Highlight
OsCSLF6 plays an important role in balance carbon metabolism and phosphate accumulation.
Key words: Cellulose, phosphate (Pi) accumulation, Pi transporters, sucrose.
Abstract
Phosphorus is an essential macronutrient for plant growth and development. However, the network that affects phosphate (Pi) accumulation in crops is not well established. It is reported here that OsCSLF6, a member of the cellulose synthase-like family (CSLF), which is found only in grasses, is involved in Pi accumulation. The oscslf6 mutants (oscslf6-1 and oscslf6-2) display Pi toxic symptoms and increased Pi accumulation in both roots and shoots under the Pi-sufficient condition, which correlate with the induced expression of Pi transporters in the knockout mutants. Consistent with the over-accumulation of Pi, a significant decrease in primary root length, adventitious root length, and adventitious root number were observed in the oscslf6 mutants when compared with the wild type (WT) under Pi-sufficient conditions. In addition, the sucrose (Suc) level was increased in the oscslf6 mutants and the expression of sucrose synthases (OsSUS4/5) and sucrose transporters (OsSUT1/2/4/OsSweet14) genes were also induced in the shoots of oscslf6 mutants, suggesting that OsCSLF6 may play a role in affecting Pi accumulation by affecting the level of carbon metabolism.
Introduction
Phosphorus is an essential macronutrient for plant growth, development, and reproduction. Despite the abundance of Pi in the soil, the form of Pi available for uptake by plants is usually present at a low level in the soil due to its precipitation with cations and conversion to organic matter (Kuo and Chiou, 2011). To cope with heterogeneous or low phosphate (Pi) availability, plants have evolved complex adaptive responses that include morphological and physiological modifications. These responses include changes in root architecture and morphology, increased Pi uptake activity, the secretion of organic acids or phosphatase, and an association with mycorrhizal fungi (López-Bucio et al., 2003; Aung et al., 2006; Chiou and Lin, 2011; Péret et al., 2011).
In recent decades, a number of genes that are involved in sensing and responding to Pi deficiency, and in their regulatory networks, have been isolated from vascular plants, including Pi transporters, phosphatases, and RNases. In Arabidopsis, a major regulatory system that involves SPX1, PHR1, SIZ1, miR399, and PHO2 in response to Pi deficiency has been identified (Fujii et al., 2005; Miura et al., 2005; Aung et al., 2006; Chiou et al., 2006; Nilsson et al., 2007; Puga et al., 2014). MYB transcription factors AtPHR1 and AtPHL1 have been identified as key regulators in the Pi-signalling pathway (Rubio et al., 2001; Bustos et al., 2010). Over-expression of AtPHR1 leads to the over accumulation of Pi in shoots and the activation of Pi starvation-induced gene expression (Nilsson et al., 2007). Puga et al. (2014) have reported that the nuclear protein SPX1 is a Pi-dependent inhibitor of PHR1 in Arabidopsis. SIZ1, a SUMO E3 ligase, is known to control PHR1 sumoylation (Miura et al., 2005). Upon Pi starvation, miR399 is up-regulated by PHR1 and is involved in the cleavage of the PHO2 mRNA, which encodes the low-Pi-responsive UBC24 (ubiquitin-conjugating E2 24) enzyme (Aung et al., 2006; Bari et al., 2006; Chiou et al., 2006). Both miR399 over-expression and mutations in PHO2 resulted in the over-accumulation of Pi and Pi toxic symptoms being exhibited in Arabidopsis (Fujii et al., 2005; Aung et al., 2006; Chiou et al., 2006).
The functional orthologue of AtPHR1 in rice (designated as OsPHR2) was also identified, and it was found that over-expression of OsPHR2 resulted in the excessive accumulation of Pi in shoots and the up-regulation of some Pi transporter genes under Pi-sufficient conditions (Zhou et al., 2008a; Liu et al., 2010). OsSPX4 has been reported to function in Pi starvation signalling and to act as a negative regulator of OsPHR2 in rice (Lv et al., 2014). OsSPX1 and OsSPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner (Wang et al., 2014). OsPHO2, the putative homologue of AtPHO2, has been shown to be involved in the Pi starvation signalling pathway mediated by OsSPX1–OsPHR2 (Bari et al., 2006; Wang et al., 2009; Liu et al., 2010).
Although our understanding of Pi starvation signalling involving SPX4, SPX1, SPX2, PHR2, miR399, and PHO2 is well established, other pathways may also be required for the Pi starvation response (PSR). For example, characterization of transcription factors such as OsPTF1 (Yi et al., 2005), OsMYB2P-1 (Dai et al., 2012), AtWRKY75 (Devaiah et al., 2007a), AtZAT6 (Devaiah et al., 2007b), AtMYB62 (Devaiah et al., 2009), AtBHLH32 (Chen et al., 2007), AtWRKY6 (Chen et al., 2009), and AtMYB2 (Baek et al., 2013) suggests that they play crucial roles in controlling the expression of the downstream genes as well as the regulation of cross-talk among different signalling pathways.
Besides these factors, sugar signalling also plays important roles in regulating plant responses to Pi starvation. Many studies have shown the importance of sugar signalling in regulating PSR, including the increased expression of PSI genes and changes in root system architecture (RSA) (Liu et al., 2005; Müller et al., 2005, 2007; Jain et al., 2007; Karthikeyan et al., 2007; Hammond and White, 2008; Zhou et al., 2008b; Chiou and Lin, 2011). Increased Suc concentrations in roots precede the induction of PSR and the inhibition of Suc biosynthesis or translocation attenuates the plant response to Pi starvation (Hammond and White, 2008, 2011). However, cross-talk between carbon metabolism and the Pi pathway in rice remains unclear.
A rice mutant that displayed Pi toxicity symptoms was identified here. Sequencing of the flanking regions of the T-DNA insertion site revealed that the insertion leads to the loss-of-function of the OsCSLF6, a gene encoding a cellulose synthase-like protein. The increased Pi content in oscslf6 mutants was associated with the increased expression of PHT genes. OsCSLF6 was also involved in Pi-dependent root architecture alteration. Moreover, the Suc level was increased and the expression of genes encoding sucrose synthases (OsSUS4/5) and sucrose transporters (OsSUT1/2/4/Sweet14) were induced in shoot of oscslf6 mutants, suggesting that OsCSLF6 may affect Pi accumulation and response through the alteration of carbon metabolism in rice.
Materials and methods
Isolation of T-DNA insertion mutants
The mutant line 04Z11DM89 [oscslf6-1; rice (Oryza sativa ssp. japonica cv. Zhonghua 11)] was obtained from the RMD database (Wu et al., 2003; Zhang et al., 2006) (http://rmd.ncpgr.cn/) and 3A-60123 (oscslf6-2; rice ssp. japonica cv. Dongjin) was ordered from the POSTECH RISD database (Jeon et al., 2000) (http://www.postech.ac.kr/life/pfg/risd/), respectively. Mutants were planted in the paddy field of Huazhong Agricultural University in the normal rice (Oryza sativa) growing season of Wuhan, China, and in a greenhouse during the winter. All transgenic plants were grown in similar growth conditions.
Hydroponic experiments
Hydroponic experiments were conducted using normal rice culture solution with 10mg l–1 Pi (Yoshida et al., 1976) and a Pi-deficient solution (0.5mg l–1 Pi). Rice seeds were surface-sterilized for 10min with ethanol (75% v/v) and for 15min with commercially diluted (1:3, v/v) NaClO, washed, and germinated for 3 d at 28 °C. The 9-d-old seedlings were transferred to nutrient solution containing 1.25mM NH4NO3, 0.35mM K2SO4, 1mM CaCl2.2H2O, 1mM MgSO4.7H2O, 0.5mM Na2SiO3.9H2O, 20mM Fe-EDTA, 20mM H3BO3, 9mM MnCl2.4H2O, 0.32mM CuSO4.5H2O, 0.77mM ZnSO4.7H2O, and 0.39mM Na2MoO4.2H2O, pH 5.5, supplemented with 10mg l–1 Pi (HP) or 0.5mg l–1 Pi (LP); 30-d-old seedlings were observed for phenotype or sampled for total P concentration measurement (Zhou et al., 2008a). The solution was refreshed every 3 d.
Vector construction and plant transformation
An 11kb genomic DNA fragment containing the entire OsCSLF6 coding region and the 2756bp upstream and 2818bp downstream sequences was isolated by digestion of the Clemson BAC clone OSJNBa0055L06 (kindly provided by R Wing, University of Arizona) and inserted into the binary vector pCAMBIA2301. An empty pCAMBIA2301 vector was used as a control. The transformation recipient was callus culture that was induced from seeds homozygous for cslf6-1.
To fuse the Os08g06380 promoter to the GUS gene, the promoter of OsCSLF6, a 1878bp fragment upstream of the ATG of Os08g06380 was amplified by PCR. The PCR product was cloned into pDONR207 by BP recombination. After sequencing, the correct clone for each gene was individually introduced into the Gateway-compatible GUS fusion vector pGWB3 (Nakagawa et al., 2007) to produce CSLF6pGUS.
All the constructs were introduced into Agrobacterium tumefaciens EHA105 and were transformed into the callus derived from the japonica cultivar Zhonghua 11 by Agrobacterium-mediated transformation as described previously (Wu et al., 2003). All primers for genotyping and vector construction are listed in Supplementary Table S1 at JXB online.
Scanning electron microscopy
Samples were prepared according to the method previously reported by Mou et al. (2000), with some modifications. In brief, the mature culms were excised with a blade and immediately placed in 70% ethanol, 5% acetic acid, and 3.7% formaldehyde for 24h. Then samples were critical-point dried, sputter-coated with gold, and observed with a scanning electron microscope (S570, Hitachi, Tokyo, Japan).
Histochemical staining
Cellulose staining was assayed according to the method described by Li et al. (2003). Fresh hand-cut sections (~20 μm thick) from rice culms were stained with a 0.005% aqueous solution of Calcofluor (fluorescent brightener 28; Sigma) for 2min and visualized with a fluorescent microscope (Leica, Wetzlar, Germany).
GUS staining
GUS staining was performed as previously described by Jefferson et al. (1987). Samples were transferred to a solution of 200mM sodium phosphate buffer, pH 7.0, 12.5mM potassium ferricyanide, 12.5mM potassium ferrocyanide, 0.3% Triton X-100, 20% methanol, and 38.3mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide and were kept overnight at 37 °C. The stained samples were then washed with 75% ethanol overnight. The cleared samples were observed by light microscopy.
Measurement of total Pi
Dry samples (approximately 0.2g) were used for the determination of total Pi as previously described by Zhou et al. (2008a).
The Pi uptake rate was measured based on the rate of depletion of the nutrient from solution over 24h (Liu et al., 2010). Thirty-day-old plants were used. Before measurement, the plants were moved into a solution culture without Pi for 3 d and then transferred to a pot with four plants per litre of fresh solution (10mg l–1 Pi). A 1ml aliquot of solution was removed from each pot at 24, 48, and 72h time points for phosphorus concentration analysis by the phosphomolybdenum blue reaction. The roots of the plants in each pot were harvested and oven-dried, and the Pi uptake rate was calculated as depletion of the Pi in the solution per gram of dried root biomass.
RT-PCR and real-time PCR
Total RNA was extracted from rice using an RNA extraction kit (TRIzol reagent; Invitrogen) according to the manufacturer’s instructions. The first-strand cDNA was synthesized using 3 μg of RNA and 200U of M-MLV reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. Real-time PCR was performed using an optical 96-well plate in an ABI Stepone plus PCR system (Applied Biosystems) by using SYBR Premix reagent F-415 (Thermo Scientific). The expression measurements were obtained using the relative quantification method (Livak and Schmittgen, 2001). All the primers used for RT-PCR and real-time PCR are listed in Supplementary Tables S1 and S2 at JXB online.
Quantitative analysis of sugar contents
Twelve-day-old seedlings were cut into shoot and root parts and thoroughly ground in liquid nitrogen. Soluble sugar was extracted in 225 μl methanol, 120 μl CHCl3, and 240 μl ddH2O at 70 °C for 15min. Samples were centrifuged at 12 000rpm for 10min, transferred to 200 μl supernatant and then dried at 80 °C. For methoximation, 40 μl of methoxyamine hydrochloride in pyridine (20mg ml–1) was added at 30 °C for 90min. After, 60 μl of N-methyl-N-trimethylsily-trifluoroacetamide was added, and the mixture was incubated at 37 °C for 30min. The derivatives were analysed by gas chromatography–mass spectrometry on a Thermo DSQII mass spectrometer using a DB-5ms column. A temperature programme was implemented as follows: initially at 70 °C, followed by heating to 300 °C at 5 °C min–1, and then held at 300 °C for a further 3min. Myoinositol was used as an internal standard.
Results
Isolation and characterization of the oscslf6 mutants in rice
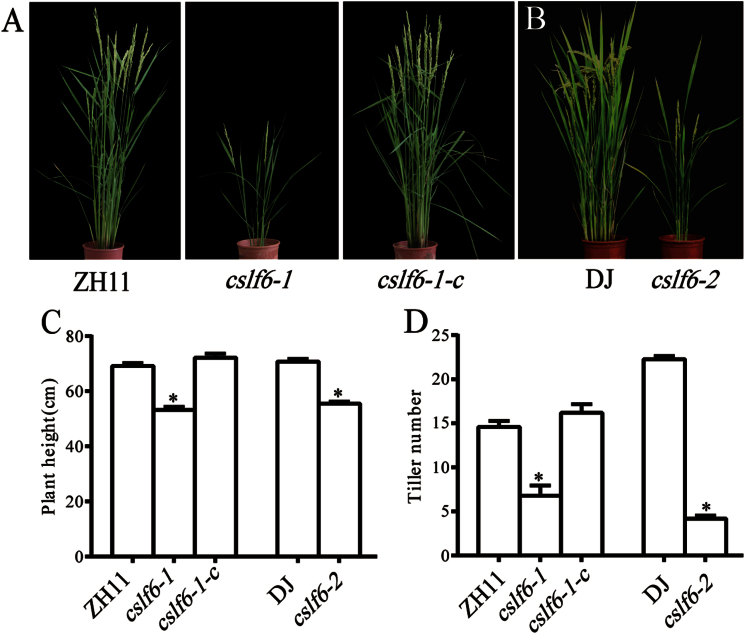
A rice mutant with greatly reduced height and tiller number was isolated in screening transgene lines after Agrobacterium tumifaciens-mediated T-DNA insertion (Wu et al., 2003; Zhang et al., 2006). Sequencing of the flanking regions of the T-DNA insertion site indicated that the T-DNA was inserted in the first intron of OsCSLF6 (see Supplementary Fig. 1A at JXB online). The mutant, designated as oscslf6-1, exhibited dwarfism throughout growth and development (Fig. 1A). At the mature stage, the height of mutant plants was about 70% of that of the WT (ZH11) plants (Fig. 1C). In addition to dwarfism, oscslf6-1 showed a marked decrease in tiller number (Fig. 1D). The Rice Functional Genomic Express database (http://signal.salk.edu/cgi-bin/RiceGE) was searched using the OsCSLF6 sequence and an allelic mutant PFG_3A-60123.L, was found which was named oscslf6-2. The T-DNA insertion site was located at 204bp upstream of ATG of LOC_Os08g06380 (see Supplementary Fig. 1A at JXB online). The Oscslf6-2 mutant also displayed similar phenotypes including reduced plant height and a decrease in tiller number (Fig. 1B–D).
Fig. 1.
Phenotypes of wild type (WT) and oscslf6 mutants grown in the field for 3 months. (A) The phenotypes of ZH11 (Zhonghua11) and oscslf6-1 and cslf6-1-complemented (cslf6-1-c) mutant plants at maturity. (B) The phenotypes of DJ (Dongjin) and oscslf6-2 mutant plants at maturity. (C) Height of ZH11, oscslf6-1, cslf6-1-c, DJ, and oscslf6-2 mutant plants. (D) Tiller number of ZH11, oscslf6-1, cslf6-1-c, DJ, and oscslf6-2 mutant plants. Error bars indicate SD (n=10). Asterisks indicate the significance of differences between WT and cslf6 mutant plants as determined by Student’s t test: *P ≤0.05. (This figure is available in colour at JXB online.)
Reverse transcription-PCR analysis indicated that oscslf6-1 and oscslf6-2 are knockout lines since no OsCSLF6 transcript could be detected in plants homozygous for the insertions (see Supplementary Fig. 1B at JXB online). To verify further that the mutant phenotype is the result of the loss-of-function of OsCSLF6, an 11kb BamH1–BamH1 fragment harbouring the entire OsCSLF6 coding region, a 2.7kb upstream and a 2.8kb downstream region were introduced into the oscslf6-1 mutant background. As expected, tillers and height defects of the complemented oscslf6-1 (cslf6-1-c) mutant were rescued (Fig. 1A–D).
The oscslf6 mutants display overaccumulation of Pi
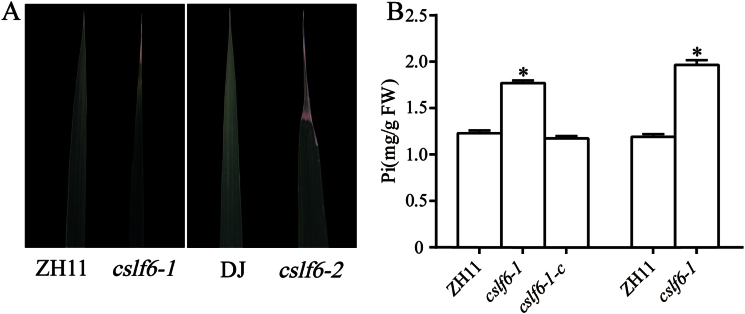
Necrotic spots were observed in the mature leaves of oscslf6-1 and oscslf6-2 plants in Pi-sufficient soil (Fig. 2A). The leaf toxic symptoms and growth retardation of the oscslf6 mutants are similar to that of Pi over-accumulation plants such as OsPHR2-overexpression plants (OsPHR2(O)) and ospho2 mutants (Zhou et al., 2008a; Hu et al., 2011).
Fig. 2.
Pi content in shoots of oscslf6 mutants. (A) Leaf tip necrosis of WT and oscslf6 mutants plants grown in the field for 3 months. (B) Shoot Pi content in 3-month-old field-grown ZH11, oscslf6-1, cslf6-1-c, DJ, and oscslf6-2 mutant plants. Error bars indicate SD (n=10). Asterisks indicate the significance of differences between WT and oscslf6 mutant plants as determined by Student’s t test: *P ≤0.05. (This figure is available in colour at JXB online.)
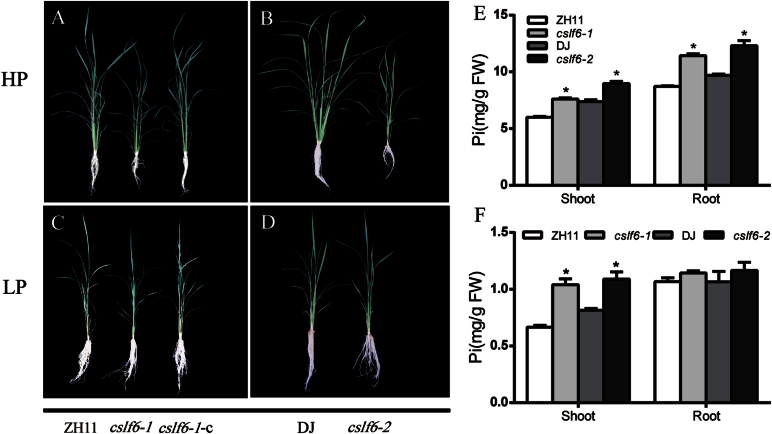
To explore whether the phenotype of the oscslf6 mutants is related to Pi overaccumulation, Pi concentrations in the shoots of the oscslf6 and cslf6-1-c plants were examined in Pi-sufficient soil. The results indicated Pi concentrations in shoots of oscslf6-1 and oscslf6-2 mutant plants were significantly higher than those of WT plants (Fig. 2B). The Pi concentrations of cslf6-1-c plants were rescued to the level of their WT counterparts (Fig. 2B). To determine whether an excess of Pi was responsible for the leaf toxic symptoms and growth retardation phenotype of oscslf6 mutants, oscslf6, cslf6-1-c, and WT plants were grown in nutrient solutions that were supplied with either a high level of Pi (HP, 0.323mM) or a low level of Pi (LP, 0.032mM) for phenotype observation.
In the HP condition, both knockout lines of oscslf6 (oscslf6-1 and oscslf6-2) displayed toxic leaf symptoms and growth retardation similar to that of the OsPHR2-overexpressing plants [PHR2(O)], the OsPT2-overexpressing plants [OsPT2(O)], and the rice ltn1 (ospho2) mutant plants (Fig. 3A, B) (Zhou et al., 2008a; Wang et al., 2009; Liu et al., 2010; Hu et al., 2011), as indicated by small plants, reduced tiller number, and decreased tiller number and decreased shoot and root biomass of the oscslf6 mutant lines (Fig. 3A–D; Table 1). Moreover, Pi concentrations in both shoots and roots of the oscslf6 mutant plants grown in HP conditions were higher than those in the WT plants (Fig. 3E).
Fig. 3.
Phenotypes of oscslf6 mutant plants under HP and LP conditions. (A) Phenotype of WT (ZH11), oscslf6-1 mutant, and cslf6-1-c plants after being grown under HP conditions (0.3mM Pi) for 30 d. The Pi toxic phenotype shows a smaller plant, reduced tiller number, and decreased shoot and root length. (B) Phenotype of WT (DJ) and oscslf6-2 mutants after being grown under HP conditions (0.3mM Pi) for 30 d. (C) Phenotype of WT (ZH11), oscslf6-1, and cslf6-1-c plants after being grown under LP conditions (0.015mM Pi) for 30 d. (D) Phenotype of the wild type (DJ) and oscslf6-2 mutants after being grown under LP conditions (0.015mM Pi) for 30 d. (E) Pi contents in shoots and roots of the wild type (ZH11), oscslf6-1, cslf6-1-c, DJ, and oscslf6-2 mutant plants under Pi-sufficient conditions. (F) Pi contents in shoots and roots of WT (ZH11), oscslf6-1, cslf6-1-c, WT (DJ), and oscslf6-2 mutant plants under Pi-deficient conditions. Plants were pregerminated in water for 7 d and then grown hydroponically for 30 d under HP and LP conditions. Error bars indicate SD (n=10). Asterisks indicate the significance of differences between WT and oscslf6 mutant plants as determined by Student’s t test: *P ≤0.05. DW, dry weight. (This figure is available in colour at JXB online.)
Table 1.
Plant height, tiller number, dry shoot biomass, and dry root biomass of WT (ZH11 and DJ), cslf6-1-complemented and oscslf6 mutants
The 9-d-old seedlings were transferred to HP or LP medium for 30 d and then the plants were sampled for the measurements. The values are means ±SD of three independent experiments, with 10 seedlings being used in each experiment. Asterisks indicate the significance of differences between the wild type and oscslf6 mutant plants as determined by Student’s t test analysis: *0.01≤ P ≤0.05, **P <0.01.
| Genotype | Plant height (cm) | Tillering number | Shoot biomass (g DW) | Root biomass (g DW) |
|---|---|---|---|---|
| HP | ||||
| ZH11 | 73.533±1.784 | 8.167±0.307 | 1.660±0.137 | 0.327±0.030 |
| cslf6-1 | 59.117±1.282** | 4.333±0.211** | 0.784±0.042** | 0.180±0.013** |
| cslf6-1-c | 74.433±0.914 | 7.500±0.224 | 1.636±0.026 | 0.340±0.010 |
| DJ | 68.433±0.878 | 7.400±0.245 | 1.604±0.077 | 0.340±0.014 |
| cslf6-2 | 53.200±0.683** | 3.400±0.245** | 0.556±0.043** | 0.168±0.017** |
| LP | ||||
| ZH11 | 51.867±0.630 | 3.167±0.307 | 0.652±0.026 | 0.294±0.010 |
| cslf6-1 | 42.133±0.455** | 2.833±0.167* | 0.455±0.014* | 0.220±0.013** |
| cslf6-1-c | 51.700±0.599 | 3.667±0.211 | 0.749±0.026 | 0.320±0.004 |
| DJ | 44.817±0.549 | 3.000±0.000 | 0.357±0.008 | 0.239±0.012 |
| cslf6-2 | 42.850±0.809 | 2.833±0.167 | 0.363±0.011 | 0.224±0.011 |
The tiller number, shoot and root biomass of oscslf6-1 mutants decreased under Pi-sufficient conditions and this was alleviated under Pi-deficient conditions (Fig. 3C, D; Table 1). Similarly, those parameters in oscslf6-2 mutant were recovered to almost the same levels as in WT plants (Fig. 3C, D; Table 1). Furthermore, leaf necrosis in the oscslf6 mutants wasnot seen when those plants were grown under Pi-deficient conditions. In LP conditions, higher Pi concentrations in the shoots of oscslf6-1 and oscslf6-2 mutants were also observed, whereas, in roots, the Pi concentration was similar to the WT (Fig. 3F) although the absolute concentration level was much lower than that at the HP level.
To confirm further that excessive Pi accumulation of oscslf6 mutants is indeed caused by the loss-of-function of OsCSLF6, cslf6-1-c lines were also investigated using 30-d-old plants grown under both HP and LP conditions (Fig. 3A, C; Table 1). The results showed the Pi concentration and growth retardation of cslf6-1-c plants were almost rescued to the levels of the WT plants, suggesting strongly that phenotypes of oscslf6-1 mutants are caused by the loss-of-function of OsCSLF6 (Fig. 3A, C; Table 1).
To explore whether the phenotype of oscslf6 mutants is related to nitrogen (N) accumulation, N concentrations in the shoots of the oscslf6 plants were also examined. The result showed that the N content was slightly increased in the shoots of the oscslf6-1 mutants but was not changed in the oscslf6-2 mutants compared with the WT (see Supplementary Fig. 2A at JXB online). However, expression of most nitrate transporters and ammonium transporters were not significantly changed in the shoots of oscslf6 mutants (see Supplementary Fig. 2B at JXB online). Furthermore, no alteration of the phenotypes was found in mutants under different N supply conditions (Supplementary Fig. 2C at JXB online). The phenotypes of toxic leaf symptoms and growth retardation were still observed in oscslf6-1 mutants under high-nitrogen (+10N;12.5mM NH4NO3) and N-deficient (no NH4NO3) conditions (see Supplementary Fig. 2C at JXB online). These results suggested that nitrogen was not the reason for the phenotypes of oscslf6 mutants.
Taken together, these results indicate that OsCSLF6 is involved in the regulation of shoot and root Pi accumulation.
Knockout of OsCSLF6 affects development of primary and adventitious roots
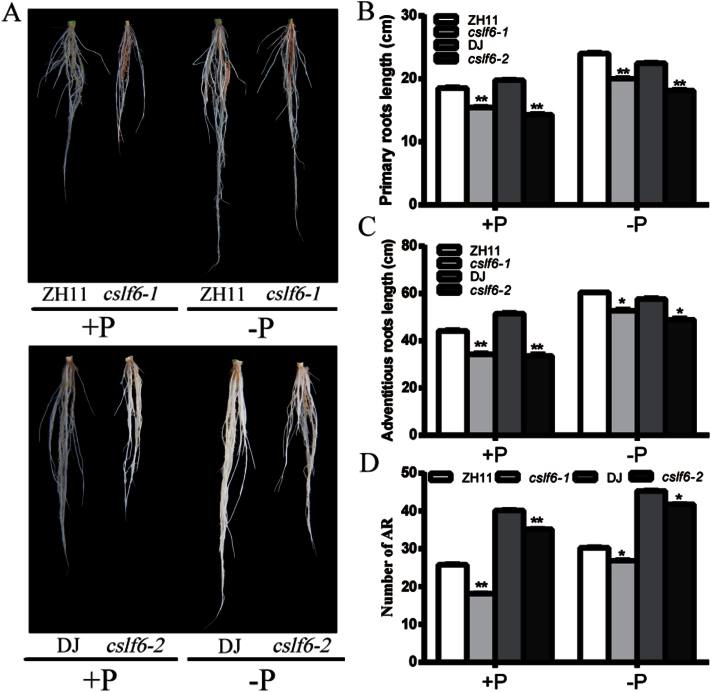
A suite of studies have shown that plants adjust their root structure and morphology to increased Pi availability (Niu et al., 2013). The root architecture system is sensitive to Pi starvation, as indicated by the elongation of primary and adventitious roots in rice (Wissuwa, 2003; Yi et al., 2005). To test whether OsCSLF6 is involved in root architecture alteration in response to Pi concentration, plants grown under Pi-sufficient or Pi-deficient hydroponic culture conditions for 14 d were used to compare the primary root length, adventitious root (AR) number, and total length of the three longest adventitious roots (Fig. 4A). Under Pi-sufficient conditions, primary and adventitious roots in the oscslf6 mutants were shorter than those in wild-type plants (Fig. 4A–C). There was a significant decrease in the number of adventitious roots of the mutant under both Pi-sufficient and Pi-deficient conditions compared with the WT (Fig. 4D). The primary roots, adventitious roots, and adventitious root number were more highly induced in mutants than in the WT under the LP condition (Fig. 4A–D). Consistently, Pi contents in oscslf6 roots were also rescued to the WT level under Pi starvation (Fig. 3F). Taken together, these results suggested that OsCSLF6 is involved in Pi-dependent root architecture alteration possibly by affecting the Pi contents in roots.
Fig. 4.
Effects of Pi availability in the medium on root architecture in WT (ZH11), oscslf6-1, WT (DJ), and oscslf6-2 mutant plants. (A) Root performance of 14-d-old seedlings of WT (ZH11), oscslf6-1, WT (DJ), and oscslf6-2 mutant plants under Pi-sufficient and Pi-deficient conditions. (B–D) The length of primary roots (B), the three longest adventitious roots (C), and the number of adventitious roots (AR) (D) of WT (ZH11), oscslf6-1, WT (DJ), and oscslf6-2 mutant plants under Pi-sufficient or Pi-deficient conditions. Error bars indicate SD (n=10). Asterisks indicate the significance of differences between WT and cslf6 mutant plants as determined by Student’s t test: *0.01≤ P ≤0.05, **P <0.01.
Knockout of OsCSLF6 affects the expression of Pi transporters
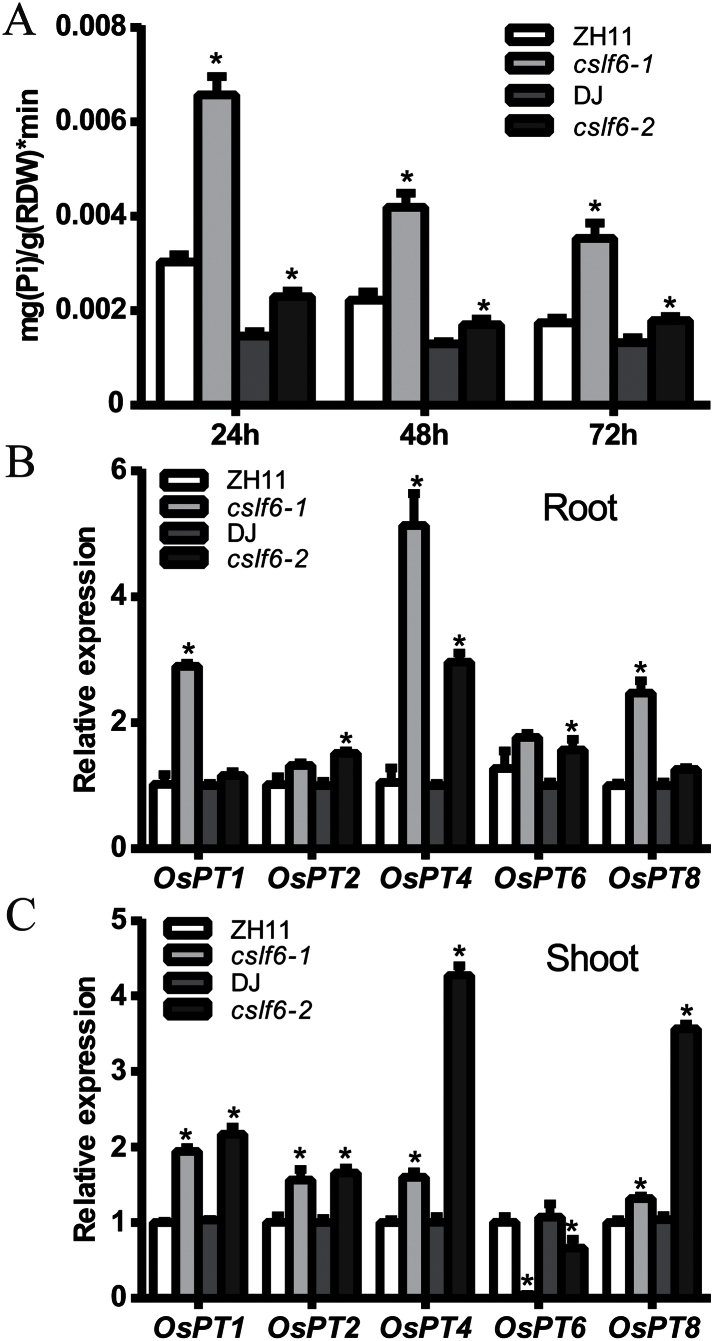
A Pi uptake experiment was then performed to explore whether Pi over-accumulation in oscslf6 was due to increased Pi uptake. The Pi uptake rates of oscslf6 mutants were significantly higher than that of WT at 24, 48, and 72h (Fig. 5A). The result indicates that knockout of OsCSLF6 leads to an increase in Pi uptake, which may result in the accumulation of excess Pi under abundant Pi conditions.
Fig. 5.
Pi uptake of WT (ZH11), cslf6-1, WT (DJ), and oscslf6-2 mutant plants. (A) Pi uptake of WT (ZH11), oscslf6-1, WT (DJ), and oscslf6-2 mutant plants under HP conditions. (B–C) Expression of Pi transporter genes in roots (B) and shoots (C) of WT (ZH11 and DJ) and oscslf6 mutant plants under HP conditions. Expression was normalized to that of ubiquitin. Error bars indicate SD (n=3). Asterisks indicate the significance of differences between WT and oscslf6 mutant plants as determined by Student’s t test: *P ≤0.05.
A number of putative high-affinity Pi transporter genes that function in Pi uptake, translocation, and homeostasis have been identified in rice (Goff et al., 2002; Ai et al., 2009; Jia et al., 2011; Sun et al., 2012). To determine whether the improved Pi uptake and increased Pi accumuation in oscslf6 mutants were associated with the induction of expression of genes encoding Pi transporters, the expression of OsPT1, OsPT2, OsPT4, OsPT6, and OsPT8 were analysed. Under Pi-sufficient conditions, the expression of OsPT1, OsPT2, OsPT4, and OsPT8 was significantly increased in both shoots and roots of oscslf6 mutants (Fig. 5B, C). OsPT6 was up-regulated in roots but was repressed in shoots of oscslf6 mutants (Fig. 5B,C). These results indicate that enhanced Pi uptake in the shoots and roots of oscslf6 mutants, which results in increased Pi accumulation at HP levels, are positively correlated with the increased expression of these OsPHT genes in these tissues.
Knockout of OsCSLF6 affects the expression of PSR genes
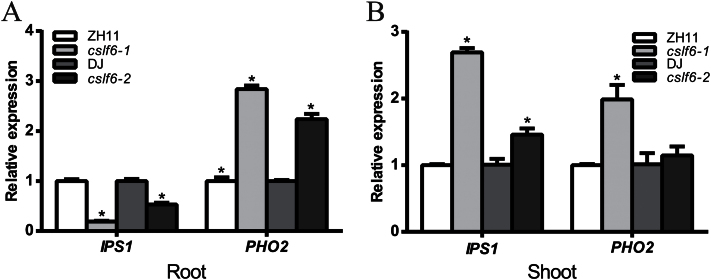
To test whether the genes related to the Pi starvation-response were also affected by loss-of-function of OsCSLF6, the expression of two genes (OsIPS1 and OsPHO2) were examined by qRT-PCR in oscslf6 mutants under Pi-sufficient conditions. OsIPS1 and OsPHO2 have been shown to be involved in the Pi starvation signalling pathway. Our results show that the expression of OsIPS1 was significantly increased in shoots but dramatically repressed in roots of oscslf6 mutants (Fig. 6A, B). The expression of OsPHO2 was significantly increased in the roots or shoots of oscslf6 mutants (Fig. 6A, B). These results implied that OsCSLF6 affects the phosphate starvation signalling pathway in rice.
Fig. 6.
Expression of Pi signalling pathway genes in the WT and oscslf6 mutants. (A) Expression of OsIPS1 and OsPHO2 in roots of WT (ZH11), oscslf6-1, WT (DJ), and oscslf6-2 mutant plants under HP conditions. (B) Expression of OsIPS1 and OsPHO2 in shoots of WT (ZH11), oscslf6-1, WT (DJ), and oscslf6-2 mutant plants under HP conditions. Expression was normalized to that of ubiquitin. Error bars indicate SD (n=3). Asterisks indicate the significance of differences between WT and oscslf6 mutant plants as determined by Student’s t test: *P ≤0.05.
Knockout of OsCSLF6 increases Suc level
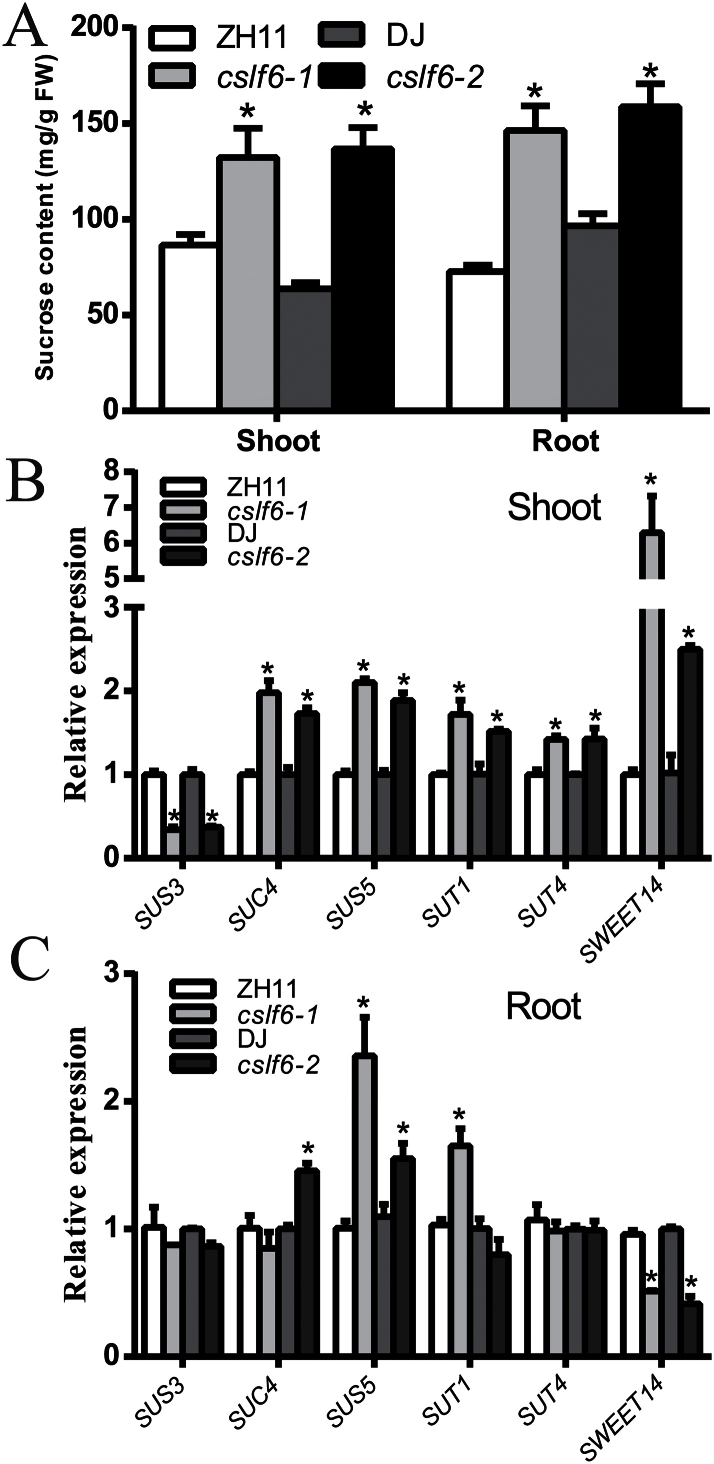
It has been reported that Suc signalling is involved in plant responses to Pi starvation (Chiou and Lin, 2011) and knockout of OsCSLF6 caused an obvious change in cell wall monosaccharide composition (Vega-Sanchez et al., 2012). The Suc content in the oscslf6 mutants was then examined. The result showed that the accumulation of Suc in both shoots and roots of both mutants (oscslf6-1 and oscslf6-2) seedlings were much higher than in the WT plants (Fig. 7A). The expression was also examined of Suc synthases (SUSs) and Suc transporters (SUTs) that are involved in Suc synthesis and transport. QRT-PCR analyses showed that the expression of SUS4, SUS5, SUT1, and SUT4 was dramatically enhanced although SUS3 was repressed in shoots of the oscslf6 mutants (Fig. 7B, C). OsSweet14, a gene that functions as Suc transporters in the HEK293T cell line and Xenopus oocytes, was also significantly increased in shoots and decreased in roots of the oscslf6 mutants compared with the WT (Fig. 7B, C) (Chen et al., 2010, 2012). These results demonstrated that knockout of OsCSLF6 results in increased Suc accumulation in rice, possibly through the activation of SUS and SUT genes. Together, these results suggest that OsCSLF6 may have a role in affecting the Pi pathway by changing the level of Suc.
Fig. 7.
Accumulation of Suc in the oscslf6 mutants. (A) Soluble sugars were extracted from 12-d-old WT and oscslf6 mutants. Suc contents were determined by gas chromatography–mass spectrometry. (B–C) Expression of SUS3, SUS4, SUT1, SUT4, and Sweet14 in both shoots (B) and roots (C) of 12-d-old WT and mutants. Expression was normalized to that of ubiquitin. Error bars indicate SD (n=3). Asterisks indicate the significance of differences between WT and oscslf6 mutant plants as determined by Student’s t test: *P ≤0.05.
Oscslf6 is defective in cell walls
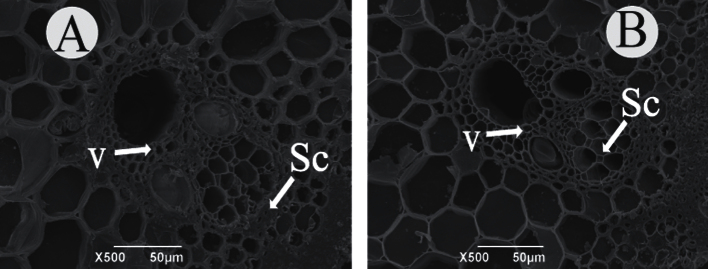
Cell wall morphology was also examined with scanning electron microscopy. Scanning electron microscopy observations revealed that the WT sclerenchyma cell walls were heavily thickened and that the cells were nearly completely filled at the mature stages of culms (Fig. 8A), in striking contrast to those of oscslf6-1 mutant plants (Fig. 8B). To investigate cell wall composition of the oscslf6 mutants, transverse sections of the culms of the WT and mutant were histochemically stained with Calcofluor solutions. Calcofluor showed much stronger fluorescent signals in the sclerenchyma cells, parenchyma cells, and vascular bundles of the WT than in the oscslf6-1 mutant (see Supplementary Fig. 4A, B at JXB online), demonstrating a significantly high level of cellulose in wild-type plants. This finding is consistent with the scanning electron microscopy observations and indicates that the oscslf6 mutant is deficient in the cell walls.
Fig. 8.
Scanning electron analysis in ZH11 and oscslf6-1 mutant plants. (A, B) Scanning electron in cross culms sections of ZH11 (A) and oscslf6-1 (B) plants. Sc, sclerenchyma cells; V, vascular bundles. Bars=50 μm.
Expression pattern of OsCSLF6
For histochemical analysis of the expression pattern of OsCSLF6, the promoter (a 1.8-kb fragment upstream of ATG) of OsCSLF6 was fused to the GUS reporter gene and transformed into WT rice. GUS activity was preferentially detected in the vascular tissues in the tissues examined, such as panicles, leaf lamina joints, root s, and culms (see Supplementary Fig. 3A–I at JXB online), and the highest GUS activity was detected in the roots (see Supplementary Fig. 3F–I at JXB online). Total RNA was extracted from flag leaves, mature leaves, roots, leaf lamina joints, panicles, and culms, and qRT-PCR analyses were performed. The result showed OsCSLF6 was expressed in all the organs examined, with expression being lowest in mature leaves and highest in roots (see Supplementary Fig. 3J at JXB online), which was consistent with the GUS analysis.
Discussion
Proteins encoded by the CSLs super-family of genes are known to be involved in the biosynthesis of cell-wall polymers and in polysaccharide biosynthesis (Richmond and Somerville, 2000). Members of the CSLF super-family, which are only found in grasses (Hazen et al., 2002), are believed to control the synthesis of mixed-linkage glucan (β-1,3; 1,4, glucan) (Burton et al., 2006). The novel function of OsCSLF6, a member of this family, in affecting Pi accumulation and root development, possibly through the alteration of carbon metabolism in rice, is reported here.
OsCSLF6 regulates Pi uptake or transport by affecting the expression of PHT genes
The oscslf6 mutants displayed excessive Pi accumulation and toxic leaf symptoms and growth retardation similar to that of OsPHR2(O), OsPT2(O), and ospho2 mutants (Liu et al., 2010; Hu et al., 2011). These phenotypes suggest that Pi absorption and translocation may be altered in oscslf6 mutants. Pi transporters are directly responsible for Pi acquisition and transport in plants (Harrison et al., 2002; Misson et al., 2004; Seo et al., 2008), and plants up-regulate the expression of Pi transporters to enhance Pi uptake and transport efficiency (Liu et al., 1998; Karthikeyan et al., 2002; Xiao et al., 2006; Hu et al., 2011). Our results showed that the expression of OsPT1/2/4/8 was significantly increased in both roots and shoots of oscslf6 mutants compared with WT plants, whereas OsPT6 was dramatically repressed in oscslf6 shoots under Pi-sufficient conditions. The differential response of PHT genes in both roots and shoots of oscslf6 mutants suggested that Pi uptake or transport in the oscslf6 mutant were affected. Increased expression of OsPT1/2/4/8 in roots may be related to Pi uptake and Pi transport from the roots to the shoots in the oscslf6 mutants. In addition, the expression of OsPT6 was dramatically repressed in shoots of ospho2 which may lead to defective Pi distribution and mobilization (Hu et al., 2011). Similarly, the significantly reduced OsPT6 expression in oscslf6 shoots may also result in defective Pi mobilization in these plants. Taken together, our results strongly indicate that OsCSLF6 is involved in Pi accumulation by affecting the expression of some PHT genes in rice.
OsCSLF6 is involved in regulating root development
Root architecture, the spatial configuration of a root system in the soil, has been shown to be important for plant P acquisition (Lynch, 1995). In Arabidopsis, plants show dramatic changes in root architecture, including reduced primary root growth and the increased formation of lateral roots and root hairs under low Pi availability (Williamson et al., 2001; López-Bucio et al., 2003). Different from Arabidopsis, the elongation of rice primary and adventitious roots are the typical traits stimulated by Pi starvation (Wissuwa, 2003; Zhou et al., 2008a). To investigate whether OsCSLF6 is involved in this process, the effect of Pi availability on root architecture alteration was analysed in WT and oscslf6 mutant plants. In our observations, consistent with the over-accumulation of Pi, a significant decrease in primary root length, and adventitious root length and number were observed in oscslf6 mutants compared with the WT under Pi-sufficient conditions. However, those parameters were alleviated under Pi-deficient conditions. Meanwhile, the Pi content in roots of the oscslf6 mutants was increased in the oscslf6 mutants under Pi-sufficient conditions but no significant difference was observed compared with the WT under Pi starvation conditions (Fig. 3F). Taken together, these results indicated that OsCSLF6 may affect Pi-dependent root architecture by affecting the Pi status in roots.
OsCSLF6 may play a role in the balance between carbon metabolism and phosphate accumulation
Increased carbohydrates have been associated in many plant species with low P availability. Over-accumulation of Suc in roots precedes the induction of PSR and the inhibition of Suc biosynthesis or translocation attenuates the plant response to Pi starvation (Hammond and White, 2008, 2011). In Arabidopsis, over-expression of SUC2 resulted in the increased translocation of Suc in the phloem and enhanced sensitivity to Pi starvation (Lei et al., 2011). The authors argued that Suc is a global regulator of P-starvation and elevated levels of Suc can directly alter the expression of a large number of PSI genes (Lei et al., 2011). It has also been reported that over-expression of ZmPTF1 improves low phosphate tolerance of maize by regulating carbon metabolism (Li et al., 2011). This evidence suggests a role of carbohydrates in communicating the plant responses to low Pi availability. In our results, knockout of OsCSLF6 increased the Pi content and enhanced the expression of some PSI genes such OsPT1/2/6/8. Increased Suc levels were also observed in the oscslf6 mutants compared with WT plants. Therefore, it is possible that increased Pi accumulation in oscslf6 mutants result from the alteration of carbon metabolism and carbohydrate allocation.
IPS1 is a PSI gene that negatively affects plant uptake (Franco-Zorrilla et al., 2007) and over-expression of AtIPS1 results in the increased accumulation of PHO2 mRNA and reduced shoot Pi content in Arabidopsis. OsIPS1 (Hou et al., 2005) and OsPHO2 (Hu et al., 2011), the two rice homologue genes to AtIPS1 and PHO2, respectively, function similarly to their Arabidopsis counterparts in response to Pi starvation. It was observed that the knockout of OsCSLF6 suppressed OsIPS1 in roots but induced OsIPS1 in shoots, while the expression of OsPHO2 was increased in both shoots and roots of the oscslf6 mutants. Similar results were also obtained in the Arabidopsis pho3 mutant that had a defective Suc transporter 2 (SUC2) (Lei et al., 2011). In this mutant, substantially reduced transportation of SUC from the shoot to the root and decreased expression of PSR genes in the root, but increased expression in the shoots, were observed (Lloyd and Zakhleniuk, 2004; Lei et al., 2011). These contrasting expression patterns of PSI genes (OsIPS1 and OsPHO2) may be correlated with the altered accumulation patterns of Suc in the oscslf6 mutants.
It was also found that the loss of OsCSLF6 affected the biosynthesis of secondary cell walls and resulted in altered cellulose content (Fig. 8C; see Supplementary Fig. 4A–C at JXB online). It is possible that the alteration in cell wall composition or structure affects the expression of PSI genes, such as OsIPS1 and OsPHO2, that are involved in Pi signalling through the plant vasculature. Xylem and phloem provide high-speed pathways for long-distance transportation in plants (Turnbull and Lopez-Cobollo, 2013; Lin et al., 2014). Various kinds of molecules, such as inorganic nutrients, phytohormones, and other metabolites are distributed by the xylem and phloem throughout the plants (Ye, 2002; Lough and Lucas, 2006; Turnbull and Lopez-Cobollo, 2013; Lin et al., 2014). Therefore, the possible effect of altered cell wall composition/structure on Pi assimilation could not be ruled out.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Molecular features of OsCSLF6.
Supplementary Fig. S2. Phenotypes of oscslf6-1 mutant plants under high-nitrogen and nitrogen-deficient conditions.
Supplementary Fig. S3. OsCSLF6 expression profile.
Supplementary Fig. S4. Staining of cellulose in ZH11 and oscslf6-1 mutant plants.
Supplementary Table S1. Primers used in this study.
Supplementary Table S2. Primers for real-time PCR analysis.
Acknowledgements
This work was supported by the Major State Basic Research Development Program of China (973 Program) (No. 2013CB127001), the National High Technology R&D Program of China (863 Program) (No. 2012AA10A303), the National Natural Science Foundation of China (No. 31070267), and the Program for New Century Excellent Talents in University of the Ministry of Education in China (NCET-09-0401).
References
- Ai P, Sun S, Zhao J, et al. 2009. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. The Plant Journal 57, 798–809. [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. 2006. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiology 141, 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Kim MC, Chun HJ, et al. 2013. Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis . Plant Physiology 161, 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. 2006. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology 141, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. 2006. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science 311, 1940–1942. [DOI] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Perez-Perez J, Solano R, Leyva A, Paz-Ares J. 2010. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genetics 6, e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, et al. 2010. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB. 2012. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. [DOI] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH. 2009. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. The Plant Cell 21, 3554–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Nimmo G, Jenkins G, Nimmo H. 2007. BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochemical Journal 405, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL. 2006. Regulation of phosphate homeostasis by microRNA in Arabidopsis. The Plant Cell 18, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. 2011. Signaling network in sensing phosphate availability in plants. Annual Review of Plant Biology 62, 185–206. [DOI] [PubMed] [Google Scholar]
- Dai X, Wang Y, Yang A, Zhang WH. 2012. OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiology 159, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. 2007. a . WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiology 143, 1789–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. 2009. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Molecular Plant 2, 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. 2007. b . Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiology 145, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics 39, 1033–1037. [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. 2005. A miRNA involved in phosphate-starvation response in Arabidopsis. Current Biology 15, 2038–2043. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Hammond JP, White PJ. 2008. Sucrose transport in the phloem: integrating root responses to phosphorus starvation. Journal of Experimental Botany 59, 93–109. [DOI] [PubMed] [Google Scholar]
- Hammond JP, White PJ. 2011. Sugar signaling in root responses to low phosphorus availability. Plant Physiology 156, 1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J. 2002. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. The Plant Cell 14, 2413–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Scott-Craig JS, Walton JD. 2002. Cellulose synthase-like (CSL) genes of rice. Plant Physiology 128, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Wu P, Jiao F, Jia Q, Chen H, Yu J, Song X, Yi K. 2005. Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant, Cell and Environment 28, 353–364. [Google Scholar]
- Hu B, Zhu C, Li F, Tang J, Wang Y, Lin A, Liu L, Che R, Chu C. 2011. LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiology 156, 1101–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, Murphy AS, Raghothama KG. 2007. Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiology 144, 232–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, et al. 2000. T-DNA insertional mutagenesis for functional genomics in rice. The Plant Journal 22, 561–570. [DOI] [PubMed] [Google Scholar]
- Jia H, Ren H, Gu M, Zhao J, Sun S, Zhang X, Chen J, Wu P, Xu G. 2011. The phosphate transporter gene OsPht1; 8 is involved in phosphate homeostasis in rice. Plant Physiology 156, 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Jain A, Held MA, Carpita NC, Raghothama KG. 2007. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225, 907–918. [DOI] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Mukatira UT, D’Urzo MP, Damsz B, Raghothama KG. 2002. Regulated expression of Arabidopsis phosphate transporters. Plant Physiology 130, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HF, Chiou TJ. 2011. The role of microRNAs in phosphorus deficiency signaling. Plant Physiology 156, 1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. 2003. The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology 6, 280–287. [DOI] [PubMed] [Google Scholar]
- Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D. 2011. Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiology 156, 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Qian O, Zhou YH, et al. 2003. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. The Plant Cell 15, 2020–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZX, Gao Q, Liu YZ, He CM, Zhang XR, Zhang JR. 2011. Overexpression of transcription factor ZmPTF1 improves low phosphate tolerance of maize by regulating carbon metabolism and root growth. Planta 233, 1129–1143. [DOI] [PubMed] [Google Scholar]
- Lin WY, Huang TK, Leong SJ, Chiou TJ. 2014. Long-distance call from phosphate: systemic regulation of phosphate starvation responses. Journal of Experimental Botany 65, 1817–1827. [DOI] [PubMed] [Google Scholar]
- Liu C, Muchhal US, Uthappa M, Kononowicz AK, Raghothama KG. 1998. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiology 116, 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang Z, Ren H, Shen C, Li Y, Ling HQ, Wu C, Lian X, Wu P. 2010. OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. The Plant Journal 62, 508–517. [DOI] [PubMed] [Google Scholar]
- Liu J, Samac DA, Bucciarelli B, Allan DL, Vance CP. 2005. Signaling of phosphorus deficiency‐induced gene expression in white lupin requires sugar and phloem transport. The Plant Journal 41, 257–268. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV. 2004. Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. Journal of Experimental Botany 55, 1221–1230. [DOI] [PubMed] [Google Scholar]
- Lough TJ, Lucas WJ. 2006. Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annual Review of Plant Biology 57, 203–232. [DOI] [PubMed] [Google Scholar]
- Lv Q, Zhong Y, Wang Y, et al. 2014. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. The Plant Cell 26, 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. 1995. Root architecture and plant productivity. Plant Physiology 109, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH. 2007. Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiology 143, 156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Nilsson L, Nielsen LK, Hamborg Nielsen T. 2005. Interaction between phosphate starvation signalling and hexokinase‐independent sugar sensing in Arabidopsis leaves. Physiologia Plantarum 124, 81–90. [Google Scholar]
- Misson J, Thibaud M-C, Bechtold N, Raghothama K, Nussaume L. 2004. Transcriptional regulation and functional properties of Arabidopsis Pht1; 4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Molecular Biology 55, 727–741. [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA. 2005. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proceedings of the National Academy of Sciences, USA 102, 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, He Y, Dai Y, Liu X, Li J. 2000. Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. The Plant Cell 12, 405–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. 2007. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Muller R, Nielsen TH. 2007. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana . Plant, Cell and Environment 30, 1499–1512. [DOI] [PubMed] [Google Scholar]
- Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS. 2013. Responses of root architecture development to low phosphorus availability: a review. Annals of Botany 112, 391–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T. 2011. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends in Plant Science 16, 442–450. [DOI] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, et al. 2014. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, 14947–14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. 2000. The cellulose synthase superfamily. Plant Physiology 124, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes & Development 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HM, Jung Y, Song S, Kim Y, Kwon T, Kim DH, Jeung SJ, Yi YB, Yi G, Nam MH, Nam J. 2008. Increased expression of OsPT1, a high-affinity phosphate transporter, enhances phosphate acquisition in rice. Biotechnology Letters 30, 1833–1838. [DOI] [PubMed] [Google Scholar]
- Sun SB, Gu MA, Cao Y, Huang XP, Zhang X, Ai PH, Zhao JN, Fan XR, Xu GH. 2012. A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiology 159, 1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull CG, Lopez-Cobollo RM. 2013. Heavy traffic in the fast lane: long-distance signalling by macromolecules. New Phytologist 198, 33–51. [DOI] [PubMed] [Google Scholar]
- Vega-Sanchez ME, Verhertbruggen Y, Christensen U, et al. 2012. Loss of cellulose synthase-like f6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiology 159, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ying S, Huang H, Li K, Wu P, Shou H. 2009. Involvement of OsSPX1 in phosphate homeostasis in rice. The Plant Journal 57, 895–904. [DOI] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM. 2001. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiology 126, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M. 2003. How do plants achieve tolerance to phosphorus deficiency? Small causes with big effects. Plant Physiology 133, 1947–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Li X, Yuan W, Chen G, Kilian A, Li J, Xu C, Li X, Zhou DX, Wang S. 2003. Development of enhancer trap lines for functional analysis of the rice genome. The Plant Journal 35, 418–427. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Ruan WY, Shi J, et al. 2014. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proceedings of the National Academy of Sciences, USA 111, 14953–14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Liu J, Dewbre G, Harrison M, Wang ZY. 2006. Isolation and characterization of root‐specific phosphate transporter promoters from Medicago truncatula . Plant Biology 8, 439–449. [DOI] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. 2005. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiology 138, 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH. 2002. Vascular tissue differentiation and pattern formation in plants. Annual Review of Plant Biology 53, 183–202. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. 1976. Laboratory manual for physiological studies of rice , 3rd edn Manila: International Rice Research Institute. [Google Scholar]
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S. 2006. RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Research 34, D745–D748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao FC, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. 2008. a . OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiology 146, 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Yamagishi M, Osaki M, Masuda K. 2008. b . Sugar signalling mediates cluster root formation and phosphorus starvation-induced gene expression in white lupin. Journal of Experimental Botany 59, 2749–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.