Highlight
When contemplating the elemental content of plant tissues, should we try to understand the dynamics of each element individually, or try to understand the dynamics of integrated groups of elements?
Key words: Elemental profiling, environment, G×E, genetics, ionomics, plant nutrition.
Abstract
It has been more than 10 years since the concept of the ionome, all of the mineral nutrients in a cell tissue or organism, was introduced. In the intervening years, ionomics, high throughput elemental profiling, has been used to analyse over 400 000 samples from at least 10 different organisms. There are now multiple published examples where an ionomics approach has been used to find genes of novel function, find lines or environments that produce foods with altered nutritional profiles, or define gene by environmental effects on elemental accumulation. In almost all of these studies, the ionome has been treated as a collection of independent elements, with the analysis repeated on each measured element. However, many elements share chemical properties, are known to interact with each other, or have been shown to have similar interactions with biological molecules. Accordingly, there is strong evidence from ionomic studies that the elements of the ionome do not behave independently and that combinations of elements should be treated as the phenotypes of interest. In this review, I will consider the evidence that we have for the interdependence of the ionome, some of its causes, methods for incorporating this interdependence into analyses and the benefits, drawbacks, and challenges of taking these approaches.
Introduction
It has been more than 10 years since the concept of the ionome, all of the mineral nutrients in a cell tissue or organism, was introduced (Lahner et al., 2003). In the intervening years, ionomics, high throughput elemental profiling, has been used to analyse over 400 000 samples from at least 10 different organisms (Broadley et al., 2008; Wu et al., 2008; Chen et al., 2009; Sankaran et al., 2009; Yu et al., 2012; Ziegler et al., 2013; Baxter et al., 2014; Bus et al., 2014; Malinouski et al., 2014; Zhang et al., 2014). There are now multiple published examples where an ionomics approach has been used to find genes of novel function, find lines or environments that produce foods with altered nutritional profiles, or define gene by environmental effects on elemental accumulation. In almost all of these studies, the ionome has been treated as a collection of independent elements, with the analysis repeated on each measured element. However, many elements share chemical properties, are known to interact with each other, or have been shown to have similar interactions with biological molecules. Accordingly, there is strong evidence from ionomic studies that the elements of the ionome do not behave independently and that combinations of elements should be treated as the phenotypes of interest. In this review, I will consider the evidence that we have for the interdependence of the ionome, some of its causes, methods for incorporating this interdependence into analyses and the benefits, drawbacks, and challenges of taking these approaches.
Evidence and cause
The strongest evidence that the elements of the ionome are interconnected is the identification of mutants in multiple organisms with multi-element phenotypes (Lahner et al., 2003; Eide et al., 2005; Chen et al., 2009; Yu et al., 2012; Ziegler et al., 2013; Malinouski et al., 2014). In Arabidopsis, many of these phenotypes have been shown to segregate as a single locus and quite a few have been cloned, verifying that a single genetic polymorphism can affect multiple elements (Fig. 1). The nature of the genes underlying these multi-element phenotypes reveals the wide variety of mechanisms that can affect multiple elements.
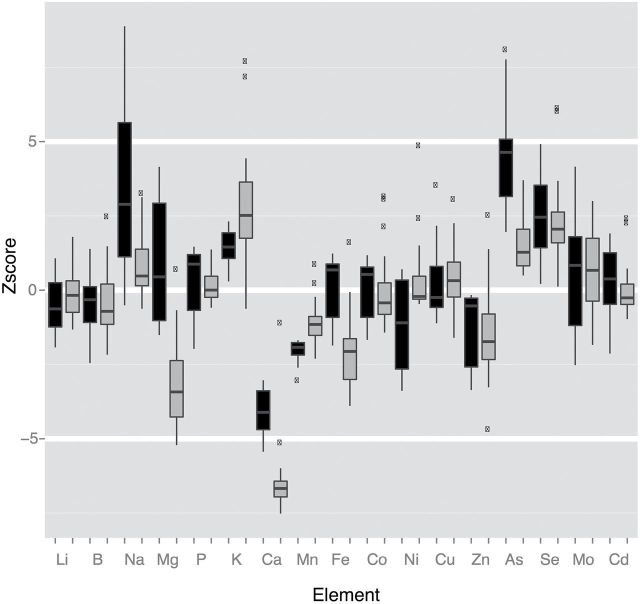
Fig. 1.
Multi-element phenotype for the esb1-2 mutant. Boxplots showing the distributions of Z-scores (number of standard deviations from the Col-0 mean) from trays 1095 (black) and 1146 (grey) at www.ionomicshub.org (last accessed: 15 December 2014). Note that Na, K, Ca, Mn, Zn, As, and Se are different in both trays, while Mg and Fe are only different in tray 1146.
Transporters
Transport of elements across membranes is a critical step in their movement and regulation. While some transporters appear to be specific for single elements, studies of plant transporter mutants or heterologous expression of plant transporters in yeast have demonstrated multi-element specificity. Examples include the Fe/Mn/Cd specificity of IRT1 and the Fe/Co specificity of FPN2 (Korshunova et al., 1999; Connolly et al., 2002; Morrissey et al., 2009). There are several examples where the plants will express a transporter that is less specific in order to increase the uptake of a scarce elemental nutrient, leading to the accumulation of deleterious elements (Rogers et al., 2000; Zhao et al., 2009, 2010; Mitani-Ueno et al., 2011). Variation in FRD3, which encodes a citrate transporter, affects the interplay between Fe and Zn homeostasis (Pineau et al., 2012). This suggests that transporters of chelators, which have multi-element specificities, can affect multiple elements by controlling the quantity and localization of the chelators.
Alterations of plant physiological properties
Several mutants have been cloned that affect the physiological properties of a plant and thereby have a significant effect on the elemental content. One of the first ionomic mutants to be cloned was the esb1 gene (Baxter et al., 2009), which affects the structure of the casparian strip (Hosmani et al., 2013). Across dozens of independent experiments, as many as nine elements were significantly different between the mutant and the wild type. Further analysis of other mutants that modify the structure of the casparian strip has confirmed that the permeability of this key structure has a major effect on the balance of elements that make it to the shoot (Pfister et al., 2014). For example, loss of the SGN3/GASSHO1 receptor-like kinase, which is critical for assembling the microdomains to build the casparian strip, leads to a K/Mg phenotype, although, surprisingly, most other elements are unaffected. Altering root structure (Hermans et al., 2010) and phloem (Tian et al., 2010) function have both been shown to affect multiple elements.
The co-ordinated response of several processes to an environmental stimulus can create a multi-element response in the ionome. Both low-Fe and low-P concentrations in the growth medium induced multi-element signatures that could be used to predict the soil condition. Mutants with a constitutive low-Fe response have the same multi-element profile as plants grown in low-Fe conditions. Mutants can also alter several processes to create a multi-element phenotype. The TSC10A mutant, for example, alters sphingolipid biosynthesis, resulting in changes to endodermal suberin deposition (and probably casparian strip function) and Fe homeostasis that lead to a seven element phenotype that overlaps with the signatures of those perturbations (Chao et al., 2011).
Studies of quantitative genetics populations provide additional, if indirect, evidence that the elements of the ionome are not independent of each other. Studies in multiple species have shown that many significant correlations between elements are observed across the lines of a population (Broadley et al., 2008; Ghandilyan et al., 2009a, b; Sankaran et al., 2009; Buescher et al., 2010; Bus et al., 2014; Zhang et al., 2014; Pinson et al., 2015). However, which elements are correlated with each other changes across environments, populations, and species, suggesting that different processes are involved depending on the genotypes and environments involved. These studies have also identified quantitative trait loci (QTLs) for different elements that co-locate to the same region of the genome, suggesting that the same causal gene variant is driving the phenotypes. With the number of elements measured and the number of genes underlying a QTL, it is possible but not likely that these co-localizations are due to random chance. This question will be resolved in the next few years as studies utilizing association mapping studies, with much narrower genetic resolution, are likely to be published.
Several other processes, including interactions with symbiotic organisms, alteration of the rhizosphere pH, and alterations of cell wall biochemistry, are likely to affect multiple ions. As the total number of cloned mutants is small, little can be inferred from the absence of mutants affecting these processes. However, an alternative hypothesis would be that there are mechanisms to buffer changes in some elements, resulting in no phenotype or single element phenotypes. Indeed nothing presented here should be taken as evidence that the elements never act independently. There are several clear examples where polymorphisms in a gene affect a single element, even when the affected process, such as cell size in the root or the pleiotropic response to perceived pathogen attack, might reasonably be expected to affect multiple elements (Borghi et al., 2011; Chao et al., 2013).
Solutions
Given all of these examples, we are left with the question of how best to conceptualize and analyse the ionome: as independent elements, or as an interdependent network?
There are several advantages to treating the elements independently, the simplest being that it reflects their fundamental chemical properties and the way that they are measured using modern spectroscopy techniques. This approach also allows for simple comparisons across tissues, experiments, and species. Given that sometimes the elements do act independently, we should continue to analyse the elements independently in every experiment.
We should not, however, assume that we are getting the full understanding of the underlying biology if we limit our analysis to the independent element frame. The examples detailed above demonstrate that there are many cases where several elements are acting in a dependent manner, and treating them as independent can muddle the thinking and interpretation. There is a temptation to attempt to define ‘the phenotype’ and then use statistical tests to see if the stated phenotype repeats. As a result, the phenotype can be defined down to those elements that are most reproducible, while eliminating elements that are more responsive to genetic or environmental variation and might provide novel insights into function. A more integrated understanding of the ionome might enable the detection of gene by environment interactions that modify the elemental signatures.
Another reason to move beyond thinking of individual elements is that multi-element approaches might allow for more sensitivity in detecting significant variation in the ionome. The change in a process that has small effects on four elements might not be detectable in any single element, but a pooled trait could boost the signal-to-noise ratio to allow detection. Combining the elements can also allow for the detection and removal of artefacts such as weighing or machine errors that affect multiple elements, improving the remaining signal for each element.
When thinking of combining elemental traits, there are two possible broad approaches, those that use directed knowledge of chemical or biological relatedness and those that use an undirected approach. Which approach is appropriate depends on the questions being asked and the level of prior knowledge about the elemental relationships in the relevant system.
The simplest way of combining elements is to create ratios to see where two elements differ, or average their normalized concentrations to determine where they are acting in concert. Elemental ratios have been used to diagnose issues in crop nutrition (Walworth and Sumner, 1987), and appear to be important signifiers of adapting to serpentine soils (Bradsahw, 2005). When this approach gets expanded beyond the small number of obvious ratios to the whole ionome, however, it greatly expands the number of traits that need to be tracked. Just 10 elements can have 45 dual element ratios and up to 405 (non-independent) amalgamated ratios, a problem that gets even worse when dealing with >20 elements (Parent et al., 2013b). There is also a danger when the measured ionome gets expanded to include trace elements that ratios of well-measured elements to elements around the limit of detection could be prone to artefacts.
In contrast to the ratio approaches, undirected approaches such as principal component analysis (PCA) can act to reduce the dimensionality of the ionome. PCA identifies orthogonal vectors through the n-dimensional elemental space that account for major components of the variation in the dataset examined. The PCAs can reflect artefacts, environmental variables or processes affecting the ionome. Since it is undirected, there is an opportunity to identify hidden factors that might not have been found in a directed approach. Indeed there are at least two unpublished examples of PCA statistics leading to the identification of strong QTLs affecting the ionome in genomic regions where no signal was detected for individual elements (Asaro and Baxter, unpublished data: BP Dilkes, personal communication). The limitations of this approach include that the PCAs are calculated within the universe defined by a given dataset, so comparison with other datasets, either within or across species, is problematic, making follow-up and interpretation difficult.
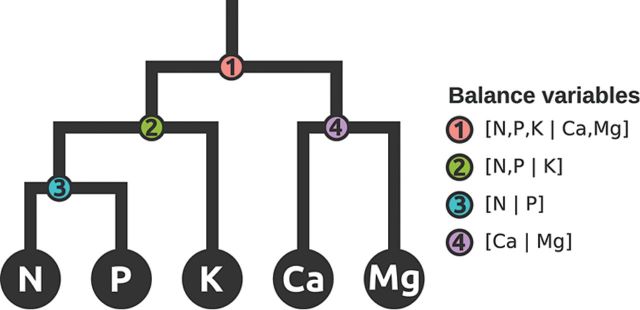
An intriguing new approach uses the nutrient balance concept to create hierarchical sets of relationships between elements based on prior knowledge (Parent et al., 2013a, b). This approach allows for an element to have different specified relationships with different elements and to test these balances in the context of all the other relationships. A simple example of this approach is detailed in Parent et al. (2013a, b), where they model four balances for five elements as shown in Fig. 2. An obvious disadvantage of this approach is that it relies on prior knowledge to define the relationships, but the authors state that unbiased ways of defining the balances are possible and under development.
Fig. 2.
Mobile-and-fulcrums at mass equilibration point illustrates four hierarchically nested balances that represent a subcomposition or subspace of nutrients in the ionome. Fig. 1 from Parent et al. (2013a; The plant ionome revisited by the nutrient balance concept. Frontiers in Plant Science 4, 39, doi: 10.3389/fpls.2013.00039).
Each dataset of 20+ elemental traits is already difficult to comprehend. Attempting to take all of these approaches would transform them into dozens if not hundreds of composite traits, making interpretation of the data within a biological context intractable. So we are left with the question of how to sort through these approaches to prioritize those that are most useful. From my perspective, genetics holds the key to resolving these questions. The approaches that we should use going forward are those that produce heritable results, which will allow for the detection of novel loci, leading to the identification of new genes and gene by environment interactions underlying elemental accumulation. The other important principle is that we should publish, curate, and maintain repositories of ionomic data so that researchers, statisticians, and bioinformaticians can develop, test, and publish their new ideas for analysing the data. The data exchange at www.ionomicshub.org was designed with this exact purpose in mind and contains >250 000 samples worth of data.
Conclusion
I hope that this review has demonstrated that the ionome is not simply a collection of 20+ independent elements. There are clear relationships between the elements, some with known causes and some that are yet to be discerned. While there are several promising approaches, and more under development, we have certainly not arrived at the best solution, and different questions may require different solutions.
References
- Baxter I, Hosmani PS, Rus A, Lahner B, Borevitz JO, Muthukumar B, Mickelbart MV, Schreiber L, Franke RB, Salt DE. 2009. Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genetics 5, e1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter IR, Ziegler G, Lahner B, Mickelbart MV, Foley R, Danku J, Armstrong P, Salt DE, Hoekenga OA. 2014. Single-kernel ionomic profiles are highly heritable indicators of genetic and environmental influences on elemental accumulation in maize grain (Zea mays). PLoS One 9, e87628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi M, Rus A, Salt DE. 2011. Loss-of-function of Constitutive Expresser of Pathogenesis Related Genes5 affects potassium homeostasis in Arabidopsis thaliana . PLoS One 6, e26360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw HD. 2005. Mutations in CAX1 produce phenotypes characteristic of plants tolerant to serpentine soils. New Phytologist 167, 81–88. [DOI] [PubMed] [Google Scholar]
- Broadley MR, Hammond JP, King GJ, et al. 2008. Shoot calcium and magnesium concentrations differ between subtaxa, are highly heritable, and associate with potentially pleiotropic loci in Brassica oleracea . Plant Physiology 146, 1707–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher E, Achberger T, Amusan I, et al. 2010. Natural genetic variation in selected populations of Arabidopsis thaliana is associated with ionomic differences. PLoS One 5, e11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bus A, Korber N, Parkin IA, Samans B, Snowdon RJ, Li J, Stich B. 2014. Species- and genome-wide dissection of the shoot ionome in Brassica napus and its relationship to seedling development. Frontiers in Plant Science 5, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Gable K, Chen M, et al. 2011. Sphingolipids in the root play an important role in regulating the leaf ionome in Arabidopsis thaliana . The Plant Cell 23, 1061–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao D-Y, Dilkes B, Luo H, Douglas A, Yakubova E, Lahner B, Salt DE. 2013. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 341, 658–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Watanabe T, Shinano T, Osaki M. 2009. Rapid characterization of plant mutants with an altered ion-profile: a case study using Lotus japonicus . New Phytologist 181, 795–801. [DOI] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML. 2002. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. The Plant Cell 14, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide DJ, Clark S, Nair TM, Gehl M, Gribskov M, Guerinot ML, Harper JF. 2005. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae . Genome Biology 6, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandilyan A, Barboza L, Tisné S, Grainer C, Reymond M, Koorneef M, Schat H, Aarts MGM. 2009. a . Genetics analysis identifies quantitative trait loci controlling rosette mineral concentrations in Arabidopsis thaliana under drought. New Phytologist 184, 180–192. [DOI] [PubMed] [Google Scholar]
- Ghandilyan A, Ilk N, Hanhart C, et al. 2009. b . A strong effect of growth medium and organ type on the identification of QTLs for phytate and mineral concentrations in three Arabidopsis thaliana RIL populations. Journal of Experimental Botany 60, 1409–1425. [DOI] [PubMed] [Google Scholar]
- Hermans C, Porco S, Verbruggen N, Bush DR. 2010. Chitinase-like protein CTL1 plays a role in altering root system architecture in response to multiple environmental conditions. Plant Physiology 152, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE. 2013. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proceedings of the National Academy of Sciences, USA 110, 14498–14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. 1999. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Molecular Biology 40, 37–44. [DOI] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, et al. 2003. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana . Nature Biotechnology 21, 1215–1221. [DOI] [PubMed] [Google Scholar]
- Malinouski M, Hasan NM, Zhang Y, Seravalli J, Lin J, Avanesov A, Lutsenko S, Gladyshev VN. 2014. Genome-wide RNAi ionomics screen reveals new genes and regulation of human trace element metabolism. Nature Communications 5, 3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani-Ueno N, Yamaji N, Zhao FJ, Ma JF. 2011. The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. Journal of Experimental Botany 62, 4391–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J, Baxter IR, Lee J, Li L, Lahner B, Grotz N, Kaplan J, Salt DE, Guerinot ML. 2009. The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. The Plant Cell 21, 3326–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent SE, Parent LE, Egozcue JJ, et al. 2013. a . The plant ionome revisited by the nutrient balance concept. Frontiers in Plant Science 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent SE, Parent LE, Rozane DE, Natale W. 2013. b . Plant ionome diagnosis using sound balances: case study with mango (Mangifera indica). Frontiers in Plant Science 4, 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister A, Barberon M, Alassimone J, et al. 2014. A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. Elife 3, e03115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau C, Loubet S, Lefoulon C, Chalies C, Fizames C, Lacombe B, Ferrand M, Loudet O, Berthomieu P, Richard O. 2012. Natural variation at the FRD3 MATE transporter locus reveals cross-talk between Fe homeostasis and Zn tolerance in Arabidopsis thaliana . PLoS Genetics 8, e1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson SRM, Tarpley L, Yan W, Yeater K, Lahner B, Yakubova E, Huang X-Y, Zhang M, Guerinot ML, Salt DE. 2015. Worldwide genetic diversity for mineral element concentrations in rice grain. Crop Science 55, 294–311. [Google Scholar]
- Rogers EE, Eide DJ, Guerinot ML. 2000. Altered selectivity in an Arabidopsis metal transporter. Proceedings of the National Academy of Sciences, USA 97, 12356–12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran RP, Huguet T, Grusak MA. 2009. Identification of QTL affecting seed mineral concentrations and content in the model legume Medicago truncatula . Theoretical and Applied Genetics 119, 241–253. [DOI] [PubMed] [Google Scholar]
- Tian H, Baxter IR, Lahner B, Reinders A, Salt DE, Ward JM. 2010. Arabidopsis NPCC6/NaKR1 is a phloem mobile metal binding protein necessary for phloem function and root meristem maintenance. The Plant Cell 22, 3963–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth JL, Sumner ME. 1987. The diagnosis and recommendation integrated system (DRIS). Advances in Soil Science 6, 149–188. [Google Scholar]
- Wu J, Yuan Y, Zhang X, et al. 2008. Mapping QTLs for mineral accumulation and shoot dry biomass under different Zn nutritional conditions in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant and Soil 310, 25–40. [Google Scholar]
- Yu D, Danku JM, Baxter I, Kim S, Vatamaniuk OK, Vitek O, Ouzzani M, Salt DE. 2012. High-resolution genome-wide scan of genes, gene-networks and cellular systems impacting the yeast ionome. BMC Genomics 13, 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Pinson SR, Tarpley L, Huang XY, Lahner B, Yakubova E, Baxter I, Guerinot ML, Salt DE. 2014. Mapping and validation of quantitative trait loci associated with concentrations of 16 elements in unmilled rice grain. Theoretical and Applied Genetics 127, 137–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FJ, Ma JF, Meharg AA, McGrath SP. 2009. Arsenic uptake and metabolism in plants. New Phytologist 181, 777–794. [DOI] [PubMed] [Google Scholar]
- Zhao XQ, Mitani N, Yamaji N, Shen RF, Ma JF. 2010. Involvement of silicon influx transporter OsNIP2;1 in selenite uptake in rice. Plant Physiology 153, 1871–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler G, Terauchi A, Becker A, Armstrong P, Hudson K, Baxter I. 2013. Ionomic screening of field-grown soybean identifies mutants with altered seed elemental composition. The Plant Genome 6, No. 2. [Google Scholar]