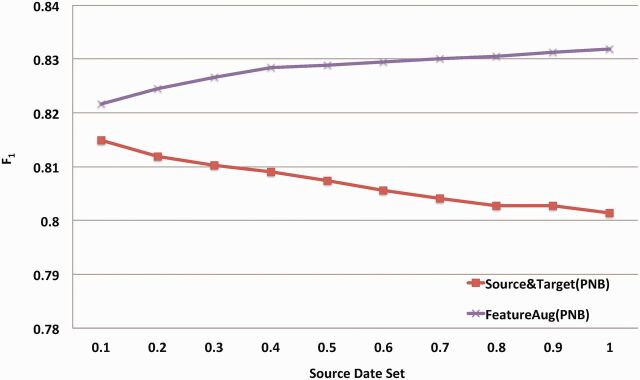
Figure 4 shows learning curves that plot the F1 value on the combined SRL task with increasing percentages of MiPACQ samples used for training when PropBank, NomBank, and BioProp were combined as DS. The Source Only baseline is a horizontal line. For other methods, increasing the sample size of the target domain (MiPACQ) leads to a consistent performance enhancement. However, the domain adaptation method (FeatureAug) clearly shows better performance than baselines of Target Only and Source & Target. Similarly, Figure 5 shows the learning curves obtained by increasing the source domain (PNB) sample size for training. Without domain adaptation, increasing the sample size of DS progressively decreased the performance. Nevertheless, a monotone increasing curve is clear when augmented with the domain adaptation algorithm (FeatureAug).
Figure 4:
Learning curves of the SRL systems that used all three sources (PNB), with increasing percentage of the target domain dataset. The x-axis denotes the percentage of target domain instances that are used for training. The y-axis denotes the averaged combined F1 value using 10-fold cross-validation. “Target Only” denotes the baseline of using only the target domain dataset for training. “Source Only” denotes the baseline of using only the source dataset for training. “FeatureAug” denotes the SRL system implemented with the FeatureAug domain adaptation algorithm.
Figure 5:
Learning curves of the SRL systems with increasing percentage of the combined source domain dataset (PNB). The x-axis denotes the percentage of source domain instances that are available during training. The y-axis denotes the averaged F1 value, using 10-fold cross-validation. “Source & Target” denotes the method that simply combines source and target domain corpora. “FeatureAug” denotes the feature augmentation domain adaptation algorithm.
Table 4 displays the SRL performance of optimized domain adaptation methods for each source, for overlapping versus nonoverlapping predicates or verb versus nominal predicates, respectively. The scores listed in parentheses in the last column of Table 4 indicate the improvements in F-measures between the baseline (Target Only) and domain adaptation methods, which are calculated in the same way as those in Table 3. As illustrated in Table 4, the employed domain adaptation algorithms improved SRL on not only overlapping predicates (PropBank 1.48%, NomBank 1.66%, and BioProp 0.83%), but also nonoverlapping predicates (PropBank 1.14%, NomBank 1.44%, and BioProp 0.47%). Our results also suggested that the employed domain adaptation algorithms improved SRL performance on not only verb predicates, but also nominal predicates (Table 4). For example, when FeatureAug was used for PropBank (containing verb predicates only), the F1 value of nominal predicates in MiPACQ was also improved by 1.16%.
Table 4:
Combined SRL Performance of verb and nominal predicates using MiPACQ only, FeatureAug with PropBank, FeatureAug with NomBank, and InstancePrune with BioProp (%)
| MiPACQ | PropBank_FeatureAug |
|||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| Overlapping verb | 83.89 | 83.11 | 83.50 | 84.97 | 84.52 | 84.74 (+1.48) |
| Nonoverlapping verb | 86.72 | 86.87 | 86.80 | 87.90 | 87.68 | 87.79 (+1.14) |
| Nominal | 69.50 | 70.13 | 69.81 | 70.31 | 70.94 | 70.62 (+1.16) |
| MiPACQ | NomBank_FeatureAug |
|||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| Overlapping Nominal | 68.85 | 69.24 | 69.04 | 69.55 | 70.84 | 70.19 (+1.66) |
| Nonoverlapping nominal | 71.36 | 72.74 | 72.04 | 72.20 | 73.98 | 73.08 (+1.44) |
| Verb | 84.53 | 83.96 | 84.24 | 84.86 | 83.97 | 84.41 (+0.20) |
| MiPACQ | BioProp_InstancePrune |
|||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| Overlapping verb | 77.90 | 78.13 | 78.01 | 80.42 | 76.99 | 78.66 (+0.83) |
| Nonoverlapping verb | 84.63 | 84.04 | 84.34 | 85.15 | 84.34 | 84.74 (+0.47) |
| Nominal | 69.50 | 70.13 | 69.81 | 70.32 | 70.52 | 70.42 (+0.87) |
DISCUSSION
In this study, we leverage existing annotation corpora from newswire and biomedical literature to improve the performance of clinical SRL by using domain adaptation algorithms. Our results showed that domain adaptation algorithms such as FeatureAug could improve the SRL task in the clinical domain by utilizing existing open domain corpora such as PropBank. In addition, we demonstrated that combining multiple sources from external domains further improved clinical SRL systems. To the best of our knowledge, this is the first study that has compared different domain adaptation algorithms for SRL in the medical domain.
The performance enhancement and cost reduction by domain adaptation for the clinical domain SRL task are illustrated more explicitly in Figure 4. It is not surprising that increasing the MiPACQ dataset for training constantly enhances the performance. An F1 value of 81.67% was be achieved using 50% of MiPACQ dataset with domain adaptation (FeatureAug on the PNB corpora). This was higher than the baseline method (81.53%), which used 90% MiPACQ dataset for training (via 10-fold cross-validation), indicating a 40% reduction in annotation cost.
The SRL performance of the Source Only baseline indicates the direct influence of source domain datasets on MiPACQ without any domain adaptation algorithm. As illustrated in Table 2, the Source Only performance of all three source domain datasets was much lower than the Target Only performance. Specifically, the recall of Source Only with NomBank, which shared 265 nominal predicates with MiPACQ, was extremely low, with only 11.03% for AI and 6.65% for the combined task. On the other hand, BioProp achieved better recalls than NomBank using the Source Only method, with recalls of 30.62% and 16.25% for AI and the combined task, respectively. A possible explanation for the low recall with NomBank is that the high overlap of predicates between source and target domain datasets does not necessarily guarantee a high recall for SRL. The similarity of PASs between source domain and target domain datasets makes the major contribution.1,31,57 As illustrated in Table 4, the recall of the combined SRL task for the nonoverlapping predicates was also improved by leveraging the source domain datasets. Another possible reason for the low recall when NomBank was used in the Source Only method was that the PAS distribution of nominal predicates in MiPACQ is low. MiPACQ only contains 2795 (22.22%) PASs of noun predicates, in contrast to 9780 (77.78%) PASs of verb predicates.
We noticed that when a single source domain dataset is employed, domain adaptation algorithms performed differently for different source domain datasets. For example, although the dataset size of BioProp is much smaller than PropBank/NomBank, it achieved better performance than PropBank/NomBank using InstancePrune (Combined task F1: BioProp 81.95%, PropBank 81.26%, NomBank 80.52%); whereas PropBank/NomBank outperformed BioProp using the FeatureAug algorithm (Combined task F1: BioProp 81.63%, PropBank 82.83%, NomBank 81.86%). This is probably attributable to the essential difference among the DS. PropBank/NomBank are built from the general English domain, while BioProp is built from biomedical literature. Based on our manual analysis of PASs in BioProp and MiPACQ, instance-level domain adaptation algorithms work better for BioProp, probably because the instances of BioProp selected by the InstancePrune algorithm have similar PASs, with a portion of instances in the target domain (MiPACQ). Feature-level algorithms have a better tolerance for domain gap,28 which is probably why FeatureAug worked best for PN. In addition, PropBank and NomBank are much larger and have higher dimensions of features, which provide the potential to contribute more useful features, as determined by the feature weighting mechanism in FeatureAug. The impact of BioProp seems trivial (PNB + MiPACQ versus PN + MiPACQ) or even negative (PB + MiPACQ versus PropBank + MiPACQ) when multiple source domain datasets are combined directly for domain adaptation. It is necessary to further investigate how to select source data instances efficiently when multiple source domain datasets are used. These findings may provide valuable insights for selecting source domain datasets and domain adaptation algorithms.
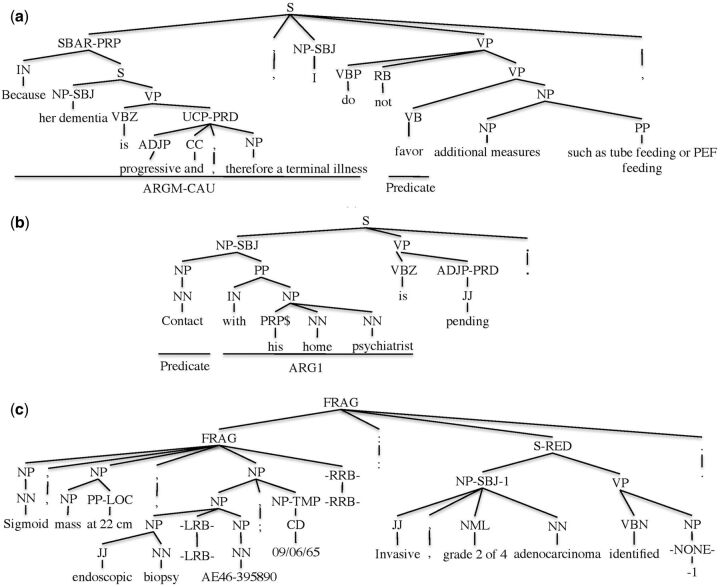
To identify which knowledge was transferred from the source domains to improve the performance, we examined the SRL results of individual instances. We found that most of the improvement was obtained by syntactic structural information learned from the source domains. As illustrated in Figure 6(a), due to the complex syntactic structure, the MiPACQ-only baseline failed to recognize “Because her dementia is progressive and, therefore, a terminal illness” as the ARGM-CAU (cause) argument of the predicate “favor” in the argument recognition stage. This argument was recognized correctly with the effort of domain adaptation. Taking the sentence in Figure 6(b) as another example, the prepositional phrase “with his home psychiatrist” should be a core argument ARG1 (entity contacted) of the nominal predicate “contact.” However, the MiPACQ-only baseline labeled its role as an adjunctive argument, “ARGM-ADJ” (adjective). Although the PropBank dataset only annotated PAS for verb predicates, the similar syntactic structure for the verb “contact” still transferred successfully and correctly labeled the semantic role as ARG1. This explains the reason why the performance of nominal predicates in MiPACQ was also improved by PropBank.
Figure 6:
Syntactic parsing results of three sample instances.
Although knowledge transferred from source domains can be adapted to the clinical domain, the unique characteristics of clinical text require domain specific resources and solutions for further SRL improvement. One type of salient attribute is the clinical lexicons and semantic relations between them. In the phrase “an advanced oropharynx cancer treated with radiation therapy and chemotherapy,” “an advanced oropharynx cancer” is annotated as ARG2 (illness or injury) in the gold standard. However, it is labeled as ARG1 by our SRL system. Additionally, in the phrase “erectile dysfunction,” “erectile” is annotated as ARG1 (job, project) of “dysfunction,” which is mistakenly labeled as ARGM-ADJ (adjective). Clinical domain knowledge needs to be employed to precisely interpret these semantic relations. Another unique characteristic of clinical text is the high frequency of fragments; that is, grammatically incomplete sentences. Figure 6(c) illustrates the syntactic parsing result of the fragment sentence “Sigmoid, mass at 22 cm, endoscopic biopsy (AE46-395890; 09/06/65): Invasive, grade 2 of 4 adenocarcinoma identified.” In this sentence, there is no semantic relation between “biopsy” and the temporal phrase “09/06/65,” which is difficult to identify even when using the current open-domain state-of-the art features. Clinical domain features like specific syntactic patterns of CNs need to be further explored.
State-of-the-art SRL systems usually employ a rich feature set57 and/or a global inference phase to further refine the output with global constraints.61 To verify the effects of global inference and domain adaptation, we developed a rule-based global inference module following important constraints defined in Punyakanok et al.61 The global inference phase improved SRL performance: the baseline Target Only method was improved from 81.53% to 82.12%. We then integrated the best domain adaption method FeatureAug with PNB with the improved SRL system (with global inference). Our results showed that FeatureAug with PNB further improved the SRL performance, with a combined F1 of 83.88% (compared with the improved baseline of 82.12%). This indicates that domain adaptation and global inference are complementary. For optimized SRL systems with a global inference phase, domain adaptation methods may further improve the performance. The performance of integrating the global inference constraints with each of the methods implemented in our study can be found in Supplementary Appendix Table S1. Common features used in current state-of-the-art open domain SRL systems were adopted in our study.31,57 The previous work of domain adaptation on the BioProp dataset (Dahlmeier and Ng, 2010)45 also employed a similar feature set to our work. In this study, we built our SRL system by following the study in Dahlmeier and Ng.45 The current performance of our SRL system on PropBank is not state-of-the-art. Using the same datasets as in Punyakanok et al.,61 our SRL system achieved an F1 of 82.77%, which is lower than the state-of-the-art performance of 86.81% F1 in Punyakanok et al.,61 probably due to the different feature sets, machine learning algorithms, and global constraints used in the study. We can further optimize the SRL performance by feature engineering57 in our future work. It is notable that domain adaptation made another contribution to significantly reduce the data annotation cost of the target dataset to achieve a comparable performance. As illustrated in Figure 4, it required <50% of training samples in the target dataset to achieve a comparable performance to the target-only baseline using 90% of the target dataset.
One potential limitation of our work is the coverage in the target domain dataset. MiPACQ is built from Mayo Clinical CN and Mayo Clinical pathology notes related to colon cancer, which may contain limited clinical findings. In addition, clinical text consists of diverse narrative types, such as discharge summaries and clinic visit notes. Therefore, the SRL systems built on MiPACQ may need further adaptation for use in other clinical subdomains. In this study, we employed common features used in current state-of-the-art open domain SRL systems for the SRL task. However, the contribution of each feature type needs to be further examined for the SRL tasks in the clinical domain. Furthermore, instead of directly combining multiple source domain datasets, we plan to investigate more sophisticated multisource domain adaptation algorithms, such as weighting on source datasets with different distributions,62,63 which may allow us to more effectively employ multisource datasets.
CONCLUSIONS
Our study investigates domain adaptation techniques for SRL in clinical text. Three state-of-the-art domain adaptation algorithms were employed and evaluated by using existing SRL resources built from newswire, biomedical literature, and clinical text. Experimental results showed that domain adaptation significantly improved the SRL performance on clinical text, indicating its potential to reduce annotation costs when building clinical SRL systems.
FUNDING
This study was supported in part by National Institute of General Medical Sciences grant 1R01GM102282, National Library of Medicine grant R01LM010681, National Cancer Institute grant R01CA141307, and Cancer Prevention and Research Institute of Texas grant R1307.
COMPETING INTERESTS
None.
CONTRIBUTORS
The work presented here was carried out in collaboration among all authors. Y.Z., B.T., and H.X. designed methods and experiments. M.J. and J.W. carried out the experiments. Y.Z., B.T., and H.X. analyzed the data, interpreted the results, and drafted the article. All authors have been attributed to, seen, and approved the manuscript.
Supplementary Material
Acknowledgments
We would like to thank the anonymous reviewers for their constructive comments.
REFERENCES
- 1. Pradhan SS, Ward WH, Hacioglu K, et al. Shallow semantic parsing using support vector machines. In: Proceedings of Human Language Technology Conference of the North American Chapter of the Association for Computational Linguistics .Boston, Massachusetts, USA: Association for Computational Linguistics; July 21-26, 2004:233–240. [Google Scholar]
- 2. Allen J. Natural Language Understanding. 2nd ed.Menlo Park, CA: Benjamin/Cummings; 1995. [Google Scholar]
- 3. Surdeanu M, Harabagiu S, Williams J, et al. Using predicate-argument structures for information extraction. In: Proceedings of the 41st Annual Meeting of the Association for Computational Linguistics. Sapporo, Japan: Association for Computational Linguistics; July 7-12, 2003: 8–15. [Google Scholar]
- 4. Akane Y, Yusuke M, Tomoko O, et al. Automatic construction of predicate-argument structure patterns for biomedical information extraction. In: Proceedings of the 2006 Conference on Empirical Methods in Natural Language Processing. Seattle, Washington, USA: Association for Computational Linguistics; July 22-23, 2006:284–292. [Google Scholar]
- 5. Yakushiji A, Miyao Y, Tateisi Y, et al. Biomedical information extraction with predicate-argument structure patterns. In: Proceedings of the First International Symposium on Semantic Mining in Biomedicine. Hinxton, Cambridge, UK: European Bioinformatics Institute; April 10-13, 2005:60–69. [Google Scholar]
- 6. Nguyen NTH, Miwa M, Tsuruoka Y, et al. Open information extraction from biomedical literature using predicate-argument structure patterns. In: Proceedings of the 5th International Symposium on Languages in Biology and Medicine. Zurich, Switzerland: December 12-13, 2013. [Google Scholar]
- 7. Wattarujeekrit T, Shah PK, Collier N. PASBio: predicate-argument structures for event extraction in molecular biology. BMC Bioinformatics 2004;5(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kogan Y, Collier N, Pakhomov S, et al. Towards semantic role labeling & IE in the medical literature. AMIA Symposium October 22-26, 2005:410–414. [PMC free article] [PubMed] [Google Scholar]
- 9. Shah PK, Bork P. LSAT: learning about alternative transcripts in MEDLINE. Bioinformatics 2006;22(7):857–865. [DOI] [PubMed] [Google Scholar]
- 10. Bethard S, Lu Z, Martin JH, et al. Semantic role labeling for protein transport predicates. BMC Bioinformatics 2008;9:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barnickel T, Weston J, Collobert R, et al. Large scale application of neural network based semantic role labeling for automated relation extraction from biomedical texts. PloS One 2009;4(7):e6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paek H, Kogan Y, Thomas P, et al. Shallow semantic parsing of randomized controlled trial reports. AMIA Symposium November 11-15, 2006:604–608. [PMC free article] [PubMed] [Google Scholar]
- 13. Albright D, Lanfranchi A, Fredriksen A, et al. Towards comprehensive syntactic and semantic annotations of the clinical narrative. J Am Med Inform Assoc. 2013;20(5):922–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y, Pakhomov S, Melton GB. Predicate argument structure frames for modeling information in operative notes. Stud Health Technol Inform. 2013;192:783–787. [PMC free article] [PubMed] [Google Scholar]
- 15. Meyers A, Reeves R, Macleod C, et al. Annotating noun argument structure for NomBank. In: Proceedings of the Language Resources and Evaluation Conference. Lisbon, Portugal: European Language Resources Association; May 26-28, 2004:803–806. [Google Scholar]
- 16. Meyers A, Reeves R, Macleod C, et al. The NomBank Project: an interim peport. In: Proceedings of the HLT-NAACL 2004 Workshop: Frontiers in Corpus Annotation. Boston, Massachusetts, USA: Association for Computational Linguistics; May 6, 2004:24–31. [Google Scholar]
- 17. Tsai RT-H, Chou W-C, Su Y-S, et al. BIOSMILE: a semantic role labeling system for biomedical verbs using a maximum-entropy model with automatically generated template features. BMC Bioinformatics 2007;8:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palmer M, Gildea D, Kingsbury P. The proposition bank: an annotated corpus of semantic roles. Comput Linguist. 2005;31(1):71–106. [Google Scholar]
- 19. Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36(6):462–477. [DOI] [PubMed] [Google Scholar]
- 20. Kilicoglu H, Shin D, Fiszman M, et al. SemMedDB: a PubMed-scale repository of biomedical semantic predications. Bioinformatics 2012;28(23):3158–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sager N. Natural Language Information Processing. UK; Addison-Wesley, 1981. [Google Scholar]
- 22. Chen ES, Hripcsak G, Xu H, et al. Automated acquisition of disease–drug knowledge from biomedical and clinical documents: an initial study. J Am Med Inform Assoc. 2008;15(1):87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simpson MS, Demner-Fushman D. Biomedical text mining: a survey of recent progress. In: Aggarwal CC, Zhai C, eds. Mining Text Data. USA; Springer; 2012:465–517. [Google Scholar]
- 24. Harris ZS, Harris Z. A Theory of Language and Information: a Mathematical Approach. Oxford: Clarendon Press; 1991. [Google Scholar]
- 25. Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. J Biomed Inform. 2002;35(4):222–235. [DOI] [PubMed] [Google Scholar]
- 26. Cohen KB, Palmer M, Hunter L. Nominalization and alternations in biomedical language. PLoS One 2008;3(9):e3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17(1):19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schuler KK. VerbNet: A Broad-Coverage, Comprehensive Verb Lexicon. ProQuest Paper AAI3179808, 2005. [Google Scholar]
- 29. Ruppenhofer J, Ellsworth M, Petruck MRL, et al. FrameNet II: extended theory and practice. http://framenet.icsi.berkeley.edu/ Accessed 10 March 2014. [Google Scholar]
- 30. Levin B. English Verb Classes and Alternations: a Preliminary Investigation. Chicago, USA: University of Chicago Press; 1993. [Google Scholar]
- 31. Gildea D, Jurafsky D. Automatic labeling of semantic roles. Comput Linguist. 2002;28(3):245–288. [Google Scholar]
- 32. Shen D, Lapata M. Using semantic roles to improve question answering. In: Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL). Prague, Czech Republic: Association for Computational Linguistics; June 28-30, 2007:12–21. [Google Scholar]
- 33. McCord MC, Murdock JW, Boguraev BK. Deep parsing in Watson. IBM J Res Dev. 2012;56(3.4):3:1–3:15. [Google Scholar]
- 34. Carreras X, Màrquez L. Introduction to the CoNLL-2005 shared task: semantic role labeling. In: Proceedings of the Ninth Conference on Computational Natural Language Learning (CoNLL-2005). Ann Arbor, Michigan: Association for Computational Linguistics; June 29-30, 2005:152–164. [Google Scholar]
- 35. Surdeanu M, Johansson R, Meyers A, et al. The CoNLL-2008 shared task on joint parsing of syntactic and semantic dependencies. In: Proceedings of the Twelfth Conference on Computational Natural Language Learning. Stroudsburg, PA, USA: Association for Computational Linguistics; August 16-17, 2008:159–177. [Google Scholar]
- 36. Xue N, Palmer M. Calibrating features for semantic role labeling. In: Proceedings of the 2004 Conference on Empirical Methods in Natural Language Processing. Barcelona, Spain: Association for Computational Linguistics; July 25-26, 2004:88–94. [Google Scholar]
- 37. Merlo P, Musillo G. Semantic parsing for high-precision semantic role labelling. In: Proceedings of the Twelfth Conference on Computational Natural Language Learning. Stroudsburg, PA, USA: Association for Computational Linguistics; August 16-17th, 2008:1–8. [Google Scholar]
- 38. Meza-Ruiz I, Riedel S. Jointly identifying predicates, arguments and senses using Markov logic. In: Proceedings of Human Language Technologies: The 2009 Annual Conference of the North American Chapter of the Association for Computational Linguistics. Stroudsburg, PA, USA: Association for Computational Linguistics; May 31-June 5, 2009:155–163. [Google Scholar]
- 39. Pan SJ, Yang Q. A survey on transfer learning. IEEE T Knowl Data Eng. 2010;22(10):1345–1359. [Google Scholar]
- 40. Wang Y, Pakhomov S, Burkart NE, et al. A study of actions in operative notes. AMIA Symposium November 3-7, 2012:1431–1440. [PMC free article] [PubMed] [Google Scholar]
- 41. Jiang J. Multi-task transfer learning for weakly-supervised relation extraction. In: Proceedings of the 47th Annual Meeting of the ACL and the 4th IJCNLP of the AFNLP August 2-7, 2009:1012–1020. [Google Scholar]
- 42. Titov I, Klementiev A. Crosslingual induction of semantic roles. In: Proceedings of the 50th Annual Meeting of the Association for Computational Linguistics. Stroudsburg, PA, USA: Association for Computational Linguistics; July 8-14, 2012:647–656. [Google Scholar]
- 43. Li Q. Literature Survey: Domain Adaptation Algorithms for Natural Language Processing. Technique Report. USA; The City University of New York; 2012. [Google Scholar]
- 44. Xu Q, Yang Q. A survey of transfer and multitask learning in bioinformatics. J Comput Sci Eng. 2011;5(3):257–268. [Google Scholar]
- 45. Dahlmeier D, Ng HT. Domain adaptation for semantic role labeling in the biomedical domain. Bioinformatics 2010;26(8):1098–1104. [DOI] [PubMed] [Google Scholar]
- 46. Ferraro JP, Daumér H, Duvall SL, et al. Improving performance of natural language processing part-of-speech tagging on clinical narratives through domain adaptation. J Am Med Inform Assoc. 2013;20(5):931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laippala V, Viljanen T, Airola A, et al. Statistical parsing of varieties of clinical Finnish. Artif Intell Med. 2014;61(3)131–136. [DOI] [PubMed] [Google Scholar]
- 48. Savova GK, Coden AR, Sominsky IL, et al. Word sense disambiguation across two domains: biomedical literature and clinical notes. J Biomed Inform. 2008;41(6):1088–1100. [DOI] [PubMed] [Google Scholar]
- 49. Demner-Fushman D, Mork JG, Shooshan SE, et al. UMLS content views appropriate for NLP processing of the biomedical literature vs. clinical text. J Biomed Inform. 2010;43(4):587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Johansson R, Nugues P. Dependency-based syntactic-semantic analysis with PropBank and NomBank. In: Proceedings of the Twelfth Conference on Computational Natural Language Learning. Manchester, UK: August 16-17, 2008:183–187. [Google Scholar]
- 51. Kim JD, Ohta T, Tateisi Y, et al. GENIA corpus–semantically annotated corpus for bio-textmining. Bioinformatics 2003;19 (Suppl 1):i180–i182. [DOI] [PubMed] [Google Scholar]
- 52. Jiang J, Zhai C. Instance weighting for domain adaptation in NLP. In: Proceedings of the 45th Annual Meeting of the Association of Computational Linguistics. Prague, Czech Republic: Association for Computational Linguistics; June 25-27, 2007:264–271. [Google Scholar]
- 53. McClosky D, Charniak E. Self-training for biomedical parsing. The 46th Annual Meeting of the Association of Computational Linguistics. Manchester, UK: Association for Computational Linguistics; June 16-18, 2008:101–104. [Google Scholar]
- 54. Xu R, Xu J, Wang X. Instance level transfer learning for cross lingual opinion analysis. In: Proceedings of the Second Workshop on Computational Approaches to Subjectivity and Sentiment Analysis. Stroudsburg, PA, USA: Association for Computational Linguistics; June 24, 2011:182–188. [Google Scholar]
- 55. Dai W., Yang Q., Xue G., et al. Boosting for transfer learning. In: Proceedings of the 24th International Conference on Machine Learning. Corvallis, OR, USA: ACM; June 20-24, 2007:193–200. [Google Scholar]
- 56. Daume H., III Frustratingly easy domain adaptation. In: Proceedings of the 45th Annual Meeting of the Association of Computational Linguistics. Prague, Czech Republic: Association for Computational Linguistics; June 25-27, 2007:256–263. [Google Scholar]
- 57. Pradhan S, Hacioglu K, Krugler V, et al. Support vector learning for semantic argument classification. J Mach Learn. 2005;60(1–3):11–39. [Google Scholar]
- 58. Fan R-E, Chang K-W, Hsieh C-J, et al. LIBLINEAR: a library for large linear classification. J Mach Learn Res. 2008;9:1871–1874. [Google Scholar]
- 59. Joachims T. Text categorization with support vector machines: learning with many relevant features. In: Proceedings of the 10th European Conference on Machine Learning. London, UK: Springer-Verlag; April 21-23, 1998:137–142. [Google Scholar]
- 60. Woolson RF. Wilcoxon signed-rank test. Wiley Encyclopedia of Clinical Trials. New York, NY, USA: John Wiley & Sons; 2007. [Google Scholar]
- 61. Punyakanok V, Roth D, Yih WT. The importance of syntactic parsing and inference in semantic role labeling. Comput Linguist. 2008;34(2):257–287. [Google Scholar]
- 62. Gao J, Fan W, Jiang J, et al. Knowledge transfer via multiple model local structure mapping. In: Proceedings of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Las Vegas, Nevada, USA: ACM; August 24-27, 2008:283–291. [Google Scholar]
- 63. Ge L, Gao J, Ngo H, et al. On handling negative transfer and imbalanced distributions in multiple source transfer learning. SIAM Conference on Data Mining. Austin, Texas, USA: Society for Industrial and Applied Mathematics; May 2-4, 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.