Abstract
Receptor activator of nuclear factor kappa-B ligand (RANKL) is an essential mediator of osteoclast formation, function and survival. In patients with solid tumor metastasis to the bone, targeting the bone microenvironment by inhibition of RANKL using denosumab, a fully human monoclonal antibody (mAb) specific to RANKL, has been demonstrated to prevent tumor-induced osteolysis and subsequent skeletal complications. Recently, a prominent functional role for the RANKL pathway has emerged in the primary bone tumor giant cell tumor of bone (GCTB). Expression of both RANKL and RANK is extremely high in GCTB tumors and denosumab treatment was associated with tumor regression and reduced tumor-associated bone lysis in GCTB patients. In order to address the potential role of the RANKL pathway in another primary bone tumor, this study assessed human RANKL and RANK expression in human primary osteosarcoma (OS) using specific mAbs, validated and optimized for immunohistochemistry (IHC) or flow cytometry.
Our results demonstrate RANKL expression was observed in the tumor element in 68% of human OS using IHC. However, the staining intensity was relatively low and only 37% (29/79) of samples exhibited≥10% RANKL positive tumor cells. RANK expression was not observed in OS tumor cells. In contrast, RANK expression was clearly observed in other cells within OS samples, including the myeloid osteoclast precursor compartment, osteoclasts and in giant osteoclast cells. The intensity and frequency of RANKL and RANK staining in OS samples were substantially less than that observed in GCTB samples. The observation that RANKL is expressed in OS cells themselves suggests that these tumors may mediate an osteoclastic response, and anti-RANKL therapy may potentially be protective against bone pathologies in OS. However, the absence of RANK expression in primary human OS cells suggests that any autocrine RANKL/RANK signaling in human OS tumor cells is not operative, and anti-RANKL therapy would not directly affect the tumor.
Abbreviations: APC, allophycocyanin; ATCC, American Type Culture Collection; cDNA, complementary deoxyribonucleic acid; ELISA, enzyme linked immunosorbent assay; FACS, fluorescence-activated cell sorting; FBS, fetal bovine serum; FFPE, formalin-fixed, paraffin-embedded; GCTB, giant cell tumor of bone; IgG1, immunoglobulin G1; IHC, immunohistochemistry; ISH, in situ hybridization; LN, lymph node; mAb, monoclonal antibody; mRNA, messenger ribonucleic acid; OS, osteosarcoma; RANK, receptor activator of nuclear factor kappa-B; RANKL, receptor activator of nuclear factor kappa-B ligand; RNA, ribonucleic acid; RT-PCR, reverse transcriptase polymerase chain reaction
Keywords: RANK, RANKL, Human osteosarcoma, Antibodies, Protein expression, Immunohistochemistry
1. Introduction
Osteosarcoma (OS) is the most common primary malignant tumor of the bone. This neoplasm is defined histologically by osteoid deposition by the malignant mesenchymal cells [1]. In OS, current evidence supports an osteoblastic population as the cell of origin [2], although the distinct histological subtypes (e.g. osteoblastic, fibroblastic, chondroblastic and telangiectactic) indicate potential for heterogeneous origins. In association with varying degrees of bone matrix deposition, OS is characterized by local bone destruction and frequent lung metastasis. Ten-year survival outcomes for patients with localized OS are approximately 65% and outcomes, along with the standard medical treatment, have not changed substantially in recent years. Patients with recurrent OS have a poor prognosis and there is great desire to develop improved therapies [3].
In solid tumors which have metastasized to bone or in giant cell tumor of bone (GCTB), lytic bone destruction is mediated by osteoclasts. Osteoclasts are highly specialized cells derived from the monocyte/macrophage lineage necessary for the degradation of the organic and inorganic matrices of bone. Receptor activator of nuclear factor kappa-B ligand (RANKL), a tumor necrosis factor ligand superfamily member, is essential for the formation, activation, and function of osteoclasts. RANKL is expressed by cells of the osteoblast lineage in the bone stroma as well as osteocytes and acts via a paracrine mechanism, binding to its cognate receptor RANK expressed on osteoclasts and osteoclast precursors [4]. Denosumab, a fully human monoclonal antibody (mAb) specific to RANKL, inhibits osteoclastogenesis and osteoclast-mediated bone destruction. In clinical studies, denosumab reduced tumor-induced bone resorption and skeletal complications of metastatic bone disease [5], [6], [7], including delaying the development of bone metastasis in men with castrate-resistant prostate cancer [8], [9].
Osteoclasts have been observed in OS at sites of bone resorption, either at the tumor/bone interface or within the tumor tissue at sites of neoplastic osteoid [10]. Cortical destruction and extension of the tumor mass into the soft tissue is frequently evident in OS patients, suggesting involvement of osteoclasts in associated bone pathologies. The human OS cell line SaOS-2 has been shown to support osteoclastogenesis via RANKL production on the surface of OS cells [11] and RANKL expression has been reported on primary feline, canine [12], and human OS cells [13] with variable frequency. In preclinical studies, animal models of OS have also indicated that RANKL levels increase in tumor-involved bone [14], [15]. Pharmacologic inhibition of RANKL has been shown to prevent increased osteolysis, reduce skeletal tumor growth and reduce lung metastases (often associated with an increased survival) in these models [16], [17], [18]. Osteoclast inhibition, achieved with either RANKL blockade or bisphosphonates, results in similar antitumor and bone-protective effects in these models [19]. These studies support the notion that RANKL, produced within the reactive bone stroma and potentially within OS cells themselves, contributes to OS-mediated bone degradation/lysis. In addition to potential alterations in RANKL in OS, RANK expression has been reported in mouse and human OS cell lines [20], [21] and in primary human OS [20], [22]. The prevalence of RANKL and RANK expression, as well as any associated prognostic significance, varies considerably in these published reports of human OS.
In a recent characterization of another primary bone tumor, GCTB, we and others have confirmed significant RANKL expression within the GCTB stroma while RANK is expressed within the myeloid-derived giant cell component [23]. In patients with recurrent or unresectable GCTB, treatment with denosumab, was associated with tumor regression and reduced tumor-associated bone lysis [24], [25]. The objective of the present study was to ascertain human RANKL and RANK protein expression in primary human OS using specific mAbs validated and optimized for immunohistochemistry (IHC).
2. Materials and methods
2.1. Tissue samples
Osteosarcoma tissue microarrays (1.5 mm [diameter] cores) were obtained from US Biomax (Rockville, MD). Large tissue sections (slides; 20 mm [diameter] cores) from anonymized OS samples were obtained from Asterand (Detroit, MI) and processed at the Amgen Tissue Bank. One limitation of the present study is the degree of chemotherapy pretreatment prior to sample acquisition or other sampling preparation variables, such as time to sample fixation, are unknown. Any effect of these variables on RANK or RANKL IHC performance is unknown.
2.2. Cell lines and expression analyses
The human cancer cell lines H1299 and COLO205 were obtained from the American Type Culture Collection (ATCC) and cultured in Roswell Park Memorial Institute media supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, and glutamine (Invitrogen). These cells are subsequently referred to as H1299-parental cells and COLO205-parental cells. The human cancer cell lines Karpas, H929, and Ramos were also obtained from ATCC and cultured as described above. Parental cells were then transduced with a retroviral LZRS-pBMNZ vector containing a full-length version of the RANK. Surface expression of RANK was confirmed by flow cytometry analysis using mAbs against human RANK (Amgen mAbs N-2B10, N-1H8, and M331) and a goat, antimouse secondary antibody conjugated to APC. Retroviral transduction methods and the generation of MDA-231-parental and MDA-231-RANK cells are described in Blake et al. [26]. Tumor xenografts were generated in female athymic (R-Foxn1<nu>) nude mice [26].
For RANK mRNA expression analysis, total RNA was isolated from tumor cell lines using the RNAeasy™ Kit (Qiagen, Valencia, CA) and transcribed into cDNA using random hexamer priming and TaqMan™ Reverse Transcription Reagents (Applied Biosystems, Inc., Foster City, CA). Ten nanograms of tumor cell cDNA was analyzed for RANK expression using Assay on Demand™ primer probe set Hs00187189_m1 (Applied Biosystems, Inc.). Quantitative real-time reverse transcriptase polymerase chain reaction (RT-PCR) was performed using the ABI Prism® 7900HT Sequence Detection System (Applied Biosystems, Inc.). Gene expression was calculated relative to β-actin expression (control) and reactions were performed in triplicate. Surface expression of RANK and RANKL was determined by flow cytometry after incubation with either 1 mg/ml of mouse anti-RANK antibody (clones N-1H8, N-2B10, or M331 as indicated), 20 µg/mL mouse anti-human RANKL antibody M366 (Amgen Inc.) or isotype control (BD Biosciences) in 2% FBS followed by APC-conjugated anti-mouse secondary antibody. Fluorescence was assessed using a FACScan sorter (BD Biosciences). For RANK surface antigen density measurement, the quantitative indirect immunofluorescence kit (QIFIkit, Dako, Denmark) using anti-RANK mAb N1-H8 was performed as described [27]. For Western blotting of huRANKL, cells were lysed in K-Lysis buffer (Cell Signaling Technology), total protein quantified using BCA (Thermo Scientific), and analyzed using SDS-PAGE and mAb M366, as described in Blake, 2014. For in situ hybridization (ISH) of huRANKL, antisense and sense control transcripts were radiolabeled and synthesized with 33P-UTP (Amersham; labeling isotope) as described [28]. Slides were counterstained with hematoxylin and eosin (H & E) and imaged using light and darkfield illumination. For IHC of L929 cells (parental or transfected with human RANKL cDNA), cells were embedded in collaplugs, formalin-fixed and paraffin embedded. Four microns sections were cut and antigen retrieval was performed in a pressure cooker via citra buffer prior to staining with 3 µg/mL of M366. Slides were developed using Romulin AEC (Biocare).
2.3. Generation and optimization of anti-huRANK and anti-huRANKL mAbs for IHC
A soluble form of the extracellular huRANK (amino acids 1 to 213 including the signal sequence) was fused to the Fc portion of human immunoglobulin G1 (IgG1). The fusion protein was expressed and purified after transfection of CHO cells, according to standard techniques [29]. Recombinant RANK-Fc was emulsified in Complete Freund’s Adjuvant (Pierce™) and immunized into Balb/c and 129xBL/6 F1 mice (The Jackson Laboratory™). Second and third immunizations were performed at 3-week intervals using the huRANK-antigens suspended in RIBI adjuvant (Sigma™). Ten days following the third immunization, blood samples were collected. Serum and hybridoma supernatants were screened for RANK-Fc binding by ELISA and the top 96 mAbs were expanded in culture and the supernatants collected for purification. Eighty IgGs were tested on FFPE control sections including positive and negative control tumor xenografts (H1299-parental and H1299-RANK) and clinical GCTB samples according to methods as summarized below. Nine of 80 IgGs specifically stained the FFPE positive control xenografts (H1299-RANK) and GCTB osteoclasts without any detectable staining to negative controls (H1299-parental xenografts). From this pool of nine mAbs, binding to membrane expressed huRANK was confirmed by FACS using the MDA-MB-231-ATCC LUCI-parental and the MDA-MB-231-ATCC LUCI-RANK cell lines described [30]. Anti-RANK mAbs which demonstrated specific binding to surface RANK were then epitope binned according to antibody competition ELISA. Two candidate antibodies (N-1H8 and N-2B10) were selected as they represented distinct epitope binning characteristics (as defined by antibody competition ELISA) and were confirmed to bind RANK by western blots, performed as described [26]. Specificity of anti-RANK mAbs N-1H8 and N-2B10 was confirmed by positive IHC staining to an additional positive control, FFPE xenograft tissue (COLO205-RANK) and negative IHC staining to multiple negative controls, FFPE xenograft tissues (COLO205-parental, Karpas, H929, and Ramos). The specific staining pattern observed with both N-1H8 and N-2B10 by IHC correlated with the detection of surface RANK by flow cytometry using the same antibodies. Finally, the positive and negative expression patterns revealed by N-1H8 and N-2B10 on IHC and flow cytometry on multiple positive (H1299-RANK, COLO205-RANK) and negative controls (H1299-parental and COLO205-parental) were concordant with the flow cytometry pattern for a distinct anti-huRANK mAb (M331) [31]. The anti-huRANKL mAb M366 has been described previously [23].
IHC analysis was carried out on FFPE samples using automated staining and optimized methods as described [23]. To assess expression for each analyte within tumor cells, the H-score approach was used. Briefly, percent of immunostained tumor cells and staining intensity, 0 (negative), 1+ (weak), 2+ (moderate) and 3+ (strong) were scored; an H-score was calculated using the following formula: H-score=(percentage of cells of weak×1)+(percentage of cells of moderate×2)+(percentage of cells of strong×3). The maximum H-score would be 300, corresponding to 100% of cells with strong intensity (3+). The staining of RANKL and RANK within normal bone or infiltrating (non-tumor) cells was determined without quantitative scoring. Microscopic images were photographed using a Nikon™ Eclipse E600 microscope with a Nikon™ DXM1200 digital camera. Resulting images were white-balanced using Adobe™ Photoshop CS software but no additional image modifications were employed.
3. Results
3.1. Development and optimization of anti-RANKL and anti-RANK mAbs for IHC applications
Our strategy for substantiating antibody specificity for both analytes, RANKL and RANK, was achieved by demonstrating concordant signals between the antibody used for IHC application with independent expression analyses using multiple methodologies, as outlined below. For validation and optimization of the anti-RANKL antibody M366 for IHC applications, specificity controls were first performed in mouse L929 cells and analyzed by IHC, western blotting and flow cytometry. Upon M366 analysis, a positive signal for each independent assay was demonstrated in L929 cells transfected with human RANKL cDNA (L929 huRANKL); an undetectable signal was observed in untransfected cells (L929-parental; negative control; Fig. 1).
Fig. 1.
huRANKL antibody validation for IHC methods. (A) Anti-huRANKL mAb M366 IHC reveals specific signal in mouse L929 cells transduced with huRANKL cDNA (L929 huRANKL), but not parental L929 cells (L929 control). (B) Similarly, analysis of dissociated proteins on western blot using mAb 366 of L929 huRANKL cells detected a protein of approximately 45 kDa, the predicted size for full-length human RANKL [28]. Positions of molecular weight markers are illustrated on left (kDa). (C) Determination of RANKL cell surface protein expression on L929 cells was performed using flow cytometry. Expression of RANKL was detected using the M366 mAb and a goat antimouse secondary antibody conjugated to APC. M366 staining on huRANKL transduced L929 cells is indicated with the red line and on parental L929 cells (L929 control) a solid black line. The M366 anti-huRANKL antibody detects a signal by IHC, western blot, and flow cytometry specifically in L929 cells transduced with huRANKL and not parental L929 cells. APC, allophycocyanin; cDNA, complementary deoxyribonucleic acid; IHC, immunohistochemistry; mAb, monoclonal antibody; RANKL, receptor activator of nuclear factor kappa-B ligand.
We also performed IHC on FFPE sections of human lymph node (LN), a tissue known to express RANKL [28], and demonstrated a RANKL signal detectable in the draining cortex. The positive IHC pattern for RANKL overlapped with that using an independent method (in situ hybridization [ISH]) and conversely, regions of the LN without a detectable ISH signal for RANKL were also negative for RANKL staining by IHC (Supplemental Fig. 1). Finally, we utilized GCTB as controls for each RANKL IHC run on test cases, since GCTB is a complex tissue where RANKL is expressed within the stromal element of GCTB but excluded from giant osteoclast cells [23], thus providing an internal positive and negative control for the RANKL IHC (Fig. 2).
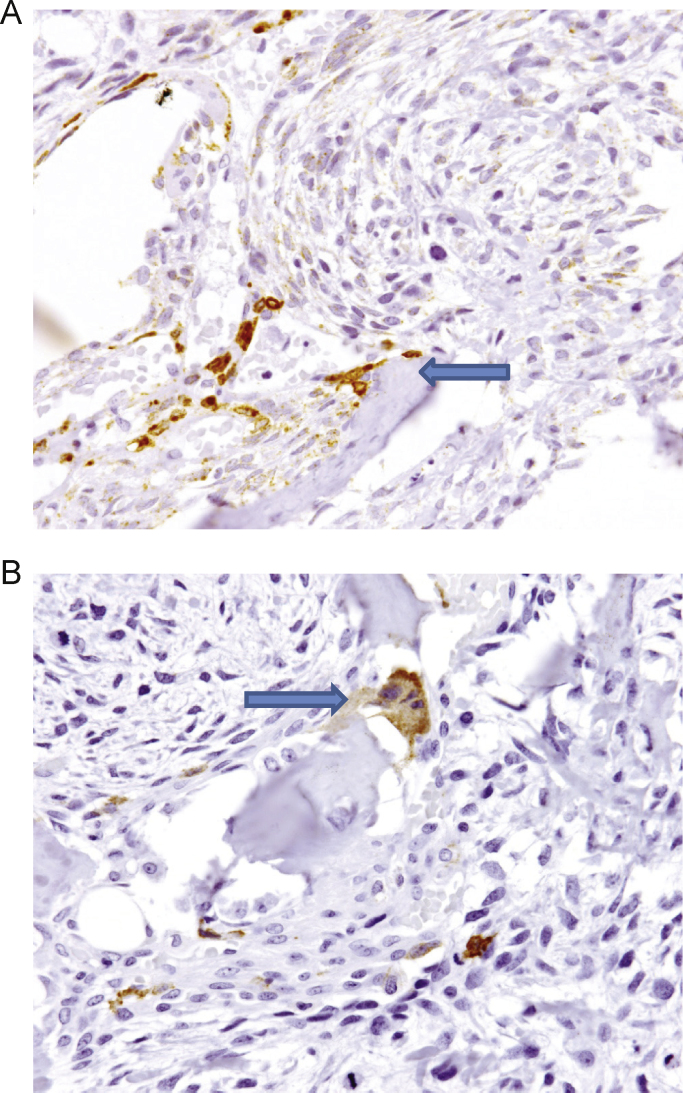
Fig. 2.
RANKL and RANK IHC of GCTB. IHC performed on FFPE section of GCTB, which serves as an internal positive and negative control since RANKL and RANK are expressed in distinct compartments [25]. GCTB is composed of osteoclast-like giant cells and myeloid giant cell precursors that express RANK and stromal tumor cells that express RANKL. (A) The RANKL IHC using M366 clearly indicates stromal tumor cells but is excluded from the giant cell component. (B) Conversely, the RANK IHC using N-2B10 recognizes giant cells and myeloid osteoclast precursors but is excluded from the stromal compartment. FFPE, formalin-fixed, paraffin-embedded; GCTB, giant cell tumor of bone; IHC, immunohistochemistry; RANK, receptor activator of nuclear factor kappa-B; RANKL, RANK ligand.
The development of anti-RANK mAbs suitable for IHC applications relied on screening mAbs directly on formalin-fixed tissues with pre-defined antigen expression, as described in Materials and methods. Specifically, we utilized multiple independent methods (e.g. flow cytometry and RT-PCR) to define human tumor cell lines with undetectable RANK expression (negative control; parental cell lines) and then transfected these cell lines with human RANK cDNA to create positive control cell lines. In order to mimic the procedure and tissue preparation used for IHC application of the antibody in surgical samples, FFPE tissues isolated from tumor xenografts derived from the parental and RANK-transfected cells grown in mice were used. Positive staining of known, RANK-positive cells in clinical samples of GCTB was also required for screening of potential anti-RANK mAbs. In order to proceed to antibody optimization in IHC, we also demonstrated that candidate antibodies had no detectable IHC staining in negative control tumor xenografts and in RANK-negative cells within GCTB (data not shown).
After initial screening steps, several anti-RANK mAbs were identified which demonstrated reproducible, specific and selective staining of human RANK in FFPE tumor tissues. Two high affinity, anti-RANK mAb candidates (clones N-1H8 and N-2B10), were selected for final optimization and represent diverse epitope binding within the RANK extracellular domain (data not shown). Both N-1H8 and N-2B10 mAbs demonstrated specific IHC staining in FFPE sections from two independent, RANK-transfected tumor xenografts (H1299-RANK and COLO205-RANK), while FFPE sections from the negative xenograft controls (untransfected H1299-parental or COLO205-parental; Fig. 3A) or Karpas, H929 or Ramos (Supplemental Fig. 2) did not show any RANK staining. Specificity and selectivity of RANK IHC staining using N-1H8 or N-2B10 mAb were further substantiated by the detection of RANK using independent analyses, including mRNA detection (RT-PCR; Supplemental Table 1) and surface protein detection (flow cytometry; Fig. 3B and Supplemental Fig. 3). As additional tests for specificity, we demonstrated that N-1H8 and N-2B10 mAbs revealed the same pattern of RANK staining on flow cytometry as compared with that observed with an anti-RANK mAb distinct from those used for IHC applications (Fig. 3B). RANK staining by IHC was only observed in xenografts from cells which were shown to be RANK positive by flow cytometry or by RT-PCR. No RANK IHC signal was detected in cells which had undetectable RANK levels by RT-PCR or flow cytometry (summarized in Supplemental Table 1).
Fig. 3.
huRANK antibody validation for IHC methods. (A) RANK IHC was performed on FFPE samples of xenografts of the specified human cancer cells. Parental cells did not detectably express RANK as confirmed by multiple independent methods. There was no detectable signal with the isotype control antibody. Both antibodies against huRANK (N-1H8 and N-2B10) provide a similar staining pattern, essentially cross-validating one another. (B) The same cells used for the above tumor xenografts were grown in vitro and processed for flow cytometry. For anti-RANK staining, the pattern of staining by both test antibodies N-1H8 and N-2B10 was compared with a previously identified mAb (M331) useful for flow cytometry applications (Armstrong [31]). Solid grey line: Unstained; Red line: Secondary control=Goat antimouse APC; Blue line: Isotype control 4D2 (anti-AGP3 muIgG1), 1 μg/mL; Purple line: M331 (anti-huRANK muIgG1), 1 μg/mL; Green line: N-1H8 (anti-huRANK muIgG1), 1 μg/mL; Black line: N-2B10 (anti-huRANK muIgG1), 1 μg/mL. The RANK signal by IHC is concordant with the signal by flow cytometry. APC, allophycocyanin; FFPE, formalin-fixed, paraffin-embedded; IHC, immunohistochemistry; mAb, monoclonal antibody; RANK, receptor activator of nuclear factor kappa-B.
Demonstration of very specific RANK IHC staining of osteoclasts in decalcified FFPE human bone sections (data not shown) provided an additional confirmation of reagent and method specificity. While IHC positive for RANK staining, the positive controls for RANK expression (H1299-RANK and COLO205-RANK) expressed very high levels of protein due to the stable transfection of RANK cDNA and were not a rigorous test of antibody sensitivity. To address sensitivity of RANK IHC with mAbs N-1H8 and N-2B10, we identified two breast cancer cell lines (HCC70 and H1954) which expressed much lower levels of RANK (estimated to express between 20- and 100-fold lower RANK expression by FACS and RT-PCR measures [Supplemental Table 1 and Supplemental Fig. 3]); quantitation of surface protein using a flow cytometry based method confirmed that these cell lines expressed low levels of surface RANK expression (1800±365 receptors/cell for H1954 and 9120±2826 receptors/cell for HCC70 cells). Despite these low levels of RANK expression, RANK protein was readily detectable in FFPE xenograft tissues samples from either HCC70 or H1954 cells using mAbs N-1H8 or N-2B10 (Supplemental Fig. 4 and summarized in Supplemental Table 1) indicating the sensitivity of this IHC approach. While both N-1H8 and N-2B10 anti-RANK mAbs gave identical staining patterns, we proceeded with N-1H8 for most IHC applications as it had slightly greater sensitivity while retaining good signal to noise ratio (Fig. 3).
3.2. Expression analysis of RANKL and RANK in OS
Expression of RANKL and RANK was determined by IHC in primary OS samples, including a TMA of small core biopsy specimens (1.5 mm [diameter]) representing 40 individual patients and also in large tissue sections (20 mm [diameter]) from a smaller study set (n=16). Incidence was scored as a positive IHC signal (any intensity) while the H-score method captured the heterogeneity and intensity of IHC signals; the maximum H-score is 300 (Material and methods). The patient characteristics are summarized in Table 1.
Table 1.
Patient characteristics.
| Age (years) | Range | 11–64 |
| Mean | 32.1 | |
| Median | 32 | |
| Sex;n(%) | Male | 36 (64%) |
| Female | 20 (36%) | |
| Site of primary tumor;n(%) | Femur | 24 (43%) |
| Tibia | 6 (11%) | |
| Humerus | 7 (13%) | |
| Rib/scapula | 5 (9%) | |
| Other/unidentified bone site | 14 (25%) | |
| Stage;n(%) | IA | 7 (12%) |
| IB | 4 (7%) | |
| IIA | 14 (25%) | |
| IIB | 22 (39%) | |
| III | 3 (5%) | |
| IV | 5 (9%) | |
| Unidentified | 1 (2%) |
Analysis of RANKL and RANK was determined within distinct compartments, including tumor tissue, infiltrating cells, and adjacent normal bone tissue when present. The H-score distributions for RANKL or RANK selectively within OS tumor cells are shown in Fig. 4. The staining distributions for RANKL or RANK within normal bone or infiltrating (nontumor) cells were noted without quantitative scoring. OS, osteosarcoma; RANK, receptor activator of nuclear factor kappa-B; RANKL, RANK ligand.
There was a wide range of RANKL expression within the OS tumor cell component of both sample sets examined, with many samples negative for RANKL expression while other samples had high levels of RANKL expression (Fig. 4, Fig. 5).
Fig. 4.
RANKL and RANK H-score distribution in OS. The H score incorporates intensity (scale of 0–3) and percentage of tumor cells stained positive, giving a range of 0–300. RANK IHC was performed using N-1H8 mAb on the small core TMA samples and confirmed using N-2B10 mAb on the larger OS tumor sections. RANKL IHC was performed using M366 mAb on both the TMA and large tissue sections. The H-score distribution for both analytes are depicted for both (A) the small TMA core samples and (B) the larger tissue sections. IHC, immunohistochemistry; OS, osteosarcoma; RANK, receptor activator of nuclear factor kappa-B; RANKL, RANK ligand.
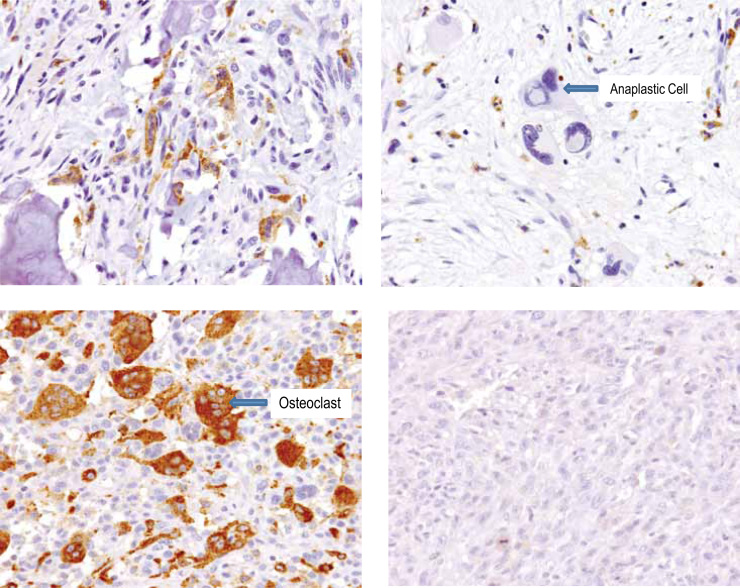
Fig. 5.
Range of RANKL protein expression in OS. RANKL IHC performed with mAb M366. Representative IHC images from four separate OS tumor samples are shown. Arrows indicate RANKL-positive tumor cells and tumor anaplastic cells. In addition, RANKL-negative osteoclasts are also clearly delineated (arrow). The four images demonstrate a range of RANKL positivity, as indicated by the different H-scores. The H-score for each image was as follows: upper left panel, H-score=260; upper right panel, H-score=210; lower left panel, H-score=180; lower right panel, H-score=20. IHC, immunohistochemistry; OS, osteosarcoma; RANK, receptor activator of nuclear factor kappa-B ligand.
Within the OS TMA, RANKL-positive tumors (any tumor cells positive) were observed for the majority of samples (65%; 26/40) but the H-scores were typically less than 100 (mean=23.4). RANKL expression within OS was not uniformly observed in all tumor cells and only a few tumor samples (20%; 8/40) exhibited>10% positive OS cells. In large OS tissue sections, the incidence of RANKL-positive tumors (any tumor cells positive) was 75% (12/16) and H-scores were higher (mean H-score=73.5) than the TMA. Fifty percent of large OS tissue sections exhibited>10% cells positive for RANKL. The quantitative and qualitative expression patterns for RANKL were similar between the two independent sample sets, with generally higher H-scores for RANKL observed in the larger tissue sections, as expected given the larger tissue surface area. The specificity of the RANKL staining was supported by the distinct expression within occasional tumor cells and tumor anaplastic cells adjacent to RANKL-negative osteoclasts (Fig. 5) as well as the expected positive RANKL expression frequently observed within normal stroma at the bone interface (Fig. 6A).
Fig. 6.
RANKL and RANK expression within adjacent bone tissue. RANKL and RANK IHC performed with mAb N-1H8 and M366, respectively. (A) RANKL-expressing stroma at bone interface. (B) RANK expression in bone osteoclast. IHC, immunohistochemistry; mAb, monoclonal antibody; RANK, receptor activator of nuclear factor kappa-B; RANKL, RANK ligand.
Analysis of RANK IHC in the large OS tissue sections and the small core biopsy specimens did not indicate any substantial RANK expression in OS tumor cells (Fig. 4, Fig. 7). Only 2.5% (1/40) samples from the TMA had RANK staining potentially in OS tumor cells. In this single case, the faint signal and low percentage of positive cells precluded a clear distinction between the spindeloid (somewhat anaplastic) OS cells or the mononucleated myeloid cells. This is in marked contrast to the high RANK signal observed within the myeloid osteoclast precursor compartment present in 65% (26/40) of tumors and the obvious RANK staining within any giant cell osteoclasts embedded in tumor (Fig. 7). Analysis of the large OS tissue sections was performed with a second anti-RANK antibody (N-2B10) with a distinct RANK epitope binding characteristics to confirm the above findings and demonstrated similar results, RANK staining within the myeloid osteoclast precursor compartment and RANK staining within any giant cell osteoclasts present; no OS tumor cells stained positive for RANK (see Fig. 4 for summary of H-scores). The fidelity, sensitivity and specificity of the staining is supported within any individual tissue sample by the distinct expression of RANK observed as expected in normal osteoclasts, either within the tumor mass (Fig. 7) or in samples where the tumor/bone interface was represented (Fig. 6B), which contrasts with the absence of signal within OS tumor cells (Fig. 7).
Fig. 7.
RANK protein expression in OS is limited to myeloid osteoclasts and osteoclast precursors. RANK IHC performed with mAb N-1H8. Representative IHC images from four separate OS tumor samples are shown. Upper left panel shows RANK-positive normal osteoclasts at the tumor–bone interface. Upper right panel shows tumor anaplastic cells were RANK-negative. Lower left panel shows abundant RANK-positive tumor-associated osteoclasts were present within the tumor in some tumors. Lower right panel shows no RANK-positive cells were present in some tumors. The H-score was zero for all images for RANK. RANK expression was observed in osteoclasts and myeloid osteoclast precursors in a majority of samples but not in the sarcoma component. IHC, immunohistochemistry; mAb, monoclonal antibody; OS, osteosarcoma; RANK, receptor activator of nuclear factor kappa-B.
4. Discussion
In the present study, we developed and optimized methodologies and monoclonal anti-RANK and anti-RANKL antibodies for IHC applications and expression analysis in human OS. We incorporated a number of independent specificity controls for each analyte. The positive and negative IHC signal patterns for the anti-RANKL mAb were very precisely correlated to the western blotting and flow cytometry signals for the control cells. Moreover, the specific pattern of RANKL staining observed by IHC with mAb M366 overlapped with RANKL in situ mRNA analysis. For the RANK expression analysis, a concordant expression pattern was observed using mAbs in IHC on a variety of positive and negative controls compared with expression analysis tested using other methods, such as flow cytometry using mAbs which bind RANK via distinct epitopes and RT-PCR. Each RANKL and RANK IHC test of OS samples included positive and negative controls, including clinical GCTB samples. These robust specificity controls provide evidence for the reliability of RANKL and RANK expression analysis reported herein.
Utilizing IHC, the majority of OS samples were positive for RANKL (68%), with RANKL staining detectable in OS tumor cells and tumor anaplastic cells. However, a wide range of RANKL expression within the OS component was observed and many samples had low intensity RANKL signal or demonstrated no detectable RANKL staining. Clearly RANKL expression was not uniformly observed within an OS tumor, as only a few samples (16/56) demonstrated more than 10% of the tumor cells positive for RANKL signal. The intensity and frequency of RANKL staining in OS samples observed in this study were substantially less than that observed in GCTB samples using the same antibody for IHC evaluation [23]. The overall incidence of RANKL staining observed in the present study of human OS (68%) is similar to that observed by Lee et al. [34] using a goat polyclonal RANKL antisera (75%) but differs substantially to the 9% RANKL positivity observed by Bago-Horvath [22]. The relatively small sample size (n=56) and limited patient follow-up data precluded any prognostic association with RANKL incidence or expression levels in the present study. We conclude that RANKL is observed in the majority of OS cases although the fraction of positive tumor cells and staining intensity is generally very low.
In contrast to the RANKL expression, there was essentially no RANK expression detected in OS tumor cells using IHC. Only one OS sample (1.9%, 1/52) demonstrated RANK staining potentially within OS tumor cells while RANK expression was observed as expected within myeloid osteoclast precursors, giant cell osteoclasts within a tumor mass, and normal osteoclasts at the bone/tumor interface whenever these cells were present. In fact, due to the very faint nature of the RANK staining in this single sample, it was impossible to clearly distinguish between RANK positivity within the spindeloid and somewhat anaplastic OS cells or mononucleated myeloid cells. We conclude that RANK protein expression is not observed in tumor cells from primary human OS to any prominent degree.
The absence of RANK expression in OS tumor cells observed in the present study is in marked contrast to the two reports that RANK is highly expressed in the majority of primary human OS [20], [22]. Mori et al. (2007) reported RANK expression to be homogeneously expressed (in 100% of tumor cells) in 57% of OS biopsy specimenss and Bago-Horvath [22] reported moderate or strongly positive RANK expression in 69% of OS tumor samples. Given that the IHC scoring systems used in all studies were not normalized, one cannot make an unbiased comparison of RANK expression incidence and intensity levels observed in those two studies with the current study. However, it is important to point out distinct features of the present study with the two previously published studies of RANK expression in human OS to address the very high incidence observed in those studies. Firstly, both previously published reports relied on commercially available anti-RANK antibodies and did not provide any evidence for the specificity and reproducibility of staining for IHC applications nor confirmation of RANK expression using independent methods or reagents. Secondly, the use of mAbs for expression analysis in the present study avoids the technical pitfalls of run-to-run reproducibility inherent in polyclonal antibodies. Thirdly, neither previously-published study documented the expected RANK staining within normal myeloid cells in the tumor mass or at the bone/tumor interface within any OS sample to document RANK expression in the expected cell types and provide context for the signal to noise for the reagents and methods used. The validation of the RANK antibodies used for IHC demonstrated concordant positive (and negative) signals between multiple independent methodologies and addressed sensitivity down to fewer than 1800 receptors/cell. Furthermore, in our analysis of OS samples, two independent anti-RANK mAbs, each recognizing distinct epitope binding sites demonstrated an identical positive and negative staining pattern, essentially cross-validating one another. Different alternatively-spliced variants of the human RANK gene have been described which alter exons 7, 8, or 9 encoding in either the transmembrane or cytoplasmic regions of RANK [32], potentially impacting IHC detection. However, the antibodies N-1H8 and N-2B10 bind to the extracellular portion of RANK common to each variant, thus the IHC results reported here would be inclusive of any rare, alternatively-spliced forms. Altogether, these specificity and sensitivity controls, along with the observed expression of RANK in the expected osteoclasts and related cells, substantiates the observation that OS tumor cells do not express RANK to any major degree. It would seem likely that technical limitations to these previously published studies may have accounted for the reportedly high incidence and high expression levels of RANK within OS tumors.
Aside from distinct technical approaches used, the well-established RANKL and RANK expression patterns in the bone further substantiates the expression patterns defined in the current study of OS. RANKL and RANK play major roles in bone metabolism due to the critical role of this pathway in osteoclastogenesis [4]. That is, RANK is expressed with the hematopoietic myeloid compartment contributing to myeloid-derived osteoclasts and their precursors consistent with the observed compartmentalization of RANK expression within giant cells, osteoclasts and osteoclast precursors found within OS samples in the present study. Given that OS tumors may arise from a mesenchymal-osteoblast origin [1], [2], it is unlikely that RANK would be expressed in the tumor cells while expression of RANKL in OS tumor cells is perhaps not surprising. In normal and pathologic bone, RANKL expression is confined to the cells of the osteoblast lineage, including osteocytes and has also been observed in certain tumor cells [33]. The observation herein of RANKL within reactive bone stroma in OS tumors as well as many OS cells themselves suggests that RANKL may stimulate osteoclast differentiation and activation potentially via multiple sources. Molyneux et al. [35] has reported that human and mouse OS with reduced expression of the gene encoding the regulatory subunit α (RIa) of PKA have high RANKL levels suggesting that pathways regulating RANKL expression may be dysfunctional in some OS. For any RANKL-positive OS, it remains unclear why RANKL is not expressed uniformly in all OS tumor cells as observed in the present study, suggesting that perhaps some element of local regulation observed in normal osteoblast-lineage cells (e.g. responsiveness to PTH1r) is retained by OS tumor cells.
In mouse models of OS, pharmacologic inhibition of RANKL is protective against bone destruction and also decreases tumor burden [18], [19], [20]. This is likely related to osteoclast inhibition, as treatment with bisphosphonates lead to essentially similar responses [21]. The reduction in skeletal tumor burden observed with osteoclast inhibitors results from interruption of the vicious cycle in which decreased osteoclastic bone resorption and subsequent reduction in localized bone matrix and growth factors indirectly reduces tumor growth and survival. These pharmacology observations demonstrating similar anti-tumor activity of bisphosphonates and RANKL inhibitors are consistent with the absence of RANK on OS tumor cells and a contribution of RANK-positive osteoclasts associated with OS tumors (this study and Avnet et al. [10]) to the bone pathologies observed in OS and potential indirect feedback to the skeletal tumor. The observation that RANKL is expressed in OS cells themselves suggests that these tumors may mediate an osteoclastic response independently of (or in addition to) RANKL within the normal or tumor-reactive bone stroma. However, the absence of RANK expression in OS tumor cells indicates that an autocrine RANKL/RANK response in human OS tumor cells is unlikely to be operative. While anti-RANKL therapy may influence the bone microenvironment and may be protective against bone pathologies in OS, the lack of RANK expression in tumor cells suggests that this approach would not directly affect the tumor.
5. Funding
This study was funded by Amgen Inc.
6. Conflicts of interest
William C. Dougall is an employee and shareholder of Amgen Inc. Daniel Branstetter, Kathy Rohrbach, Li-Ya Huang, Mark Tometsko, Allison Jacob, Michelle Blake and Rosalia Soriano are former employees and shareholders of Amgen Inc.
Acknowledgments
The authors would like to acknowledge Aaron Winters and Martine Roudier for technical assistance with antibody development and validation. Albert Y. Rhee (Amgen Inc.) provided editorial assistance.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jbo.2015.06.002.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Klein M.J., Siegal G.P. Osteosarcoma: anatomic and histologic variants. Am J Clin Pathol. 2006;125:555–581. doi: 10.1309/UC6K-QHLD-9LV2-KENN. [DOI] [PubMed] [Google Scholar]
- 2.Mutsaers A.J., Walkley C.R. Cells of origin in osteosarcoma: mesenchymal stem cells or osteoblast committed cells? Bone. 2014;62:56–63. doi: 10.1016/j.bone.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Botter S.M., Neri D., Fuchs B. Recent advances in osteosarcoma. Curr Opin Pharmacol. 2014;16:15–23. doi: 10.1016/j.coph.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Lacey D.L., Boyle W.J., Simonet W.S., Kostenuik P.J., Dougall W.C., Sullivan J.K. Bench to bedside: elucidation of the OPG-RANK-RANKL pathway and the development of denosumab. Nat Rev Drug Discov. 2012;11:401–419. doi: 10.1038/nrd3705. [DOI] [PubMed] [Google Scholar]
- 5.Stopeck A.T., Lipton A., Body J.J., Steger G.G., Tonkin K., de Boer R.H. Denosumab compared with zoledronic acid for the treatment of bone metastases in patients with advanced breast cancer: a randomized, double-blind study. J Clin Oncol. 2010;28:5132–5139. doi: 10.1200/JCO.2010.29.7101. [DOI] [PubMed] [Google Scholar]
- 6.Henry D.H., Costa L., Goldwasser F., Hirsh V., Hungria V., Prausova J. Randomized, double-blind study of denosumab versus zoledronic acid in the treatment of bone metastases in patients with advanced cancer (excluding breast and prostate cancer) or multiple myeloma. J Clin Oncol. 2011;29:1125–1132. doi: 10.1200/JCO.2010.31.3304. [DOI] [PubMed] [Google Scholar]
- 7.Fizazi K., Carducci M., Smith M., Damiao R., Brown J., Karsh L. Denosumab versus zoledronic acid for treatment of bone metastases in men with castration-resistant prostate cancer: a randomised, double-blind study. Lancet. 2011;377:813–822. doi: 10.1016/S0140-6736(10)62344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith M.R., Saad F., Coleman R., Shore N., Fizazi K., Tombal B. Denosumab and bone-metastasis-free survival in men with castration-resistant prostate cancer: results of a phase 3, randomised, placebo-controlled trial. Lancet. 2012;379:39–46. doi: 10.1016/S0140-6736(11)61226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith M.R., Saad F., Oudard S., Shore N., Fizazi K., Sieber P. Denosumab and bone metastasis-free survival in men with nonmetastatic castration-resistant prostate cancer: exploratory analyses by baseline prostate-specific antigen doubling time. J Clin Oncol. 2013;31:3800–3806. doi: 10.1200/JCO.2012.44.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avnet S., Longhi A., Salerno M., Halleen J.M., Peru F., Granchi D. Increased osteoclast activity is associated with aggressiveness of osteosarcoma. Int J Oncol. 2008;33:1231–1238. [PubMed] [Google Scholar]
- 11.Itoh K., Udagawa N., Matsuzaki K., Takami M., Amano H., Shinki T. Importance of membrane- or matrix-associated forms of M-CSF and RANKL/ODF in osteoclastogenesis supported by SaOS-4/3 cells expressing recombinant PTH/PTHrP receptors. J Bone Miner Res. 2000;15:1766–1775. doi: 10.1359/jbmr.2000.15.9.1766. [DOI] [PubMed] [Google Scholar]
- 12.Barger A.M., Fan T.M., de Lorimier L.P., Sprandel I.T., O’Dell-Anderson K. Expression of receptor activator of nuclear factor kappa-B ligand (RANKL) in neoplasms of dogs and cats. J Vet Intern Med. 2007;21:133–140. doi: 10.1892/0891-6640(2007)21[133:eoraon]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.A., Jung J.S., Kim D.H., Lim J.S., Kim M.S., Kong C.B. RANKL expression is related to treatment outcome of patients with localized, high-grade osteosarcoma. Pediatr Blood Cancer. 2011;56:738–743. doi: 10.1002/pbc.22720. [DOI] [PubMed] [Google Scholar]
- 14.Lamoureux F., Richard P., Wittrant Y., Battaglia S., Pilet P., Trichet V. Therapeutic relevance of osteoprotegerin gene therapy in osteosarcoma: blockade of the vicious cycle between tumor cell proliferation and bone resorption. Cancer Res. 2007;67:7308–7318. doi: 10.1158/0008-5472.CAN-06-4130. [DOI] [PubMed] [Google Scholar]
- 15.Molyneux S.D., Di Grappa M.A., Beristain A.G., McKee T.D., Wai D.H., Paderova J. Prkar1a is an osteosarcoma tumor suppressor that defines a molecular subclass in mice. J Clin Invest. 2010;120:3310–3325. doi: 10.1172/JCI42391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akiyama T., Dass C.R., Shinoda Y., Kawano H., Tanaka S., Choong P.F. Systemic RANK-Fc protein therapy is efficacious against primary osteosarcoma growth in a murine model via activity against osteoclasts. J Pharm Pharmacol. 2010;62:470–476. doi: 10.1211/jpp.62.04.0009. [DOI] [PubMed] [Google Scholar]
- 17.Akiyama T., Choong P.F., Dass C.R. RANK-Fc inhibits malignancy via inhibiting ERK activation and evoking caspase-3-mediated anoikis in human osteosarcoma cells. Clin Exp Metastasis. 2010;27:207–215. doi: 10.1007/s10585-010-9319-y. [DOI] [PubMed] [Google Scholar]
- 18.Lamoureux F., Picarda G., Rousseau J., Gourden C., Battaglia S., Charrier C. Therapeutic efficacy of soluble receptor activator of nuclear factor-kappa B-Fc delivered by nonviral gene transfer in a mouse model of osteolytic osteosarcoma. Mol Cancer Ther. 2008;7:3389–3398. doi: 10.1158/1535-7163.MCT-08-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heymann D., Ory B., Blanchard F., Heymann M.F., Coipeau P., Charrier C. Enhanced tumor regression and tissue repair when zoledronic acid is combined with ifosfamide in rat osteosarcoma. Bone. 2005;37:74–86. doi: 10.1016/j.bone.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 20.Mori K., Le Goff B., Berreur M., Riet A., Moreau A., Blanchard F. Human osteosarcoma cells express functional receptor activator of nuclear factor-kappa B. J Pathol. 2007;211:555–562. doi: 10.1002/path.2140. [DOI] [PubMed] [Google Scholar]
- 21.Beristain A.G., Narala S.R., Di Grappa M.A., Khokha R. Homotypic RANK signaling differentially regulates proliferation, motility and cell survival in osteosarcoma and mammary epithelial cells. J Cell Sci. 2012;125:943–955. doi: 10.1242/jcs.094029. [DOI] [PubMed] [Google Scholar]
- 22.Bago-Horvath Z., Schmid K., Rossler F., Nagy-Bojarszky K., Funovics P., Sulzbacher I. Impact of RANK signalling on survival and chemotherapy response in osteosarcoma. Pathology. 2014;46:411–415. doi: 10.1097/PAT.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 23.Branstetter D.G., Nelson S.D., Manivel J.C., Blay J.Y., Chawla S., Thomas D.M. Denosumab induces tumor reduction and bone formation in patients with giant-cell tumor of bone. Clin Cancer Res. 2012;18:4415–4424. doi: 10.1158/1078-0432.CCR-12-0578. [DOI] [PubMed] [Google Scholar]
- 24.Thomas D., Henshaw R., Skubitz K., Chawla S., Staddon A., Blay J.Y. Denosumab in patients with giant-cell tumour of bone: an open-label, phase 2 study. Lancet Oncol. 2010;11:275–280. doi: 10.1016/S1470-2045(10)70010-3. [DOI] [PubMed] [Google Scholar]
- 25.Chawla S., Henshaw R., Seeger L., Choy E., Blay J.Y., Ferrari S. Safety and efficacy of denosumab for adults and skeletally mature adolescents with giant cell tumour of bone: interim analysis of an open-label, parallel-group, phase 2 study. Lancet Oncol. 2013;14:901–908. doi: 10.1016/S1470-2045(13)70277-8. [DOI] [PubMed] [Google Scholar]
- 26.Blake M.L., Tometsko M., Miller R., Jones J.C., Dougall W.C. RANK expression on breast cancer cells promotes skeletal metastasis. Clin Exp Metastasis. 2014;31:233–245. doi: 10.1007/s10585-013-9624-3. [DOI] [PubMed] [Google Scholar]
- 27.Smith K.B., Ellis S.A. Standardisation of a procedure for quantifying surface antigens by indirect immunofluorescence. J Immunol Methods. 1999;228:29–36. doi: 10.1016/s0022-1759(99)00087-3. [DOI] [PubMed] [Google Scholar]
- 28.Lacey D.L., Timms E., Tan H.L., Kelley M.J., Dunstan C.R., Burgess T. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 29.Anderson D.M., Maraskovsky E., Billingsley W.L., Dougall W.C., Tometsko M.E., Roux E.R. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390:175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 30.Blake M.G., Boccia M.M., Krawczyk M.C., Baratti C.M. Hippocampal alpha7-nicotinic cholinergic receptors modulate memory reconsolidation: a potential strategy for recovery from amnesia. Neurobiol Learn Mem. 2013;106:193–203. doi: 10.1016/j.nlm.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Armstrong A.P., Miller R.E., Jones J.C., Zhang J., Keller E.T., Dougall W.C. RANKL acts directly on RANK-expressing prostate tumor cells and mediates migration and expression of tumor metastasis genes. Prostate. 2008;68:92–104. doi: 10.1002/pros.20678. [DOI] [PubMed] [Google Scholar]
- 32.Papanastasiou A.D., Sirinian C., Kalofonos H.P. Identification of novel human receptor activator of nuclear factor-kB isoforms generated through alternative splicing: implications in breast cancer cell survival and migration. Breast Cancer Res. 2012;14:R112. doi: 10.1186/bcr3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dougall W.C., Holen I., Gonzalez Suarez E. Targeting RANKL in metastasis. Bonekey Rep. 2014;3:519. doi: 10.1038/bonekey.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J.A., Jung J.S., Kim D.H., Lim J.S., Kim M.S., Kong C.B., Song W.S., Cho W.H., Jeon D.G., Lee S.Y., Koh J.S. RANKL expression is related to treatment outcome of patients with localized, high-grade osteosarcoma. Pediatr Blood Cancer. 2011;56(5):738–743. doi: 10.1002/pbc.22720. [DOI] [PubMed] [Google Scholar]
- 35.Molyneux S.D., Di Grappa M.A., Beristain A.G., McKee T.D., Wai D.H., Paderova J., Kashyap M., Hu P., Maiuri T., Narala S.R., Stambolic V., Squire J., Penninger J., Sanchez O., Triche T.J., Wood G.A., Kirschner L.S., Khokha R. Prkar1a is an osteosarcoma tumor suppressor that defines a molecular subclass in mice. J Clin Invest. 2010;120(9):3310–3325. doi: 10.1172/JCI42391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material