Highlight
NO generated by nitrate reductase plays a pivotal role in improving N-use efficiency by increasing lateral root initiation and inorganic N uptake, representing a strategy for rice to adapt to fluctuating nitrate supply.
Abstract
Increasing evidence shows that partial nitrate nutrition (PNN) can be attributed to improved plant growth and nitrogen-use efficiency (NUE) in rice. Nitric oxide (NO) is a signalling molecule involved in many physiological processes during plant development and nitrogen (N) assimilation. It remains unclear whether molecular NO improves NUE through PNN. Two rice cultivars (cvs Nanguang and Elio), with high and low NUE, respectively, were used in the analysis of NO production, nitrate reductase (NR) activity, lateral root (LR) density, and 15N uptake under PNN, with or without NO production donor and inhibitors. PNN increased NO accumulation in cv. Nanguang possibly through the NIA2-dependent NR pathway. PNN-mediated NO increases contributed to LR initiation, 15NH4 +/15NO3 – influx into the root, and levels of ammonium and nitrate transporters in cv. Nanguang but not cv. Elio. Further results revealed marked and specific induction of LR initiation and 15NH4 +/15NO3 – influx into the roots of plants supplied with NH4 ++sodium nitroprusside (SNP) relative to those supplied with NH4 + alone, and considerable inhibition upon the application of cPTIO or tungstate (NR inhibitor) in addition to PNN, which is in agreement with the change in NO fluorescence in the two rice cultivars. The findings suggest that NO generated by the NR pathway plays a pivotal role in improving the N acquisition capacity by increasing LR initiation and the inorganic N uptake rate, which may represent a strategy for rice plants to adapt to a fluctuating nitrate supply and increase NUE.
Introduction
Nitrogen (N) nutrition affects all levels of plant functions, including metabolism resource allocation, growth, and development (Crawford, 1995; Stitt, 1999). Plants have the potential to adapt to dramatic fluctuations in N availability by modulating their nutrient acquisition capacity and by altering their metabolism and morphology, such as their root architecture (Crawford, 1995; López-Bucio et al., 2003; Song et al., 2013a ; Manoli et al., 2014). A dual effect of external nitrate on root system development has been demonstrated in the model plant Arabidopsis thaliana: (i) localized stimulation of lateral root (LR) elongation at the site of contact with a nitrate-rich supply and (ii) systemic inhibition of lateral primordia by uniformly high nitrate concentrations during the post-emergence stage (Zhang and Forde, 1998; Zhang et al., 1999; Linkohr et al., 2002; Zhang et al., 2007; Ruffel et al., 2011; Forde, 2014). Several known pathways involve microRNAs, transcription factors, hormonal signals, and nitrate transporters with a dual affinity for nitrate and auxin (Remans et al., 2006; Chiou, 2007; Gifford et al., 2008; Krouk et al., 2010; Vidal et al., 2010; Gojon et al., 2011; Ruffel et al., 2011; Trevisan et al., 2011, 2012a, b ; Xu et al., 2011; Zhao et al., 2011, 2012; Nischal et al., 2012; Zhao et al., 2013; Alvarez et al., 2014; Begheldo et al., 2014; Manoli et al., 2014; Zeng et al., 2014). However, our understanding of how plants sense external nitrate conditions and the signal transduction system that influences root system development and nutrient acquisition capacity remains limited.
Nitric oxide (NO) is a signalling molecule involved in many physiological processes during plant development and nutrient assimilation. Several lines of evidence suggest that NO is involved in the growth and development of the root system (Pagnussat et al., 2003; Correa-Aragunde et al., 2004,2006; Lombardo et al., 2006; Stöhr and Stremlau, 2006; Guo et al., 2008; Chen et al., 2010; Wang et al., 2010; Fernández-Marcos et al., 2011; Meng et al., 2012; Niu et al., 2013; Wang et al., 2013). Correa-Aragunde et al. (2004) suggested that auxin and NO are involved in a linear signalling pathway during the LR formation process in tomato. Several recent reports highlighted a role for NO in N assimilation and suggested that it is an important regulator of the first steps in the nitrate-sensing pathway (Du et al., 2008; Jin et al., 2009; Rosales et al., 2010; Sanz-Luque et al., 2013; Frungillo et al., 2014). For example, nitrate reductase (NR) in the leaves of wheat was inhibited by NO (Rosales et al., 2010), but was activated in cabbage (Du et al., 2008). Indeed, the inhibition or activation of NR by NO in tomato roots may depend on the nitrate concentration (Jin et al., 2009). However, the mechanisms of NO-regulated N uptake in response to nitrate fluctuation remain poorly understood.
NO synthase (NOS) and NR are two potential enzymatic sources of NO production in plants. Plant NOS has not yet been identified (Crawford et al., 2006; Moreau et al., 2008, 2010; Gas et al., 2009; Gupta et al., 2011), although experiments using inhibitors of the animal NOS enzyme have provided some evidence of the role of the L-arginine pathway in NO production (Zhao et al., 2007). NR is one of the most important sources of NO in plants, and the nitrate concentration in the rooting medium can affect the amount of NO via mediation of NR activity (Yamasaki et al., 1999; Meyer et al., 2005; Yamasaki, 2005). Studies on Arabidopsis and Vicia faba confirmed that the relative and absolute levels of nitrate and nitrite were key determinants of NR-induced production of NO (Vanin et al., 2004). Moreover, the localization of mRNA for NR in the root apex corresponded to the major sites of NO accumulation, as seen in Arabidopsis (Stöhr and Stremlau, 2006), suggesting that these NR genes represent pivotal elements of a finely tuned NO homeostasis and signalling system. NO is a nitrate-related signal generated by the NR pathway to regulate root system architecture (Zhao et al., 2007; Manoli et al., 2014; Trevisan et al., 2014). However, the role of NO in the nitrate signalling pathway requires further investigation.
Rice (Oryza sativa L.), one of the most important staple food crops globally, is traditionally cultivated under flood conditions. Although ammonium (NH4 +) is preferred over nitrate as the form of N as a nutrient in rice, rice roots are exposed to partial nitrate nutrition (PNN) due to nitrification in the rhizosphere (Li et al., 2008). There is increasing evidence that PNN improves rice growth by inducing N uptake and assimilation, as well as the formation of adventitious roots and LRs (Qian et al., 2004; Duan et al., 2007a, b ; Cao et al., 2008; Li et al., 2008; Zhang et al., 2011; Song et al., 2011a, b , 2013a , b ). In previous studies, seven rice cultivars with high N-use efficiency (NUE) and three with low NUE were selected from 177 japonica rice cultivars to study their responsiveness to PNN (Duan et al., 2007a ; Zhang et al., 2008; Song et al., 2011a ). Under hydroponic conditions, five of the seven rice cultivars with high NUE were sensitive to PNN (Duan et al., 2007b ), and three rice cultivars with low NUE were insensitive to PNN (Song et al., 2011a ). Moreover, experiments using rice cultivars with contrasting NUE demonstrated that the increase in NUE caused by PNN is attributable to improved N uptake (Duan et al., 2007b ), suggestive of a possible relationship between PNN and NUE. It was hypothesized that NO plays a key role in the improved NUE of rice as an evolutionary adaptation to nitrate nutrition based on the following observations: (i) significant induction by PNN of nitrate concentrations in rice cultivars with high rather than low NUE relative to treatment with NH4 + alone (Song et al., 2011a , b); and (ii) significant induction by PNN of auxin accumulation, and formation of adventitious roots and LRs in rice cultivars with high NUE (Song et al., 2011b, 2013a). Auxin showed common stages with NO in the signal transduction cascade, which resulted in auxin-induced adventitious root and LR formation (Pagnussat et al., 2003; Correa-Aragunde et al., 2004). In this study, the role of NO in the regulation of rice LR formation and N-uptake capacity in response to PNN was examined in two rice cultivars with contrasting nitrate responses and N-uptake efficiencies. It was proposed that a high NUE in rice plants was attributable to NO generated by NR, which induces LR formation and inorganic N uptake.
Materials and methods
Plant materials and growth
Two rice cultivars (cvs Nanguang and Elio) were selected from 177 japonica rice cultivars based on their similar growth patterns and differential responses to N application in field trials conducted in 2003 and 2004 (Zhang et al., 2007, 2008). Cv. Nanguang was identified as a high-nitrate-response and high-NUE cultivar, while cv. Elio was a low-nitrate-response and low-NUE cultivar (Duan et al., 2007a, b; Zhang et al., 2009).
Cvs Nanguang and Elio were grown in a greenhouse under natural light at day/night temperatures of 30 °C/18 °C. Seven-day-old seedlings of uniform size and vigour were transplanted into holes in a lid placed over the top of pots (four holes per lid and three seedlings per hole). Nutrient solutions varying from a quarter to half-strength were applied for 2 d, after which full-strength nutrient solution was applied for an additional 14 d. Seedlings were subjected to two NH4 +-N/NO3 –-N ratios, namely 100/0 (NH4 +) and 75/25 (PNN), by adding 1.43mM in the form of (NH4)2SO4 or a mixture of (NH4)2SO4 and NH4NO3. The chemical composition of the International Rice Research Institute (IRRI) nutrient solution was (mM): 0.3 KH2PO4, 0.35 K2SO4, 1.0 CaCl2, 1.0 MgSO4·7H2O, 0.5 Na2SiO3, and (μM) 20.0 Fe-EDTA, 9.0 MnCl2, 0.39 (NH4)6Mo7O24, 20.0 H3BO3, 0.77 ZnSO4, and 0.32 CuSO4; pH 5.5. A nitrification inhibitor (dicyandiamide, 7.0 μM) was added to each pot to prevent NH4 + oxidation. The nutrient solution was replaced daily with fresh solution. Nitrate was not detected in medium containing NH4 + alone. Rice plants were harvested 14 d after treatments. Each treatment consisted of four replicates arranged in a completely randomized design to avoid edge effects. In addition, all experiments included three independent biological replicates.
Preliminary experiments were conducted to determine the final amount of pharmacological application for cvs Nanguang and Elio. Pharmacological responsiveness to sodium nitroprusside (SNP) and tungstate (Tu) differed between the two rice cultivars (Supplementary Figs S1, S2 available at JXB online). For example, application of 1 μM or 2.5 μM SNP in addition to sole NH4 + nutrition induced an LR density of cv. Nanguang to a similar level to PNN treatment. However, the application of SNP (≥5 μM) induced LR density in cv. Elio comparable with that of sole NH4 + nutrition. Similarly, application of 50 μM Tu in addition to PNN decreased the LR density of cv. Nanguang to a similar level to sole NH4 + treatment. However, application of 100 μM Tu decreased the LR density of cv. Elio compared with PNN. Thus, different concentrations of SNP (2.5 μM for cv. Nanguang and 5 μM for cv. Eilo) and Tu (50 μM for cv. Nanguang and 100 μM for cv. Eilo) were applied in subsequent experiments. In addition, 80 μM 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) was applied to the plant growth media.
Measurement of LR density and LR primordia number
As reported previously, PNN increased the formation of adventitious roots and LRs in cv. Nanguang (Song et al., 2011b ). During shorter durations of the experiments (14 d), LR formation increased significantly but adventitious root formation was unaffected (Song et al., 2013a ). Thus, LR density and primordia were selected for subsequent, detailed analyses.
In this study, stages of LR development followed Malamy and Benfey (1997), with stages I–XII grouped here as unemerged primordia. The primordia of LRs were classified into unemerged and emerged primordia. An emerged LR primordium longer than 0.5mm (visible to the naked eye) was considered an LR, and was referred to as being activated (Song et al., 2013a ). To visualize the development of LRs, pDR5::GUS transgenic rice plants were exploited. After the roots were stained in GUS (β-glucuronidase) buffer solution, it was simple to count the number of primordia and LRs. The scaleplate on the stereomicroscope (Olympus Optical Co. Ltd, Tokyo, Japan) simplifies determination of the length of emerged primordia and LRs. Seminal root length was measured with a ruler, and LR density was calculated as LR number divided by seminal root length.
Measurements of NO in the roots
NO was assayed using DAF-FM DA (diaminofluorescein-FM diacetate) and epifluorescence microscopy. Roots were loaded with 10 μM DAF-FM DA in 20mM HEPES-NaOH buffer (pH 7.5). After incubating in darkness for 30min, the roots were washed three times with fresh buffer and immediately visualized (OLYMPUS MVX10 stereomicroscope, with a colour CCD camera, excitation at 488nm, emission at 495–575nm). Signal intensities of green fluorescence in the images were quantified according to the method of Jin et al. (2011) using Photoshop software (Adobe Systems). Data are presented as mean fluorescence intensities.
Determination of total N concentration and nitrate reductase activity (NRA)
The total N concentration in plants was determined using the Kjeldahl method (Li et al., 2008). Maximum and active NR activities (NRAmax and NRAact) were measured in fresh roots using the method described by Li et al. (2008).
Determination of 15N uptake rate
The 15N influx rate was assayed as described previously (Orsel et al., 2006) for hydroponically grown plants. Seedlings were grown in the presence of 1.43mM NH4 + for 14 d and then N starved for 3 d. The plants were transferred to 0.1mM CaSO4 for 1min, then to a complete nutrient solution with the same N concentration (1.43mM) containing 15NH4 +, 15NH4 +/15NO3 – (PNN), 15NH4 ++SNP (2.5 μM, cv. Nanguang; 5 μM, cv. Eilo), and 15NH4 +/15NO3 – (PNN)+cPTIO (80 μM for two cultivars) for 10min, and finally to 0.1mM CaSO4 for 1min. The 15N abundance in each fraction was determined using a MAT25 isotope mass spectrometer. NH4 + was labelled by [atom% 15N: (15NH4)2SO4, 40%] and PNN was labelled by [atom% 15N: (15NH4)2SO4, 40%/15NH4 15NO3, 40% (50/50)] during the treatments.
Quantitative reverse transcription–polymerase chain reaction (qRT–PCR)
Total RNA was isolated from the roots of rice seedlings. RNA extraction, reverse transcription, and qRT–PCR were performed as described previously (Tang et al., 2012). Amplification of real-time quantitative PCR products was performed with a single-colour Real-Time PCR Detection System (MyiQ Optical Module; Bio-Rad) in a reaction mixture of 20 μl of SYBR Green master mix (SYBR Premix Ex Tag TMII; TaKaRa Bio; http://www.takara-bio.com) according to the manufacturer’s instructions (TaKaRa Biotechnology). The choice of the reference gene (OsACT) was supported by Tang et al. (2012). Primers and gene locus numbers for the NIA1, NIA2, NOA, AMT1, AMT2, AMT3, NRT2, and NAR2 genes are listed in Supplementary Tables S1 and S2 at JXB online.
Data analysis
Experimental data were pooled to calculate means and standard errors (SE), and were analysed by one-way analysis of variance (ANOVA) followed by least significant difference (LSD) to determine the significance of differences between individual treatments. All statistical procedures were conducted using SPSS ver. 11.0 (SPSS Inc., Chicago, IL, USA). In all analyses, P<0.05 was considered to indicate statistical significance.
Results
LR density and NO increased in cv. Nanguang in response to PNN relative to sole NH4 + nutrition
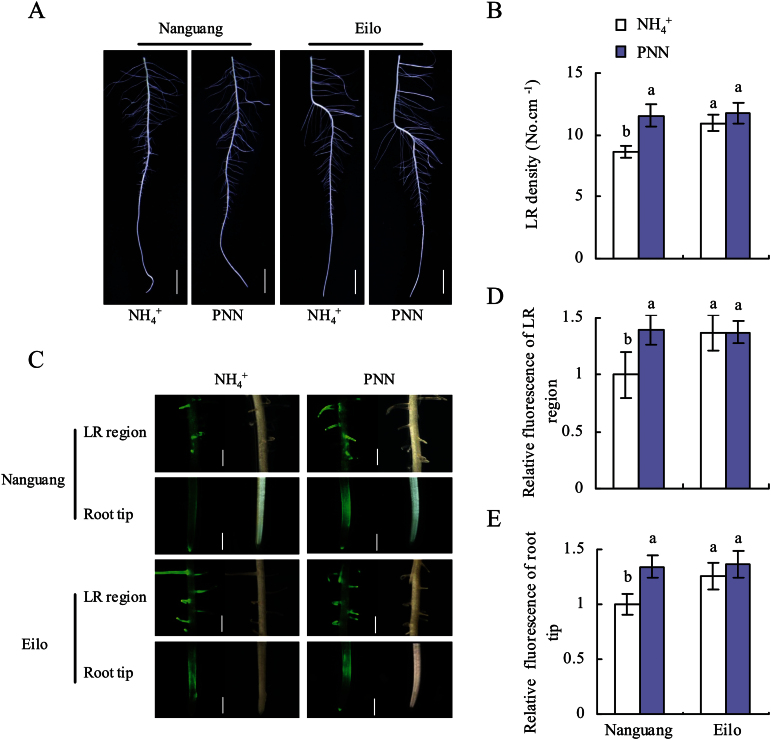
As reported previously (Song et al., 2011a, b, 2013a ), LR density of the seminal root in the high-nitrate-response cultivar Nanguang, but not the low-nitrate-response cultivar Elio, increased significantly under PNN conditions compared with NH4 + alone (Fig. 1A, B). The NO-associated green fluorescence in both the root tip and LR regions of cv. Nanguang increased under PNN treatment, but no difference was observed in cv. Elio (Fig. 1C). Quantification of the fluorescence signal intensities (Fig. 1D, E) indicated an ~34% increase in the NO content in the LR region and the root tip of cv. Nanguang under PNN conditions compared with NH4 + treatment. These results indicated that NO production in the root tip and LR regions of cv. Nanguang under PNN conditions was higher than that with NH4 + treatment.
Fig. 1.
Lateral root (LR) development and NO accumulation in the tip and LR regions of seminal roots in cvs Nanguang and Elio. (A) LR morphology in the seminal root; scale bar=1cm. (B) LR density in the seminal root. (C) NO production, as indicated by green fluorescence, in representative roots. Scale bar=1mm. (D and E) NO production expressed as the fluorescence intensity relative to the same root region of Nanguang under NH4 + supply. Seedlings were grown under NH4 + and partial nitrate nutrition (PNN) for 14 d in hydroponic media. Data are means ± standard error (SE), and bars with different letters indicate significant differences at P<0.05, as determined by ANOVA.
NO participated in PNN-induced LR formation in cv. Nanguang but not in cv. Eilo
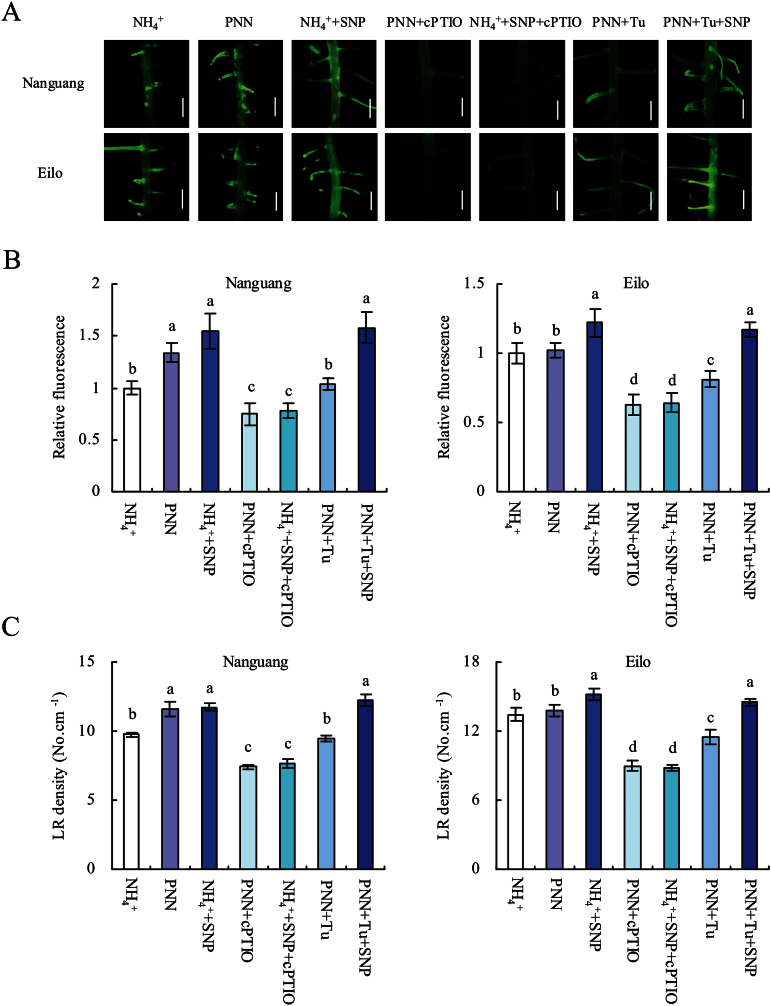
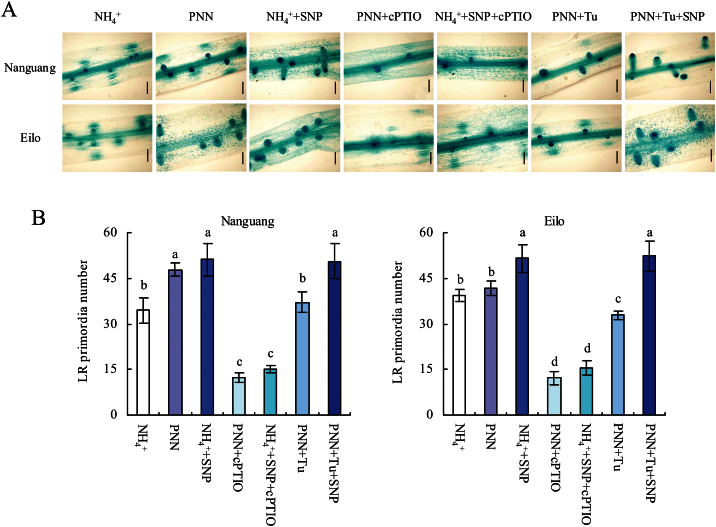
The effects of an NO donor and scavenger (SNP and cPTIO) on LR density and primordia were examined to determine whether the effect of PNN on the LR density in cv. Nanguang was mediated by NO. In cv. Nanguang, LR density increased upon application of 1–10 μM SNP. In cv. Elio, LR density increased slightly upon application of up to 5 μM SNP, and increased significantly with concentrations ≥5 μM (Supplementary Fig. S1 at JXB online). Thus, different SNP concentrations (2.5 μM, cv. Nanguang; 5 μM, cv. Elio) were used in subsequent analyses (Figs 2, 3). Application of SNP markedly induced NO accumulation and LR density and primordia in both rice cultivars (Fig. 3). Treatment with the NO scavenger cPTIO resulted in inhibition of PNN- and SNP-induced LR density and primordia in cv. Nanguang to a similar level. In cv. Elio, cPTIO application markedly decreased NO accumulation and LR density and primordia (Figs 2, 3). These results suggested that NO participated in LR formation in both rice cultivars and that NO participated in PNN-induced LR formation in cv. Nanguang but not in cv. Eilo.
Fig. 2.
Lateral root (LR) development and NO accumulation in the LR region of seminal roots in cvs Nanguang and Elio. (A) NO production in the LR region is shown as green fluorescence. (B) NO production expressed as the fluorescence intensity relative to the LR region of cvs Nanguang and Elio under NH4 + supply. (C) LR density in the seminal root. Seedlings were grown under NH4 + and partial nitrate nutrition (PNN) with or without application of SNP (2.5 μM, cv. Nanguang; 5 μM, cv. Eilo), cPTIO (80 μM for the two cultivars), or tungstate (Tu; 50 μM, cv. Nanguang; 100 μM, cv. Eilo) for 14 d in hydroponic media. Scale bar=1mm. Data are means ± standard error (SE), and bars with different letters indicate significant differences at P<0.05, as determined by ANOVA.
Fig. 3.
Lateral root (LR) primordia number in cvs Nanguang and Elio. Seedlings were grown under NH4 + and partial nitrate nutrition (PNN) with or without application of SNP (2.5 μM, cv. Nanguang; 5 μM, cv. Eilo), cPTIO (80 μM for the two cultivars), or tungstate (Tu; 50 μM, cv. Nanguang; 100 μM, cv. Eilo) for 14 d in hydroponic media. Scale bar=200 μm. Data are means ± standard error (SE), and bars with different letters indicate significant differences at P<0.05, as determined by ANOVA.
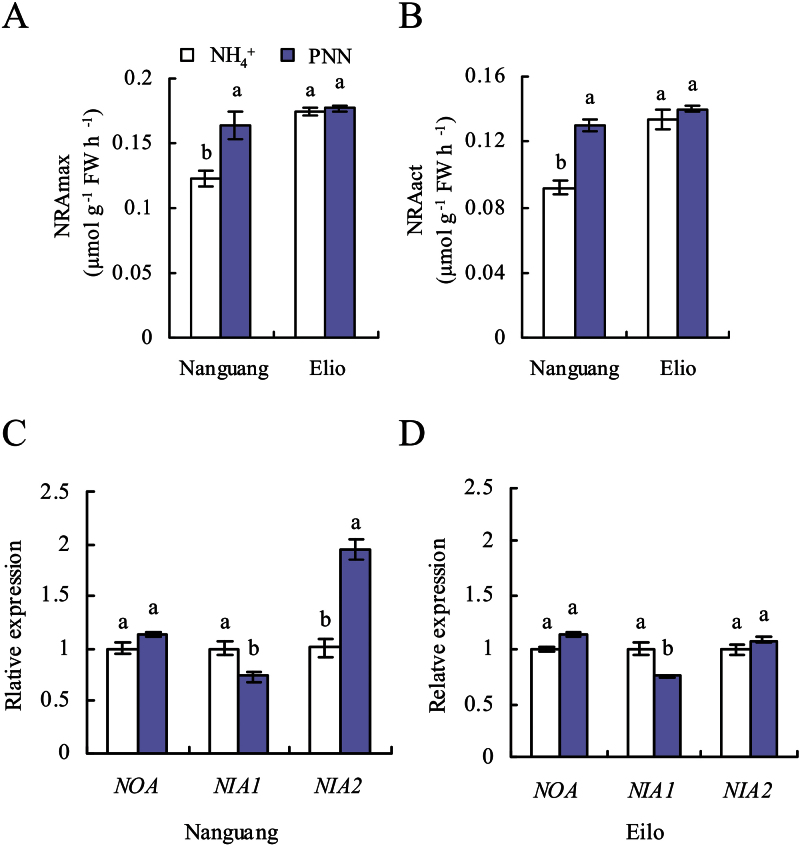
The PNN-induced NO accumulation is derived mainly from an NIA2-dependent NR source
NRAmax and NRAact levels in the roots of cv. Nanguang increased by 33% and 41% under PNN compared with NH4 + treatment, while no differences were observed in cv. Elio (Fig. 4A, B). Cao et al. (2008) reported that NIA2 expression was highly induced by nitrate in rice cultivars. NIA2 expression in the root of cv. Nanguang increased ~2-fold under PNN treatment compared with NH4 + treatment, consistent with the findings of Cao et al. (2008) (Fig. 4C). No difference in NIA2 expression was observed in the roots of cv. Elio. However, NIA1 expression decreased slightly in the roots of both rice cultivars (Fig. 4C, D). This change in NIA2 expression is in agreement with that of NRAmax and NRAact, indicating that PNN affected NR at the transcriptional level. Upon treatment of cv. Nanguang with the NR inhibitor Tu under PNN conditions, the NO-associated green fluorescence and the LR density and primordia decreased to levels similar to those under NH4 + treatment (Figs 2, 3). Similarly, Tu application significantly decreased DAF fluorescence, and LR density and primordia in cv. Elio. Furthermore, NOA expression in the two rice cultivars did not differ between the two N treatments (Fig. 4C, D). These results suggested that NIA2-dependent NR-derived NO is a key signal in PNN-induced LR formation in cv. Nanguang.
Fig. 4.
Maximum and active nitrate reductase activities (NRAmax and NRAact), and qRT–PCR analysis of OsNOA, OsNIA1, and OsNIA2 expression in the roots of cvs Nanguang and Elio. (A and B) NRAmax and NRAact in cvs Nanguang and Elio. (C and D) OsNOA, OsNIA1, and OsNIA2 expression levels in cvs Nanguang (C) and Elio (D). Seedlings were grown under NH4 + and partial nitrate nutrition (PNN) for 14 d (A and B) and 2 d (C and D) in hydroponic media. Relative mRNA levels were normalized to OsACT. Data are means ± standard error (SE), and bars with different letters in the four treatments (A and B) and the same genes (C and D) indicate significant differences at P<0.05, as determined by ANOVA.
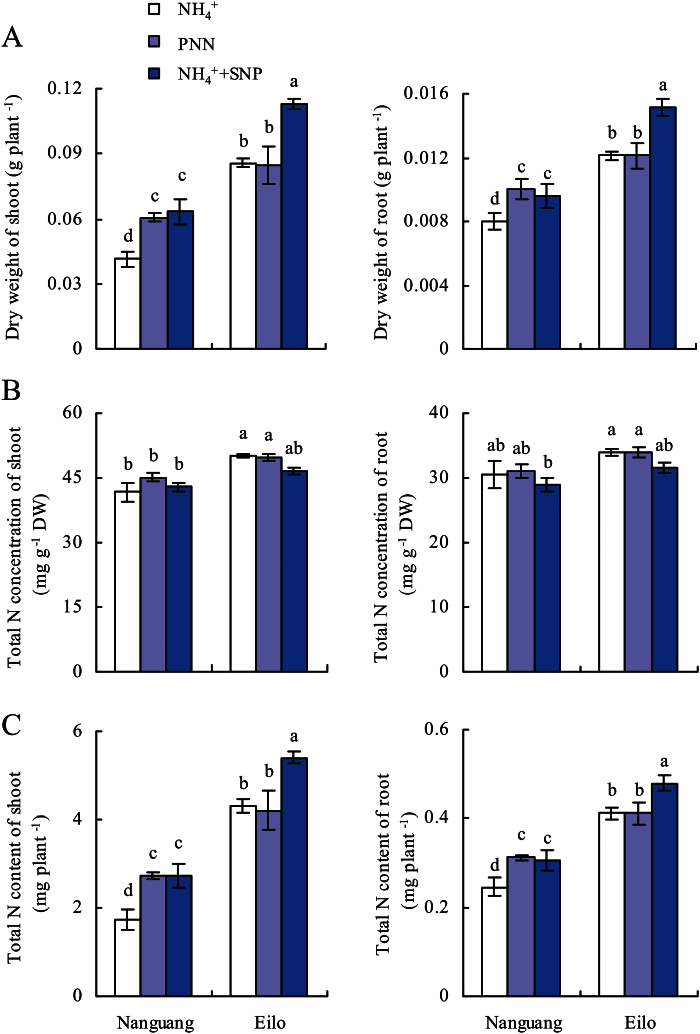
Role of NO in PNN-enhanced biomass and N accumulation
As reported previously (Duan et al., 2007a, b; Cao et al., 2008; Song et al., 2013b ), PNN treatment increased the dry weight and N accumulation of cv. Nanguang, but not cv. Eilo. For example, the dry weights of shoots and roots increased by 46% and 25%, respectively, in cv. Nanguang with PNN treatment compared with NH4 + treatment (Fig. 5A, B). However, no differences were recorded for cv. Elio. Interestingly, SNP application in addition to NH4 + nutrition increased the dry weights of shoots and roots in rice cultivars. The total N concentration did not differ in each rice cultivar under the three treatments (Fig. 5B). However, the total N contents of shoots and roots increased under SNP application in both cvs Nanguang and Eilo. These results indicated that NO might be involved in N accumulation of cv. Nanguang under PNN treatment. For example, PNN treatment increased the total N contents of shoots and roots in cv. Nanguang by ~57% and 27%, respectively, and SNP application by ~57% and 25%, respectively (Fig. 5C). In addition, application of SNP induced the total N contents of shoots and roots in cv. Elio. These results supported the involvement of NO as a key signal in PNN-induced N accumulation in rice.
Fig. 5.
Growth and N accumulation of cvs Nanguang and Elio in response to partial nitrate nutrition (PNN) and SNP. Dry weight (A), total N concentration (B), and total N content (C) in the shoots and roots of cvs Nanguang and Elio. Rice seedlings were grown under NH4 + or PNN for 14 d in hydroponic media with or without SNP (2.5 μM, cv. Nanguang; 5 μM, cv. Eilo). Data are means ± standard error (SE), and bars with different letters indicate significant differences at P<0.05, as determined by ANOVA.
Role of NO in PNN-enhanced N uptake rate
PNN treatment increased the rate of 15NH4 + uptake in cv. Nanguang, but not cv. Elio (Duan et al., 2007b ). To explore the effects of NO on NH4 + and NO3 – influx into intact plants, seedlings of both rice cultivars were incubated in solutions containing 15NH4 +, 15NH4 +/15NO3 – (PNN), 15NH4 ++SNP, and 15NH4 +/15NO3 – (PNN)+cPTIO for 10min. Figure 6 shows that PNN treatment increased 15N influx in cv. Nanguang, but not cv. Eilo. Meanwhile, application of SNP in addition to NH4 + nutrition increased the 15N influx, and cTPIO application in addition to PNN reduced the 15N influx in both rice cultivars (Fig. 6). These results supported the involvement of NO as a key signal in PNN-induced N uptake rate in rice.
Fig. 6.
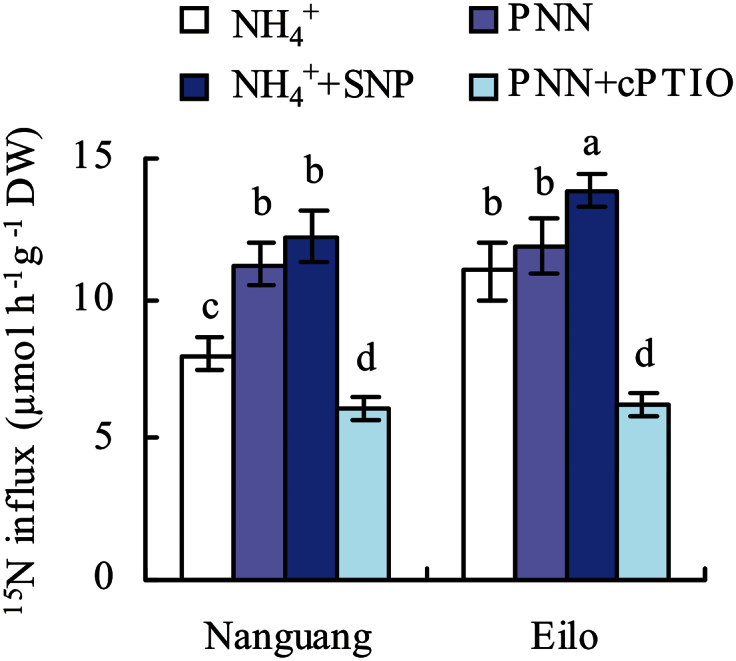
15N influx rates of the roots of cvs Nanguang and Eilo as measured by 15NH4 + and 15NO3 –. Seedlings were grown under 1.43mM NH4 + for 14 d and then N starved for 3 d, before being resupplied with 15NH4 +, 15NH4 +/15NO3 – (75/25, PNN), 15NH4 ++SNP (2.5 μM, cv. Nanguang; 5 μM, cv. Eilo), and 15NH4 +/15NO3 – (75/25, PNN)+cPTIO (80 μM for the two cultivars) for 10min before harvest. Data are means ± standard error (SE), and bars with different letters indicate significant differences at P<0.05, as determined by ANOVA.
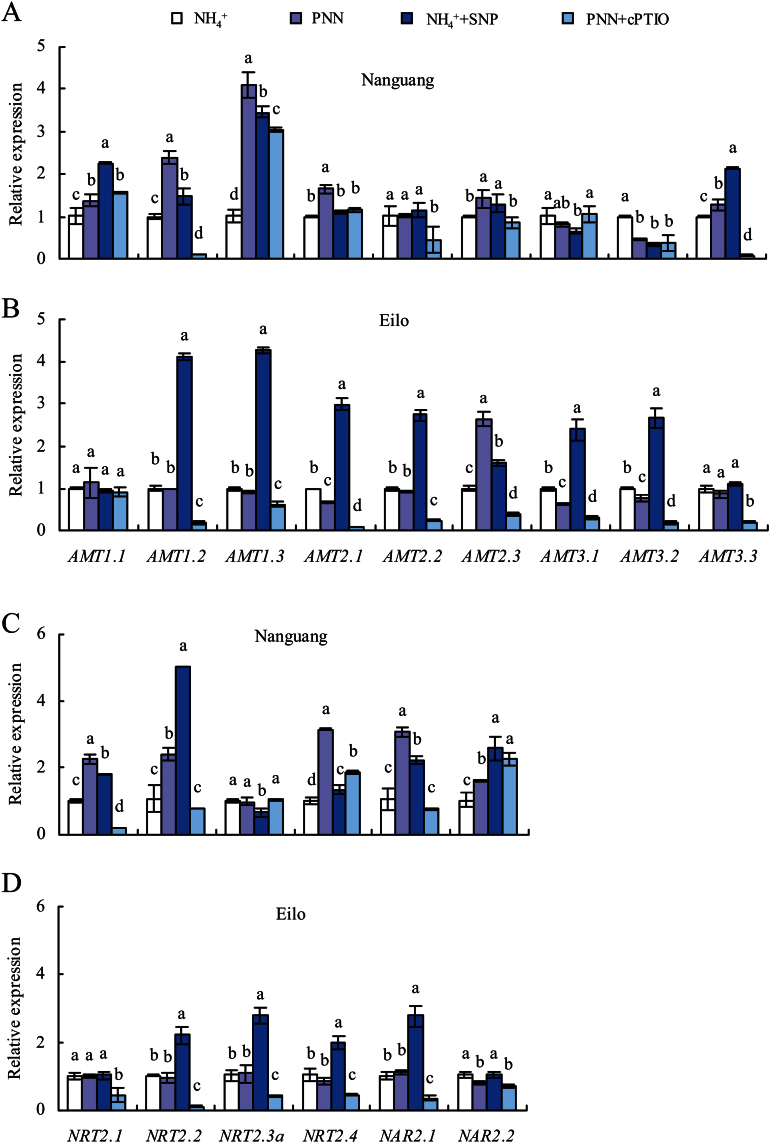
NO donor and scavenger affected OsAMT1–3, OsNRT2, and OsNAR2 gene expression
The expression levels of AMT, NRT2, and NAR2 genes were analysed using qRT–PCR (Fig. 7). The expression of AMT1.1–3, AMT2.1, AMT2.3, AMT3.3, NRT2.1–2, NRT2.4, and NAR2.1–2 were significantly increased under PNN compared with NH4 + in cv. Nanguang (Fig. 7A, C). SNP application under NH4 + conditions induced transcript levels of most genes in cv. Nanguang. Conversely, the application of cPTIO under PNN conditions decreased the expression levels of most genes in cv. Nanguang, with the exception of AMT3.1, NRT2.3a, and NAR2.2 (Fig. 7A, C). These results indicated that NO increased the N uptake rate under PNN treatment by up-regulating the expression of most nitrate and ammonium transporter genes.
Fig. 7.
qRT–PCR analysis of AMT1–3, NRT2, and NAR2 family genes in cvs Nanguang (A and C) and Elio (B and D) rice seedlings. Seedlings were grown in nutrient solution containing NH4 + only and partial nitrate nutrition (PNN) with or without SNP (2.5 μM, cv. Nanguang; 5 μM, cv. Eilo) and cPTIO (80 μM for tthe two cultivars) for 2 d in hydroponic media. Relative mRNA levels were normalized to OsACT. Data are means ± standard error (SE), and bars with different letters indicate significant differences at P<0.05 as determined by ANOVA.
Compared with cv. Nanguang, the two N treatments affected the expression levels of few genes in cv. Elio. Only the expression of AMT2.3 was significantly increased by PNN in cv. Eilo compared with the NH4 + treatment (Fig. 7B). Application of SNP under NH4 + conditions increased the expression of AMT1.2–3, AMT2.1–3, AMT3.1–2, NRT2.2–4, and NAR2.1 in cv. Eilo compared with NH4 + treatment alone (Fig. 7B, D). Meanwhile, the application of cPTIO decreased the expression levels of most genes in cv. Elio, with the exception of AMT1.1. These results showed that, for cv. Elio, PNN did not increase the N uptake rate and did up-regulate the expression level of AMT2.3. NO also increased the expression levels of most nitrate and ammonium transporters.
Discussion
Nitrogen is a major limiting element of plant growth, and crops depend on intense fertilization, which has potential negative effects on the environment (Xu et al., 2012). The situation is worse in China due to the added pressure of a large population (Zhang et al., 2009). The identification of crop cultivars with improved nutrient acquisition efficiencies in low-input farming systems continues to be an important goal for plant scientists (Robertson and Vitousek, 2009; Xu et al., 2012). Due to its lower 1000-grain weight, the rice cv. Nanguang, a high-NUE and high-nitrate-response cultivar, had less biomass accumulation than cv. Elio, a low-NUE and low-nitrate-response cultivar, during the early growth stage. However, a higher growth rate was recorded in cv. Nanguang, resulting in higher biomass accumulation and grain yield than in cv. Elio during the maturity stage (Duan et al., 2007b ; Zhang et al., 2007; Zhang et al., 2009). Previous studies have shown that PNN increased the N uptake efficiency, mainly by increasing root growth and the NH4 + uptake rate, which was attributed to high NUE in cv. Nanguang relative to sole NH4 + nutrition (Duan et al., 2007a, b; Li et al., 2008; Zhang et al., 2011; Song et al., 2011a , b, 2013a ). However, the mechanism(s) by which rice senses external nitrate conditions and the corresponding signal transduction pathway remains unclear.
Together with its role as an essential nutrient, nitrate acts as a signal to trigger a number of molecular and physiological events that lead to the overall response of a plant to N availability (Gojon et al., 2011). More recently, molecular NO has been reported to be a key signal in nitrate sensing as an early response to nitrate supply in maize roots (Trevisan et al., 2011; Manoli et al., 2014). The nitrate supply caused consistent increases in DAF fluorescence during the first minutes. The role of NOA during NO synthesis remains controversial. It has been demonstrated that Arabidopsis NOA1 does not possess NOS activity, being instead a GTPase (Moreau et al., 2008). However, Schlicht et al. (2013) identified noa1 mutants that contained lower NO levels in the root relative to the wild type. In this study, the NOA gene, a homologue of NOA1 in Arabidopsis, showed a similar transcript level in the two rice cultivars between the two N treatments (Fig. 4C, D). It is possible that PNN did not increase NO levels through the NOS pathway; alternatively, NOA may not play a role in NO synthesis. Moreover, the application of chemicals that interfere with NO biosynthesis and scavenging provided further evidence that NO was produced by NR rather than by NOS.
Nitrate reductase is also involved in NO production in response to biotic and abiotic stresses in several upland plants (Modolo et al., 2005; Srivastava et al., 2009; Zhao et al., 2009; Freschi et al., 2010; Kolbert et al., 2010). The observations that NIA1 (but not NIA2) expression was sensitive to hormonal and developmental cues (Yu et al., 1998), and the role of NIA1 in NO production in guard cells (Bright et al., 2006) and in response to cold acclimation (Zhao et al., 2009), support that NIA1 is a key component of NR-mediated NO production. It is known that nitrate rapidly induces the expression of NR genes in plants. However, the NR genes responsible for NR-mediated NO production in response to nitrate remain unclear.
In this study, it was shown that NO production was generated by NR in response to nitrate nutrition in rice roots. First, NRAmax and NRAact were induced significantly in response to nitrate supply in cv. Nanguang, but not cv. Elio, compared with in the presence of NH4 + nutrition. The NRAact to NRAmax ratio remained constant under the two N treatments, indicating that PNN did not regulate NR at the post-translational level. The significant increase in NO accumulation in the root tip and LR zone of rice plants was consistent with the pattern of NR activity. Secondly, for rice plants, the transcript level of NIA2 was considerably higher than that of NIA1 (Fan et al., 2007; Cao et al., 2008), consistent with the results in Arabidopsis (Wilkinson and Crawford, 1991). The expression of NIA2 in cv. Nanguang was markedly induced by the nitrate supply and correlated well with tissue NR activity, consistent with the results of Cao et al. (2008). However, different responses of NIA1 mRNA levels to PNN in rice plants were recorded in the present study and by Cao et al. (2008), possibly resulting from the fact that ammonium is the predominant N supply form for this species. Although a strong correlation between NIA2 mRNA levels and NRA was observed, further experiments are required to evaluate the effects of PNN-induced changes. Thirdly, application of the NR inhibitor Tu in addition to PNN reduced DAF fluorescence in both cvs Nanguang and Eilo, suggestive of the involvement of NR in NO generation in rice roots.
The most important example of plant plasticity with regard to N availability is their ability to rearrange their root architecture and maximize nutrient capture (Lόpez-Bucio et al., 2003; Zhang et al., 2007; Forde, 2014). Nitrate affects root development by regulating the growth of LRs in a manner dependent on its localization and external concentration (Zhang and Forde, 1998; Zhang et al., 1999; Linkohr et al., 2002; Zhang et al., 2007; Ruffel et al., 2011; Mounier et al., 2013; Forde, 2014). Due to the previous results showing PNN-induced LR density during a 14 d experimental stage with no change in seminal and adventitious roots relative to sole NH4 + nutrition, the effects of nitrate on NO generation in the root tip and LR zone were examined (Fig. 1C). These results showed marked and specific induction of LR initiation in seminal roots of rice seedlings supplied with NH4 ++SNP relative to those supplied with NH4 + alone, and considerable inhibition upon the application of cPTIO or Tu with the PNN. This result is in agreement with the change in relative NO fluorescence in the two rice cultivars. These results suggest that the NO generated by NR contributes to LR initiation in response to PNN.
Nitrate acts both as a nutrient and as a signal that regulates plant N acquisition and metabolism (Crawford, 1995; Stitt, 1999; Forde, 2002). Early studies on nitrate signalling showed that nitrate induced expression of NR and nitrate transporters in the nitrate assimilation pathway (Zielke and Filner, 1971; Somers et al., 1983; Stitt, 1999; Wang et al., 2000). Studies on nitrate as a nutrient suggested that its ability to prevent acidification of the root medium plays an important role in maintaining the plasma membrane potential and therefore the physiological patterns of NH4 + uptake (Babourina et al., 2007). Previous reports have shown that PNN induced increased N accumulation by enhancing the N-uptake efficiency in rice cultivars with high NUE (Duan et al., 2007b ). In agreement with this, N accumulation in cv. Nanguang increased by 53% by PNN or SNP in addition to NH4 + nutrition, but was enhanced by SNP treatment only in cv. Elio. These effects were probably due to NO induction of N uptake and assimilation. NR activity in cabbage roots was consistently higher following SNP treatment than in the control, while the NR protein content was unaffected by SNP (Du et al., 2008), indicating that NO stimulated NR at the post-translational level. Similarly, in beech seedlings, the NO-mediated increase in the ammonium uptake rate was mediated at the level of protein modification rather than gene expression (Simon et al., 2009). However, no such phenomenon was detected in Scots pine seedlings (Simon et al., 2013), suggesting that the effects of NO on N uptake differ among plant species. The present short-duration 15N labelling experiments showed a significant induction of 15N influx in rice seedlings supplied with PNN (15NH4 +/15NO3 –) or 15NH4 ++SNP relative to NH4 + nutrition alone, and a marked inhibition upon application of cPTIO in addition to the PNN supply. Interestingly, expression of high-affinity ammonium transporters (AMT1.2–1.3) and high-affinity nitrate transporters (NRT2.2 and NAR2.1) in both rice cultivars was induced significantly by SNP, but reduced by cPTIO application. This result is in agreement with the changes in 15N influx, suggesting that NO regulated N uptake at the transcriptional level.
Taken together, these results suggest that NO generated by NR plays a pivotal role in improving N acquisition capacity by modulating LR initiation and the N-uptake rate. This may represent a strategy by which rice plants adapt to variations in nitrate supply and increase their NUE.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Figure S1. Effect of the NO donor SNP on LR density of seminal roots.
Supplementary Figure S2. Effect of the NR inhibitor tungstate on LR density of seminal roots.
Supplementary Table S1. The primers for qRT–PCR of OsNOA, OsNIA1, and OsNIA2 genes.
Supplementary Table S2. The primers for qRT–PCR of OsAMT1–3, OsNRT2, and OsNAR2 genes.
Acknowledgements
This work was funded by the Ministry of Science and Technology of China (no. 2011CB100302), the National Nature Science Foundation of China (nos 31071846, 31172022, and 31471936), the Innovative Research Team Development Plan of the Ministry of Education of China (no. IRT1256), the 111 Project (no. 12009), PAPD in Jiangsu Province of China, China Scholarship Council (CSC), and Innovative Plan of Jiangsu Province of China (CXLX13_280 and 568). The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, http://www.textcheck.com/certificate/ubiBjX.
References
- Alvarez JM, Riveras E, Vidal EA, et al. 2014. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant Journal 80, 1–13. [DOI] [PubMed] [Google Scholar]
- Babourina O, Voltchanskii K, McGann B, Newman I, Renge Z. 2007. Nitrate supply affects ammonium transport in canola roots. Journal of Experimental Botany 58, 651–658. [DOI] [PubMed] [Google Scholar]
- Begheldo M, Nonis A, Trevisan S, Ruperti B, Quaggiotti S. 2014. The dynamic regulation of microRNAs circuits in plant adaptation to abiotic stresses: a survey on molecular, physiological and methodological aspects. Environmental and Experimental Botany (in press). [Google Scholar]
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ. 2006. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. The Plant Journal 45, 113–122. [DOI] [PubMed] [Google Scholar]
- Cao Y, Fan X, Sun S, Xu G, Hu J, Shen Q. 2008. Effect of nitrate on activities and transcript levels of nitrate reductase and glutamine synthetase in rice. Pedosphere 18, 664–673. [Google Scholar]
- Chen WW, Yang JL, Qin C, Jin CW, Mo JH, Ye T, Zheng SJ. 2010. Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis. Plant Physiology 154, 810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ. 2007. The role of microRNAs in sensing nutrient stress. Plant, Cell and Environment 30, 323–332. [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N, Graziano M, Chevaller C, Lamattina L. 2006. Nitric oxide modulates the expression of cell cycle regulatory genes during lateral root formation in tomato. Journal of Experimental Botany 57, 581–588. [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N, Graziano M, Lamattina L. 2004. Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218, 900–905. [DOI] [PubMed] [Google Scholar]
- Crawford NM. 1995. Nitrate: nutrient and signal for plant growth. The Plant Cell 7, 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM. 2006. Mechanisms for nitric oxide synthesis in plants. Journal of Experimental Botany 57, 471–478. [DOI] [PubMed] [Google Scholar]
- Du S, Zhang Y, Lin X, Wang Y, Tang C. 2008. Regulation of nitrate reductase by nitric oxide in Chinese cabbage pakchoi (Brassica chinensis L.). Plant, Cell and Environment 31, 195–204. [DOI] [PubMed] [Google Scholar]
- Duan YH, Zhang YL, Wang SW, Shen QR. 2007. a Effect of NH4 + to NO3 – ratio (NH4 +/NO3 –) on biological characteristics of rice with different nitrogen use efficiency. Journal of Nanjing Agricultural University 30, 73–77. [Google Scholar]
- Duan YH, Zhang YL, Ye LT, Fan XR, Xu GH, Shen QR. 2007. b Responses of rice cultivars with different nitrogen use efficiency to partial nitrate nutrition. Annals of Botany 99, 1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XR, Jia LJ, Li YL, Smith SJ, Miller AJ, Shen QR. 2007. Comparing nitrate storage and remobilization in two rice cultivars that differ in their nitrogen use efficiency. Journal of Experimental Botany 58, 1729–1740. [DOI] [PubMed] [Google Scholar]
- Fernández-Marcos M, Sanz L, Lewis DR, Muday GK, Lorenzo O. 2011. Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proceedings of the National Academy of Sciences, USA 108, 18506–18511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG. 2002. The role of long-distance signaling in plant responses to nitrate and other nutrients. Journal of Experimental Botany 53, 39–43. [PubMed] [Google Scholar]
- Forde BG. 2014. Nitrogen signaling pathways shaping root system architecture: an update. Current Opinion in Plant Biology 21, 30–36. [DOI] [PubMed] [Google Scholar]
- Freschi L, Rodrigues MA, Domingues DS, Purgatto E, Van Sluys MA, Magalhaes JR, Kaiser WM, Mercier H. 2010. Nitric oxide mediates the hormonal control of crassulacean acid metabolism expression in young pineapple plants. Plant Physiology 152, 1971–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frungillo L, Skelly MJ, Loake GJ, Spoel SH, Salgado I. 2014. S-nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nature Communications 5, 5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gas E, Flores-Pérez U, Sauret-Güeto S, Rodríguez-Concepción M. 2009. Hunting for plant nitric oxide synthase provides new evidence of a central role for plastids in nitric oxide metabolism. The Plant Cell 21, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. 2008. Cell-specific nitrogen responses mediate developmental plasticity. Proceedings of the National Academy of Sciences, USA 105, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A, Krouk G, Perrine-Walker F, Laugier E. 2011. Nitrate transceptor(s) in plants. Journal of Experimental Botany 62, 2299–2308. [DOI] [PubMed] [Google Scholar]
- Guo K, Xia K, Yang Z. 2008. Regulation of tomato lateral root development by carbon monoxide and involvement in auxin and nitric oxide. Journal of Experimental Botany 59, 3443–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KJ, Fernie AR, Kaiser WM, van Dongen JT. 2011. On the origins of nitric oxide. Trends in Plant Science 16, 160–168. [DOI] [PubMed] [Google Scholar]
- Jin CW, Du ST, Zhang YS, Lin XY, Tang CX. 2009. Differential regulatory role of nitric oxide in mediating nitrate reductase activity in roots of tomato (Solanum lycocarpum). Annals of Botany 104, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbert Z, Ortega L, Erdei L. 2010. Involvement of nitrate reductase (NR) in osmotic stress-induced NO generation of Arabidopsis thaliana L. roots. Journal of Plant Physiology 167, 77–80. [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, et al. 2010. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Developmental Cell 18, 927–937. [DOI] [PubMed] [Google Scholar]
- Li YL, Fan XR, Shen QR. 2008. The relationship between rhizosphere nitrification and nitrogen-use efficiency in rice plants. Plant, Cell and Environment 31, 73–85. [DOI] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser O. 2002. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. The Plant Journal 29, 751–760. [DOI] [PubMed] [Google Scholar]
- Lombardo MC, Graziano M, Polacco JC, Lamattina L. 2006. Nitric oxide functions as a positive regulator of root hair development. Plant Signaling and Behavior 1, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramı′rez A, Herrera-Estrella L. 2003. The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology 6, 280–287. [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Manoli A, Begheldo M, Genre A, Lanfranco L, Trevisan S, Quaggiotti S. 2014. NO homeostasis is a key regulator of early nitrate perception and root elongation in maize. Journal of Experimental Botany 65, 185–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng ZB, Chen LQ, Suo D, Li GX, Tang CX, Zheng SJ. 2012. Nitric oxide is the shared signalling molecule in phosphorus- and iron-deficiency-induced formation of cluster roots in white lupin (Lupinus albus). Annals of Botany 109, 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Lea US, Provan F, Kaiser WM, Lillo C. 2005. Is nitrate reductase a major player in the plant NO (nitric oxide) game? Photosynthesis Research 83, 181–189. [DOI] [PubMed] [Google Scholar]
- Modolo LV, Augusto O, Almeida IMG, Magalhaes JR, Salgado I. 2005. Nitrite as the major source of nitric oxide production by Arabidopsis thaliana in response to Pseudomonas syringae. FEBS Letters 579, 3814–3820. [DOI] [PubMed] [Google Scholar]
- Moreau M, Lee G, I, Wang Y, Crane BR, Klessig DF. 2008. At NOS/A1 is a functional Arabidopsis thaliana cGTPase and not a nitric oxide synthase. Journal of Biological Chemistry 283, 32957–32967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Lindermayr C, Durner J, Klessig D. 2010. NO synthesis and signaling in plants—where do we stand? Physiologia Plantarum 138, 372–383. [DOI] [PubMed] [Google Scholar]
- Mounier E, Pervent M, Ljung K, Gojon A, Nacry P. 2013. Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant, Cell and Environment 37, 162–174. [DOI] [PubMed] [Google Scholar]
- Nischal L, Mohsin M, Khan I, Kardam H, Wadhwa A, Abrol Y P, Iqbal M, Ahmad A. 2012. Identification and comparative analysis of microRNAs associated with low-N tolerance in rice genotypes. PLoS One 7, e50261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS. 2013. Responses of root architecture development to low phosphorus availability: a review. Annals of Botany 112, 391–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ. 2006. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis: physiology and protein–protein interaction. Plant Physiology 142, 1304–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Lanteri ML, Lamattina L. 2003. Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiology 132, 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian XQ, Shen QR, Xu GH, Wang JJ, Zhou MY. 2004. Nitrogen form effects on yield and nitrogen uptake of rice crop grown in aerobic soil. Journal of Plant Nutrition 27, 1061–1076. [Google Scholar]
- Remans T, Nacry P, Pervent M, et al. 2006. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proceedings of the National Academy of Sciences, USA 103, 19206–19211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GP, Vitousek PM. 2009. Nitrogen in agriculture: balancing the cost of an essential resource. Annual Review of Environment and Resources 34, 97–125. [Google Scholar]
- Rosales EP, Iannone MF, Groppa MD, Benavides MP. 2010. Nitric oxide inhibits nitrate reductase activity in wheat leaves. Plant Physiology and Biochemistry 49, 124–130. [DOI] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. 2011. Nitrogen economics of root foraging: transitive closure of the nitrate–cytokinin relay and distinct systemic signaling for N supply vs. demand. Proceedings of the National Academy of Sciences, USA 108, 18524–18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Luque E, Ocaña-Calahorro F, Llamas A, Galvan A, Fernandez E. 2013. Nitric oxide controls nitrate and ammonium assimilation in Chlamydomonas reinhardtii . Journal of Experimental Botany 64, 3373–3383. [DOI] [PubMed] [Google Scholar]
- Schlicht M, Müller JL, Burbach C, Volkmann D, Baluska F. 2013. Indole-3-butyric acid induces lateral root formation via peroxisome-derived indole-3-acetic acid and nitric oxide. New Phytologist 200, 473–482. [DOI] [PubMed] [Google Scholar]
- Simon J, Dong F, Buegger F, Rennenberg H. 2013. Rhizospheric NO affects N uptake and metabolism in Scots pine (Pinus sylvestris L.) seedlings depending on soil N availability and N source. Plant, Cell and Environment 36, 1019–1026. [DOI] [PubMed] [Google Scholar]
- Simon J, Stoelken G, Rienks M, Rennenberg H. 2009. Rhizospheric NO interacts with the acquisition of reduced N sources by the roots of European beech (Fagus sylvatica L.). FEBS Letters 583, 2907–2910. [DOI] [PubMed] [Google Scholar]
- Somers DA, Kuo TM, Kleinhofs A, Warner BL, Oaks A. 1983. Synthesis and degradation of barley nitrate reductase. Plant Physiology 72, 949–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava N, Gonugunta VK, Puli MR, Raghavendra AS. 2009. Nitric oxide production occurs downstream of reactive oxygen species in guard cells during stomatal closure induced by chitosan in abaxial epidermis of Pisum sativum. Planta 229, 757–765. [DOI] [PubMed] [Google Scholar]
- Stöhr C, Stremlau S. 2006. Formation and possible roles of nitric oxide in plant roots. Journal of Experimental Botany 57, 463–470. [DOI] [PubMed] [Google Scholar]
- Song W, Jin J, Ha L, Tu E, Shen Q, Zhang Y. 2011. a Response of rice plants different in response to nitrate to enhanced nitrate supply in root growth at the seedling stage. Acta Pedologica Sinica 48, 1006–1012. [Google Scholar]
- Song W, Li J, Sun H, Huang S, Gong X, Ma Q, Zhang Y, Xu G. 2013. b Increased photosynthetic capacity in response to nitrate is correlated with enhanced cytokinin levels in rice cultivar with high responsiveness to nitrogen nutrients. Plant and Soil 373, 981–993. [Google Scholar]
- Song W, Makeen K, Wang D, et al. 2011. b Nitrate supply affects root growth differentially in two rice cultivars differing in nitrogen use efficiency. Plant and Soil 343, 357–368. [Google Scholar]
- Song W, Sun H, Li J, Gong X, Huang S, Zhu X, Zhang Y, Xu G. 2013. a Auxin distribution is differentially affected by nitrate in roots of two rice cultivars differing in responsiveness to nitrogen nutrients. Annals of Botany 112, 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M. 1999. Nitrate regulation of metabolism and growth. Current Opinion in Plant Biology 2, 178–186. [DOI] [PubMed] [Google Scholar]
- Stöhr C, Stremlau S. 2006. Formation and possible roles of nitric oxide in plant roots. Journal of Experimental Botany 57, 463–470. [DOI] [PubMed] [Google Scholar]
- Tang Z, Fan XR, Li Q, Feng HM, Miller AJ, Shen QR, Xu GH. 2012. Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiology 160, 2052–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisan S, Begheldo M, Nonis A, Quaggiotti S. 2012. b The miRNA-mediated post-transcriptional regulation of maize response to nitrate. Plant Signaling and Behavior 7, 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisan S, Manoli A, Begheldo M, Nonis A, Enna M, Vaccaro S, Caporale G, Ruperti B, Quaggiotti S. 2011. Transcriptome analysis reveals coordinated spatiotemporal regulation of hemoglobin and nitrate reductase in response to nitrate in maize roots. New Phytologist 192, 338–352. [DOI] [PubMed] [Google Scholar]
- Trevisan S, Manoli A, Quaqqiotti S. 2014. NO signaling is a key componet of root growth response to nitrate in Zea mays L. Plant Signaling and Behavior 9, e28290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisan S, Nonis A, Begheldo M, Manoli A, Palme K, Caporale G, Ruperti B, Quaggiotti S. 2012. a Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant, Cell and Environent 35, 1137–1155. [DOI] [PubMed] [Google Scholar]
- Vanin AF, Svistunenko DA, Minkoyan VD, Serezhenkov VA, Fryer MJ, Baker NR, Cooper CE. 2004. Endogenous superoxide production and the nitrite/nitrate ratio control the concentration of bioavailable free nitric oxide in leaves. Journal of Biological Chemistry 279, 24100–24107. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutierrez RA. 2010. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 107, 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BL, Tang XY, Cheng LY, et al. 2010. Nitric oxide is involved in phosphorus deficiency-induced cluster-root development and citrate exudation in white lupin. New Phytologist 187, 1112–1123. [DOI] [PubMed] [Google Scholar]
- Wang H, Xiao W, Niu Y, Jin C, Chai R, Tang C, Zhang Y. 2013. Calcium is involved in nitric oxide- and auxin-induced lateral root formation in rice. Planta 237, 137–144.22990909 [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM. 2000. Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. The Plant Cell 12, 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Crawford NM. 1991. Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. The Plant Cell 3, 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ. 2012. Plant nitrogen assimilation and use efficiency. Annual Review of Plant Biology 63, 153–182. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhong S, Li X, Li W, Rothstein SJ, Zhang S, Bi Y, Xie C. 2011. Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS One 6, e28009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H. 2005. The NO world for plants: achieving balance in an open system. Plant, Cell and Environment 28, 78–84. [Google Scholar]
- Yamasaki H, Sakihama Y, Takahashi S. 1999. An alternative pathway for nitric oxide production in plants: new features of an old enzyme. Trends in Plant Science 4, 128–129. [DOI] [PubMed] [Google Scholar]
- Yu X, Sukumaran S, Márton L. 1998. Differential expression of the Arabidopsis Nia1 and Nia2 genes. Plant Physiology 116, 1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Wang G, Hu X, Wang H, Du L, Zhu Y. 2014. Role of microRNAs in plant responses to nutrient stress. Plant and Soil 374, 1005–1021. [Google Scholar]
- Zhang H, Rong H, Pilbeam D. 2007. Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana . Journal of Experimental Botany 58, 2329–2338. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Forde BG. 1998. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Jennings A, Barlow PW, Forde BG. 1999. Dual pathways for regulation of root branching by nitrate. Proceedings of the National Academy of Sciences, USA 96, 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lv H, Wang D, Deng J, Song W, Makeen K, Shen Q, Xu G. 2011. Partial nitrate nutrition amends photosynthetic characteristics in rice (Oryza sativa L. var. japonica) differing in nitrogen use efficiency. Plant Growth Regulation 63, 235–242. [Google Scholar]
- Zhang YH, Fan JB, Zhang YL, Wang DS, Huang QW, Shen QR. 2007. N accumulation and translocation in four japonica rice cultivars at different N rates. Pedosphere 17, 792–800. [Google Scholar]
- Zhang YL, Fan JB, Duan YH, Wang DS, Ye LT, Shen QR. 2008. Variation of nitrogen use efficiency of rice different in genotype and its evaluation. Acta Pedologica Sinica 45, 267–273. [Google Scholar]
- Zhang YL, Fan JB, Wang DS, Shen QR. 2009. Genotypic differences in grain yield and physiological nitrogen use efficiency among rice cultivars. Pedosphere 19, 681–691. [Google Scholar]
- Zhao M, Chen L, Zhang L, Zhang W. 2009. Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiology 151, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Ding H, Zhu JK, Zhang F, Li WX. 2011. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytologist 190, 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Tai H, Sun S, Zhang F, Xu Y, Li WX. 2012. Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS One 7, e29669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Tian QY, Zhang WH. 2007. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiology 144, 206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xu Z, Mo Q, Zou C, Li W, Xu Y, Xie C. 2013. Combined small RNA and degradome sequencing reveals novel miRNAs and their targets in response to low nitrate availability in maize. Annals of Botany 112, 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke HR, Filner P. 1971. Synthesis and turnover of nitrate reductase induced by nitrate in cultured tobacco cells. Journal of Biological Chemistry 246, 1772–1779. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.