Abstract
Objective Many tasks in natural language processing utilize lexical pattern-matching techniques, including information extraction (IE), negation identification, and syntactic parsing. However, it is generally difficult to derive patterns that achieve acceptable levels of recall while also remaining highly precise.
Materials and Methods We present a multiple sequence alignment (MSA)-based technique that automatically generates patterns, thereby leveraging language usage to determine the context of words that influence a given target. MSAs capture the commonalities among word sequences and are able to reveal areas of linguistic stability and variation. In this way, MSAs provide a systemic approach to generating lexical patterns that are generalizable, which will both increase recall levels and maintain high levels of precision.
Results The MSA-generated patterns exhibited consistent F1-, F.5-, and F2- scores compared to two baseline techniques for IE across four different tasks. Both baseline techniques performed well for some tasks and less well for others, but MSA was found to consistently perform at a high level for all four tasks.
Discussion The performance of MSA on the four extraction tasks indicates the method’s versatility. The results show that the MSA-based patterns are able to handle the extraction of individual data elements as well as relations between two concepts without the need for large amounts of manual intervention.
Conclusion We presented an MSA-based framework for generating lexical patterns that showed consistently high levels of both performance and recall over four different extraction tasks when compared to baseline methods.
Keywords: information extraction, natural language processing, text mining
OBJECTIVE
Many tasks in natural language processing (NLP) utilize lexical pattern-matching techniques, including information extraction (IE), negation identification, and syntactic parsing. Existing pattern formulation constructs, such as regular expressions, enable the generation of lexical patterns that can precisely capture specific sequences of tokens while enabling approximate matching through the use of wildcards and other flexible mechanisms. Though much NLP research has focused on applying machine learning techniques, the declarative and transparent nature of lexical patterns, which makes them easy to understand and maintain, can be very advantageous for processing large-scale data both accurately and robustly for real-world applications. A recent study has shown that commercial IE systems are mostly rule-based, and very few have adopted machine learning methods, because they must deal with dynamic requirements that demand the interpretability of rule-based systems.1 However, the main disadvantage of using rule-based systems is that crafting high-quality and high-performing patterns can be both challenging and labor-intensive. Thus, methodologies that automate the pattern generation process will become increasingly important as the need to process and analyze large-scale textual data continues to grow.
Current automated pattern generation systems typically leverage static windows of tokens (eg, two to four words) and the resulting patterns can often handle some degree of variation between each token. However, the variation built into these patterns may not reflect the language usage in the text of documents being processed. Phrases of interest within documents may exhibit a high level of variation in some areas and consistency in others, and patterns used to identify these phrases must be designed to account for these differing regions of variation and stability. In this paper, we present an automated pattern generation methodology that uses multiple sequence alignments (MSAs) to capture the commonalities and disparities between groups of phrases in order to systematically identify areas of conservation and variability. We show that this technique is able to generate high-quality lexical patterns using minimal manually annotated data over several IE tasks when compared with patterns generated using static token windows.
BACKGROUND AND SIGNIFICANCE
This work is closely related to NLP research focusing on characterizing the context of words or phrases using lexical, syntactic, and semantic features. A simple, and often effective, way of defining context is using a bag of words that surround the target. This technique has been successfully applied to several NLP problems, including word sense disambiguation,2 text classification,3 and information retrieval.4 To recover information lost by ignoring word order, n-grams have been utilized as a straightforward method of including sequential characteristics in models of context.5 Information retrieval systems represent contexts as vectors in word or n-gram space and calculate similarity between contexts using a standard distance metric (eg, cosine).6 To more fully capture word order, sequence models are often used to characterize the language generation process and the relationships between tokens and previously occurring information. These methods often take the form of Hidden Markov Models7 or Conditional Random Fields8 for modeling language semantics as probabilistic transitions between states. Other machine learning algorithms utilize a full range of features to generate statistical models of contexts. These features range from the word itself, to various lexical features of the words (eg, capitalization, word endings), to part of speech tags, to semantic labels from a specified ontology.9,10
Much work has been done on IE methodologies and systems over the past several decades. Early systems were mostly rule-based, and methodologies focused on automating the rule generation process, while later systems utilized machine learning or hybrid approaches that integrate aspects of both techniques.11,12 There has also been much work in the medical informatics domain on IE systems for clinical documents. Approaches mirror those of general IE and range from rule-based13–17 to machine learning18–21 to hybrid systems.22–24
The techniques that are most relevant to this work are IE systems that automatically generate extraction rules for individual concepts or relations. An earlier seminal work on automatically generating rules utilized annotated examples for training and syntactic clausal boundaries to determine the window of tokens.25 More recently, Riloff26 describes a methodology for generating extraction rules for relations by identifying pairs of concepts that are known to have a target relation and generating patterns based on words that lie between any two instances of known concepts. Yang and Cardie27 presents a rule-based IE system that uses a window of two to four words around the target to generate candidate rules. The rules are then scored using partially labeled data and approximately matched against text to perform extraction. Gupta et al.28 generates candidate rules using a seed of target entities and uses the presence of trigger words to determine the window of words for generating rules. The rules are eventually codified as finite automata for the matching process.
Though sharing the same overall goals with the previously mentioned work, the method described in this paper differs in that it automatically captures context as a window of words surrounding a target by leveraging patterns of language use. Instead of relying on phrasal boundaries based on syntactic analysis or utilizing a static window of words, the methodology generates dynamic contexts based on commonly used phrases. These commonalities are summarized within MSAs that reveal areas of variation and/or stability within groups of similar phrases.
MATERIALS AND METHODS
Determining Context
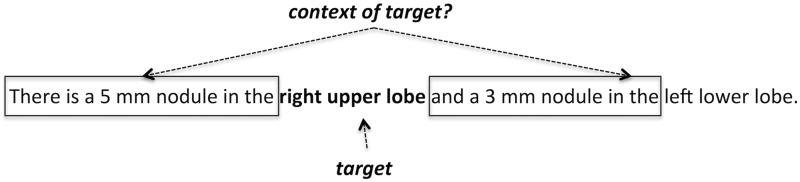
The usage-based theory of language is a formalism for modeling language acquisition and states that the meaning of language is highly dependent on how it is used.29 Thus, to determine the meaning of a target word or phrase, it is necessary to characterize its usage. Fundamentally, this takes the form of identifying the target’s context, which are the words and phrases that influence and shape the target’s meaning. In general, accurately identifying this context is a difficult task. Lexical proximity may provide some general guidance, but identifying the specific words and phrases that determine the meaning of a target is a nontrivial task. Figure 1 shows a simple example where lexical proximity alone would not provide a definitive determination of the context for the phrase “right upper lobe.” Our hypothesis is that particular word sequences that frequently occur with a target will most influence its meaning, and robustly identifying these phrases will lead to accurate delineations of the target’s context. If the same phrase is continually co-located in proximity to targets across several documents, MSAs can be used to model the basic form of the phrase as well as any lexical variations that might occur (eg, addition/deletion of one or two words). This work extends and further generalizes previous research using MSAs for IE as well as other NLP tasks.30–33
Figure 1:
Determining the words that make up the context of a target is a nontrivial task. Using distance does not favor one context over the other for the example target.
Generating Multiple Sequence Alignments
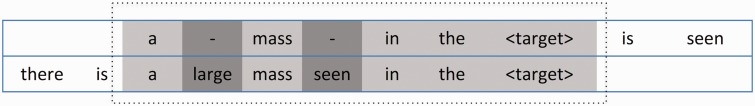
A brief description of our MSA generation algorithm is given here, with additional details provided in the supplementary materials. We employed a simple algorithm that progressively constructs MSAs by incrementally adding the results of the pair-wise local alignment of sentences. The basic idea is that after a pair-wise local alignment has been formed, the aligned sequences are added to an MSA that shares the same common tokens. The tokens that are common among all sequences within an alignment will be referred to as base tokens. We used the Smith-Waterman approximate local alignment algorithm to perform the pair-wise local alignments with a trivial scoring (or substitution) matrix that returns a positive value (eg, 1.0) for all exact matches between two tokens and 0.0 for everything else.34 To ensure that targets are always included in alignments, their match score was set to be much higher than the match score for other tokens (eg, 100.0). Semantic distance between tokens in general was not codified within the scoring matrix for the work reported in this paper and is left to future research. The gap penalty was set to a small negative number (eg, −0.01) to give the algorithm freedom to insert a limited number of gaps. Figure 2 shows the sequences of a pair-wise local alignment between two sentences using the Smith-Waterman algorithm, where gaps have been inserted into the top sequence to optimize the overall alignment. The alignment itself is shown within the dotted rectangle and tokens shown in light gray boxes are base tokens, tokens shown in dark gray show areas of variation, and tokens shown in white were not included in the pair-wise alignment. A maximum allowable number for gaps inserted into the pair-wise local alignment can be enforced to maintain a desired level of similarity among phrases clustered within the same MSA.
Figure 2:
Pair-wise alignment of two sentences, where the light gray shaded boxes represent matched tokens (base tokens) while the dark gray shaded boxes represent tokens that have been inserted. White boxes indicate tokens that do not participate in the alignment.
Once the Smith-Waterman algorithm generates an alignment, a search is run for an existing MSA that contains rows with the same set of base tokens as the alignment. If such an MSA is identified, both sequences in the pair-wise alignment are added to the MSA. Adding a sequence to an MSA involves inserting gaps into either the sequence that is being added and/or into the MSA’s existing rows in order to preserve the alignment of the base tokens. This process is repeated for both sequences in the pair-wise alignment. If there is no MSA that matches the pairwise alignment’s base tokens, a new MSA is created using the sequences in the alignment as the initial rows.
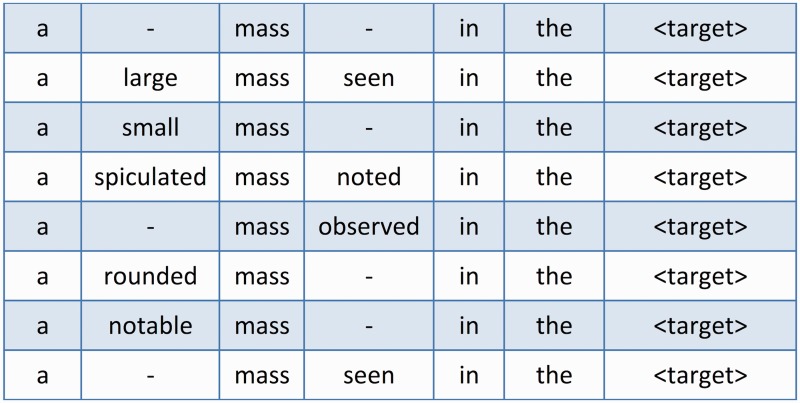
The MSA-generation algorithm uses a set of sentences that contain a specific type of target, such as a number, date, or anatomy phrase, as input. The type of target being used will depend on the task this technique is being used to accomplish. In the case of numbers, the possible task could be the extraction of tumor sizes from chest radiology reports, because numbers are the most generic form of measurements. Note that a sentence may contain more than one instance of a target and, thus, the target could occur multiple times in the sentence set and focus on a different target instance for each occurrence. Pair-wise, the algorithm locally aligns each sentence with every other sentence in the set and builds MSAs using the process described above. The end result of the algorithm’s run will be a set of MSAs that each represent a cluster of similar phrases that commonly occur in proximity to the target of interest (eg, numbers). Figure 3 shows an example MSA in which several sequences with the same set of base tokens (a, mass, in, the, <target>) have been aligned together.
Figure 3:
An example MSA generated from the common tokens of several different sentences that share the same target. The MSA clearly shows the areas of stability and variation among the different sentences.
Certain types of tokens or phrases were given generalized symbols in place of the original text in order to reduce variation and increase normalization, such as “<number>” for numbers and “<target>” for the target. Care must be taken when using these types of generalizations, because potentially vital semantic information can be lost (eg, indications of temporality when verb tenses are ignored). Also, all tokens were preserved and no stop words were removed.
Lexical Patterns from MSAs
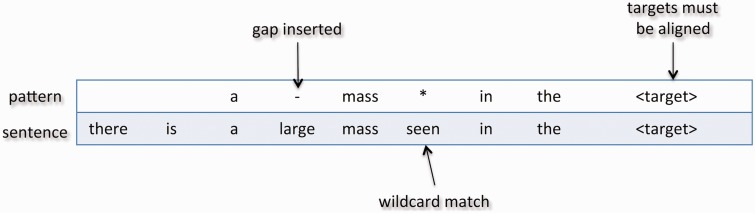
The generated MSAs now form the basis for the lexical patterns that can be utilized for extracting targets or relations involving targets. Each column of an MSA can be summarized using either a token for stable columns (base tokens) or a wildcard (asterisks) if the column contains variation, and summary information from all columns concatenated together determines the MSA’s lexical pattern. Matching an MSA-based lexical pattern with other text is basically a pair-wise local alignment between the two. The previously described alignment mainly differs in that lexical patterns have wildcards that can match any token. In addition, every base token in the lexical pattern is required to match a corresponding one within the text. The reason for this is that MSA-based patterns are assumed to represent phrases that are complete semantic units, and any modification of the base token sequence may alter the meaning of the phrase. Similar to the approximate local alignment described previously, targets from both sequences must align, and a maximum number of inserted gaps allowed can be enforced. An example of a lexical pattern being aligned with a sentence is shown in Figure 4. Note that gaps can be inserted into the lexical pattern as long as all base tokens have been aligned. Table 1 shows three extraction patterns generated for extracting relations between nodule sizes and locations (numeric values representing sizes are targets).
Figure 4:
Matching a pattern with a sentence is a pair-wise alignment with the inclusion of wildcards that can be matched against any token. In addition, to be considered a successful match, all tokens within the pattern must correspond with a token in the.
Table 1:
Example Extraction Patterns for Relations Between Nodule Size and Anatomy Location
| there be * <target> * mm * <nodule> in the <anatomy> |
| <anatomy> * <number> mm * previously <target> |
| <nodule> * in * the * <anatomy> * measure <number> x <target> mm |
Particular types of tokens, such as numbers (<number>), nodule types, including tumor, lesion, nodule, and opacity (<nodule>), and anatomy phrases (<anatomy>), were replaced by generic symbols. Target phrases are represented by <target> and wildcards are represented by asterisks. Words have been normalized to their root forms.
Clinical Application
The clinical application of this research is the automatic determination of lung cancer patient status based on the RECIST 1.1 criteria35 using information gathered from radiology reports. In order to automatically evaluate patient status using RECIST, specific data elements need to be obtained from the medical record, including tumor size, tumor location, the existence of metastases, etc. RECIST tracks changes in size of one or more target tumors over time and determines whether the patient’s disease is stable, worsening, or improving. In terms of facilitating IE, RECIST provides a structured framework that clearly indicates the data elements that need to be extracted.
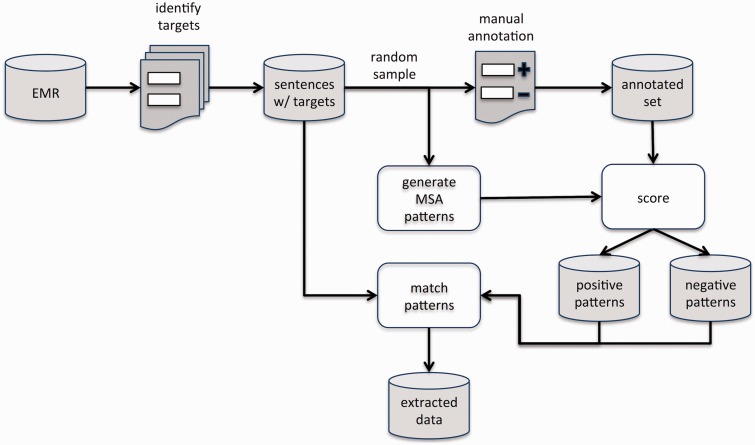
Pattern Generation Workflow
The overall workflow of the pattern generation methodology is similar to typical processes for generating IE patterns. As shown in Figure 5, targets such as anatomy concepts, dates, or numbers were first identified within the documents. Next, for a given target, a random subsample of target instances is drawn from the full set of instances, and each is manually annotated for a specific extraction task (eg, determining the anatomical location of lung nodules). The annotated instances are then used both to score patterns, to determine positive or negative cases, as well as to validate the performance of the extracted patterns after they are scored. The patterns themselves are automatically generated using MSAs and are divided into positive and negative, depending on the score assigned using the manually annotated subsample.
Figure 5:
System workflow. Sentences containing targets are first identified from documents and a subsample is set aside for both generating and scoring patterns. Scoring is based on a manually annotated set of instances. The scored patterns are then matched against sentences containing yet-to-be-extracted targets.
Extraction Tasks
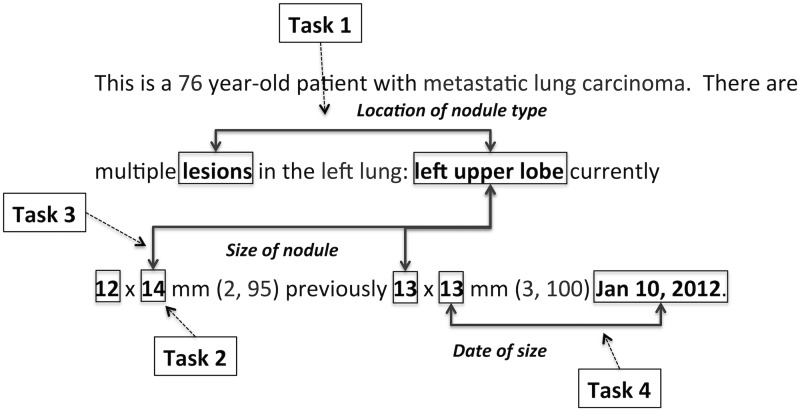
The system was trained to extract lung nodule type, anatomic location, and size from within chest radiology reports, using four extraction tasks: 1) extracting relations between lung anatomy phrases and nodule types; 2) identification of numeric values that are measurements or sizes; 3) extracting relations between sizes and lung anatomy locations; and 4) extracting relations between sizes and dates. Figure 6 shows the four extraction tasks using a specific example, where some markings have been left out to preserve clarity.
Figure 6:
Extraction tasks. Task 1 extracts the relation between nodule types and anatomy locations, Task 2 extracts nodule sizes, Task 3 extracts the relation between sizes and anatomy locations, and Task 4 extracts the relation between sizes and dates. Some markings have been left out, to preserve clarity.
Document Set and Preprocessing
We gathered a set of 7365 chest radiology reports from the UCLA healthcare system’s electronic medical records and processed them using a GATE NLP pipeline36 that included annotators for tokenization, sentence splitting, part of speech tagging, morphological analysis, detection of dates and numbers, and the MetaMap Unified Medical Language System concept mapping tool.37 The resulting set of over 6.5 million annotations covering 11 annotation types was stored in a MySQL relational database to facilitate further processing by other applications.
Gold Standard Annotation
One thousand targets were randomly sampled for each of the four extraction tasks, then manually annotated. For tasks that identified relations between the target and another concept, the target was labeled with the concept being related. Thus, all lung anatomic locations were labeled with nodule types (eg, nodule, tumor, or lesion) for Task 1, numeric values representing nodule measurements were labeled with lung anatomic locations for Task 3, and dates were labeled with nodule sizes, if they described the size’s temporality, for Task 4. For Task 2, numeric values were labeled either as “yes” or “no” for indicating a measurement. As an example of the annotation process, in the sentence, “The left upper lobe mass has increased in size from 12 × 13 mm on 1/1/2014 to 14 × 14 mm on 4/1/2013,” the lung anatomic phrase “left upper lobe” would be labeled with the word “mass” as the nodule type. The numeric values “12” and “13” would be labeled with “left upper lobe” as their associated anatomic location, to indicate that they are nodule sizes, as well as the date “1/1/2014,” to indicate their temporality.
Evaluation Procedure
We evaluated the methodology using the following process for each task. MSAs were generated by the previously described algorithm using sentences containing target instances that were not included in the gold standard. Extraction patterns were derived from the generated MSAs and were ordered from longest to shortest length (measured by number of words). The 1000 manually annotated target instances from the gold standard were divided into a scoring set (400 instances) and a validation set (600 instances). The extraction patterns were scored as positive or negative based on their accuracy (determined by using the scoring set). For scoring, each pattern was matched against every instance of a target in the scoring set, and the precision of the pattern was calculated. If the precision exceeded a predefined threshold (eg, 80%), the pattern would be labeled positive, and, otherwise, it was labeled negative. Each sentence containing a target instance in the validation set was then matched against the ordered list of extraction patterns. The best-matching pattern was determined to be the first (longest) pattern that matched all of its base tokens while maintaining the number of gaps below the predefined maximum. If a sentence matched both a positive and negative pattern, the target instance would be labeled as negative, if the negative pattern was a proper superset of the positive pattern. The F1-score, F.5-score, and F2-score were used as performance metrics. We compared our MSA-based technique with a baseline window-based pattern generation methodology – Bwin. Bwin patterns were generated by varying a window that extends from 1 to 11 words on either side of the target and generating an extraction pattern for each possible configuration of the window. For instance, a pattern was generated for one token to the left and one token to the right of the target, and another was generated for one token to the left and two tokens to the right, etc. The patterns generated by Bwin contain no wildcards, but approximate matching is implemented by inserting gaps during the matching process. For extraction Tasks 1, 3, and 4, which identify relations, a second baseline method, which extracts relations with targets by finding concepts with the shortest distance from the target (Bdist), was also compared to the MSA-based technique. Multiple evaluation runs were made by varying the values for the positive/negative pattern scoring threshold and the maximum number of gaps allowed when matching against text. The positive/negative threshold affects the precision of positive patterns, and, if set to high values (eg, 0.99), high levels of precision can be achieved. Higher numbers of gaps allowed in the alignments increases the flexibility of the patterns and improves overall recall, because this enables more approximate matches between extraction patterns and text. See supplementary materials for a more detailed explanation of the baseline methods.
RESULTS
Table 2 shows the results for all extraction tasks comparing the F1, F.5, and F2 scores of MSA and Bwin. For Tasks 1, 3, and 4, the scores were also compared with Bdist. The highest values are shown in bold, and values with an asterisk indicate that MSA is statistically significantly better based on approximate randomization.38
Table 2:
Results of MSA Compared with Bwin and Bdist for F1, F.5, and F2-scores with Precision and Recall Shown in Parentheses (P, R)
| Task | MSA | Bwin | Bdist |
|---|---|---|---|
| Task1 | |||
| F1-score | 0.940 (0.919, 0.962) | 0.880* (0.907, 0.855) | 0.935 (0.892, 0.983) |
| F.5-score | 0.935 (0.943, 0.907) | 0.916 (0.946, 0.812) | 0.909 |
| F2-score | 0.963 (0.882, 0.985) | 0.865* (0.907, 0.855) | 0.963 |
| Task 2 | |||
| F1-score | 0.987 (0.987, 0.987) | 0.987 (0.993, 0.981) | – |
| F.5-score | 0.991 (0.993, 0.981) | 0.991 (0.993, 0.981) | – |
| F2-score | 0.978 (0.987, 0.987) | 0.986 (0.981, 0.987) | – |
| Task 3 | |||
| F1-score | 0.940 (0.896, 0.988) | 0.893* (0.918, 0.870) | 0.920 (0.886, 0.956) |
| F.5-score | 0.934 (0.949, 0.877) | 0.908 (0.918, 0.870) | 0.900* |
| F2-score | 0.970 (0.883, 0.994) | 0.879* (0.918, 0.870) | 0.941 |
| Task 4 | |||
| F1-score | 0.924 (0.963, 0.889) | 0.922 (0.981, 0.871) | 0.747* (0.599, 0.991) |
| F.5-score | 0.955 (1.00, 0.838) | 0.967 (1.00, 0.855) | 0.650* |
| F2-score | 0.919 (0.849, 0.939) | 0.918 (0.918, 0.918) | 0.876 |
Bold values indicate the highest score for each row and an asterisk shows that MSA’s result is statistically significant over this value (p <.05).
Table 3 shows the number of candidate patterns generated for each method
Table 3:
Number of Patterns Generated by MSA vs. Bwin for Each Extraction Task.
| Task | MSA | Bwin |
|---|---|---|
| 1) Nodule location | 115 175 | 539 686 |
| 2) Nodule size | 10 194 | 196 984 |
| 3) Nodule size location | 61 307 | 305 552 |
| 4) Nodule size date | 2041 | 34 923 |
DISCUSSION
The overall results show that MSA performed the most consistently across the four tasks and attests to the versatility of the pattern generation process. The F1, F.5, and F2-scores were chosen as comparison metrics, because they test different importance weightings of precision and recall relative to one another, where F1 balances them equally, F.5 emphasizes precision, and F2 emphasizes recall.
Bwin is an exhaustive enumeration of word sequences surrounding a target type within the training set (up to 11 tokens on either side) and is a greatly expanded and more complete version of typical current IE pattern generation techniques. Thus, it was expected that Bwin’s precision would be very high and that recall would also be reasonable, because some approximate matching to generalize the patterns would be allowed. As seen in the results, Bwin did indeed perform well for all metrics, and the precision for the F.5 scores was always relatively high, but Bwin ran into some difficulty on Task 3 on which it was outperformed by both MSA and Bdist. For its simplicity, Bdist performed surprisingly well, achieving competitive scores for most of the tasks. It mainly suffered from low precision levels, because choosing the concepts with shortest distance does not take linguistic content into account at all, thereby ignoring a great deal of vital information. Bdist’s relatively good performance on Tasks 1 and 3 attests to the inherent regularity of language when radiologists state anatomy location with respect to nodule types and sizes. However, Task 4’s language pattern was the least favorable to this simple method, and Bdist’s performance was significantly worse on this task. The fact that MSA’s performance was always at or near the top causes us to believe that the technique is able to generate patterns that are generally superior to both baseline methods. This is especially true in the case of Bwin, in which exhaustive enumeration over a training set of 600 instances is still not sufficient for producing extraction patterns that perform at a high level over all four tasks. This shows that generating patterns must take language usage into consideration and that areas of regularity and variation within patterns must be chosen based on linguistic content.
As seen in Table 3, MSA also generates far fewer candidate patterns than Bwin, sometimes more than an order of magnitude less. This can be a significant gain in terms of the time and computational power spent to score patterns and makes MSA an attractive option even if the two methods exhibit similar performance levels.
Currently, the patterns contain only tokens, wildcards, and some generalized symbols, which provide for some level of flexibility but may be too limited for more complex cases. In the current implementation, the generic symbols need to be configured by domain experts who are able to determine the impact of the generalizations on the performance of the system. One future direction for research would be to develop techniques to automatically generalize patterns using multiple semantic levels, including generic symbols, parts of speech, semantic classes, and actual tokens from the original text.
From our analysis, some of the most difficult cases for the system to extract were relationships between concepts that were lexically distant from one another. This distance usually resulted from multiple phrases that were inserted between concepts. An example of this can be seen in the following sentence: “Multiple nodules seen in the left upper lobe 11 × 14 mm, right lower lobe 9 × 8 mm, and right upper lobe 10 × 11 mm.” Here, when the system tries to relate the word “nodules” with the phrase “right upper lobe,” the distance and variation could not be handled by most extraction patterns. One possible solution would be to represent groups of tokens that have already been semantically identified by the system using a generic symbol. In this example, if “left upper lobe” has already been identified as a nodule location along with its size (“11 × 14 mm”), it could be represented using the generic symbol <tumor-location-size>. This would reduce the complexity for identifying “right lower lobe” as a nodule location, and it would also be represented as a generic symbol, ultimately enabling the extraction of “right upper lobe.”
CONCLUSION
We presented a methodology for automatically generating patterns for IE systems within the clinical domain. This technique addresses one of the major drawbacks of using rule-based IE systems, which is the difficulty and labor-intensive nature of manually crafting high performing rules. We demonstrated that our MSA-based pattern generation technique was able to generate extraction rules that performed consistently across four different extraction tasks compared to two baseline methods. MSA also generated far fewer candidate patterns compared to a sliding window method, making it more computationally efficient. Overall, MSAs show much promise as a utility for systematically determining variation and stability for NLP lexical patterns.
FUNDING
This work was supported by the following National Institutes of Health grants: R01 CA1575533 (National Cancer Institute), R01 NS076534 (National Institute of Neurological Disorders and Stroke), and R01 LM011333 (National Library of Medicine).
COMPETING INTERESTS
None.
CONTRIBUTORS
F.M. was responsible for the conception, design, implementation, and evaluation of this work. C.M. participated in the design and interpretation of the results and revision of the draft.
PROVENANCE AND PEER REVIEW
None commissioned; externally peer reviewed.
Acknowledgments
The authors wish to thank Prof. William Hsu (UCLA) for recommending data sources for use in this study and Dr. Louis Fiore (MAVERIC) for providing clinical guidance.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1.Chiticariu L, Li Y, Reiss F. Rule-Based Information Extraction is Dead! Long Live Rule-Based Information Extraction Systems! EMNLP. 2013:827–832. [Google Scholar]
- 2.Yarowsky D. Unsupervised word sense disambiguation rivaling supervised methods. In: proceedings of the 33rd annual meeting on Association for Computational Linguistics (ACL '95). Association for Computational Linguistics; 1995:189–196; Stroudsburg, PA, USA [Google Scholar]
- 3.Ko Y. A study of term weighting schemes using class information for text classification. In: proceedings of the 35th international ACM SIGIR conference on Research and development in information retrieval (SIGIR '12). ACM; 2012:1029–1030; New York, NY, USA. [Google Scholar]
- 4.Carrillo M, López-López A. Concept based representations as complement of bag of words in information retrieval. AIAI, volume 339 of IFIP Advances in Information and Communication Technology, Springer; 2010:154–161. [Google Scholar]
- 5.Tandon N, de Melo G. Information Extraction from Web-Scale N-Gram Data (2010). In: Proc. Web N-gram Workshop at SIGIR 2010:59-63;Association for Computing Machinery (ACM). [Google Scholar]
- 6.Salton G, Wong A, Yang CS. A vector space model for automatic indexing. Commun ACM. 1975;18(11):613–620. [Google Scholar]
- 7.Skounakis M, Craven M, Ray S. Hierarchical hidden Markov models for information extraction. In: proceedings of the 18th international joint conference on Artificial intelligence (IJCAI'03). Morgan Kaufmann Publishers Inc.; 2003:427–433; San Francisco, CA, USA. [Google Scholar]
- 8.Peng F, McCallum A. Information extraction from research papers using conditional random fields. Inf Process Manage. 2006;42(4):963–979. [Google Scholar]
- 9.Ireson N, Ciravegna F, Califf ME, Freitag D, Kushmerick N, Lavelli A. Evaluating machine learning for information extraction. In: proceedings of the 22nd International Conference on Machine learning (ICML '05). ACM; 2005:345–352; New York, NY, USA. [Google Scholar]
- 10.Téllez-Valero A, Montes-y-Gómez M, Villaseñor-Pineda L. A Machine Learning Approach to Information Extraction Computational Linguistics and Intelligent Text Processing . 2005:539–547. [Google Scholar]
- 11.Chang C-H, Kayed M, Girgis MR, Shaalan KF. A Survey of Web Information Extraction Systems. IEEE Trans Knowl Data Eng. 2006;18(10):1411–1428. [Google Scholar]
- 12.Piskorski J, Yangarber R. Information extraction: past, present and future. In: Poibeau T, Saggion H, Piskorski J, Yangarber R, eds. Multi-source, Multilingual Information Extraction and Summarization. Berlin/New York, NY: Springer; 2013. [Google Scholar]
- 13.Bejan CA, Wei WQ, Denny JC. Assessing the role of a medication-indication resource in the treatment relation extraction from clinical text. J Am Med Inform Assoc. 2014:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben Abacha A, Zweigenbaum P. Automatic extraction of semantic relations between medical entities: a rule based approach. J Biomed Semantics. 2011;2(Suppl 5):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garvin JH, DuVall SL, South BR, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. JAMIA 2012;19(5):859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17(1):19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc. 2011;18(Suppl 1):i116–i1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Avolio LW, Nguyen TM, Goryachev S, Fiore LD. Automated concept-level information extraction to reduce the need for custom software and rules development. J Am Med Inform Assoc. 2011;18(5):607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esuli A, Marcheggiani D, Sebastiani F. An enhanced CRFs-based system for information extraction from radiology reports. J Biomed Inform. 2013;46(3):425–435. [DOI] [PubMed] [Google Scholar]
- 20.Patrick JD, Nguyen DH, Wang Y, Li M. A knowledge discovery and reuse pipeline for information extraction in clinical notes. J Am Med Inform Assoc. 2011;18(5):574–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rink B, Harabagiu S, Roberts K. Automatic extraction of relations between medical concepts in clinical texts. J Am Med Inform Assoc. 2011;18(5):594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovacevic A, Dehghan A, Filannino M, Keane JA, Nenadic G. Combining rules and machine learning for extraction of temporal expressions and events from clinical narratives. J Am Med Inform Assoc. 2013;20(5):859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang B, Wu Y, Jiang M, Chen Y, Denny JC, Xu H. A hybrid system for temporal information extraction from clinical text. J Am Med Inform Assoc. 2013;20(5):828–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang YC, Dai HJ, Wu JC, Chen JM, Tsai RT, Hsu WL. TEMPTING system: a hybrid method of rule and machine learning for temporal relation extraction in patient discharge summaries. J Biomed Inform. 2013;46(Suppl):S54–S62. [DOI] [PubMed] [Google Scholar]
- 25.Riloff E. Automatically generating extraction patterns from untagged text. In: proceedings of the Thirteenth National Conference on Artificial Intelligence - Volume 2 (AAAI'96), Vol. 2. AAAI Press; 1996:1044–1049. [Google Scholar]
- 26.Riloff Yang B, Cardie C. Extracting opinion expressions with semi-Markov conditional random fields. In: proceedings of the 2012 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL '12). Association for Computational Linguistics: Stroudsburg, PA, USA; 2012:1335–1345. [Google Scholar]
- 27.Gupta S, MacLean DL, Heer J, Manning CD. Induced lexico-syntactic patterns improve information extraction from online medical forums. J Am Med Inform Assoc. 2014;21:902–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talukdar PP, Brants T, Liberman M, Pereira F. A context pattern induction method for named entity extraction. In: proceedings of the Tenth Conference on Computational Natural Language Learning (CoNLL-X '06). Association for Computational Linguistics: Stroudsburg, PA, USA; 2006:141–148. [Google Scholar]
- 29.Tomasello M. Constructing a Language: a Usage-Based Theory of Language Acquisition. Harvard University Press: Stroudsburg, PA, USA; 2005. [Google Scholar]
- 30.Barzilay R, Lee L. Learning to paraphrase: an unsupervised approach using multiple-sequence alignment. In: proceedings of the 2003 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology, Vol. 1. 2003:16–23. [Google Scholar]
- 31.Hakenberg J, Plake C, Royer L, Strobelt H, Leser U, Schroeder M. Gene mention normalization and interaction extraction with context models and sentence motifs. Genome Biol. 2008;9(Suppl 2):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeganova L, Kim W, Comeau DC, Wilbur WJ. Finding biomedical categories in Medline®. J Biomed Semantics. 2012;3 (Suppl 3):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng F, Morioka CA, El-Saden S. Determining word sequence variation patterns in clinical documents using multiple sequence alignment. AMIA Annual Symposium Proceedings. 2011:934–943. [PMC free article] [PubMed] [Google Scholar]
- 34.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147(1):195–197. [DOI] [PubMed] [Google Scholar]
- 35.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. [DOI] [PubMed] [Google Scholar]
- 36.Cunningham H, Maynard D, Bontcheva K, Tablan V. GATE: an architecture for development of robust HLT applications. In: proceedings of the 40th Annual Meeting on Association for Computational Linguistics (ACL '02). Association for Computational Linguistics, Stroudsburg, PA, USA; 2002:168–175. [Google Scholar]
- 37.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;1(32):D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeh A. More accurate tests for the statistical significance of result differences. In: proceedings of the 18th conference on Computational linguistics - Volume 2 (COLING '00), Vol. 2. Association for Computational Linguistics, Stroudsburg, PA, USA; 2000:947–953. [Google Scholar]