Highlight
Cell–microbe interactions that have evolved over millions of years enable the acquisition of novel developmental functions. The underlying themes of different symbioses, that are suggestive of a common symbiotic architecture, are reviewed.
Key words: Arbuscular mycorrhizal root interactions, evolution of root architecture, nodulation, plant–microbe signalling, rhizosphere biology, SYM symbiosis genes.
Abstract
Plant root rhizosphere interactions with mutualistic microbes are diverse and numerous, having evolved over time in response to selective pressures on plants to attain anchorage and nutrients. These relationships can be considered to be formed through a combination of architectural connections: molecular architecture interactions that control root–microbe perception and regulate the balance between host and symbiont and developmental architecture interactions that enable the microbes to be ‘housed’ in the root and enable the exchange of compounds. Recent findings that help to understand the common architecture that exists between nodulation and mycorrhizal interactions, and how this architecture could be re-tuned to develop new symbioses, are discussed here.
Evolution of plant below-ground tissues and contact with soil microbes
Plants must interact with their environment both above-ground (in the phyllosphere) and below-ground (in the rhizosphere). The rhizosphere represents a particularly complex interactive matrix since it consists of multivariate organism–environment relationships, involving both physical and molecular plant–microbe–soil interactions. Together with studying soil biodiversity and soil health, the rhizosphere has gained increased interest in recent years due to the availability of new visualization techniques (Downie et al., 2012; Mairhofer et al., 2012) together with advances in sequencing of inhabiting microbial communities (Bulgarelli et al., 2012; Lundberg et al., 2012; Schlaeppi et al., 2014). This review aims to use such recent findings to discuss aspects of the architecture connecting mutualistic plant–microbe interactions.
Underlying the colonization of land by plant species, several important processes occurred (Fig. 1). These includes development of multicellular specialized tissues such as roots—below-ground organs that help plants to acquire water and nutrients for development. During plant evolution the gradual shift from aquatic environments towards moist terrestrial habitats (Becker and Marin, 2009) can be hypothesized as the selective pressure that led to rhizoid development in plants. These rhizoid features provided the opportunity for anchorage of the plant as well as water and nutrient uptake (Delaux et al., 2012). Plant colonization of drier soil landscapes, the development of specialized rhizoid tissues, and more widespread root system architecture opened up the possibility of increased interaction of these early roots with soil microbes. However, cell–microbe interaction was not new. Even before the appearance of multi-cellular organisms, primitive cell–cell interaction events had already occurred. Eukaryotic cells themselves are the product of endosymbiosis between ancestral prokaryotic cell types (Margulis, 1970; reviewed in Archibald, 2011), suggesting that cells have molecular responses to allow or block cell–cell interactions between different organisms. This situates the context of cell–cell interactions (and thus symbiosis) in a broader and more ancestral scenario (Fig. 1). Pathogenesis, symbiosis, and intermediate interactions could all originate from a type of cell–cell interaction. The comparison of current innate immune signalling pathways in plants (reviewed in Dodds and Rathjen, 2010; Dangl et al., 2013; Wirthmueller et al., 2013) and animals suggests that they have a different evolutionary origin (reviewed in Ausubel, 2005). However, it could also be that certain components have a common origin, but that it diverged in an early premature eukaryotic stage and that both animals and plants evolved into highly specialized and differentiated multicellular organisms.
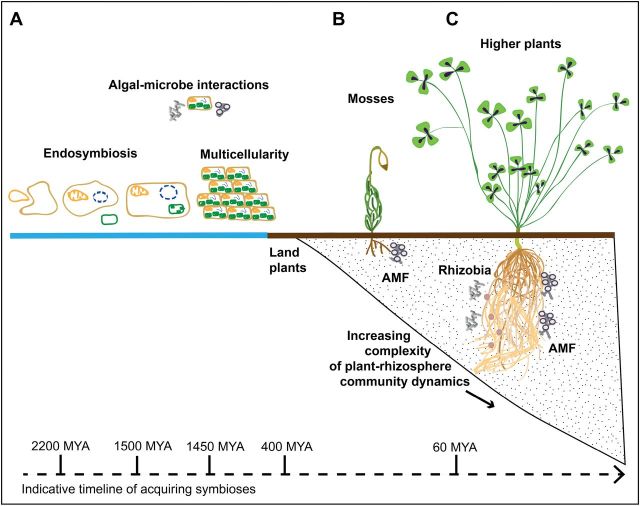
Fig. 1.
Accumulation of symbioses. Schematic representation of the evolution of molecular and biochemical interactions related to symbiosis over an indicative time-scale. (A) Ancient cell–cell interactions together with the development of molecular recognition mechanisms allowed the establishment of endosymbiotic events. This facilitated higher energy efficiency in eukaryotic cells, underpinning the ability to form multicellular tissues. (B) Land colonization of plants required the development of highly complex multicellular tissues including rhizoids and roots to achieve water and nutrient uptake. Early land plants formed primitive plant–microbe mutualistic interactions. Such interactions are also evident in single cell algal–microbe interactions, for example as found in lichens. (C) Within the rhizosphere, microbial diversity together with more specialized recognition genetic signalling pathways in roots has enabled increased symbiosis with higher plants. In legumes (such as M. truncatula), plants interact with AM fungi and rhizobia, enabling more efficient nutrient exchange between plants and the soil. This enables plasticity of the root system to colonize new rhizosphere niches.
Cell–cell interactions between different organisms, controlled both by evolutionary and environmental pressures, have shaped the types of interactions that we currently know about. Interestingly, it has been found recently that simple environmental changes can induce free-living organisms to be mutualistic without requiring adaptive co-evolution. However, the degree of success on these mutualisms seems to be controlled by species-specific traits (Aanen and Bisseling, 2014; Hom and Murray, 2014). To address this subject much more would need to be understood about the environmental, molecular, and biochemical profile of species interactions.
Soil and rhizosphere diversity provides a complex interactive environment for roots
Soil is a complex matrix containing mineral nutrients present in a broad range of chemical species and states, together with water and diverse different-sized physical particles. Another enormous component of soil is a vast diversity of living organisms including microbes. From what we know, soil microbes represent the largest reservoir of biodiversity on earth (Berendsen et al., 2012). It is estimated that 1g of soil could contain 1 billion bacterial cells and 200 million fungal hyphae as well as many other types and species of organisms. Although there is not a large difference in the total amount of microbe cells between forest and agricultural soils, there is, in fact, a dramatic difference in the number and type of taxa present. It has been found that certain forest soils are the richest in microbe biodiversity (Helgason et al., 1998; Roesch et al., 2007; Wagg et al., 2014). Soil microbe biodiversity, in turn, influences soil properties including the physical structure and nutrient composition (de Vries et al., 2013) and ultimately affects soil–root interactions and thus the environment where plants develop and grow.
Recent technical advances have allowed the exploration of natural soil bacterial communities and the characterization of the rhizosphere microbiota (Bulgarelli et al., 2012, 2013; Knief et al., 2012; Lundberg et al., 2012;). These studies have enabled progress to be made in understanding the different microbe–root interactions that can potentially be established within a complex community, from endophytic to symbiotic and pathogenic, across a wide range of intermediate states. This work has also shown that plant genotype contributes to the biodiversity of the root microbiota (reviewed in Bulgarelli et al., 2013), probably due to the composition of root exudates ‘rhizodeposition’ (reviewed in Bednarek et al., 2010; Bulgarelli et al., 2013).
In the soil environment, plant roots are therefore surrounded by a large variety of soil microbes and they can establish different types of molecular and physical interactions with them. This then defines a wide range of root–microbe associations where beneficial or pathogenic interactions can be considered to be the most extreme lifestyles, but many others can remain in intermediate associative states. Some microbes can colonize root host tissue and remain neutral, without causing any damage or disease symptoms in a so-called co-existence. The natural abundance of this type of plant–microbe interaction is generally unknown since they are hard to detect. Other microbial species could also be beneficial for the plant, acting as mutualists, for example, promoting growth and impacting function via production of plant regulators such as auxin, cytokinin or gibberellins (reviewed in Reinhold-Hurek and Hurek, 2011). As an example of growth promotion, the endophyte fungus Piriformospora indica, establishes interactions with a broad range of plant species with the consequence of enhancing plant growth and enabling biotic and abiotic stress resistance without causing disease symptoms in the host (Schäfer et al., 2007; Qiang et al., 2012). However, there are cases in which endophytes could turn into more saprophytic or pathogenic stages during their whole life cycle (reviewed in Slippers and Wingfield, 2007; Behie and Bidochka, 2014).
Evolution of a molecular architecture supporting plant–mutualist interactions
It can be hypothesized that the establishment of current root–microbe interactions, with pathogenic or mutualistic as the most extreme, was an extremely long evolutionary transition driven by many selective pressures. This includes the accordance of a ‘molecular language’ between the microbe and the plant (Bulgarelli et al., 2013; Oldroyd, 2013) and the co-ordination of plant microbe responses (Zamioudis and Pieterse, 2011; Doehlemann et al., 2014). Since plants and microbes establish a broad range of interactions, it can be considered that there were many attempts at communication between these organisms, thereby generating a spectrum of interactions. If a successful mutualistic interaction was established, positive selective pressure could have helped to maintain it. This new mutualist pathway enabling microbe entry would then have been susceptible for future evolutionary changes and new symbiotic interactions, but also susceptible to attack by pathogenic microbes.
Legume–rhizobia (reviewed in Suzaki and Kawaguchi, 2014) and plant–mycorrhizal interactions (reviewed in Bonfante and Genre, 2010; Gutjahr and Parniske, 2013) are both well-studied examples of higher plant root interactions with mutualistic microbes. They both involve recognition of microbial signatures followed by microbial entry into roots, concomitant with plant developmental changes and hormone signalling, all carefully orchestrated in specific cell types.
Most likely all plants form interactions with soil fungi, and at least 90% form interactions with mycorrhizal fungi (Wang and Qiu, 2006). Among the seven different types of mycorrhizal fungi, Arbuscular Mycorrhizal Fungi (AMF) are the best studied, and it has been found that they form interactions with >80% of higher plants (reviewed in Behie and Bidochka, 2014). All AMF belong to the Glomeromycota phyla, and the levels of specificity among host plants and fungal species are fairly low. Many species of AMF have a cosmopolitan distribution (reviewed in Richardson et al., 2000), consequently allowing the ubiquity of this interaction.
The interaction between AMF and plant roots has the important benefit of enabling plants to obtain the highly immobile and limiting nutrient phosphorus (P) (Lynch and Brown, 2008). In exchange, the fungus is provided with carbon compounds (Helgason and Fitter, 2005; Brundrett, 2009). P can occur as a range of negatively and positively charged or uncharged chemical species in the soil solution, the distribution of which is much dependent on the pH and on the concentration of metal cations such as Ca, Fe, and Al and organic and inorganic ligands (Helgason and Fitter, 2005). Consequently, the chemical speciation of P in soil is complex but it is mostly found in low mobile precipitates in soil that are not directly available for the plant (Shen et al., 2011). AMF are able to take up P and provide it directly to the plant, bypassing the metabolically inefficient direct P uptake at the plant epidermis (Helgason and Fitter, 2005).
Fossil evidence suggests that AMF interactions similar to current interactions already existed around 400 million years ago (MYA) and it is thought that this interaction aided plants to colonize land at the beginning of root system development (reviewed in Remy et al., 1994) (Fig. 1). Primitive mycorrhizal associations with early land plants were probably also in water-rich environments such as bogs (Helgason and Fitter, 2005). Supporting this, it is known that AMF also form associations with mosses, one of the closest existing relatives of the first plants that colonized land (Zhang and Guo, 2007). These AMF structures have been detected in stems and leaves since mosses lack proper roots (Zhang and Guo, 2007). Furthermore there is also evidence of aquatic and semi-aquatic plant species that form mycorrhizal symbiosis (Beck-Nielsen and Vindbæk Madsen, 2001).
It is known from classical studies that mycorrhization predates the evolution of nodulation (see below), and analysis of genetic and molecular regulators has established that a large part of the signalling machinery for nodulation is shared, or derived simultaneously with the pathways for AM formation (Op den Camp et al., 2011; Delaux et al., 2013, 2014). Genetic and phylogenetic analyses place gain of nodulation function occurring around 58 MYA (Sprent, 2007). During nodule development, rhizobia-derived signal molecules (Nod factors) are sensed in root epidermal cells and, concomitantly with bacterial entry, cortical cell division is induced, enabling the formation of a niche to host the bacteria (reviewed in Oldroyd, 2013). Inside a functional nodule, rhizobia fix atmospheric nitrogen into a form usable by the plant and, in exchange, the plant supplies the bacteria with a mixture of carbon compounds and amino acids (reviewed in Lodwig et al., 2003; Oldroyd et al., 2011 b; Oldroyd, 2013). This interaction normally occurs in plant roots, but there are some exceptions where nodules are developed in stems such as in the case of Sesbania (Becker and George, 1995), although this stem nodulation appears to be a unique evolutionary gain of function event.
Pathogenesis and mutualism: a genetic and molecular shift from foe to friend?
One hypothesis suggests that pathogenic interactions derived from a mutualistic plant–microbe relationship (reviewed in Stukenbrock and McDonald, 2008). An alternative, non-mutually-exclusive hypothesis suggests that mutualistic root–microbe interactions evolved from early pathogenic interactions, since both require prevention of host defence programmes for microbe survival (Pel and Pieterse, 2013). Several lines of evidence suggest that the initial perception of beneficial microbes activates the plant immune system (Helgason and Fitter, 2005; Zamioudis and Pieterse, 2011) and then, in later stages of symbiotic associations, the expression of defence-related genes is down-regulated (Zamioudis and Pieterse, 2011). This suggests that suppression of the host immune system enables symbiosis to occur. There is a tight interplay between pathogen and beneficial microbe recognition by host plants. For example, in the legume L. japonicus, defence responses induced by the Microbe-Associated Molecular Pattern (MAMP) flg22 inhibit rhizobial symbiosis (Lopez-Gomez et al., 2011). In addition, more recent work suggests that non-legumes respond to the rhizobial-produced Nod Factor, suppressing plant innate immunity and rendering plants more susceptible to pathogen attack (Liang et al., 2013).
The potential shift from a pathogenic interaction to a symbiotic one could be partially due to the presence of more specialized LysMs with small alterations in their protein domains (Nakagawa et al., 2011) that enable different microbes including pathogens and mutualists to be distinguished. In legumes, the entire LysM family duplicated 60 MYA, potentially enabling the divergence of family members for roles in AMF signalling and also rhizobial symbiosis (reviewed in Young and Bharti, 2012). This was corroborated by Delaux et al. (2014) who carried out phylogenetic and phenotypic analysis across plants with differing levels of abilities to form AMF interactions and who found a highly interrelated evolutionary relationship between AMF interactions and host plant symbiosis regulatory genes (Delaux et al., 2014).
Molecular evidence suggests that the duplication of LysM receptor genes could have generated two closely related genes, NFP (for nodulation) and LYK related 1 (LYR1, for mycorrhization) that now exist in M. truncatula (Young et al., 2011). This supports the idea that the emergence of nodulation in legumes was associated with the whole genome duplication event (reviewed in Young and Bharti, 2012). The fact that Nod factors and Myc factors are highly similar lipo-chitooligosaccharide molecules also supports this theory (Maillet et al., 2011). Most recently, work on the impact of symbiotic associations on the host genome has shown that the evolutionary loss of many genes of the ‘symbiotic toolkit’ makes it impossible for non-hosts to establish symbiotic interactions (Delaux et al., 2014). The specific loss of mycorrhization genes in some legumes such as those from Lupinus is suggested as an explanation of why they are able to nodulate but not form AMF interactions. The establishment of these species in extreme soil conditions (Lambers and Teste, 2013) could possibly have been a selective pressure for the specific loss of mycorrhization genes but not the genes involved in nodulation (Delaux et al., 2014).
Common genetic and molecular architecture underlying both nodulation and mycorrhization pathways
Rhizobia-derived lipo-chitooligosaccharides (LCOs) or Nod factors are recognized by plasma membrane-localized Lysine Motif-Receptor-Like Kinases (LysMs) (Limpens et al., 2003; Oldroyd and Long, 2003). These include NFP and LYK3 in Medicago truncatula (Amor et al., 2003) or NFR1 and NFR5 in Lotus japonicus (Madsen et al., 2003). Structural variations in the rhizobia-derived Nod factors, together with variations in the plant-derived-signal flavonoids, enable species-specificity in nodulation (reviewed in Denarie et al., 1996). During plant-AMF recognition, AMF-derived LCOs (or Myc factors) are thought to be perceived by Myc LCO receptors. Their precise identity is still unknown in most species although they are postulated to have a similar structure and membrane-bound location to Nod factor-recognizing LysMs (Maillet et al., 2011; Op den Camp et al., 2011; Gust et al., 2012).
Nodulation is associated with legume species, but is not restricted to them, since plant species from the non-legume genus Parasponia can also form nodules through interaction with rhizobia (Trinick, 1973; Akkermans et al., 1978) (Fig. 2). Parasponia nodules differ from legume nodules in that they are modified lateral roots with a central vascular bundle and infected cells in the peripheral zone. This is in comparison to legume nodules that have a peripheral vasculature with a central zone of infected cells (Pawlowski and Sprent, 2008). These differences in nodule structure and development in Parasponia, together with its close relationship to the non-nodulating genus Trema, suggest that Parasponia gained nodulation ability in an independent and more recent evolutionary event to legumes (Streng et al., 2011) (Fig. 2). By contrast with the whole genome duplication event giving rise to nodulation genes in legumes, in Parasponia andersonii, the same LysM-type receptor has been found to enable both mycorrhizal and rhizobia recognition. This dual nature could explain why Parasponia interacts with a broad range of rhizobial species, resulting in differences in nitrogen fixation efficiency (Op den Camp et al., 2012). The dual function of LysM seems also to render Parasponia susceptible to pathogen attack since some rhizobial interactions with Parasponia can become parasitic (Op den Camp et al., 2012). The dual nature of the LysM receptor in P. andersonii contrasts with the function of LysMs in the model legumes M. truncatula and L. japonicus, in which mutations in the Nod factor receptor have no influence on AMF symbiosis (Limpens et al., 2003; Madsen et al., 2003; Oldroyd and Long, 2003). It would be interesting to identify how calcium spiking and signal transduction occurs in Parasponia, since, in this species, the same molecular pathway seems to be utilized by both rhizobia and AMF.
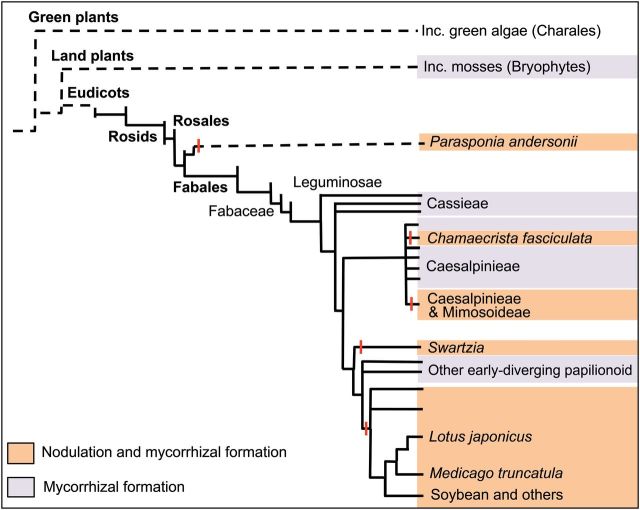
Fig. 2.
Overview of Leguminosae, highlighting multiple origins of nodulation. Relative/indicative (not to scale) phylogenetic positions of species that form AMF interactions (purple shading) and those that form AMF and rhizobial interactions (orange shading). Putative origins of nodulation are indicated with vertical red lines. Indicative positions of land plants including mosses, and green plants including green algae, are also shown. The Tree is drawn from information in the Tree of Life Project (http://tolweb.org/tree/), and for Leguminosae following Sprent (2007).
Common SYM genes pre-date nodulation and AMF interactions
Downstream of the symbiotic receptors, transcription factor-mediated signalling induces calcium spiking, involving proteins associated with the nuclear membrane (extensively reviewed, for example, in Oldroyd et al., 2011 a). In M. truncatula, for example, DMI1 and DMI2 are involved in initiating calcium spiking to activate both nodulation and AM formation. This results in the activation of calcium/calmodulin-dependent protein kinase (CCaMK) and subsequent nodule organogenesis (Kosuta et al., 2008; Capoen et al., 2009). DMI1, DMI2 and CCaMK are three members of a defined set of common symbiosis ‘SYM’ regulatory genes (Fig. 3). Upon spore germination, AMF perception involves regulation of SYM genes, calcium spiking, starch accumulation in roots, and also lateral root formation prior to colonization (Chabaud et al., 2011). It has been hypothesized that the frequency of epidermal calcium spiking codes for microbe specificity (Oldroyd and Downie, 2008) since, in response to rhizobia these spikes are highly regular, whereas in response to AMF they can be highly irregular (Chabaud et al., 2011). However, other studies show that the calcium spiking between different types of microbial interactions are indistinguishable and that differences in calcium spiking are a reflection of the stage of the establishment of the symbiosis (Sieberer et al., 2012). Another alternative explanation is that rhizobia and AMF might have different binding requirements for calmodulin to bind to CCaMK (Shimoda et al., 2012).
Fig. 3.
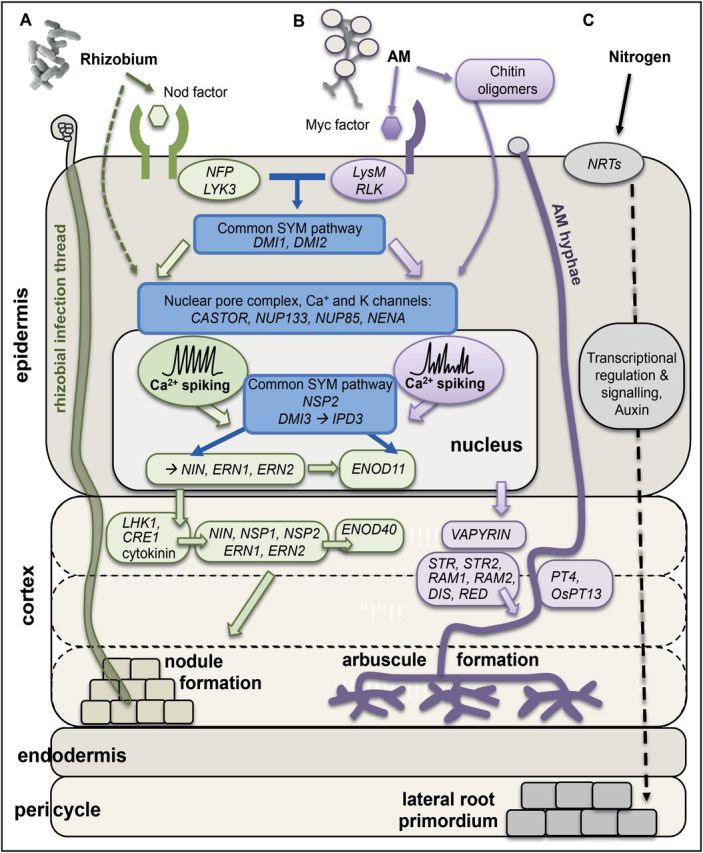
Comparative overview of (A) nodulation, (B) mycorrhizal formation, and (C) lateral root development in response to nitrogen. Rhizobia- and AM-derived small molecules are perceived by LysM receptors on the epidermal plasma membrane. Root nodules form under nitrogen-deprived conditions. Common SYM genes (blue) lead to the activation of calcium spiking in the nucleus and gene regulatory signalling between cell types (green=key nodulation genes; purple=key mycorrhizal interaction genes) that trigger nodule or arbuscule establishment in the inner cortex. Nitrogen has a complex effect on lateral root development, both activating and repressing genes that control primordium initiation and subsequent emergence (reviewed in Vidal et al., 2008). In brief, lateral root primordia develop in the pericycle, regulated by signalling downstream of Nitrate Transporters (NRTs), transcriptional regulation, and auxin signalling. Currently nine common SYM genes have been identified and characterized: SYMRK=DMI2, POLLUX=DMI1, CCaMK=DMI3 and CYCLOPS=IPD3, CASTOR, NUP85, NUP133, NENA, and NSP2 (when characterized in two species, Lotus japonicus names are given first then Medicago truncatula names: as discussed in Oldroyd, 2013). Within the plant, a number of regulators including VAPYRIN, STR1*, STR2*, RAM1, RAM2, DIS, RED, PT4, and OsPT13* identified in M. truncatula (bold), L. japonicus (underlined) or rice (*) control formation of the mature arbuscule (reviewed by Gutjahr and Parniske, 2013).
SYM genes are functionally conserved in rice, which suggests their existence prior to the diversification of angiosperms (reviewed by Gutjahr and Parniske, 2013). Homologues are also found in bryophytes and in the green algae order Charales, which raises the possibility that the common symbiosis genes evolved prior to the colonization of land by plants (reviewed in Delaux et al., 2013).
AMF interaction, nodulation, and lateral root development pathways: diversification but conservation during evolution
Genetic evidence shows that, aside from the commonalities with AMF interaction, nodule development integrates pre-existing plant regulatory pathways that are related to lateral root organogenesis (Mathesius, 2003). Regulation of lateral root development itself shares parallels with root–mutualist interactions, although, rather than microbes, these plant pathways are regulated by environmental conditions including nitrogen and phosphorus form and availability (reviewed in Giehl et al., 2014). Lateral root development and nodulation share a more specific common environmental regulation—they are both tightly regulated by nitrogen (although nodulation is also rhizobia-dependent). In addition, the development of the two organs can be seen to be fundamentally similar since both nodules and lateral roots are formed by the reactivation of cell division in differentiated cells in specific developmental zones (Malamy and Benfey, 1997; Stougaard, 2000) and are controlled spatially and quantitatively by hormone flux (De Smet et al., 2007, 2008; Tirichine et al., 2007; van Noorden et al., 2007). This similarity is even more relevant in Parasponia, where the architecture of a nodule is more similar to a lateral root (Pawlowski and Sprent, 2008). Although lateral roots and nodules are formed from cell division in different root cell types (pericycle and cortex, respectively), they both involve cortical cell division. In the case of lateral roots, these cortical divisions are orientated to repair consequential damage to the cortex and to ensure structural integrity to the new lateral root (Mathesius, 2003). There is also considerable overlap between the genes whose expression changes in lateral root and nodule development that could be related to the common requirement for the regulation of cell division and differentiation (Hirsch and LaRue, 1997). In the case of AMF–plant interactions, although a new organ does not develop, there is stimulation of root growth as well as differentiation and re-orientation of root cortical cells (Harrison, 1999). Possibly due to this common regulation of cell growth and development, nodulation has been found to increase the likelihood of mycorrhizal interaction establishment on the same plant, and vice versa (Xie et al., 1995, 1998). In M. truncatula it has been found that Nod factors and Myc factors, both stimulate root branching via lateral root development (Olah et al., 2005; Maillet et al., 2011). These interactions occur both at the level of symbiont/environment recognition and more directly between developmental pathways (reviewed in Olah et al., 2005; Mathesius, 2003).
Lateral root development and nodulation are genetically linked in legumes. For example, in M. truncatula, RNAi knockdown of the cytokinin receptor MtCRE1 results in a reduction in nodule number but an increase in lateral root number (Gonzalez-Rizzo et al., 2006). Suggestive of a shared genetic pathway linking lateral root and nodule development, lateral root organ defective (latd/nip) mutants in M. truncatula are unable to form active nodules or to complete lateral root formation with consequential effects on primary root development (Bright et al., 2005). The LATD gene is a high affinity nitrate transporter, with additional developmental functions likely as have already been found for other nitrate transporters (Bagchi et al., 2012).
Nitrogen levels regulate both nodule and lateral root number, and high levels of nitrogen in the soil or in the plant inhibit the formation of nodules via autoregulation of nodulation (AON) mechanisms. This is controlled by the Supernumary Nodules (MtSUNN) gene in M. truncatula (Schnabel et al., 2005), and the probably orthologous Hypernodulation Aberrant Root formation (LjHAR1) gene in L. japonicus (Wopereis et al., 2000). In the presence of rhizobia the har1 mutant has an overabundance of nodules (termed hypernodulation) and drastically reduced root growth. In the absence of rhizobia, har1 does not nodulate but there are effects on root growth. In the har1 mutant root pericycle layer, there is a significantly higher level of mitotic activity, resulting in higher lateral root density compared with the wild type. Together this suggests that the LjHAR1/SUNN gene is a common developmental regulator of lateral root and nodule formation.
At low levels of nitrogen, both Nod factor and Myc factor stimulate lateral root development via a DMI1 and DMI2-regulated signalling pathway (Olah et al., 2005; Maillet et al., 2011). However, it has been found that the presence of rhizobia at high levels of nitrogen that inhibit nodulation, actually represses lateral root development. We have discovered that this appears to act via signalling from the AON mechanism (Bonyadi Pour et al., unpublished data).
Together the nodule–lateral root developmental and molecular connections support the received hypothesis that the nodule structure arose from a lateral root blueprint (discussed in Mathesius, 2003) whilst also ‘co-opting’ or utilizing genetic interactions involved in AMF interactions. This suggests tight co-regulation of lateral root and nodule formation, with the connection of gene regulatory pathways a probable mechanism underlying this. These regulatory connections appear to be conserved in non-symbiotic species. For example, it has recently been found that the putative orthologue of M. truncatula NSP2 in Arabidopsis (At4g08540) has a different lateral root phenotype depending on nitrate availability (B Lagunas et al., unpublished data). Both genes are GRAS transcription factors and appear to mediate GA-controlled developmental root processes in specific root cell types that form nodules or lateral roots.
Future prospects
Understanding how plants manage and balance interactions with different microorganisms is more important than ever with a rapidly increasing world population (UN, 2013). Sustainable solutions are, therefore, needed to ensure food security. The importance of investigating root–environment interactions is achieving increasing recognition as a critical area in this global challenge. In this direction, transferring nodulation ability into crops such as rice and wheat would be one sustainable solution to decrease human dependence on fertilizer usage. To approach this goal, it is necessary to gain a deeper knowledge of the symbiotic mechanisms in current model symbiotic systems. This would include carrying out a molecular cost–benefit analysis to quantify the carbon requirements of nodulation but also the effects of introducing a new symbiosis upon those already existing in a plant species. It is also important to consider how to develop an efficient technical strategy for transferring it to crops and to determine which species will be best to transfer it into (see comments in Rogers and Oldroyd, 2014). Clues for this can come from studying the evolutionary history of nodulation and AMF interactions, since the gene families in existence today define the range and scale of symbiosis. We could also assess how to integrate novel aspects of the nitrogen cycle. For example free-living nitrogen fixing bacteria could mitigate low N levels in particular agricultural soil types (Bashan et al., 2014).
We are still discovering to what extent symbiotic and pathogenic mechanisms are linked and will need to know whether increasing or enabling symbiotic interactions has an effect on the ability of pathogens to infect plants. One avenue to study these connections could be to analyse pathogen susceptibility of the single-LysM-receptor species Parasponia andersonii. We must further begin to model symbioses together in order to define a multi-scale model of Parasponia plant–mutualist interactions. For this, our analysis can be informed by examining the expression patterns and mode of regulation for orthologous symbiosis-controlling genes. By connecting information about the regulation of different pathways at the phenotypic, molecular, and biophysical levels, it will be possible to gain new insight. For example, by integrating individual mathematical models describing the regulation of shoot growth and architecture by day-length, light intensity, and temperature, with carbon partitioning and gene regulatory network dynamics, the combined multi-model had much greater power for testing novel hypotheses with agronomically relevant outputs (Chew et al., 2014). These multi-scale approaches are particularly relevant for understanding developmental processes with a common origin and hold potential for us, in future, to identify the regulatory connections between developmental pathways and species.
Acknowledgements
This work was supported by a Biotechnology and Biological Sciences Research Council New Investigator BB/H109502/1 grant and a BBSRC BB/H019502/1 grant to MLG.
References
- Aanen DK, Bisseling T. 2014. Microbiology. The birth of cooperation. Science 345, 29–30. [DOI] [PubMed] [Google Scholar]
- Akkermans ADL, Abdulkadir S, Trinick MJ. 1978. Nitrogen-fixing root nodules in Ulmaceae. Nature 274, 190–190.26885 [Google Scholar]
- Amor BB, Shaw SL, Oldroyd GED, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C. 2003. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. The Plant Journal 34, 495–506. [DOI] [PubMed] [Google Scholar]
- Archibald J. 2011. Origin of eukaryotic cells: 40 years on. Symbiosis 54, 69–86. [Google Scholar]
- Ausubel FM. 2005. Are innate immune signaling pathways in plants and animals conserved? Nature Immunology 6, 973–979. [DOI] [PubMed] [Google Scholar]
- Bagchi R, Salehin M, Adeyemo OS, Salazar C, Shulaev V, Sherrier DJ, Dickstein R. 2012. Functional assessment of the Medicago truncatula NIP/LATD protein demonstrates that it is a high-affinity nitrate transporter. Plant Physiology 160, 906–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron C, Zambryski PC. 1995. The plant response in pathogenesis, symbiosis, and wounding: variations on a common theme? Annual Review of Genetics 29, 107–129. [DOI] [PubMed] [Google Scholar]
- Bashan Y, de-Bashan LE, Prabhu SR, Hernandez J-P. 2014. Advances in plant growth-promoting bacterial inoculant technology: formulations and practical perspectives (1998–2013). Plant and Soil 378, 1–33. [Google Scholar]
- Beck-Nielsen D, Vindbæk Madsen T. 2001. Occurrence of vesicular–arbuscular mycorrhiza in aquatic macrophytes from lakes and streams. Aquatic Botany 71, 141–148. [Google Scholar]
- Becker B, Marin B. 2009. Streptophyte algae and the origin of embryophytes. Annals of Botany 103, 999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, George T. 1995. Nitrogen fixation response of stem- and root-nodulating Sesbania species to flooding and mineral nitrogen. Plant and Soil 175, 189–196. [Google Scholar]
- Bednarek P, Kwon C, Schulze-Lefert P. 2010. Not a peripheral issue: secretion in plant–microbe interactions. Current Opinion in Plant Biology 13, 378–387. [DOI] [PubMed] [Google Scholar]
- Behie SW, Bidochka MJ. 2014. Nutrient transfer in plant–fungal symbioses. Trends in Plant Science 19, 734–740. [DOI] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM. 2012. The rhizosphere microbiome and plant health. Trends in Plant Science 17, 478–486. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Genre A. 2010. Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nature Communications 1 48. [DOI] [PubMed] [Google Scholar]
- Bright L, Liang Y, Mitchell DM, Harris JM. 2005. The LATD gene of Medicago truncatula is required for both nodule and root development. Molecular Plant–Microbe Interactions 18, 521–532. [DOI] [PubMed] [Google Scholar]
- Brundrett M. 2009. Mycorrhizal associations and other means of nutrition of vascular plants: understanding the global diversity of host plants by resolving conflicting information and developing reliable means of diagnosis. Plant and Soil 320, 37–77. [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, et al. 2012. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annual Review of Plant Biology 64, 807–838. [DOI] [PubMed] [Google Scholar]
- Capoen W, Den Herder J, Sun J, Verplancke C, De Keyser A, De Rycke R, Goormachtig S, Oldroyd G, Holsters M. 2009. Calcium spiking patterns and the role of the calcium/calmodulin-dependent kinase CCaMK in lateral root base nodulation of Sesbania rostrata . The Plant Cell 21, 1526–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, Genre A, Sieberer BJ, Faccio A, Fournier J, Novero M, Barker DG, Bonfante P. 2011. Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytologist 189, 347–355. [DOI] [PubMed] [Google Scholar]
- Chew YH, Wenden B, Flis A, et al. 2014. Multiscale digital Arabidopsis predicts individual organ and whole-organism growth. Proceedings of the National Academy of Sciences, USA 111, E4127–E4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. 2013. Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis . Development 134, 681–690. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, et al. 2008. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594–597. [DOI] [PubMed] [Google Scholar]
- de Vries FT, Thébault E, Liiri M, et al. 2013. Soil food web properties explain ecosystem services across European land use systems. Proceedings of the National Academy of Sciences, USA 110, 14296–14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux P-M, Sejalon-Delmas N, Becard G, Ane J-M. 2013. Evolution of the plant–microbe symbiotic ‘toolkit’. Trends in Plant Science 18, 298–304. [DOI] [PubMed] [Google Scholar]
- Delaux P-M, Varala K, Edger PP, Coruzzi GM, Pires JC, Ané J-M. 2014. Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genetics 10, e1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux P-M, Xie X, Timme RE, Puech-Pages V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon-Delmas N. 2012. Origin of strigolactones in the green lineage. New Phytologist 195, 857–871. [DOI] [PubMed] [Google Scholar]
- Denarie J, Debelle F, Prome JC. 1996. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annual Review of Biochemistry 65, 503–535. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. 2010. Plant immunity: towards an integrated view of plant–pathogen interactions. Nature Reviews Genetics 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Doehlemann G, Requena N, Schaefer P, Brunner F, O’Connell R, Parker JE. 2014. Reprogramming of plant cells by filamentous plant-colonizing microbes. New Phytologist 204, 803–814. [DOI] [PubMed] [Google Scholar]
- Downie H, Holden N, Otten W, Spiers AJ, Valentine TA, Dupuy LX. 2012. Transparent soil for imaging the rhizosphere. PLoS One 7, e44276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RF, Gruber BD, von Wiren N. 2014. It’s time to make changes: modulation of root system architecture by nutrient signals. Journal of Experimental Botany 65, 769–778. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. 2006. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti . The Plant Cell 18, 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, Willmann R, Desaki Y, Grabherr HM, Nurnberger T. 2012. Plant LysM proteins: modules mediating symbiosis and immunity. Trends in Plant Science 17, 495–502. [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Parniske M. 2013. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annual Review of Cell and Developmental Biology 29, 593–617. [DOI] [PubMed] [Google Scholar]
- Harrison MJ. 1999. Molecular and cellular aspects of the abuscular mycorrhizal symbiosis. Annual Review of Plant Physiology and Plant Molecular Biology 50, 361–389. [DOI] [PubMed] [Google Scholar]
- Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW. 1998. Ploughing up the wood-wide web? Nature 394, 431–431. [DOI] [PubMed] [Google Scholar]
- Helgason T, Fitter A. 2005. The ecology and evolution of the arbuscular mycorrhizal fungi. Mycologist 19, 96–101. [Google Scholar]
- Hirsch AM, LaRue TA. 1997. Is the legume nodule a modified root or stem or an organ sui generis? Critical Reviews in Plant Science 16, 361–392. [Google Scholar]
- Hom EF, Murray AW. 2014. Plant–fungal ecology. Niche engineering demonstrates a latent capacity for fungal–algal mutualism. Science 34, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, von Mering C, Vorholt JA. 2012. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. International Society for Microbial Ecology Journal 6, 1378–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S, Hazledine S, Sun J, Miwa H, Morris RJ, Downie JA, Oldroyd GED. 2008. Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proceedings of the National Academy of Sciences, USA 105, 9823–9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Teste FP. 2013. Interactions between arbuscular mycorrhizal and non-mycorrhizal plants: do non-mycorrhizal species at both extremes of nutrient availability play the same game? Plant, Cell and Environment 36, 1911–1915. [DOI] [PubMed] [Google Scholar]
- Liang Y, Cao Y, Tanaka K, Thibivilliers S, Wan J, Choi J, Kang Ch, Qiu J, Stacey G. 2013. Nonlegumes respond to rhizobial nod factors by suppressing the innate immune response. Science 341, 1384–1387. [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. 2003. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302, 630–633. [DOI] [PubMed] [Google Scholar]
- Lodwig EM, Hosie AHF, Bourdes A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS. 2003. Amino-acid cycling drives nitrogen fixation in the legume–Rhizobium symbiosis. Nature 422, 722–726. [DOI] [PubMed] [Google Scholar]
- Lopez-Gomez M, Sandal N, Stougaard J, Boller T. 2011. Interplay of flg22-induced defence responses and nodulation in Lotus japonicus . Journal of Experiemental Botany 63, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, et al. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Brown KM. 2008. Root strategies for phosphorus acquisition. In: White PJ, Hammond JP, eds. The ecophysiology of plant–phosphorus interactions. Dordrecht, The Netherlands: Springer, 83–116. [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, et al. 2003. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640. [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, Andre O, et al. 2011. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–63. [DOI] [PubMed] [Google Scholar]
- Mairhofer S, Zappala S, Tracy SR, Sturrock C, Bennett M, Mooney SJ, Pridmore T. 2012. RooTrak: automated recovery of three-dimensional plant root architecture in soil from X-ray microcomputed tomography images using visual tracking. Plant Physiology 158, 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana . Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Margulis L. 1970. Origin of eukaryotic cells. New Haven: Yale University Press. [Google Scholar]
- Mathesius U. 2003. Conservation and divergence of signalling pathways between roots and soil microbes: the Rhizobium–legume symbiosis compared to the development of lateral roots, mycorrhizal interactions and nematode-induced galls. Plant and Soil 255, 105–119. [Google Scholar]
- Nakagawa T, Kaku H, Shimoda Y, Sugiyama A, Shimamura M, Takanashi K, Yazaki K, Aoki T, Shibuya N, Kouchi H. 2011. From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume–Rhizobium symbiosis. The Plant Journal 65, 169–180. [DOI] [PubMed] [Google Scholar]
- Olah B, Briere C, Becard G, Denarie J, Gough C. 2005. Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. The Plant Journal 44, 195–207. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews Microbiology 11, 252–263. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA. 2008. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annual Review of Plant Biology 59, 519–546. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Long SR. 2003. Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiology 131, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS, Downie JA. 2011. The rules of engagement in the legume–rhizobial symbiosis. Annual Review of Genetics 45 119–144. [DOI] [PubMed] [Google Scholar]
- Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, Ammiraju JSS, Kudrna D, Wing R, Untergasser A. 2011. LysM-type mycorrhizal receptor recruited for Rhizobium symbiosis in the non-legume Parasponia . Science 331, 909–912. [DOI] [PubMed] [Google Scholar]
- Op den Camp RHM, Polone E, Fedorova E, Roelofsen W, Squartini A, Op den Camp HJM, Bisseling T, Geurts R. 2012. Non-legume Parasponia andersonii deploys a broad Rhizobium host range strategy resulting in largely variable symbiotic effectiveness. Molecular Plant–Microbe Interactions 25, 954–963. [DOI] [PubMed] [Google Scholar]
- Pawlowski K, Sprent JI. 2008. Comparison between actinorhizal and legume symbiosis. In: Pawlowski K, Newton W, eds. Nitrogen-fixing actinorhizal symbioses. The Netherlands: Springer, 261–288. [Google Scholar]
- Pel MJC, Pieterse CMJ. 2013. Microbial recognition and evasion of host immunity. Journal of Experimental Botany 64, 1237–1248. [DOI] [PubMed] [Google Scholar]
- Qiang X, Weiss M, Kogel K-H, Schäfer P. 2012. Piriformospora indica—a mutualistic basidiomycete with an exceptionally large plant host range. Molecular Plant Pathology 13, 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Hurek T. 2011. Living inside plants: bacterial endophytes. Current Opinion in Plant Biology 14, 435–443. [DOI] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H. 1994. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proceedings of the National Academy of Sciences, USA 91, 11841–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DM, Allsopp N, D’Antonio CM, Milton SJ, Rejmánek M. 2000. Plant invasions—the role of mutualisms. Biological Reviews 75, 65–93. [DOI] [PubMed] [Google Scholar]
- Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. International Society for Microbial Ecology Journal 1, 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Oldroyd GED. 2014. Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. Journal of Experimental Botany 65, 1939–1946. [DOI] [PubMed] [Google Scholar]
- Schäfer P, Khatabi B, Kogel K-H. 2007. Root cell death and systemic effects of Piriformospora indica: a study on mutualism. FEMS Microbiology Letters 275, 1–7. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. 2014. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proceedings of the National Academy of Sciences, USA 111, 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J. 2005. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Molecular Biology 58, 809–822. [DOI] [PubMed] [Google Scholar]
- Shen J, Yuan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F. 2011. Phosphorus dynamics: from soil to plant. Plant Physiology 156, 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda Y, Han L, Yamazaki T, Suzuki R, Hayashi M, Imaizumi-Anraku H. 2012. Rhizobial and fungal symbioses show different requirements for calmodulin binding to calcium calmodulin–dependent protein kinase in Lotus japonicus . The Plant Cell 24, 304–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Fournier J, Timmers ACJ, Barker DG. 2012. A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula . The Plant Journal 69, 822–830. [DOI] [PubMed] [Google Scholar]
- Slippers B, Wingfield MJ. 2007. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biology Reviews 21, 90–106. [Google Scholar]
- Sprent JI. 2007. Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytologist 174, 11–25. [DOI] [PubMed] [Google Scholar]
- Stougaard J. 2000. Regulators and regulation of legume root nodule development. Plant Physiology 124, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streng A, op den Camp R, Bisseling T, Geurts R. 2011. Evolutionary origin of rhizobium Nod factor signaling. Plant Signaling & Behavior 6, 1510–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, McDonald BA. 2008. The origins of plant pathogens in agro-ecosystems. Annual Review of Phytopathology 46, 75–100. [DOI] [PubMed] [Google Scholar]
- Suzaki T, Kawaguchi M. 2014. Root nodulation: a developmental program involving cell fate conversion triggered by symbiotic bacterial infection. Current Opinion in Plant Biology 21C, 16–22. [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. 2007. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315, 104–107. [DOI] [PubMed] [Google Scholar]
- Trinick MJ. 1973. Symbiosis between Rhizobium and the non-legume, Trema aspera . Nature 244, 459–460. [Google Scholar]
- UN. 2013. World population prospects: the 2012 revision. Population Division of the Department of Economic and Social Affairs of the United Nations Secretariat. [Google Scholar]
- van Noorden GE, Kerim T, Goffard N, Wiblin R, Pellerone FI, Rolfe BG, Mathesius U. 2007. Overlap of proteome changes in Medicago truncatula in response to auxin and Sinorhizobium meliloti . Plant Physiology 144, 1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA. 2008. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Current Opinion in Plant Biology 11, 521–529. [DOI] [PubMed] [Google Scholar]
- Wagg C, Bender SF, Widmer F, van der Heijden MGA. 2014. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proceedings of the National Academy of Sciences, USA 111, 5266–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Qiu YL. 2006. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16, 299–363. [DOI] [PubMed] [Google Scholar]
- Wirthmueller L, Maqbool A, Banfield MJ. 2013. On the front line: structural insights into plant–pathogen interactions. Nature Reviews Microbiology 11, 761–776. [DOI] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang Q, Gresshoff PM, De Bruijn FJ, Stougaard J, Szczyglowski K. 2000. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. The Plant Journal 23, 97–114. [DOI] [PubMed] [Google Scholar]
- Xie Z-P, Muller J, Wiemken A, Broughton WJ, Boller T. 1998. Nod factors and tri-iodobenzoic acid stimulate mycorrhizal colonization and affect carbohydrate partitioning in mycorrhizal roots of Lablab purpureus . New Phytologist 139, 361–366. [Google Scholar]
- Xie ZP, Staehelin C, Vierheilig H, Wiemken A, Jabbouri S, Broughton WJ, Vogeli-Lange R, Boller T. 1995. Rhizobial nodulation factors stimulate mycorrhizal colonization of nodulating and nonnodulating soybeans. Plant Physiology 108, 1519–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Bharti AK. 2012. Genome-enabled insights into legume biology. Annual Review of Plant Biology 63, 283–305. [DOI] [PubMed] [Google Scholar]
- Young ND, Debelle F, Oldroyd GED, et al. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis C, Pieterse CMJ. 2011. Modulation of host immunity by beneficial microbes. Molecular Plant–Microbe Interactions 25, 139–150. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Guo L-D. 2007. Arbuscular mycorrhizal structure and fungi associated with mosses. Mycorrhiza 17, 319–325. [DOI] [PubMed] [Google Scholar]