ABSTRACT
The gene encoding the neural cell adhesion molecule Contactin-6 (Cntn6 a.k.a. NB-3) has been implicated as an autism risk gene, suggesting that its mutation is deleterious to brain development. Due to its GPI-anchor at Cntn6 may exert cell adhesion/receptor functions in complex with other membrane proteins, or serve as a ligand. We aimed to uncover novel phenotypes related to Cntn6 functions during development in the cerebral cortex of adult Cntn6−/− mice. We first determined Cntn6 protein and mRNA expression in the cortex, thalamic nuclei and the hippocampus at P14, which decreased specifically in the cortex at adult stages. Neuroanatomical analysis demonstrated a significant decrease of Cux1+ projection neurons in layers II-IV and an increase of FoxP2+ projection neurons in layer VI in the visual cortex of adult Cntn6−/− mice compared to wild-type controls. Furthermore, the number of parvalbumin+ (PV) interneurons was decreased in Cntn6−/− mice, while the amount of NPY+ interneurons remained unchanged. In the hippocampus the delineation and outgrowth of mossy fibers remained largely unchanged, except for the observation of a larger suprapyramidal bundle. The observed abnormalities in the cerebral cortex and hippocampus of Cntn6−/− mice suggests that Cntn6 serves developmental functions involving cell survival, migration and fasciculation. Furthermore, these data suggest that Cntn6 engages in both trans- and cis-interactions and may be involved in larger protein interaction networks.
KEYWORDS: ASD, autism, CAMs, cell adhesion molecule, Cntn6, contactin, hippocampus, NB-3, visual cortex
Introduction
Cell adhesion molecules (CAMs) play an important role in the development of the nervous system by mediating multiple processes that rely on cell-cell contact and –signaling. Cell adhesion is regulated by the trans-interactions of an expressed collection of CAMs on specific subsets of cells. Cell adhesion functions facilitate neuronal migration, lamination, neurite outgrowth and axon guidance, and synaptogenesis.1-3 Furthermore, CAMs may serve receptor properties and thereby regulate neuronal apoptosis or survival.4,5
Contactin-6 (Cntn6 a.k.a. NB-3) is a member of the contactin family of immunoglobulin (Ig) cell adhesion molecules (IgCAMs) and is specifically expressed in the central nervous system. The Cntn family consists of six members which are extracellularly attached on the cell surface by a glycosylphosphatidylinositol link (GPI), and contain six Ig and four fibronecin type III domains (FNIII).6,7 CNTN6 is of particular interest, since studies have indicated that copy number variants in this gene contribute to several neuropsychiatric disorders.8-14 This indicates that CNTN6 proteins exert functions that are required for normal neurodevelopment.
In the murine brain, the Cntn6 gene is expressed in the cortex, hippocampus, thalamus and cerebellum,15-17 reaching a maximum at P14 and thereafter declining to a constant level in adulthood.15 The notion that loss of Cntn6 results in inappropriate neurodevelopment is strengthened by the phenotypes identified in null-mutants. A number of observations have pointed to a role of Cntn6 in synapse formation and axonal outgrowth. In particular, Cntn6−/− mice exhibited a significant reduction the density of parallel fiber glutamatergic synaptiC-terminals in the cerebellum.18 Similarly, a significant reduction of glutamatergic synapses was found in the hippocampus of Cntn6−/− mice, whereas there was no difference in the amount of GABAergic synapses.17 Cntn6 was previously found to regulate neurite outgrowth in vitro,19 and this property was reflected by the delayed formation of the corticospinal tract in Cntn6−/− mice.20,21 These known neurobiological processes exerted by Cntn6 can be attributed to the cell adhesion properties of Cntn6.
Such properties of Cntn6 may be the basis for the development of neuropsychiatric disorders in case of loss-of-function mutations of CNTN6. Studies on the visual cortex in Cntn6−/− mice demonstrated the significance of Cntn6 in cortical development. Cntn6 is expressed in a low-rostral to high-caudal gradient in the cortex, with the highest expression levels in the visual cortex, where deep layer pyramidal neurons develop axonal and dendritic processes.21,22 A substantial alteration in the orientation of apical dendrites of pyramidal neurons in layer V of the visual cortex of one month old Cntn6−/− mice was found.22
The aim of this study was to examine phenotypes in Cntn6−/− mice that could further inform about the roles Cntn6 plays during neurodevelopment. Compromised functions of IgCAMs can critically affect the laminar organization of the cerebral cortex,23,24 while the mossy fiber tracts in the hippocampus are sensitive to fasciculation defects.25-27 For this reason we examined phenotypes in the visual cortex and the hippocampus of Cntn6−/− mice. Here we describe abnormalities in Cntn6−/− mice which indicate that Cntn6 functions in processes that regulate the numbers of subtype-specific neurons and interneurons in the cortex and in the formation of mossy fiber tracts in the hippocampus. These results suggest that Cntn6 might be involved in cell survival, migration and fasciculation by engaging both trans- and cis-interactions with itself or other molecules.
Results
Differential spatiotemporal expression of Cntn6 in the brain
We first determined the expression of the Cntn6 gene in the forebrain, since it has been reported that Cntn6 mRNA levels in whole mouse brain peak around P7.15 In situ hybridization was performed on P14 and adult C57BL/6 wild-type mouse brains. In the P14 sections, Cntn6 was most prominently expressed in the anterodorsal (AD) nucleus of the thalamus and in layer V of the motor cortex, somatosensory cortex, and visual cortex (Fig. 1A–B). In the adult brain, the expression in the AD nucleus remained constant and expression was also visible in the anteroventral (AV) nucleus of the thalamus. Furthermore, the overall expression in the cortex was decreased, most notably in the visual cortex. In the P14 and adult hippocampus, weak Cntn6 mRNA was observed in the cornu ammonis 1 (CA1), the dentate gyrus (DG) and in the hilus of the DG (Fig. 1C).
Figure 1.
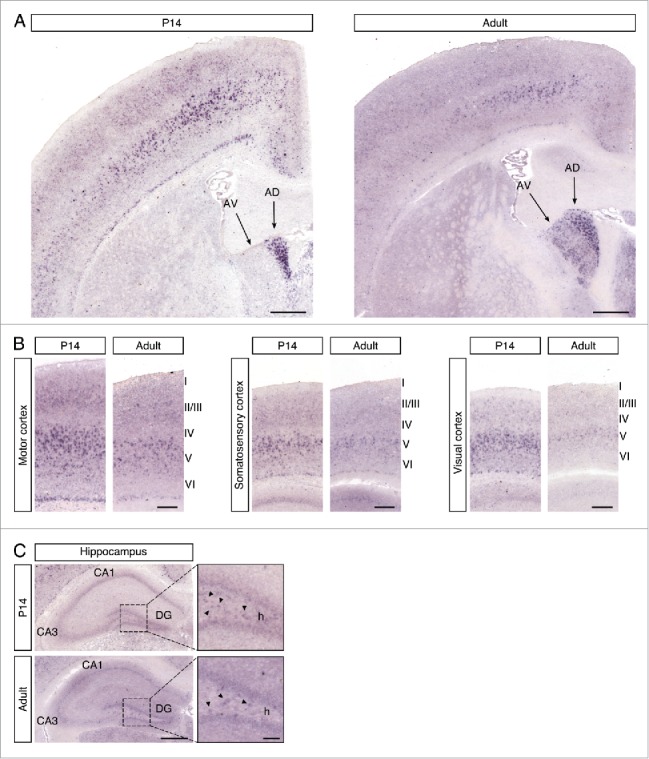
Cntn6 mRNA expression in the murine brain. (A) A low magnification of Cntn6 mRNA expression in coronal sections of P14 and adult mouse brain. Cntn6 was highly expressed the anterodorsal (AD) nucleus of the thalamus at P14. In the adult brain, both the AD and anteroventral (AV) nucleus of the thalamus expressed Cntn6. The scale bar represents 500 μm. (B) Cntn6 mRNA was present at high levels in the motor cortex, somatosensory cortex, and visual cortex at P14. The expression was lower in adult brains, most notably in the visual cortex. The scale bar represents 250 μm. (C) In P14 and adult hippocampi, Cntn6 mRNA was present in the cornu ammonis 1 (CA1) and in the dentate gyrus (DG). Cntn6 expression was visible in cell bodies in the hilus (h) of the DG as indicated by the arrows in the dashed squares, which are the magnified adjacent images. CA3, cornu ammonis 3. The scale bar represents 500 μm. The scale bar in the magnified image is 100 μm.
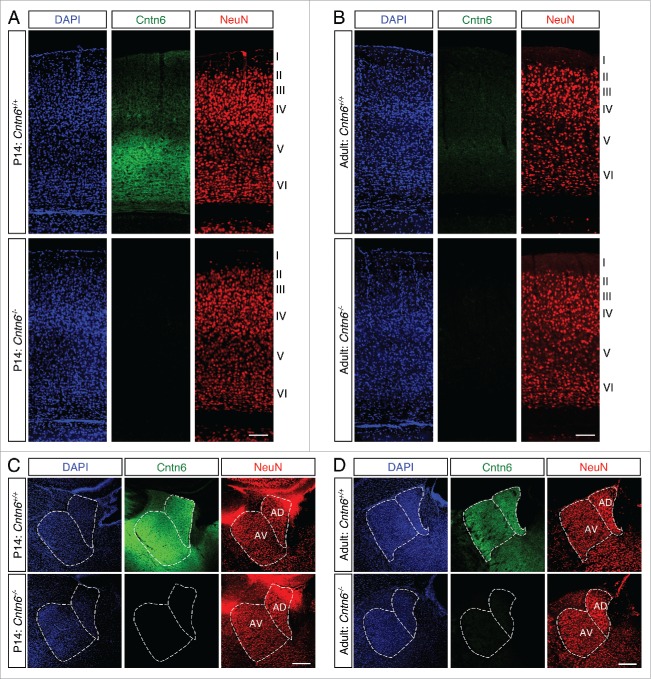
Next, immunostaining was performed to determine if the decreased expression of Cntn6 mRNA in the adult brain also affected the Cntn6 protein levels. Immunohistochemistry using a Cntn6 antibody showed strong staining in layer V of the visual cortex of young wild-type mice (Fig. 2A). This agrees with the reported expression of Cntn6 in the visual cortex in early postnatal stages.22 In line with the in situ data, staining in the visual cortex was very weak in adult wild-type mice (Fig. 2B). In addition, Cntn6 staining was observed in the AD and AV nuclei of both P14 and adult mice (Fig. 2C–D). Immunostaining signal was absent in tissues from Cntn6−/− mice (Fig. 2A–B), demonstrating the specificity of the Cntn6 antibody.
Figure 2.
Cntn6 protein sexpression in the adult mouse brain. (A-B) Immunohistochemistry for Cntn6 (green) and NeuN (red) in the visual cortex of P14 and adult wild-type and Cntn6−/− mice. Specific Cntn6 immunoreactivity was only observed in layer V of the P14 wild-type cortex and was strongly decreased in the adult. DAPI is in blue. The scale bar represents 250 μm. (C-D) Cntn6 (green) and NeuN (red) staining in the thalamic region of P14 and adult wild-type and Cntn6−/− mice. The staining revealed the presence of Cntn6 protein in the AD and AV nuclei of the thalamus in both P14 and adult wild-type mice but not in Cntn6−/− mice. DAPI is in blue. The scale bar represents 250 μm.
It has been reported that apical dendrites of cortical pyramidal neurons were misoriented in one month old Cntn6−/− mice.22 The patterning of axodendritic projections of layer V pyramidal neurons is regulated by molecular guidance cues in the extracellular environment.28 Dendritic misorientation has been seen in several mouse lines deficient for neuronal guidance cues like semaphoring-3A (Sema3A), close homolog of L1 (Chl1), and protein tyrosine phosphatase α (Ptpra).22,29,30 This raises the question whether the defects observed in Cntn6−/− mice points to properties of Cntn6 in cell-cell interaction. These may also become apparent in migration of neurons and lamination of the adult cortex.
Normal cortical thickness in Cntn6−/− mice
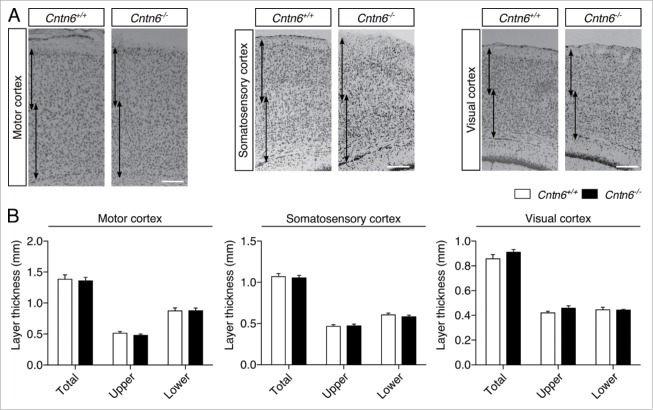
We first reevaluated the gross cortical morphology between adult wild-type and Cntn6−/− mice. Therefore, we measured the cortical thickness of the motor cortex, the somatosensory cortex and the visual cortex in adult mice (Fig. 3A). A distinction was made between the superficial and deeper layers of the cortices (i.e. layers I-IV and V-VI respectively), which were measured separately and in combination. There were no differences found between wild-type and Cntn6−/− mice in any of the measured cortices (Fig. 3B), consistent with previous reports.16 These results suggest that Cntn6 is not involved in the regulation of gross cortical morphology but rather in the fine-tuning of specific features of cortical development, such as the orientation of dendritic outgrowth. Previous reports suggest that Cntn6 does not affect the migration of developing pyramidal neurons.22 However, other processes that are involved in cortical organization might still be affected.31
Figure 3.
Cortical thickness in Cntn6-deficient mice. (A) Nissl-stained sections of the motor cortex, somatosensory cortex and visual cortex of adult wild-type and Cntn6−/− mice. Arrows indicate upper and lower layer thickness. The scale bar indicates 250 μm. (B) Quantitative analysis of the represented upper and lower cortical areas demonstrate equal cortical thickness between the genotypes. Analysis was performed on at least two sections per brain from wild-type and Cntn6−/− adult mice (n = 5 per genotype) using unpaired Student's t test and one-way ANOVA. Data are presented as mean ±SEM.
Altered numbers of projection neurons in the visual cortex of adult Cntn6−/− mice
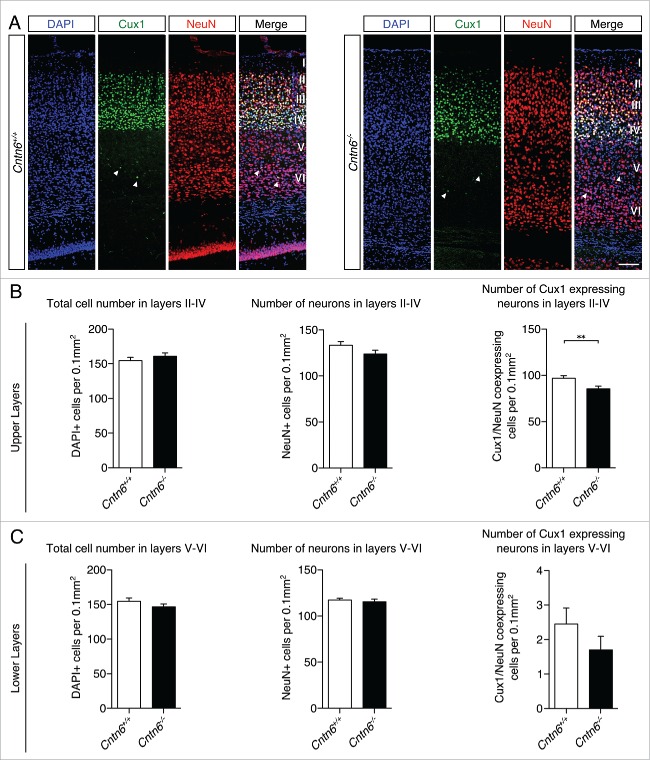
Next, we focused on the laminar positioning and numbers of specific cortical projection neurons in the visual cortex of wild-type and Cntn6−/− mice. The location and density of upper and lower layer projection neurons were investigated by using Cux1 antibodies, which marks cells destined for layers II-IV in adult wild-type and Cntn6−/− mice (Fig. 4A). Cell counting revealed no differences in the total numbers of cells and neurons, which are DAPI+ and NeuN+ cells respectively, in the upper layers between genotypes. However, the number of Cux1+ neurons in the upper layers was significantly decreased in Cntn6−/− mice (Fig. 4B). Measurements revealed no statistical difference in the total number of cells, neurons, and the number of Cux1+ displaced neurons in the lower layers of Cntn6−/− mice (Fig. 4C).
Figure 4.
Upper layer projection neurons in Cntn6-deficient mice. (A) Representative images of Cux1 expression (green) in layers II-IV of the visual cortex together with NeuN (red) in adult wild-type and Cntn6−/− mice. DAPI is in blue. The arrowheads show displaced Cux1+ cells in lower cortical layers. The scale bar represents 100 μm. (B) Quantitative analysis of the upper layers (layers II-IV) did not show differences in total cell number and total neuron number, but did show a significant decrease in the number of Cux1+ neurons in adult Cntn6−/− mice compared to wild-type mice. (C) Quantitative analysis of the lower layers (layers V-VI) did not show differences in total number of cells and neurons, and in the Cux1+ neurons in adult Cntn6−/− mice compared to wild-type mice. Analysis was performed on at least two sections per brain from wild-type and Cntn6−/− adult mice (n = 5 per genotype) using unpaired Student's t test and one-way ANOVA. Data are presented as mean ±SEM. **, p < 0.01.
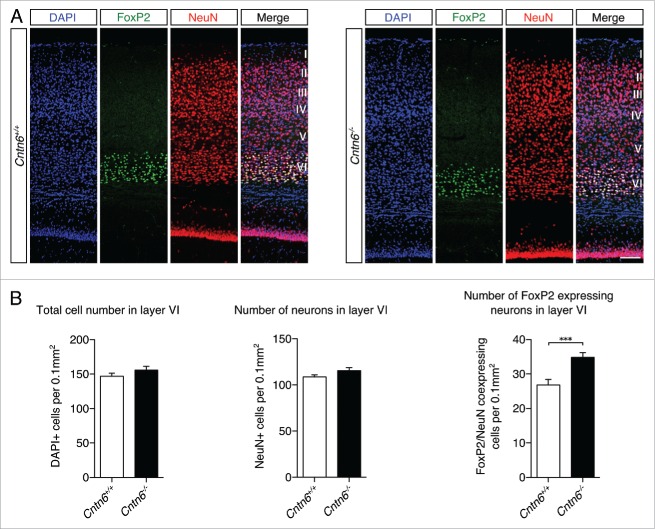
FoxP2 staining was used to visualize projection neurons in layer VI (Fig. 5A). Cell counting showed no statistical difference in the total number of cells and neurons in layer VI between genotypes. However, an increase in FoxP2+ neurons in layer VI was found in the Cntn6−/− mice (Fig. 5B). These observations indicated a shift in the numbers of Cux1+ and FoxP2+ neurons in their respective layers of the visual cortex in Cntn6−/− mice. This shift in the numbers of projection neurons may relate to changes in survival, apoptosis or migration, which are processes commonly affected by CAMs.
Figure 5.
Lower layer projection neurons in Cntn6-deficient mice. (A) Representative images of FoxP2 (green) expression, NeuN expression and DAPI staining in layer VI of the visual cortex in the visual cortex of adult wild-type and Cntn6−/− mice. The scale bar represents 100 μm. (B) Quantitative analysis of layer VI did not show differences in the total numbers of cells and neurons, and did show a significant increase in the number of FoxP2+ neurons in adult Cntn6−/− mice compared to wild-type mice. Analysis was performed on at least two sections per brain from wild-type and Cntn6−/− adult mice (n = 5 per genotype) using unpaired Student's t test and one-way ANOVA. Data are presented as mean ±SEM. **, p < 0.01.
Reduced number of subtype-specific interneurons in Cntn6−/− mice
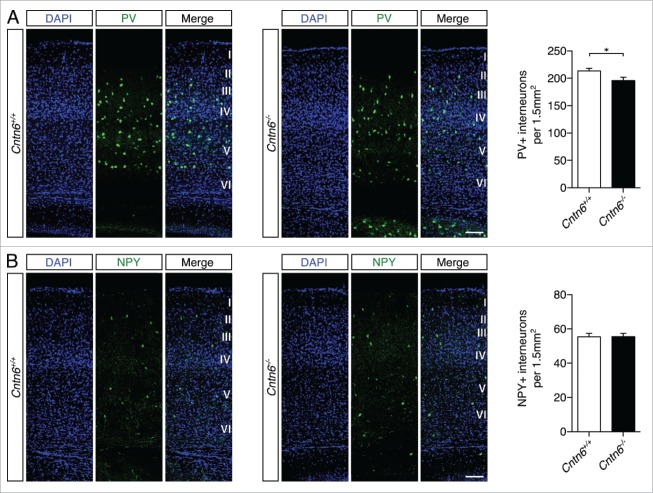
GABAergic interneuron dysfunction was previously associated with autism and epilepsy32 and was also found in the cortex of contactin-associated protein-2 (Cntnap2) knockout mice.24 Defects in migration may cause these dysfunctions. The indication that Cntn6 exerts a number of CAM functions like axon guidance and survival,18,22 prompted us to examine the migration of GABAergic interneurons. We quantified two non-overlapping populations of interneurons in the visual cortex of wild-type and Cntn6−/− mice, namely interneurons positive for parvalbumin (PV) or neuropeptide Y (NPY).33 We observed a significant decrease in the number of PV+ interneurons in Cntn6−/− mice (Fig. 6A), while the amount of NPY+ interneurons remained unchanged (Fig. 6B). These results indicate that Cntn6 exerts functions in a process that determine the number of PV+ interneurons in the visual cortex.
Figure 6.
Parvalbumin- and NPY-positive interneuron numbers in the cortex of Cntn6-deficient mice. (A) Representative images of PV+ interneurons (green) across the visual cortex of adult wild-type and Cntn6−/− mice. DAPI is in blue. Quantification of PV+ cells showed a significant decrease in Cntn6−/− mice. (B) NPY+ interneurons (green) were stained in the visual cortex, with DAPI in blue. Quantifications of NPY+ cells revealed no difference between wild-type and Cntn6−/− mice. The scale bars represent 100 μm. Analysis was performed on at least two sections per brain from wild-type and Cntn6−/− adult mice (n = 7 for PV+ quantifications and n = 5 for NPY+ quantifications per genotype) using unpaired Student's t test and one-way ANOVA. Data are presented as mean ±SEM. *, p < 0.05.
Abnormal suprapyramidal bundle in Cntn6−/− mice
In order to investigate if Cntn6 is necessary for axonal fasciculation, we determined the integrity of hippocampal mossy fiber projections in the absence of Cntn6. The fasciculation of efferents of this system is sensitive to alterations in axon guidance molecules and CAMs.26,27,34-36 In the hippocampus of P14 and adult mice, there was low Cntn6 mRNA expression in the pyramidal neurons of CA1 and CA3, in the granular neurons of the DG and in the dispersed cell bodies inside the hilus of the DG (Fig. 1C). Similarly, Cntn6 protein expression was faint in CA1 and CA3 at this age (Fig. 7A–B). Upon closer inspection of the DG, Cntn6 expression surrounded the granular neurons of the DG and Cntn6 was clearly expressed in the stratum lacunosum-moleculare (slm) and on cell bodies inside the hilus of the DG (Fig. 7C).
Figure 7.
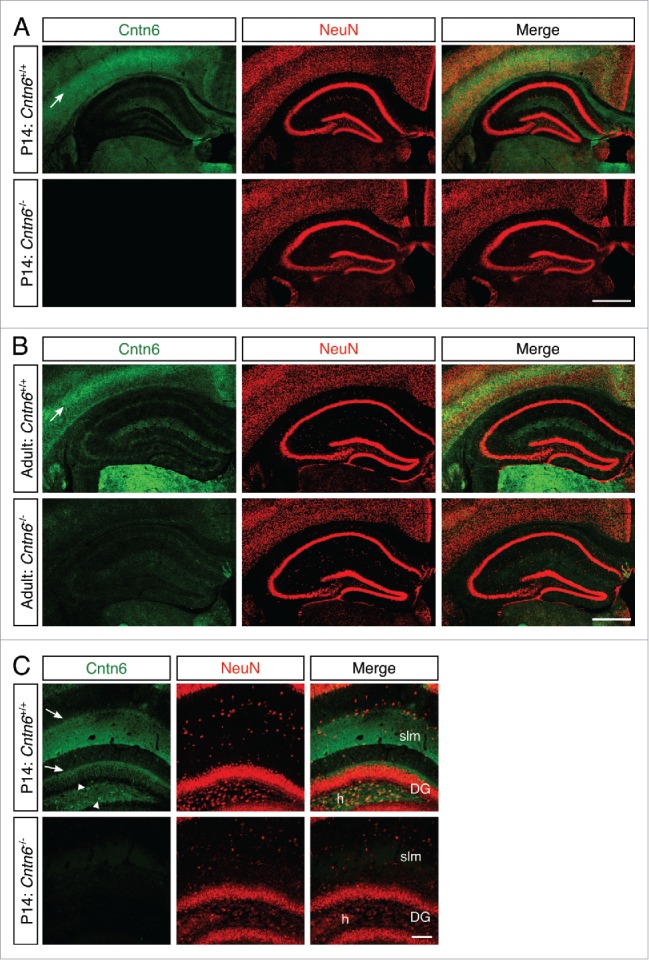
Cntn6 protein expression in the hippocampus. (A-B) Immunohistochemistry for Cntn6 (green) and NeuN (red) showed Cntn6 expression in the hippocampus of P14 and adult wild-type mice. No significant background staining was found in Cntn6−/− adult mice. Specific Cntn6-staining was also observed in the cortex of wild-type mice (arrows). (C) A higher magnification of Cntn6 (green) and NeuN (red) immunostaining of the hippocampus of P14 wild-type and Cntn6−/− mice. The arrowheads show Cntn6 expression at cell bodies in the hilus (h) of the dentate gyrus (DG) and the arrows indicate Cntn6 expression adjacent to the granule layer of the DG and at the stratum lacunosum-moleculare (slm) of wild-type P14 mice. All scale bars represent 100 μm.
The expression of Cntn6 protein in the hippocampus at P5 was described before and confirms our results.17 However, another study reported strong Cntn6 mRNA expression in the CA1 region without staining of other hippocampal structures in P7 mouse pups.15 The difference between results may be caused by the spatiotemporal expression of Cntn6.
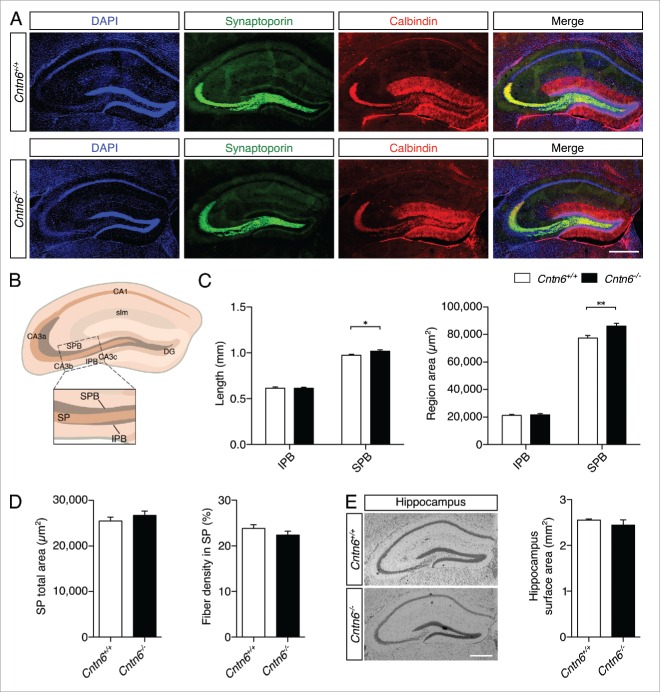
Immunostaining with two specific markers was used to outline the organization of hippocampal mossy fibers and their terminals (Fig. 8A). Synaptoporin was expressed in the mossy fiber system in the hilus of the DG, and staining shows the suprapyramidal bundle (SPB) and the infrapyramidal bundle (IPB).37 Staining for the calcium-binding protein calbindin revealed DG granular cells and mossy fiber axons.38,39 The density of mossy fibers crossing the stratum pyramidale (SP) of the CA3 region was measured, which emanate from the DG, bifurcate and segregate into the SPB and IPB. Both bundles are located on either side of the SP, which is the layer containing pyramidal neuron somata. Additionally, the lengths and areas sizes of the SPB, IPB, and the area size of the SP were measured (Fig. 8B). The length and area size of the IPB remained unchanged in Cntn6−/− mice, while these parameters were significantly increased in the SPB (Fig. 8C). The area size of the SP and the number of mossy fibers crossing the SP were unaffected in Cntn6−/− mice. Although an increase in area size was found in the SPB, no difference was found in total hippocampal surface area between the genotypes (Fig. 8D–E). These results indicated that Cntn6 is required for the normal integrity of lamina-restricted projections of mossy fibers forming the SPB.
Figure 8.
Hippocampal mossy fiber distribution in Cntn6-deficient mice. (A) Representative image of synaptoporin (green) and calbindin (red) expression in adult wild-type and Cntn6−/− mice. DAPI is in blue. The scale bars represent 250 μm. (B) Schematic representation of the adult mouse hippocampus. The rectangle indicates the area and location used for quantification of mossy fiber crossings in the SP of the CA3. Abbreviations: CA1, cornu ammonis 1; CA3a-c, cornu ammonis 3a-c; DG, dentate gyrus; SPB, suprapyramidal bundle; IPB, infrapyramidal pundle; SP, stratum pyramidale; slm, stratum lacunosum-moleculare. (C) Quantification of the length (left panel) and area size (right panel) of the IPB and SPB in wild-type and Cntn6−/− mice only showed a significant increase of both parameters on the SPB. (D) Quantification of total area size (left panel) and percentage of mossy fibers crossing the SP (right panel) did not reveal a difference between wild-type and Cntn6−/− mice. Analysis was performed on at least three sections per brain from wild-type and Cntn6−/− adult mice (n = 7 per genotype) using unpaired Student's t test and one-way ANOVA. Data are presented as mean ±SEM. *, p < 0.05, **, p < 0.01. (E) Nissl-stained sections of adult wild-type and Cntn6−/− mice did not demonstrate a difference in hippocampal surface areas between genotypes. Analysis was performed on at least two sections per brain from wild-type and Cntn6−/− adult mice (n = 5 per genotype) using unpaired Student's t test and one-way ANOVA. Data are presented as mean ±SEM.
Discussion
We here report that Cntn6 contributes to the appropriate development of cortical layers and hippocampal mossy fiber tracts. The numbers of projection neurons and interneurons were changed in the visual cortex of Cntn6−/− mice. Additionally, the size of the SPB in the hippocampus was altered in these mice. We interpret these impairments as signs that Cntn6 affects the adequate numbers of subtype-specific projection neurons and interneurons in the visual cortex. We speculate that Cntn6 could regulate survival and apoptosis, migration and the fasciculation of axons. Earlier reports on axon guidance functions of Cntn6 in the cortex demonstrated misoriented apical dendrites of cortical pyramidal neurons and a delay in corticospinal tract formation in Cntn6−/− mice.21,22 Together with these data, these results point to functions of Cntn6 in neuronal migration, guidance and survival. The underlying mechanism requires Cntn6 to engage cis- and trans-interactions with membrane bound proteins. We do not have evidence that Cntn6 employs homophilic interactions. Therefore, the present data suggests that the interactions involve other, partly unknown proteins. Several partners of Cntn6 have been described in literature,7 but lack functional evidence so far. Increasing evidence indicate that other Cntns engage adhesive trans-interactions, for example the role of Cntn2 in the construction of the node of Ranvier,40 and a role in signaling by cis-interaction, for example by the heterodimeric Cntn4-APP complex.41
It has been made apparent that CAMs are important for the construction of a normal cerebral cortex from numerous studies in mice deficient of CAMs.23,24,26 Such roles were demonstrated for Cntn6 in the regulation of cortical development after the observation of aberrant dendritic orientation of pyramidal neurons in layer V of the visual cortex in Cntn6−/− mice.22 The visual cortex of Chl1−/− mice showed a similar effect and this particular phenotype was enhanced in mice heterozygous for both Cntn6 and Chl1.22 Close examination of the visual cortex in Chl1−/− mice revealed impairments of neuronal migration in this area.42 These impairments were not observed in Cntn6−/− mice.22 Other phenotypes in cortical organization are generally implicated,31 including the numbers of projection neurons and inhibitory interneurons.
Our conclusion that Cntn6 regulates numbers of projection neurons in the visual cortex is based on the observation of the altered numbers of subtype-specific neurons in the visual cortex of Cntn6−/− mice. In these mice, the number of Cux1+ neurons in the upper layers was significantly reduced, while the amount of FoxP2+ neurons in layer VI was increased in the visual cortex. The decrease of the number of Cux1+ neurons might be a direct effect of a decrease of the total number of neurons in layer II-IV in the visual cortex of Cntn6−/− mice. Although the decrease of the total number of neurons was not significant, it was indeed slightly reduced. A similar observation was made for the lower projection neurons. The significant increase in the number of FoxP2+ neurons might be a direct effect of, although not significant, a slight increase of the total number of neurons in layer VI of the visual cortex in Cntn6−/− mice. Simultaneously, the absence of Cntn6 might have a beneficial effect on neuronal survival specifically in layer VI. These data indicate that Cntn6 may be involved in neurogenesis or neuronal survival in the visual cortex. The notion that Cntn6 plays a role in neuronal survival was previously demonstrated by the increased amount of apoptosis in granule cells of the cerebellum and in primary cultures derived from Cntn6−/− mouse cortex.18,20
A selective role of Cntn6 in interneuron function was derived from the observation that the number of PV+ interneurons was significantly decreased in Cntn6−/− mice, while the number of NPY+ interneurons remained unchanged. Cntn6 may serve as a ligand for receptors on PV+ interneurons and thereby contribute to migration, guidance or survival or other functions of these interneurons in the visual cortex. In this case, Cntn6 may serve as a signal to receptors on PV+ interneurons. Interestingly, PV+ interneurons in particular have been reported to play important roles in the rhythmic pacing of cortical neuronal activity.43,44 The fast-spiking PV+ interneurons play an essential role in the regulation of critical periods in the cerebral cortex.45,46 Developmentally regulated perineuronal nets mediate the initiation and termination of the critical periods, as has been found in studies on PV+ neurons in the visual cortex. In an animal model of Rett syndrome, which is a genetic neurodevelopmental disorder with ASD symptoms, an aberrant regulation of PV+ interneuron-associated perineuronal net was linked to functional deficits.47 Our results suggest that Cntn6 is crucial for maintaining the typical number of PV+ interneurons in the visual cortex. With respect to the underlying mechanism we suggest that Cntn6 may mediate contact and adhesion of migrating PV+ interneurons and of growing apical dendrites with the extracellular matrix or other cells, by providing a permissive environment or a directional cue for movement and growth. These suggestions need mechanistic studies that are beyond the scoop of this study.
We examined if Cntn6 plays a role in the fasciculation of mossy fibers in the hippocampus during development. This system is very sensitive to axon guidance defects or to absence of certain CAMs, as was demonstrated in Chl1−/− mice. Mossy fibers represent the fasciculated axonal projections of DG granule cells on pyramidal cells in the hippocampus. Their terminals form synapses in the stratum lucidum with the proximal portion of the apical dendrites of CA3 pyramidal cells. Numerous mossy fiber terminals were seen scattered within the pyramidal cell layer in Chl1−/− mice, suggesting that the terminals were formed not only on proximal dendrites of the pyramidal cells, but also on the cell bodies.26,27,36 Interestingly, NCAM-deficient animals also have demonstrated similar abnormal projections of mossy fibers in the hippocampus.25 However, we did not observe an increased mossy fiber invasion into the SP of Cntn6−/− mice, but rather an increase in length and size of mossy fiber projections in the SPB. These findings imply that Cntn6 absence from the DG might impair the fasciculation of mossy fibers innervating the SPB.
Our results provide insight in the roles of Cntn6 during neurodevelopment, which include maintaining the proper numbers of subtype-specific neurons and interneurons in the visual cortex and regulating the size of the SPB in the hippocampus. We speculate that these observations may relate to changes in cell survival, migration and fasciculation. Further investigation of the molecular pathways and protein networks of Cntn6, which exerts these functions, will provide more insight in the manner Cntn6 regulates neurodevelopment.
Materials and methods
Animals and tissue treatment
B57BL/6 and Cntn6−/− mice were obtained from Charles River and Nagaoka University,16 respectively. Mice were maintained on a 12-h light/dark cycle with ad libitum food and water in the animal facility of the Brain Center Rudolf Magnus, Utrecht. Homozygous and heterozygous Cntn6 mice and littermate contrls were obtained from heterozygous breeding. For immunohistochemistry, adult mice were anesthetized with an overdose of sodium pentobarbital (19.4 μl/gr) and were perfused intracardially with 0.9% saline, followed by 4% PFA in PBS, pH 7.5. Brains were post fixed in 4% PFA before transferred to 30% sucrose for cryopreservation. Tissue was sectioned at 40 μm sections and free-floating sections were stored in PBS with 0.02% sodium azide until immunohistochemistry was performed. For in situ hybridization, P14 mouse pups and adult mice were killed by decapitation and their brains were quickly dissected and flash-frozen in 2-methylbutane between −50 and −55°C. Brains were sliced into 16 μm sections using a cryostat and mounted onto Superfrost slides (VWR, 631–0108). For immunohistochemistry and in situ hybridization, the following stereotaxic coordinates were used for coronal sections of adult mouse brain (Atlas Franklin and Paxinos): visual cortex (−2.80 mm to bregma), thalamic nuclei (−0.82 mm to bregma) and hippocampus (between −1.5 mm and −2.5 anterior posterior (AP) from bregma).
In situ hybridization
Nonradioactive in situ hybridization was performed according to Pasterkamp et al., 1998.48 In brief, probe sequences for Cntn6 (NM_017383.3: nt 283–876) was polymerase chain reaction (PCR)-amplified from cDNA. Digoxigenin (DIG)-labeled RNA probes were generated by a RNA polymerase reaction using 10x DIG RNA labeling mix (Sigma, 11277073910). Tissue sections were post-fixed in 4% PFA in PBS, pH 7.40 at room temperature (RT) for 20 min. To enhance tissue penetration and decrease aspecific background staining, sections were acetylated with 0.25% acetic anhydride in 0.1 M triethanolamine and 0.06% HCl at RT for 10 min. Sections were prehybridized in hybridization buffer (50% formamide, 5x Denhardt's solution, 5x SSC, 250 μg/ml baker's yeast tRNA and 500 μg/ml sonicated salmon sperm DNA) at RT for 2 hr. Hybridization was performed with 2000 ng/ml denatured DIG-labeled probe diluted in hybridization buffer at 68°C for 15 hr. After hybridization, sections were first washed briefly in 2x SSC followed by incubation in 0.2x SCC at 68°C for 2 hr. Sections were adjusted to RT in 0.2x SSC for 5 min. DIG-labeled RNA hybrids were detected with anti-DIG Fab fragments conjugated to AP (Roche, 11093274910) diluted in 1:2500 in TBS (pH 7.4) at 4°C overnight. Binding of AP-labeled antibody was visualized by incubating the sections in detection buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl and 50 mM MgCl2) containing 240 μg/ml levamisole and nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphatase (NBT/BCIP) (Roche, 11681451001) at RT for 14 hr. Sections subjected to the entire in situ hybridization procedure, but with no probe or sense probe added, did not exhibit specific hybridization signals. The specificity of the in situ hybridization procedure was also inferred from the clearly distinct gene expression patterns observed. Staining was visualized using a Zeiss Axioskop 2 microscope.
Nissl staining and cortical thickness analysis
Coronal sections of mouse brain were immersed in 0.5% cresyl violet for several minutes and then subjected to the ethanol series and embedded. Analysis of cortical thickness was performed within the frontal motor cortex (+0.5 mm to bregma), primary somatosensory areas (−1.70 mm to bregma) and visual cortex (−2.80 mm to bregma) in all used brains. Of the cortices the superficial and deeper layers (i.e., layers I-IV and layers V-VI resp) were measured, which were performed on at least 2 sections of each area in wild-type and Cntn6−/− adult mice (n = 5 per genotype) using ImageJ software. A statistical analysis was carried out using unpaired Student's t test and one-way analysis of variance (ANOVA).
Immunohistochemistry
The sections were washed with PBS and incubated in blocking buffer (1% BSA, 0.2% fish skin gelatin (Sigma, G7765), 0.1% Triton X-100 in PBS) for 45 min. Sections were washed and incubated in permeabilization buffer (0.3% Triton X-100 in PBS) for 10 min before incubation with primary antibody in blocking buffer at 4°C for 2 hr. The sections were washed in PBS and pre-incubated with blocking buffer before incubating with secondary antibody at RT for 2 hr. The sections were washed again in PBS and a 10 min DAPI incubation was performed. The sections were embedded with Polyvinyl alcohol mounting medium with DABCO antifading (Fluka, 10981) onto glass slides after additional PBS wash steps. Antibodies used were sheep anti-Cntn6 (R&D systems, AF5890) 1:100; mouse anti-NeuN (Milipore, MAB377) 1:200; rabbit anti-Cux1 (Santa Cruz, sc-13024) 1:200; rabbit anti-FoxP2 (Sigma, HPA000382) 1:250; rabbit anti-Parvalbumin (PV) (ImmunoStar, 24428) 1:250; rabbit anti-Neuropeptide Y (NPY) (Abcam, ab30914) 1:1000; rabbit anti-Synaptoporin (Synaptic Systems, 102002) 1:1000; mouse anti-Calbindin (Swant, 300) 1:3000; and DAPI, followed by species-specific secondary antibodies conjugated to Alexa Fluor (Invitrogen). Images were captured by confocal laser scanning microscopy (Olympus FV1000) and a Zeiss Axioscop A1. For the quantification analysis, images from the Zeiss Axioscop A1 were used. For the quantifications of projection neurons in the visual cortex (−2.80 mm to bregma), measurements were performed on at least two sections from wild-type and Cntn6−/− adult mice (n = 5 per genotype) using ImageJ software. Cells positive for NeuN, Cux1 and DAPI were counted in images taken of layers II-IV and images taken of layers V-VI. Cells positive for NeuN, FoxP2 and DAPI were counted in images taken of layer VI. These images comprised of at least 20 randomly selected microscope fields in the designated layers (area 0.1 mm2) from the visual cortex. For the quantifications of interneurons in the visual cortex, measurements were performed on at least two sections from wild-type and Cntn6−/− adult mice (n = 7 per genotype for PV-analysis and n = 5 for NPY-analysis) using ImageJ software. Cells positive for PV and NPY were counted in images taken of the entire cortex. These images comprised of at least 20 randomly selected microscope fields (area 1.5 mm2) from the visual cortex. For the analyses of the hippocampus (between −1.5 mm and −2.5 AP from bregma), measurements were performed on at least three sections from wild-type and Cntn6−/− adult mice (n = 7 per genotype) using ImageJ software. Anti-calbindin was used to visualize mossy fibers and anti-synaptoporin was used to visualize mossy fiber synapses in the hippocampus. The lengths and area sizes of the supra- and infrapyramidal bundles (SPB and IPB) were assessed by tracing the bundles from the endpoints of the dentate gyrus (DG) blades to the last visible bundle staining at the CA3 side. The analytical region of the stratum pyramidale (SP) was assessed by placing a rectangle (Fig. 8B) from the endpoints of the DG blades to the last visible staining of the IPB at the CA3 side. The SP region in this rectangle was traced and both the area size and fiber density were measured. At least 28 randomly selected microscope fields from the hippocampus were used for measurements. Statistical analysis was carried out using unpaired Student's t test and one-way analysis of variance (ANOVA).
Ethics statement
The experiments performed in this study were approved by the Experimental Animal Committee (DEC) of Utrecht (2010.I.06.073). All animal experiments were conducted in agreement with Dutch law (Wet op de Dierproeven, 1996) and European regulations (Guideline 86/609/EEC) related to the protection of vertebrate animals used for experimental and other scientific purposes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Marloes van den Nieuwendijk, Jan Sprengers and Heleen van't Spijker for their contribution to the quantifications of cortical neurons. We thank Henk Spierenburg for performing the in situ hybridization experiments and for genotyping of the animals. We are grateful to Eljo van Battum and Julie Tastet for discussing the approach to measure the hippocampal parameters. We thank K Watanabe and Y Shimoda for the generous gift of the knock-out animals.
Funding
This study was supported by a Fellowship from the Dutch Brain Foundation nr. F2008(1)-08 (BVDZ) and by a JSPS Fellowship (AOA).
References
- [1].Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci 2007; 8:206-20; PMID:17299456; http://dx.doi.org/ 10.1038/nrn2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci 2007; 10:19-26; PMID:17189949; http://dx.doi.org/ 10.1038/nn1827 [DOI] [PubMed] [Google Scholar]
- [3].Murase S, Schuman EM. The role of cell adhesion molecules in synaptic plasticity and memory. Curr Opin Cell Biol 1999; 11:549-53; PMID:10508654; http://dx.doi.org/ 10.1016/S0955-0674(99)00019-8 [DOI] [PubMed] [Google Scholar]
- [4].Anderson AA, Kendal CE, Garcia-Maya M, Kenny AV, Morris-Triggs SA, Wu T, Reynolds R, Hohenester E, Saffell JL. A peptide from the first fibronectin domain of NCAM acts as an inverse agonist and stimulates FGF receptor activation, neurite outgrowth and survival. J Neurochem 2005; 95:570-83; PMID:16135080; http://dx.doi.org/ 10.1111/j.1471-4159.2005.03417.x [DOI] [PubMed] [Google Scholar]
- [5].Naus S, Richter M, Wildeboer D, Moss M, Schachner M, Bartsch JW. Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease-disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. J Biol Chem 2004; 279:16083-90; PMID:14761956; http://dx.doi.org/ 10.1074/jbc.M400560200 [DOI] [PubMed] [Google Scholar]
- [6].Shimoda Y, Watanabe K. Contactins: emerging key roles in the development and function of the nervous system. Cell Adh Migr 2009; 3:64-70; PMID:19262165; http://dx.doi.org/ 10.4161/cam.3.1.7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zuko A, Kleijer KTE, Oguro-Ando A, Kas MJH, van Daalen E, van der Zwaag B, Burbach JPH. Contactins in the neurobiology of autism. Eur J Pharmacol 2013; 719:63-74; PMID:23872404; http://dx.doi.org/ 10.1016/j.ejphar.2013.07.016 [DOI] [PubMed] [Google Scholar]
- [8].van Daalen E, Kemner C, Verbeek NE, van der Zwaag B, Dijkhuizen T, Rump P, Houben R, van't Slot R, de Jonge MV, Staal WG, et al.. Social Responsiveness Scale-aided analysis of the clinical impact of copy number variations in autism. Neurogenetics 2011; 12:315-23; PMID:21837366; http://dx.doi.org/ 10.1007/s10048-011-0297-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al.. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010; 466:368-72; PMID:20531469; http://dx.doi.org/ 10.1038/nature09146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fernandez TV, García-González IJ, Mason CE, Hernández-Zaragoza G, Ledezma-Rodríguez VC, Anguiano-Alvarez VM, E'Vega R, Gutiérrez-Angulo M, Maya ML, García-Bejarano HE, et al.. Molecular characterization of a patient with 3p deletion syndrome and a review of the literature. Am J Med Genet 2008; 146A:2746-52; PMID:18837054; http://dx.doi.org/ 10.1002/ajmg.a.32533 [DOI] [PubMed] [Google Scholar]
- [11].Wang K, Zhang H, Bloss CS, Duvvuri V, Kaye W, Schork NJ, Berrettini W, Hakonarson H. Price Foundation Collaborative Group. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol Psych 2011; 16:949-59; http://dx.doi.org/ 10.1038/mp.2010.107 [DOI] [PubMed] [Google Scholar]
- [12].Hu J, Liao J, Sathanoori M, Kochmar S, Sebastian J, Yatsenko SA, Surti U. CNTN6 copy number variations in 14 patients: a possible candidate gene for neurodevelopmental and neuropsychiatric disorders. J Neurodev Disord 2015; 7:26; PMID:26257835; http://dx.doi.org/ 10.1186/s11689-015-9122-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kashevarova AA, Nazarenko LP, Schultz-Pedersen S, Skryabin NA, Salyukova OA, Chechetkina NN, Tolmacheva EN, Rudko AA, Magini P, Graziano C, et al.. Single gene microdeletions and microduplication of 3p26.3 in three unrelated families: CNTN6 as a new candidate gene for intellectual disability. Mol Cytogenet 2014; 7:97; PMID:25606055; http://dx.doi.org/ 10.1186/s13039-014-0097-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kashevarova AA, Nazarenko LP, Skryabin NA, Salyukova OA, Chechetkina NN, Tolmacheva EN, Sazhenova EA, Magini P, Graziano C, Romeo G, et al.. Array CGH analysis of a cohort of Russian patients with intellectual disability. Gene 2014; 536:145-50; PMID:24291026; http://dx.doi.org/ 10.1016/j.gene.2013.11.029 [DOI] [PubMed] [Google Scholar]
- [15].Lee S, Takeda Y, Kawano H, Hosoya H, Nomoto M, Fujimoto D, Takahashi N, Watanabe K. Expression and regulation of a gene encoding neural recognition molecule NB-3 of the contactin/F3 subgroup in mouse brain. Gene 2000; 245:253-66; PMID:10717476; http://dx.doi.org/ 10.1016/S0378-1119(00)00031-7 [DOI] [PubMed] [Google Scholar]
- [16].Takeda Y, Akasaka K, Lee S, Kobayashi S, Kawano H, Murayama S, Takahashi N, Hashimoto K, Kano M, Asano M, et al.. Impaired motor coordination in mice lacking neural recognition molecule NB-3 of the contactin/F3 subgroup. J Neurobiol 2003; 56:252-65; PMID:12884264; http://dx.doi.org/ 10.1002/neu.10222 [DOI] [PubMed] [Google Scholar]
- [17].Sakurai K, Toyoshima M, Takeda Y, Shimoda Y, Watanabe K. Synaptic formation in subsets of glutamatergic terminals in the mouse hippocampal formation is affected by a deficiency in the neural cell recognition molecule NB-3. Neurosci Lett 2010; 473:102-6; PMID:20176085; http://dx.doi.org/ 10.1016/j.neulet.2010.02.027 [DOI] [PubMed] [Google Scholar]
- [18].Sakurai K, Toyoshima M, Ueda H, Matsubara K, Takeda Y, Karagogeos D, Shimoda Y, Watanabe K. Contribution of the neural cell recognition molecule NB-3 to synapse formation between parallel fibers and Purkinje cells in mouse. Dev Neurobiol 2009; 69:811-24; PMID:19672956; http://dx.doi.org/ 10.1002/dneu.20742 [DOI] [PubMed] [Google Scholar]
- [19].Mercati O, Danckaert A, André-Leroux G, Bellinzoni M, Gouder L, Watanabe K, Shimoda Y, Grailhe R, De Chaumont F, Bourgeron T, et al.. Contactin 4, −5 and −6 differentially regulate neuritogenesis while they display identical PTPRG binding sites. Biol Open 2013; 2:324-34; PMID:23519440; http://dx.doi.org/ 10.1242/bio.20133343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huang X, Sun J, Zhao T, Wu KW, Watanabe K, Xiao ZC, Zhu LL, Fan M. Loss of NB-3 aggravates cerebral ischemia by impairing neuron survival and neurite growth. Stroke 2011; 42:2910-6; PMID:21817151; http://dx.doi.org/ 10.1161/STROKEAHA.110.609560 [DOI] [PubMed] [Google Scholar]
- [21].Huang Z, Yu Y, Shimoda Y, Watanabe K, Liu Y. Loss of neural recognition molecule NB-3 delays the normal projection and terminal branching of developing corticospinal tract axons in the mouse. J Comp Neurol 2012; 520:1227-45; PMID:21935948; http://dx.doi.org/ 10.1002/cne.22772 [DOI] [PubMed] [Google Scholar]
- [22].Ye H, Tan YLJ, Ponniah S, Takeda Y, Wang S-Q, Schachner M, Watanabe K, Pallen CJ, Xiao ZC. Neural recognition molecules CHL1 and NB-3 regulate apical dendrite orientation in the neocortex via PTP α. EMBO J 2008; 27:188-200; PMID:18046458; http://dx.doi.org/ 10.1038/sj.emboj.7601939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bruining H, Matsui A, Oguro-Ando A, Kahn RS, Van't Spijker HM, Akkermans G, Stiedl O, van Engeland H, Koopmans B, van Lith HA, et al.. Genetic Mapping in Mice Reveals the Involvement of Pcdh9 in Long-Term Social and Object Recognition and Sensorimotor Development. Biol Psychiat 2015; 78(7):485-95; PMID:25802080 [DOI] [PubMed] [Google Scholar]
- [24].Peñagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, et al.. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 2011; 147:235-46; http://dx.doi.org/ 10.1016/j.cell.2011.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cremer H, Chazal G, Goridis C, Represa A. NCAM is essential for axonal growth and fasciculation in the hippocampus. Mol Cell Neurosci 1997; 8:323-35; PMID:9073395; http://dx.doi.org/ 10.1006/mcne.1996.0588 [DOI] [PubMed] [Google Scholar]
- [26].Heyden A, Angenstein F, Sallaz M, Seidenbecher C, Montag D. Abnormal axonal guidance and brain anatomy in mouse mutants for the cell recognition molecules close homolog of L1 and NgCAM-related cell adhesion molecule. Neuroscience 2008; 155:221-33; PMID:18588951; http://dx.doi.org/ 10.1016/j.neuroscience.2008.04.080 [DOI] [PubMed] [Google Scholar]
- [27].Montag-Sallaz M, Schachner M, Montag D. Misguided axonal projections, neural cell adhesion molecule 180 mRNA upregulation, and altered behavior in mice deficient for the close homolog of L1. Mol Cell Biol 2002; 22:7967-81; PMID:12391163; http://dx.doi.org/ 10.1128/MCB.22.22.7967-7981.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science 1996; 274:1123-33; PMID:8895455; http://dx.doi.org/ 10.1126/science.274.5290.1123 [DOI] [PubMed] [Google Scholar]
- [29].Ye H, Zhao T, Tan YLJ, Liu J, Pallen CJ, Xiao Z-C. Receptor-like protein-tyrosine phosphatase α enhances cell surface expression of neural adhesion molecule NB-3. J Biol Chem 2011; 286:26071-80; PMID:21622556; http://dx.doi.org/ 10.1074/jbc.M110.214080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sasaki Y, Cheng C, Uchida Y, Nakajima O, Ohshima T, Yagi T, Taniguchi M, Nakayama T, Kishida R, Kudo Y, et al.. Fyn and Cdk5 mediate semaphorin-3A signaling, which is involved in regulation of dendrite orientation in cerebral cortex. Neuron 2002; 35:907-20; PMID:12372285; http://dx.doi.org/ 10.1016/S0896-6273(02)00857-7 [DOI] [PubMed] [Google Scholar]
- [31].Rubenstein JLR. Annual Research Review: Development of the cerebral cortex: implications for neurodevelopmental disorders. J Child Psychol Psychiatry 2011; 52:339-55; PMID:20735793; http://dx.doi.org/ 10.1111/j.1469-7610.2010.02307.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Levitt P, Eagleson KL, Powell EM. Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders. Trends Neurosci 2004; 27:400-6; PMID:15219739; http://dx.doi.org/ 10.1016/j.tins.2004.05.008 [DOI] [PubMed] [Google Scholar]
- [33].Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci 2006; 7:687-96; PMID:16883309; http://dx.doi.org/ 10.1038/nrn1954 [DOI] [PubMed] [Google Scholar]
- [34].Suto F, Tsuboi M, Kamiya H, Mizuno H, Kiyama Y, Komai S, Shimizu M, Sanbo M, Yagi T, Hiromi Y, et al.. Interactions between plexin-A2, plexin-A4, and semaphorin 6A control lamina-restricted projection of hippocampal mossy fibers. Neuron 2007; 53:535-47; PMID:17296555; http://dx.doi.org/ 10.1016/j.neuron.2007.01.028 [DOI] [PubMed] [Google Scholar]
- [35].Suto F, Ito K, Uemura M, Shimizu M, Shinkawa Y, Sanbo M, Shinoda T, Tsuboi M, Takashima S, Yagi T, et al.. Plexin-a4 mediates axon-repulsive activities of both secreted and transmembrane semaphorins and plays roles in nerve fiber guidance. J Neurosci 2005; 25:3628-37; PMID:15814794; http://dx.doi.org/ 10.1523/JNEUROSCI.4480-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Van Battum EY, Gunput RAF, Lemstra S, Groen EJN, Yu KL, Adolfs Y, Zhou Y, Hoogenraad CC, Yoshida Y, Schachner M, et al.. The intracellular redox protein MICAL-1 regulates the development of hippocampal mossy fibre connections. Nat Commun 2014; 5:4317; PMID:25007825; http://dx.doi.org/ 10.1038/ncomms5317 [DOI] [PubMed] [Google Scholar]
- [37].Singec I, Knoth R, Ditter M, Hagemeyer CE, Rosenbrock H, Frotscher M, Volk B. Synaptic vesicle protein synaptoporin is differently expressed by subpopulations of mouse hippocampal neurons. J Comp Neurol 2002; 452:139-53; PMID:12271488; http://dx.doi.org/ 10.1002/cne.10371 [DOI] [PubMed] [Google Scholar]
- [38].Scharfman HE, Goodman JH, Sollas AL. Granule-like neurons at the hilar/CA3 border after status epilepticus and their synchrony with area CA3 pyramidal cells: functional implications of seizure-induced neurogenesis. J Neurosci 2000; 20:6144-58; PMID:10934264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tóth K, Freund TF. Calbindin D28k-containing nonpyramidal cells in the rat hippocampus: their immunoreactivity for GABA and projection to the medial septum. Neuroscience 1992; 49:793-805; http://dx.doi.org/ 10.1016/0306-4522(92)90357-8 [DOI] [PubMed] [Google Scholar]
- [40].Poliak S, Peles E. The local differentiation of myelinated axons at nodes of Ranvier. Nat Rev Neurosci 2003; 4:968-80; PMID:14682359; http://dx.doi.org/ 10.1038/nrn1253 [DOI] [PubMed] [Google Scholar]
- [41].Osterhout JA, Stafford BK, Nguyen PL, Yoshihara Y, Huberman AD. Contactin-4 mediates axon-target specificity and functional development of the accessory optic system. Neuron 2015; 86:985-99; PMID:25959733; http://dx.doi.org/ 10.1016/j.neuron.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Demyanenko GP, Schachner M, Anton E, Schmid R, Feng G, Sanes J, Maness PF. Close homolog of L1 modulates area-specific neuronal positioning and dendrite orientation in the cerebral cortex. Neuron 2004; 44:423-37; PMID:15504324; http://dx.doi.org/ 10.1016/j.neuron.2004.10.016 [DOI] [PubMed] [Google Scholar]
- [43].Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 2009; 459:698-702; PMID:19396159; http://dx.doi.org/ 10.1038/nature07991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Takada N, Pi HJ, Sousa VH, Waters J, Fishell G, Kepecs A, Osten P. A developmental cell-type switch in cortical interneurons leads to a selective defect in cortical oscillations. Nat Commun 2014; 5:5333; PMID:25354876; http://dx.doi.org/ 10.1038/ncomms6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Morishita H, Cabungcal JH, Chen Y, Do KQ, Hensch TK. Prolonged Period of Cortical Plasticity upon Redox Dysregulation in Fast-Spiking Interneurons. Biol Psychiatry 2015; 78:396-402; PMID:25758057; http://dx.doi.org/ 10.1016/j.biopsych.2014.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nowicka D, Soulsby S, Skangiel-Kramska J, Glazewski S. Parvalbumin-containing neurons, perineuronal nets and experience-dependent plasticity in murine barrel cortex. Eur J Neurosci 2009; 30:2053-63; PMID:20128844; http://dx.doi.org/ 10.1111/j.1460-9568.2009.06996.x [DOI] [PubMed] [Google Scholar]
- [47].Krishnan K, Wang BS, Lu J, Wang L, Maffei A, Cang J, Huang ZJ. MeCP2 regulates the timing of critical period plasticity that shapes functional connectivity in primary visual cortex. Proc Natl Acad Sci 2015; 112:E4782-91; PMID:26261347; http://dx.doi.org/ 10.1073/pnas.1506499112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pasterkamp RJ, De Winter F, Holtmaat AJGD, Verhaagen J. Evidence for a Role of the Chemorepellent Semaphorin III and Its Receptor Neuropilin-1 in the Regeneration of Primary Olfactory Axons. J Neurosci 1998; 18:9962-76; PMID:9822752 [DOI] [PMC free article] [PubMed] [Google Scholar]