Abstract
The RNA exosome is a conserved complex for RNA degradation with two ribonucleolytic subunits, Dis3 and Rrp6. Rrp6 is a 3′–5′ exonuclease, but it also has a structural role in helping target RNAs to the Dis3 activity. The relative importance of the exonuclease activity and the targeting activity probably differs between different RNA substrates, but this is poorly understood. To understand the relative contributions of the exonuclease and the targeting activities to the degradation of individual RNA substrates in Schizosaccharomyces pombe, we compared RNA levels in an rrp6 null mutant to those in an rrp6 point mutant specifically defective in exonuclease activity. A wide range of effects was found, with some RNAs dependent mainly on the structural role of Rrp6 (“protein-dependent” targets), other RNAs dependent mainly on the catalytic role (“activity-dependent” targets), and some RNAs dependent on both. Some protein-dependent RNAs contained motifs targeted via the RNA-binding protein Mmi1, while others contained a motif possibly involved in response to iron. In these and other cases Rrp6 may act as a structural adapter to target specific RNAs to the exosome by interacting with sequence-specific RNA-binding proteins.
Keywords: Rrp6, Dis3, exosome, pombe, Mmi1, iron
INTRODUCTION
The RNA exosome is a large complex conserved from archaea to higher eukaryotes. It functions in surveillance, processing, and degradation of RNAs including rRNA, tRNA, and small RNAs (for review, see Schmid and Jensen 2008; Lykke-Andersen et al. 2009; Chlebowski et al. 2010). In eukaryotes, the complex comprises a nine-subunit core of RNA-binding proteins associated with two ribonucleases, Rrp6 and Dis3 (Liu et al. 2006; Dziembowski et al. 2007). Dis3 is essential and has endonuclease activity and 3′–5′ exonuclease activity. Rrp6 has only 3′–5′ exonuclease activity, and is found in the nuclear but not the cytoplasmic form of the exosome (Allmang et al. 1999).
Crystal structures of exosome–RNA complexes reveal that Rrp6 and Dis3 are found at opposite ends of the exosome (Wasmuth and Lima 2012; Makino et al. 2013, 2015; Wasmuth et al. 2014). The exonuclease domain of Rrp6 is solvent-exposed even when associated with the exosome. In contrast, the exonuclease domain of Dis3 faces the inner core of the exosome, through which the RNA substrate is threaded. Although earlier studies showed that Rrp6 and Dis3 have distinct RNA targets (Kiss and Andrulis 2010), recent studies have shown that Rrp6 can regulate the catalytic activity of Dis3 in vitro (Wasmuth and Lima 2012). This may be because Rrp6 associates with the top of the exosome where the RNA enters, and possibly regulates RNA threading through the exosome core (Wasmuth and Lima 2012; Makino et al. 2013, 2015). Strikingly, even a catalytically inactive Rrp6 is able to enhance RNA degradation by Dis3 in vitro (Wasmuth and Lima 2012). These studies show that, at least in vitro, Rrp6 has a structural role in the exosome in addition to its catalytic role.
In Schizosaccharomyces pombe, the RNA exosome plays a crucial role in regulating meiosis. Many genes are up-regulated during the switch from vegetative growth to meiosis (Mata et al. 2002). During vegetative growth, some early meiotic genes are constitutively transcribed. The RNA-binding protein Mmi1 binds these early meiotic mRNAs and targets them for degradation to keep meiosis off (Harigaya et al. 2006). Upon meiotic entry, Mmi1 is inactivated, allowing early meiotic transcripts to accumulate and be translated. Further studies on the mechanism of Mmi1-mediated RNA turnover showed the exosome degrades Mmi1-bound RNAs in vegetative cells, and is aided by polyadenylation of these RNAs (St-Andre et al. 2010; Yamanaka et al. 2010; Chen et al. 2011). Previous studies have shown both rrp6 and dis3 mutations interfere with degradation of Mmi1 targets (Yamanaka et al. 2010; Chen et al. 2011). Some Mmi1 targets, such as rec8 mRNA, undergo Mmi1-dependent hyperadenylation and thus become a substrate for Rrp6. However, a small fraction of the rec8 mRNA is a read-through transcript that is targeted by Dis3 (Chen et al. 2011). In summary, both Dis3 and Rrp6 keep many early meiotic mRNAs off in vegetative cells (Chen et al. 2011).
Delivery of the mRNAs bound by Mmi1 to the exosome involves a multiprotein complex called NURS or MTREC (Sugiyama and Sugioka-Sugiyama 2011; Sugiyama et al. 2012; Yamashita et al. 2013; Egan et al. 2014; Zhou et al. 2015). NURS/MTREC complex bridges Mmi1 and the exosome; it interacts with Mmi1 through Iss10 (Egan et al. 2014), and it interacts with Rrp6 through Mtl1 (Egan et al. 2014) and Red1 (Zhou et al. 2015).
In S. pombe, it is currently believed that Rrp6 directly degrades Mmi1-bound RNAs. This is based on the observation that rrp6Δ mutants accumulate Mmi1-target RNAs. However, dis3 mutants also accumulate Mmi1-target RNAs (Yamanaka et al. 2010; Chen et al. 2011). One possibility is that Rrp6 and Dis3 are each partly responsible for degrading Mmi1-bound RNAs. Alternatively, loss of Rrp6 protein could result in defective recruitment of Mmi1-bound RNAs for degradation by Dis3. This led us to the question of whether Rrp6 has distinct catalytic and “structural” functions. The structural functions could include both an enhancement of Dis3 activity, and a recruitment activity via protein–protein interactions. To distinguish which RNAs are degraded by Rrp6, and which are somehow targeted to Dis3 via Rrp6, we made catalytic (rrp6-cat), null (rrp6Δ), and rrp6-cat dis3-4 (ts) mutants and measured the genome-wide abundance of RNAs.
RESULTS AND DISCUSSION
Analysis of phenotypes of exosome exonuclease mutants
To examine the role of the Rrp6 exonuclease activity, we designed an allele of rrp6 to abolish its 3′–5′ exonuclease activity (rrp6-cat). When compared across species, the DEDD-Y exonuclease domain of Rrp6 is highly conserved (Supplemental Fig. S1A). We constructed rrp6-cat by mutating residues D243, E245, D303, Y366, and D370 to alanine. We selected these residues based on reports showing these residues are required for processing 5.8S rRNAs, some snRNAs and snoRNA in yeast (Briggs et al. 1998; Phillips and Butler 2003). We mutated all of them to increase the likelihood of obtaining a catalytically dead Rrp6 protein. All these amino acids are located inside the catalytic core of Rrp6 as shown in a homology model (Supplemental Fig. S1B).
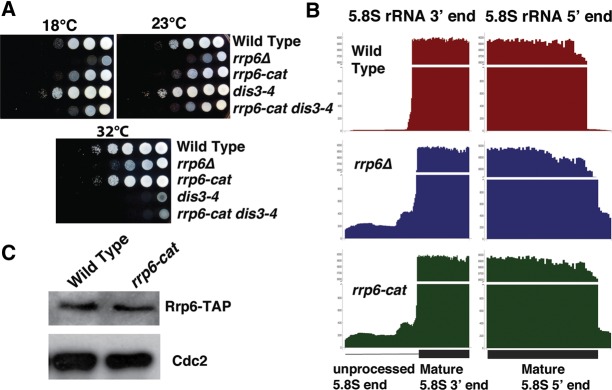
The rrp6-cat mutant had a slight growth defect compared to wild-type at all temperatures (Fig. 1A). In contrast, the rrp6Δ mutant showed severe growth defects at all temperatures (Fig. 1A). This shows that Rrp6-cat protein retains some functions of the wild-type protein.
FIGURE 1.
Phenotypes of Rrp6 and Dis3 mutants. (A) Growth assay for rrp6 and dis3 mutants on YES media at different temperatures. (B) Bar chart showing RNA-seq reads at one 5.8S rRNA locus (SPRRNA.51) at the 5′ and 3′ ends (direction of transcription is from right to left). The y-axis was compressed to clearly show the unprocessed reads. (C) Western blot showing the levels of Rrp6-TAP (strain JLP1840) and Rrp6-cat-TAP (strain JLP1841).
To further characterize the rrp6-cat mutant, we attempted to make double mutants with dis3, the core nuclease of the exosome. We first crossed to a cold-sensitive mutant, dis3-54 (Kinoshita et al. 1991). Despite dissecting 40 tetrads, we were unable to find any rrp6-cat dis3-54 double mutants among the 11 complete tetrads (P < 0.005), suggesting that this double mutant is inviable even at the dis3-54 permissive temperature. This is consistent with the idea that rrp6-cat lacks exonuclease activity. We then crossed rrp6Δ to dis3-4 (ts), a different allele of dis3 (Yamanaka et al. 2010), but were again unable to recover any double mutants, presumably because the rrp6Δ dis3-4 (ts) double mutant is also inviable. However, we succeeded in generating the rrp6-cat dis3-4 (ts) double mutant. This rrp6-cat dis3-4 double mutant was temperature sensitive because of the dis3-4 allele and also slightly cold sensitive like the rrp6-cat mutant (Fig. 1A). This indicated that both Rrp6 and Dis3 functions were impaired in the rrp6-cat dis3-4 double mutant.
The dis3-4 allele was generated by error-prone PCR (Yamanaka et al. 2010), and is not well characterized. We sequenced it, and found three mutations: D307G, L642S, and F954S. These lie in the OB RNA-binding region, the catalytic RNB region, and the S1 RNA-binding region, respectively. Thus, the molecular basis of the dis3-4 defect is unclear.
Previous studies showed that Rrp6 is important for processing the 3′ end of a fraction of 5.8S rRNA (Briggs et al. 1998; Allmang et al. 2000). To test if rrp6-cat is defective for catalysis, we analyzed 5.8S rRNA processing by sequencing total RNA from wild-type, rrp6Δ, and rrp6-cat mutants. We made the libraries by ligating a sequencing adapter to the 3′ end of unfragmented total RNA (see Materials and Methods) to determine 3′ processing changes. We found that the rrp6-cat and rrp6Δ mutants each contained a substantial and similar amount of unprocessed 5.8S rRNA at both the 5′ and 3′ ends (Fig. 1B); little or no such unprocessed 5.8S rRNA was seen in the WT strain (Fig. 1B). Nevertheless, the total amount of mature 5.8S rRNA was similar in all three strains (Fig. 1B, y-axis). This result shows that mutating the five amino acids in the exonuclease domain did indeed abolish catalytic activity of Rrp6, since, for this phenotype, rrp6-cat phenocopies rrp6Δ. It also suggests that there are other, redundant mechanisms for producing the mature 5.8S rRNA.
The levels of untagged, wild-type Rrp6 protein and mutant Rrp6-cat protein were the same, as assayed with a rabbit polyclonal serum to native Rrp6 (data not shown), and the levels of WT Rrp6-TAP and mutant Rrp6-cat-TAP (i.e., with a C-terminal TAP-tag) were also the same (Fig. 1C). Furthermore, a C-terminal TAP-tag did not affect the growth phenotypes of either the wild-type or rrp6-cat strain (data not shown).
Rrp6 mostly plays a structural role in degrading its mRNA targets
We used RNA-seq to determine the genome-wide RNA targets of Rrp6 and Dis3. The RNA-seq libraries were made using poly(A) selected RNA from wild-type and mutants rrp6Δ, rrp6-cat, dis3-4, and rrp6-cat dis3-4, with two biological replicates for each strain. The strains with dis3-4 were grown at permissive temperature (25°C) and then shifted to restrictive temperature (30°C) for 1 h and RNA was isolated and sequenced from both conditions. We mapped the RNA-seq reads to the S. pombe transcriptome and computed fragments per kilobase per million (FPKM) as a measure of RNA levels for each gene using cuffnorm and cuffdiff (Trapnell et al. 2013). The replicate experiments were highly correlated (Supplemental Fig. S2), having a correlation coefficient greater than 0.92 for all replicates.
We compared the transcriptomes of the wild-type and the rrp6Δ mutant using cuffdiff to determine which RNAs were significantly different. Many RNAs accumulated in the rrp6Δ mutant while few RNAs had lower levels (data not shown). This is consistent with the idea that deleting an exosome subunit should lead to RNA accumulation. We defined Rrp6 RNA targets as those RNAs that accumulated significantly (P < 0.01) in the rrp6Δ mutant with respect to wild-type (Supplemental Table S1). According to this definition, Rrp6 targets included 157 mRNAs (Supplemental Table S1A) and 103 noncoding RNAs (Supplemental data, Rrp6_targets.xlsx). (Note that since there are about 5000 protein-coding genes in S. pombe, with a two-tailed P-value threshold of 0.01, there could be around 25 false positives among the 157 mRNAs accumulated.) We plotted the WT/rrp6Δ FPKM ratios of these Rrp6 targets as a frequency distribution and found that the ratios were distributed between 0 and 0.5 (Fig. 2A); i.e., all the targets were increased twofold or more in the rrp6Δ mutant.
FIGURE 2.
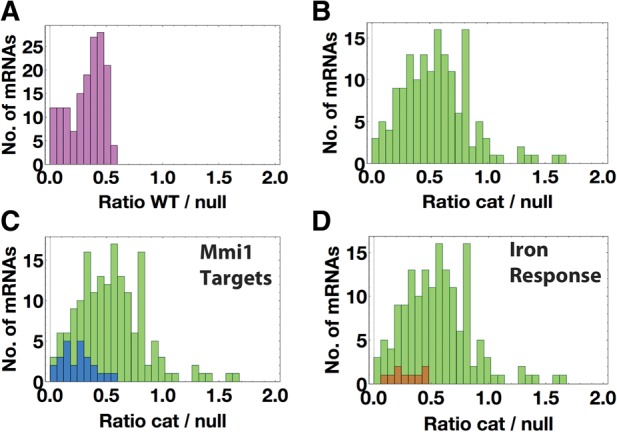
Target mRNAs of Rrp6 protein. (A) Frequency distribution of WT/rrp6Δ FPKM ratios of mRNA targets of Rrp6, defined as accumulating significantly by Cuffdiff (P < 0.01). The bin width is 0.05. (B) Frequency distribution of rrp6-cat/rrp6Δ FPKM ratios of mRNA targets of Rrp6, defined as accumulating significantly by Cuffdiff (P < 0.01). (C) Histogram showing the distribution of rrp6-cat/rrp6Δ FPKM ratios of Mmi1 target mRNAs (blue) and all mRNA targets of Rrp6 (green). (D) Frequency histogram showing the distribution of rrp6-cat/rrp6Δ FPKM ratios of iron homeostasis mRNAs (brown) and all mRNA targets of Rrp6 (green).
To compare the effect of the Rrp6 catalytic mutant to the null mutant for each mRNA, we plotted the rrp6-cat/rrp6Δ FPKM ratio for each Rrp6 target RNA as a frequency distribution (Fig. 2B; Supplemental Fig. S3B). At one extreme, if the mRNA is not stabilized at all by the catalytic mutation, but largely stabilized by the null mutation, this ratio could be as low as zero (i.e., very little mRNA in the catalytic mutant, a large amount of mRNA in the null mutant). At the other extreme, if degradation depends entirely on the Rrp6 exonuclease activity, so that the RNA is equally stabilized by the catalytic mutation and the null mutation, then the ratio could be as high as one (ratios higher than one are likely due to noise). We found a roughly normal distribution of rrp6-cat/rrp6Δ ratios between 0 and 1, with a peak around 0.5, suggesting that for a large majority of Rrp6 targets, the catalytic mutation had a weaker effect on abundance than the null mutation (Fig. 2B). On average, over all mRNAs, the catalytic mutation had only about half the effect of the null mutation. In other words, overall, only about half of the effect of Rrp6 on mRNA degradation is due to its exonuclease activity.
Rrp6 exonuclease activity could be distributed equally among its mRNA targets, or Rrp6 exonuclease could show a preference in degrading some targets over others. When we compared the rrp6-cat/rrp6Δ FPKM ratios of Rrp6 targets in the two replicate experiments, we found that they were highly correlated (Supplemental Fig. S3A). The correlation was smaller for noncoding RNA targets, likely due to poly(A) selection (Supplemental Fig. S3C). The fact that each mRNA gave essentially the same result in replicates 1 and 2 strongly suggests that the catalytic/null ratios in Figure 2B are accurate characterizations of each individual mRNA; that is, some mRNAs are “protein-dependent” targets—their degradation depends on the Rrp6 protein, but not its activity; while other mRNAs are “activity-dependent” targets—their degradation depends on Rrp6 catalytic activity.
As further evidence that different mRNAs have different dependencies on the catalytic activity of Rrp6, we analyzed the replicate data shown in Figure 2B and Supplemental Fig. S3A using ANOVA to test whether the mRNAs all come from a single population. ANOVA rejected this null hypothesis (P < 10−31), showing that different mRNAs have different dependencies on the catalytic activity. Thus, there are some mRNAs efficiently degraded directly by Rrp6 (“activity dependent”), and other mRNAs that are not, but still require Rrp6 for degradation for some indirect reason (“protein dependent”) (e.g., targeting to Dis3 exonuclease).
Protein-dependent targets of Rrp6 include Mmi1 targets and iron homeostasis mRNAs
To try to understand why degradation of some mRNAs might depend on the Rrp6 protein, but not its exonuclease activity, we applied Gene Ontology (GO) analysis (Ashburner et al. 2000) to the “protein dependent” targets. For this purpose, we defined “protein dependent” targets as those 121 mRNAs with a catalytic/null ratio of 0.75 or less, which corresponds to the first (leftmost) 15 bars of Figure 2B (Supplemental Table S1A). This analysis revealed two functional groups. The first significant category of genes was involved in meiosis (Table 1; Supplemental data, Rrp6_GO.txt), and the genes were immediately recognizable as those coding for early meiotic mRNAs targeted by Mmi1 (Fig. 2C—blue). The second and nonoverlapping group contained genes involved in iron ion homeostasis, mostly iron transport (Fig. 2D; Table 1; Supplemental data, Rrp6_GO.txt).
TABLE 1.
Functions of 121 protein dependent mRNA targets of Rrp6

Previously we identified a set of 30 mRNAs targeted by the RNA-binding protein Mmi1 for degradation by the exosome during vegetative growth (Chen et al. 2011). In our current experiments, we obtained no sequence reads for four of them, mcp6, mug1, mcp5, and SPAC6C3.05. When we calculated the rrp6-cat/rrp6Δ FPKM ratios of the remaining 26 Mmi1 targets, we found that they accumulated much more in rrp6Δ than in rrp6-cat (Figs. 2C, blue, 3A,B; Table 2); that is, they had very low cat/null ratios. Thus, the Rrp6 protein may be channeling Mmi1 target-mRNAs to Dis3 for degradation. We speculate that this occurs because Rrp6 is indirectly bound to the RNA-binding protein Mmi1, and Mmi1 is bound to these particular mRNAs (Egan et al. 2014; Zhou et al. 2015). We suggest that in wild-type cells, the exosome is recruited to the Mmi1-bound RNAs through the NURS/MTREC–Rrp6 interaction (Egan et al. 2014; Zhou et al. 2015). The exosome then degrades these RNAs using Dis3 (Fig. 4A). However, as indicated by the dotted line, Rrp6 also has some catalytic role given that the Mmi1 targets accumulated somewhat (about fourfold) in rrp6-cat (Fig. 3B). Our results are also consistent with structural studies showing that the presence of Rrp6 protein in the exosome enhances RNA degradation by the exosome (Wasmuth and Lima 2012).
FIGURE 3.
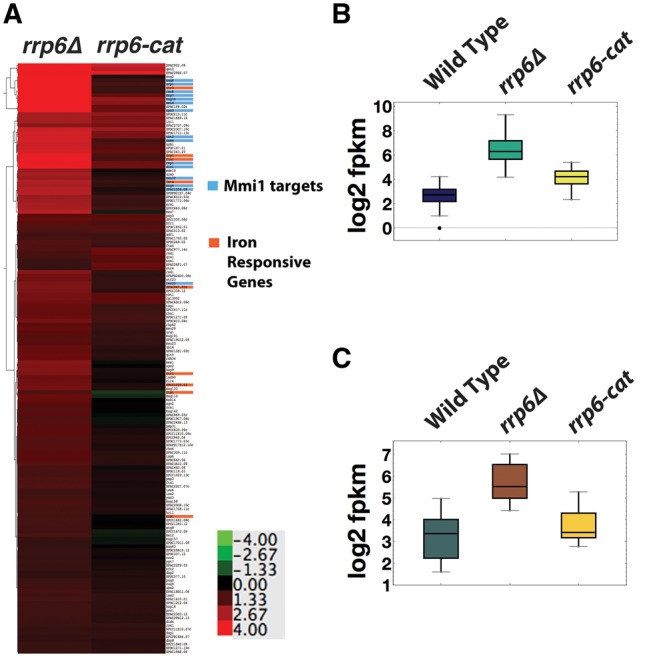
RNA accumulation of Rrp6 target mRNAs in rrp6 deletion and catalytic mutants. (A) Hierarchical clustering of the log2 (mutant/WT) FPKM ratio of the 157 Rrp6 target mRNAs, in rrp6Δ and rrp6-cat. Mmi1 target RNAs are highlighted in blue and iron homeostasis mRNAs are highlighted in orange. (B) Median log2 FPKM values for Mmi1 target mRNAs and (C) iron response mRNAs in wild-type and rrp6 mutants.
TABLE 2.
Effect of Rrp6 null and catalytic mutations on Mmi1 target RNAs
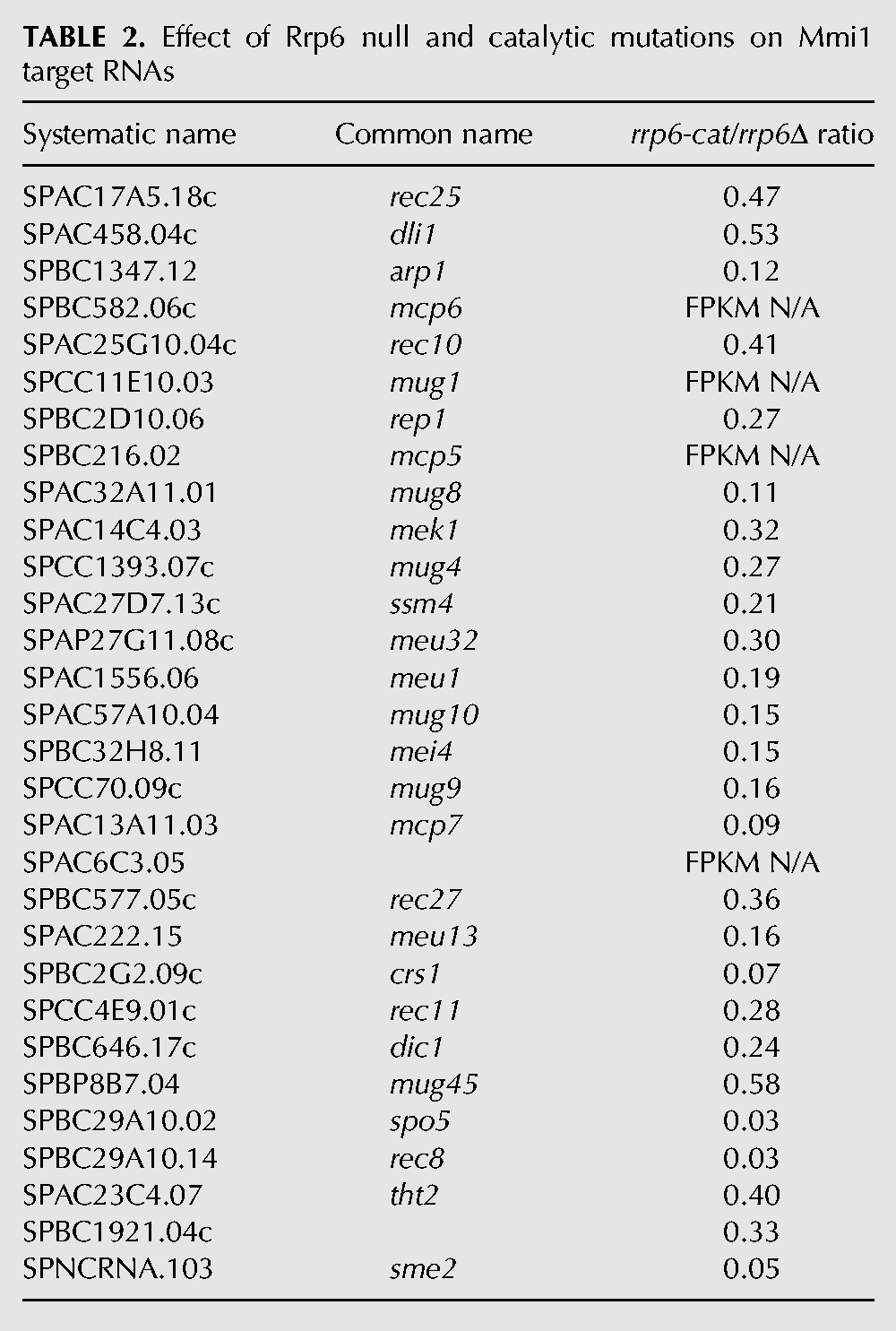
FIGURE 4.
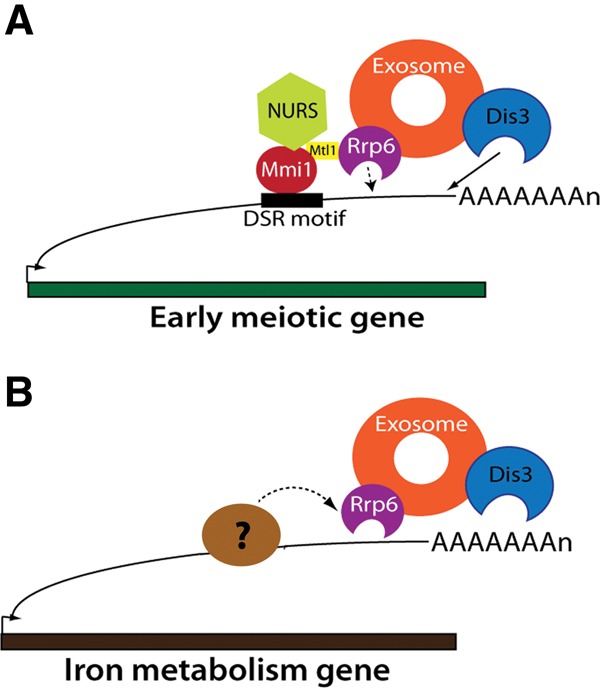
A schematic model for the role of Rrp6 as a structural adapter protein. (A) Rrp6 associates with the NURS complex through its interaction with Mtl1. NURS complex associates with Mmi1 bound to early meiosis RNAs during mitosis. Therefore, exosome recruitment to these RNAs is possible and these RNAs are degraded largely by Dis3. Rrp6 degrades only some of the Mmi1 target RNA (depicted by dotted line). (B) Rrp6 facilitates the degradation of iron homeostasis mRNAs during high-iron conditions. It does so presumably by interacting with as yet unidentified RNA-binding proteins that bind to these mRNAs.
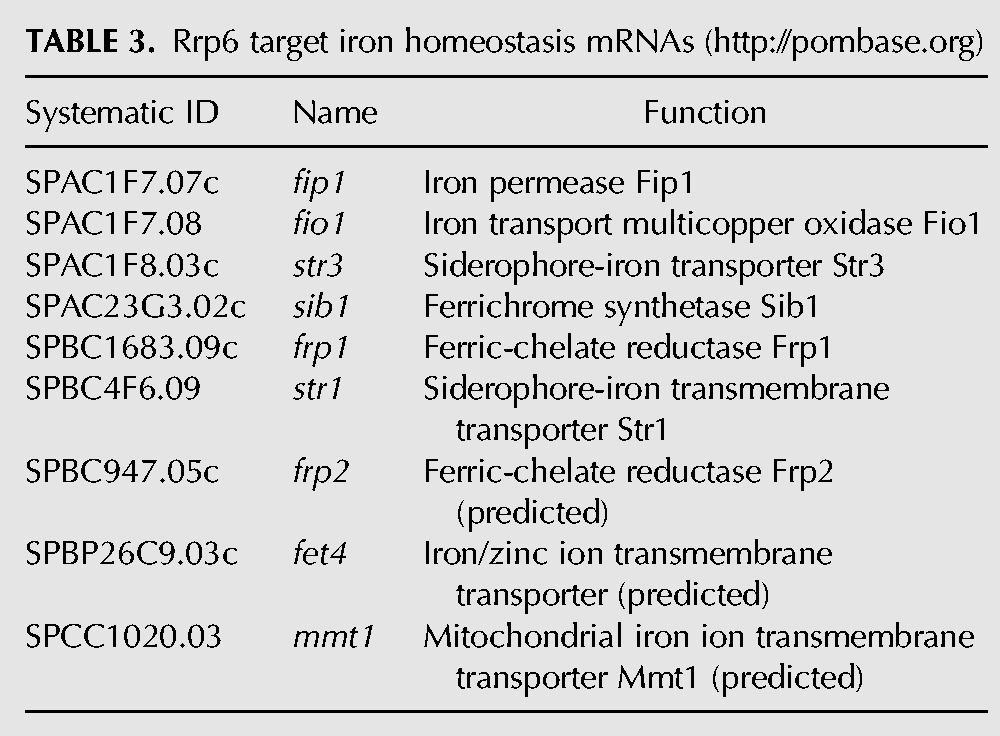
Similarly, degradation of a set of nine iron response mRNAs was dependent on the Rrp6 protein, but not on its catalytic activity (Figs. 2D, brown, 3A,C; Table 1). The results from RNA-seq were confirmed by qPCR for several of the genes (Supplemental Fig. S4A). The genes in this category and their functions are listed in Table 3 and their expression levels are in the Supplemental data, “iron.txt.” Most of the genes code for cell surface transporters of iron.
TABLE 3.
Rrp6 target iron homeostasis mRNAs (http://pombase.org)
Prior studies on iron metabolism genes suggest that they are up-regulated during iron depletion, but repressed when iron is present in the growth medium (Roman et al. 1993; Pelletier et al. 2003; Mercier et al. 2006). Since we grew cells in YES (nutrient-rich) medium, it is unlikely that these genes were up-regulated due to iron depletion. Thus, our results suggest that in addition to transcriptional regulation, these genes are post-transcriptionally regulated by exosome-mediated RNA degradation during high iron conditions. In mammals and budding yeast, iron-responsive elements (IREs) in the mRNAs of iron metabolism genes control either stability or translation of those mRNAs (Hentze et al. 1987, 1989; Rouault et al. 1988).
Analogous to the Mmi1 pathway, in S. pombe there may be one or more RNA-binding proteins bound to these mRNAs during high-iron conditions that targets them to the exosome using Rrp6 as an adapter (Fig. 4B). This idea suggests that these mRNAs could contain a nucleotide motif for recognition by this hypothetical RNA-binding protein. Motif analysis using MEME found that eight of the nine iron homeostasis Rrp6 targets had a total of 30 copies of a purine-rich motif (Supplemental Fig. S4B) with an E-value of 0.015. An alternative model would be analogous to the regulation of iron starvation mRNAs in budding yeast by the Cth1, Cth2 RNA-binding proteins (Martinez-Pastor et al. 2013). Under high iron conditions, Rrp6 processes the 3′ end of the CTH2 pre-mRNA resulting in mature CTH2 mRNA (Ciais et al. 2008). Cth2 recognizes AU-rich elements in the 3′ UTR of iron starvation mRNAs and targets them for degradation (Puig et al. 2005, 2008). However, Cth1 and Cth2 have no homologs in S. pombe.
Of the 121 protein-dependent mRNAs, 15 are targets of Mmi1, and nine are probable targets of a putative RNA-binding protein involved in iron homeostasis (Table 1). Thus there are an additional 97 mRNAs that may be targeted to the exosome via Rrp6 and unknown RNA-binding proteins (Table 1), and subsequently degraded largely by Dis3.
Effects of dis3-4
As described above, we were able to make rrp6-cat dis3-4 double mutants, but not rrp6Δ dis3-4, rrp6-cat dis3-54, or rrp6Δ dis3-54 double mutants. We investigated mRNA levels at permissive and restrictive temperatures in the dis3-4 mutant and the rrp6-cat dis3-4 double mutant. However, by RNA-seq, these had about the same mRNA levels at both temperatures (Supplemental Fig. S5), suggesting that the biochemical activity of Dis3-4 is not highly dependent on temperature.
We clustered the mRNA abundances from RNA-seq in the rrp6Δ, rrp6-cat, dis3-4, and rrp6-cat dis3-4 mutants (Supplemental Fig. S6A). We found that the Mmi1 targets accumulated at a higher level in dis3-4 than in rrp6-cat (Supplemental Fig. S6A,B). However, this result is difficult to interpret because the specific defect of Dis3-4 mutant protein is uncharacterized.
MATERIALS AND METHODS
Yeast strains, media, and growth conditions
S. pombe culture methods were used as described in Forsburg and Rhind (2006). When required, cells were grown in Edinburgh Minimal Media (EMM) with supplements. For RNA isolation and spot assays, cells (except dis3-4 mutants) were grown in liquid YES at 32°C, until they reached OD600 = 0.5–0.6. Temperature-sensitive mutants (dis3-4) were grown at 25°C before shifting to 30°C for 1h. For RNA isolation, cells were harvested at 4°C, washed with ice-cold water, frozen in liquid nitrogen, and stored at −80°C. For spot assays, wild-type (CHP1365), rrp6Δ (JLP1704), and rrp6-cat (JLP1708) strains were grown in liquid YES at 32°C, and dis3-4 (JT449), and rrp6-cat dis3-4 (JLP1863) were grown in liquid YES at 25°C until they reached an optical density (600 nm) of 0.6. Cells were then suspended in water at a density of 2 × 106 cells/mL. Tenfold serial dilutions were made and 5 µL of each dilution was spotted on YES-agar plates and incubated at the temperatures indicated. For transformation, cells were grown in liquid YES at 32°C overnight and then diluted in an equal volume of fresh YES media. Cells were grown another 3 h before harvesting. All strains used in this study are listed in Supplemental Table S2.
Cloning and site-directed mutagenesis of rrp6 exonuclease domain
A region containing the exonuclease domain of rrp6 was PCR amplified using primers rrp6-exon2F and rrp6-exon2R. The PCR product was T-A cloned and the resulting plasmid, pKM-rrp6-ex2, was mutagenized by site-directed mutagenesis to give plasmid pKM-rrp6-cat. Site-directed mutagenesis was performed using the Stratagene QuikChange Multi Site-Directed Mutagenesis kit according to instructions. Primers used are listed in Supplemental Table S3.
Construction of rrp6Δ, rrp6-cat, and rrp6-cat dis3-4 mutants
The rrp6 gene was disrupted using ura5-lys7 cassette as described in Mudge et al. (2012). ura5-lys7 was PCR amplified, with 250 nt overhangs homologous to exon2 of rrp6, from plasmid pUL57. This PCR product was transformed into CHP1365 (kindly provided by Dr. Charles Hoffman) using a protocol described in Bahler et al. (1998). Cells were plated on EMM with leucine, adenine, and histidine to select for ura+ lys7+ colonies resulting in the rrp6::ura5-lys7 (rrp6Δ) strain, JLP1704. JLP1704 was then transformed with a PCR product from pKM-rrp6-cat generated using primers rrp6-exon2F and rrp6-exon2R. Transformants were counter-selected on EMM media containing 0.1% 5-FOA. Mutants were screened by colony PCR, and sequencing the rrp6 gene. The resulting rrp6-cat mutant, JLP1708, was crossed to the dis3-4 (ts) mutant (JT449, kindly provided by Dr. Masayuki Yamamoto, (Yamanaka et al. 2010)) and tetrads were dissected to obtain the rrp6-cat dis3-4 mutant (JLP1863).
Construction of TAP-tagged rrp6 and rrp6-cat alleles
TAP-tagged Rrp6 and Rrp6-cat were made by transforming strains CHP1365 and JLP1708 with a C-terminal TAP>>KanMX cassette, integrated at the 3′ end of the wild-type rrp6 and mutant rrp6-cat allele resulting in strains JLP1840 and JLP1841.
RNA isolation and Illumina RNA-seq
RNA was extracted from cells using RiboPure Yeast kit (Ambion) according to manufacturer's instructions. Twenty micrograms of total RNA was DNase-treated using Turbo DNase (Ambion). One microliter of DNase-treated RNA was used for PCR using srp7F and srp7R primers to ascertain whether there was no genomic DNA contamination. The integrity of total RNA was measured using Agilent Bioanalyzer 2100.
RNA-seq library preparation for total RNA using 3′ ligation
One microgram of unfragmented total RNA was end repaired using polynucleotide kinase. A modified oligonucleotide adapter from the Illumina Art-Seq/Riboprofile kit was ligated to the 3′ end. First strand cDNA was made using reagents provided with this kit. Second strand cDNA was made using reagents from the NuGen Ovation Universal RNA-seq System according to manufacturer's instructions. cDNA was sonicated using a Misonix 3000 sonicator at amplitude 60, for 10 min on ice. Following sonication, the libraries were prepared using reagents and instructions from the NuGen Ovation Universal RNA-seq system but with the following modification. Only 5′ adapter was ligated to the fragmented, end-repaired cDNA. The libraries were PCR amplified using forward and reverse primers provided in the Illumina Art-Seq/Riboprofile kit. The libraries were purified using 0.8× Ampure XP beads and sequenced on Illumina MiSeq.
RNA-seq library preparation for poly(A) selected RNA
Thirty micrograms of DNase-free RNA was used for poly(A) selection using the Poly(A)Purist kit (Ambion). One hundred nanograms of poly(A) selected RNA was used as starting material for making strand-specific cDNA-seq libraries using the Ovation Universal RNA-Seq Library System (NuGen) according to manufacturer's instructions. cDNA was sonicated using a Misonix 3000 sonicator at amplitude 60, for 10 min on ice. Strand-specific cDNA was PCR-amplified for 12 cycles and sequenced on Illumina MiSeq by paired-end sequencing. All RNA-seq data from 3′ end sequencing and poly(A) selected RNA sequencing are available in GEO (accession number: GSE74411). FPKMs for all genes were generated using cuffdiff as described below, and are listed in the Supplemental data, fpkm_cuffdiff.txt.
Computational methods
Clustal alignment
The Rrp6 exonuclease domain amino acid sequences from Saccharomyces cerevisiae, S. pombe, Drosophila melanogaster, and Homo sapiens was aligned by Clustal X multiple sequence alignment using default parameters (Thompson et al. 2002). Jalview was used to visualize and annotate the results (Waterhouse et al. 2009).
Homology modeling
A homology model for S. pombe Rrp6 was created based on the crystal structure of S. cerevisiae Rrp6 using http://swissmodel.expasy.org (Arnold et al. 2006). Pymol was used to visualize and annotate the Rrp6 homology model.
Bowtie
Reads from total RNA libraries made by end-ligation were aligned using Bowtie2. Reads mapping to the chromosome coordinates corresponding to SPRRNA.51 (5.8S rRNA) were extracted using Samtools mpileup and plotted as a bar chart.
Tophat
Reads were aligned to the S. pombe transcriptome (ASM294v2.26) using Tophat2 (Kim et al. 2013). Discordant mapping of pairs was not allowed and minimum and maximum intron lengths were set to 29 and 1000, respectively (all Tophat options used will be provided upon request). The mapping did not allow for discovery of novel intron junctions. Mapped reads were sorted and indexed using Samtools (Li et al. 2009) and visualized on the Integrative Genomics Viewer (IGV) (Robinson et al. 2011; Thorvaldsdottir et al. 2013).
Cuffquant, Cuffnorm, and Cuffdiff
For differential gene expression, Cuffquant was used to estimate abundances for each transcript. To estimate correlation between replicate experiments, Cuffnorm was used to generate normalized fragments per kilo base per million (FPKM) by considering each replicate as an independent sample (Trapnell et al. 2012). To determine targets of Rrp6, we used Cuffdiff (Trapnell et al. 2013) to compare the transcriptomes of wild-type and rrp6Δ mutant by pooling the replicate data for each sample. Cuffdiff provided the expression level (FPKM) as well as a measure of differential expression, for each gene in wild-type and rrp6Δ.
Analysis of rrp6 mutants’ transcriptome and RNA target determination
From the Cuffdiff analysis, the RNAs that accumulated significantly in rrp6Δ/WT (P < 0.01) were considered Rrp6 target RNAs. Genes with 0 FPKM in all samples were removed from our analysis. A pseudocount of 1 was added to FPKM of all genes in all conditions to enable log2 transformation and ratio calculation. For the Rrp6 targets, we determined their Rrp6-cat dependence by calculating their FPKM ratio in rrp6-cat/rrp6Δ. All plots were generated using Mathematica.
Cluster
(log2 FPKM mutant − log2 FPKM WT) for each gene, was clustered by hierarchical clustering with Euclidian distance as the similarity metric using Cluster 3.0 (de Hoon et al. 2004). The clusters were visualized and analyzed by Java TreeView (Saldanha 2004).
GO analysis was done using http://go.princeton.edu/cgi-bin/GOTermFinder as described in Ashburner et al. (2000).
MEME
The motif search in iron assimilation mRNAs was done using MEME motif finder. We allowed for multiple repetitions of motifs in each sequence. Only the CDS (including UTRs) was used for the analysis. A similar search using the shuffled sequences of the same genes gave a motif with a significantly higher E-value (not shown).
Western blot
Strains JLP1840 and JLP1841 were lysed according to the procedure mentioned in Forsburg and Rhind (2006) and equal amounts of protein from each sample were resolved by 10% SDS-PAGE. Rrp6-TAP was detected by chemiluminescence using a 1:5000 dilution of HRP-conjugated peroxidase anti-peroxidase antibody.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Charles Hoffman at Boston College for providing strains CHP1364 and CHP1365. We also thank Dr. Masayuki Yamamoto (NIBB, Japan) for providing us strain JT449. This work was supported by National Institutes of Health (NIH) grant P01 GM088297 to B.F.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.051490.115.
REFERENCES
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. 1999. The yeast exosome and human PM-Scl are related complexes of 3′ --> 5′ exonucleases. Genes Dev 13: 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Mitchell P, Petfalski E, Tollervey D. 2000. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res 28: 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem 273: 13255–13263. [DOI] [PubMed] [Google Scholar]
- Chen HM, Futcher B, Leatherwood J. 2011. The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PLoS One 6: e26804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A, Tomecki R, Lopez ME, Seraphin B, Dziembowski A. 2010. Catalytic properties of the eukaryotic exosome. Adv Exp Med Biol 702: 63–78. [PubMed] [Google Scholar]
- Ciais D, Bohnsack MT, Tollervey D. 2008. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res 36: 3075–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B. 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22. [DOI] [PubMed] [Google Scholar]
- Egan ED, Braun CR, Gygi SP, Moazed D. 2014. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. RNA 20: 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL, Rhind N. 2006. Basic methods for fission yeast. Yeast 23: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. 2006. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Caughman SW, Rouault TA, Barriocanal JG, Dancis A, Harford JB, Klausner RD. 1987. Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science 238: 1570–1573. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Rouault TA, Harford JB, Klausner RD. 1989. Oxidation-reduction and the molecular mechanism of a regulatory RNA-protein interaction. Science 244: 357–359. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N, Goebl M, Yanagida M. 1991. The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol Cell Biol 11: 5839–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss DL, Andrulis ED. 2010. Genome-wide analysis reveals distinct substrate specificities of Rrp6, Dis3, and core exosome subunits. RNA 16: 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Brodersen DE, Jensen TH. 2009. Origins and activities of the eukaryotic exosome. J Cell Sci 122: 1487–1494. [DOI] [PubMed] [Google Scholar]
- Makino DL, Baumgartner M, Conti E. 2013. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 495: 70–75. [DOI] [PubMed] [Google Scholar]
- Makino DL, Schuch B, Stegmann E, Baumgartner M, Basquin C, Conti E. 2015. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature 524: 54–58. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor M, Vergara SV, Puig S, Thiele DJ. 2013. Negative feedback regulation of the yeast CTH1 and CTH2 mRNA binding proteins is required for adaptation to iron deficiency and iron supplementation. Mol Cell Biol 33: 2178–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147. [DOI] [PubMed] [Google Scholar]
- Mercier A, Pelletier B, Labbe S. 2006. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell 5: 1866–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge DK, Hoffman CA, Lubinski TJ, Hoffman CS. 2012. Use of a ura5+–lys7+ cassette to construct unmarked gene knock-ins in Schizosaccharomyces pombe. Curr Genet 58: 59–64. [DOI] [PubMed] [Google Scholar]
- Pelletier B, Beaudoin J, Philpott CC, Labbe S. 2003. Fep1 represses expression of the fission yeast Schizosaccharomyces pombe siderophore-iron transport system. Nucleic Acids Res 31: 4332–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S, Butler JS. 2003. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA 9: 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120: 99–110. [DOI] [PubMed] [Google Scholar]
- Puig S, Vergara SV, Thiele DJ. 2008. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab 7: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman DG, Dancis A, Anderson GJ, Klausner RD. 1993. The fission yeast ferric reductase gene frp1+ is required for ferric iron uptake and encodes a protein that is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Mol Cell Biol 13: 4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA, Hentze MW, Caughman SW, Harford JB, Klausner RD. 1988. Binding of a cytosolic protein to the iron-responsive element of human ferritin messenger RNA. Science 241: 1207–1210. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. 2008. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci 33: 501–510. [DOI] [PubMed] [Google Scholar]
- St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F. 2010. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J Biol Chem 285: 27859–27868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Sugioka-Sugiyama R. 2011. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J 30: 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Sugioka-Sugiyama R, Hada K, Niwa R. 2012. Rhn1, a nuclear protein, is required for suppression of meiotic mRNAs in mitotically dividing fission yeast. PLoS One 7: e42962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2: 3. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Lima CD. 2012. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol Cell 48: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Januszyk K, Lima CD. 2014. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature 511: 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. 2010. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J 29: 2173–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Takayama T, Iwata R, Yamamoto M. 2013. A novel factor Iss10 regulates Mmi1-mediated selective elimination of meiotic transcripts. Nucleic Acids Res 41: 9680–9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu J, Schermann G, Ohle C, Bendrin K, Sugioka-Sugiyama R, Sugiyama T, Fischer T. 2015. The fission yeast MTREC complex targets CUTs and unspliced pre-mRNAs to the nuclear exosome. Nat Commun 6: 7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.