Abstract
We present a multiphoton microscopy approach with clearing optimized for pathology evaluation producing image quality comparable to traditional histology. Use of benzyl alcohol/benzyl benzoate with 4',6-diamidino-2-phenylindole and eosin in an optimized imaging setup results in optical sections nearly indistinguishable from traditionally-cut sections. Application to human renal tissue demonstrates diagnostic-level image quality can be maintained through 1 millimeter of tissue. Three dimensional perspectives reveal changes of glomerular capsule cells not evident on single sections. Collagen-derived second harmonic generation can be visualized through entire biopsies. Multiphoton microscopy with clearing has potential for increasing the yield of histologic evaluation of biopsy specimens.
OCIS codes: (180.0180) Microscopy, (180.4315) Nonlinear microscopy, (180.6900) Three-dimensional microscopy
1. Introduction
Despite the substantial clinical utility of standard histology techniques, the traditional process of tissue embedding, sectioning, and staining suffers from significant drawbacks including low sensitivity for sparse lesions, difficult quantitation, a limited two-dimensional perspective, and visual artifacts of preparation. Multiphoton microscopy (MPM) has been proposed as an alternative for clinical histologic evaluation that can be performed in vivo [1] or ex vivo [2] with the potential to address many of these shortcomings. However, in their standard implementation optical sectioning techniques are themselves limited to relatively shallow depths, which is further restricted to about 100 µm in tissue fixed for preservation by formaldehyde [3]. Recently, this limitation has been addressed by the advancement of chemical clearing. Clearing minimizes refractive index differences responsible for the high scattering of tissue samples so that the imaging depth (and three dimensional capability) of optical sectioning methods can be dramatically improved. The potential histologic importance of this 3D perspective can be seen in published investigative efforts that have resorted to serial sectioning of tissue samples, including renal tissue [4]. This manual analysis has shown that changes in renal histology not noted on individual slices are evident with serial sectioning, albeit at the cost of significant additional work, precluding its use in routine clinical evaluation. With these efforts in mind, we have sought to extend the technique of MPM with clearing to human tissue for investigative and clinical renal disease evaluation with the aim of producing images that are of sufficient quality for definitive pathologist evaluation while yielding the additional 3D perspective.
Although fluorescence-based MPM is expected to produce a volumetric view, development of clearing-MPM for primary diagnostic clinical evaluation by pathologists, i.e. as a substitute for traditional techniques, requires generation of images that are at least comparable in contrast and resolution to slide-based microscopy. Much of routine histologic evaluation by pathologists is performed with a 20x objective, but primary diagnosis is generally not performed without access to at least a 40x objective. Thus, definitive diagnosis typically requires a numerical aperture of approximately 0.65 to 0.95, corresponding to resolution ranging from 420 nm down to 290 nm. Further image clarity is ensured by use of slices that are as thin as possible. Target thickness is typically 5 μm or less, as thicker sections lead to blurry and overlapping nuclear and other cytologic features that are critical for accurate morphologic assessment. Optimally, images from optical sectioning microscopy would also recreate the stain characteristics that the clinicians are familiar with, allowing pathologists to employ and build on the accumulated knowledge of traditional pattern recognition in histologic evaluation that has evolved over more than a century of field practice. Finally, a practical method for clinical use requires low toxicity components and simple processing. Such features are not trivially achieved. Although a rigorous analysis of resolution is not available for most clearing approaches, many are likely to lack the resolution possible with 40x or even 20x light microscopy on tissue slices, particularly when evaluating depths greater than tens of microns. For example, recently developed clearing agents that have been applied for deep optical slicing such as Scale, a urea-based solution [5]; SeeDB, based on saturated fructose solutions [6]; and ClearT, using formamide [7], are not good candidates for clinical use due to either very slow processing times (up to weeks) and/or refractive indices lower than 1.51 resulting in limited index-matching and incomplete clearing. While these may be suitable for studies of large-scale structure at low magnification, obtaining optical sections suitable for histology requires the best possible matching between tissue and clearing medium. Residual differences in refractive index lead to scattering and aberrations that degrade the point-spread function at depth. Axial resolution is affected most strongly, particularly when using high numerical aperture objectives, reducing clarity of optical sections and three dimensional reconstructions [8]. The alternative clearing method known as Clarity that employs hydrogel scaffolds and electrophoresis to remove lipids [9] affords excellent index matching, but is too labor intensive and tissue altering for practical clinical histology. Also, implementations of clearing microscopy have typically lacked the specific combination of nuclear and protein channels that produce images analogous to hematoxylin and eosin (H&E) stained sections, making them less easily interpretable by pathologists.
Another recently developed clearing approach using tetrahydrofuran (THF) and dibenzyl ether known as 3DISCO has improved index matching [10]. It was developed mainly to address reports of reduced fluorescence of genetically encoded fluorescence proteins with the clearing agent solution of benzyl alcohol/benzyl benzoate in a 1:2 ratio (BABB), itself a modification by Murray [11] of a clearing technique that dates back 100 years to Spalteholz [12]. However, BABB has properties of high refractive index, relatively rapid clearing, and compatibility with H&E-like fluorescent dye combinations, as well as a benign safety profile (compared to THF peroxide risk) that make it an excellent option for clinical application. In this article, we describe an approach for clearing-MPM that is optimized for pathologist evaluation of tissue based on a combination of fluorescent stain and clearing agent choice, optical system selection, and post-imaging processing that results in images that are essentially indistinguishable from traditional H&E. In addition, we present new observations of renal tissue injury that are made possible only through three-dimensional reconstructions, with implications for enhanced evaluation of kidney disease.
2. Methods
2.1 Tissue samples and histology
Biopsy-sized tissue portions were extracted from fixed human autopsy renal specimens and kidney resections using a 1.2 mm diameter biopsy needle. Tissues had been previously fixed in formalin for periods ranging from 24 hours to 12 months. Samples were processed for clearing using a simplified two-step process comprised of an approximately four-hour methanol dehydration followed by at least 1 hour immersion in benzyl alcohol/benzyl benzoate in a 1:2 ratio (BABB), with both steps performed under agitation and heating to 42°C. Fluorescent staining was performed during the dehydration step. A combination of Eosin Y (Thermo Fisher Scientific, Waltham, MA), concentration 0.5% by volume, and 4',6-diamidino-2-phenylindole (DAPI) dilactate (Biotium, Freemont CA), concentration 90 mM, was used for images presented here.
2.2 Imaging system and image processing
Samples were imaged on a home-built multiphoton microscope based on a Ti-Sapphire laser (80MHz repetition rate, <100 fs pulse length) with galvanometer scanning. Image acquisition used Scanimage [13] software modified for automated tiling with a motorized stage, and a Leica objective (HCX Apo L20x/0.95 IMM) designed for immersion in BABB. Use of a high NA objective is critical to obtain thin optical sections, but necessitates using a lens and immersion fluid matched to the clearing solution, as the axial resolution is otherwise severely degraded by spherical aberration. An objective equipped with a correction collar can also yield good results, but requires repeated adjustment when imaging thick samples. Data was collected at a pixel size of approximately 0.29 µm. Excitation wavelength was either 780 or 800 nm. Second harmonic generation was detected in transmission. Single slices could be imaged at a rate of approximately 1 mm2 per minute, while imaging an entire biopsy sample with a slice separation of 5 microns required 30 hours.
Image processing consisted of normalization of contrast and brightness, stitching, and pseudo-coloring of fluorescence. The dyes used were a combination of a nuclear dye DAPI (analogous to hematoxylin) and a general protein dye (eosin). After normalization, images were pseudo-colored by conversion of fluorescence intensity values to RGB optical densities (inverse color deconvolution), as previously described [2]. ImageJ based FIJI software plugins and custom python routines were employed. For reconstructions of Bowman’s capsule, images were obtained at 1 µm z-steps. Signal corresponding to glomerulus was deleted for clear visualization, by manually drawing a contour on each slice using ImageJ. Due to the empty space within the capsule, segmentation is relatively straightforward and tolerant of imprecision, although the process still requires approximately 30 minutes per capsule. A z-step size of 5 µm was used for complete specimen 3D reconstructions. Visualization was done with ImageJ and Drishti software [14].
Traditional H&E section images were collected on an Olympus BX-2 microscope equipped with a 100x PLAN 1.25 oil immersion objective and using a SPOT Insight 4 megapixel camera.
3. Results
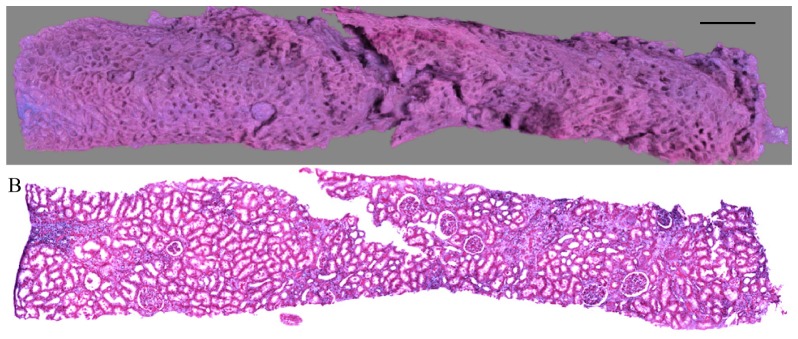
An entire, un-sectioned renal biopsy sample could be imaged using MPM with BABB clearing, corresponding to an imaging depth of greater than 1 mm (Fig. 1(a)). Diagnostic-quality images were collected throughout the uncut and un-embedded specimen using the MPM/clearing technique with the combination of nuclear and protein staining and subsequent processing using the pseudo-coloring algorithm (Fig. 1(b)).
Fig. 1.
A) 3D renal biopsy reconstruction using MPM with clearing. Scale bar = 500 µm. B) Corresponding multiphoton image obtained at depth of 400 μm, pseudo-colored to mimic H&E.
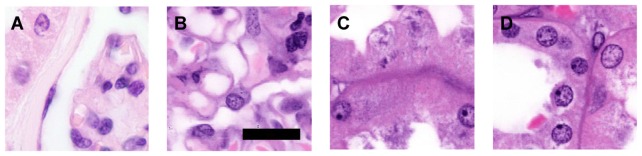
Contrast and resolution were equivalent to traditionally processed H&E sections, while avoiding folding or microtome cutting artifacts and preserving the entire tissue for additional analyses. Compared to traditional slide preparation, which typically uses 3-5 µm tissue sections, the optical sections of approximately 2 µm thickness provide superior clarity and contrast (Fig. 2). Full magnification images permit detection of fine structures such as glomerular capillary walls, cytoplasmic granules, and nuclear detail including membranes, nucleoli, and chromatin patterns (Fig. 2(b)-2(d)).
Fig. 2.
A) Traditionally processed kidney H&E section using 100x lens. B-D) Pseudo-colored MPM with clearing. Range of fine features recognizable include B) glomerular capillary walls, C) cytoplasmic granules, and D) nuclear membranes and chromatin patterns. Scale bar – 30 µm.
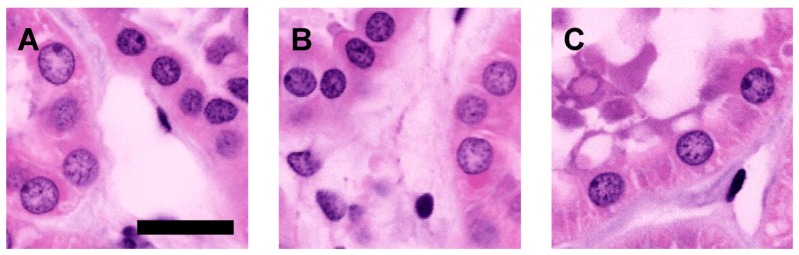
Image quality is not noticeably reduced throughout an imaging depth of 1 mm (Fig. 3). High magnification SHG images of collagen filaments at a depth of 1 mm were used to determine the optical resolution of the system. The measured values (FWHM) were 440 nm laterally and 2.1 microns axially, close to the theoretical values [15] of 370 nm and 1.7 microns for this numerical aperture and imaging wavelength.
Fig. 3.
Pseudo-colored MPM images on cleared renal tissue show preservation of clarity and contrast throughout entire core renal biopsies. Depths - A) 200 µm, B) 600 µm, C) 1000 µm. Scale bar – 40 µm.
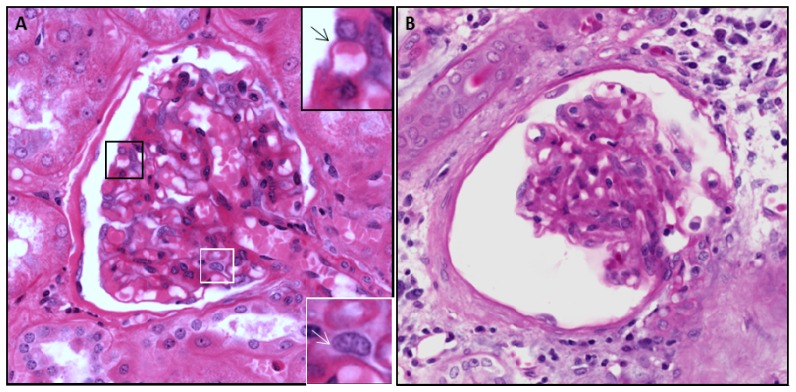
In addition to being able to closely mimic H&E staining, the digital two-channel images offer the ability for individual channel selection, color variation, contrast enhancement, and gamma adjustment that can highlight specific features of diagnostic interest such as mesangial cells, distinguishable by their gray cytoplasm (white inset, Fig. 4(a)), and typically very difficult to visualize with standard H&E. The resolution is also sufficient to allow assessment of thickness of capillary loops (black inset/arrow, Fig. 4(a)), normally less than 400 nm in thickness, and a component of routine renal biopsy evaluation requiring high resolution.
Fig. 4.
A). Renal glomerulus using MPM/clearing at 300 µm depth. Black inset shows capillary loop (black arrow) with luminal RBC. White inset shows visualization of mesangial cell (white arrow) facilitated by thin optical sectioning and high color contrast. B) Atubular glomerulus cross-section using MPM/clearing. Glomerular changes and surrounding connective tissue and edema are noted, but the functional status based on parietal epithelial cells and tubular connection cannot be assessed in a single section. Dimensions – 350 x 350 µm.
The three dimensional perspective also allowed visualization of glomerular capsules in a manner that is not possible with traditional two-dimensional sectioning histology. There has been considerable recent interest among renal disease investigators in characterizing cells known as parietal epithelial cells (PEC) that line the peri-glomerular capsule (Bowman’s capsule) [16]. However, single two-dimensional sections do not reliably reveal distinctive differences in the capsular lining cells of diseased glomeruli (Fig. 4(b)). By contrast, comparison of three-dimensional reconstructions of a normal glomerular capsule and a damaged glomerular capsule demonstrated marked morphologic changes in the distribution and morphology of PEC nuclei on the surface of the glomerular capsule (Fig. 5). Furthermore, although the existence of glomeruli without tubular connection (atubular glomeruli) has been described in carefully executed manual serial sectioning experiments [1], research into their prevalence and pathophysiologic role has been hampered by the lack of accessible tools for routine visualization, which is made possible by the 3D reconstructions.
Fig. 5.
Reconstructions in 3D of a normal (A) and atubular (B) capsule showing changes in the nuclei of the parietal epithelial cells lining the capsule that are not apparent on two dimensional histologic sections. Glomeruli have been digitally removed. Sections correspond to Fig. 4(a) and 4(b), respectively. Block dimensions are approximately 400x400x150μm. Arrows in (A) indicate proximal tubule connection.
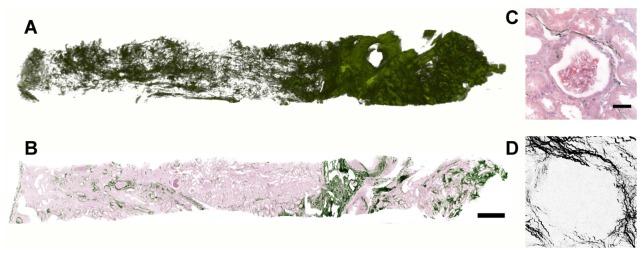
In addition, simultaneous collection of second harmonic generation signals allowed characterization of the collagen matrix organization throughout the entire specimen (Fig. 6(a)), a feature of significant potential diagnostic importance. Although heterogeneous patterns of collagen are recognizable even from a single section (Fig. 6(b)), full thickness imaging offers advantages for analysis and visualization. As collagen is sparse in many regions (Fig. 6(c)), averaging multiple slices will improve accuracy of quantitation. Similarly, patterns which are difficult to recognize from a single slice may become more apparent when viewed as a stack, 3D rendering or projection (Fig. 6(d)).
Fig. 6.
A). 3D reconstruction of collagen in human kidney biopsy using MPM/SHG in cleared tissue. B) Single MPM section showing two dimensional collagen arrangement (green) with modified background pseudo H&E coloring. (Scale bar – 500 µm) C) Higher magnification view of B showing sparse collagen SHG around glomerulus. (Scale bar – 50 µm) D) Z-projection over depth of 200 µm within the same glomerulus as in C.
4. Discussion
The promise of optical sectioning microscopy for pathologist evaluation in clinical settings depends critically on the production of high quality images at depth. While much recent emphasis has been placed on in vivo/in situ evaluation of tissue for microscopic visualization, the inability to physically alter the optical properties of live tissue remains a barrier to contrast and resolution. As a result, images created with available in vivo approaches are of insufficient quality for adequate definitive diagnosis, thus necessitating secondary evaluation by traditional wax embedding. On the other hand, the use of optical clearing and application of extrinsic dyes enables contrast and resolution with ex vivo samples that in some ways surpasses that achievable with traditional histology preparations. Notably, clearing techniques differ in their capability to produce these high resolution images, primarily due to refractive index differences. The data presented here using the combination of MPM with BABB clearing illustrates that images of sufficient clarity for primary diagnostic evaluation are accessible without wax-embedding or tissue sectioning. Key elements are inferred to be proper matching of the clearing agent refractive index to endogenous structures, and adequate matching of the optical chamber and imaging system to the clearing fluid. These result in the highest possible lateral and axial resolution, the latter of which is of paramount importance for best clarity of images.
In this application to human renal tissue, evaluating entire renal biopsy specimens augments considerably the number of glomeruli that can be visually inspected, an aspect of important practical consequence where guidelines for a minimum number of glomeruli for evaluation often necessitate a biopsy of additional tissue. The virtual serial sectioning also ensures that sparse lesions of potential diagnostic importance are identified. It is commonly appreciated by renal pathologists that glomerular lesions characteristic of diseases such as diabetes (e.g. Kimmelstein-Wilson nodules) can be sparse and may be missed on evaluation of single biopsy slices. Likewise, inflammatory aggregates and increased fibrosis may be restricted to uncut portions of a biopsy, but visible with complete virtual sectioning. In addition, the high resolution three dimensional reconstructions of glomerular capsules show changes in the pattern of parietal epithelial cells of atubular glomeruli not recognizable on single sections. Although this association must be further evaluated through detailed clinical/anatomic studies, it illustrates the added capability of the MPM/clearing technique in revealing abnormalities not evident on single sections.
Furthermore, the combination of clearing with SHG shows significant promise for additional qualitative and more quantitative assessment of collagen patterns. As a large volume extension of prior work describing the clinical importance of SHG for collagen assessment [17], this technique could address recognized limitations in the evaluation of collagen fibrosis in renal and other tissue specimens.
While the results presented here are limited to renal tissue, we have found MPM/clearing to produce good results in a broad variety of tissue types. Some reduction in clarity with depth has been observed in axonally dense tissue such as spinal cord, likely due to incomplete index-matching, while opaque particles in samples such as bone marrow can create shadowed regions. Remaining challenges for clinical implementation relate primarily to speed. Although a total preparation time of approximately 6 hours is comparable to traditional processing, improvements in both processing time and imaging time are likely to be important for practical integration in a pathology laboratory workflow.
While diffusion of fixative, dyes, and clearing agents sets a limit on the processing time for thick samples, several avenues exist for decreasing imaging time. Speed can likely be increased by an order of magnitude through optimization of the imaging software, improved motorized stage scanning, and incorporation of technologies such as resonant galvonometer [18] or polygon scanners [19]. Utility may also be increased by adopting alternative scanning strategies. Current imaging was carried out either by exhaustive scanning of the sample, or by “live” scanning followed by imaging of a small volume. Alternatively, a number of entire sections of a sample separated by 50-100 microns could be imaged in a short (30-60 minute) session at intermediate or low resolution, followed by human or automated review to locate structures of interest which are then imaged at full resolution.
In conclusion, we propose that the use of MPM with BABB clearing, SHG, and a combination of nuclear and protein contrast dyes, coupled with an index-matched objective provides images with equivalent or superior contrast and resolution to traditionally processed kidney samples, while adding a three dimensional perspective that offers additional insights, opening an avenue for investigational studies and clinical assessment of these and other biopsy samples.
Disclosure
RT and MJL have an ownership interest in Applikate Technologies, a company developing technology for histology of un-embedded specimens.
References and links
- 1.Zipfel W. R., Williams R. M., Christie R., Nikitin A. Y., Hyman B. T., Webb W. W., “Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation,” Proc. Natl. Acad. Sci. U.S.A. 100(12), 7075–7080 (2003). 10.1073/pnas.0832308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres R., Vesuna S., Levene M. J., “High-resolution, 2- and 3-dimensional imaging of uncut, unembedded tissue biopsy samples,” Arch. Pathol. Lab. Med. 138(3), 395–402 (2014). 10.5858/arpa.2013-0094-OA [DOI] [PubMed] [Google Scholar]
- 3.Centonze V. E., White J. G., “Multiphoton excitation provides optical sections from deeper within scattering specimens than confocal imaging,” Biophys. J. 75(4), 2015–2024 (1998). 10.1016/S0006-3495(98)77643-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcussen N., “Atubular glomeruli and the structural basis for chronic renal failure,” Lab. Invest. 66(3), 265–284 (1992). [PubMed] [Google Scholar]
- 5.Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., Noda H., Fukami K., Sakaue-Sawano A., Miyawaki A., “Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain,” Nat. Neurosci. 14(11), 1481–1488 (2011). 10.1038/nn.2928 [DOI] [PubMed] [Google Scholar]
- 6.Ke M.-T., Fujimoto S., Imai T., “SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction,” Nat. Neurosci. 16(8), 1154–1161 (2013). 10.1038/nn.3447 [DOI] [PubMed] [Google Scholar]
- 7.Kuwajima T., Sitko A. A., Bhansali P., Jurgens C., Guido W., Mason C., “ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue,” Development 140(6), 1364–1368 (2013). 10.1242/dev.091844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young P. A., Clendenon S. G., Byars J. M., Dunn K. W., “The effects of refractive index heterogeneity within kidney tissue on multiphoton fluorescence excitation microscopy,” J. Microsc. 242(2), 148–156 (2011). 10.1111/j.1365-2818.2010.03448.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung K., Deisseroth K., “CLARITY for mapping the nervous system,” Nat. Methods 10(6), 508–513 (2013). 10.1038/nmeth.2481 [DOI] [PubMed] [Google Scholar]
- 10.Ertürk A., Becker K., Jährling N., Mauch C. P., Hojer C. D., Egen J. G., Hellal F., Bradke F., Sheng M., Dodt H. U., “Three-dimensional imaging of solvent-cleared organs using 3DISCO,” Nat. Protoc. 7(11), 1983–1995 (2012). 10.1038/nprot.2012.119 [DOI] [PubMed] [Google Scholar]
- 11.Dent J. A., Polson A. G., Klymkowsky M. W., “A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus,” Development 105(1), 61–74 (1989). [DOI] [PubMed] [Google Scholar]
- 12.W. Spalteholz, Über das Durchsichtigmachen von menschlichen und tierischen Präparaten und seine theoretischen Bedingungen, nebst Anhang: Über Knochenfärbung (S. Hirzel, 1914). [Google Scholar]
- 13.Pologruto T. A., Sabatini B. L., Svoboda K., “ScanImage: flexible software for operating laser scanning microscopes,” Biomed. Eng. Online 2(1), 13 (2003). 10.1186/1475-925X-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. Limaye, “Drishti - Volume Exploration and Presentation Tool” (2006), retrieved https://sf.anu.edu.au/Vizlab/drishti/index.shtml.
- 15.Zipfel W. R., Williams R. M., Webb W. W., “Nonlinear magic: multiphoton microscopy in the biosciences,” Nat. Biotechnol. 21(11), 1369–1377 (2003). 10.1038/nbt899 [DOI] [PubMed] [Google Scholar]
- 16.Shankland S. J., Smeets B., Pippin J. W., Moeller M. J., “The emergence of the glomerular parietal epithelial cell,” Nat. Rev. Nephrol. 10(3), 158–173 (2014). 10.1038/nrneph.2014.1 [DOI] [PubMed] [Google Scholar]
- 17.Chen X., Nadiarynkh O., Plotnikov S., Campagnola P. J., “Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure,” Nat. Protoc. 7(4), 654–669 (2012). 10.1038/nprot.2012.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen Q. T., Callamaras N., Hsieh C., Parker I., “Construction of a two-photon microscope for video-rate Ca(2+) imaging,” Cell Calcium 30(6), 383–393 (2001). 10.1054/ceca.2001.0246 [DOI] [PubMed] [Google Scholar]
- 19.Kim K. H., Buehler C., So P. T., “High-speed, two-photon scanning microscope,” Appl. Opt. 38(28), 6004–6009 (1999). 10.1364/AO.38.006004 [DOI] [PubMed] [Google Scholar]