Highlight
A host-induced gene-silencing strategy for controlling potato late blight is presented, a plant disease that conventionally requires regular application of fungicides at high rates.
Key words: Late blight, oomycete, Phytophthora infestans, potato, RNA interference, small RNA.
Abstract
Phytophthora infestans is an oomycete that causes severe damage to potato, and is well known for its ability to evolve rapidly in order to overcome resistant potato varieties. An RNA silencing strategy was evaluated here to clarify if small interfering RNA homologous to selected genes in P. infestans could be targeted from the plant host to reduce the magnitude of the infection. As a proof-of-concept, a hairpin RNA (hp-RNA) construct using the GFP marker gene was designed and introduced in potato. At 72 hpi, a 55-fold reduction of the signal intensity of a corresponding GFP expressing P. infestans strain on leaf samples of transgenic plants, compared with wild-type potato, was detected. This suggests that an RNA interference construct in the potato host could be processed and target a transcript of the pathogen. Three genes important in the infection process of P. infestans, PiGPB1, PiCESA2, and PiPEC, together with PiGAPDH taking part in basic cell maintenance were subsequently tested using an analogous transgenic strategy. Out of these gene candidates, the hp-PiGPB1 targeting the G protein β-subunit (PiGPB1) important for pathogenicity resulted in most restricted disease progress. Further, Illumina sequencing of inoculated transgenic potato leaves revealed sRNAs of 24/25 nt size homologous to the PiGPB1 gene in the transgenic plants indicating post-transcriptional silencing of the target gene. The work demonstrates that a host-induced gene-silencing approach is functional against P. infestans but is highly dependent on target gene for a successful outcome. This finding broadens the arsenal of control strategies to this important plant disease.
Introduction
Phytophthora infestans is the oomycete pathogen responsible for the late blight disease on its potato host (Solanum tuberosum) inciting the worldwide most severe potato losses (Haverkort et al., 2008; Forbes, 2012). Enormous breeding efforts to produce new varieties with improved resistance have been ongoing for more than 100 years. Exploitation of resistance genes from wild Solanum species started with S. demissum (Reddick, 1928, 1934) and has continued ever since (Vleeshouwers et al., 2011). Besides pyramiding dominant resistance genes, the emphasis has been on introducing quantitative resistance traits to reduce the short ‘shelf life’ of qualitative resistance genes. However, finding durable resistance gene combinations without negative trait drag from donor species such as, for example, late maturity, remains a challenge. The success of P. infestans as a pathogen originates from its effective reproduction in both asexual and sexual forms. Under ideal conditions, the life cycle can be completed on foliage in about five days, where one lesion can generate up to hundreds of thousands of new sporangia (Fry, 2008). Furthermore, the genome of P. infestans is one of the largest among oomycetes (240Mb), containing vast numbers of transposable elements (TE) (Haas et al., 2009). There are also hundreds of predicted genes coding for disease-promoting effector proteins, predominantly located in the TE-rich genomic regions which together drive the diversification process, all leading to an exceptional high potential to adapt to new control strategies of the potato crop.
Effector proteins in P. infestans belong mainly to two groups that target distinct sites in the host plant. The apoplastic effectors are secreted into the plant extracellular space, whereas cytoplasmic effectors are translocated inside the plant cell where they target different subcellular compartments (Armstrong et al., 2005; Kamoun, 2006; Whisson et al., 2007). Both classes of effectors are modular proteins with cleavable amino-terminal secretion signals. Cytoplasmic effectors carry an additional domain after the signal peptide that mediates translocation inside host cells and is defined by conserved motifs, such as the RXLR amino acid sequence. The discovery of many potential effectors in the genomic sequence of P. infestans has enabled high-throughput analysis for additional resistance gene candidates in a variety of species in the Solanaceae (Vleeshouwers et al., 2008; Oh et al., 2009).
Gene silencing or RNA interference (RNAi) is a master regulatory mechanism with diverse roles such as the control of gene expression at transcriptional and post-transcriptional levels and chromatin organization of eukaryotic organisms. Central players in RNA silencing are small RNAs (sRNAs), often ranging in size from 19–40 nucleotides, and divided into different classes with diverse roles (Ghildiyal and Zamore, 2009; Malone and Hannon, 2009; Ruiz-Ferrer and Voinnet, 2009). Generally, sRNAs are generated from long double stranded RNA precursors which are digested by the type III RNase called Dicer or Dicer-like (Dcl) into short duplexes known as siRNA (short interfering RNA). The duplex is unwound and the antisense strand incorporated to an Argonaute (Ago) protein, which then binds to homologous mRNA and degrades it or inhibits its translation. This intramolecular hybridization of self-complementary RNA based on hairpin or stem-loop RNA is a key step in RNA silencing. Another important protein associated with RNAi is RNA-dependent RNA polymerase which is responsible for the conversion of RNA into double-stranded RNA and for the amplification of the silencing signal through the generation of secondary siRNA.
P. infestans possesses the components of canonical gene silencing pathways similar to those of other eukaryotes but the involved proteins display unusual protein domain organization (Vetukuri et al., 2011a). First, only one Dicer-like protein, having the expected dual RNase III domains but lacking other typical domains for Dicers, was identified. A second Dicer-like protein (PiDcl2) was later found in the genome trace archive (Fahlgren et al., 2013). Small RNAs of approximately 21 nt have been associated with partial silencing in P. infestans (Ah-Fong et al., 2008) and 40 nt sRNAs were associated with TE silencing (Vetukuri et al., 2011b). Based on deep sequencing of sRNAs from P. infestans, distinct classes of sRNAs at 21, 25/26, and 32 nt were found where the biogenesis of 21 nt sRNAs was shown to be PiDcl1-dependent while longer sRNAs were PiAgo-dependent (Vetukuri et al., 2012). Furthermore, P. infestans lacks DNA methylation associated proteins known to be ubiquitous in plants.
Plants utilize post-transcriptional gene silencing to protect themselves against invasive nucleic acids such as transposons, viruses, and transgenes (Cogoni and Macino, 2000). Knowledge on the basic mechanisms of gene silencing provides new opportunities to explore plant–pathogen interactions and potential strategies for novel disease control. In this area there are promising reports where RNAi-based constructs in plants were designed to target fungal plant pathogens (Nowara et al., 2010; Koch et al., 2013; Ghag et al., 2014). In order to expand the toolbox for potato breeders not just to rely on dominant resistance genes from various Solanum species, it would be interesting to know if a similar gene-silencing approach driven by the plant host could be a functional strategy to target this oomycete plant pathogen.
The main objective of this study was to investigate the possibility of exploiting RNA silencing to target selected genes in P. infestans via the host plant and, thereby, reduce its capacity to initiate and develop disease on potato. The work showed that sRNAs targeting a pathogen gene, generated in the host plant during infection, incites host-induced gene silencing of the corresponding pathogen transcript. However, the choice of target genes and precursor hp-RNA is crucial for a successful outcome of this strategy.
Materials and methods
P. infestans strains and plant inoculation
The eGFP strain under the promoter of Ham34 (Avrova et al., 2008) and the wild-type 88069 strain were cultured as described earlier by Vetukuri et al. (2011a). Each P. infestans strain at a concentration of 5×104 spores ml–1 water was used for leaf inoculations (Grenville-Briggs et al., 2005) of potato plants at 4 weeks post transfer to soil from in vitro conditions.
Potato transformation and cultivation
For transformation, in vitro potato plants of cv. Desiree were grown at 22 °C with a 16h photoperiod on Murashige–Skoog (MS) medium (Duchefa Biochemie B.V., Amsterdam, Netherlands), supplemented with 2% (w/v) sucrose and 0.3% (w/v) gelrite (Duchefa). The internodes of the plants were used as explants for transformation (Millam, 2007) using the Agrobacterium tumefaciens strain C58. Ten potential transgenic shoots per construct were grown on MS media with 50 μg ml–1 kanamycin. Plantlets were transferred to soil and grown under the same light and temperature conditions in a culture chamber as for the plants grown in vitro. Validation of transgenic plants was done by PCR (the primers are listed in Supplementary Table S1 at JXB online) and Sanger sequencing.
hp-RNA constructs for potato transformation
All the gene-silencing constructs were made with the ubiquitin1 (UBQ1) promoter for constitutive expression and the heat shock protein 18.2 (HSP) terminator from Arabidopsis thaliana. To enable a hairpin (hp) formation of selected sequences, a 71bp intron from the Ste20-like PiC20 gene of P. infestans (Tani et al., 2004) was used. cDNA of 88069 and the Ham34:eGFP strains of P. infestans were used as templates in the gene PCR amplification step. Besides the green fluorescence protein (GFP) marker gene, sequences of the following genes were used: G-protein β-subunit (PiGPB1) (Latijnhouwers and Govers, 2003), cellulose synthase A2 (PiCESA2) (Grenville-Briggs et al., 2008), pectinesterase (PiPEC) (Ospina-Giraldo et al., 2010a), and the constitutive glyceraldehyde 3-phosphate dehydrogenase (PiGAPDH) gene. The choice of target genes relied on published data on PiCESA1 and PiGAPDH (Zeng et al., 2006; Grenville-Briggs et al., 2008) together with our analysis of PiGPB1 and PiPEC in infected potato plants (see Supplementary Fig. S1 at JXB online). The specificity of each target gene was determined by BLAST and, using sequences from highly conserved nucleotides, from CLUSTAL-W alignments within each gene family. The BP and LR reactions were performed by using MultiSite Gateway® Pro Technology (Invitrogen, Carlsbad, CA, USA), and recombined into the binary vector pGWB1 (Nakagawa et al., 2007). All primers used for plasmid construction are listed in Supplementary Table S2 at JXB online, and detailed cloning steps are outlined in Supplementary Fig. S2 at JXB online. All constructs were confirmed by sequencing (Macrogen Inc. Seol, Korea).
Quantitative real-time PCR (qRT-PCR)
Leaf samples from wild-type, cv. Desiree, and transgenic plants were collected at 24, 48, and 72h post-inoculation (hpi), snap-frozen in liquid nitrogen, and stored at –70 °C. The leaf materials were used for RNA extraction, cDNA synthesis, and SYBR green qRT-PCR assays (Vetukuri et al., 2011a). To check the transcript level for each gene of interest, specific primers were designed to anneal outside the hairpin sequence. Primers are listed in Supplementary Table S3 at JXB online. Calculations and statistical analyses were carried out according to Avrova et al. (2003). Primers for hairpin expression analysis are listed in Supplementary Table S4 at JXB online. DNA quantification of P. infestans and calculation was as described earlier by Llorente et al. (2010). At least three biological replicates were used for each individual qRT-PCR analysis.
Phenotypic analysis of transgenic potato plants
Colonization and disease progression were monitored up to 30 d after inoculations. The effect of PiGPB1 silencing on spore formation was assayed by analysing the shape and number of sporangia using an epifluorescence microscope (Leica DMI 4000). Sporangia were collected at 24, 48, 72, and 96 hpi from detached leaves and counted 1h after leaf detachment as described by Lehtinen et al. (2009).
Confocal microscopy
The GFP expression levels on leaves of the transgenic and wild-type plants inoculated by the GFP-expressing Ham34:eGFP P. infestans strain were analysed by confocal microscopy (Zeiss LSM510). GFP was excited at 488nm,and detected at 505–530nm. Images were analysed using the LSM 510 software.
Northern blot hybridization
To detect sRNA molecules homologous to the PiGPB1 gene, Northern blot hybridization was performed (Vetukuri et al., 2011b). Primers used for the generation of PiGPB1-specific riboprobes were: Spe-Fw ATATACTAGTAGT TCTCAGCCAATCTTCG and Not-Rev ATATGCGGCCGCTTCAACTTGGT CTAGTTTCCAT.
Western blotting
Total GFP proteins from the GFP-tagged P. infestans strain colonized on hp-GFP transgenic plants, were resolved on SDS-containing 9% polyacrylamide gel. The proteins were transferred to nitrocellulose membrane and incubated with an anti-GFP monoclonal antibody, following the procedure described previously by Moschou et al. (2008). The membranes were subsequently incubated with a chemi-luminescent substrate (ECL Western blotting system kit, GE Healthcare, Uppsala) and exposed using a Fuji Phosphorimager. Ponceau staining of the membrane (Bannur et al., 1999) was used as a loading control.
Small RNA sequencing
Total RNA was extracted from hp-PiGPB1 transgenic and wild-type potato leaves at 24, 48, and 72 hpi infected by the 88069 strain, the 88069 mycelia control, and non-infected transgenic plants using the mirVanaTM miRNA isolation kit (Ambion, Austin, TX, USA). Eight sRNA libraries were generated using the Illumina small RNA sample preparation kit and sequenced using Illumina HiSeq 2500 at SciLifeLab, Stockholm, Sweden.
Bioinformatic analysis
All sRNA reads were trimmed at any base with a phred-scaled quality score below 10 and adapter sequences were removed using Cutadapt v1.3 (Martin, 2011). Reads shorter than 18 bases in length were excluded from all further analysis. The resulting set of filtered reads for each sample was aligned to the P. infestans reference genome (http://www.broadinstitute.org/annotation/genome/phytophthora_infestans/MultiDownloads.html) using Bowtie2 v2.1.0 (Langmead and Salzberg, 2012) with one mismatch allowed per seed (-N 1), a seed length of 18 (-L 18), and the remainder of alignment options set to their default values. The SAM format alignments produced by Bowtie2 were converted to BAM files, sorted, and indexed using SAMtools v0.1.19 (Li et al., 2009). The number, locations, lengths, and sense/antisense orientations of reads mapping to the PiGPB1 gene in P. infestans were obtained using a combination of the SAMtools view command and custom python scripts.
Results
Generation and validation of transgenic potato harbouring hp-RNA constructs
A series of gene-cloning steps were performed to accomplish binary vectors containing the target gene sequence in a sense and antisense orientation together with an intron (I) to enable a short hp-RNA formation (see Supplementary Fig. S2 at JXB online). This construction work resulted in five binary vectors: UBQ1:GFP-I-GFP; UBQ1:PiGPB1-I-PiGPB1; UBQ1:PiCesA2-I-PiCesA2; UBQ1:PiPEC-I-PiPEC; and UBQ1:PiGAPDH-I-PiGAPDH, that were used for A. tumefaciens-mediated transformation. Ten transgenic potato plants from each construct were made, and three individual lines L1, L2, and L3 were used in subsequent analysis. The transgenic status of the plant materials was validated by PCR amplification of the sequences matching individual constructs in DNA samples from the different transgenic plants (see Supplementary Fig. S3A at JXB online) and by performing Sanger sequencing to check that each construct was intact in the plants, followed by analysing the transcription levels of each gene by qRT-PCR (see Supplementary Fig. S3B at JXB online). Further, in order to verify that the silencing of the target gene was not an effect of the transformation procedure it self, the PiGPB1 transcript level was analysed in hp-GFP plants and compared with hp-PiGPB1 and wild-type potato plants infected with P. infestans. The PiGPB1 transcript level in the hp-GFP plants compared with wild-type plants remained intact at 24, 48, and 72 hpi while the transcript level was gradually decreasing with time in hp-PiGPB1 plants as expected (see Supplementary Fig. S3C at JXB online).
A GFP-expressing P. infestans strain can be targeted from the host plant
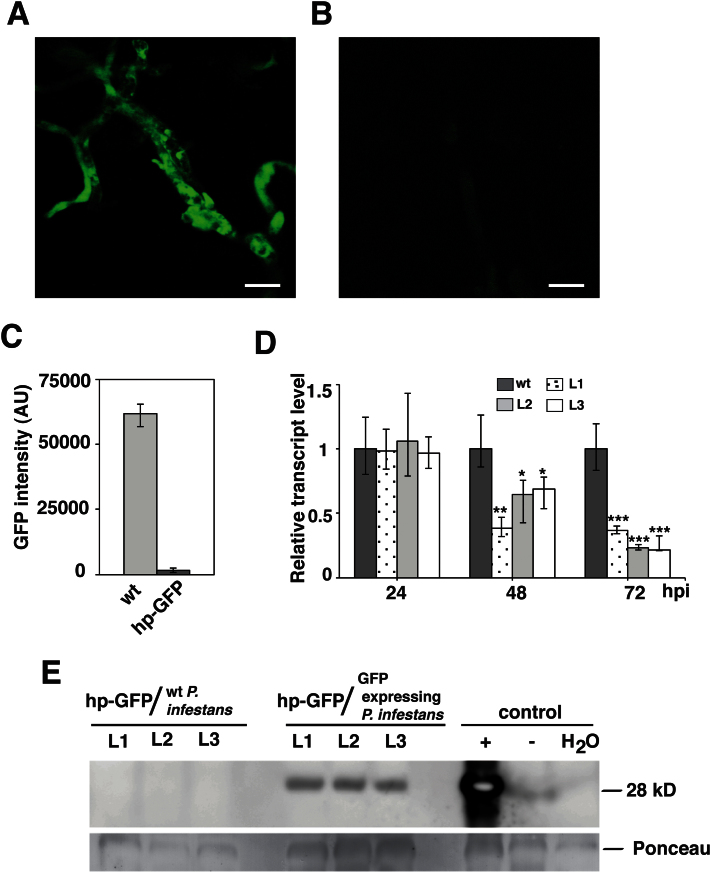
Three transgenic potato lines (hp-GFPL1, hp-GFPL2, and hp-GFPL3) harbouring the hp-RNA construct matching a GFP-tagged P. infestans strain were analysed first. The intensity of the GFP signal in the P. infestans-colonizing transgenic potato leaves was monitored and quantified using confocal microscopy. There was a marked reduction in the GFP signal at 72 hpi in all three transgenic lines compared with wild-type plants (Fig. 1A, B; see Supplementary Figs S4 and S5 at JXB online). At 72 hpi, as much as a 55-fold reduction of the signal intensity of the GFP-expressing P. infestans strain was recorded on leaf samples of transgenic plants compared with wild-type potato (Fig. 1C). In order to monitor the GFP-silencing in P. infestans further, transcript levels were checked. A significant reduction in relative transcript levels of the GFP gene in P. infestans growing on transgenic plants at 48 and 72 hpi was detected compared with wild-type potato (Fig. 1D). To check if the protein level in the hp-GFP potato lines was altered, a Western blot was run (Fig. 1E). A reduction of the GFP protein was seen in the potato transgenic lines compared with wild-type plants upon infection with the GFP-tagged P. infestans isolate. Together, these data suggest that hp-RNA in the potato host could be processed and target a transcript of the invading P. infestans pathogen.
Fig. 1.
Silencing of GFP in P. infestans by hp-RNA. Confocal laser scanning microscopy of P. infestans transformants (Ham34:eGFP) expressing green fluorescent protein. GFP expression in mycelia grown on (A) wild-type and (B) hp-GFPL1 (UBQ:GFP-I-GFP) transgenic potato. Bars=25 μm. (C) Fold change of total intensity of GFP in mycelia grown on hp-GFP plants compared with wild-type plants. AU=arbitrary unit. (D) Transcript abundance of GFP in P. infestans transformants grown on wild-type and transgenic plants at 24, 48, and 72 hpi, quantified by qRT-PCR. Data are normalized to P. infestans actinA mRNA levels and represent means ±SE (n=3 pooled leaves of 3 plants). Asterisks indicate significant difference to the wild type (Student’s t test; *P <0.05; **P <0.01; ***P <0.001). (E) Western blot analysis of GFP protein. hp-GFP plant lines inoculated with: wild type (wt) P. infestans (88069), the GFP-tagged P. infestans, wild-type plants inoculated with: the GFP-tagged P. infestans (+), 88069 (–), and water. Ponceau staining was used as loading control.
Monitoring RNA-mediated inhibition of disease progression
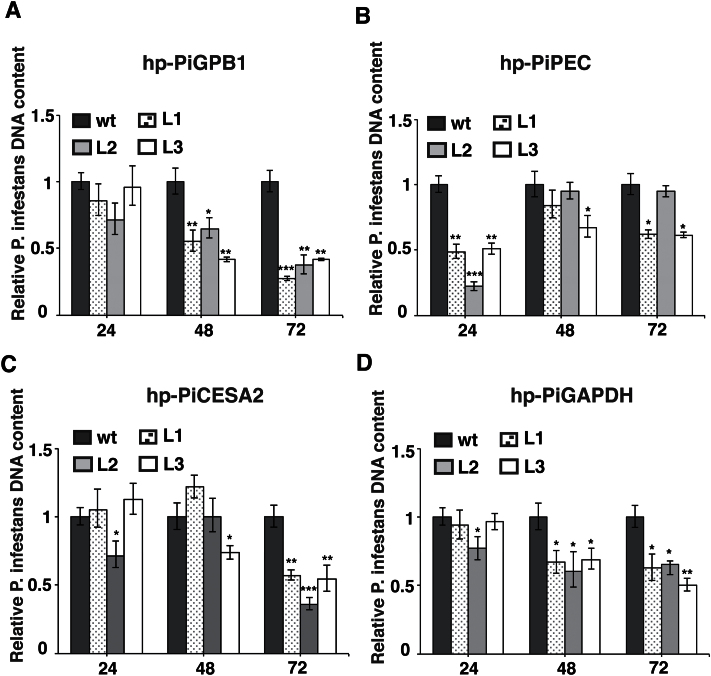
Encouraged by the data on GFP silencing in P. infestans, the next step was to monitor the effects of host-mediated silencing of the PiGPB1, PiCESA2, PiPEC, and PiGAPDH genes. A reduction of P. infestans DNA content over time on these transgenic plant leaves compared with the wild type was generally found but that trend was most obvious in the hp-PiGPB1 lines (Fig. 2A). In the case of the hp-PiPEC plants, a rather rapid decrease of P. infestans DNA was already detected at 24 hpi (Fig. 2B). By contrast, the DNA levels were lowest at 72 hpi compared with earlier time-points in the hp-PiCESA2 plants but not reaching the level seen in the hp-PiGPB1 lines (Fig. 2C). In the leaf samples from the hp-PiGAPDH transgenic lines, the growth of P. infestans seemed to be less at 48 and 72 hpi compared with 24 hpi (Fig. 2D). For all these transgenic plants, colonization by P. infestans occurred as necrotic lesions on the leaves to a limited extent compared with the wild-type plants (see Supplementary Fig. S6 at JXB online).
Fig. 2.
P. infestans DNA in leaves of the wild type and three individual transgenic lines L1, L2, and L3 of (A) hp-PiGPB1, (B) hp-PiPEC, (C) hp-PiCESA2, and (D) hp-PiGAPDH potato plants quantified by qRT-PCR at 24, 48, and 72 hpi. Data are normalized to potato EF1 DNA levels and represent means ±SE (n=3 pooled leaves of 3 plants). Asterisks indicate significant difference to the wild type. (Student’s t test; *P <0.05; **P <0.01; ***P <0.001).
Reduced transcript level in hp-PiGPB1 and hp-PiGAPDH plants
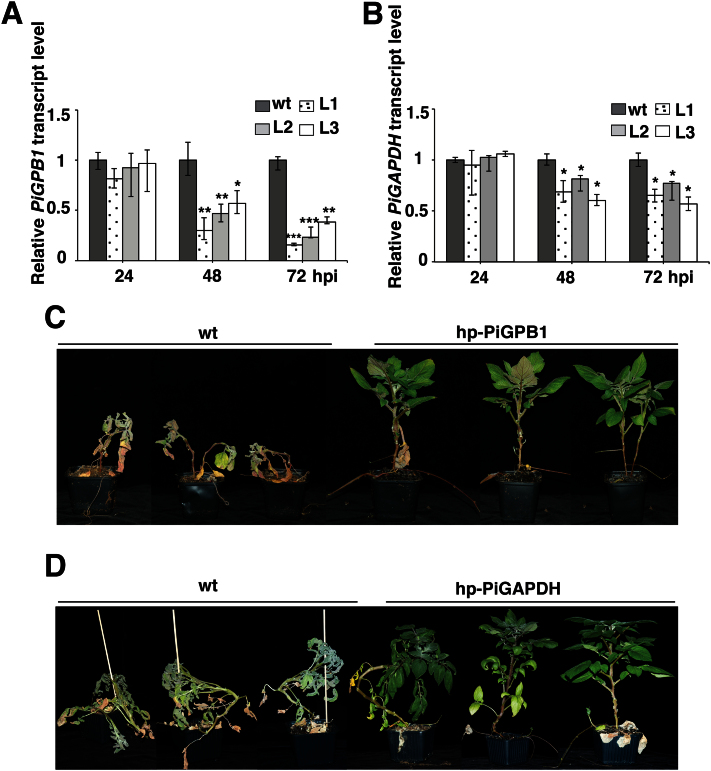
Further studies were done on the hp-PiGPB1 plants where a gene important for sporangia formation was targeted with promising effects on the pathogen. The basic cell maintenance gene in the hp-PiGAPDH plants was included for comparison. In order to monitor the silencing of PiGPB1 and PiGAPDH in P. infestans, transcript levels of these genes were checked in the transgenic material. The relative transcript levels of PiGPB1 at 48 and 72 hpi were significantly down-regulated in all three transgenic lines compared with inoculated wild-type plants (Fig. 3A). In the case of the hp-PiGAPDH plants, a reduced transcript level of PiGAPDH was particularly seen at 48 hpi and 72 hpi (Fig. 3B). These results were in line with a reduced P. infestans DNA content detected in infected leaves. To record overall plant performance, the plants were allowed to grow for a month. After 30 d, the inoculated wild-type plants were almost dead, whereas transgenic hp-PiGPB1 plants showed greatly reduced disease spread disease symptoms (Fig. 3C). A similar performance was seen for the hp-PiGAPDH transgenic plants (Fig. 3D), although the effect was not as prominent compared with the hp-PiGPB1 plants.
Fig. 3.
Analysis of transgenic potato harbouring a hp-PiGPB1 or hp-PiGAPDH construct. (A) Transcript abundance of the PiGPB1 in wild-type and three lines of hp-PiGPB1 plants. (B) Transcript abundance of the PiGAPDH gene in wild-type and hp-PiGAPDH plants inoculated with P. infestans at 24, 48, and 72 hpi quantified by qRT-PCR. Data are normalized to P. infestans actinA mRNA levels and represent means ±SE (n=3 pooled leaves of 3 plants). Asterisks indicate significant difference to the wild type (Student’s t test; *P <0.05: **P <0.01; ***P <0.001). Overall performance of (C) wild-type and hp-PiGPB1 transgenic plants, and (D) wild-type and hp-PiGAPDH transgenic plants at 30 dpi.
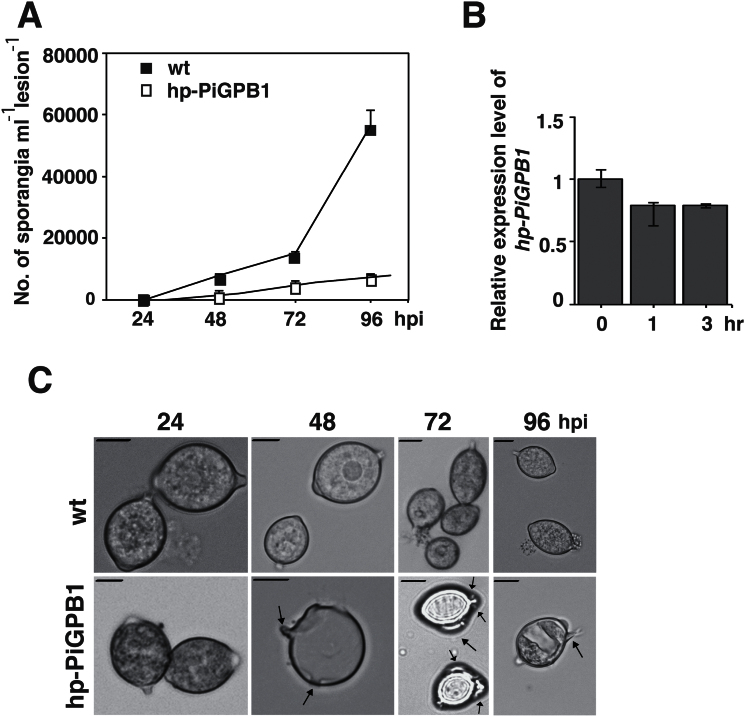
Host induced silencing of PiGPB1 gene affects spore formation
PiGPB1 is a gene known to be involved in phosphatase-mediated signalling in mycelia leading to the formation of sporangia (Kasahara et al., 1997; Latijnhouwers and Govers, 2003). Silencing of this gene causes P. infestans to produce abundant aerial mycelia and very few defective sporangia. In the present study, no sporangia were found on leaves of the hp-PiGPB1 plants. In order to assay any phenotypic changes whatsoever, a detached leaf assay was used where inoculated leaves were taken at 24, 48, 72, and 96 hpi from the hp-PiGPB1 plants. Leaving the leaves detached for 1h in a plastic box under humid conditions promoted sporangia formation. A condition that by time favoured the pathogen over the plant host with declining hp-PiGPB1 expression (Fig. 4B). Yet, a 6-fold reduction in the number of sporangia on colonized transgenic leaves was observed compared with the wild-type at 96 hpi (Fig. 4A). Sporangia produced by P. infestans at 24 hpi, grown on both control and hp-PiGPB1 leaves, were normal with the expected ‘lemon shapes’ and single hyphae. By contrast, sporangia formed on hp-PiGPB1 plants at 48, 72, and 96 hpi were abnormal with an outgrowth of multiple hyphae and the loss of the characteristic phenotype at 48 hpi (Fig. 4C). This reduction in number of sporangia and sporangia with multiple hyphal outgrowths are in agreement with the phenotype of the PiGPB1-silenced mutants of P. infestans observed earlier by Latijnhouwers and Govers (2003).
Fig. 4.
Effects on spore formation in transgenic hp-PiGPB1 plants. (A) Numbers of sporangia produced by the 88069 strain of P. infestans on wild-type and transgenic plants at 24, 48, 72, and 96 hpi. Data represent means ±SE (n=3 pooled samples of 3 plants). (B) Relative expression of hp-PiGPB1 construct in hp-PiGPB1 plants inoculated with P. infestans at 0, 1, and 3h after leaf detachment. Data are normalized to P. infestans actinA mRNA levels and represent means ±SE (n=3 pooled leaves of 3 plants) (C) Phenotypes of sporangia collected at 24, 48, 72, and 96 hpi from wild-type and hp-PiGPB1 transgenic plants. Bars =10 μm.
Detection of sRNAs homologous to PiGPB1 gene in P. infestans
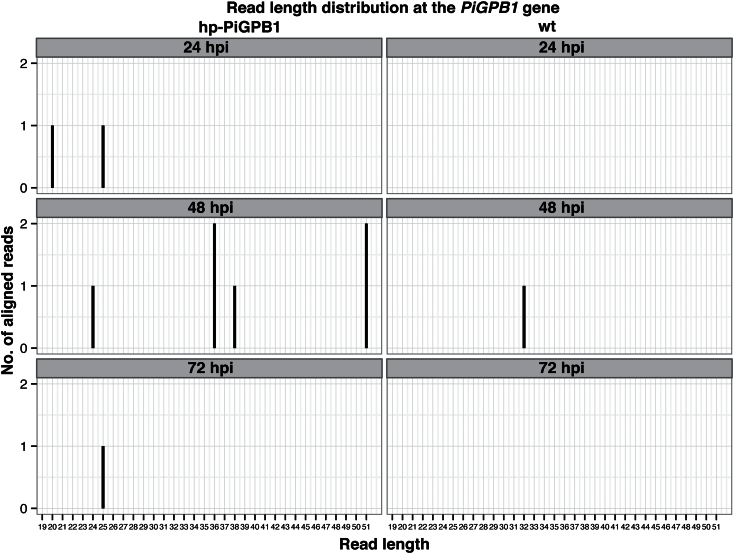
In order to confirm the presence of sRNAs homologous to hp-RNA constructs in transgenic plants, Northern blot hybridizations were used. Probes specific to PiGPB1 detected antisense sRNAs, ranging from 21, 25, 26, 27, and 28 nt in size (see Supplementary Fig. S7 at JXB online). Subsequently, Illumina sequencing of sRNAs generated from wild-type and hp-PiGPB1 transgenic plants resulted in unique but few 24/25 nt size class sRNAs only mapping to the PiGPB1 gene in the transgenic plants and not seen in control plants (Fig. 5). These data suggest that siRNAs are generated from the hp-RNA constructs in transgenic plants during the infection process.
Fig. 5.
Small RNAs complementary to the PiGPB1 gene. Distribution of small RNA reads homologous to the PiGPBI gene in transgenic hp-PiGPB1 and wild-type potato plants at 24, 48, and 72 hpi based on Illumina sequencing.
Discussion
This work demonstrated the possibility that hp-RNA constructs in potato could affect the host colonization and invasion by P. infestans to various degrees. Three genes of importance in the first stages of infection, together with a gene involved in basic cell maintenance, were examined. The current understanding of key genes and signals in early plant infection phases is meagre. It could be anticipated that various receptor classes are main players in these interactions, not least G protein-coupled receptors common in eukaryotes. Heterotrimeric G proteins are known to be key regulators in numerous signalling pathways impacting arrays of downstream targets. However distinct differences are known between animals and plants (Urano and Jones, 2014). P. infestans has only a single α (PiGPA1) and one β (PiGPB1) subunit gene. PiGPB1 is differentially expressed in various stages of the life cycle of P. infestans (Laxalt et al., 2002; Latijnhouwers and Govers, 2003; Dong et al., 2004). Primarily, PiGPB1 is important for sporangia formation and is not strongly expressed in mycelia. When the infection progresses from a biotrophic to a necrotrophic mode at 36–48 hpi, the continuous formation of sporangia is required (Judelson and Blanco, 2005), a process which is impaired in the hp-PiGPB1 transgenic plants. Fewer sporangia, and at the same time abnormal phenotypes, impose difficulties to infect new host cells leading to reduced colonization and spread of the pathogen.
Oomycetes have cell walls composed of cellulose, not chitin as fungi do, but both pathogenic organism groups have evolved pectinases to break down plant cell walls, enzymes aiding in the penetration and subsequent establishment of the infection. Phytophthora species encode large sets of different cell wall-degrading enzymes (Ospina-Giraldo et al., 2010b). Thus, influencing cell wall-associated genes, such as hp-PiCESA2 and hp-PiPEC, was also of great interest. In the hp-PiCESA2 plants, P. infestans is supposed to have impaired the formation of appressoria (Grenville-Briggs et al., 2008), leading to attenuated colonization but such an effect was not consistent over time. On the hp-PiPEC plants, P. infestans encountered difficulty in establishing the infection process but the few spores that managed to penetrate the mesophyll cells continued to colonize the leaves. Some of this ‘incomplete’ inhibition could be attributed to redundancy since CESA, PEC, and GAPDH belong to gene families of varying size in P. infestans. The challenge here was to design hp-gene constructs that specifically targeted P. infestans and not the host plant.
In the Arabidopsis–Phytophthora parasitica interaction, host-driven gene-silencing targeting GFP and the PnPMA gene encoding for a plasma membrane H+ ATPase were not reported to be successful and was explained by the lack of proper machinery required for the uptake of silencing signals in the oomycete pathogen (Zhang et al., 2011). This is contrary to what is already a well-established methodology in oomycete research; experiments where the treatment of protoplasts with hp-RNA results in transient RNA silencing, which shows beyond doubt that oomycetes do have the machinery needed for the uptake of silencing molecules (Whisson et al., 2005; Vetukuri et al., 2011a). Nevertheless, the mechanism of RNA uptake from the plant host during the infection process into P. infestans cells has not been well studied. It was shown recently that the fungus Botrytis cinerea delivers sRNAs to hijack the host RNAi machinery and selectively silences host immunity genes during the infection process of Arabidopsis (Weiberg et al., 2013). Notably, out of the 73 B. cinerea sRNAs with the potential to silence plant genes, 52 were derived from transposable elements (TE) of the fungal genome. If sRNAs are included in the vast list of P. infestans effectors, this implies thousands of new candidates because of its TE-saturated genome constitution, and adds to the enormous capacity to overcome host resistance. The Illumina sequencing data revealed sRNAs of 24/25 nt size homologous to the PiGPB1 gene in the transgenic plants, indicating post-transcriptional silencing of the target gene. The number of sRNA reads detected were considerably less than expected which could be attributed to unknown timing for the required cellular processes, which thereby leads to non-optimal timing of sample collection. However, based on hairpin RNA expression studies in plants, it can be presumed that the sRNAs are initially formed on the plant side (Mansoor et al., 2006).
The process of effector protein translocation between potato and P. infestans has been explored for many years and, hitherto, led to inconclusive results (Wawra et al., 2013). Similarly, the sRNA translocation process is poorly understood. No homologous proteins to SID1 and SID2, earlier identified in C. elegans to be responsible for the intercellular communication of the RNA-induced silencing signal, and also environmental RNA interference (Winston et al., 2002, 2007) are neither found in P. infestans nor in potato or Arabidopsis genomes. In the case of the barley–Blumeria graminis interaction, exosomes have been speculated to be involved in the translocation of the silencing signal (Nowara et al., 2010) but this critical question remains to be answered.
Future potential
The hp-RNA-mediated gene silencing of P. infestans clearly contributes to reduce disease progress in potato and should be regarded as an additional strategy to complement resistance gene deployment. Broader resistance would most likely be achieved by combining several RNAi transgenes, preferably targeting single copy genes of importance in the infection process. Understanding potential sRNA transport from P. infestans to the host plant opens up alternative revenues to control this important plant pathogen. During the processing of this paper one work on the interaction between B. lcatucae and lettuce was published (Govindarajulu et al., 2014).
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Data on in planta expression of PiGPB1 and PiPEC gene.
Supplementary Fig. S2. Schematic representation of plasmid constructs.
Supplementary Fig. S3. Data on hp-RNA constructs.
Supplementary Fig. S4. Silencing of GFP in P. infestans by hp-RNA.
Supplementary Fig. S5. Mycelia growth in hp-GFP transgenic potato lines.
Supplementary Fig. S6. Overall picture of necrotic lesions on leaves.
Supplementary Fig. S7. Northern blot analysis.
Supplementary Table S1. PCR primers for transgene detection.
Supplementary Table S2. Primers for plasmid constructs.
Supplementary Table S3. Primers for qRT-PCR.
Supplementary Table S4. Primers for hp-RNA expression analyses.
Acknowledgements
The authors would like to acknowledge support from the Science for Life Laboratory, the National Genomics Infrastructure, and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure, and to Dr S Whisson (James Hutton Institute, Dundee, UK) for the eGFP P. infestans strain. The work was supported by grants from the Swedish Research Councils Formas and VR, the Farmers Foundation for Agricultural Research (SLF) and Board of Agriculture (SJV).
References
- Ah Fong AMV, Bromann-Chung CA, Judelson HS. 2008. Optimization of transgene mediated silencing in Phytophthora infestans and its association with small-interfering RNAs. Fungal Genetics and Biology 45, 1197–1205. [DOI] [PubMed] [Google Scholar]
- Armstrong MR, Whisson SC, Pritchard L, et al. 2005. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proceedings of the National Academy of Sciences, USA 102, 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrova AO, Boevink PC, Young V, Grenville-Briggs LJ, van West P, Birch PRJ, Whisson SC. 2008. A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cellular Microbiology 10, 2271–2284. [DOI] [PubMed] [Google Scholar]
- Avrova AO, Venter E, Birch PR, Whisson SC. 2003. Profiling and quantifying differential gene transcription in Phytophthora infestans prior to and during the early stages of potato infection. Fungal Genetics and Biology 40, 4–14. [DOI] [PubMed] [Google Scholar]
- Bannur SV, Kulgod SV, Metkar SS, Mahajan SK, Sainis JK. 1999. Protein determination by Ponceau S using digital color image analysis of protein spots on nitrocellulose membranes. Analytical Biochemistry 267, 382–389. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. 2000. Post-transcriptional gene silencing across kingdoms. Current Opinion in Genetics & Development 10, 638–643. [DOI] [PubMed] [Google Scholar]
- Dong W, Latijnhouwers M, Jiang RH, Meijer HJ, Govers F. 2004. Downstream targets of the Phytophthora infestans Gα subunit PiGPA1 revealed by cDNA-AFLP. Molecular Plant Pathology 5, 483–494. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Bollmann SR, Kasschau KD, et al. 2013. Phytophthora have distinct endogenous small RNA populations that include short interfering and microRNAs. PLoS ONE 8, e77181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes GA. 2012. Using host resistance to manage potato late blight with particular reference to developing countries. Potato Research 55, 205–216. [Google Scholar]
- Fry W. 2008. Phytophthora infestans: the plant (and R gene) destroyer. Molecular Plant Pathology 9, 385–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghag SB, Shekhawat UKS, Ganapathi TR. 2014. Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnology Journal 12, 541–553. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nature Reviews Genetics 10, 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenville-Briggs LJ, Anderson VL, Fugelstad J, et al. 2008. Cellulose synthesis in Phytophthora infestans is required for normal appressorium formation and successful infection of potato. The Plant Cell 20, 720–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenville-Briggs LJ, Avrova AO, Bruce CR, Williams A, Whisson SC, Birch PR, van West P. 2005. Elevated amino acid biosynthesis in Phytophthora infestans during appressorium formation and potato infection. Fungal Genetics and Biology 42, 244–256. [DOI] [PubMed] [Google Scholar]
- Govindarajulu M, Epstein L, Wroblewski T, Michelmore RW. 2014. Host-induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnology Journal, doi:10.1111/pbi.12307. [DOI] [PubMed]
- Haas BJ, Kamoun S, Zody MC, et al. 2009. Genome sequencing and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Haverkort AJ, Boonekamp PM, Hutten R, Jacobsen E, Lotz LAP, Kessel GJT, Visser RGF, van der Vossen EAG. 2008. Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Research 51, 47–57. [Google Scholar]
- Judelson HS, Blanco FA. 2005. The spores of Phytophthora: weapons of the plant destroyer. Nature Reviews Microbiology 3, 47–58. [DOI] [PubMed] [Google Scholar]
- Kamoun S. 2006. A catalogue of the effector secretome of plant pathogenic oomycetes. Annual Review of Phytopathology 44, 41–60. [DOI] [PubMed] [Google Scholar]
- Kasahara S, Nuss DL. 1997. Targeted disruption of a fungal G-protein β subunit gene results in increased vegetative growth but reduced virulence. Molecular Plant–Microbe Interaction 10, 984–993. [DOI] [PubMed] [Google Scholar]
- Koch A, Kumar N, Weber L, Keller H, Imani J, Kogel KH. 2013. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. Proceedings of the National Academy of Sciences, USA 110, 19324–19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers M, Govers F. 2003. A Phytophthora infestans G-Protein β subunit is involved in sporangium formation. Eukaryotic Cell 2, 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxalt AM, Latijnhouwers M, van Hulten M, Govers F. 2002. Differential expression of G protein α and β subunit genes during development of Phytophthora infestans . Fungal Genetics and Biology 36, 137–146. [DOI] [PubMed] [Google Scholar]
- Lehtinen A, Andersson B, Le VH, et al. 2009. Aggressiveness of Phytophthora infestans on detached potato leaflets in four Nordic countries. Plant Pathology 58, 690–702. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Bravo-Almonacid F, Cvitanich C, Orlowska E, Torres HN, Flawia MM, Alonso GD. 2010. A quantitative real-time PCR method for in planta monitoring of Phytophthora infestans growth. Letters in Applied. Microbiology 51, 603–610. [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. 2009. Small RNAs as guardians of the genome. Cell 136, 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- Millam S. 2007. Potato (Solanum tuberosum L.). Methods in Molecular Biology 344, 25–35. [DOI] [PubMed] [Google Scholar]
- Mansoor S, Amin I, Hussain M, Zafar Y, Briddon RW. 2006. Engineering novel traits in plants through RNA interference. Trends in Plant Science 11, 559–565. [DOI] [PubMed] [Google Scholar]
- Moschou PN, Paschalidis KA, Delis ID, Andriopoulou AH, Lagiotis GD, Yakoumakis DI, Roubelakis-Angelakis KA. 2008. Spermidine exodus and oxidation in the apoplast induced by abiotic stress is responsible for H2O2 signatures that direct tolerance responses in tobacco. The Plant Cell 20, 1708–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, et al. 2007. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Bioscience, Biotechnology and Agrochemistry 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nowara D, Gay A, Lacommen C, Shaw J, Ridout C, Douchkow D, Hensel G, Kumlehn J, Schweizer P. 2010. HIGS: host-induces gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis . The Plant Cell 22, 3130–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SK, Young C, Lee M, et al. 2009. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. The Plant Cell 21, 2928–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina-Giraldo MD, Griffith JG, Laird EW, Mingora C. 2010. b . The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora . BMC Genomics 11, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina-Giraldo MD, McWalters J, Seyer L. 2010. a . Structural and functional profile of the carbohydrate esterase gene complement in Phytophthora infestans . Current Genetics 56, 495–506. [DOI] [PubMed] [Google Scholar]
- Reddick D. 1928. Blight resistant potatoes. Phytopathology 18, 483–502. [Google Scholar]
- Reddick D. 1934. Elimination of potato late blight from North America. Journal of Phytopathology 24, 555–557. [Google Scholar]
- Ruiz-Ferrer V, Voinnet O. 2009. Roles of plant small RNAs in biotic stress responses. Annual Review of Plant Biology 60, 485–510. [DOI] [PubMed] [Google Scholar]
- Tani S, Yatzkan E, Judelson HS. 2004. Multiple pathways regulate the induction of genes during zoosporogenesis in Phytophthora infestans . Molecular Plant–Microbe Interactions 17, 330–337. [DOI] [PubMed] [Google Scholar]
- Urano D, Jones AM. 2014. Heterotrimeric G protein-coupled signaling in plants. Annual Review of Plant Biology 65, 365–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetukuri RR, Åsman AK, Tellgren-Roth C, et al. 2012. Evidence for small RNAs homologous to effector-encoding genes and transposable elements in the oomycete Phytophthora infestans . PLoS ONE 7, e51399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetukuri RR, Avrova AO, Grenville-Briggs LJ, Van West P, Söderbom F, Savenkov EI, Whisson SC, Dixelius C. 2011. a . Evidence for involvement of dicer-like, argonaute, and histone deacetylase proteins in gene silencing in Phytophthora infestans . Molecular Plant Pathology 12, 772–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetukuri RR, Tian Z, Avrova AO, Savenkov EI, Dixelius C, Whisson SC. 2011. b . Silencing of the PiAvr3a effector-encoding gene from Phytophthora infestans by transcriptional fusion to a short interspersed element. Fungal Biology 115, 1225–1233. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VG, Rietman H, Krenek P, et al. 2008. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE 3, e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers VGAA, Raffaele S, Vossen J, et al. 2011. Understanding and exploiting late blight resistance in the age of effectors. Annual Review of Phytopathology 49, 507–531. [DOI] [PubMed] [Google Scholar]
- Wawra S, Djamei A, Albert I, Nurnberger T, Kahmann R, van West P. 2013. In vitro translocation experiments with RxLR-reporter fusion proteins of Avr1b from Phytophthora sojae and AVR3a from Phytophthora infestans fail to demonstrate specific autonomous uptake in plant and animal cells. Molecular Plant–Microbe Interactions 26, 528–536. [DOI] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H. 2013. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson SC, Avrova AO, van West P, Jones JT. 2005. A method for double-stranded RNA-mediated transient gene silencing in Phytophthora infestans . Molecular Plant Pathology 6, 153–163. [DOI] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, et al. 2007. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP. 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459. [DOI] [PubMed] [Google Scholar]
- Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP. 2007. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proceedings of the National Academy of Sciences, USA 104, 10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Wang Y, Shen G, Zheng X. 2006. A Phytophthora sojae gene of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) induced in host infection and its anti-oxidative function in yeast. Chinese Science Bulletin 51, 1316–1323. [Google Scholar]
- Zhang M, Wang Q, Xu K, et al. 2011. Production of dsRNA sequences in the host plant is not sufficient to initiate gene silencing in the colonizing oomycete pathogen Phytophthora parasitica . PLoS ONE 6, e28114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.