Highlight
Phosphate transporters AtPHT1;8 and AtPHT1;9, but not AtPHT1;1, discriminate between phosphite and phosphate. Phosphate-starvation-responsive transcript profiles show altered kinetics with phosphite, hence allowing further dissection of phosphorus signalling networks.
Key words: Arabidopsis thaliana, phosphate-starvation response, phosphate transport, phosphite, phosphonate, phosphorous acid, phosphorus signalling networks, PSR genes, transcriptional regulation.
Abstract
Phosphite is a less oxidized form of phosphorus than phosphate. Phosphite is considered to be taken up by the plant through phosphate transporters. It can mimic phosphate to some extent, but it is not metabolized into organophosphates. Phosphite could therefore interfere with phosphorus signalling networks. Typical physiological and transcriptional responses to low phosphate availability were investigated and the short-term kinetics of their reversion by phosphite, compared with phosphate, were determined in both roots and shoots of Arabidopsis thaliana. Phosphite treatment resulted in a strong growth arrest. It mimicked phosphate in causing a reduction in leaf anthocyanins and in the expression of a subset of the phosphate-starvation-responsive genes. However, the kinetics of the response were slower than for phosphate, which may be due to discrimination against phosphite by phosphate transporters PHT1;8 and PHT1;9 causing delayed shoot accumulation of phosphite. Transcripts encoding PHT1;7, lipid-remodelling enzymes such as SQD2, and phosphocholine-producing NMT3 were highly responsive to phosphite, suggesting their regulation by a direct phosphate-sensing network. Genes encoding components associated with the ‘PHO regulon’ in plants, such as At4, IPS1, and PHO1;H1, generally responded more slowly to phosphite than to phosphate, except for SPX1 in roots and MIR399d in shoots. Two uncharacterized phosphate-responsive E3 ligase genes, PUB35 and C3HC4, were also highly phosphite responsive. These results show that phosphite is a valuable tool to identify network components directly responsive to phosphate.
Introduction
Phosphite (H2PO3 –, Phi) is a less oxidized form of phosphorus (P) than phosphate (H2PO4 –, Pi). Phi is highly water soluble and less prone than Pi to adsorb to soil particles, which makes it more accessible to plants (Ruthbaum and Baille, 1964). Phi competes with the essential macronutrient Pi for uptake by plants, most probably through both high- and low-affinity transport systems (d’arcy-Lameta and Bompeix, 1991; Danova-Alt et al., 2008). Phi uptake is strongly and competitively inhibited in the presence of Pi (Pratt et al., 2009). Within the plant, Phi can be translocated, and it preferentially accumulates in sink tissues (Nartvaranant et al., 2004).
Phosphite was once abundant in the oceans, but it has been oxidized over time (Pasek et al., 2013). Many microbes have retained the ability to oxidize Phi to Pi, and even use it as a reducing agent, namely for sulphate reduction (Poehlein et al., 2013). Plants, however, are not able to metabolize Phi (McDonald et al., 2001). Instead, P-limited plants are highly sensitive to Phi and display toxicity symptoms such as leaf chlorosis and stunted growth (McDonald et al., 2001; Ratjen and Gerendas, 2009; Thao and Yamakawa, 2009). Other detrimental effects caused by Phi are the arrest of primary root growth, yellowing of the leaf lamina of young leaves, and a patchy accumulation of anthocyanins in older leaves (Varadarajan et al., 2002). Pratt et al. (2009) also showed that respiration rates declined upon Phi treatment of P-limited sycamore cells. It was recently found that the accumulation of Phi impacts on metabolism in Arabidopsis thaliana, leading to changes in the levels of several central metabolites (Berkowitz et al., 2013).
Phi also triggers broad-spectrum resistance against pathogens with a (hemi)biotrophic lifestyle, such as oomycetes, fungi, and nematodes (Smillie et al., 1989; Hofgaard et al., 2010; Dias-Arieira et al., 2013; Percival and Banks, 2014). Phi has been suggested to act as a priming agent of plant defence responses in a number of plant–pathogen interactions (Machinandiarena et al., 2012; Massoud et al., 2012; Dalio et al., 2014). However, it is unclear how the primary recognition of Phi takes place, and which molecular pathways are altered within the plant subsequently to induce this primed state of heightened defence. Given that Phi is transported by Pi transporters, these primary molecular interactions could trigger changes in signal perception (Schothorst et al., 2013).
Phi accumulates in both the cytosol and organelles, while the presence of Pi enhances Phi sequestration in the vacuole (Danova-Alt et al., 2008). This is probably why plants with an adequate P status can tolerate moderate Phi exposure without visible toxicity symptoms (Thao and Yamakawa, 2009). Conversely, Phi inhibits the efflux of Pi from the vacuole, which could exacerbate Pi-starvation symptoms (Pratt et al., 2009) and lead to accelerated plant death (Singh et al., 2003). Interestingly, the combined concentrations of Phi plus Pi within roots and shoots of A. thaliana were remarkably constant, regardless of their ratio in the growth medium, demonstrating that plants sense both Pi and Phi and adjust their uptake and allocation accordingly (Berkowitz et al., 2013).
Due to its physical similarity to Pi and non-metabolizable nature, Phi has been used as a tool to understand Pi-dependent signalling networks in plants. In several studies, Phi in fact seemed to mimic Pi effectively. Brassica nigra seedlings germinated on low-Pi media in the presence of high (1–10mM) Phi concentrations had reduced activation of Pi-starvation-induced phosphoenolpyruvate phosphatase and pyrophosphate-dependent phosphofructokinase compared with P-limited control plants (Carswell et al., 1996). While Phi did not affect the total adenylate pool in P-limited Brassica napus suspension cells in the same way as Pi, it did cause changes in the in vivo phosphorylation status of a number of proteins (Carswell et al., 1997). In A. thaliana, Ticconi et al. (2001) observed that Phi prevented the induction of transcripts from the Pi-starvation-responsive (PSR) genes ACP5, At4, and PT2 upon 14 d exposure of P-sufficient seedlings to a medium lacking Pi, but containing high concentrations of Phi. The same plants showed reduced in vitro activities of PSR ribonucleases RNS1 and RNS2 and of an acid phosphatase. Within 1 d of transfer of P-sufficient A. thaliana seedlings to a medium lacking Pi, Phi suppressed the typical root hair formation and transcript accumulation of purple acid phosphatase PAP1 and Pi transporters PT1 and PT2 that occur upon Pi withdrawal (Varadarajan et al., 2002). Exposure of A. thaliana to Phi prevented not only PSR MGD2 and MGD3 expression, but also changes in glycerolipid profiles that accompany P-limited growth (Kobayashi et al., 2006). In P-limited tomato seedlings, Phi mimicked Pi in promoting proteolytic turnover of purple acid phosphatases (Bozzo et al., 2004). In rice, long-term exposure (5–7 d) to Phi suppressed the Pi-starvation-induced expression of OsIPS1 and OsIPS2 (Hou et al., 2005). In tobacco BY-2 cells, Phi caused the reversion of autophagic protein turnover triggered by Pi deprivation (Tasaki et al., 2014).
The first evidence suggesting that Phi and Pi have discrete effects on P signalling networks came from work by Stefanovic et al. (2007), who showed that transcripts of PHO1 and its close paralogue PHO1;H1 differentially accumulated in plants treated with Pi or Phi. The PHR1-dependent induction of PHO1;H1 under P-limiting conditions was attenuated by Phi, while the PHR1-independent induction of PHO1 was not. This effect does not directly depend on the MYB transcription factor PHR1, because, unlike for PHO1;H1, the induction of another PHR1-regulated paralogue, PHO1;H10, was not affected by Phi (Ribot et al., 2008). Interestingly, both PHO1 and PHO1;H1 transcripts were less abundant in the P-limited pho2 mutant and more strongly induced in the P-limited pdr2 mutant compared with those in the wild type (Stefanovic et al., 2007). Disruption of the gene encoding endoplasmic reticulum (ER)-resident P5-type ATPase PDR2 affected local Pi-sensing networks and heightened the sensitivity and amplitude of metabolic responses to P limitation (Ticconi et al., 2004). The conditional pdr2 short-root phenotype was reversible by Phi. These observations strongly suggest that Phi mimics Pi in local signalling networks, irrespective of the plant’s P status.
Studies have so far addressed the question of whether Phi can prevent the long-term accumulation of PSR gene transcripts. In this study, the question of whether the shorter term kinetics of Phi suppression were similar to those of Pi was addressed (Müller et al., 2004; Morcuende et al., 2007). Organ-level accumulation of both Pi and Phi in P-limited seedlings in A. thaliana accession Col-0 and three PHT1 transporter mutants was therefore determined. Root growth and anthocyanin accumulation as well as gene expression profiles in response to Phi treatment or Pi resupply were monitored in P-limited Col-0 seedlings over a time-course from 1 d to 7 d.
Materials and methods
Plant material and growth conditions
Seeds of A. thaliana (L.) Heynh. Col-0 and homozygous T-DNA insertion lines for pht1;1–2 (SALK 088568C) (Shin et al., 2004), pht1;8 (SALK 056529, Lapis-Gaza et al., 2014), and pht1;9-1 (SALK 050730) (Remy et al., 2012) were surface-sterilized for 2min in 70% (v/v) ethanol and 5min in 5% (v/v) NaOCl, before being rinsed five times in sterile water. Seeds were resuspended in sterile 0.1% (w/v) agar and stratified in the dark for 24–48h at 4 °C. Seedlings (12 per plate) were grown vertically on 10×10cm plates containing 50ml of nutrient solution [1mM Ca(NO3)2, 2mM KNO3, 0.5mM MgSO4, 0.25mM KH2PO4, 40 μM Fe-EDTA, 25 μM H3BO3, 2 μM MnCl2, 2 μM ZnSO4, 0.5 μM CuSO4, 0.075 μM (NH4)6Mo7O24, 0.15 μM CoCl2, 50 μM KCl, pH 5.8] with 0.5% (w/v) 2-(N-morpholino)ethanesulphonic acid and 1% (w/v) sucrose, and solidified with 0.7% (w/v) agar (Plant TC Agar, cat.#A111, PhytoTechnology Laboratories, Shawnee Mission, KS, USA). Plates were sealed with 3M™ Micropore medical tape (Intouch Direct, Springwood, Australia). Seedlings were grown in a 10/14h day/night cycle with 200 μmol m–2 s–1 photosynthetically active radiation (PAR) at 21 °C (day), 19 °C (night), and 65% relative humidity. The plant-available Pi present in the agar added another 5 μM to the medium. This amount is within the range of Pi concentrations across gelling agents (Jain et al., 2009). Preliminary experiments showed that concentrations of Pi ranging from 250 μM to 1mM do not limit seedling growth in this system (data not shown). For the experiment, seedlings grown on a medium with 250 μM Pi for 5 d were grown for 4 d on plates without Pi supplementation (containing 250 μM KCl instead) before being transferred to plates containing minimal Pi (5 μM residual Pi in agar), or equimolar concentrations (250 μM) of either Pi or Phi. The Phi solution was prepared from a fresh batch of phosphorous acid (99%, Sigma Aldrich, Castle Hill, Australia) as a filter-sterilized 250mM stock. The pH was adjusted to pH 5.8 with KOH. There was <0.1% oxidation of Phi to Pi in this solution during 1 month storage at 4 °C.
At harvest, the 12 seedlings on each plate were pooled into one sample. Roots were rinsed in MilliQ water for 5min. Roots and shoots were blotted dry and shock-frozen in liquid nitrogen. Harvesting started 3h after the beginning of the light period in synchrony with the experimental time-course to ensure that plants were at a comparable physiological state.
Root growth analysis and microscopy
After emergence of the radicle or transfer to a new plate, the position of the primary root tip was marked at 24h intervals. Prior to transfer or harvest, the seedlings were scanned at 600 dpi resolution to determine root and root hair length, growth rate, and lateral root number (LSM Image Browser v4.2; Carl Zeiss Microscopy GmbH, Jena, Germany).
For microscopy (Axioplan Universal microscope; Carl Zeiss Microscopy GmbH), roots were mounted onto slides in water under glass cover slips. Images were electronically processed (AxioVision4; Carl Zeiss Microscopy GmbH).
Metabolite quantification
Fifteen volumes of 1% (v/v) acetic acid were added to frozen plant powder (30–50mg) and homogenized for three cycles of 45 s at 5000rpm in the presence of two ceramic beads (ø 2mm, Precellys 24 Tissue Disruptor; Bertin Technologies, Montigny-le-Bretonneux, France). After incubation for 15min on ice, the homogenization process was repeated once. Cleared supernatants were used to determine organ Pi concentrations via the reduction of a phosphomolybdate complex by ascorbic acid (Ames, 1966). Phi concentrations were determined using the same extracts in a high-throughput enzymatic fluorescence assay (Berkowitz et al., 2011).
Anthocyanins in leaf samples were determined using a pH-differential method (Wrolstad et al., 2005). Concentrations were calculated using the molar absorptivity of cyanidin-3-glucoside (ε=26 900 l mol–1 cm–1), the predominant anthocyanin in A. thaliana leaves (Tohge et al., 2005).
Relative quantification of transcript abundance
mRNA was captured from tissue homogenates using oligo(dT)25-coated magnetic beads (Dynabeads, Life Technologies Australia Pty Ltd, Mulgrave, Australia) and converted to cDNA as previously described (Jost et al., 2007). Aliquots of 0.5ng of cDNA were amplified in a 10 μl reaction volume containing 0.3 μM of each primer and PCR master mix (Power SYBR® Green, Applied Biosystems, Scoresby, Australia). Quantitative PCR and threshold cycle (Ct) determination were performed using a fluorescence baseline setting of 0.3 (7500 FAST Real-Time PCR System, Applied Biosystems, Scoresby, Australia). Data were normalized against PP2AA3 (formerly PDF2) and UBC9 reference genes (Czechowski et al., 2005). PCR efficiencies for each primer pair were determined using the LinReg algorithm (Ruijter et al., 2009) (Supplementary Table S1 available at JXB online). Data were expressed either relative to normalized Ct values in control samples (ΔΔCt) or as 40–ΔCt values that correlate with the relative transcript expression of the gene of interest (Bari et al., 2006). The detection limit of the assay was calculated to be a 40–ΔCt value of 25.7±0.1.
Statistical analysis
Statistically significant differences between treatments were determined using analysis of variance (ANOVA) and defined as P≤0.05 (SigmaStat v. 12.3, Systat Software Inc., San Jose, CA, USA). Two-way ANOVA followed by Tukey’s post-hoc test was used to separate means. Hierarchical clustering was performed using squared Euclidean distance and complete linkage [J-Express 2012, Norwegian Bioinformatics Platform and Norwegian Microarray Consortium (http://www.molmine.com)] http://jexpress.bioinfo.no/site/ (last accessed 27 January 2015) (Dysvik and Jonassen, 2001).
Results
Phosphite strongly reduced plant biomass production
A vertical growth system was used for A. thaliana Col-0 seedlings that allowed a direct comparison of the effects of Phi versus Pi on the repression of Pi-starvation responses without the confounding effects of competition between Pi and Phi. Using this system, P-limited seedlings were subjected to continued Pi deprivation, Pi resupply, or Phi treatment. Plant biomass did not differ significantly among the treatments within the first 2 d of transfer (Fig. 1). After 3 d of treatment, both the P-limited seedlings and those resupplied with Pi had greater root and shoot biomass than seedlings at days 1 and 2, while the biomass of the Phi-treated seedlings was unchanged. Over the next 4 d, seedlings resupplied with Pi recovered from P limitation with a proportional increase in both root and shoot biomass that maintained the root-to-shoot ratio at 0.30±0.01. P-limited seedlings preferentially allocated resources to roots over shoots, leading to a final root-to-shoot ratio of 0.47±0.03. Despite the greater partitioning of biomass to roots, the root biomass after 7 d of further P limitation was only 84% of that in the Pi-resupplied seedlings. The shoot biomass of the P-limited seedlings was only 53% of that of the Pi-resupplied seedlings. By contrast, the Phi treatment slowed seedling growth much more severely. After 7 d exposure to Phi, the final root and shoot biomass of seedlings was only 26% and 38%, respectively, of that in Pi-resupplied seedlings. While there was a 55% increase in shoot biomass over time, the root biomass of Phi-treated seedlings did not change. Since this severe inhibition of root growth contrasted with both P-limited and P-sufficient seedling growth, the kinetics of root elongation were examined in more detail.
Fig. 1.
Accumulation of root and shoot biomass. Seeds were germinated on media containing 250 μM phosphate (Pi) on vertical plates as described in the Materials and methods. Five-day-old seedlings were transferred to a low-Pi medium for 4 d before being transferred to plates containing minimal Pi (5 μM, white bars), high Pi (250 μM, black bars), or phosphite (250 μM, grey bars) media. Root and shoot biomass was determined at 1, 2, 3, and 7 d after transfer (mean ±SE, n=4 replicates with 12 seedlings each). Statistically significant differences between time points as determined by Tukey’s HSD for each treatment at P<0.001 are indicated by an asterisk. Differences between treatments were significant for both organs only at 7 d after transfer (P<0.001).
Phosphite strongly inhibited primary root elongation
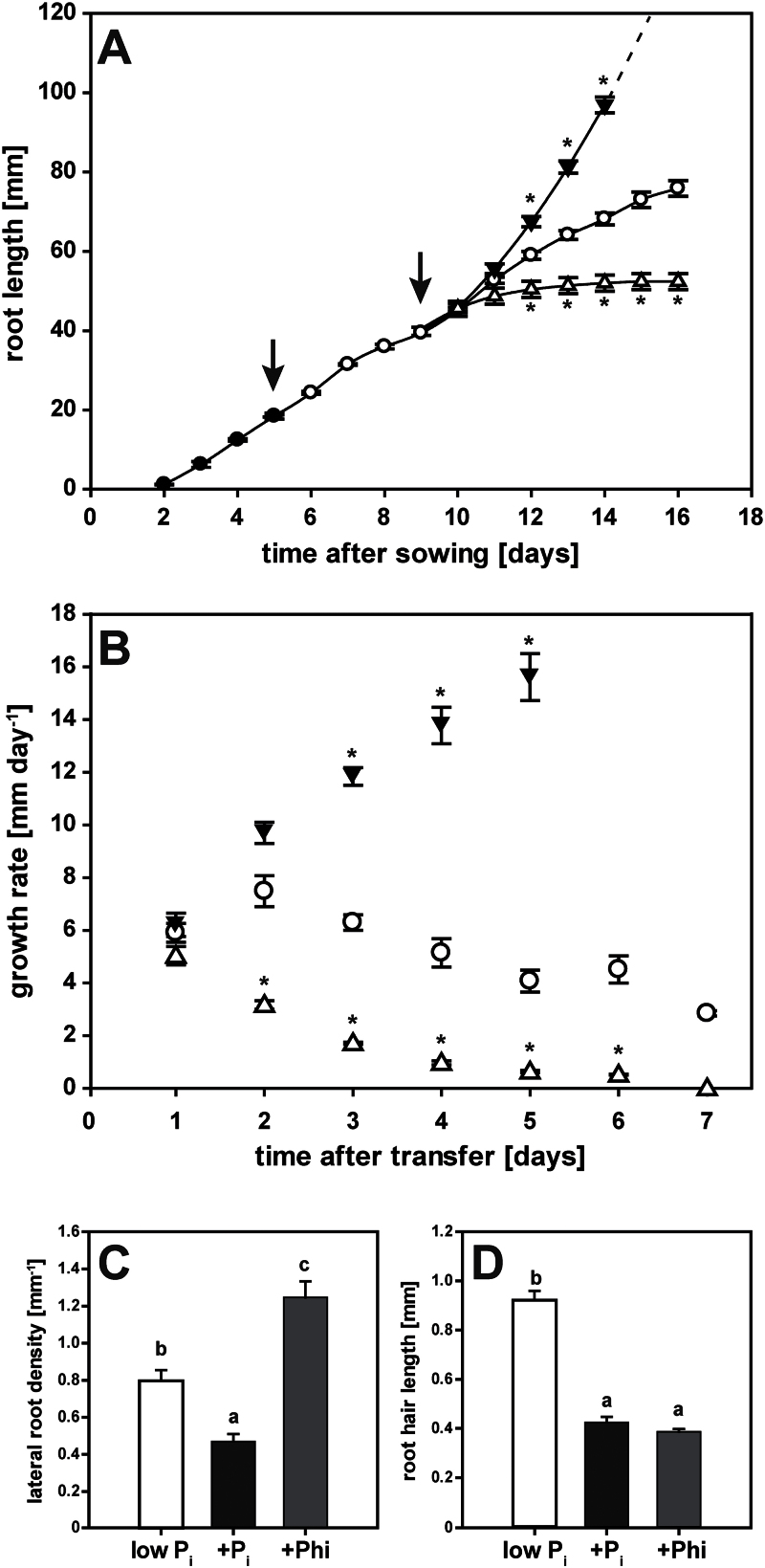
Seedlings germinated on high-Pi media showed steady primary root growth from 2 d after sowing (Fig. 2A). Root growth was initially maintained when the seedlings were transferred to a Pi-deficient medium 5 d after sowing. Imposing Pi resupply or Phi treatments after 4 d of Pi withdrawal did not affect root elongation during the first day (Fig. 2A). Two days after the transfer to the final medium (day 11), roots of both P-limited and Pi-resupplied seedlings grew at similar rates (Fig. 2B). In contrast, primary roots of Phi-treated seedlings showed much lower growth rates during this period, and elongation ceased completely within the next 48h. Root growth in Pi-resupplied seedlings accelerated exponentially during this same time period (Fig. 2B), with roots reaching the bottom of the 10-cm plate by 6 d after imposing the treatment. Root growth in P-limited seedlings decelerated by 2%, resulting in a final total root length that was almost 30% shorter than in Pi-resupplied seedlings. These results show that, unlike Pi resupply, Phi treatment accentuated the reduction in root growth caused by Pi depletion.
Fig. 2.
Changes in root architecture in response to phosphate (Pi) resupply and phosphite (Phi) treatment. (A) Primary root growth over the course of the experiment. Seedlings were germinated on media containing 250 μM Pi (filled circles). After 5 d, they were transferred to media containing minimal Pi (5 μM, open circles) before being transferred to plates with minimal Pi (5 μM, open circles), high Pi (250 μM, filled triangles), or Phi (250 μM, open triangles). Arrows indicate transfer to new plates. (B) Root growth rates in response to treatments. Symbols are the same as in (A). Shown in (A, B) are means ±SE, n=16 (four seedling roots each were measured individually from four separate plates). (C) Lateral root density in seedlings harvested 7 d after transfer to minimal Pi (white bars), high Pi (black bars), or 250 μM Phi (grey bars). Emerging lateral roots were counted in root segments that were formed 3 d after transfer. Shown are means ±SE, n=10 (five seedlings each from two plates). (D) Root hair length of seedlings harvested 7 d after transfer to minimal Pi (white bars), high Pi (black bars), or 250 μM Phi (grey bars). Shown are means ±SE, n=30 (3 root hairs×5 seedlings×2 plates). Statistically significant differences across time points in (A, B) were determined by Tukey’s HSD for treatments relative to Pi-limited seedlings at P<0.001. In (C) and (D), pairwise multiple comparisons between treatments identified statistically significant differences at P<0.005.
Phosphite altered seedling root architecture
At the end of the time-course experiment, high-resolution scans of primary root segments initiated on day 3 after the final transfer were used to analyse the effects of the three treatments on root development (Fig. 2C, D). The chosen root segment was proximal to the root apex, at the beginning of the root branching zone (Dubrovsky and Forde, 2012). The short-root phenotype caused by Phi resulted in an almost 2-fold greater lateral root density than in P-limited seedlings in this newly formed section of the root (Fig. 2C). Remarkably, the number of lateral roots per segment in Phi-treated seedlings (2.3±0.2) was 2-fold lower than that in Pi-limited (4.7±0.3) and Pi-resupplied (5.0±0.5) segments. Primary root growth in Pi-resupplied seedlings decreased lateral root density by nearly 2-fold compared with P-limited seedlings. While lateral roots elongated similarly under both Pi limitation and Pi resupply, emergence of lateral roots was inhibited in the presence of Phi. This phenomenon was also observed in a hydroponics growth system, where transfer to different nutrient solutions is less damaging to roots (Supplementary Fig. S1 at JXB online; note that in order to compensate for slower uptake of Phi over Pi, 1mM Phi was used in this experiment).
Root hairs of Phi-treated seedlings were 57% shorter than in P-limited seedlings (Fig. 2D). This shortening was similar to the 52% reduction observed for seedlings resupplied with Pi. A concomitant reduction in root hair density by 38% for Phi-treated seedlings (14±1mm–1) compared with P-limited seedlings (22±1mm–1) was also very similar to the 44% reduction observed in Pi-resupplied seedlings (13±1mm–1). Hence this local response to Pi resulting in fewer and shorter root hairs appears to be mimicked by Phi.
Anthocyanin accumulation in P-limited seedlings was repressed by both Pi and Phi
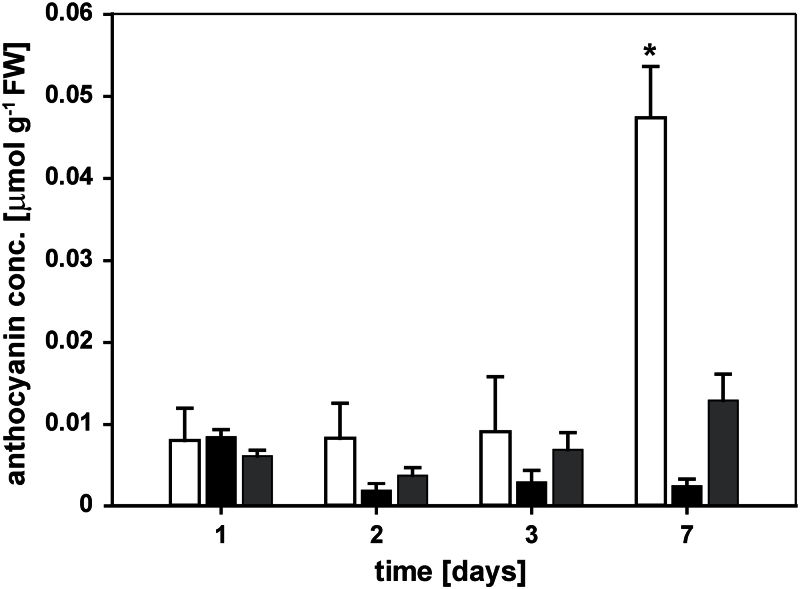
Anthocyanins accumulated to significant levels in leaves of seedlings after a total of 11 d of growth on minimal Pi media (4 d Pi withdrawal+7 d treatment; Fig. 3). This slow accumulation indicates that the seedlings were not highly stressed by the Pi deprivation imposed during the early stage of the experiment, and were probably accessing and gradually depleting P reserves that accumulated during the initial 5-d growth on Pi-containing medium. Seedlings resupplied with Pi after a starvation period of 4 d had lower levels of anthocyanins within 2 d of treatment. Phi-treated seedlings also had reduced leaf anthocyanin levels within the first 2 d of treatment, but not as low as in Pi-resupplied seedlings. In Phi-treated seedlings, the leaf anthocyanin concentration was higher at day 7 than in Pi-supplied seedlings, but was 72% lower than in P-limited seedlings. Therefore, Phi attenuated anthocyanin accumulation in P-limited plants that was completely suppressed by Pi resupply.
Fig. 3.
Anthocyanin accumulation in leaves of phosphorus-limited seedlings. Five-day-old seedlings were grown on low-phosphate (Pi) medium for 4 d before being transferred to plates with minimal Pi- (5 μM, white bars), high Pi- (250 μM, black bars), or phosphite- (250 μM, grey bars) containing media. Leaf anthocyanin concentrations were determined at day 1, 2, 3, and 7 after transfer (mean ±SE, n=3 replicates with 12 seedlings each). Statistically significant differences between time points and treatments were determined by Tukey’s HSD at P<0.001. Differences within the low-Pi series and between the low-Pi and the other two treatments were significant only at 7 d after transfer (P<0.001).
Root-to-shoot transport favoured Pi over Phi
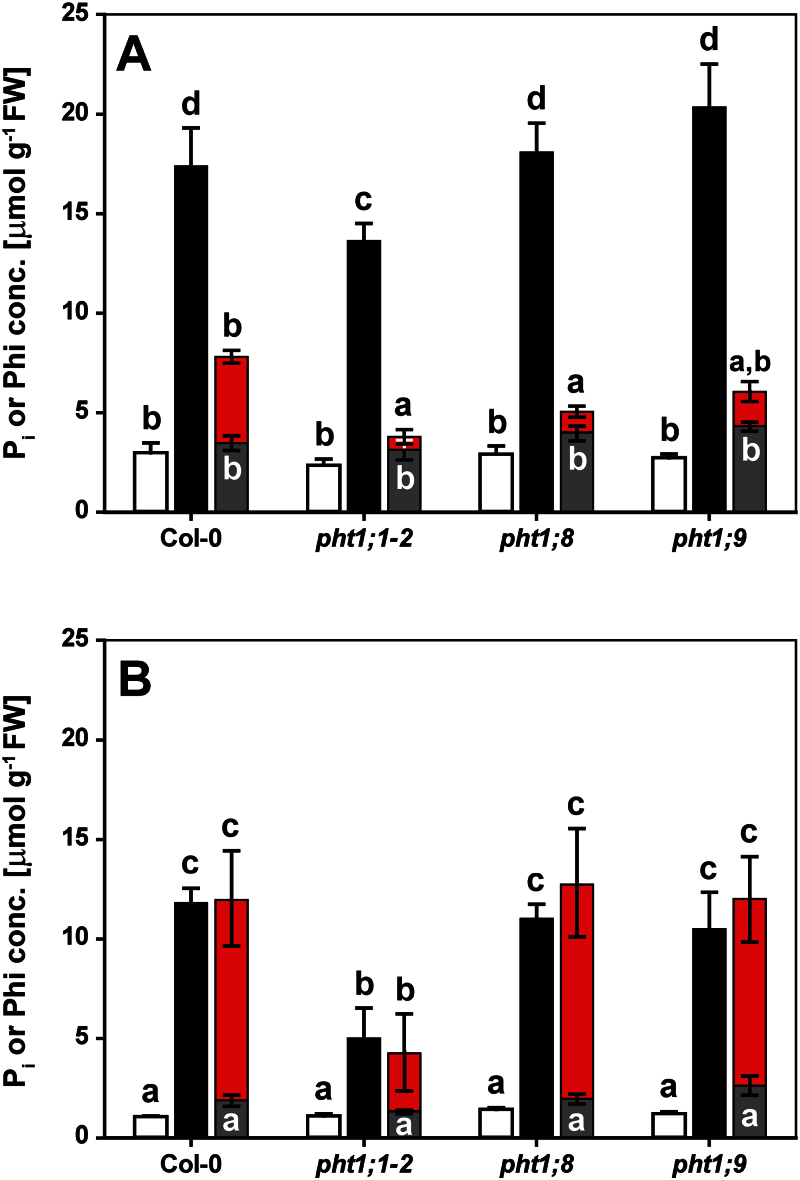
To appreciate fully the differences in the physiological and molecular responses to Phi compared with Pi, the accumulation of both anions in roots and shoots was determined over time. While roots accumulated both Pi and Phi equally within 1 d of exposure, there was a delay in the accumulation of Phi relative to that of Pi in the shoot (Fig. 4). Shoot Pi concentrations reached ~13 μmol g–1 fresh weight (FW) within 1 d of resupply (Fig. 4A) which was greater than the level of free Pi in seedlings continuously grown on a sufficient Pi supply (~5 μmol g–1 FW). The shoot Pi concentration nearly doubled over the next 6 d to a final concentration of ~21 μmol g–1 FW. In roots, Pi levels increased to 9 μmol g–1 FW within 1 d of resupply, matching the Pi concentration found in roots of seedlings continuously receiving Pi (~10 μmol g–1 FW). Pi concentrations remained at this level for several days, before dropping to 6 μmol g–1 FW by day 7 (Fig. 4B). The drop in Pi was probably due to a combination of depletion from the medium, continued export to the shoot, conversion to organic P compounds, and internal dilution by root growth. In roots of Phi-treated seedlings, Phi accumulated to similar levels as Pi within 1 d, and remained high for the 7 d of the experiment, with a final concentration of 12 μmol g–1 FW. In shoots of Phi-treated seedlings, Phi concentrations were lower than the Pi concentrations in Pi-resupplied seedlings at the two earliest time points (3 μmol g–1 FW; Fig. 4A). After 3 d, the shoot Phi concentration of 10 μmol g–1 FW caught up with the shoot Pi concentration found after only 1 d of Pi resupply. At the final harvest, the shoot Phi concentration of 27 μmol g–1 FW in Phi-treated seedlings was higher than that of the free Pi concentration in resupplied seedlings, probably due to metabolic conversion of Pi but not Phi into organic compounds. In shoots of P-limited seedlings, the Pi concentration tended to decline over the course of the experiment to a final concentration of 1.5 μmol g–1 FW. The Pi concentration in roots of P-limited plants (2 μmol g–1 FW) was constant over the time-course. In roots and shoots of Phi-treated seedlings, the Pi concentration (3 μmol and 4 μmol Pi g–1 FW, respectively) was constant over time, and Pi concentrations at the final harvest were higher than those in P-limited organs, most probably due to Phi-induced Pi retention in the vacuole (Pratt et al., 2009).
Fig. 4.
Kinetics of phosphate (Pi) and phosphite (Phi) accumulation in seedling organs. Five-day-old seedlings were depleted of Pi for 4 d before being transferred to plates for the different treatments as indicated. (A) Shoot and (B) root accumulation of Pi in phosphorus-limited seedlings (white bars), upon Pi resupply (black bars), or with Phi treatment (grey bars). Phi accumulation in Phi-treated seedlings is shown as red bars. Shown are means ±SE, n=3 or n=4 replicates with 12 seedlings each. Statistically significant differences between time and treatments are indicated by different letters according to Tukey’s HSD at P<0.001.
Phosphite tissue accumulation was differentially affected among a set of pht1 mutants
To gather direct evidence that Phi is transported by Pi transporters of the PHT1 family, the Phi accumulation in roots and shoots of homozygous T-DNA insertion lines was analysed in the Col-0 background lacking either PHT1;1, one of the major Pi transporters at the root–soil interface (Shin et al., 2004), PHT1;8, or PHT1;9. The latter two PHT1 transporters are involved in translocation of Pi to the shoot (Lapis-Gaza et al., 2014). Seedlings were grown on vertical plates and depleted of Pi as described above, before supplying them with either 250 μM Pi or 250 μM Phi for 24h prior to harvest. Pi starvation led to similar residual organ Pi concentrations across genotypes (Fig. 5). Compared with the corresponding wild-type Col-0, the pht1;1–2 mutant accumulated 58% less Pi in roots and 22% less Pi in shoots of Pi-resupplied seedlings over the 24-h period (Fig. 5). The effect of this mutation on Phi uptake by P-limited seedlings was significantly more pronounced, leading to 71% less Phi in roots and 84% less Phi in shoots of the mutant than in the wild type. Knocking out PHT1;8 or PHT1;9 had no effect on either root or shoot Pi accumulation. In contrast to pht1;1–2, Phi concentrations in roots of both pht1;8 and pht1;9-1 were similar to those in the wild type, but Phi accumulation in shoots was reduced by 76% for pht1;8 and by 60% for pht1;9-1 compared with Col-0, the same extent as seen in pht1;1–2. The basal organ Pi concentrations in Phi-treated seedlings were similar across mutants. The same trends in organ Pi and Phi concentrations were observed after 2 d of treatment, although differences between Col-0 and the three mutants were diminished by day 7 (Supplementary Fig. S2 at JXB online). Throughout the time-course, root and shoot biomass accumulation was largely unaffected by the lack of individual PHT1 proteins (Supplementary Fig. S3).
Fig. 5.
Phosphate (Pi) and phosphite (Phi) accumulation in P-limited Col-0 and pht1 mutant organs after 1 d of Pi resupply or Phi treatment. Five-day-old seedlings were depleted of Pi for 4 d before being treated as indicated. (A) Shoot and (B) root accumulation of Pi in P-limited (white bars), Pi-resupplied (black bars), and Phi-treated (grey bars) seedlings and accumulation of Phi (red bars). Shown are means ±SE, n=3 replicates with 12 seedlings each. Genotypes and treatments with a letter in common are not significantly different according to Tukey’s HSD at P<0.05.
Phosphite altered transcript accumulation for a subset of Pi-responsive genes
The short-term effect of Phi on PSR gene expression was assessed by quantitative reverse-transcriptase PCR (qRT-PCR) for a set of well-documented PSR genes representing various metabolic and regulatory steps within plant P signalling networks (Hammond et al., 2003; Wu et al., 2003; Misson et al., 2005; Morcuende et al., 2007; Woo et al., 2012). If Phi was a true Pi analogue and sensed in the same way as Pi by as yet unidentified cellular signalling components, one would expect the effect of the two chemicals on transcript profiles to be similar; that is, lower transcript levels for Pi-starvation-induced genes and higher transcript abundance for genes involved in organophosphate biosynthesis or encoding negative regulators such as the E2 ubiquitin conjugase PHO2 (Aung et al., 2006; Bari et al., 2006) or the F-box protein FBX2 and transcription factor BHLH32 (Chen et al., 2008).
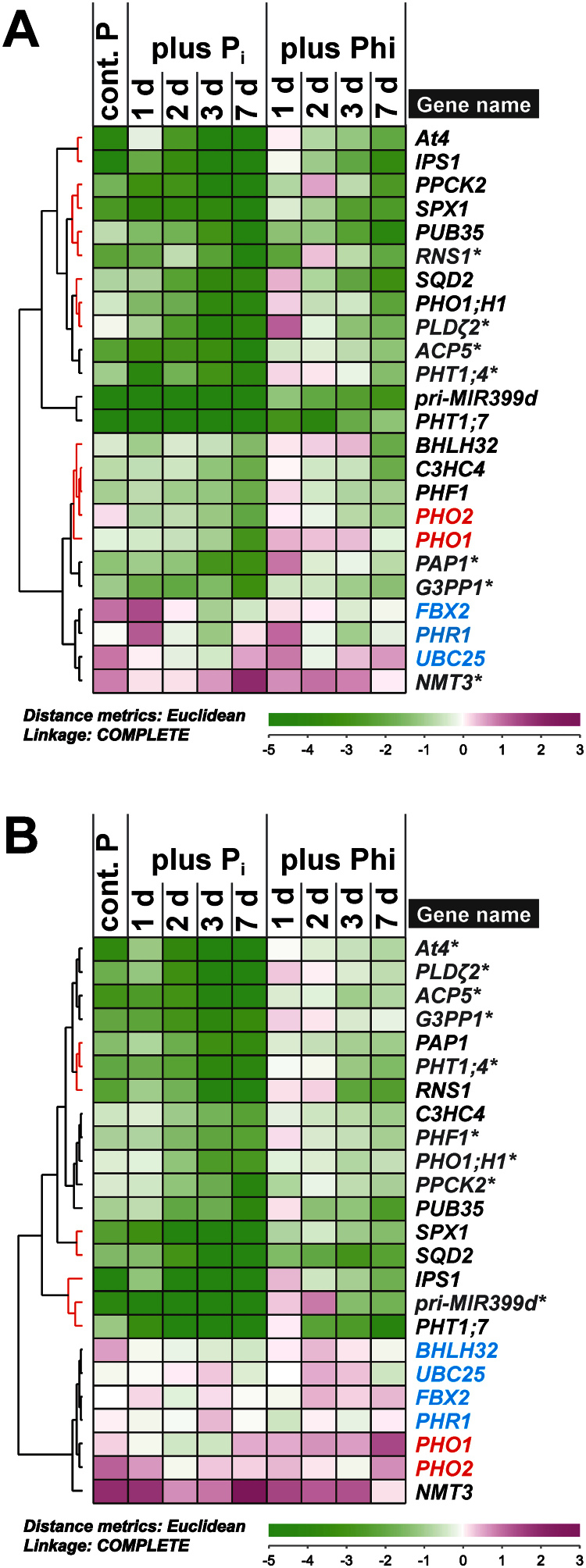
The selected PSR genes showed the previously documented expression changes within 1 d of Pi resupply (Fig. 6). Surprisingly, 33% of the target genes showed no significant change in transcript abundance in response to Phi in shoots of P-limited plants over the 7-d treatment period (Fig. 6A, grey transcript names). Within this non-responsive group were the Pi transporter gene PHT1;4, as well as genes involved in Pi metabolism (ACP5, G3PP1, NMT3, PAP1, PLD ζ2, and RNS1). In shoots, the majority of PSR genes tested showed an attenuated response to Phi treatment with a 1 d or 2 d delay compared with Pi resupply. This set included genes encoding regulatory components such as At4, IPS1, PHO1;H1, and SPX1, as well as genes encoding protein kinase PPCK2 and sulpholipid synthase SQD2 (Fig. 6A, red clusters). In contrast, other genes responded strongly to Phi, as they did to Pi resupply. These responses included an 8-fold suppression within 24h of Phi treatment for PHT1;7 transcript amounts, with a further 16-fold drop within 2 d of treatment. Similarly, transcripts encoding U-box-containing E3 ligase PUB35 were less abundant in shoots within 24h of Phi treatment. A milder suppression compared with Pi was observed for the primary transcript of regulatory microRNA miR399d. Transcripts encoding transcription factor BHLH32, E3 ubiquitin ligase C3HC4, and transport facilitator PHF1 responded more slowly but similarly to both Pi resupply and Phi treatment, with a >4-fold lower abundance than in shoots of P-limited seedlings at the end of the experiment. PHO2 transcripts showed an unexpected profile in shoots, with 2- to 4-fold lower levels in Pi-resupplied over P-limited seedlings. Phi treatment triggered a similar 2-fold decline in PHO2 transcripts within 3 d of treatment.
Fig. 6.
Effect of phosphate (Pi) and phosphite (Phi) on transcript abundance in P-limited seedlings. Hierarchical cluster analysis of a time-course on relative transcript abundance in P-limited Arabidopsis thaliana (A) shoots and (B) roots in response to Pi resupply or Phi treatment. Mean log2 expression ratios (–ΔΔCt) relative to the normalized expression in P-limited plants with three biological replicates for each sample are shown. Raw data were normalized against the transcript abundance of PP2AA3 and UBC9 reference genes. Clusters that contain Phi-responsive transcripts are highlighted by red lines in the tree. Transcripts in black change abundance in response to both Pi and Phi treatment, while those in grey (*) are unresponsive to Phi treatment (P≤0.05). PHO1 and PHO2 transcripts are highlighted in red. Transcripts in blue show no significant change in abundance across treatments. Details on individual transcript expression patterns and statistical analysis can be found in Supplementary Table S2 at JXB online.
Despite the fact that Phi accumulated as quickly as Pi in roots, 43% of the tested P-responsive transcripts did not respond to Phi in this organ (Fig. 6B, grey transcript names). Transcripts from ACP5, G3PP1, PLDζ2, and PHT1;4 were among those that were also identified as being non-responsive to Phi in shoots. In roots, Phi-non-responsive transcripts included those from At4, PHO1;H1, PHF1, and PPCK2, all of which responded to Phi to some extent in shoots. On the other hand, transcripts encoding phosphatase PAP1 and ribonuclease RNS1 were more responsive to Phi in roots compared with shoots. As in shoots, NMT3 transcript abundance in roots increased 2-fold in response to Phi within 24h of treatment, but transcript levels did not continue to increase and were 11-fold lower compared with roots of Pi-resupplied plants on day 7 (Supplementaty Table S2 at JXB online). PHT1;7, PUB35, SPX1, and SQD2 were highly Phi responsive in roots as well as in shoots. However, the response was relatively delayed in roots for PHT1;7 and PUB35, while SPX1 and SQD2 transcripts were more quickly suppressed in roots than in shoots. C3HC4, IPS1, and pri-MIR399d transcript abundance showed a weaker response to Phi in roots compared with shoots. In contrast to shoots, PHO1 transcript abundance did not respond to Pi resupply in roots. Curiously, within 48h of Phi treatment, PHO1 transcript abundance was ~2-fold greater than that in roots of P-limited plants and continued to increase throughout the time-course. PHO2 transcript abundance in roots did not respond to either Pi or Phi treatment.
It has to be noted that seedlings were not severely P starved at the beginning of the experiment. Evidence for this was the small changes in transcript abundance in organs of P-limited control plants at day 1 of the experiment compared with transcript levels in plants continuously supplied with Pi (blue bar in Supplementary Table S2 at JXB online). As a consequence, transcript levels of the target Pi-starvation-induced genes continued to increase over the time-course in P-limited control plants. This was also the case for those transcripts that did not show a response to Phi in Phi-treated seedlings.
In contrast to the gradual response to Pi deprivation, Pi resupply led to the suppression of Pi-starvation-induced genes within 24h (Supplementary Table S2 at JXB online). Thereafter, transcript abundance remained at the newly established lower levels for the rest of the time-course. Exceptions to this expression profile were those of microRNA antagonists IPS1 and At4, which showed a more gradual response to Pi resupply in both roots and shoots. In shoots, PHT1;4 transcripts also showed this gradual decrease in abundance in response to Pi. Unlike all other target genes, transcripts from both IPS1 and PHT1;4 decreased in abundance to below the level observed in shoots of seedlings that were continuously supplied with Pi. In roots, PHO2 transcript levels tended to increase transiently within 24h of Pi resupply, rather than showing a sustained increase over P-limited plants. PHO2 transcripts did not respond to Pi resupply in shoots.
Discussion
Phi has been demonstrated to suppress the induction of Pi-starvation responses. This conclusion was drawn from a series of experiments where P-sufficient plants were transferred to Pi-containing or Pi-free media supplemented with increasing Phi concentrations, or where seeds were germinated on these media (Carswell et al., 1996; Ticconi et al., 2001; Varadarajan et al., 2002; Berkowitz et al., 2013; Eshraghi et al., 2014). Thus, these studies focused on the ability of Phi to interfere with the induction of PSR genes in response to Pi removal or the lack of Pi supply. The experimental set-up used in this study allowed direct comparison of Phi and Pi effects on the suppression of Pi-starvation responses through monitoring plant growth, Pi anion and anthocyanin accumulation, as well as PSR gene expression. The experimental set-up has several advantages. (i) Withdrawal of Pi from the medium prior to Phi treatment avoids competition between the two anions for uptake. (ii) A direct comparison of Phi and Pi effects on the suppression of PSR genes can be conducted. (iii) Phi accumulation in the cytosol and organelles should be favoured over the vacuole under these conditions, so that more direct effects on metabolism and gene regulatory networks can be observed. (iv) The kinetic dependences of these effects on the accumulation of both P anions in roots and shoots can be determined.
Discrimination between Pi and Phi by PHT1 transporters
The differential movement of Phi and Pi into the shoots of plants suggests different affinities for these molecules within their transport routes. Measurements of transport kinetics in different systems have concluded that Pi transporters are able to transport Phi, albeit with a lower affinity than for Pi (d’arcy-Lameta and Bompeix, 1991; Pratt et al., 2004; Danova-Alt et al., 2008; Basheer et al., 2011). This means that Phi can bind to Pi transporter proteins without inducing the same conformational changes necessary for efficient transport (Basheer et al., 2011). It is unknown if all plant PHT transporters interact with Phi with the same affinity or whether some discriminate more strongly against Phi. In this study, the more pronounced delay in root-to-shoot transport of Phi in the pht1;8 and pht1;9-1 mutants than in wild-type seedlings, without a delay in Phi uptake, suggests that the encoded transporters discriminate more strongly against Phi than PHT1;1. The fact that discrimination is stronger in the absence of either PHT1;8 or PHT1;9 could mean that the two only partially complement each other (Lapis-Gaza et al., 2014) which would slow down transport even further. Alternatively, a third transport process, perhaps involving the Pi exporter PHO1 (Arpat et al., 2012), could be implicated in the stronger discrimination between Pi and Phi in both mutants. The alleviation of the Phi discrimination phenotype over time is most probably due to remobilization processes between sink and source organs involving other PHT transporters, such as PHT1;5 (Nagarajan et al., 2011).
Differential recognition of Phi by different PHT proteins may modulate not only transport activity, but also signalling events associated with this activity (Schothorst et al., 2013). It is unclear whether such a ‘transceptor’ function applies to the plant PHT family, but complex post-translational regulation has already been shown. Bayle et al. (2011) showed that some high-affinity PHT1 proteins undergo complex post-translational modifications, including protein phosphorylation. PHT1 protein abundance is also controlled by ubiquitin-mediated protein degradation (Lin et al., 2013; Park et al., 2014). Both PHT1;8 and PHT1;9 proteins can be distinguished from other family members by the presence of a PEST [proline, glutamic acid (E), serine, threonine] domain that mediates phosphorylation-dependent protein degradation in many systems (Rechsteiner and Rogers, 1996), for example the high- and low-affinity Pi transporters in yeast (Lagerstedt et al., 2004; Estrella et al., 2008).
Differential expression of ‘PHO regulon’ genes in response to local Pi signalling in roots and shoots
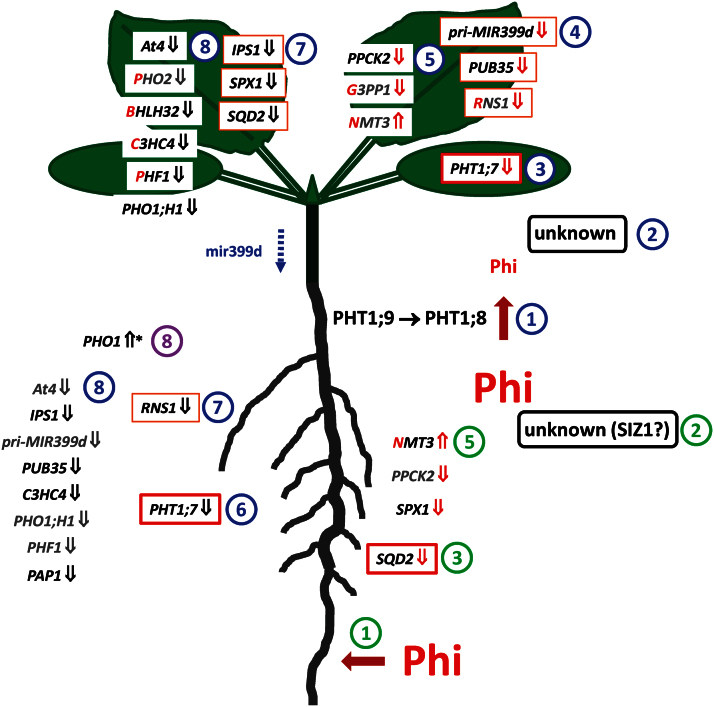
There is mounting evidence that the local and systemic control of PSR gene expression is governed by different signalling circuits in roots and shoots, and that different circuits within each organ respond either to the direct perception of Pi or to a more indirect process involving downstream metabolites or other as yet unidentified signals (Müller et al., 2004; Bari et al., 2006; Thibaud et al., 2010; Woo et al., 2012; Rojas-Triana et al., 2013). The discrimination between Pi and Phi by PHT1;8 and PHT1;9 shown in this study leads to a delayed accumulation of Phi in shoots. This delayed accumulation of Phi may hence be an elegant tool for dissecting direct sensing of Pi from other potential signals of P status in the shoot. PHT1;7 and pri-MIR399d transcripts in the present study were suppressed earlier in shoots than in roots and responded before Phi accumulated to significant levels. This would place them into an early-response circuit more directly connected to a Pi-specific sensor in the root-to-shoot transport route. PHT1;7 and pri-MIR399d expression was deregulated in the pht1;9-1 (Lapis-Gaza et al., 2014) and the phr1 mutant, but not the pho2 mutant (Bari et al., 2006). Slower shoot accumulation of Phi correlated with an attenuated down-regulation of a select subset of PSR genes closely associated with the ‘PHO regulon’, such as At4, IPS1, SPX1, PHF1, and PHO1;H1. This would support their response to local Pi or, in this case, Phi availability in the shoot. Interestingly, these genes were also deregulated in both P-limited phr1 and Pi-resupplied pho2 mutants (Bari et al., 2006). These findings may indicate that early Pi- and Phi-responsive genes are more directly connected to PHR1, possibly through a SIZ1-co-ordinated network in roots (Miura et al., 2005) and an unknown signalling component in shoots (Fig. 7) (Klecker et al., 2014). Only very few locally responsive PSR genes in the shoot seem to be PHO2 dependent (Pant et al., 2015). In a split-root system, genes that were systemically regulated in roots showed a strong enrichment of the P1BS element for PHR1 binding in their promoter regions (Thibaud et al., 2010). This may indicate differences in signal perception between roots and shoots. A clear distinction of regulatory groups of genes according to their responsiveness to Pi and Phi in space and time would therefore be useful to define individual response circuits further.
Fig. 7.
A model for the sequence of changes in phosphate-starvation-responsive (PSR) gene expression observed in roots and shoots of phosphorus-limited Arabidopsis thaliana seedlings in response to phosphite (Phi) treatment. Blue pathway: the discrimination of Phi by PHT1;9 and subsequently by PHT1;8 during xylem loading (1) may indicate the recognition by a receptor that signals the availability of phosphate (Pi) and Phi to the shoot, possibly involving SIZ1 (2). This sequence of events may primarily affect PHT1;7 expression in the shoot (3), followed by the consecutive suppression of other PSR genes within 24h (4+5) or later after 3 d when Phi finally started to accumulate in the shoot (7+8). Green pathway: in roots, early local recognition of Phi is possibly restricted to the suppression of SQD2 within 24h (3) and of the less responsive SPX1 and PPCK2 as well as to the induction of NMT3 (5). Compared with shoots, PHT1;7, RNS1 and a couple of transcripts in group 8 (in grey) responded more slowly in roots, most probably indicating that their expression in roots is regulated by PHO2 and relies on systemic signalling, perhaps through reduced levels of mir399d in the phloem (blue dotted arrow). Curiously, PHO1 expression in roots increased within 3 d of Phi treatment, which may indicate its connection to independent regulatory networks (purple 8) that directly respond to the overall P status of the plant or the growth inhibition triggered by Phi. Note that the number of genes responding equally well to either Pi or Phi (red first letter in gene name) was greater in shoots than in roots. Gene names in black indicate a 2-fold expression change in response to Phi over P-limited controls (Fig. 6). An orange border indicates a 4-fold expression change. A bold red border indicates an 8-fold change. Grey names indicate non-significant changes. Red arrows following gene names indicate suppression (↓) or induction (↑) within 24h of Phi exposure, while black arrows indicate a response within 3 d of treatment. An asterisk indicates a Phi-specific expression change that was not observed in Pi-resupplied seedlings.
PHO1 transcripts encoding a Golgi-localized Pi exporter (Arpat et al., 2012) showed a contrasting expression profile in roots to that of the other PSR genes tested in this study: instead of being suppressed by either Pi or Phi addition, they were more abundant in roots of Phi-treated compared with P-limited plants and did not respond to Pi resupply. In shoots, Pi resupply caused the down-regulation of PHO1, while Phi treatment caused a transient increase in PHO1 transcript levels similar to its effect in roots. PHO1 is therefore the only PSR gene tested that responded to the more severe depletion of local cytosolic Pi pools that is expected in the presence of Phi (Pratt et al., 2009). Alternatively, PHO1 expression may be triggered by the strong inhibition of seedling growth in the presence of Phi. In this context, it is interesting to note that shoot growth in transgenic lines with reduced PHO1 expression is uncoupled from the actual P status of the shoot (Rouached et al., 2011). PHO1-associated signalling components could therefore integrate growth stimuli and P status.
PHO1;H1 transcript accumulation was suppressed by Pi in both roots and shoots, with a strong suppression by Phi in shoots. These results confirm the findings of Stefanovic et al. (2007) showing that PHR1-dependent PHO1;H1 expression is Phi responsive, while PHR1-independent PHO1 expression is not. PHO1 and PHO1;H1 are SPX (SYG1, Pho81, and XPR1) domain proteins (Secco et al., 2012). Transcripts encoding another SPX domain protein, SPX1, responded to Phi in both roots and shoots. SPX1 is a competitive inhibitor of PHR1 binding to the P1BS element in PSR gene promoters (Puga et al., 2014). Its interaction with PHR1 is also highly dependent on the presence of either Pi or Phi. In contrast to most PSR genes in the present study, it responded much more quickly to Phi in roots. Both PHO1;H1 and SPX1 are regulated in a PHR1- and PHO2-dependent manner (Bari et al., 2006), but SPX1 is also controlled by SIZ1 (Duan et al., 2008). The latter may explain its more direct response to local Phi concentrations in the root (Miura et al., 2011). This would put SIZ1 into a position close to the local Pi- and Phi-sensing module in roots (Fig. 7). Surprisingly, SPX1 is also systemically regulated in P-limited roots in a split-root system (Thibaud et al., 2010).
PHO2 transcripts encoding an E2 ubiquitin conjugase (Aung et al., 2006; Bari et al., 2006) accumulated transiently in Pi-resupplied roots, but were largely unresponsive to Phi treatment. This suggests that PHO2 is connected to a signalling circuit that responds very sensitively to changes in overall P status, perhaps through monitoring concentrations of a downstream P metabolite (Klecker et al., 2014; Pant et al., 2015). In support of this interpretation, P-sufficient pht1;9 mutants showed a stronger accumulation of At4 and pri-MIR399d transcripts, and lower transcript accumulation of PHO2 in shoots which did not correlate with Pi concentrations in pht1;9 roots or shoots (Lapis-Gaza et al., 2014). The decline in PHO2 transcripts over the treatment period could therefore be an early response to the Pi depletion of the media resulting in lower levels of a downstream P metabolite. This Pi depletion after 7 d of treatment would also explain the observed lower Pi concentration and the higher transcript abundance for PHO1 and SPX1 in roots as well as increasing transcript levels for PHT1;7 and pri-MIR399d in shoots of Pi-resupplied seedlings. In shoots of Pi-resupplied seedlings, PHO2 expression was even lower than that in P-limited seedlings. Since At4 and IPS1 transcript levels were significantly lower in shoots in response to either Pi or Phi treatment, the late increase in pri-MIR399d transcript abundance, which underlies PHO2 repression, might explain the further drop in PHO2 transcript amounts in the shoot. In contrast to roots, this response was mimicked by Phi to some extent, again highlighting the differences in Pi perception between the two organs.
All the genes mentioned in this section respond very quickly to changes in P status. However, there is a clear distinction in the regulation of SPX1 that responds very early in roots, PHO1, which seems to respond to signals associated with growth, PHO2 which responds to unknown downstream P signals, and all other components of the ‘PHO regulon’ that do show strong responses to both Pi and Phi, especially in shoots. It is possible that the first perception of Pi takes place during root-to-shoot transport or within the shoot itself. Conversely, PSR gene expression in the root largely responds to secondary, shoot-derived signals as previously demonstrated (Bari et al., 2006; Lin et al., 2008; Thibaud et al., 2010).
Phosphite-dependent expression changes in roots affect transcripts for local lipid-remodelling pathways
In roots, transcripts encoding sulpholipid synthase SQD2 that catalyses the last step in sulpholipid biosynthesis and phosphoethanolamine N-methyltransferase NMT3 that synthesizes the head group of the phospholipid phosphatidylcholine responded very quickly to both Pi and Phi, while their response was slower in shoots. By contrast, PLDζ2 transcripts encoding a phospholipase D isoform showed a response to Pi, but not to Phi. NMT3 is one of the few genes that respond to Pi independently of PHR1 and PHO2 in A. thaliana seedlings (Bari et al., 2006). In the study of Woo et al. (2012), many lipid-remodelling genes such as PLDζ2 and SQD2 were among the group of genes that specifically responded to Pi in both roots and shoots. Their response to Pi was PHR1-dependent, but undisturbed in pho2 seedlings (Bari et al., 2006). They were also systemically regulated in a split-root system, but their induction was attenuated compared with that in P-limited control roots (Thibaud et al., 2010). These genes were also highly responsive to Phi in an earlier acclimation study (Berkowitz et al., 2013). Kobayashi et al. (2006) demonstrated that the promoter of another lipid-remodelling gene, MGD2, responds very strongly to Phi in roots and shoots, and that Phi is able to cause the modification of shoot lipid profiles in a similar fashion to Pi, including lower proportions of galactolipids and sulpholipids, and higher proportions of phospholipids. These findings indicate that direct sensing of Pi affects the lipid-remodelling pathway through PHR1-dependent and PHR1-independent signalling cascades, with a more rapid local perception of Pi in roots.
Phosphite effects on root architecture may be caused by altered accumulation of transcripts encoding proteins involved in protein turnover and vesicle trafficking
In this study, a U-box/ARM-repeat E3 ligase gene, PUB35, was responsive to the plant’s P status and was among a small group of PSR genes that were highly responsive to Phi, especially in roots. U-box and RING-finger E3 ligases, such as PUB35 and C3HC4, that responded to both Pi and Phi in the present study, are highly responsive to the plant’s P status, with many of them showing PHR1/PHL1-dependent regulation (Rojas-Triana et al., 2013). The U-box E3 ligase, PUB9, has recently been implicated in linking auxin-dependent and Pi-regulated lateral root emergence with vesicle trafficking. PUB9 interacts with the S-domain receptor kinase ARK2 that is implicated in P-derived signal recognition (Deb et al., 2014). The ark2-1/pub9-1 double mutant features shorter primary roots under low Pi supply, thus mimicking the Phi-induced phenotype in this study. While it has been demonstrated that several PUB E3 ligases can interact with ARK2 in vitro (Samuel et al., 2008), this has yet to be demonstrated for PUB35.
What makes this potential link between Pi signalling and Phi recognition particularly intriguing is the fact that U-box proteins have also been implicated in triggering plant immunity (Gonzalez-Lamothe et al., 2006; Trujillo et al., 2008). Many of the 64 predicted U-box-containing proteins in A. thaliana are associated with mono-ubiquitination and proteasomal degradation of signalling components during stress responses that trigger cell death (Yee and Goring, 2009).
Indirect effects of Phi treatment on plant growth
In P-limited cell suspension cultures, Phi exacerbates Pi starvation by inhibiting vacuolar efflux of Pi (Pratt et al., 2009). This could explain the arrest in primary and lateral root growth observed upon Phi treatment in this study and upon longer term Phi exposure (Berkowitz et al., 2013; Eshraghi et al., 2014). In both instances, the growth arrest was much more severe than the slowing of primary root elongation observed upon Pi withdrawal alone. However, plants in the present study were not experiencing severe Pi starvation, given that anthocyanin levels in leaves of P-limited controls only started to increase towards the end of the experiment. Also, a significant reduction of both root and shoot biomass in the presence of Phi compared with that of both continued P-limited growth and Pi resupply was only observed on the third day of treatment, at the time when Phi first significantly accumulated in shoots. The inhibitory effect that Phi has on organ growth might thus be directly triggered by the accumulation of Phi within the cytosol and organelles of the shoot (Danova-Alt et al., 2008; Pratt et al., 2009). As would be expected from a mildly cytotoxic agent, Phi then seems to affect both root and shoot growth in a similar fashion. This is very different from the opposing hormonal effects on root and leaf development (King et al., 1995; Werner et al., 2003).
These observations would imply that whilst Phi is a great tool to tease apart direct, Pi-triggered effects on P signalling networks from those further downstream, care has to be taken to interpret longer term effects due to its immediate toxicity on many Pi-dependent metabolic pathways.
Conclusion
The present results indicate that Phi is perceived as Pi and suggest that this perception is stronger in shoots than in roots. The perception of Phi most probably affects distinct regulatory circuits in both organs, and is more closely associated with factors that interact with PHR1-associated networks, such as SIZ1 (Miura et al., 2011). The strong root architectural changes induced by Phi in P-limited plants are most probably invoked by its interference with local signalling components that affect lipid remodelling (PLDζ2, SQD2, NMT3) and protein turnover (PUB35, C3HC4). In the longer term, Phi severely affects plant growth, most probably by inhibiting vital Pi-dependent metabolic pathways. Several of these pathways have the potential to trigger the priming of plant defences. Used with caution, Phi can be a useful tool in further disentangling these complex interactions.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Root phenotypic responses in phosphorus-limited plants to phosphate resupply or phosphite treatment.
Figure S2. Phosphate and phosphite accumulation in roots and shoots of Col-0 and pht1 mutants over time.
Figure S3. Biomass accumulation in roots and shoots of Col-0 and pht1 mutants over time.
Table S1. Information on target genes and primers used in qRT-PCR analyses.
Table S2. Time-course of relative transcript abundance of known phosphate-responsive genes in phosphorus-limited A. thaliana seedlings in response to either phosphate resupply or phosphite treatment.
Acknowledgements
This work was supported by the Australian Research Council (LP0776252 to HL and PMF), an Endeavour Fellowship from the Australian Government (to MP), an Academic Recharging Fellowship from the Indonesian Directorate of Higher Education (to MP), and a Scholarship for International Research Fees from the University of Western Australia (to HRL-G).
Glossary
Abbreviations:
- P
phosphorus
- Phi
phosphite
- Pi
inorganic phosphorus/phosphate/H2PO4–
- PSR
phosphate-starvation-responsive.
References
- Ames BN. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods in Enzymology 8, 115–118. [Google Scholar]
- Arpat AB, Magliano P, Wege S, Rouached H, Stefanovic A, Poirier Y. 2012. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. The Plant Journal 71, 479–491. [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. 2006. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiology 141, 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Pant BD, Stitt M, Scheible WR. 2006. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology 141, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basheer S, Samyn D, Hedstrom M, Thakur MS, Persson BL, Mattiasson B. 2011. A membrane protein based biosensor: use of a phosphate–H(+) symporter membrane protein (Pho84) in the sensing of phosphate ions. Biosensors and Bioelectronics 27, 58–63. [DOI] [PubMed] [Google Scholar]
- Bayle V, Arrighi JF, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L. 2011. Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. The Plant Cell 23, 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz O, Jost R, Kollehn DO, Fenske R, Finnegan PM, O’Brien PA, Hardy GE, Lambers H. 2013. Acclimation responses of Arabidopsis thaliana to sustained phosphite treatments. Journal of Experimental Botany 64, 1731–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz O, Jost R, Pearse SJ, Lambers H, Finnegan PM, Hardy GES, O’Brien PA. 2011. An enzymatic fluorescent assay for the quantification of phosphite in a microtiter plate format. Analytical Biochemistry 412, 74–78. [DOI] [PubMed] [Google Scholar]
- Bozzo GG, Singh VK, Plaxton WC. 2004. Phosphate or phosphite addition promotes the proteolytic turnover of phosphate-starvation inducible tomato purple acid phosphatase isozymes. FEBS Letters 573, 51–54. [DOI] [PubMed] [Google Scholar]
- Carswell MC, Grant BR, Plaxton WC. 1997. Disruption of the phosphate-starvation response of oilseed rape suspension cells by the fungicide phosphonate. Planta 203, 67–74. [DOI] [PubMed] [Google Scholar]
- Carswell C, Grant BR, Theodorou ME, Harris L, Niere JO, Plaxton WC. 1996. The fungicide phosphonate disrupts the phosphate-starvation response in Brassica nigra seedlings. Plant Physiology 110, 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Jenkins GI, Nimmo HG. 2008. Identification of an F-box protein that negatively regulates Pi starvation responses. Plant and Cell Physiology 49, 1902–1906. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalio RJD, Fleischmann F, Humez M, Osswald W. 2014. Phosphite protects Fagus sylvatica seedlings towards Phytophthora plurivora via local toxicity, priming and facilitation of pathogen recognition. PLoS One 9, e87860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danova-Alt R, Dijkema C, De Waard P, Köck M. 2008. Transport and compartmentation of phosphite in higher plant cells—kinetic and P-31 nuclear magnetic resonance studies. Plant, Cell and Environment 31, 1510–1521. [DOI] [PubMed] [Google Scholar]
- d’arcy-Lameta A, Bompeix G. 1991. Systemic transport of tritiated phosphonate in tomato plantlets (Lycopersicon esculentum Mill). Pesticide Science 32, 7–14. [Google Scholar]
- Deb S, Sankaranarayanan S, Wewala G, Widdup EE, Samuel M. 2014. The S-domain receptor kinase AtARK2 and the U-box/ARM-repeat-containing E3 ubiquitin ligase AtPUB9 module mediates lateral root development under phosphate starvation in Arabidopsis. Plant Physiology 165, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias-Arieira CR, de Melo Santana-Gomes S, Higashi Puerari H, Fernanda Fontana L, Martins Ribeiro L, Mattei D. 2013. Induced resistance in the nematodes control. African Journal of Agricultural Research 8, 2312–2318. [Google Scholar]
- Duan K, Yi KK, Dang L, Huang HJ, Wu W, Wu P. 2008. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. The Plant Journal 54, 965–975. [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Forde BG. 2012. Quantitative analysis of lateral root development: pitfalls and how to avoid them. The Plant Cell 24, 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dysvik B, Jonassen I. 2001. J-Express: exploring gene expression data using Java. Bioinformatics 17, 369–370. [DOI] [PubMed] [Google Scholar]
- Eshraghi L, Anderson JP, Aryamanesh N, McComb JA, Shearer B, Hardy GE. 2014. Suppression of the auxin response pathway enhances susceptibility to Phytophthora cinnamomi while phosphite-mediated resistance stimulates the auxin signalling pathway. BMC Plant Biology 14, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella LA, Krishnamurthy S, Timme CR, Hampsey M. 2008. The Rsp5 E3 ligase mediates turnover of low affinity phosphate transporters in Saccharomyces cerevisiae . Journal of Biological Chemistry 283, 5327–5334. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JDG. 2006. The U-Box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. The Plant Cell 18, 1067–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ. 2003. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiology 132, 578–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofgaard IS, Ergon A, Henriksen B, Tronsmo AM. 2010. The effect of potential resistance inducers on development of Microdochium majus and Fusarium culmorum in winter wheat. European Journal of Plant Pathology 128, 269–281. [Google Scholar]
- Hou XL, Wu P, Jiao FC, Jia QJ, Chen HM, Yu J, Song XW, Yi KK. 2005. Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant, Cell and Environment 28, 353–364. [Google Scholar]
- Jain A, Poling MD, Smith AP, Nagarajan VK, Lahner B, Meagher RB, Raghothama KG. 2009. Variations in the composition of gelling agents affect morphophysiological and molecular responses to deficiencies of phosphate and other nutrients. Plant Physiology 150, 1033–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost R, Berkowitz O, Masle J. 2007. Magnetic quantitative reverse transcription PCR: a high-throughput method for mRNA extraction and quantitative reverse transcription PCR. Biotechniques 43, 206–211. [DOI] [PubMed] [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB. 1995. A mutation altering auxin homeostasis and plant morphology in Arabidopsis. The Plant Cell 7, 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecker M, Gasch P, Peisker H, Dörmann P, Schlicke H, Grimm B, Mustroph A. 2014. A shoot-specific hypoxic response of Arabidopsis thaliana sheds light on the role of the phosphate-responsive transcription factor PHR1. Plant Physiology 165, 774–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Masuda T, Takamiya KI, Ohta H. 2006. Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. The Plant Journal 47, 238–248. [DOI] [PubMed] [Google Scholar]
- Lagerstedt JO, Voss JC, Wieslander Å, Persson BL. 2004. Structural modeling of dual-affinity purified Pho84 phosphate transporter. FEBS Letters 578, 262–268. [DOI] [PubMed] [Google Scholar]
- Lapis-Gaza HR, Jost R, Finnegan PM. 2014. Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biology 14, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ. 2008. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiology 147, 732–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WY, Huang TK, Chiou TJ. 2013. NITROGEN LIMITATION ADAPTATION, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. The Plant Cell 25, 4061–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machinandiarena MF, Lobato MC, Feldman ML, Daleo GR, Andreu AB. 2012. Potassium phosphite primes defense responses in potato against Phytophthora infestans . Journal of Plant Physiology 169, 1417–1424. [DOI] [PubMed] [Google Scholar]
- Massoud K, Barchietto T, Le Rudulier T, Pallandre L, Didierlaurent L, Garmier M, Ambard-Bretteville F, Seng JM, Saindrenan P. 2012. Dissecting phosphite-induced priming in Arabidopsis infected with Hyaloperonospora arabidopsidis . Plant Physiology 159, 286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald AE, Grant BR, Plaxton WC. 2001. Phosphite (phosphorous acid): its relevance in the environment and agriculture and influence on plant phosphate starvation response. Journal of Plant Nutrition 24, 1505–1519. [Google Scholar]
- Misson J, Raghothama KG, Jain A, et al. 2005. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proceedings of the National Academy of Sciences, USA 102, 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Gong QQ, Ma SS, Jin JB, Yoo CY, Miura T, Sato A, Bohnert HJ, Hasegawa PM. 2011. SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiology 155, 1000–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, et al. 2005. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proceedings of the National Academy of Sciences, USA 102, 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, et al. 2007. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant, Cell and Environment 30, 85–112. [DOI] [PubMed] [Google Scholar]
- Müller R, Nilsson L, Krintel C, Nielsen TH. 2004. Gene expression during recovery from phosphate starvation in roots and shoots of Arabidopsis thaliana . Physiologia Plantarum 122, 233–243. [Google Scholar]
- Nagarajan VK, Jain A, Poling MD, Lewis AJ, Raghothama KG, Smith AP. 2011. Arabidopsis PHT1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiology 156, 1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nartvaranant P, Hamill S, Leonardi J, Whiley AW, Subhadrabandhu S. 2004. Seasonal effects of foliar application of phosphonate on phosphonate translocation, in vitro pollen viability and pollen germination in ‘Hass’ avocado (Persea americana Mill.). Journal of Horticultural Science and Biotechnology 79, 91–96. [Google Scholar]
- Pant B-D, Pant P, Erban A, Huhman D, Kopka J, Scheible W-R. 2015. Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus-limitation. Plant, Cell and Environment 38, 172–187. [DOI] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua N-H. 2014. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. The Plant Cell 26, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasek MA, Harnmeijer JP, Buick R, Gull M, Atlas Z. 2013. Evidence for reactive reduced phosphorus species in the early Archean ocean. Proceedings of the National Academy of Sciences, USA 110, 10089–10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival GC, Banks JM. 2014. Evaluation of plant defence activators for the potential control of Pseudomonas syringae pv. aesculi . Arboricultural Journal 36, 76–88. [Google Scholar]
- Poehlein A, Daniel R, Schink B, Simeonova DD. 2013. Life based on phosphite: a genome-guided analysis of Desulfotignum phosphitoxidans . BMC Genomics 14, 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt J, Boisson AM, Gout E, Bligny R, Douce R, Aubert S. 2009. Phosphate (Pi) starvation effect on the cytosolic Pi concentration and Pi exchanges across the tonoplast in plant cells. An in vivo 31P-NMR study using methylphosphonate as a Pi analogue. Plant Physiology 151, 1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt JR, Mouillon JM, Lagerstedt JO, Pattison-Granberg J, Lundh KI, Persson BL. 2004. Effects of methylphosphonate, a phosphate analogue, on the expression and degradation of the high-affinity phosphate transporter Pho84, in Saccharomyces cerevisiae . Biochemistry 43, 14444–14453. [DOI] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, et al. 2014. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, 14947–14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratjen AM, Gerendas J. 2009. A critical assessment of the suitability of phosphite as a source of phosphorus. Journal of Plant Nutrition and Soil Science – Zeitschrift für Pflanzenernährung und Bodenkunde 172, 821–828. [Google Scholar]
- Rechsteiner M, Rogers SW. 1996. PEST sequences and regulation by proteolysis. Trends in Biochemical Sciences 21, 267–271. [PubMed] [Google Scholar]
- Remy E, Cabrito TR, Batista RA, Teixeira MC, Sa-Correia I, Duque P. 2012. The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytologist 195, 356–371. [DOI] [PubMed] [Google Scholar]
- Ribot C, Wang Y, Poirier Y. 2008. Expression analyses of three members of the AtPHO1 family reveal differential interactions between signaling pathways involved in phosphate deficiency and the responses to auxin, cytokinin, and abscisic acid. Planta 227, 1025–1036. [DOI] [PubMed] [Google Scholar]
- Rojas-Triana M, Bustos R, Espinosa-Ruiz A, Prat S, Paz-Ares J, Rubio V. 2013. Roles of ubiquitination in the control of phosphate starvation responses in plants. Journal of Integrative Plant Biology 55, 40–53. [DOI] [PubMed] [Google Scholar]
- Rouached H, Stefanovic A, Secco D, Bulak Arpat A, Gout E, Bligny R, Poirier Y. 2011. Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. The Plant Journal 65, 557–570. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthbaum HP, Baille WJH. 1964. The use of red phosphorus as a fertilizer. Part 4. Phosphite and phosphate retention in soil. New Zealand Journal of Science 7, 446–451. [Google Scholar]
- Samuel MA, Mudgil Y, Salt JN, Delmas F, Ramachandran S, Chilelli A, Goring DR. 2008. Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiology 147, 2084–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schothorst J, Kankipati H, Conrad M, Samyn D, Zeebroeck G, Popova Y, Rubio-Texeira M, Persson B, Thevelein J. 2013. Yeast nutrient transceptors provide novel insight in the functionality of membrane transporters. Current Genetics 59, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Wang C, Arpat BA, Wang ZY, Poirier Y, Tyerman SD, Wu P, Shou HX, Whelan J. 2012. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytologist 193, 842–851. [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. 2004. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. The Plant Journal 39, 629–642. [DOI] [PubMed] [Google Scholar]
- Singh VK, Wood SM, Knowles VL, Plaxton WC. 2003. Phosphite accelerates programmed cell death in phosphate-starved oilseed rape (Brassica napus) suspension cell cultures. Planta 218, 233–239. [DOI] [PubMed] [Google Scholar]
- Smillie R, Grant BR, Guest D. 1989. The mode of action of phosphite—evidence for both direct and indirect modes of action on 3 Phytophthora spp in plants. Phytopathology 79, 921–926. [Google Scholar]
- Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J, Belbahri L, Delessert S, Poirier Y. 2007. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. The Plant Journal 50, 982–994. [DOI] [PubMed] [Google Scholar]
- Tasaki M, Asatsuma S, Matsuoka K. 2014. Monitoring protein turnover during phosphate starvation-dependent autophagic degradation using a photoconvertible fluorescent protein aggregate in tobacco BY-2 cells. Frontiers in Plant Science 5, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao HTB, Yamakawa T. 2009. Phosphite (phosphorous acid): fungicide, fertilizer or bio-stimulator? Soil Science and Plant Nutrition 55, 228–234. [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. 2010. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. The Plant Journal 64, 775–789. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Abel S. 2001. Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiology 127, 963–972. [PMC free article] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S. 2004. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. The Plant Journal 37, 801–814. [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, et al. 2005. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. The Plant Journal 42, 218–235. [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. 2008. Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Current Biology 18, 1396–1401. [DOI] [PubMed] [Google Scholar]
- Varadarajan DK, Karthikeyan AS, Matilda PD, Raghothama KG. 2002. Phosphite, an analog of phosphate, suppresses the coordinated expression of genes under phosphate starvation. Plant Physiology 129, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. 2003. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. The Plant Cell 15, 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J, Macpherson CR, Liu J, Wang H, Kiba T, Hannah M, Wang XJ, Bajic VB, Chua NH. 2012. The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biology 12, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrolstad RE, Durst RW, Lee J. 2005. Tracking color and pigment changes in anthocyanin products. Trends in Food Science and Technology 16, 423–428. [Google Scholar]
- Wu P, Ma LG, Hou XL, Wang MY, Wu YR, Liu FY, Deng XW. 2003. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiology 132, 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D, Goring DR. 2009. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. Journal of Experimental Botany 60, 1109–1121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.