Highlight
The cargo receptor cornichon, located in the endoplasmic reticulum, interacts with the low-affinity Na+ transporter OsHKT1;3 for its delivery to the Golgi apparatus.
Key words: Cornichon, endoplasmic reticulum, Golgi, OsHKT1;3, protein–protein interaction.
Abstract
Membrane proteins are synthesized and folded in the endoplasmic reticulum (ER), and continue their path to their site of residence along the secretory pathway. The COPII system has been identified as a key player for selecting and directing the fate of membrane and secretory cargo proteins. Selection of cargo proteins within the COPII vesicles is achieved by cargo receptors. The cornichon cargo receptor belongs to a conserved protein family found in eukaryotes that has been demonstrated to participate in the selection of integral membrane proteins as cargo for their correct targeting. Here it is demonstrated at the cellular level that rice cornichon OsCNIH1 interacts with OsHKT1;3 and, in yeast cells, enables the expression of the sodium transporter to the Golgi apparatus. Physical and functional HKT–cornichon interactions are confirmed by the mating-based split ubiquitin system, bimolecular fluorescence complementation, and Xenopus oocyte and yeast expression systems. The interaction between the two proteins occurs in the ER of plant cells and their co-expression in oocytes leads to the sequestration of the transporter in the ER. In the yeast cornichon mutant erv14, OsHKT1;3 is mistargeted, preventing the toxic effects of sodium transport in the cell observed in wild-type cells or in the erv14 mutant that co-expressed OsHKT1;3 with either OsCNIH1 or Erv14p. Identification and characterization of rice cornichon as a possible cargo receptor opens up the opportunity to improve our knowledge on membrane protein targeting in plant cells.
Introduction
Membrane transport proteins have to be selectively targeted to specific membranes to control ion and metabolite fluxes into and within the cell. However, very little is known about the sorting mechanisms of plant transporters. During evolution, eukaryotic cells developed multiple systems to target proteins to their respective compartments. Once membrane proteins are synthesized and folded in the endoplasmic reticulum (ER), they are directed by specific protein–protein interactions through the secretory pathway to the site where they will finally dwell. Transport of membrane and secretory cargo proteins is mediated by COPII vesicles that help to select these cargoes from ER-resident proteins. In yeast, humans, and plants, there is evidence that selection of cargo proteins is mediated specifically by the Sec24 subunit of the COPII complex (Miller et al., 2002; Mossessova et al., 2003; Mancias and Goldberg, 2008; Faso et al., 2009; Conger et al., 2011). Conserved domains involved in cargo selection have been identified (Miller et al., 2003; Mossessova et al., 2003). Accessory proteins known as cargo participate in the selection of specific cargo that can be either soluble or membrane proteins destined for different organelle or vesicular membranes (Dancourt and Barlowe, 2010). Correct targeting of membrane proteins, in particular ion transporters, is important for controlling ion homeostasis within the cell. Participation of cargo receptors is proposed to increase the specificity of the secretory pathway, with a particular receptor recognizing a specific cargo to be delivered to a precise cellular site of residence (Dancourt and Barlowe, 2010). In yeast, the role of Erv proteins (endoplasmic reticulum vesicle proteins) as cargo receptors is indicated by their association with COPI and COPII vesicles, their cycling between the ER and Golgi, and the selective inhibition of cargo transport with the deletion of a particular Erv cargo receptor (Belden and Barlowe, 2001). For example, Erv29p has been demonstrated to select soluble secretory proteins such as carboxypeptidase Y and proteinase A (Belden and Barlowe, 2001; Otte and Barlowe, 2004), while Erv26p is involved in the export of membrane proteins such as alkaline phosphatase and mannosyltransferase (Bue et al., 2006; Bue and Barlowe, 2009). In contrast, Erv14p participates in the ER export of membrane proteins through ER lumen-localized sorting signals (Powers and Barlowe, 1998, 2002). Yeast Erv14p is a well conserved protein found in eukaryotes, initially identified in Drosophila as cornichon (Roth et al., 1995), and later in mammals where it was found to be associated with glutamate receptors of the AMPA subtype (Schwenk et al., 2009). In plants, there is little information on the possible role of supplementary cargo receptors in the secretory pathway, with the exception of vacuolar cargo receptor proteins directly involved in the selection of soluble proteins for their delivery to the vacuole lumen (Sanderfoot et al., 1998; Tse et al., 2004; Wang et al., 2011). However, it has been demonstrated in plant transporters that the presence of diacidic (Mikosch et al., 2006; Dunkel et al., 2008) or dileucine motifs (Komarova et al., 2012; Wolfenstetter et al., 2012) or loops between transmembrane domains (Komarova et al., 2012) is important for the correct delivery to the target membrane.
While the acronym HKT for high-affinity K+ transporter implies that members of the family transport K+, the majority of these proteins play a more significant role in low-affinity Na+ transport and have been identified as key players in salt tolerance (Plett et al., 2010; Munns et al., 2012; Schroeder et al., 2013). Yet, knowledge of the regulation of these transporters is limited. HKT transporters form two distinct subfamilies. Members of subfamily 1 are associated with Na+ tolerance based on their expression in the parenchyma cells of the xylem where they remove Na+ from the root xylem (Uozumi et al., 2000; Ren et al., 2005; Sunarpi et al., 2005) or the leaf sheath (Cotsaftis et al., 2012; Munns et al., 2012), preventing its transport to the leaf blade. Alternatively, they may participate in Na+ recirculation from the leaves to the root along the phloem (Berthomieu et al., 2003). The specific overexpression of AtHKT1;1 in the root stele of Arabidopsis thaliana (Møller et al., 2009) or rice (Oryza sativa) (Plett et al., 2010) caused a decreased Na+ transport to the shoot, with a consequent lower accumulation of Na+ in the leaves and higher fresh weight, and resulting in an increase in salinity tolerance. Members of subfamily 2 probably participate in ion absorption (Rubio et al., 1995; Horie et al., 2001, 2007; Yao et al., 2010), as indicated by their localization at the plasma membrane (Horie et al., 2007, 2011; Lan et al., 2010; Xue et al., 2011).
In A. thaliana, only a single member of the HKT family, AtHKT1;1, is present (Uozumi et al., 2000), while the rice genome contains nine HKT genes, one of them probably a pseudo-gene (Garciadeblás et al., 2003). The presence of more than one HKT gene in rice, with members from the two subfamilies, indicates that the corresponding proteins must play particular roles in the physiology of the plant, either by expression in a particular tissue or organelle, or by presenting unique transport properties. Previous studies have confirmed this view, with members from the two subfamilies showing differences in ion selectivity (Horie et al., 2001, 2011; Golldack et al., 2002; Jabnoune et al., 2009; Yao et al., 2010; Sassi et al., 2012) and differential gene expression profiles in plant tissues (Golldack et al., 2002; Kader et al., 2006; Jabnoune et al., 2009).
OsHKT1;3 functions as a highly selective Na+ transporter and its transcript is expressed in the vascular tissue of roots and leaves. The high expression in leaf adaxial epidermal bulliform cells has been used to propose its involvement in turgor changes for rolling and unrolling of leaves in response to environmental variations (Jabnoune et al., 2009). Of particular interest is the high Na+ selectivity of OsHKT1;3 which, if not unique among HKTs, raises the question of the role of such a mechanism, in view of the toxic effects that sodium has on glycophytes, such as rice, and its widespread expression in the plant (Jabnoune et al., 2009). To study the regulation of OsHKT1;3, interacting proteins that might regulate its activity/trafficking were searched for employing a large-scale protein–protein interaction screen based on the mating-based split ubiquitin system (mbSUS) (Lalonde et al., 2010; Chen et al., 2012; Jones et al., 2014). In this screen, a plant homologue of the putative cargo receptor, cornichon, was identified. Here the characteristics of rice cornichon are reported at the cellular level, together with its interaction with OsHKT1;3 as a cargo protein, and the intracellular localization of OsHKT1;3 to the Golgi system where it may function as the shunt conductance for the H+ pumps that acidify this organelle.
Materials and methods
Mating-based split ubiquitin system (mbSUS)
For PCR amplification of the open reading frame (ORF) for OsHKT1;3 and OsCNIH1 genes without their stop codon, the primers described in Supplementary Table S1 available at JXB online were employed. Entry clones were generated with the pENTR-TOPO vector/plasmid following the manufacturer’s instructions (Invitrogen). The integrity of gene insertion was confirmed by sequencing and by digestion with PvuII (Roche). The LR clonase (Invitrogen) was employed to transfer the OsHKT1;3 and OsCNIH1 genes to the pMETYC_GW (Cub clones) and pXN32_GW (Nub clones) vectors, respectively. Yeast media were prepared as previously described (Lalonde et al., 2010). The THY.AP4 (MATa ura3, leu2, lexA::LacZ::trp1 lexA::HIS3 lexA::ADE2) and THY.AP5 (MATα URA3, leu2, trp1, his3 loxP::ade2) yeast strains were transformed with the pMETYC_GW and pXN32_GW vectors, respectively (Obrdlik et al., 2004), employing the LiAc protocol previously described (Lalonde et al., 2010). Cub clones were pre-screened to identify false-positive and false-negative fusions. Those fusions that did not interact with the soluble NubWT, that has a strong affinity for the Cub domain, were considered as false negatives; while those that did interact with the NubG (with reduced affinity for Cub) corresponded to false positives. Those Cub fusions that passed the pre-screening were used with the A. thaliana membrane-linked interactome protein library Nub clones. Positive interactors were chosen by the similar growth shown to the soluble NubWT in IS-500 medium (Lalonde et al., 2010).
Transitory expression of OsHKT1;3 and OsCNIH1 tagged with florescent proteins in Nicotiana benthamiana leaf epidermis
For the transitory transformation of N. benthamiana leaves, plants were grown in the glasshouse from seeds in pots containing Metromix 500 soil (SunGro) at 25 °C under natural light conditions. Leaves from 4-week-old plants were infiltrated with Agrobacterium tumefaciens harbouring either the pOsHKT1;3-EYFP, pOsHKT1;3-mCherry, or pOsCNIH1-mCherry constructs under the control of the 35S promoter and/or the indicated subcellular marker gene constructions (Nelson et al., 2007). For bimolecular fluorescence complementarion (BiFC) experiments, leaf infiltration was done using the constructs pYFC43-OsHKT1;3, pYFC43-OsCNIH1, pYFC43-AtPIP2A, pYFN43-OsHKT1;3, pYFN43-OsCNIH1, and pYFN43-AtPIP2A (Belda-Palazón et al., 2012). Agrobacterium tumefaciens containing each construct were grown in 30ml of LB with rifampicin (50 μg ml–1) and spectinomycin (50 μg ml–1) or kanamycin (5050 μg ml–1) at 28 °C at an OD600 between 0.3 and 0.5. Leaves were infiltrated with a solution containing sodium phosphate buffer pH 7, 0.1mM acetosyringone (Sigma), 28mM glucose, and bacterial culture with an OD600 of 0.3. To obtain the expression clone for each gene, an LR gateway-based recombination reaction (Invitrogen) with either pX-EYFP-GW or pX-Cherry-GW for C-terminal translational fusions, or pYFC43 and pYFN43 for BiFC assays, were achieved. Transformed electrocompetent cells of the strain GV3101 of A. tumefaciens with each of these constructions were used for expression in tobacco. Image J software (http://imagej.nih.gov/ij/) was used to analyse the confocal images for co-localization. Pearson’s coefficient and scatter plots were obtained using the JACoP Plug-in (Bolte and Cordelières, 2006).
Functional expression of OsHKT1;3 in Xenopus oocytes
The two-electrode voltage-clamp technique was used to record the activity of OsHKT1;3 and OsCNIH1, individually or co-expressed in oocytes 2 d after cRNA injection as described (Ortiz-Ramirez et al., 2011). For all measurements, oocytes were clamped at their free running membrane potential. The reported results are the means ±SD of 5–10 oocytes. For expressing OsHKT1;3 and OsCNIH1 in Xenopus oocytes, both genes were cloned into the pOO2 oocyte expression vector (Ludewig et al., 2002) employing the primers described in Supplementary Table S1 at JXB online. An OsHKT1;3–EGFP fusion protein was made by adding the ORF of the enhanced green fluorescent protein (pEGFP-C1; Clontech) at the C-terminus of OsHKT1;3 which was then subcloned into pOO2. Clones were verified by sequencing. Capped cRNA was transcribed in vitro by SP6 RNA polymerase using the mMessage mMachine kit (Ambion), after linearization of the plasmid with Bbr P1 (Roche).
Heterologous expression of OsHKT1;3 in yeast
OsHKT1;3 was amplified by PCR employing the primers described in Supplementary Table S1 at JXB online. The PCR-amplified OsHKT1;3 gene was inserted into the YEp352-NHA1-URA and pGRU1-NHA1-URA vectors (previously digested with PstI and PvuII, respectively) behind the Saccharomyces cerevisiae NHA1 promoter (Kinclová et al., 2001) using homologous recombination in S. cerevisiae BW31a cells (MATa leu2-3/122 ura3-1 trp1-1 his3-11/15 ade2-1 can1-100 GAL SUC2 mal10 ena1-4Δ::HIS3 nha1::LEU2). The resulting vectors were pYEp352-OsHKT1;3 and pGRU1-OsHKT1;3. The correct constructions were confirmed by sequencing using oligos described in Supplementary Table S1. OsCNIH1 and ERV14 cloned in pENTR-TOPO were transferred to pDR-F1-GW-LEU by an LR reaction (Invitrogen). pDR-F1-OsCNIH1 and pDR-F1-ERV14 constructs are under control of the PMA1 promoter. Constructions were verified by PvuII digestion (Thermo Scientific). The S. cerevisiae BY4741 strain (MATa his3Δleu2Δmet15Δura3Δ) was used to determine NaCl sensitivity in drop-test assays. Yeast cells were grown aerobically at 30 °C in standard YPD (to prepare competent cells for transformation) or Yeast Nitrogen Base (YNB) media without amino acids (to select and maintain transformants), with 2% glucose as carbon source and appropriate auxotrophic supplements. The growth phenotype of cells was estimated in drop tests on solid YNB medium supplemented with NaCl after 4 d. The ERV14 gene was deleted by homologous recombination using the KanMX marker gene and the Cre–loxP system-producing strain BY4741Δerv14 (Supplementary Fig. S1). Primers used are described in Supplementary Table S2.
Fluorescence microscopy
Fluorescence from EYFP was visualized by excitation with an Argon laser at 514nm with the spectral detector set between 540nm and 30nm for the emission. The wavelengths employed for Citrine and mCherry were 488nm and 543nm (excitation), and 515/30nm and 630/60nm (emission), respectively. Abaxial epidermal peels of mature leaves were placed onto microscope slides in water, covered with a cover slide, and observed by fluorescence microscopy using an inverted multiphoton confocal microscope (Olympus FV1000) equipped with a ×60 oil immersion objective. Results are representative images from >10 cells from at least four different independent transformations.
Results
Analysis of protein–protein interactions between OsHKT1;3 and the Arabidopsis membrane interactome
In order to obtain additional information on the functioning of OsHKT1;3, the possible regulation of the transporter through protein–protein interactions was studied by employing the mbSUS (Obrdlik et al., 2004; Lalonde et al., 2010). Screening an A. thaliana membrane-linked interactome protein library as prey (Nub clones), with OsHKT1;3 protein used as bait (Cub clone) (Lalonde et al., 2010; Jones et al., 2014), 19 possible interactions were identified, as indicated by growth of diploid yeast cells under selective conditions (Table 1). These interactions were common to two other rice HKTs, OsHKT1;1 and OsHKT2;1 (Table 1).
Table 1.
Arabidopsis proteins interacting with OsHKT1;3
Arabidopsis proteins identified with the mbSUS employing OsHKT1;3 as bait (Cub clone) and the Arabidopsis interactome (Nub clones) (Jones et al., 2014) as prey.
| OsHKT1;3 (Cub) | AGs Arabidopsis (Nub) | Description |
|---|---|---|
| Os02g07830 | AT3G17000 | Ubiquitin-conjugating enzyme 32 |
| AT3G25805 | Unknown protein | |
| AT1G17280 | Ubiquitin-conjugating enzyme 34 | |
| AT1G21240 | Cell wall-associated kinase | |
| AT1G34640 | Unknown protein | |
| AT1G47640 | Unknown protein | |
| AT1G63110 | Cell division cycle protein-related | |
| AT2G26180 | IQ-domain 6 (IQD6) | |
| AT3G08040 | Member of the MATE family | |
| AT3G10640 | VPS60 vesicle-mediated transport | |
| AT3G12180 | Cornichon family protein | |
| AT3G28220 | Meprin and TRAF homology domain-containing protein | |
| AT4G30850 | Heptahelical transmembrane protein homologous to human adiponectin receptors and progestin receptors | |
| AT5G06100 | myb family of transcription factors (MYB33) | |
| AT5G06320 | Tobacco hairpin-induced gene (HIN1) | |
| AT5G10450 | 14-3-3 gene family | |
| AT5G37050 | Unknown protein | |
| AT5G49540 | Unknown protein | |
| AT5G52240 | Similar to progesterone-binding proteins in animals. |
OsHKT1;3 interacts with OsCNIH1 in yeast
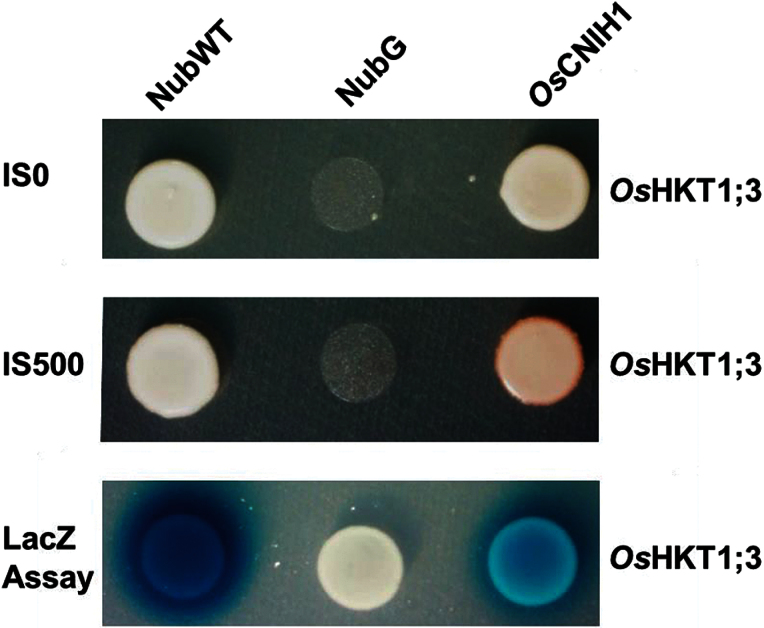
After analysing the 19 interacting proteins, it was decided to focus on a protein homologous to Erv14p in yeast and cornichon in Drosophila and mammals, which has been shown to direct the trafficking of membrane proteins from the ER to the Golgi apparatus (Gillingham et al., 2004), or to the plasma membrane (Schwenk et al., 2009; Herzig et al., 2012). Confirmation of the interaction between OsHKT1;3 and rice cornichon (OsCNIH1; Os06g04500) was obtained by employing OsHKT1;3 and OsCNIH1 as Cub and Nub clones, respectively, in the mbSUS. Growth of diploid yeast cells in selective medium confirmed the interaction between OsHKT1;3 and OsCNIH1 (Fig. 1; IS0), and the strength of the interaction was demonstrated by growth of the diploid cells in the presence of 0.5mM methionine which acts as a repressor of the pMETYC promoter (Grefen et al., 2007; Fig. 1, IS500). As expected, OsHKT1;3 did not interact with soluble NubG, but it did with soluble NubWT, that were used as false-positive and false-negative controls, respectively (Fig. 1). Additional confirmation of the interaction between OsHKT1;3 and OsCNIH1 was derived from analysing LacZ activity with X-Gal as a substrate, whereby clear blue signals in the diploid cells harbouring OsHKT1;3 and OsCNIH1 clones were observed. The blue precipitate was also observed with the positive control (soluble NubWT), but not with soluble NubG, the negative control (Fig. 1, LacZ).
Fig. 1.
Homologous interaction between OsHKT1;3 and OsCNIH1. Homologous interaction between OsHKT1;3 and OsCNIH1 confirmed with the mbSUS in yeast with selective medium in the absence (IS0) or in the presence of methionine (0.5mM; IS500). Corroboration of the interaction between OsHKT1;3 and OsCNIH1 was demonstrated by activation of LacZ and revealed with X-Gal as a substrate. Representative results of three different assays are shown.
Sequence analysis of rice cornichon
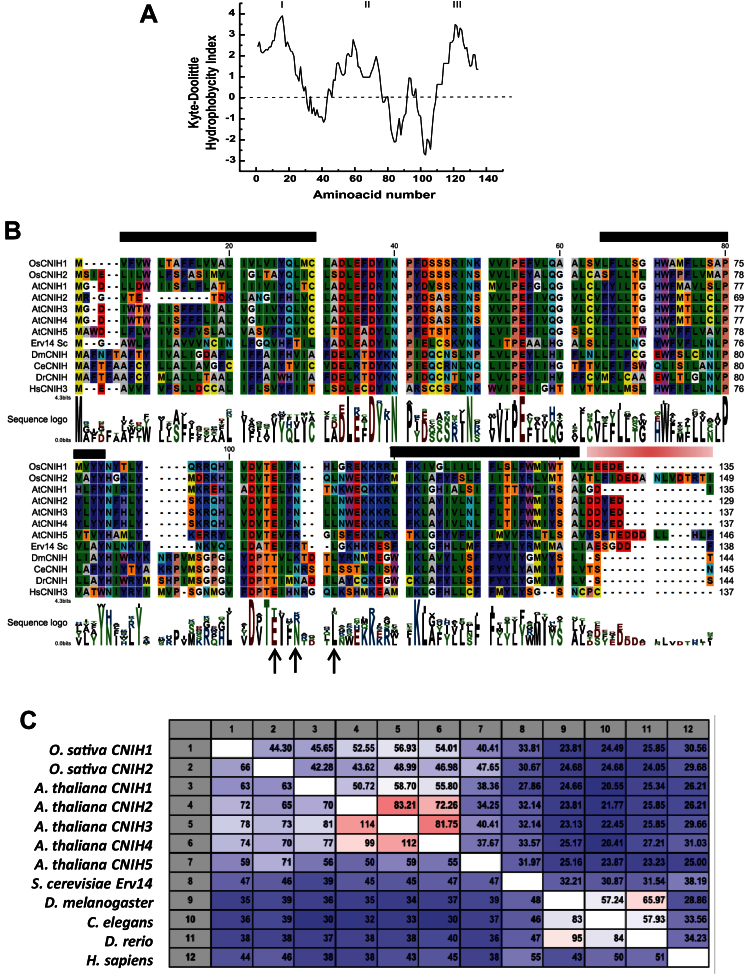
The rice genome contains two cornichons, OsCNIH1 (Os06g04500), a small hydrophobic protein of 135 amino acids (Fig. 2A, B), and OsCNIH2; (Os12g32180), which encodes a slightly larger protein (149 amino acids; Fig. 2B). The two isoforms share 44% identity at the amino acid level. Cornichon belongs to a family of membrane proteins unique to eukaryotes that is predicted to have three membrane-spanning α-helixes (Fig. 2A, Kyte–Doolittle; Fig. 2B, TMHMM, black lines), with the N-terminus towards the cytoplasm according to the predicted positive-inside rule (von Heijne, 1992), and the C-terminus located in the ER lumen. All plant cornichon homologues possess an acidic domain in the luminal C-terminus (Fig. 2B, red line). The similarity between plants, yeast, fly, worm, zebra fish, and human cornichons is ~40%, with OsCNIH1 showing higher identity with the Arabidopsis homologues AtCNIH2, AtCNIH3, and AtCNIH4 (Fig. 2C).
Fig. 2.
OsCNIH1 sequence analysis. (A) A Kyte–Doolittle hydropathy plot shows three potential transmembrane domains (I, II, and III; values >0) in OsCNIH1. (B) Sequence alignment of OsCNIH1 with OsCNIH2, AtCNIH1, AtCNIH2, AtCNIH3 AtCNIH4, AtCNIH5, ScErv14, and cornichon homologues from Drosophila (Drosophila melanogaster), worm (Caenorhabditis elegans), zebra fish (Danio rerio), and human isoform 4 (Homo sapiens). Accession numbers are Os06g04500, Os12g32180, At4g12090, At1g12340, At1g12390, At1g62880, At3g12180, YGL054C Erv14, NP_477068, CAB01516, NP_001028278, and NP_001264129.1, respectively. Black bars indicate putative transmembrane domains; arrows indicate conserved residues involved in binding to COPII in yeast (I96, F97, and L100); the grey bar denotes an acidic domain. (C) Pairwise comparison between the cornichon proteins listed in (B) shows the percentage identity (upper right values) and number of identical residues (bottom left values). (This figure is available in colour at JXB online.)
Split-YFP analyses demonstrate that OsHKT1;3 and OsCNIH1 interact in the ER
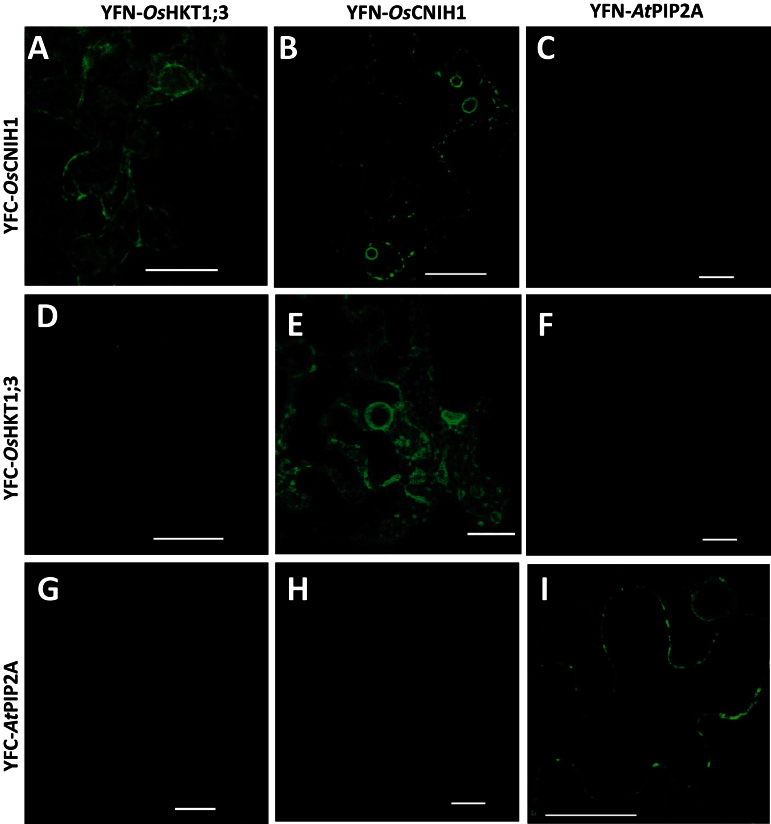
To confirm the interaction between OsHKT1;3 and OsCNIH1 in planta, and to identify the site(s) of interaction, the split-YFP (yellow fluorescent protein) system was employed by fusing the N-terminal half of EYFP to the N-terminal half of OsHKT1;3 (YFN-OsHKT1;3) and the C-terminus of EYFP to the N-terminus of OsCNIH1 (YFC-OsCNIH1). Co-expressing these constructs in tobacco leaves led to fluorescence recovery of EYFP (Fig. 3A), indicating the association between OsHKT1;3 and OsCNIH1 at the ER, according to the reticulated structure highlighted by EYFP. The co-localization of the two proteins was confirmed with fusions of the N- and C-terminal halves of EYFP to the N-terminus of OsCNIH1 (YFN–OsCNIH1) or OsHKT1;3 (YFC–OsHKT1;3), respectively (Fig. 3E). As a positive control for the BiFC assay, the well-known tetramerization of the plasma membrane aquaporin AtPIP2A was used. For this, the N- or C-terminal halves of EYFP were fused to the N-terminus of the aquaporin AtPIP2A (YFN–AtPIP2A and YFC–AtPIP2A). As expected, oligomerization of the aquaporin was observed at the plasma membrane (Fig. 3I). Co-expression of YFN-OsCNIH1 and YFC-AtPIP2A did not lead to reconstitution of EYFP fluorescence (Fig. 3C, H). Additional analyses demonstrated fluorescence reconstitution by expressing the C- and N-termini of YFP fused independently to the N-terminus of OsCNIH1, indicating the oligomerization of the protein in intracellular structures that resembled the Golgi apparatus (Fig. 3B). In contrast, similar studies with OsHKT1;3 failed in reconstituting YFP fluorescence, suggesting that this transporter does not oligomerize (Fig. 3D). No interactions were observed between the aquaporin AtPIP2A and OsHKT1;3 or OsCNIH1 (Fig. 3C, F–H). These results demonstrated that the interaction between OsHKT1;3 and OsCNIH1 occurs at the ER, and indicated the possible oligomerization of rice cornichon. Moreover, the results suggested that BiFC did not seem to be a result of overexpression of the proteins in the same membrane, as indicated by failure in reconstituting EYFP associated with OsHKT1;3.
Fig. 3.
BiFC confirms the interaction of OsCNIH1 with OsHKT1;3 and demonstrates the likely oligomerization of OsCNIH1 in transiently transfected tobacco leaves. (A, E) Reciprocal interaction between YFC–OsCNIH1/YFN–OsHKT1;3 and YFN–OsCNIH1/YFC–OsHKT1;3 confirms the interaction of the two proteins in the ER. (B) Co-expression of YFC–OsCNIH1 and YFN–-OsCNIH1 indicates the possible oligomerization of cornichon in the ER. Absence of a fluorescence complementation signal indicates that: OsCNIH1 does not interact with AtPIP2A (C, H); OsHKT1;3 does not form oligomers (D); and the transporter and the aquaporin do not interact (F, G). (I) Oligomerization of AtPIP2A. Scale bar=25 μm.
Cornichon co-localizes with OsHKT1;3 and resides in the ER and Golgi
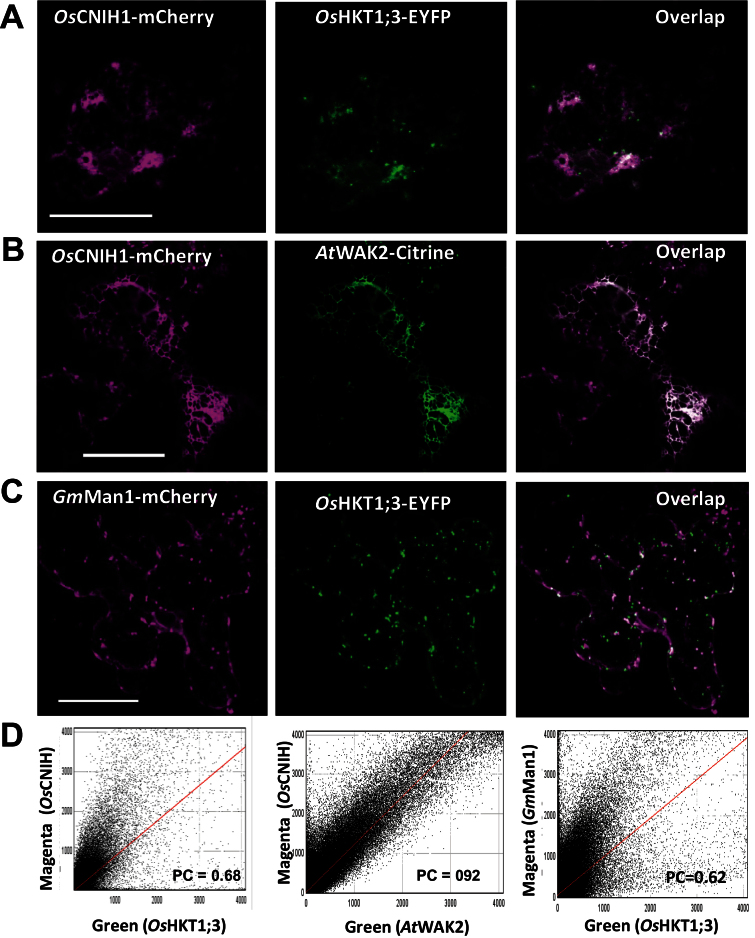
Further support for the interaction between OsHKT1;3 and OsCNIH1 was obtained by analysing tobacco leaf epidermis co-transformed with OsCNIH1-mCherry and OsHKT1;3-EYFP. Expression of OsCNIH1–mCherry was generally observed as a reticulated structure (Fig. 4A, left), while that of OsHKT1;3–EYFP mainly appeared as bright puncta (Fig. 4A, centre). Overlapping both images showed that the puncta associated with OsHKT1;3–EYFP superimposed the reticula highlighted by OsCNIH1–mCherry (Fig. 4A, right, white dots). By plotting the pixel number for OsHKT1;3–EYFP against that from OsCNIH1–mCherry, pixel distribution was demonstrated to occur along a straight line (Fig. 4D, left), and, by applying Costes’ method (Costes et al., 2004) for image analysis using ImageJ (JACoP Plug-in), a Pearson’s coefficient (PC) and P-value of 0.68 and 100%, respectively, were calculated (Fig. 4D, left) (Bolte and Cordelières, 2006). These findings confirmed that OsCNIH1 and OsHKT1;3 partially co-localized in the ER. In view of these results, it was important to determine and confirm the site(s) of residence for OsCNIH1 and OsHKT1;3 individually, and for this co-expression studies were carried out employing several well-characterized membrane markers fused to fluorescent proteins. OsCNIH1–mCherry highlighted a reticulated structure, strongly suggesting its expression at the ER (Fig. 4B, left); this was confirmed by the comparable expression observed for the ER marker AtWAK2–Citrine (Fig. 4B, centre) (Nelson et al., 2007) and by the overlapping of signals from these two markers (Fig. 4B, right, white signal). Image analysis demonstrated a high degree of co-localization between OsCNIH1 and AtWAK2 (PC=0.82; P=100%; Fig. 4D, centre). Localization of OsHKT1;3–EYFP emphasized small, highly motile punctate structures in the cytoplasm, indicating localization to potentially vesicular structures (Fig. 4C, centre). To identify this compartment, OsHKT1;3–EYFP was co-expressed with the Golgi marker GmMan1–mCherry (Nelson et al., 2007), and similar punctate cytoplasmic signals were observed for both proteins (Fig. 4C, left and centre), that when overlapped showed partial co-localization (Fig. 4C, right, white areas). Applying Costes’ method, a calculated PC of 0.62 with a P- value of 100% was obtained, confirming the co-localization of the two proteins (Fig. 4D, right) which indicated that OsHKT1;3–EYFP partially localized to the Golgi.
Fig. 4.
Intracellular co-localization of OsCNIH1 with OsHKT1;3 in plants. (A) Expression of OsCNIH1–mCherry (left) and OsHKT1;3–EYFP (centre), and co-localization of the two proteins (right) in tobacco leaves. (B) Expression of OsCNIH1–mCherry (left) and the ER marker AtWAK2–Citrine (centre), and co-localization of the two proteins (right). (C) Expression of OsHKT1;3–EYFP (centre) and the Golgi marker GmMan1–mCherry (left), and co-localization of the two proteins (right). (D) Scatter plots of pixel distribution of the magenta (y-axis) and green (y-axis) channels employing the Costes algorithm for images shown in (A, left), (B, centre), and (C, right). Scale bar=25 μm.
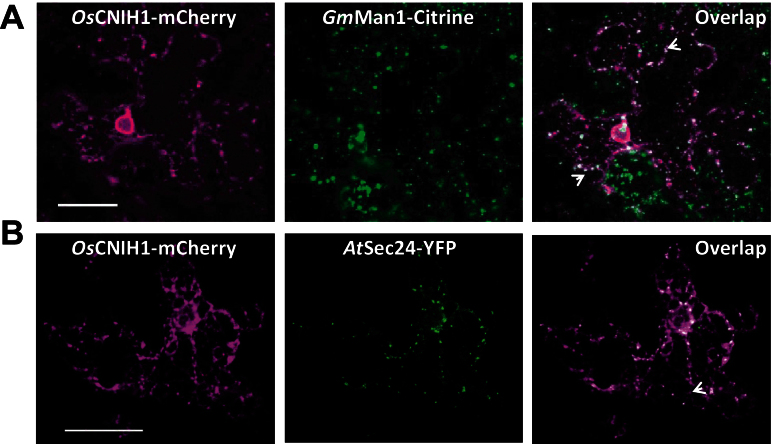
Work in yeast and mammals hase demonstrated that cornichon homologues did not locate exclusively to one specific compartment, but rather circulated actively between the Golgi and the ER, in association with COPII vesicles (Powers and Barlowe, 1998; Harmel et al., 2012). To identify more clearly the plant cornichon location, GmMan1–Citrine was used as a Golgi marker and AtSec24–YFP as a marker for ER exit sites (ERES)/COPII (Hanton et al., 2009; Langhans et al., 2012). Co-expression of OsCNIH1–mCherry (Fig. 5A, left) with GmMan1–Citrine (Fig. 5A, centre) showed that both proteins highlighted punctate structures with properties similar to that of the Golgi, with OsCNIH1–mCherry also highlighting the network associated with the ER, particularly surrounding the nucleus (Fig. 5A, left). Quantitative analyses of the overlapped images (Fig. 5A, right) showed a relatively low correlation (PC=0.40, P=100%; Supplementary Fig. S2A at JXB online), indicating that OsCNIH1 was also present in the Golgi apparatus. When OsCNIH1 was co-expressed with AtSec24–YFP, OsCNIH1 labelled the ER (Fig. 5B, left), while AtSec24 appeared as bright puncta dispersed throughout the cytoplasm (Fig. 5B, centre). AtSec24–YFP co-localized with the reticulate OsCNIH1–mCherry fluorescence (Fig. 5B, right) as indicated by the calculated PC=0.66 and P=100% (Supplementary Fig. S2B), corresponding to the proposed structure for the ERES/COPII compartment. Together, these imaging data indicated that rice cornichon located to the Golgi and ER.
Fig. 5.
OsCNIH1 is also located at the Golgi and ERES. (A) Expression of OsCNIH1–mCherry (left) and the Golgi marker GmMan1–Citrine (centre), and the co-localization of the two proteins (right). (B) Expression of OsCNIH1–mCherry (left) and the ERES/COPII marker AtSec24–YFP (centre), and overlapping of the two images showing the co-localization of the two proteins (right). Arrows indicate the punctate sites of co-localization (white signal). Scale bar=25 μm.
Subcellular localization of OsHKT1;3
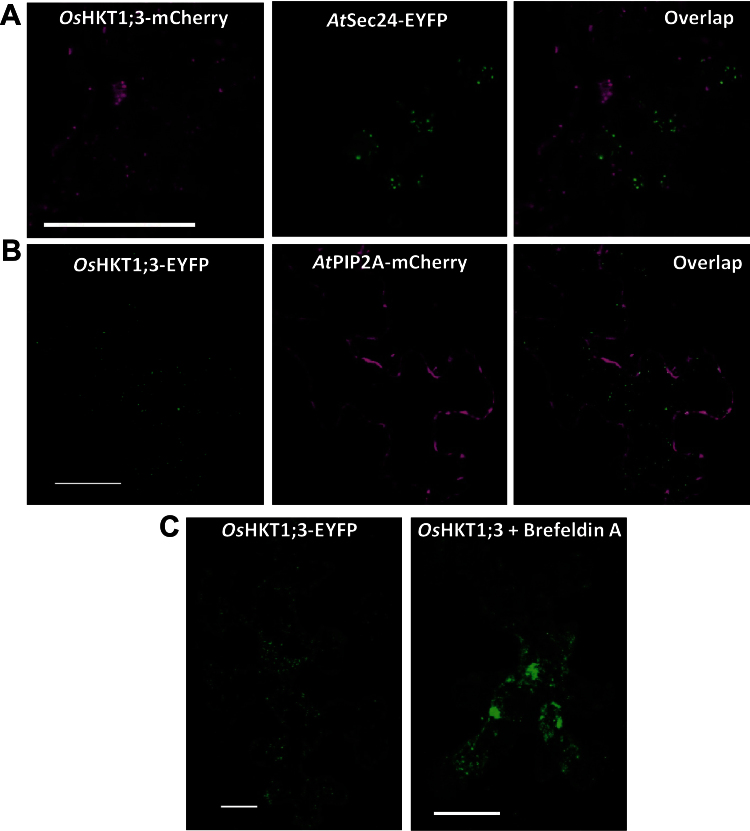
The intracellular localization of OsHKT1;3 (Fig. 4A, C) differed from that of plasma membrane-localized HKT isoforms (Horie et al.., 2007, 2011; Lan et al., 2010; Xue et al., 2011). To explore the localization of OsHKT1;3 in more detail, co-localization analysis was performed with the same membrane markers used for OsCNIH1 (Figs 4, 5). Co-expression of OsHKT1;3–mCherry with AtSec24–YFP demonstrated that both proteins were visualized as punctate structures that did not locate to the same compartment (Fig. 6A). Confirmation that co-localization of OsHKT1;3–mCherry and AtSec24–YFP was low (Fig. 6A, right) was supported by the low PC of 0.32 (P=100%; Supplementary Fig. S2C at JXB online). Co-expression of OsHKT1;3 and AtPIP2A–mCherry in tobacco leaf epidermal cells verified that the puncta corresponding to OsHKT1;3–EYFP localized to the cytoplasm (Fig. 6B, left), while expression of AtPIP2A–mCherry was limited to the plasma membrane (Fig. 6B, centre). Merging of the two channels yielded few co-localization points (Fig. 6B, right); quantitative analysis revealed no co-localization between OsHKT1;3 and AtPIP2A, as indicated by the low PC of 0.14 (P=100%; Supplementary Fig. S2D). Additional evidence supporting the Golgi as the site of residence for OsHKT1;3 was obtained with use of the fungal toxin brefeldin A (BFA). BFA blocks the activation of the GTPase Arf1 by causing a non-productive complex with its Sec7 GTP exchange factor (Chardin and McCormick, 1999), resulting in the formation of Golgi aggregates described as BFA bodies (Ritzenthaler et al., 2002). Incubation of tobacco leaf epidermis transformed with OsHKT1;3-EYFP for 15min with 25 μM BFA caused the appearance of relatively large fluorescent aggregates (Fig. 6C, right) that resembled BFA bodies, in comparison with the smaller puncta from OsHKT1;3–EYFP in the untreated epidermis (Fig. 6C, left). Measurement of the rapid movement of the fluorescent puncta associated with OsHKT1;3–EYFP gave a mean velocity of 0.14±0.02 μm s–1 (mean ±SD, n=9; Supplementary Fig. S3, Supplementary Video S1), similar to the rates that have been reported for the highly motile Golgi apparatus (Boevink et al., 1998). Although some puncta did not move, corresponding to stationary Golgi stacks (Lerich et al., 2012), others moved at a lower speed that varied between 0.081 μm s–1 and 0.061 μm s–1. All these observations provided further evidence that OsHKT1;3–EYFP locates to the Golgi apparatus of plant cells.
Fig. 6.
OsHKT1;3 does not localize to the ERES or the plasma membrane and forms aggregates upon exposure to brefeldin A. (A) Expression of OsHKT1;3–mCherry (left) and the ERES/COPII marker AtSec24–EYFP (centre), and overlapping of the two images (right). (B) Co-localization analysis of OsHKT1;3–EYFP (left) with the plasma membrane marker AtPIP2A–mCherry (centre), and overlapping of the two images (right). The intracellular localization of OsHKT1;3–EYFP, seen as fluorescent puncta distributed throughout the cell (C, left), was modified after incubation of the epidermis with brefeldin A at 25 μM for 15min, resulting in the formation of aggregated bodies (C, right). Scale bar=25 μm.
Co-expression of OsCNIH1 and OsHKT1;3 in Xenopus oocytes modified the cellular localization of the transporter and inhibited its activity
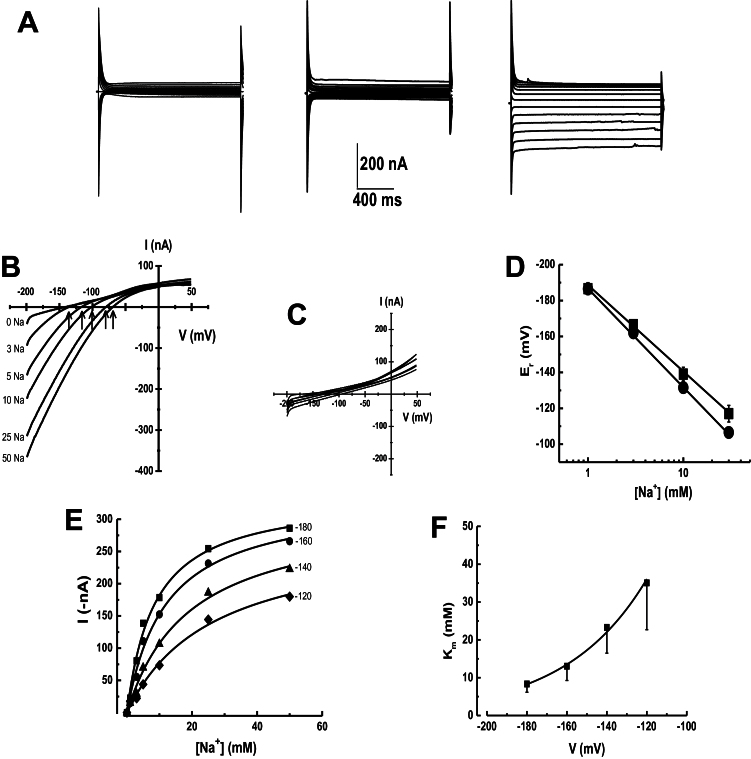
In view of the results that indicated the interaction and co-localization of OsCNIH1 and OsHKT1;3 in plant cells, the oocyte expression system was employed to study the potential effects of OsCNIH1 on the transport activity of OsHKT1;3. Initially, the properties of the transporter reported as an Na+-selective transport mechanism were confirmed (Jabnoune et al., 2009). Oocytes injected with OsHKT1;3 cRNA showed the activation of inward currents when exposed to Na+ solutions (Fig. 7A, right), but not when the cation was absent (Fig. 7A, centre). A control water-injected oocyte exposed to sodium failed to activate any measurable currents (Fig. 7A, left). Confirmation that the inward currents corresponded to Na+ movement was derived from the increasing current magnitude recorded with increasing concentrations of Na+, together with clear shifts in the reversal potential (E r) (Fig. 7B, arrows) activated by voltage ramps. Similar experiments with a water-injected oocyte did not show changes in current magnitude or in E r with variable Na+ concentrations (Fig. 7C). Plotting the E r values against external Na+ concentrations gave a linear relationship with a slope of 54.2 mV per decade Na+ (Fig. 7D, circles). A similar relationship was observed for E r values obtained in the presence of 1mM K+ (Fig. 7D, squares). These results clearly indicated that OsHKT1;3 functions as an Na+-selective transporter/channel and that OsHKT1;3 does not function as an Na+/K+ symporter, as has been reported for some members of the subfamily 2 of HKT transporters (Rubio et al., 1995; Gassmann et al., 1996; Horie et al., 2001). Kinetic properties of the transporter were obtained by plotting current magnitude from voltage ramps against external Na+ concentrations at different holding potentials, observing saturation at concentrations >50mM (Fig. 7E). Data analysis with the Michaelis–Menten equation showed that the apparent transport affinity constant (K m) for Na+ increased at less negative holding potentials, with values of 6.4, 9.6, 17.1, and 42.6mM obtained at –180, –160, –140, and –120 mV, respectively (Fig. 7E). Assuming a single binding site for sodium, Equation 1 was employed to evaluate the voltage dependence of K m, where δ is the fractional electrical distance, e is the elementary charge, V is the membrane potential, k is Boltzmann’s constant, and T is the absolute temperature (Woodhull, 1973).
Fig. 7.
Transport properties of OsHKT1;3. (A) Original traces of currents activated by voltage pulses between –200 mV and 50 mV in 20 mV steps from a control water-injected oocyte exposed to 30mM NaCl (left), or expressing OsHKT1;3 exposed to the bath solution either without (centre) or with 30mM NaCl (right). (B) I–V plot from currents recorded in an oocyte expressing OsHKT1;3 and exposed to different concentrations of NaCl (mM); E r (arrows). (C) I–V plot from currents recorded from a control oocyte and exposed to the same NaCl solutions as in (B). (D) Plot showing the linear relationship between E r and extracellular Na+ concentrations from oocytes expressing OsHKT1;3 in the absence (circles) or presence of 1mM KCl (squares). Lines are least square linear regressions fits with a slope of 54.2 mV per decade. (E) Sodium transport kinetics were voltage dependent. Lines are fits to the Michaelis–Menten equation at the corresponding voltages with r 2 ≥0.9. (F) The affinity (K m) of OsHKT1;3 for Na+ was voltage dependent. The line is a fit to Equation 1. Data are from more than five oocytes from 3–4 different frogs and correspond to the mean ±SD.
| (1) |
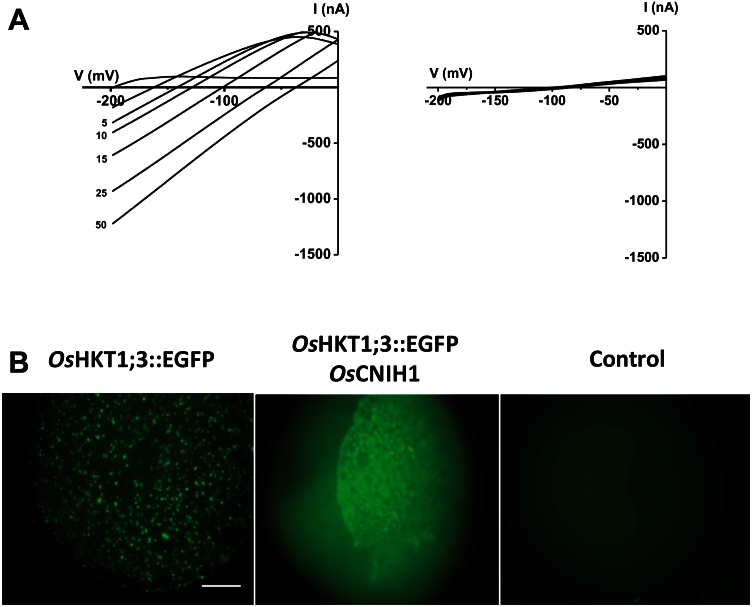
From this analysis, the putative Na+ binding site was calculated to be located at (δ) 65% within the membrane electrical field (Fig. 7F, fitted line). Having confirmed the basic properties of OsHKT1;3 (Jabnoune et al., 2009), the effects of co-expressing OsHKT1;3 together with OsCNIH1 in the oocyte system were then analysed. Voltage-clamp recordings showed that the activity of the transporter was inhibited by the presence of OsCNIH1, as indicated by the absence of inward currents at all extracellular Na+ concentrations tested (Fig. 8A, right). In comparison, an oocyte injected with only the cRNA for OsHKT1;3, activated large inward currents in response to voltage ramps, similar to those previously observed (Fig. 8A, left; see also Fig. 7D). The inhibition of OsHKT1;3 by co-injection of OsCNIH1 in the oocytes could be the result either of direct inhibition of the transporter activity or of the interaction between the two proteins preventing the default targeting of OsHKT1;3 to the oocyte membrane. To distinguish between these two possibilities, EGFP was fused to the C-terminus of OsHKT1;3 and then the construct was expressed in albino Xenopus oocytes, individually or together with OsCNIH1, and the expression was observed under epifluorescence microscopy. When expressed alone, OsHKT1;3–EGFP was observed as punctate structures at the membrane of the oocyte (Fig. 8B, left). In contrast, when co-injected with OsCNIH1, OsHKT1;3–EGFP fluorescence was observed in the interior of the oocyte as a reticulated structure, resembling the ER (Fig. 8B, centre). Autofluorescence from a control water-injected oocyte was minimal (Fig. 8B, right).
Fig. 8.
Co-expression of OsHKT1;3 and OsCNIH1 in Xenopus oocytes caused retention of the transporter in the ER, preventing the activation of Na+ currents. (A) Sodium inward currents activated by voltage ramps from an oocyte expressing OsHKT1;3 (left). Co-expression of OsHKT1;3 and OsCNIH1 (cRNA ratio injected 1:1) in a Xenopus oocyte (right). (B) Expression of OsHKT1;3–EGFP was observed as puncta at the plasma membrane of a Xenopus oocyte (left); upon co-expression with OsCNIH1, fluorescence was observed exclusively in a reticulated structure (centre). Autofluorescence is from a water-injected oocyte (right). Scale bar=250 μm. (This figure is available in colour at JXB online.)
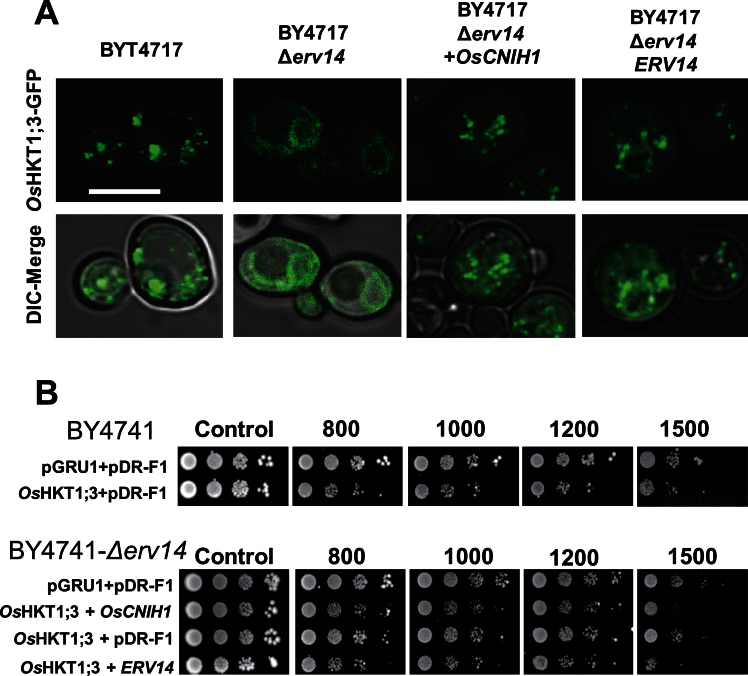
Deletion of ERV14 in yeast modifies the intracellular location of OsHKT1;3 which is restored upon expression of OsCNIH1 or ERV14
To gain further insight into the interaction between OsCNIH1 and OsHKT1;3, the effect of mutating ERV14, the yeast cornichon homologue, on the heterologous expression of OsHKT1;3 tagged with GFP (pGRU1-OsHKT1;3) in BY4741 yeast cells was tested. Transformation of BY4741 yeast cells with the pGRU1-OsHKT1;3 vector showed that localization of OsHKT1;3 was intracellular, highlighting several round bodies that indicated the presence of the transporter at the Golgi apparatus (Fig. 9A; BY4741) (Losev et al., 2006; Matsuura-Tokita et al., 2006). A different localization for OsHKT1;3–GFP was observed in the BY4741Δerv14 mutant cells, as indicated by the diffused fluorescence observed around the nucleus and throughout most of the cytoplasm, a distribution that has been associated with the ER (Fig. 9A; BY4741Δerv14) (Manford et al., 2012). Confirmation that localization of OsHKT1;3 at the Golgi apparatus depended on the presence of OsCNIH1 was obtained by co-transforming BY4741Δerv14 cells with OsCNIH1 and OsHKT1;3-GFP and observing the localization of the latter as round structures corresponding to the Golgi apparatus (Fig. 9A; BY4741Δerv14+OsCNIH1). Similar results were observed when rice cornichon was replaced with ERV14 in the co-transformation of the BY4741Δerv14 cells (Fig. 9A; BY4741Δerv14+ERV14), indicating that both rice cornichon and yeast Erv14p can direct the transporter to the Golgi apparatus. Additional evidence was gathered by investigating the effects of expressing OsHKT1;3 in BY4741 and BY4741Δerv14 yeast cells upon salt stress. Figure 9B (top) shows that transformation of BY4741 cells with OsHKT1;3 and the empty pDR-F1 vector (lower row) reduced cell growth between 0.8M and 1.5M NaCl, when compared with the parental cells transformed with the two empty vectors pGRU1 and pDR-F1 (Fig. 9B, upper row). Co-transformation of BY4741Δerv14 cells with OsHKT1;3 and the empty vector pDR-F1, in contrast, did not affect cell growth in the presence of NaCl (Fig. 9B, bottom, third row). Co-transformation of the BY4741Δerv14 mutant cells with OsCNIH1 and OsHKT1;3 partially restored the sensitivity of the cells towards NaCl (Fig. 9B, bottom, second row), a similar response to that caused by the co-transformation of the BY4741Δerv14 cells with OsHKT1;3 and ERV14 (Fig. 9B, bottom, fourth row).
Fig. 9.
OsCNIH1 restores the intracellular expression of OsHKT1;3 in the yeast mutant BY4741Δerv14 and sensitivity to NaCl. (A) Fluorescence and DIC images of living BY4741 and BY4741Δerv14 cells expressing OsHKT1;3–GFP observed by confocal fluorescence microscopy and co-expressing either OsCNIH1 or ERV14p. Scale bar=5 μm. (B) Drop-test assay on yeast strains BYT4741 (top) and BYT45Δerv14 (bottom) grown in YNB solid medium with different Na+ concentrations and transformed with pGRU1 and pDR-F1, OsHKT1;3 and pDR-F1, OsHKT1;3 and OsCNIH1, or OsHKT1;3 and ERV14. Representative results of at least three different experiments are shown.
Discussion
The putative cargo receptor OsCNIH1 interacts with the Na+ transporter OsHKT1;3
Protein–protein interactions play an essential role in cell structure and function, and have been shown to be important for protein function, regulation, and targeting (Lee et al., 2009; Geiger et al., 2010). The mbSUS with the Arabidopsis interactome (Lalonde et al., 2010; Chen et al., 2012; Jones et al., 2014) was employed to identify proteins that might modulate the activity of OsHKT1;3 by direct protein–protein interactions. A total of 19 potential membrane protein interactions from Arabidopsis were identified (Table 1), and, from these, a protein which shows homology to Drosophila and human cornichon (CNIH) (Bökel et al., 2006; Schwenk et al., 2009) and yeast Erv14p (Powers and Barlowe, 1998, 2002) was selected for further analysis. In all these biological systems, the cornichon homologues have been described as cargo receptors for membrane proteins that may (Bökel et al., 2006; Schwenk et al., 2009) or may not (Powers and Barlowe, 1998, 2002; Herzig et al., 2012) remain attached to the cargo. However, as far as could be assessed from the literature, there is no knowledge of the cornichon homologues in plants.
The interaction between the rice homologue OsCNIH1 and the Golgi membrane Na+ transport protein OsHKT1;3 was identified. Evidence for the interaction between OsHKT1;3 and OsCNIH1 was derived from growth of diploid yeast cells on selective media (IS0 and IS500) and activation of the reporter LacZ (Fig. 1). This interaction was confirmed in planta by bimolecular complementation of EYFP in tobacco cells where intracellular fluorescence signals were observed from reticulated structures reminiscent of the ER (Fig. 3A, E). This result was observed with either the C- or N-terminus of EYFP fused to the N-terminus of either OsCNIH1 or OsHKT1;3 (Fig. 3A, E). These observations were corroborated by co-localization studies between OsCNIH1 and OsHKT1;3 which also demonstrated the intracellular occurrence of both proteins in co-transformed tobacco leaves, where OsHKT1;3 was observed as small puncta superimposed on the ER highlighted by OsCNIH1 (Fig. 4A). Together, these results may be used to propose that direct interaction between OsCNIH1 and OsHKT1;3 occurs at the ER (Fig. 3A, E) and is required to direct the Na+ transporter to the Golgi (Fig. 4C). The absence of fluorescence complementation when tobacco leaves expressed the YFN–OsCNIH1 and YFN–AtPIP2A protein chimeras indicated that the aquaporin does not interact with rice cornichon (Fig. 3C), and suggests that the interaction observed between OsCNIH1 and OsHKT1;3 was specific and not a general occurrence between cornichon and membrane proteins. Moreover, lack of EYFP fluorescence complementation by co-expression of YFN–OsHKT1;3 and YFC–OsHKT1;3 (Fig. 4D) indicates that overexpression of the protein at the same membrane does not, by itself, lead to reconstitution of the fluorescent protein, discarding the possibility that the results obtained with the interaction between OsCNIH1 and OsHKT1;3 are an artefact. This result was also supported by the absence of interaction observed with the mbSUS that indicated that OsHKT1;3 does not oligomerize (Supplementary Fig. S4 at JXB online). Corroboration of the interaction of OsCNIH1 with OsHKT1;3 was obtained independently by employing Xenopus oocytes co-expressing the two proteins, resulting in the retention of the transporter in internal structures (Fig. 8B, centre), and the inability to measure OsHKT1;3-dependent Na+ currents (Fig. 8A, right). In the absence of OsCNIH1, the transporter followed the default pathway for exogenous proteins to the oocyte plasma membrane, similarly to what was observed for other heterologous expressed organelle-localized proteins, including aquaporins from plants. This response resembles the unregulated transport of the glutamate receptor (AMPAR) to the plasma membrane observed in the Caenorhabditis elegans cnih1 mutant (Brockie et al., 2013), and may be a result of an imbalance in the assembly of the COPII system caused by the lack of cornichon which leads to the uncontrolled targeting of OsHKT1;3 to the oocyte plasma membrane. Only when the cargo receptor is present is the transporter retained in the endomembrane system (Fig. 8B, centre). Alterations in COPII-mediated ER membrane protein export have also been observed upon overexpression of Sec12p and were proposed to be a result of Sar1p titration (D’Enfert et al., 1989; DaSilva et al., 2004). Yet another result that further supported the interaction between OsCNIH1 and OsHKT1;3 as being responsible for the proper targeting of the transporter to the Golgi membrane was the restoration of the location of OsHKT1;3 in the yeast mutant BY4741Δerv14 upon co-expression with either of the cornichon homologues, OsCNIH1 or Erv14p (Fig. 9A). Parallel to these results, restoration of salt sensitivity to the BY4741Δerv14 yeast mutant by co-expression of the rice sodium transporter together with either of the two cornichon homologues (Fig. 9B) further supported the dependence of OsHKT1;3 on OsCNIH1 for the correct targeting of the transporter to its membrane of residence, the Golgi. These results strongly indicated that in the absence of Erv14p, OsHKT1;3 was not delivered to the Golgi membrane and, thus, did not disturb the functioning of the cell, allowing yeast growth in the presence of Na+.
The presence of OsCNIH1 in the ER and the Golgi apparatus (Figs 4, 5) is comparable with that reported for the homologues in yeast (Powers and Barlowe, 1998) and mammals (Harmel et al., 2012). CNIH’s homologues are associated with the transport of membrane proteins, including Axl2p in yeast (Powers and Barlowe, 1998, 2002; Gillingham et al., 2004), Gurken in Drosophila (Bökel et al., 2006), and GluRAo in mammals (Schwenk et al., 2009; Kato et al., 2010; Harmel et al., 2012; Herring et al., 2013), and, it is argued, OsHKT1;3 in rice. OsCNIH1 conserves three out of five amino acids (I96, F97, and L100; Fig. 2B, arrows) in the cytoplasmic loop between TMD2 and TMD3 that effect the binding of Erv14p to COPII vesicles (Powers and Barlowe, 2002), suggesting that in plants, transport of membrane proteins in the early secretory pathway may also involve the association of OsCNIH1 with COPII vesicles. This is supported by the co-localization of OsCNIH1 with the bona fide COPII marker AtSec24 in tobacco leaves (Fig. 5B), a site where OsCNIH1 would be functioning as a cargo receptor for OsHKT1;3 (Fig. 4A). Targeting of OsHKT1;3 to the Golgi apparatus by OsCNIH1 can be compared with the Erv14p-dependent localization of Rud3p to the Golgi in yeast that also involves the participation of the GTP-binding protein Arf1p (Gillingham et al., 2004). Recent results in yeast have revealed the central role played by Erv14p as a cargo receptor for a number of membrane proteins including the flippases Dnf1p and Dnf2p, the hexose transporters Hxt3p, Hxt4p, and Hxt5p, as well as the Na+/H+ exchanger Nha1p, among others (Herzig et al., 2012). As indicated by the BiFC results (Fig. 3B), OsCNIH1 seems to form oligomers. This feature seems to be shared with cargo receptors such as ERGIC-53 and p24 protein families (Dancourt and Barlowe, 2010) and may be important for its functioning. In particular, Emp47p, a Type I membrane protein with homology to ERGIC-53, has been demonstrated to oligomerize as a requisite for its exit from the ER (Sato and Nakano, 2003). Further studies will help to confirm this result with OsCNIH1.
In view of the evidence that cornichons may act as cargo receptors in plants, the Arabidopsis Membrane-based Interactome Network Database (MIND1; Jones et al., 2014) was analysed. It was found that the Arabidopsis cornichon AtCNIH1 (At3g12180) interacts with 535 proteins (Supplementary Table S3 at JXB online), with 30% of these corresponding to membrane transport proteins (Supplementary Fig. S5), strengthening the proposed role of CNIH as a membrane protein cargo receptor in plants.
OsHKT1;3 localizes to the Golgi system
Co-localization of OsHKT1;3–EYFP with the Golgi membrane marker GmMan1 (Fig. 4C), its apparent aggregation in BFA bodies caused by BFA (Fig. 6C; Ritzenthaler et al., 2002), and the high motility of the punctate structures in which it is observed (Supplementary Fig. S3, Supplementary Video S1 at JXB online) are evidence consistent with localization of this transporter at the Golgi membrane. OsHKT1;3 does not localize to the plasma membrane (Fig. 6B) as no co-localization with the aquaporin AtPIP2A was observed, distinguishing this HKT from other members of the gene family (Horie et al., 2007, 2011; Munns et al., 2012). Similar protein fusions of GFP to AtAMT1;1 (a plasma membrane resident transporter) and to another rice HKT, OsHKT1;4, demonstrated that these two proteins localized to the plasma membrane (unpublished results), discarding the possibility that tagging of the protein might have modified its membrane of residence (Figs 4C, 6A, C, D). In plants, transporters belonging to large gene families, as is the case for HKT, have members localized to multiple cellular locations, which can include the Golgi. In particular, NHX5 and NHX6, two Na+/H+ exchangers (Bassil et al., 2011), as well as a phosphate transporter, PHT4;6 (Cubero et al., 2009), have been localized to the Golgi, whereas other family members are either plasma membrane (i.e. NHX7, NHX8, and PHT1) or tonoplast localized (NHX1–NHX4). Moreover, members of the aquaporin family of proteins have diverse subcellular localizations, being present in several intracellular compartments (Wudick et al., 2009). Therefore, it is likely that HKT transporters may be located in different cellular compartments.
OsHKT1;3 is an Na+ transporter
Functional characterization in two different heterologous expression systems; Xenopus oocytes and yeast, demonstrated that OsHKT1;3 is an Na+ transporter (Figs 7–9). Electrophysiological data from the oocyte system showed that the reversal potential changed according to Nernst with extracellular Na+ (54.2 mV per decade) (Fig. 7D), indicative of the high selectivity of OsHKT1;3 for sodium, confirming results reported previously (Jabnoune et al., 2009). Expression of OsHKT1;3 in the yeast BY4741 rendered the colonies more susceptible to Na+ (Fig. 9B), probably by generating an ionic imbalance in the cell and further confirming the selectivity of the transporter for this cation.
Although no direct evidence is available, it is proposed that OsHKT1;3 in the Golgi membrane could catalyse the downhill transport of Na+ towards the cytoplasm and, thus, may function as an alternative shunt conductance for the H+ pumps located in this organelle (Mitsuda et al., 2001; Shimaoka et al., 2004; Dettmer et al., 2005; Strompen et al., 2005). The Golgi-located NHX5 or NHX6 Na+ exchangers (Yokoi et al., 2002; Bassil et al., 2011) would lead to Na+ accumulation within the Golgi, establishing the Na+ gradient required for the functioning of OsHKT1;3. Recently, an Na+-selective mechanism has been demonstrated to play a similar role in the lysosomal membrane of mammalian cells (Wang et al., 2012).
Together, the results demonstrate that the highly selective Na+ transporter/channel OsHKT1;3 unexpectedly is located at the Golgi membrane, targeted to this endomembrane by its interaction with a newly described cargo receptor in plants, OsCNIH1. Finding OsHKT1;3 at the Golgi raises the possibility of new functions for the HKT family in addition to those previously described. Future research would demonstrate the biological significance of OsCNIH1 and OsHKT1;3 in the rice plant.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Deletion of ERV14 in S. cerevisiae.
Figure S2. Quantification of co-localization between OsCNIH1 and OsHKT1;3 and other membrane markers.
Figure S3. Dynamics of OsHKT1;3–EYFP in tobacco epidermal cells.
Figure S4. Rice HKT transporters do not interact with each other, indicating the absence of oligomerization.
Figure S5. Classification of AtCNIH-interacting proteins.
Table S1. Primers used for gene cloning into the Gateway (TOPO), pYeP352, and pOO2 plasmids.
Table S2. Primers used for the deletion of ERV14 in S. cerevisiae.
Table S3. Arabidopsis thaliana proteins interacting with AtCNIH1.
Video S1. Dynamics of OsHKT1;3–GFP-labelled bodies that resemble the movement associated with the Golgi apparatus.
Acknowledgements
We thank Saman Parsa and Erika Valle for guidance with the mbSUS method; Pavla Herynková for her assistance with the yeast work; and Andrés Saralegui and Arturo Pimentel for their support with confocal microscopy. The HKT clones from rice were a kind gift from Dr Alonso Rodríguez-Navarro. We also thank Dr Federica Brandizzi for the AtSec24 COPII marker. Help from our technician Maria Guadalupe Munoz Garcia is also recognized. This work was supported by CONACyT grant 79191, DGAPA grant IN203112 to OP, and grant LD13037 to HS.
References
- Bassil E, Ohto M, Esumi T, Tajima H, Zhu Z, Cagnac O, Belmonte M, Peleg Z, Yamaguchi T, Blumwald E. 2011. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. The Plant Cell 23, 224–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazón B, Ruiz L, Martí E, Tárraga S, Tiburcio AF, Culiáñez F, Farràs R, Carrasco P, Ferrando A. 2012. Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS One 7, e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Barlowe C. 2001. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science 294, 1528–1531. [DOI] [PubMed] [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, et al. 2003. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO Journal 22, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. 1998. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. The Plant Journal 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Bökel C, Dass S, Wilsch-Bräuninger M, Roth S. 2006. Drosophila Cornichon acts as cargo receptor for ER export of the TGFalpha-like growth factor Gurken. Development 133, 459–470. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. Journal of Microscopy 224, 213–232. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Jensen M, Mellem JE, et al. 2013. Cornichons control ER export of AMPA receptors to regulate synaptic excitability. Neuron 80, 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bue CA, Barlowe C. 2009. Molecular dissection of Erv26p identifies separable cargo binding and coat protein sorting activities. Journal of Biological Chemistry 284, 24049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bue C, Bentivoglio C, Barlowe C. 2006. Erv26p directs pro-alkaline phosphatase into endoplasmic reticulum-derived coat protein complex II transport vesicles. Molecular Biology of the Cell 17, 4780–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin P, McCormick F. 1999. Brefeldin A: the advantage of being uncompetitive. Cell 97, 153–155. [DOI] [PubMed] [Google Scholar]
- Chen J, Lalonde S, Obrdlik P, Noorani Vatani A, Parsa SA, Vilarino C, Revuelta JL, Frommer WB, Rhee SY. 2012. Uncovering Arabidopsis membrane protein interactome enriched in transporters using mating-based split ubiquitin assays and classification models. Frontiers in Plant Science 3, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conger R, Chen Y, Fornaciari S, Faso C, Held M a, Renna L, Brandizzi F. 2011. Evidence for the involvement of the Arabidopsis SEC24A in male transmission. Journal of Experimental Botany 62, 4917–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. 2004. Automatic and quantitative measurement of protein–protein colocalization in live cells. Biophysical Journal 86, 3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsaftis O, Plett D, Shirley N, Tester M, Hrmova M. 2012. A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS One 7, e39865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero B, Nakagawa Y, Jiang X-Y, Miura K-J, Li F, Raghothama KG, Bressan RA, Hasegawa PM, Pardo JM. 2009. The phosphate transporter PHT4;6 is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Molecular Plant 2, 535–552. [DOI] [PubMed] [Google Scholar]
- D’Enfert C, Wuestehube LJ, Lila T, Schekman R. 1989. Secl2p-dependent membrane binding of the small GTP binding protein Sarlp promotes formation of transport vesicles from the ER. Journal of Cell Biology 114, 663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C. 2010. Protein sorting receptors in the early secretory pathway. Annual Review of Biochemistry 79, 777–802. [DOI] [PubMed] [Google Scholar]
- DaSilva LL, Snapp E, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F. 2004. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. The Plant Cell 16, 1753–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Schubert D, Calvo-Weimar O, Stierhof Y-D, Schmidt R, Schumacher K. 2005. Essential role of the V-ATPase in male gametophyte development. The Plant Journal 41, 117–124. [DOI] [PubMed] [Google Scholar]
- Dunkel M, Latz A, Schumacher K, Müller T, Becker D, Hedrich R. 2008. Targeting of vacuolar membrane localized members of the TPK channel family. Molecular Plant 1, 938–949. [DOI] [PubMed] [Google Scholar]
- Faso C, Chen Y-N, Tamura K, et al. 2009. A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. The Plant Cell 21, 3655–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A. 2003. Sodium transport and HKT transporters: the rice model. The Plant Journal 34, 788–801. [DOI] [PubMed] [Google Scholar]
- Gassmann W, Rubio F, Schroeder J. 1996. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. The Plant Journal 10, 869–882. [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, et al. 2010. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proceedings of the National Academy of Sciences, USA 107, 8023–8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Hin A, Tong Y, Boone C, Munro S. 2004. The GTPase Arf1 and the ER to Golgi cargo receptor Erv14p cooperate to recruit the golgin Rud3p to the cis-Golgi. Journal of Cell Biology 167, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D, Su H, Quigley F, Kamasani UR, Muñoz-Garay C, Balderas E, Popova O, V, Bennett J, Bohnert HJ, Pantoja O. 2002. Characterization of a HKT-type transporter in rice as a general alkali cation transporter. The Plant Journal 31, 529–542. [DOI] [PubMed] [Google Scholar]
- Grefen C, Lalonde S, Obrdlik P. 2007. Split-ubiquitin system for identifying protein–protein interactions in membrane and full-length proteins. Current Protocols in Neuroscience 41, 5.27:5.27.1–5.27.41. [DOI] [PubMed] [Google Scholar]
- Hanton SL, Matheson L a, Chatre L, Brandizzi F. 2009. Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. The Plant Journal 57, 963–974. [DOI] [PubMed] [Google Scholar]
- Harmel N, Cokic B, Zolles G, Berkefeld H, Mauric V, Fakler B, Stein V, Klöcker N. 2012. AMPA receptors commandeer an ancient cargo exporter for use as an auxiliary subunit for signaling. PLoS One 7, e30681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Shi Y, Suh YH, Zheng C-Y, Blankenship SM, Roche KW, Nicoll RA. 2013. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron 77, 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig Y, Sharpe HJ, Elbaz Y, Munro S, Schuldiner M. 2012. A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by Erv14. PLoS Biology 10, e1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Brodsky DE, Costa A, Kaneko T, Lo Schiavo F, Katsuhara M, Schroeder JI. 2011. K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiology 156, 1493–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung H-Y, Miyao A, Hirochika H, An G, Schroeder JI. 2007. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO Journal 26, 3003–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A. 2001. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa . The Plant Journal 27, 129–138. [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Espeout S, Mieulet D, et al. 2009. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiology 150, 1955–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Xuan Y, Xu M, et al. 2014. Border control—a membrane-linked interactome of Arabidopsis. Science 344, 711–716. [DOI] [PubMed] [Google Scholar]
- Kader A, Seidel T, Golldack D, Lindberg S. 2006. Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. Journal of Experimental Botany 57, 4257–4268. [DOI] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Ho MT, et al. 2010. Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron 68, 1082–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinclová O, Ramos J, Potier S, Sychrová H. 2001. Functional study of the Saccharomyces cerevisiae Nha1p C-terminus. Molecular Microbiology 40, 656–68. [DOI] [PubMed] [Google Scholar]
- Komarova NY, Meier S, Meier A, Grotemeyer MS, Rentsch D. 2012. Determinants for Arabidopsis peptide transporter targeting to the tonoplast or plasma membrane. Traffic 13, 1090–1105. [DOI] [PubMed] [Google Scholar]
- Lalonde S, Sero A, Pratelli R, et al. 2010. A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Frontiers in Physiology 1, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W-Z, Wang W, Wang S-M, Li L-G, Buchanan BB, Lin H-X, Gao J-P, Luan S. 2010. A rice high-affinity potassium transporter (HKT) conceals a calcium-permeable cation channel. Proceedings of the National Academy of Sciences, USA 107, 7089–7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans M, Meckel T, Kress A, Lerich A, Robinson DG. 2012. ERES (ER exit sites) and the ‘Secretory Unit Concept’. Journal of Microscopy 247, 48–59. [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. 2009. A protein kinase–phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proceedings of the National Academy of Sciences, USA 106, 21419–21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerich A, Hillmer S, Langhans M, Scheuring D, van Bentum P, Robinson DG. 2012. ER import sites and their relationship to ER exit sites: a new model for bidirectional ER–Golgi transport in higher plants. Frontiers in Plant Science 3, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losev E, Reinke C a, Jellen J, Strongin DE, Bevis BJ, Glick BS. 2006. Golgi maturation visualized in living yeast. Nature 441, 1002–1006. [DOI] [PubMed] [Google Scholar]
- Ludewig U, von Wirén N, Frommer WB. 2002. Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. Journal of Biological Chemistry 277, 13548–13555. [DOI] [PubMed] [Google Scholar]
- Mancias JD, Goldberg J. 2008. Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO Journal 27, 2918–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. 2012. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Developmental Cell 23, 1129–1140. [DOI] [PubMed] [Google Scholar]
- Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, Nakano A. 2006. Live imaging of yeast Golgi cisternal maturation. Nature 441, 1007–1010. [DOI] [PubMed] [Google Scholar]
- Mikosch M, Hurst AC, Hertel B, Homann U. 2006. Diacidic motif is required for efficient transport of the K+ channel KAT1 to the plasma membrane.Plant Physiology 142, 923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E, Antonny B, Hamamoto S, Schekman R. 2002. Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO Journal 21, 6105–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Beilharz TH, Malkus PN, Lee MCS, Hamamoto S, Orci L, Schekman R. 2003. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114, 497–509. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Enami K, Nakata M, Takeyasu K, Sato MH. 2001. Novel type Arabidopsis thaliana H(+)-PPase is localized to the Golgi apparatus. FEBS Letters 488, 29–33. [DOI] [PubMed] [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M. 2009. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. The Plant Cell 21, 2163–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Bickford LC, Goldberg J. 2003. SNARE selectivity of the COPII coat. Cell 114, 483–495. [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, et al. 2012. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nature Biotechnology 30, 360–364. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Obrdlik P, El-bakkoury M, Hamacher T, et al. 2004. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proceedings of the National Academy of Sciences, USA 101, 12242–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ramirez C, Mora SI, Trejo J, Pantoja O. 2011. PvAMT1;1, a highly selective ammonium transporter that functions as H+/NH4(+) symporter. Journal of Biological Chemistry 286, 31113–31122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte S, Barlowe C. 2004. Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nature Cell Biology 6, 1189–1194. [DOI] [PubMed] [Google Scholar]
- Plett D, Safwat G, Gilliham M, Skrumsager Møller I, Roy S, Shirley N, Jacobs A, Johnson A, Tester M. 2010. Improved salinity tolerance of rice through cell type-specific expression of AtHKT1;1. PLoS One 5, e12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Barlowe C. 1998. Transport of Axl2p depends on Erv14p, an ER–vesicle protein related to the Drosophila cornichon gene product. Journal of Cell Biology 142, 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Barlowe C. 2002. Erv14p directs a transmembrane secretory protein into COPII-coated transport vesicles. Molecular Biology of the Cell 13, 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z-H, Gao J-P, Li L-G, Cai X-L, Huang W, Chao D-Y, Zhu M-Z, Wang Z-Y, Luan S, Lin H-X. 2005. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genetics 37, 1141–1146. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler C, Nebenführ A, Movafeghi A, Stussi-Garaud C, Behnia L, Pimpl P, Staehelin LA, Robinson DG. 2002. Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. The Plant Cell 14, 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg FS, Barcelo G, Schüpbach T. 1995. Cornichon and the EGF receptor signaling process are necessary for both anterior-–posterior and dorsal–ventral pattern formation in Drosophila . Cell 81, 967–978. [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI. 1995. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270, 1660–1663. [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Ahmed SU, Marty-Mazars D, Rapoport I, Kirchhausen T, Marty F, Raikhel NV. 1998. A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots. Proceedings of the National Academy of Sciences, USA 95, 9920–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi A, Mieulet D, Khan I, Moreau B. 2012. The rice monovalent cation transporter OsHKT2;4: revisited ionic selectivity. Plant Physiology 160, 498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nakano A. 2003. Oligomerization of a cargo receptor directs protein sorting into COPII-coated transport vesicles. Molecular Biology of the Cell 14, 3055–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Delhaize E, Frommer WB, et al. 2013. Using membrane transporters to improve crops for sustainable food production. Nature 497, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, et al. 2009. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323, 1313–1319. [DOI] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki K-I, Maeshima M, Yokota A, Tomizawa K-I, Mimura T. 2004. Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana . Plant and Cell Physiology 45, 672–83. [DOI] [PubMed] [Google Scholar]
- Strompen G, Dettmer J, Stierhof Y-D, Schumacher K, Jurgens G, Mayer U, Jürgens G. 2005. Arabidopsis vacuolar H-ATPase subunit E isoform 1 is required for Golgi organization and vacuole function in embryogenesis. The Plant Journal 41, 125–32. [DOI] [PubMed] [Google Scholar]
- Sunarpi Horie T, Motoda J, et al. 2005. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. The Plant Journal 44, 928–938. [DOI] [PubMed] [Google Scholar]
- Tse Y, Mo B, Hillmer S, Zhao M. 2004. Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. The Plant Cell 16, 672–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI. 2000. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae . Plant Physiology 122, 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. 1992. Membrane hydrophobicity protein structure prediction analysis and the positive-inside rule. Journal of Molecular Biology 225, 487–494. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhuang X-H, Hillmer S, Robinson DG, Jiang L-W. 2011. Vacuolar sorting receptor (VSR) proteins reach the plasma membrane in germinating pollen tubes. Molecular Plant 4, 845–853. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang X, Dong X-P, et al. 2012. TPC proteins are phosphoinositide-activated sodium-selective ion channels in endosomes and lysosomes. Cell 151, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenstetter S, Wirsching P, Dotzauer D, Schneider S, Sauer N. 2012. Routes to the tonoplast: the sorting of tonoplast transporters in Arabidopsis mesophyll protoplasts. The Plant Cell 24, 215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhull AM. 1973. Ionic blockage of sodium channels in nerve. Journal of General Physiology 61, 687–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudick MM, Luu DD-T, Maurel C. 2009. A look inside: localization patterns and functions of intracellular plant aquaporins. New Phytologist 184, 289–302. [DOI] [PubMed] [Google Scholar]
- Xue S, Yao X, Luo W, Jha D, Tester M, Horie T, Schroeder JI. 2011. AtHKT1;1 mediates nernstian sodium channel transport properties in Arabidopsis root stelar cells. PLoS One 6, e24725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Horie T, Xue S, Leung H-Y, Katsuhara M, Brodsky DE, Wu Y, Schroeder JI. 2010. Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiology 152, 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM. 2002. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. The Plant Journal 30, 529–539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.