Abstract
Objective Extracting medical knowledge from electronic medical records requires automated approaches to combat scalability limitations and selection biases. However, existing machine learning approaches are often regarded by clinicians as black boxes. Moreover, training data for these automated approaches at often sparsely annotated at best. The authors target unsupervised learning for modeling clinical narrative text, aiming at improving both accuracy and interpretability.
Methods The authors introduce a novel framework named subgraph augmented non-negative tensor factorization (SANTF). In addition to relying on atomic features (e.g., words in clinical narrative text), SANTF automatically mines higher-order features (e.g., relations of lymphoid cells expressing antigens) from clinical narrative text by converting sentences into a graph representation and identifying important subgraphs. The authors compose a tensor using patients, higher-order features, and atomic features as its respective modes. We then apply non-negative tensor factorization to cluster patients, and simultaneously identify latent groups of higher-order features that link to patient clusters, as in clinical guidelines where a panel of immunophenotypic features and laboratory results are used to specify diagnostic criteria.
Results and Conclusion SANTF demonstrated over 10% improvement in averaged F-measure on patient clustering compared to widely used non-negative matrix factorization (NMF) and k-means clustering methods. Multiple baselines were established by modeling patient data using patient-by-features matrices with different feature configurations and then performing NMF or k-means to cluster patients. Feature analysis identified latent groups of higher-order features that lead to medical insights. We also found that the latent groups of atomic features help to better correlate the latent groups of higher-order features.
Keywords: non-negative tensor factorization, unsupervised learning, subgraph mining, natural language processing
INTRODUCTION AND RELATED WORK
One primary source of medical knowledge lies in clinical patient cases that are documented in electronic medical records (EMRs) with increasing detail. The transformation from clinical cases and experiences to knowledge is largely an expert task and faces an ongoing need for periodic labor-intensive revision. Within oncology, for example, the most recent revision of the lymphoma classification guideline by the World Health Organization (WHO) lasted >1 year, involving an eight-member steering committee and over 130 pathologists and hematologists worldwide.1 Moreover, only around 1400 cases from Europe and North America were reviewed in the context of this revision, subjecting this process to substantial selection bias. To assist with expert review, an automated approach that can cover a much broader and larger patient population and minimize selection bias is clearly needed.
Advances in machine learning have opened avenues toward more effective mining and modeling of EMRs to facilitate translational research.2,3 However, clinicians often regard existing machine learning models as hard-to-interpret black boxes. In lymphoma pathology report, immunophenotypic features may be expressed in the form of relations among medical concepts such as lymphoid cells and antigens (e.g., “[large atypical cells] express [CD30]”). We refer to the above relations as higher-order features, and the words (e.g., “large,” “cells”) as atomic features. When interpreting pathology reports and evaluating lymphoma subtypes, clinicians usually reason at the level of higher-order features (e.g., cell-antigen relations) besides atomic features (e.g., individual words). Moreover, multiple higher-order features (such as “[large atypical cells] express [CD30],”“ [large atypical cells] express [CD15],” and “[large atypical cells] have [Reed-Sternberg appearance]”) can strengthen the confidence of suspected lymphoma (Hodgkin lymphoma here). Such a group of higher-order features naturally encodes medical knowledge as in the WHO lymphoma classification guideline1 (referred to as WHO guideline later), where a panel of morphologic and immunophenotypic features are used to specify diagnostic criteria. For computational modeling, atomic features can help correlate higher-order features in order to discover medically meaningful groupings. For example, the above relations all share the words “large,” “atypical,” and “cells,” which indicates that they all describe the characteristics of tumor cells. However, extracting higher-order features is itself a difficult task and often involves manually constructed rules and domain knowledge.4–7 In addition, modeling interactions between higher-order features and atomic features are usually ignored by machine learning algorithms that mostly adopt a flat patient-by-feature matrix view (patients as rows and features as columns). Although theoretically one can add interactions as additional features or embed graphical models to account for feature interactions, the problem quickly becomes intractable for large feature dimensionality.
On the other hand, limited availability of expert annotation leads to the fact that most clinical data are still either unannotated or sparsely annotated. Thus unsupervised machine learning approaches have often been used to analyze biomedical data.8,9 Moreover, the expense of expert engineered features also argues for unsupervised feature learning instead of manual feature engineering.10–12 In particular, non-negative matrix factorization (NMF) has been a highly effective unsupervised method13 to cluster similar patients14 and sample cell lines,15 to identify subtypes of diseases16 and to learn groups of atomic features or expert engineered features such as temporal patterns from predefined events17 and genetic expression patterns.18–22 As the multi-dimension extension of NMF, non-negative tensor factorization (NTF)23–25 has recently been studied to model the genetic associations with phenotypes26–28 and interaction between cellular activities.29 However, none of these approaches model the correlations among higher-order features, and some even do not consider higher-order features. Our work is more closely related to previous works on applying NMF and NTF in text mining in the general domains such as email and security surveillance.30–33 In particular, our approach differs from the NTF based text document analysis30,33 in that we augment the NTF with subgraphs to capture relation oriented higher-order features instead of standalone entities. In addition, we adopted the Tucker tensor factorization model instead of the PARAFAC model,34 where the support for factor matrices with different group numbers better serves our application purpose.
In this paper, we develop an unsupervised framework that can generate machine learning models naturally interpretable to clinicians. The framework adopts NTF to discover groupings of subgraph encoded higher-order features, hence the name subgraph augmented non-negative tensor factorization (SANTF).
METHODS
Workflow of SANTF
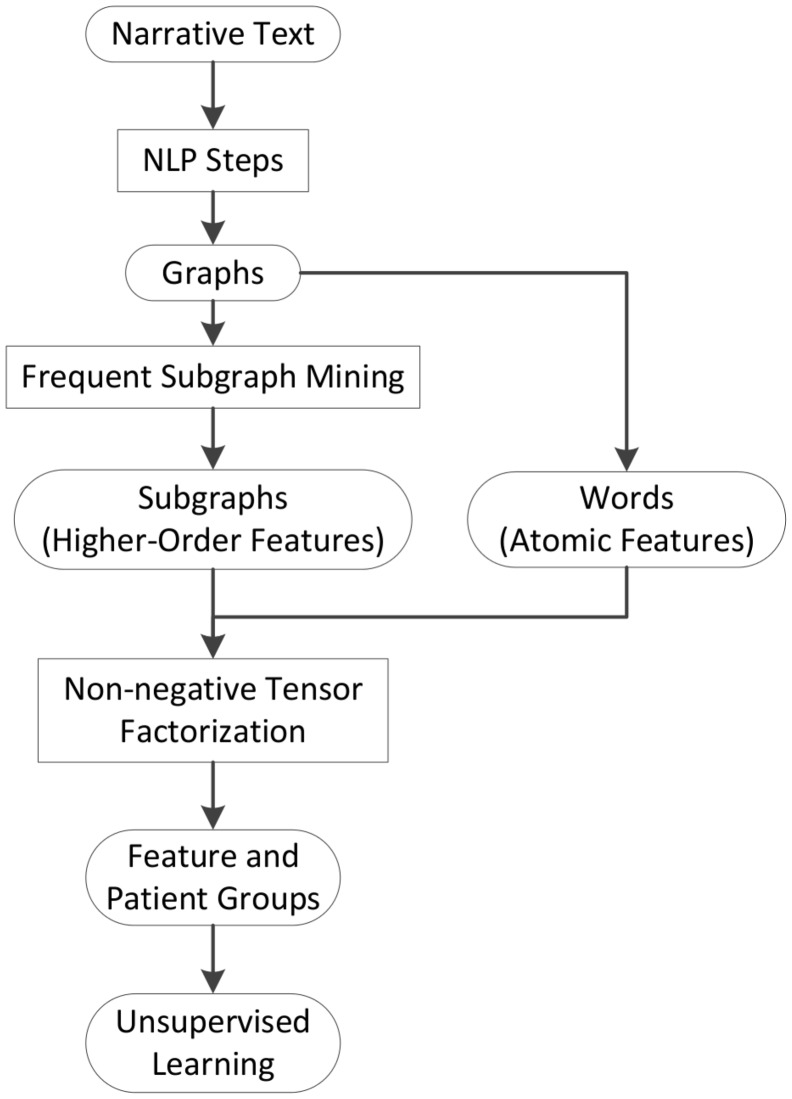
We first outline SANTF workflow in Figure 1. Narrative text sentences are first converted to graph representations. The graph representation is derived from natural language processing (NLP) steps for pathology reports as shown in Figure 2. We use frequent subgraph mining (FSM)35 tools to collect important subgraphs, which are relations among medical concepts mentioned in the sentences. Examples of higher-order features for clinical narrative text are shown in Figure 2. With such representations, subgraphs naturally encode higher-order features, and we use “subgraphs” and “higher-order features” interchangeably throughout the paper. We jointly model the higher-order features and atomic features, and apply NTF to discover groups of features and patients, and then perform unsupervised learning to identify the association between feature groups and patient groups. We next explain the tensor modeling and factorization in more detail.
Figure 1:
The workflow of subgraph augmented non-negative tensor factorization (SANTF). FSM—frequent subgraph mining; NLP—natural language processing.
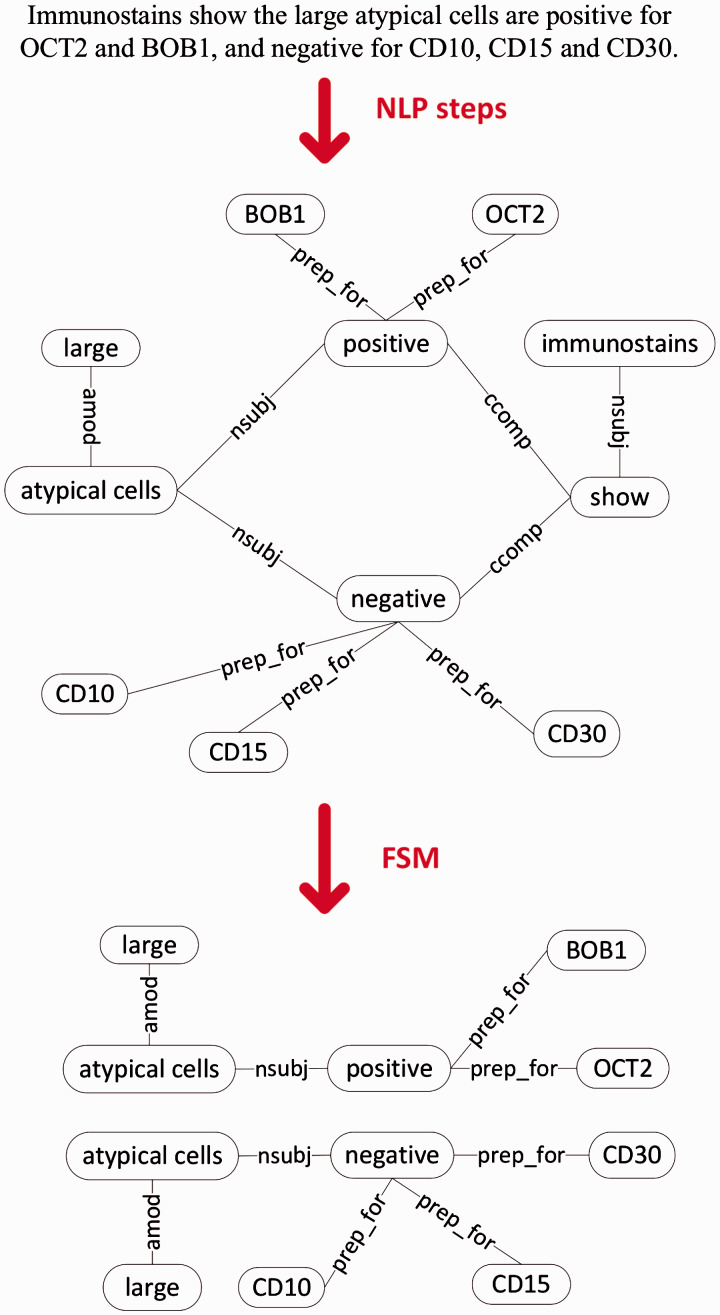
Figure 2:
Graph generation and subgraph collection in SANTF. The graph representation for the example sentence: “Immunostains show the large atypical cells are positive for OCT2 and BOB1, and negative for CD10, CD15 and CD30.” Example frequent subgraphs are shown after the frequent subgraph mining (FSM) steps.
Representing text as graphs
Figure 2 shows the steps to convert text to graphs for clinical narrative text, with an example sentence. We apply several NLP steps, including sentence breaking, tokenization, part-of-speech tagging, and a two-phase sentence parsing step that utilizes UMLS Metathesaurus,10 to convert narrative sentences into graph representation (also described in the Supplementary data). Our subgraph mining approach10 differs from previous works (e.g., 36–39) in that we extract subgraphs whose nodes usually correspond to UMLS (Unified Medical Language System) concepts instead of individual tokens in the sentence. The highly variable ways of expressing concepts in clinical narrative text favors this method. In order to generate similar representation for semantically similar but grammatically different language constructs (e.g., active voice vs. passive voice), we do not distinguish edge labels and we use the root form of verbs in the actual graph/subgraph representation. We then collect frequent subgraphs from the resultant graph corpus.
Frequent subgraph mining
We perform FSM, which is defined on the notion of graph subisomorphism. We say one graph is subisomorphic to another if all its nodes and edges coincide with part of the other one. A subgraph occurs once in a corpus whenever it is subisomorphic to a graph in that corpus. FSM identifies those subgraphs that occur in a corpus above a given threshold number of times.40,41 In this work, we use the frequent subgraph miner GASTON35 with the frequency threshold set to 5. Example frequent subgraphs from pathology report narrative text are shown in Figure 2.
Joint modeling of higher-order features and atomic features using a tensor
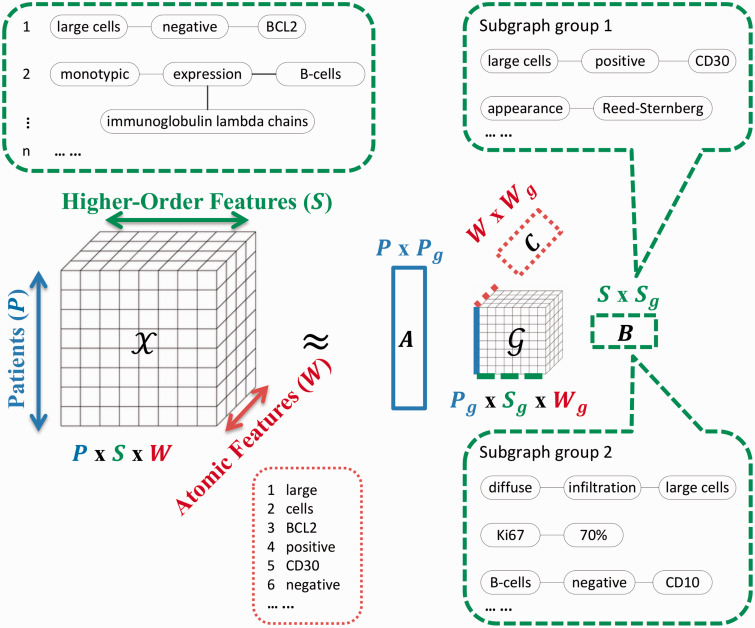
In clinical narrative text, higher-order features are often correlated with each other in medically meaningful ways. For example, the two subgraphs in Figure 2 both describe the surface markers expressed by the “large atypical cells” that are often tumor cells. However, as pointed out in the introduction, with a flat matrix view and binary feature representation, such correlations are difficult to account for. Motivated by the need to explicitly model correlations among the higher-order features, we compose a three-mode tensor, in which one mode represents the patients, a second the higher-order features (subgraphs), and a third the atomic features. Note that in tensor terminology,34 we speak of mode in place of dimension. Figure 3 shows the schematic view of tensor modeling. We select as atomic features the words that are covered by or next to a subgraph node (neighborhood window size was set to two for this work). The intuition is that subgraphs that share affiliated (covered and contextual) words are likely to be conceptually related. By taking the union over all words that are affiliated with the nodes of a sentence subgraph, we obtain the distributional representations of that sentence subgraph. Each entry of the tensor is the count of a certain combination of patient, subgraph, and word, and is non-negative (see Figure 3 for an example). We then used a generalized tf-idf weighting of co-occurrence counts of subgraph-word pairs (i.e., counting and weighting subgraph-word pairs instead of counting and weighting words), which leads to better empirical performance.
Figure 3:
Tensor modeling and factorization with distributional representations of the sentence subgraphs. In the figure, we show some higher-order features (the sentence subgraphs), as well as some atomic features (the words). The higher-order features are numbered with the first subgraph being “[large cells] – [negative] – [BCL2].” This subgraph matches the sentence “The large cells are negative for BCL2,” where the word “cells” is one of the neighboring contextual words for the node “[negative].” If the pathology report of patient 1 has a sentence “The large cells are negative for BCL2,” then subgraph 1 is associated with this patient. As the subgraph covers the word “large,” the first atomic feature, the tensor entry (1,1,1) is increased by 1. The factor matrix is the 〈patient, patient group〉 matrix, the 〈subgraph, subgraph group〉 matrix, the 〈atomic feature, atomic feature group〉 matrix. The core tensor G captures the interactions between the patient groups, subgraph groups, and atomic feature groups. We also show example subgraph group 1 and subgraph group 2. It is desirable that some subgraph groups correspond to panels of characteristic features for lymphoma subtypes. For example, subgraph group 1 includes mentions of CD30 staining and Reed-Sternberg appearance of cells, and suggests Hodgkin lymphoma; subgraph group 2 includes mentions of diffuse infiltration of large cells, moderately high Ki67 expression, and no CD10 staining, and suggests diffuse large B-cell lymphoma.
Patient and feature group discovery using SANTF
The non-negative tensor is then factorized to reduce dimensionality and obtain groups for each mode. We follow the Tucker factorization scheme,23 where the data tensor is factorized into a core tensor multiplied by factor matrices (one factor matrix for each mode, and is orthogonal in our setting). The core tensor specifies the level of interaction between groups from different modes. The column vectors in a factor matrix specify the grouping in the corresponding mode. Such groupings can capture similar patients, similar sentence subgraphs and similar words; meanwhile they allow sharing of an element among different groups as specified by its fractional weights across groups. In Figure 3, two example subgraph groups are shown. The top subgraphs in the subgraph group 1 correlate with Hodgkin lymphoma. The top subgraphs in the subgraph group 2 correlate with diffuse large B-cell lymphoma (DLBCL). Meaningful groupings will not only improve the performance of multiple machine learning tasks but also identify panels of characteristic features of patient subcategories, in the same form as specified by the diagnostic guidelines.
SANTF differs from previous NTF related works26–28 by introducing a mode that captures higher-order features. SANTF performs group discovery over sentence subgraphs based on the intuition that these higher-order features encode more aggregated information. In addition, SANTF simultaneously identifies the groups of the atomic features, which indirectly helps the group discovery for higher-order features through the core tensor. This is possible as the core tensor encodes the interactions among the groups of patients, higher-order features, and atomic features. We refer the reader to the supplement for detailed SANTF algorithm.
EXPERIMENTS AND RESULTS
We experimented with SANTF on clustering lymphoma subtypes based on pathology report narrative text. SANTF itself does not require annotated training data, but in order to verify our algorithms, we use annotated datasets for ground truth. We collected narrative text pathology reports from the Massachusetts General Hospital. We requested reports from the Research Patient Data Registry (RPDR) and obtained our patient cases by having two Massachusetts General Hospital medical oncologists and one hematopathologist review pathology reports of patients diagnosed between the years 2000 and 2010. Our dataset consists of 897 patients whose written diagnosis (in the final diagnoses section) maps to one of the following three lymphomas: Diffuse large B-cell lymphoma (DLBCL, the most common lymphoma), follicular lymphoma (the second most common lymphoma), and Hodgkin lymphoma (the most common lymphoma in young patients). The written diagnoses themselves were excluded from being processed by the feature extraction steps. The case distribution of the ground truth is shown in Table 1, where the dataset is partitioned roughly equally, and stratified by type of lymphoma, into a training set (471 cases) and a testing set (426 cases).
Table 1:
Statistics of the lymphoma subtype distribution in the pathology narrative text corpus
| Clinical Narrative Text | |||
|---|---|---|---|
| Lymphoma | All | Train | Test |
| DLBCL | 589 | 305 | 284 |
| Follicular | 184 | 101 | 83 |
| Hodgkin | 124 | 65 | 59 |
To study the impact of being able to model the interactions among multiple types of features, we establish three types of baselines for non-NMF and two configurations of k-means, a frequently used clustering method. The two configurations of k-means differ in their distance metrics used: Euclidean distance and cosine distance.42 The first type of baseline applies NMF or k-means on the 〈patient, atomic feature〉 matrices. The second baseline applies NMF or k-means on the 〈patient, higher-order feature〉 matrices. The third baseline applies NMF or k-means on the 〈patient, combined feature〉 matrices, where the combined features are generated by adjoining the atomic features and the higher-order features, because we want to exclude the possibility that the improvements of SANTF only come from simply adding features. Under orthogonality constraints, NMF is equivalent to simultaneous clustering of rows and columns of a matrix,43 and similar arguments can be made for NTF. Thus for each factorization scheme, we obtain the factor matrix of 〈patient, patient group〉, and translate this matrix into a clustering interpretation in that for each patient case, we pick the maximum column as its cluster label. For the pathology reports, recorded texts reflect results from tests and labs that are performed in order to make differential diagnoses among possible subtypes of lymphoma. Thus it is reasonable to expect that clustering based on these data will lead to patient groupings that reflect the lymphoma subtypes.
The tensor has 3773 higher-order features and 2841 atomic features. The patient group number is set to three, the same as the number of lymphoma subtypes. Because our method is unsupervised, there is no a priori mapping from patient groups to lymphoma subtypes. We therefore consider the label permutation that yields the best evaluation metrics as a parameter. For SANTF, the ideal group numbers for the higher-order features and for the atomic features are also parameters. All parameters are selected using 5-fold cross-validation on the training data and then applied to the held-out testing data.
For the evaluation metrics of clustering performance, we use the commonly adopted metrics of averaged precision, recall, f-measure, and accuracy that all apply to multi-class clustering.44 Let TP denote the number of true positives in the contingency table, FP denote the number of false positives, and FN denote the number of false negatives, the definition of precision is P = TP/(TP + FP), recall is R = TP/(TP + FN), F-measure is . Averaging computes a direct arithmetic average over classes. The accuracy computes the proportions of the sum of diagonal entries out of all entries from the multi-class contingency table. Because neither the NMF nor the NTF has a global convergence guarantee,34,45,46 we use random initialization for all factorization schemes and average the clustering evaluation metrics from 100 runs. We show the results in Table 2 for the lymphoma subtype clustering. We also perform significance testing based on the student t-test with. We see that SANTF significantly outperforms all nine baselines, and in particular, by over 10% margins in average F-measure to all baselines. Given that the classes are highly imbalanced, the results seem to suggest that improvements by SANTF come not only from the fact that more patient cases are correctly grouped (better accuracy), but also from more balanced clustering among multiple classes (better averaged precision, recall and F-measure). We refer the reader to the supplement Table 2 for detailed per-class evaluations.
Table 2:
Clustering performances for Massachusetts General Hospital lymphoma dataset
| Methods | Avg. Precision | Avg. Recall | Avg. F-measure | Accuracy |
|---|---|---|---|---|
| (1) NMF pt wd | 0.492 | 0.495 | 0.428 | 0.626 |
| (2) NMF pt sg | 0.621 | 0.765 | 0.601 | 0.605 |
| (3) NMF pt [sg wd] | 0.637 | 0.787 | 0.615 | 0.614 |
| (4) k-means (Euclidean) pt wd | 0.483 | 0.420 | 0.398 | 0.664 |
| (5) k-means (Euclidean) pt sg | 0.700 | 0.602 | 0.584 | 0.708 |
| (6) k-means (Euclidean) pt [sg wd] | 0.690 | 0.593 | 0.573 | 0.726 |
| (7) k-means (Cosine) pt wd | 0.620 | 0.694 | 0.618 | 0.617 |
| (8) k-means (Cosine) pt sg | 0.647 | 0.762 | 0.624 | 0.615 |
| (9) k-means (Cosine) pt [sg wd] | 0.648 | 0.759 | 0.626 | 0.617 |
| (10) SANTF pt sg wd | 0.720 1–9 | 0.849 1–9 | 0.743 1–9 | 0.751 1–9 |
Each factorization and clustering scheme is numbered in the “methods” column. Significant improvements (p < 05) are in bold-face and marked with superscripts indicating the baselines against which they were significantly improved from. SANTF chose by cross-validation 3 × 180 × 60 as the core tensor size for the lymphoma dataset.
FEATURE ANALYSIS
We performed feature analysis to identify groups of higher-order feature contributing to lymphoma subtype clustering. The analyzed subgraph groups correspond to the core tensor size of selected by cross-validation. We follow the standard approach of analyzing groups in factorization models,47 and make necessary adaptation to SANTF output. Based on the core tensor after factorization, we associate subgraph groups with patient clusters using the following calculation. Adopting the standard notation,34 for each slice () corresponding to a particular patient cluster, we sum over its word mode (mode 3) to get a vector whose elements correspond to the subgraph groups. We then sort the vector and investigate the top 10 subgraph groups for each patient cluster. For each subgraph group, we sort the subgraphs according to their weights in the subgraph factor matrix and display the top subgraphs, where the weight is the entry value in the matrix indexed by the corresponding subgraph and subgraph group. For each patient cluster, we select its top four subgraph groups and list them in Tables 3–5. For readability, we translated each subgraph into a partial sentence. Note that in the first DLBCL-associated subgraph group, although we have listed “cells are CD30 + , MUM1+” in order in the partial sentence, the subgraph does not distinguish the order between “CD30+” and “MUM1+” as they are both linked to “cells.” We analyze each cluster and relate them in the context of the WHO guideline,1 which reflects the current consensus knowledge.
Table 3:
Top higher-order feature groups associated with diffuse large B-cell lymphoma
| DLBCL First Subgraph Group | DLBCL Second Subgraph Group | ||
|---|---|---|---|
| 0.6640 | atypical cells | 0.0530 | atypical cells |
| 0.0929 | large lymphoid cells | 0.0293 | large lymphoid cells |
| 0.0057 | show … positive cells | 0.0240 | large cells |
| 0.0040 | large lymphoid cell with vesicular nuclei | 0.0070 | monotypic staining of immunoglobulin light chains |
| 0.0025 | show the cells are … B-cells co-expressing | 0.0059 | show large atypical cells with … vesicular nuclei |
| 0.0019 | large cells predominate | 0.0051 | B-lineage antibody PAX5 … stain … large cells |
| 0.0010 | cells are CD30+, MUM1+ | 0.0049 | associated cells |
| 0.0005 | large cells stain for CD79a | 0.0047 | a few large cells |
| 0.0005 | admixed small lymphocytes | 0.0037 | atypical cells are CD10−, BCL2−… |
| 0.0004 | large cells stain positively for CD20 | 0.0034 | infiltrate of large … cells with … scant cytoplasm |
| 0.0002 | large atypical cell with vesicular nuclei | 0.0034 | sheet of … cells |
| DLBCL Third Subgraph Group | DLBCL Fourth Subgraph Group | ||
|---|---|---|---|
| 0.0385 | diffuse infiltrate of large … cells | 0.0144 | negative for cytokeratin |
| 0.0329 | large lymphoid cells | 0.0111 | stain positively for CD20 |
| 0.0312 | large atypical cells | 0.0104 | in-situ hybridization show |
| 0.0137 | diffuse infiltrate of large … cells with … vesicular nuclei | 0.0103 | positive for immunoglobulin kappa chains |
| 0.0082 | B-lineage antibody PAX5 … stain … large cells | 0.0101 | cells show … stain |
| 0.0077 | infiltrate of large … cells with … scant cytoplasm | 0.0094 | Ki67 proliferation index is greater than 70% |
| 0.0051 | sections show … tissue with … infiltrate of … cells | 0.0086 | Ki67 proliferation index is >60% |
| 0.0041 | positive for CD20, BCL2 | 0.0075 | positive for CD79a |
| 0.0028 | cells … form | 0.0060 | stain for Ki67 |
| 0.0014 | atypical large cells … positive for CD20 | 0.0053 | large cells stain positively for CD20 |
| 0.0009 | monotypic staining with immunoglobulin lambda chains | 0.0044 | positive for cytokeratin |
Subgraphs are translated to partial sentences. In each list item, e.g., “0.0010,… cells are CD30+, MUM1+ …”, 0.0010 indicates its weight in the group. The “… cells are CD30+, MUM1+ …” is the partial sentence translated from the corresponding subgraph. Partial sentences that are not mentioned in feature analysis are grayed out. For brevity, we omit the leading and trailing “…” for partial sentences in the table.
Table 4:
Top higher-order feature groups associated with follicular lymphoma
| Follicular First Subgraph Group | Follicular Second Subgraph Group | ||
|---|---|---|---|
| 0.0308 | interstitial lymphoid aggregates | 0.0583 | nodal architecture … effaced |
| 0.0196 | predominantly small … cell | 0.0213 | B-cells co-expressing BCL2, CD10 |
| 0.0171 | paratrabecular lymphoid aggregates | 0.0201 | biopsy of lymph node |
| 0.0149 | focal | 0.0091 | sclerotic tissue |
| 0.0127 | cells in the follicles | 0.0063 | lymph node architecture effaced by … follicular proliferation |
| 0.0117 | large paratrabecular lymphoid aggregates | 0.0061 | sections show enlarged lymph nodes |
| 0.0107 | diffuse infiltrate of small lymphoid cells | 0.0059 | cell with reduced size |
| 0.0093 | infiltrate consisting of … lymphoid cells | 0.0055 | sections show … lymph nodes |
| 0.0080 | CD10+/− B-cell population | 0.0045 | residual … follicle center cells |
| 0.0062 | core needle biopsy | 0.0043 | cells stain positively for … BCL2 |
| 0.0050 | follicles contain … centroblasts | 0.0021 | flow cytometry demonstrate … population |
| Follicular Third Subgraph Group | Follicular Fourth Subgraph Group | ||
|---|---|---|---|
| 0.0829 | B-cells are negative for CD5 | 0.0642 | lymphoid infiltration |
| 0.0466 | B-cells express | 0.0269 | atypical infiltration |
| 0.0405 | CD5−, … , CD23− | 0.0267 | dense lymphoid infiltration |
| 0.0315 | negative for CD10 | 0.0133 | mucosa infiltration |
| 0.0271 | positive for CD23 | 0.0102 | small lymphoid cells |
| 0.0251 | positive for CD10 | 0.0095 | small lymphocytes |
| 0.0148 | positive for CD19, CD20, CD23 | 0.0084 | cleaved centrocytes |
| 0.0060 | containing … large atypical cells … | 0.0082 | diffuse infiltrate of small lymphoid cells |
| 0.0041 | positive for CD3 | 0.0060 | cells … in follicular dendritic pattern |
| 0.0024 | show B-cells are positive for CD3, CD20 | 0.0059 | fibroadipose tissue |
| 0.0018 | CD5−, CD10−… B-cells | 0.0044 | dense infiltrate containing lymphoid cells |
Subgraphs are translated to partial sentences. Partial sentences that are not mentioned in feature analysis are grayed out.
Table 5:
Top higher-order feature groups associated with Hodgkin lymphoma
| Hodgkin First Subgraph Group | Hodgkin Second Subgraph Group | ||
|---|---|---|---|
| 0.0362 | large cells | 0.0143 | positive for CD30 |
| 0.0312 | atypical cells | 0.0083 | large cells are negative |
| 0.0303 | large cells stain | 0.0065 | positive for CD15, CD30 |
| 0.0263 | positive for CD15 | 0.0063 | expressing PAX5 |
| 0.0196 | scattered large … cells | 0.0063 | large atypical cells |
| 0.0117 | infiltrate of large … cells with lobated nuclei | 0.0060 | large cells are negative for CD20 |
| 0.0103 | many large cells | 0.0058 | inflammatory cells |
| 0.0064 | large neoplastic cells | 0.0058 | large cells are Reed-Sternberg like |
| 0.0046 | stain positively for CD15 | 0.0049 | rare cells are … positive |
| 0.0042 | multilobated … cells | 0.0040 | histiocytes |
| 0.0027 | background contain … lymphocytes | 0.0034 | irregular nuclei |
| Hodgkin Third Subgraph Group | Hodgkin Fourth Subgraph Group | ||
|---|---|---|---|
| 0.0233 | necrosis | 0.0237 | positive for CD3 |
| 0.0142 | dense sclerosis | 0.0209 | B-cells positive for immunoglobulin lambda chains |
| 0.0106 | vaguely nodular pattern | 0.0179 | small CD3 positive lymphocytes |
| 0.0099 | collagen fibrosis | 0.0169 | CD3 positive T-cells |
| 0.0098 | mixed inflammatory cells | 0.0140 | B-cells expressing … kappa and lambda light chains |
| 0.0073 | nodular pattern | 0.0100 | expression of B-cell antigens |
| 0.0053 | atypical infiltration | 0.0053 | number of … B-cells |
| 0.0043 | collagen bands | 0.0048 | large atypical cells |
| 0.0042 | nodular lymphoid proliferation | 0.0047 | expressing CD45 |
| 0.0018 | areas of vague nodularity | 0.0025 | positive for OCT2, PAX5 |
| 0.0017 | cells … with Reed-Sternberg forms | 0.0020 | many scattered … T-cells |
Subgraphs are translated to partial sentences. Partial sentences that are not mentioned in feature analysis are grayed out.
For the DLBCL cluster as shown in Table 3, the first associated subgraph group recognizes the following histologic (light microscope-visible) facts: the cells are atypical in appearance and are large lymphoid cells with vesicular nuclei (the critical visual hallmarks of DLBCL). Immunohistochemically the group appropriately identifies staining for the B cell markers CD79a and CD20. Although the staining for CD79a, CD20 can also be seen in the scattered large lymphocyte-predominant (LP) cells in nodular LP Hodgkin lymphoma (NLPHL) (see p. 324 of the WHO guideline1), these LP cells generally lack CD30 staining. Also, the predominance of large cells helps to rule out NLPHL. Thus these features all together offer insights into the differential diagnosis of DLBCL (see Chapter 10 of the WHO guideline1). The second DLBCL associated subgraph group is again highly consistent with the current pathologic definition of DLBCL and in this group the additional feature of monotypic light chain expression is identified. This group appears to be directed toward the identification of the activated B cell-like subtype of DLBCL which is CD10 negative. The third DLBCL associated subgraph group echoes the characteristic features of DLBCL: diffuse infiltrate of neoplastic cells, expression of common B-cell lineage antibodies, and monotypic immunoglobulin expression. The second and third groups also reflect the mixed expression levels of BCL2 in DLBCL. The fourth DLBCL associated subgraph group states the following interesting facts: Ki67 proliferation index is moderately high. Note that when discretizing percentages, we choose multiple dichotomy thresholds with a step size of 10%. Thus collectively the subgraphs on Ki67 proliferation index point out that the index is moderately high in DLBCL. This in addition to the positivity of CD20 and CD79a, and the monoclonality of immunoglobulin light chains collectively associate with the differential diagnosis of DLBCL (see Chapter 10 of the WHO guideline1).
For the follicular lymphoma cluster as shown in Table 4, the first associated subgraph group is consistent with the fact that follicular lymphoma is typically composed of both centrocytes (small cells) and centroblasts, and in bone marrow biopsies the lymphoma characteristically localizes to the paratrabecular region in bone marrow and may spread into the interstitial area (see p. 222 of the WHO guideline1). The second follicular lymphoma associated subgraph group is consistent with frequent BCL2 overexpression, accompanied sclerosis, and enlargement and effacement in the architecture of lymph nodes in the setting of follicular lymphoma. The third follicular lymphoma associated subgraph group summarizes typical immunophenotypic features such as lack of expression for the cell surface marker CD5, and mixed expression levels of CD10 (together with the first and second follicular lymphoma associated subgraph groups) and CD23, all of which are consistent with Table 8.01 in the WHO guideline.1 The fourth follicular lymphoma associated subgraph group reveals characteristic morphological features including dense infiltration of small lymphoid cells, the presence of cleaved centrocytes, and the staining of cells in follicular dendritic pattern (see p. 220 of the WHO guideline1).
For the Hodgkin lymphoma cluster as shown in Table 5, the first associated subgraph group correctly identifies the morphological feature of the large neoplastic Reed-Sternberg cells that are usually multilobated and stain positively for CD15 (see p. 327 of the WHO guideline1). The second Hodgkin lymphoma associated subgraph group extracts additional essential hematopathologic features for the malignant cells of Hodgkin lymphoma: CD30 positivity, CD15 positivity, CD20 negativity, and the appearance suggestive of Reed-Sternberg cells, which often express PAX5 and occur with histiocytes (see p. 328 of the WHO guideline1). The third Hodgkin lymphoma associated subgraph group is mostly consistent with the nodular sclerosis subtype of classical Hodgkin lymphoma, where the lymphoma contains Reed-Sternberg cells as well as a microenvironment of non-neoplastic inflammatory cells, the lymph nodes show a nodular growth pattern, collagen bands often surround nodules, and necrosis may occur (see p. 330 of the WHO guideline1). The fourth Hodgkin lymphoma associated subgraph group is mostly consistent with the subtype of NLPHL, in that large neoplastic cells (LP cells) are positive for CD45, OCT2, PAX5, and immunoglobulin light (kappa and/or lambda) chains. The subgraph group is also consistent with the co-occurrence of LP cells and CD3 positive T-cells (see p. 324 of the WHO guideline1).
We note the advantage of using subgraph groups as features compared to using individual subgraphs as features. For example, in the third follicular lymphoma associated subgraph group, standalone positivity or negativity on CD5, CD10, and CD23 may not be discriminative enough, but collectively they offer medically important information favoring follicular lymphoma.
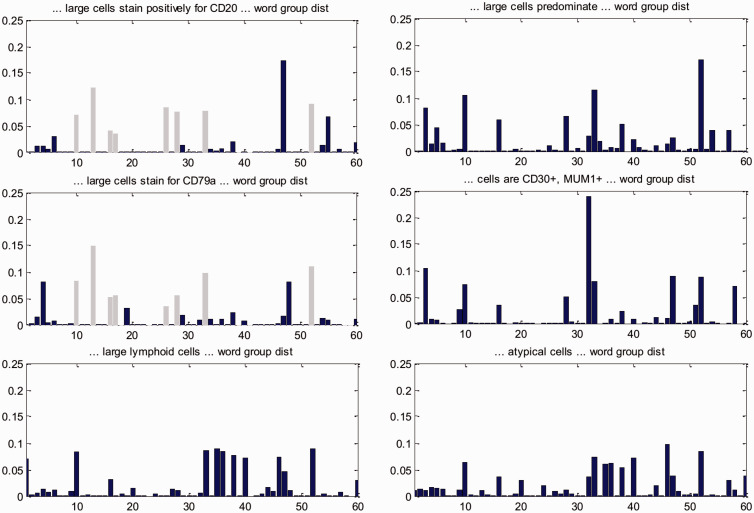
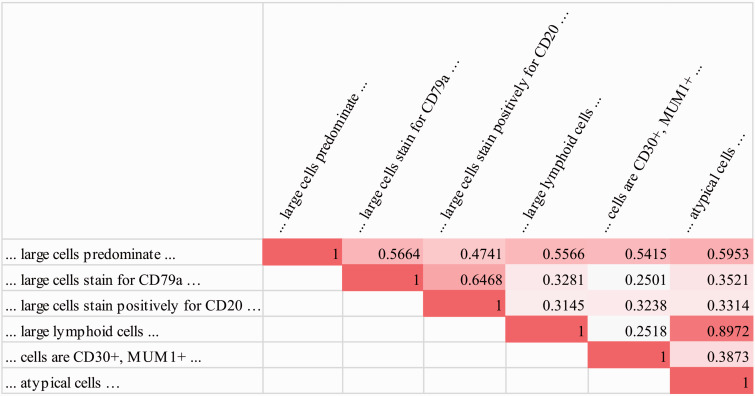
We next look into why the atomic feature groups as jointly discovered by SANTF help to better group individual subgraphs, in order to validate our intuition that exploiting interactions between both feature types is beneficial. Continuing from the analysis of important higher-order feature groups, we give an analysis on word group distributions associated with individual subgraphs. In the first DLBCL associated subgraph group in Table 3, the following subgraphs (partial sentences) are together ranked among the top subgraphs: “… large cells predominate …,” “… large cells stain for CD79a …,” “… large cells stain positively for CD20 …,” “… large lymphoid cells …,” “… cells are CD30+, MUM1+ …,” “… atypical cells …” By contrast, we did not find a similar grouping in patterns generated by those baselines that have subgraphs as features (baselines 2 and 3 in Table 2, k-means clustering does not produce subgraph groups). The positivity for the antigens CD79a and CD20 may associate with the scattered large LP cells in NLPHL, but the group includes additional positive staining for MUM1 and CD30, which favors the differential diagnosis of DLBCL. We look into the above six subgraphs and identify word groups associated with each subgraph. Intuitively, such associations are expressed in the core tensor and one can sum out the patient mode to explicitly associate a subgraph with the word groups (see SANTF algorithm section in the supplement on how to identify word groups associated with a specific subgraph from the tensor factorization results). The associated word group distribution for each subgraph is shown in Figure 4, and their correlation coefficients are shown in Figure 5. It becomes evident from Figure 5 that each of the subgraphs is correlated with at least one other subgraph with a correlation coefficient above 0.5, indicating relatively strong correlation. Figure 4 gives details on which word groups help to correlate subgraphs. For example, the word groups 10, 13, 16, 17, 26, 28, 33, and 52 help correlate subgraphs “… large cells stain positively for CD20 …” and “… large cells stain for CD79a ….” This illustrates the benefits of using word group distribution to correlate subgraphs. In summary, analysis of word groups suggests that adding the word mode (including covered and contextual words) to the tensor and jointly learning the subgraph groups and the word groups help to better capture the correlations between subgraph features.
Figure 4:
Word group distribution for six of the top subgraphs in the first DLBCL associated subgraph group. For example, the word groups 10, 13, 16, 17, 26, 28, 33, and 52 help correlate subgraphs “… large cells stain positively for CD20 …” and “… large cells stain for CD79a …”, as highlighted in light gray.
Figure 5:
Correlation between six of the top subgraphs (partial sentences) in the first DLBCL associated subgraph group, only upper triangular matrix is shown due to symmetry.
DISCUSSION AND FUTURE WORK
Currently the selection of SANTF parameters such as core tensor size relies on cross validation. We recognize the potential of using a nonparametric Bayesian approach to discover such parameters directly from data. For example, in the nonparametric Bayesian setting, each patient in a dataset can be associated with hidden variables describing groups (causes) that are responsible for generating the patient’s data. Although there can be an infinite number of possible groups to choose from, under proper prior distributions (e.g., specified using the Indian buffet process48), only a finite number of groups would be selected. Care needs to be taken when defining generative processes for multiple types of features to account for the fact that atomic features aggregate into higher-order features and to allow for an efficient inference algorithm. Clearly, the performance of SANTF depends on the nature of the relationships among the various modes of the tensor. We suspect that there is an information-theoretic analysis that can shed light on quantifying these relationships, where the suggested generative model could provide a basis for such an analysis.
SANTF applies to any medical subdomain where information can be represented as higher-order features and atomic features. For example, we recognize the potential benefits of applying SANTF to physiologic time series. Recent studies49,50 called for learning risk stratification models automatically from patient physiologic times series, for example, laboratory test values and vital measurements of patients monitored in the intensive care units. Progression of multiple physiologic variables can be summarized into temporal patterns (higher-order features) using graph representation and mining. Intuitively, similar numerical values (atomic features) of various physiologic measurements are helpful in identifying groupings of physiologic temporal trends by indicating similar states through which the patients have passed. Thus it is reasonable to expect that SANTF is also likely to improve modeling of physiologic time series in predictive tasks such as mortality risk stratification.
SANTF is currently computationally intensive. The tensor factorization on average takes 22 min on a computer with Intel Core 2 Duo P8600 and 8 GB RAM. The steps of document preprocessing including parsing, UMLS concept identification and graph/subgraph construction also take considerable amount of time. We parallel the computations into batches of 50 patients and run them on the pHPC clusters at Partners Health Care which has 600 processing cores in total and a maximum 100 core concurrency per user. The paralleled pre-processing time is within 30 min, which could be improved by parallelization into smaller batches on a larger cluster. We also plan to explore parallelization and approximation techniques such as stochastic gradient descent to speed up tensor factorization in future work.
Parsing challenges may arise with less formal clinical notes such as discharge summaries. For example, many connecting parts of speech (conjunctions, articles, prepositions) may be elided, which makes parsing dependency difficult for even statistical parsers. For less formal clinical notes, we expect a hybrid form of NLP may work better. Namely, for longer sentences, graph construction can be based on dependency parsing, while for shorter sentences, graph construction can be based on co-occurrence of concepts. Choosing the threshold of longer versus shorter sentences is non-trivial and may depend on the characteristics of clinical notes, we intend to explore such trade-offs in future work. On the other hand, different institutions may have different clinical documentation systems and styles. Such generalizability challenges are partly addressed by our clinical text subgraph mining approaches10 such as using UMLS concepts as subgraph nodes and ignoring dependency types, which can mitigate the impact of the terminology and style differences between institutions. Using atomic features to correlate higher-order features as done by SANTF also helps connect higher-order features whose differences are mainly in writing style. We are expanding the lymphoma classification project across institutions and across nations, and systematic generalizability analysis is part of our future work.
CONCLUSIONS
We proposed a novel unsupervised framework of subgraph augmented non-negative tensor factorization (SANTF), which can automatically generate machine learning models that are naturally interpretable to clinicians. SANTF can jointly model the interactions among different types of features by integrating them into the learning objective. We applied SANTF to unsupervised learning tasks on clustering lymphoma subtypes based on narrative text from pathology reports. We established nine baselines with widely used non-negative matrix factorization (NMF) and k-means clustering methods. For each of NMF or k-means configuration, the first baseline explores the atomic features. The second baseline explores the higher-order subgraph features. The third baseline explores both types of features but not their correlations. Experimental evaluation demonstrated that SANTF significantly outperforms all nine baselines, in particular, by over 10% margins in average F-measure to all baselines. A closer look at the subgraph groups that are generated by SANTF offers more clinical insights about lymphoma subtypes than atomic features or even standalone subgraphs. We also found that the atomic feature groups as jointly discovered by SANTF help to better correlate individual subgraphs, validating our intuition that exploiting interactions between different feature types is beneficial.
COMPETING FINANCIAL INTERESTS
None.
ETHICS APPROVAL
The Institutional Review Boards governing oncology care at the Massachusetts General Hospital approved this study. A waiver of informed consent was obtained. The intensive care data are from a dataset distributed under a limited data use agreement, which was approved by the Beth Israel Deaconess Hospital’s IRB.
FUNDING
The work described was supported in part by Grant Number U54LM008748 from the National Library of Medicine and by the Scullen Center for Cancer Data Analysis.
CONTRIBUTORS
YL is the primary author and was instrumental in developing the subgraph and tensor modeling, and performed data analysis. YX contributed to tensor modeling and analysis. EH provided expertise on lymphoma pathology. RJ provided input to feature analysis. OU contributed to the subgraph modeling and evaluation. PS provided expertise in machine learning and data analysis. EH and PS are the principal investigator for the grants involving the secondary use of clinical data. All co-authors reviewed and edited the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
Supplementary Material
REFERENCES
- 1. Swerdlow SH, Campo E, Harris NL, et al, eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press; 2008.
- 2. Winslow RL, Trayanova N, Geman D, Miller MI. Computational medicine: translating models to clinical care. Sci Transl Med. 2012;4:158rv11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shipp MA, Ross KN, Tamayo P, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. [DOI] [PubMed] [Google Scholar]
- 4. Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Informat. 2001;34:301–310. [DOI] [PubMed] [Google Scholar]
- 5. Hristovski D, Friedman C, Rindflesch TC, Peterlin B. Exploiting semantic relations for literature-based discovery. AMIA Ann Symp Proc. 2006;2006:349–353. [PMC free article] [PubMed] [Google Scholar]
- 6. Xu H, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Irwin JY, Harkema H, Christensen LM, et al. Methodology to develop and evaluate a semantic representation for NLP. AMIA Ann Symp Proc. 2009;2009:271. [PMC free article] [PubMed] [Google Scholar]
- 8. Gordon MM, Moser AM, Rubin E. Unsupervised analysis of classical biomedical markers: robustness and medical relevance of patient clustering using bioinformatics tools. PloS One. 2012;7:e29578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luo Y, Szolovits P, Sohani A, Hochberg E. Automatic lymphoma classification with sentence subgraph mining from pathology reports. JAMIA. 2014;21:824–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lasko TA, Denny JC, Levy MA. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PloS One. 2013;8:e66341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Norén GN, Hopstadius J, Bate A, Star K, Edwards IR. Temporal pattern discovery in longitudinal electronic patient records. Data Min Knowl Disc. 2010;20:361–387. [Google Scholar]
- 13. Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. [DOI] [PubMed] [Google Scholar]
- 14. Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nat Methods. 2013;10:1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Müller F-J, Laurent LC, Kostka D, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang F, Lee N, Hu J, Sun J, Ebadollahi S. Towards heterogeneous temporal clinical event pattern discovery: a convolutional approach. In: proceedings of the 18th ACM SIGKDD international conference on Knowledge discovery and data mining, Beijing, China; 2012:453–461. [Google Scholar]
- 18. Kim H, Park H. Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics. 2007;23:1495–1502. [DOI] [PubMed] [Google Scholar]
- 19. Brunet J-P, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA. 2004;101:4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao Y, Church G. Improving molecular cancer class discovery through sparse non-negative matrix factorization. Bioinformatics. 2005;21:3970–3975. [DOI] [PubMed] [Google Scholar]
- 21. Nik-Zainal S, Wedge DC, Alexandrov LB, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet. 2014;46:487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tucker LR. Some mathematical notes on three-mode factor analysis. Psychometrika. 1996;31:279–311. [DOI] [PubMed] [Google Scholar]
- 24. Sun J, Tao D, Papadimitriou S, Yu PS, Faloutsos C. Incremental tensor analysis: theory and applications. ACM Trans Knowl Discov Data (TKDD) 2008;2:11. [Google Scholar]
- 25. Harshman RA, Lundy ME. Uniqueness proof for a family of models sharing features of Tucker’s three-mode factor analysis and PARAFAC/CANDECOMP. Psychometrika. 1996;61:133–154. [Google Scholar]
- 26. Omberg L, Golub GH, Alter O. A tensor higher-order singular value decomposition for integrative analysis of DNA microarray data from different studies. Proc Natl Acad Sci USA. 2007;104:18371–18376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Omberg L, Golub GH, Alter O. Global effects of DNA replication and DNA replication origin activity on eukaryotic gene expression. Mol Syst Biol. 2009;5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ozcaglar C, Shabbeer A, Vandenberg S, Yener B, Bennett KP. Sublineage structure analysis of Mycobacterium tuberculosis complex strains using multiple-biomarker tensors. BMC Genomics. 2011;12:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yener B, Acar E, Aguis P, et al. Multiway modeling and analysis in stem cell systems biology. BMC Syst Biol. 2008;2:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bader BW, Puretskiy AA, Berry MW. Scenario discovery using nonnegative tensor factorization. Progress in Pattern Recognit, Image Anal Appl. 2008;5197:791–805. [Google Scholar]
- 31. Berry MW, Browne M. Email surveillance using non-negative matrix factorization. Comput Math Organ Th. 2005;11:249–264. [Google Scholar]
- 32. Shahnaz F, Berry MW, Pauca VP, Plemmons RJ. Document clustering using nonnegative matrix factorization. Inform Process Manag. 2006;42:373–386. [Google Scholar]
- 33. Bader BW, Berry MW, Browne M. Discussion tracking in Enron email using PARAFAC. Survey of Text Mining II. 2008;147–163. [Google Scholar]
- 34. Kolda TG, Bader BW. Tensor decompositions and applications. SIAM Rev. 2009;51:455–500. [Google Scholar]
- 35. Nijssen S, Kok JN. The gaston tool for frequent subgraph mining. Electron Notes Theor Comput Sci. 2005;127:77–87. [Google Scholar]
- 36. Liu H, Hunter L, Kešelj V, Verspoor K. Approximate subgraph matching-based literature mining for biomedical events and relations. PloS One. 2013;8:e60954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang C, Coenen F, Sanderson R, Zito M. Text classification using graph mining-based feature extraction. Knowledge-Based Syst. 2010;23:302–308. [Google Scholar]
- 38. Rink B, Bejan CA, Harabagiu SM. Learning textual graph patterns to detect causal event relations. FLAIRS Conference, Daytona Beach, Florida. 2010. [Google Scholar]
- 39. Liu H, Komandur R, Verspoor K. From graphs to events: a subgraph matching approach for information extraction from biomedical text. In: proceedings of the BioNLP shared task 2011 workshop, Portland, Oregon; 2011:164–172. [Google Scholar]
- 40. Chi Y, Muntz RR, Nijssen S, Kok JN. Frequent subtree mining-an overview. Fundam Inform. 2005;66:161–198. [Google Scholar]
- 41. Jiang C, Coenen F, Zito M. A survey of frequent subgraph mining algorithms. Knowl Eng Rev. 2013;28:75–105. [Google Scholar]
- 42. Manning CD, Schütze H. Foundations of Statistical Natural Language Processing. Newport Beach, CA: MIT press; 1999. [Google Scholar]
- 43. Ding CH, He X, Simon HD. On the equivalence of nonnegative matrix factorization and spectral clustering. SDM. 2005;5:606–610. [Google Scholar]
- 44. Manning CD, Raghavan P, Schütze H. Introduction to Information Retrieval. Vol. 1 Cambridge: Cambridge University Press; 2008. [Google Scholar]
- 45. Xu Y, Yin W. A block coordinate descent method for regularized multiconvex optimization with applications to nonnegative tensor factorization and completion. SIAM J Imaging Sci. 2013;6:1758–1789. [Google Scholar]
- 46. Liu J, Liu J, Wonka P, Ye J. Sparse non-negative tensor factorization using columnwise coordinate descent. Pattern Recogn. 2012;45:649–656. [Google Scholar]
- 47. Griffiths TL, Steyvers M. Finding scientific topics. Proc Natl Acad Sci USA. 2004;101:5228–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Griffiths TL, Ghahramani Z. The Indian buffet process: an introduction and review. J Mach Learn Res. 2011;12:1185–1224. [Google Scholar]
- 49. Saria S, Rajani AK, Gould J, Koller DL, Penn AA. Integration of early physiological responses predicts later illness severity in preterm infants. Sci Transl. Med. 2010;2:48–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Joshi R, Szolovits P. Prognostic physiology: modeling patient severity in intensive care units using radial domain folding. AMIA Annu Symp Proc. 2012;2012:1276. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.