Abstract
Intron patterns in plant mitochondrial genomes differ significantly between the major land plant clades. We here report on a new, clade-specific group II intron in the rps1 gene of monilophytes (ferns). This intron, rps1i25g2, is strikingly similar to rpl2i846g2 previously identified in the mitochondrial rpl2 gene of seed plants, ferns, and the lycophyte Phlegmariurus squarrosus. Although mitochondrial ribosomal protein genes are frequently subject to endosymbiotic gene transfer among plants, we could retrieve the mitochondrial rps1 gene in a taxonomically wide sampling of 44 monilophyte taxa including basal lineages such as the Ophioglossales, Psilotales, and Marattiales with the only exception being the Equisetales (horsetails). Introns rps1i25g2 and rpl2i846g2 were likewise consistently present with only two exceptions: Intron rps1i25g2 is lost in the genus Ophioglossum and intron rpl2i846g2 is lost in Equisetum bogotense. Both intron sequences are moderately affected by RNA editing. The unprecedented primary and secondary structure similarity of rps1i25g2 and rpl2i846g2 suggests an ancient retrotransposition event copying rpl2i846g2 into rps1, for which we suggest a model. Our phylogenetic analysis adding the new rps1 locus to a previous data set is fully congruent with recent insights on monilophyte phylogeny and further supports a sister relationship of Gleicheniales and Hymenophyllales.
Keywords: group II intron, RNA editing, intron transfer, reverse splicing, intron loss, monilophyte phylogeny
INTRODUCTION
Complete mitochondrial genome sequences have become available for five of the six major groups of land plants: liverworts, mosses, hornworts, lycophytes, and spermatophytes with the unique exception of monilophytes (Bock and Knoop 2012). The latter clade, which comprises horsetails, whisk ferns, and “true” ferns, has unequivocally been identified as monophyletic (Pryer et al. 2001). The major hindrance for straightforward assemblies of many vascular plant mitochondrial DNAs is their high recombinational activity, often producing complex mtDNA structures with alternative coexisting gene arrangements, made all the more difficult by the large mitochondrial genome sizes in many cases. The particular complex heavily recombining mtDNAs of some lycophytes are an example for the former (Grewe et al. 2009; Hecht et al. 2011). The mtDNAs of angiosperms like Amborella trichopoda, the Cucurbitaceae or in the genus Silene, which may easily exceed one or even 10 Mbp, and accordingly the genome sizes of most free-living bacteria, are an example for the latter (Ward et al. 1981; Alverson et al. 2010, 2011; Rodríguez-Moreno et al. 2011).
On the other hand, plant mitochondrial DNA (mtDNA) has proven to be an interesting source of phylogenetic information. The slowly evolving and highly conserved mitochondrial coding sequences of plants are of particular interest for deep-level phylogenetics and this is similarly true for the introns in plant mtDNAs (chondromes). Introns in plant mitochondrial genes are highly conserved within the major plant clades with the exception of the lycophytes. Mitochondrial introns differ, however, significantly between the major plant clades, indicating a high degree of gains and losses along the backbone of early plant phylogeny followed by a later stasis within the individual clades (Qiu et al. 1998, 2006; Pruchner et al. 2002; Qiu and Palmer 2004; Groth-Malonek et al. 2005; Volkmar et al. 2012). For example, not a single one of nearly 100 mitochondrial introns in bryophytes is shared between all three classes, i.e., the liverworts, mosses, and hornworts (Knoop 2010, 2013). In contrast, 25 of 27 mitochondrial introns in the gymnosperm Cycas taitungensis have orthologous counterparts in the mtDNAs of angiosperms (the only exception being rps3i249g2 in the rps3 gene and the chloroplast-derived trnVi36g2).
We identified group II intron rps1i25g2 as a novel, monilophyte-specific intron initially in the fern Gleichenia dicarpa. This intron is related to its paralog rpl2i846g2, which in contrast is more widely conserved among tracheophytes. Although both mitochondrial ribosomal protein genes are subject to independent nuclear gene transfer in hornworts, lycophytes, and seed plants, rps1 and rpl2 and their respective introns are widely conserved among monilophytes. We suggest that rps1i25g2 originated by retrotransposition from its more ancestral counterpart rpl2i846g2.
RESULTS
The studies reported here were initially triggered by the discovery of a novel group II intron in the mitochondrial rps1 gene (encoding the ribosomal protein 1 of the small ribosomal subunit) during the characterization of mtDNA-containing fosmids of the fern Gleichenia dicarpa. The novel group II intron in rps1 is inserted after nucleotide 25 of the rps1 reading frame (thus disrupting the ninth codon in phase 1) and is consequently named rps1i25g2 according to the nomenclature proposal for mitochondrial introns (Dombrovska and Qiu 2004; Knoop 2004). Surprisingly, the Gleichenia dicarpa rps1i25g2 intron sequence revealed strong sequence similarity with group II intron rpl2i846g2 (Fig. 1), a paralog in the rpl2 gene previously identified in the mtDNA of the lycophyte Phlegmariurus squarrosus (Liu et al. 2012), previously labeled Huperzia squarrosa and renamed by Field and Bostock (2013), and in seed plants, including the gymnosperm Cycas taitungensis (Chaw et al. 2008). The mitochondrial rpl2 gene, encoding protein 2 of the large ribosomal subunit and its intron rpl2i846g2 were recently found conserved in a wide sampling of monilophytes (Knie et al. 2015).
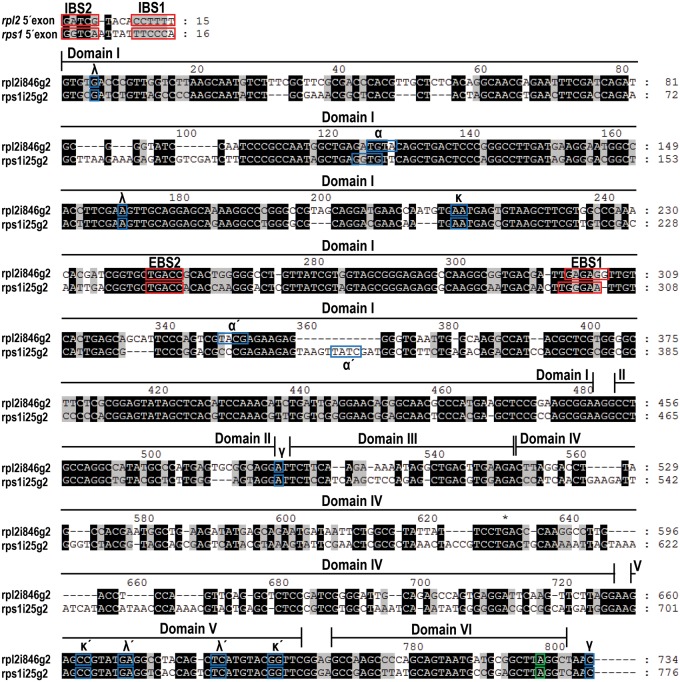
FIGURE 1.
Alignment of the rps1i25g2 and rpl2i846g2 sequences highlighting regions important for secondary structure folding. The six domains of the introns are indicated. Nucleotides shaded in black (65%) are identical, similar nucleotides explained by transitions (A/G and C/U) are shaded in gray (13%). Intron internal interaction sites (Greek letters) are marked with blue boxes, exon/intron binding sites with red boxes, and the branching adenosine with a green box. The corresponding intron binding sites in the 5′ exons are shown on top. Two indels in the EBS1 loop of rps1i25g2 versus rpl2i846g2 may have targeted the new intron to the rps1 gene as discussed in the text. The shaded sequence alignment was created with GeneDoc (http://genedoc.software.informer.com/2.7/).
We confirmed the functionality of the two mitochondrial loci in Gleichenia dicarpa at the RNA level by verifying functional splicing as predicted and detecting several events of RNA editing in the flanking exon sequences. We hypothesized that the rpl2 intron is an ancient gain in the tracheophyte lineage and later gave rise to the rps1 intron exclusively in the fern (i.e., monilophyte) lineage (Fig. 2).
FIGURE 2.
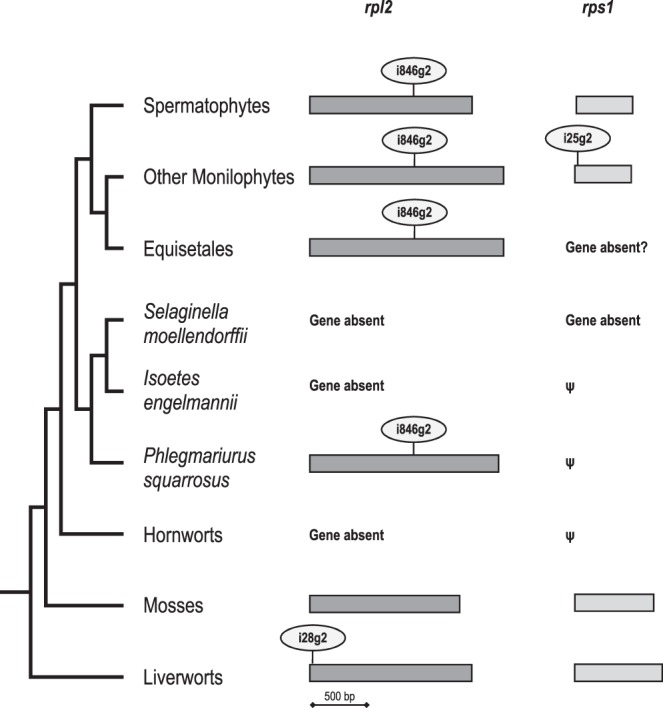
Phylogenetic overview on the mitochondrial rpl2 and rps1 genes and their respective introns in land plants. The rpl2 gene is present in the mitochondrial genomes of liverworts, mosses, the lycophyte Phlegmariurus squarrosus (Liu et al. 2012), in monilophytes (Knie et al. 2015), and in most spermatophytes (Adams et al. 2001, 2002; Adams and Palmer 2003). In hornworts (Li et al. 2009; Xue et al. 2010), lycophytes Isoetes engelmannii (Grewe et al. 2009, 2011) and Selaginella moellendorffii (Hecht et al. 2011) rpl2 genes are absent from the mtDNA, indicating transfer to the nucleus. Intron rpl2i846g2 is present in all tracheophytes where rpl2 is retained in the mtDNA with the exception of independent losses in Mimulus guttatus (Mower et al. 2012) and Equisetum bogotense (Knie et al. 2015). Liverworts carry the unrelated intron rpl2i28g2 in their mitochondrial rpl2 genes. The rps1 gene is likewise conserved in liverworts, mosses, and many spermatophytes. No homologs or only pseudogene fragments exist in hornworts and lycophytes. Monilophytes, except Equisetales, have conserved rps1 genes featuring the unique intron rps1i25g2 except for the genus Ophioglossum where the intron is secondarily lost (this study).
To explore this hypothesis, we investigated the rps1 locus in a phylogenetically wide sampling of 44 monilophyte species. The selected taxa ideally overlapped with a previous rpl2 sampling of early emerging and evolutionary old monilophyte lineages (Table 1). Whereas ribosomal protein genes are frequently subject to endosymbiotic gene transfer in angiosperms (Adams et al. 2001, 2002; Adams and Palmer 2003), we were able to consistently retrieve the rps1 gene in all cases where rpl2 is present with only one exception: Despite numerous attempts employing different primer combinations, we were unable to obtain rps1 PCR products for any of the six Equisetum species tested.
TABLE 1.
Taxa and loci
The monilophyte rps1 locus
The rps1 group II intron rps1i25g2 is highly conserved in length among the fern species (766–826 bp), except in Tmesipteris elongata which has a size of 1002 bp. The unique length extension of rps1i25g2 in Tmesipteris is due to the multiplication of two sequence motifs in the intron domain I at the orthologous position 328 of Gleichenia dicarpa (Fig. 3). The purine-rich sequence GGAGRGAAGGAGGGATAGAAGTCGGGAGAG occurs twice in tandem, separated by only 6 bp. Additionally, a 22-mer motif AGGTACACTCTCCCGACAAGAT exists in four copies and is separated by sequences of 3 bp. Otherwise, the Tmesipteris intron sequence is characterized by a (CACG)3 motif, similar to tandem repetitions of trinucleotide or tetranucleotide motifs extending intron sequences to small extents in Asplenium, Azolla, Pteris, or Salvinia. Except for these motif repetitions, the sequence divergence of rps1i25g2 is, like its length variability, rather restricted. One unique exception, however, is the rps1 sequence in the genus Anemia. Like the exon sequences, and similarly the rpl2 locus, the Anemia sequence shows a unique sequence drift resulting in long branches in the phylogenetic trees. We did not find evidence for the existence of a second copy of either the rps1 or the rpl2 gene in any of the investigated species.
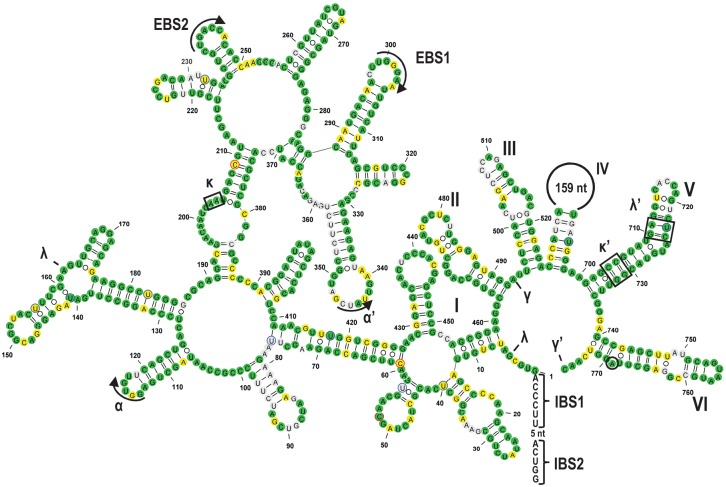
FIGURE 3.
Secondary structure model of rps1i25g2, exemplarily shown for Gleichenia dicarpa, featuring all structural elements typically conserved in group II introns (Michel et al. 1989; Michel and Ferat 1995; Qin and Pyle 1998; Toor et al. 2001; Simon et al. 2008; McNeil et al. 2016). The six intron domains are labeled with Roman numerals (I–VI) and the tertiary interaction sites with Greek letters. Exon binding sites (EBS) and corresponding intron binding sites (IBS) in the 5′ exon are indicated. All nucleotide positions that are identical to the rpl2i846g2 paralog are shown in green. Differences in nucleotide sequences that can be explained by transitions (A/G and C/U) are shown in yellow and all other nucleotides are shown in gray. Experimentally verified RNA editing sites in the intron are marked with blue (C-to-U RNA editing) or red (reverse U-to-C RNA editing) circles. The branch point adenosine for lariat formation is encircled. The intron folding was drawn with the VARNA software (Darty et al. 2009).
Despite numerous attempts with various different primer combinations, we were not able to amplify rps1 from any horsetail (Equisetum) species tested. Intriguingly, BLAST searches in the 1KP database (Matasci et al. 2014) with rps1 queries similarly did not reveal any significant hits, raising general questions about the fate of the RPS1 protein in mitochondrial ribosomes of horsetails.
The rpl2 locus
The mitochondrial rpl2 genes reveal significant variability in lengths of the encoded proteins, e.g., 464 amino acids in the moss Physcomitrella patens, 502 in the liverwort Marchantia polymorpha, 478 in the gymnosperm Cycas taitungensis, and 584 in the lycophyte Phlegmariurus squarrosus. The group II intron rpl2i846g2 in the rpl2 gene of P. squarrosus is conserved in seed plants and in monilophytes but is absent in bryophytes and accordingly most likely gained in the common ancestor of all tracheophytes (Fig. 2). The rpl2 gene is absent altogether from the mtDNAs of the lycophytes Isoetes engelmannii and Selaginella moellendorffii (Grewe et al. 2009; Hecht et al. 2011). Alignment of the flanking exon sequences with a wider sequence sampling allowed us to label the tracheophyte rpl2 intron as rpl2i846g2 to indicate the insertion site behind the upstream coding nucleotide of Marchantia polymorpha as a reference, as proposed previously (Dombrovska and Qiu 2004). We here suggest a secondary structure model for rpl2i846g2 (Supplemental Fig. 1), confirming its structural similarity with rps1i25g2.
The size of rpl2i846g2 in monilophytes is near-universally conserved between 724 bp (in Anemia) and 788 bp (in Salvinia) except for unique size extensions, mainly in domain IV, of the Ophioglossaceae, e.g., 1655 bp in Ophioglossum petiolatum (Knie et al. 2015). Like in rps1i25g2, no traces indicating a previous existence of a maturase-like reading frame are discernible in any of the fern intron rpl2i846g2 homologs. Independent losses of intron rpl2i846g2 are hitherto only found in Equisetum bogotense (Knie et al. 2015) and Mimulus guttatus (Mower et al. 2012).
RNA editing in the intron paralogs rps1i25g2 and rpl2i846g2
Several plant mitochondrial introns have been shown to be subject to RNA editing and at least some cases of intron editing are very likely required to restore the ability for splicing (Lippok et al. 1994; Carrillo and Bonen 1997; Carrillo et al. 2001; Castandet et al. 2010; Bégu et al. 2011; Farré et al. 2012; Oldenkott et al. 2014). Using a combined strategy to obtain cDNAs of intron sequences, we identified several events of RNA editing in the two introns under investigation (Fig. 3; Supplemental Fig. 1; Supplemental Table 1). In Gleichenia dicarpa we found eight sites of RNA editing in rpl2i846g2 and 15 editing sites in rps1i25g2. Many edits restore conserved A–U or G–C binding sites in stem regions. One RNA editing site in the λ′ interaction site in domain V is particularly interesting because it is present in both introns (Fig. 3; Supplemental Fig. 1).
The aligned sequences of the Gleichenia dicarpa introns rpl2i846g2 and rps1i25g2 reveal 65% identical and 13% similar nucleotides (Fig. 1). The biggest differences are mainly found in unstructured regions of domain IV and in some regions of domain I, whereas domains V and VI of the two paralogous introns are nearly identical. The secondary structure models (Fig. 3; Supplemental Fig. 1), which took into account the edited cDNA sequences, reveal a high degree of structural conservation, most notably also in the stem–loop structures of the exon binding sites (EBS).
A model for the origin of rps1i25g2
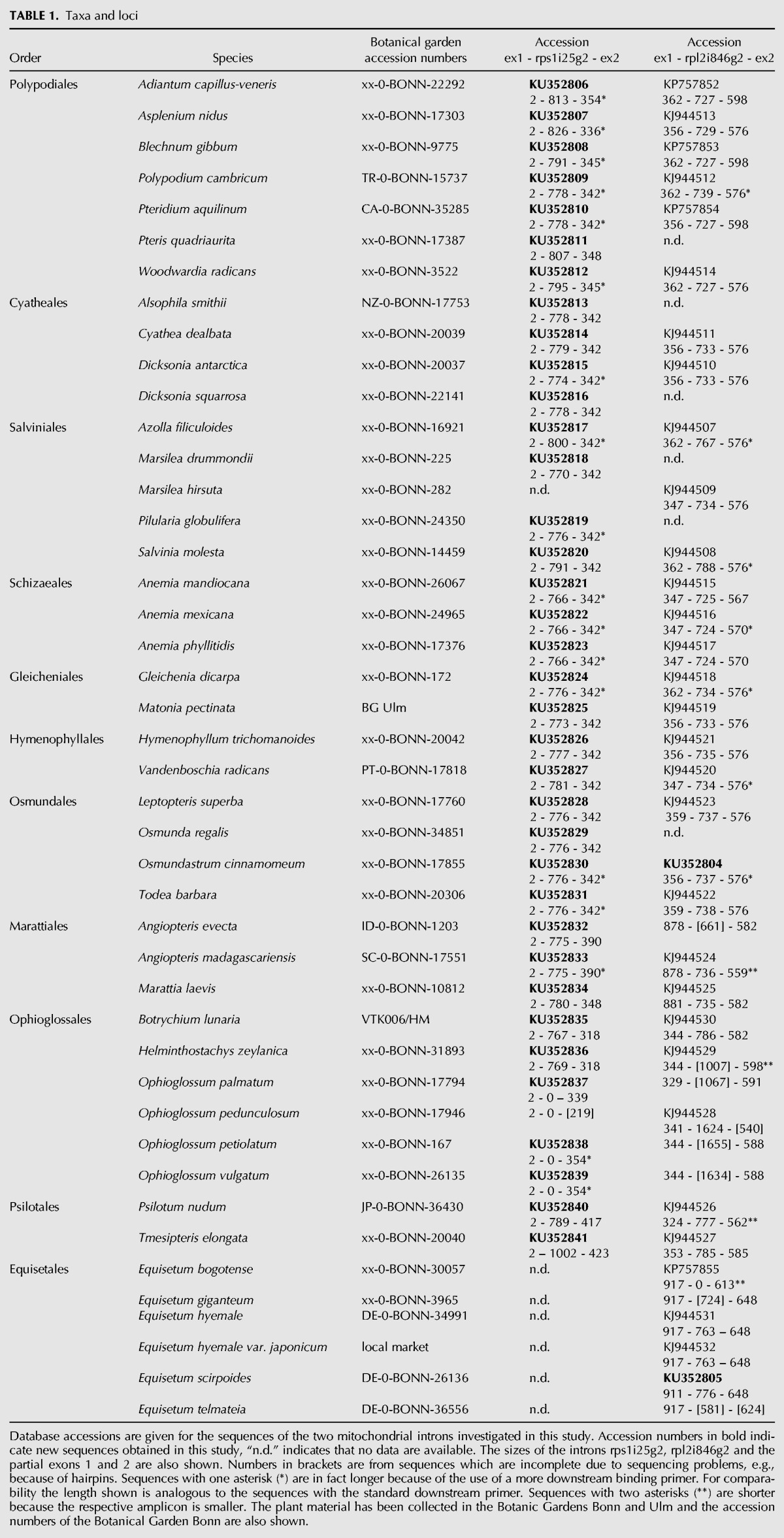
From an evolutionary perspective, intron rpl2i846g2 likely originated in the tracheophyte stem lineage and would be phylogenetically older than rps1i25g2, emerging later either in the monilophyte stem lineage or very early in monilophytes after the split from horsetails (Fig. 2). Assuming an intron-copying via reverse-splicing, this evolutionary scenario suggests that rpl2i846g2 gave rise to rps1i25g2 rather than the other way around.
Exon binding sites are very likely to play a key role in targeting new transcript regions for intron propagation via reverse splicing. Intriguingly, the EBS regions (exon binding sites) of rpl2i846g2 match the heterologous rps1 target nicely, assuming a 1-nt shift in the EBS1 loop (Figs. 1, 4). The heterologous interaction and insertion of rpl2i846g2 into the new site by reverse splicing would be facilitated by a perfect match of EBS2 and an additional base pair at the 3′-end of EBS1 with the newly emerging IBS2 and IBS1 sequences, respectively, in rps1 (Fig. 4B). We assume that the shift in the base-pairing nucleotides in EBS1 was subsequently stabilized by upstream insertion of a cytidine nucleotide (Fig. 4A,B). The deletion of the terminal guanosine nucleotide in the original EBS1 sequence may have subsequently compensated for the upstream C insertion and stabilized the EBS1 loop structure (Fig. 4B,C). Three additional nucleotide substitutions (all of them purine transitions) would ultimately have strengthened the new rps1 EBS1–IBS1 interaction and the stem of the EBS1 loop, respectively, by creating a proper G–C base pair and converting weak G–U into A–U base pairs (Fig. 4B,C). Accordingly, the ancestral EBS1 of rpl2i846g2 would have changed from 5′-GAGAGG-3′ to 5′-UGGGAA-3′ in the newly created rps1i25g2 intron in the course of evolution.
FIGURE 4.
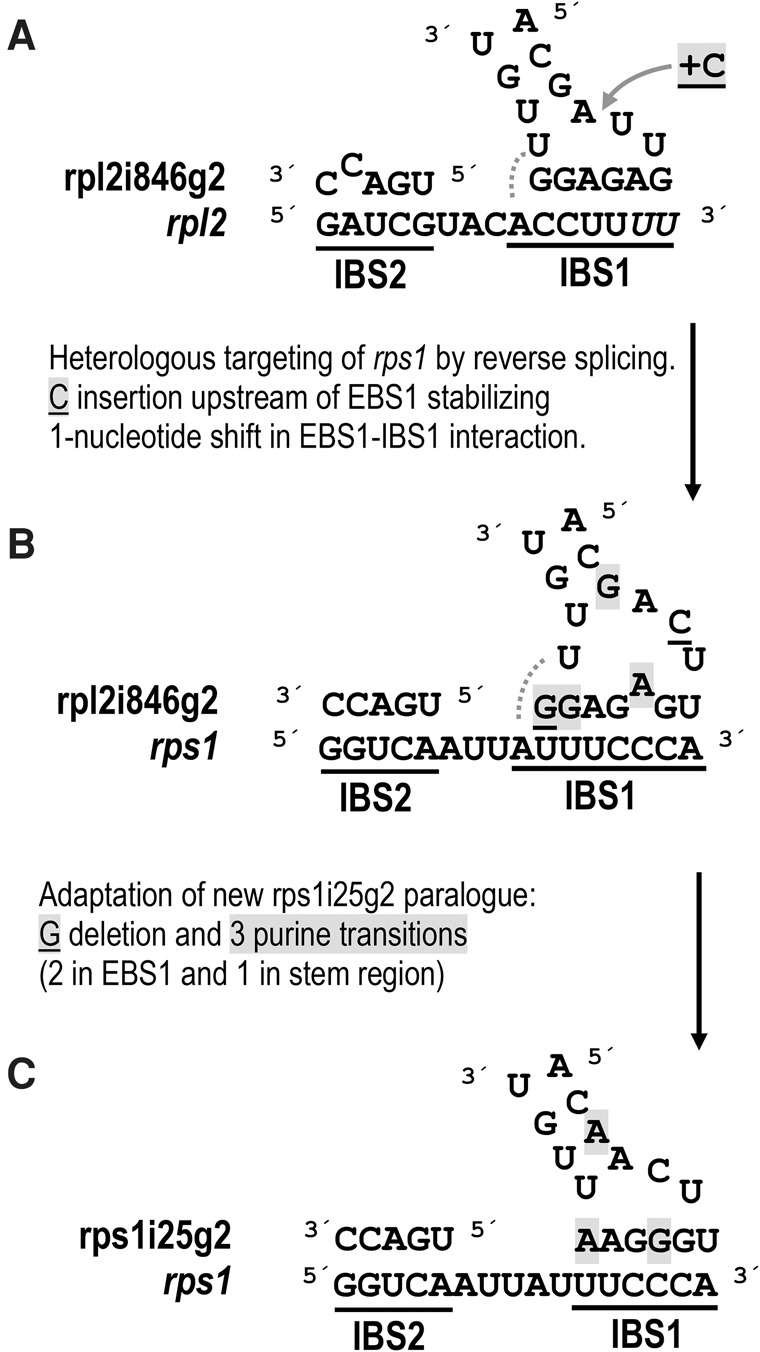
Model for an ancient retrotransposition of rpl2i846g2 into the rps1 gene creating the rps1i25g2 paralog. The transposition is mediated by permissive heterologous EBS-IBS interactions (exon/intron binding sites 1 and 2) and a subsequent adaptation of rps1i25g2 by point mutations. (A) Interaction of rpl2i846g2 EBS sequences with the orthologous rpl2 IBS sequences in the upstream exon. The two terminal positions of the upstream rpl2 exon are affected by C-to-U editing (italics). The IBS1–EBS1 interaction may be extended by a further A–U base pair at the expense of the terminal A–U base pair in the EBS1 stem (dotted line). (B) A heterologous interaction of rpl2i846g2 with the rps1 target may initially have been based on a 1-nt shift in the EBS1–IBS1 recognition subsequently stabilized by the insertion of an upstream cytidine (underlined and shaded gray under A). As in the orthologous rpl2 interaction, a further A–U base pair may extend the EBS1–IBS1 interaction at the expense of the terminal EBS1 stem base-pairing (dotted line). (C) Functional adaptation of the newly created rps1i25g2 paralog has likely been facilitated by deletion of a guanosine (gray shaded and underlined in B) to compensate for the initial upstream cytidine insertion and three purine transitions (gray shading in B and C) that stabilized the base pairings in the EBS1–IBS1 interaction and in the EBS1 stem in the course of evolution.
Losses of mitochondrial group II introns in the basal monilophyte lineages
Investigating rpl2 and rps1, we detected only two cases of intron losses in our taxon sampling: rpl2i846g2 is absent in Equisetum bogotense and rps1i25g2 is absent in the genus Ophioglossum. Initially observing the absence of rps1i25g2 in O. petiolatum we investigated three additional Ophioglossum species: O. palmatum, O. pedunculosum, and O. vulgatum. The absence of rps1i25g2 in all of them indicates an early loss, likely a molecular synapomorphy of the genus Ophioglossum. This observation is particularly interesting given that O. palmatum has alternatively been placed as Cheiroglossa palmata in a genus of its own (Hauk et al. 2003) and that the related species Helminthostachys zeylanica and Botrychium lunaria within the Ophioglossales have retained the intron rps1i25g2.
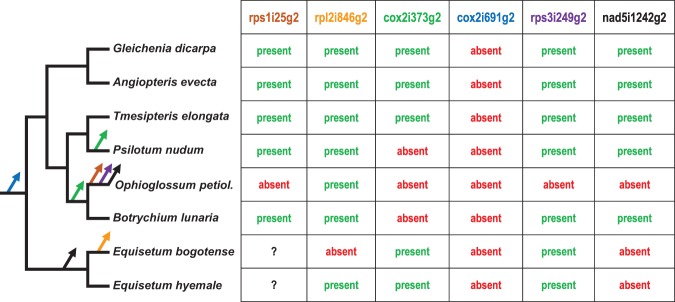
We noted that another mitochondrial intron, nad5i1242g2, present in most monilophytes, has previously also been found to be lost in Ophioglossum and Equisetum (Vangerow et al. 1999). This raised the question if intron losses may be a characteristic feature of basal (eusporangiate) fern lineages, especially in the genus Ophioglossum. We therefore exemplarily checked the intron status of three other selected mitochondrial group II introns—cox2i373g2, cox2i691g2, and rps3i249g2—by PCR analysis in Equisetum giganteum, Botrychium lunaria, Ophioglossum petiolatum, Psilotum nudum, Tmesipteris elongata, Angiopteris evecta, and Gleichenia dicarpa (Fig. 5).
FIGURE 5.
Losses of mitochondrial introns in early-branching fern lineages. The absence of the six mitochondrial introns rps1i25g2, rpl2i846g2, cox2i373g2, cox2i691g2, rps3i249g2, and nad5i1242g2 is parsimoniously explained by losses along branches as indicated by arrows in the respective colors.
Intron cox2i691g2 is absent in all fern lineages, suggesting that the loss of this intron is a molecular synapomorphy of all monilophytes. Intron cox2i373g2, however, is present in the investigated taxa including Tmesipteris elongata (Psilotales), but absent in Ophioglossum and Psilotum nudum, providing a further example of mitochondrial intron loss in Ophioglossum (and Psilotum).
Group II intron rps3i249g2 is present in Phlegmariurus squarrosus (Liu et al. 2012), Adiantum capillus-veneris (Bonavita and Regina 2016), and in gymnosperms (Ran et al. 2010; Regina and Quagliariello 2010), but it has been lost at least three times among the latter (Ran et al. 2010). The absence of this intron was also reported for Psilotum nudum (Regina and Quagliariello 2010). According to our analysis, however, rps3i249g2 is present in most ferns investigated including Psilotum. Instead, we find it lacking in Ophioglossum. To check the previously reported absence of rps3i249g2 in Psilotum, we performed a BLAST search with the annotated sequence (EU516362) and found that it reveals strong similarities to gymnosperm sequences but not to the fern homologs.
Taken together we indeed find evidence for an enhanced loss of mitochondrial introns in the early-branching monilophytes and in particular in the genus Ophioglossum (Fig. 5).
Monilophyte phylogeny incorporating rps1 as new phylogenetic locus
The rpl2 gene and its intron have recently been included in a multigene analysis together with five chloroplast (atpA, atpB, matK, rbcL, rps4) and three mitochondrial loci (atp1, nad2, nad5) to infer the phylogeny of monilophytes (Knie et al. 2015). In that study it was shown that the horsetails (Equisetales) are sister to all other ferns and that the Ophioglossales/Psilotales clade is sister to a joint clade of Marattiales and leptosporangiate ferns. This and previous studies, however, were not able to conclusively resolve the positions of the Hymenophyllales and Gleicheniales along the backbone phylogeny of monilophytes. The phylogeny of Gleicheniales, Hymenophyllales and the well-supported clade comprising the “core leptosporangiate” ferns and the Schizaeales had previously remained as an unresolved polytomy. We have now added the new rps1 data obtained here to the previous nine-gene data set. The phylogenetic analysis of the new 10-gene data set now places the Hymenophyllales and Gleicheniales as sister to each other into a joint “G(M)H” clade, i.e., Gleicheniales incl. Matoniaceae and Hymenophyllales, with a reasonable bootstrap support of 74% (Fig. 6).
FIGURE 6.
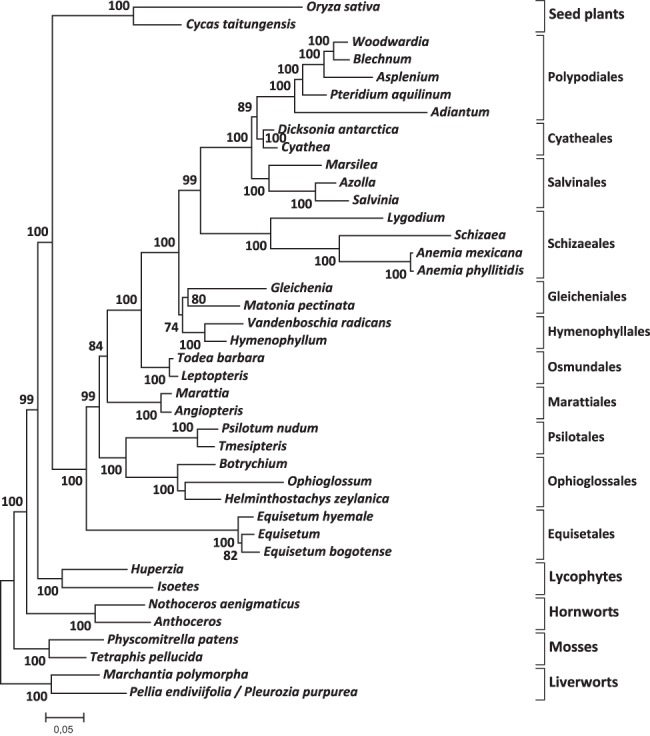
Phylogeny of monilophytes with the new rps1 locus added to a nine-gene data set (atpA-atpB-matK-rbcL-rps4-atp1-nad2-nad5-rpl2) from a previous study (Knie et al. 2015). Bootstrap values from 1000 pseudoreplicates are shown when exceeding 70.
DISCUSSION
Monilophytes (i.e., ferns sensu lato including horsetails) are the only one of the six major land plant clades still lacking a complete mitochondrial genome sequence. A fosmid cloning approach for Gleichenia dicarpa proved insufficient in our laboratory to reveal its complete chondrome and is currently being complemented with a next-generation sequencing approach (F Grewe, V Knoop, and JP Mower, unpubl.). While apparently retaining a large ancestral mitochondrial gene set, the mtDNA of Gleichenia dicarpa seems at the same time to be affected by an unparalleled amount of DNA recombination, possibly even surpassing what has been observed for the lycophytes Isoetes engelmannii and Selaginella moellendorffii (Grewe et al. 2009; Hecht et al. 2011).
Consequently, our insights into the structure of fern mitochondrial genomes are rather limited so far. Only the mitochondrial loci nad5 (Vangerow et al. 1999), atp1 (Wikström and Pryer 2005), and rpl2 (Knie et al. 2015) have previously been studied for broader samplings of ferns. Some monilophytes have also been included in a wider taxonomic sampling investigating evolution of the nad1 gene (Dombrovska and Qiu 2004). Moreover, a study in the horsetail Equisetum arvense reported on the atp9, cob, cox1, and cox2 gene structures (Bégu and Araya 2009), revealing the novel group II intron cox1i747g2 in Equisetum arvense. Another fern-specific intron is atp1i361g2 (Wikström and Pryer 2005), which may have been gained in the common ancestor of leptosporangiate ferns and the Marattiales (Grewe et al. 2013; Knie et al. 2015).
We here report on yet another novel monilophyte-specific group II intron, rps1i25g2, present in all monilophyte orders. We were unable to retrieve rps1 sequences via PCR or in the transcriptome data from horsetails (Equisetales). Hence, it remains unclear at present whether the gain of rps1i25g2 occurred in the monilophyte stem lineage or after the earliest dichotomy in the monilophyte phylogeny separating the Equisetales (Fig. 2).
One possible explanation for gain of a new mitochondrial intron could be an event of horizontal gene transfer (HGT), as previously shown for the transfer of nad1 intron nad1i477g2 into Gnetum (Won and Renner 2003) and of nad5 intron nad5i230g2 in Pinus canariensis (Wang et al. 2015). These cases, however, included flanking exon sequences creating paralogous gene copies and the affected introns seem to be nonfunctional. We did not observe any evidence for a coexistence of different copies of rpl2 or rps1 in our broad sampling of monilophytes. Yet another case of HGT are the multiple independent transfers of cox1 intron cox1i729g1, likely originating from a fungal source in diverse angiosperms (Vaughn et al. 1995; Cho et al. 1998). We did not find any evidence that a similar scenario could explain the existence of rps1i25g2 in monilophytes, but a one-time intron transfer from an as yet unidentified source very early in monilophyte evolution remains a remote possibility.
As a yet further alternative hypothesis, intron rps1i25g2 could have been gained by a recombination event placing a copy of the rpl2i846g2 intron at the 5′ end of the rps1 gene creating a new small upstream exon. However, there is no discernible sequence similarity outside of the paralogous introns. The rps1 amino terminus is highly conserved in comparison to mosses and liverworts (Supplemental Fig. 2), whereas no similarity to rpl2 upstream of rpl2i846g2 is discernible, either on nucleotide or amino acid level or after the necessary frameshift shifting the new intron from phase 0 to phase 1. Moreover, no traces of rpl2 similarity are identified in the available mitochondrial DNA sequences upstream of rps1 in Gleichenia dicarpa.
Given the striking similarity of rps1i25g2 to its paralog rpl2i846g2, we find a scenario invoking a reverse-splicing copying of the rpl2 intron into the rps1 locus much more likely. The case of rps1i25g2 and its likely donor source rpl2i846g2 reveal particularly strong similarities of two paralogous group II introns both in primary sequence and in their secondary structures. We assume that an ancient version of rpl2i846g2 with exon binding sequences promiscuously targeting the rps1 gene at the new insertion site was reverse spliced (Fig. 4), followed by reverse transcription and integration into the mtDNA via recombination. Using mobile group II introns in yeast mitochondria, it has been shown previously that restoring a conserved nucleotide in the IBS1 of a retrohoming-deficient strain led to a transfer of the mobile intron in a process of retrohoming (Eskes et al. 1997). Neither rpl2i846g2 nor rps1i25g2 reveal any evidence for intron-encoded maturases (frequently carrying reverse transcriptase domains). It is unclear at present whether nuclear-encoded maturases or other proteins could have played a role and identifying any candidates is currently still hampered by a lack of high-quality genomic or transcriptomic data for monilophytes.
While we found that the intron rps1i25g2 is well conserved among monilophytes, it is absent in the genus Ophioglossum as an exception. Observing that another mitochondrial intron, nad5i1242g2, likewise highly conserved among ferns, had also previously been found to be absent in Ophioglossum and Equisetum (Vangerow et al. 1999), we checked whether the early-branching monilophyte lineages are generally prone to mitochondrial intron losses. Indeed, we found evidence for a loss of intron rps3i249g2 in Ophioglossum and for a loss of cox2i373g2 in Ophioglossales and Psilotum nudum. Intron rpl2i846g2 has previously been lost in Equisetum bogotense (Knie et al. 2015). Taken together, the observations may indeed indicate that the earliest-branching monilophyte orders, Equisetales, Psilotales, and Ophioglossales, and in particular the genus Ophioglossum, may be particularly prone to independent losses of mitochondrial introns, similar to what has previously been observed for the genus Silene among the angiosperms (Sloan et al. 2010) or Welwitschia mirabilis among the gymnosperms (Guo et al. 2016).
MATERIALS AND METHODS
Plant material and molecular work
Monilophyte material came from the Botanical Garden of the University of Ulm (Vangerow et al. 1999) or from the University of Bonn Botanical Garden (see Table 1). Total DNA was isolated using the CTAB method (Doyle and Doyle 1990) followed by RNA digestion with RNase A (Thermo Scientific/Fermentas). The mitochondrial loci were amplified by Touchdown-PCR with initial annealing temperatures starting at 55°C or 45°C and lowered to 45°C or 42°C, respectively. Elongation time varied between 1 min 30 sec and 2 min 30 sec. PCR-products were separated by gel electrophoresis using a 0.8% agarose gel, recovered with the NucleoSpin Extract II Kit (Macherey Nagel) and cloned into the pGEM-T Easy Vector (Promega). Sequencing of plasmids was performed by Macrogen Europe or GATC Biotech AG. All new sequences obtained for this study were submitted to GenBank (Table 1). PCR primer sequences are listed in Supplemental Table 2.
RNA editing analysis
For RNA editing analysis of intron sequences, two different methods were used. Total RNA was isolated by DNase I digestion (Thermo Scientific/Fermentas) of the CTAB extracts. For routine analysis the synthesis of cDNA was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific/Fermentas) in the presence of random hexamer primers. The RT-PCRs were performed using primer pairs (Supplemental Table 2) with one primer binding in the intron and one binding to editing sites in the flanking exon in order to enrich for partially matured/edited pre-mRNAs. Two amplicons per gene with primers binding in the up- and downstream exons were analyzed to cover the full length of the introns.
In an alternative analysis, “branch” primers were designed to bridge across the adenosine branch site in domain VI and the neighboring nucleotides of the spliced out lariat. The 5′ end of these primers are reverse complementary to the 5′ end of the intron and the 3′ end binds to the branching adenosine and 6- to 8-nt upstream (Supplemental Table 2). With these lariat-specific primers, cDNA synthesis was performed with the thermostable MaximaRT (Thermo Scientific/Fermentas) and incubation temperatures between 60°C and 65°C. High temperatures were used to improve the specificity of the primer and to denaturize secondary structures. For the following RT-PCR we used the lariat-specific primer and a second primer that binds near the 5′ end of the intron. As a negative control we used DNA as a template in the RT-PCR to rule out that the lariat-specific primer may bind to unspliced mRNA during the cDNA synthesis (Supplemental Fig. 3). One species per monilophyte order was analyzed by both methods. All experimentally proven RNA editing sites are listed in Supplemental Table 1. RT-PCR attempts were only successful for the species listed.
Sequence handling and phylogenetic analyses
Sequence handling and alignment analyses were done using the alignment feature of MEGA 5.05 (Tamura et al. 2011). The new rps1 sequences were fused to a previously established nine-gene data set of five chloroplast (atpA, atpB, matK, rbcL, and rps4) and four mitochondrial (atp1, nad2, nad5, and rpl2) loci (Knie et al. 2015). Gaps and missing data in the alignment were treated with the partial deletion option and a site coverage cut-off set to 12%. With these settings, every character that was not present in at least five taxa was excluded from the phylogenetic analyses. Phylogenetic trees were obtained by the maximum likelihood method using the GTR+Γ+I substitution model (Rodríguez et al. 1990) proposed after model test runs using Bayesian information criterion (Schwarz 1978) and the corrected Akaike information criterion (Burnham and Anderson 2004). Node support was determined with bootstrapping using 1000 alignment pseudoreplicates. Group II intron secondary structures were folded manually and graphic displays were produced with the VARNA software (Darty et al. 2009).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the help of Dr. Wolfram Lobin, curator of the Bonn Botanic Garden, and his team, especially Bernd Reinken, for providing fern material and thank Monika Polsakiewicz for her expert technical assistance. We also wish to thank the two anonymous reviewers for their helpful comments to improve the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.056572.116.
REFERENCES
- Adams K, Palmer JD. 2003. Evolution of mitochondrial gene content. Gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395. [DOI] [PubMed] [Google Scholar]
- Adams KL, Ong HC, Palmer JD. 2001. Mitochondrial gene transfer in pieces: fission of the ribosomal protein gene rpl2 and partial or complete gene transfer to the nucleus. Mol Biol Evol 18: 2289–2297. [DOI] [PubMed] [Google Scholar]
- Adams KL, Qiu Y, Stoutemyer M, Palmer JD. 2002. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci 99: 9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol 27: 1436–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD. 2011. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 23: 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bégu D, Araya A. 2009. The horsetail Equisetum arvense mitochondria share two group I introns with the liverwort Marchantia, acquired a novel group II intron but lost intron-encoded ORFs. Curr Genet 55: 69–79. [DOI] [PubMed] [Google Scholar]
- Bégu D, Castandet B, Araya A. 2011. RNA editing restores critical domains of a group I intron in fern mitochondria. Curr Genet 57: 317–325. [DOI] [PubMed] [Google Scholar]
- Bock R, Knoop V, ed. 2012. Genomics of chloroplasts and mitochondria. Springer, New York. [Google Scholar]
- Bonavita S, Regina TMR. 2016. The evolutionary conservation of rps3 introns and rps19-rps3-rpl16 gene cluster in Adiantum capillus-veneris mitochondria. Curr Genet 62: 173–184. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. 2004. Model selection and multimodel inference. Springer, New York. [Google Scholar]
- Carrillo C, Bonen L. 1997. RNA editing status of nad7 intron domains in wheat mitochondria. Nucleic Acids Res 25: 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo C, Chapdelaine Y, Bonen L. 2001. Variation in sequence and RNA editing within core domains of mitochondrial group II introns among plants. Mol Gen Genet 264: 595–603. [DOI] [PubMed] [Google Scholar]
- Castandet B, Choury D, Bégu D, Jordana X, Araya A. 2010. Intron RNA editing is essential for splicing in plant mitochondria. Nucleic Acids Res 38: 7112–7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw S, Shih AC, Wang D, Wu Y, Liu S, Chou T. 2008. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol Biol Evol 25: 603–615. [DOI] [PubMed] [Google Scholar]
- Cho Y, Qiu YL, Kuhlman P, Palmer JD. 1998. Explosive invasion of plant mitochondria by a group I intron. Proc Natl Acad Sci 95: 14244–14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. 2009. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25: 1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrovska O, Qiu Y. 2004. Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol Phylogenet Evol 32: 246–263. [DOI] [PubMed] [Google Scholar]
- Doyle LL, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Eskes R, Yang J, Lambowitz AM, Perlman PS. 1997. Mobility of yeast mitochondrial group II introns: engineering a new site specificity and retrohoming via full reverse splicing. Cell 88: 865–874. [DOI] [PubMed] [Google Scholar]
- Farré J, Aknin C, Araya A, Castandet B. 2012. RNA editing in mitochondrial trans-introns is required for splicing. PLoS One 7: e52644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field A, Bostock P. 2013. New and existing combinations in Palaeotropical Phlegmariurus (Lycopodiaceae) and lectotypification of the type species Phlegmariurus phlegmaria (L.) T.Sen & U.Sen. PhytoKeys 20: 33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe F, Viehoever P, Weisshaar B, Knoop V. 2009. A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res 37: 5093–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe F, Herres S, Viehöver P, Polsakiewicz M, Weisshaar B, Knoop V. 2011. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res 39: 2890–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe F, Guo W, Gubbels EA, Hansen AK, Mower JP. 2013. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol Biol 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth-Malonek M, Pruchner D, Grewe F, Knoop V. 2005. Ancestors of trans-splicing mitochondrial introns support serial sister group relationships of hornworts and mosses with vascular plants. Mol Biol Evol 22: 117–125. [DOI] [PubMed] [Google Scholar]
- Guo W, Grewe F, Fan W, Young GJ, Knoop V, Palmer JD, Mower JP. 2016. Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol Biol Evol 33: 1448–1460. [DOI] [PubMed] [Google Scholar]
- Hauk WD, Parks CR, Chase MW. 2003. Phylogenetic studies of Ophioglossaceae: evidence from rbcL and trnL-F plastid DNA sequences and morphology. Mol Phylogenet Evol 28: 131–151. [DOI] [PubMed] [Google Scholar]
- Hecht J, Grewe F, Knoop V. 2011. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol 3: 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knie N, Fischer S, Grewe F, Polsakiewicz M, Knoop V. 2015. Horsetails are the sister group to all other monilophytes and Marattiales are sister to leptosporangiate ferns. Mol Phylogenet Evol 90: 140–149. [DOI] [PubMed] [Google Scholar]
- Knoop V. 2004. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet 46: 123–139. [DOI] [PubMed] [Google Scholar]
- Knoop V. 2010. Looking for sense in the nonsense: a short review of non-coding organellar DNA elucidating the phylogeny of bryophytes. Bryophyt Divers Evol 31: 51. [Google Scholar]
- Knoop V. 2013. Plant mitochondrial genome peculiarities evolving in the earliest vascular plant lineages. J Syst Evol 51: 1–12. [Google Scholar]
- Li L, Wang B, Liu Y, Qiu Y. 2009. The complete mitochondrial genome sequence of the hornwort Megaceros aenigmaticus shows a mixed mode of conservative yet dynamic evolution in early land plant mitochondrial genomes. J Mol Evol 68: 665–678. [DOI] [PubMed] [Google Scholar]
- Lippok B, Brennicke A, Wissinger B. 1994. Differential RNA editing in closely related introns in Oenothera mitochondria. Mol Gen Genet 243: 39–46. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang B, Cui P, Li L, Xue J, Yu J, Qiu Y. 2012. The mitochondrial genome of the lycophyte Huperzia squarrosa: the most archaic form in vascular plants. PLoS One 7: e35168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N, Hung L, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M, et al. 2014. Data access for the 1,000 Plants (1KP) project. GigaScience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil BA, Semper C, Zimmerly S. 2016. Group II introns. Versatile ribozymes and retroelements. Wiley Inerdiscip Rev RNA 7: 341–355. [DOI] [PubMed] [Google Scholar]
- Michel F, Ferat JL. 1995. Structure and activities of group II introns. Annu Rev Biochem 64: 435–461. [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. 1989. Comparative and functional anatomy of group II catalytic introns—a review. Gene 82: 5–30. [DOI] [PubMed] [Google Scholar]
- Mower JP, Case AL, Floro ER, Willis JH. 2012. Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol Evol 4: 670–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenkott B, Yamaguchi K, Tsuji-Tsukinoki S, Knie N, Knoop V. 2014. Chloroplast RNA editing going extreme: more than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 20: 1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruchner D, Beckert S, Muhle H, Knoop V. 2002. Divergent intron conservation in the mitochondrial nad2 gene: signatures for the three bryophyte classes (mosses, liverworts, and hornworts) and the lycophytes. J Mol Evol 55: 265–271. [DOI] [PubMed] [Google Scholar]
- Pryer KM, Schneider H, Smith AR, Cranfill R, Wolf PG, Hunt JS, Sipes SD. 2001. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 409: 618–622. [DOI] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM. 1998. The architectural organization and mechanistic function of group II intron structural elements. Curr Opin Struct Biol 8: 301–308. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Palmer JD. 2004. Many independent origins of trans splicing of a plant mitochondrial group II intron. J Mol Evol 59: 80–89. [DOI] [PubMed] [Google Scholar]
- Qiu YL, Cho Y, Cox JC, Palmer JD. 1998. The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature 394: 671–674. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Li L, Wang B, Chen Z, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J, et al. 2006. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci 103: 15511–15516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran J, Gao H, Wang X. 2010. Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer II and further evidence for the phylogeny of gymnosperms. Mol Phylogenet Evol 54: 136–149. [DOI] [PubMed] [Google Scholar]
- Regina TMR, Quagliariello C. 2010. Lineage-specific group II intron gains and losses of the mitochondrial rps3 gene in gymnosperms. Plant Physiol Biochem 48: 646–654. [DOI] [PubMed] [Google Scholar]
- Rodríguez F, Oliver JL, Marín A, Medina JR. 1990. The general stochastic model of nucleotide substitution. J Theor Biol 142: 485–501. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Moreno L, González VM, Benjak A, Martí MC, Puigdomènech P, Aranda MA, Garcia-Mas J. 2011. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genomics 12: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G. 1978. Estimating the dimension of a model. Ann Stat 6: 461–464. [Google Scholar]
- Simon DM, Clarke NAC, McNeil BA, Johnson I, Pantuso D, Dai L, Chai D, Zimmerly S. 2008. Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA 14: 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR. 2010. Extensive loss of RNA editing sites in rapidly evolving Silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics 185: 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Hausner G, Zimmerly S. 2001. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7: 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangerow S, Teerkorn T, Knoop V. 1999. Phylogenetic information in the mitochondrial nad5 gene of Pteridophytes: RNA editing and intron sequences. Plant Biol 1: 235–243. [Google Scholar]
- Vaughn JC, Mason MT, Sper-Whitis GL, Kuhlman P, Palmer JD. 1995. Fungal origin by horizontal transfer of a plant mitochondrial group I intron in the chimeric coxI gene of Peperomia. J Mol Evol 41: 563–572. [DOI] [PubMed] [Google Scholar]
- Volkmar U, Groth-Malonek M, Heinrichs J, Muhle H, Polsakiewicz M, Knoop V. 2012. Exclusive conservation of mitochondrial group II intron nad4i548 among liverworts and its use for phylogenetic studies in this ancient plant clade. Plant Biol (Stuttg) 14: 382–391. [DOI] [PubMed] [Google Scholar]
- Wang B, Climent J, Wang X. 2015. Horizontal gene transfer from a flowering plant to the insular pine Pinus canariensis (Chr. Sm. Ex DC in Buch). Heredity 114: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward BL, Anderson RS, Bendich AJ. 1981. The mitochondrial genome is large and variable in a family of plants (Cucurbitaceae). Cell 25: 793–803. [DOI] [PubMed] [Google Scholar]
- Wikström N, Pryer KM. 2005. Incongruence between primary sequence data and the distribution of a mitochondrial atp1 group II intron among ferns and horsetails. Mol Phylogenet Evol 36: 484–493. [DOI] [PubMed] [Google Scholar]
- Won H, Renner SS. 2003. Horizontal gene transfer from flowering plants to Gnetum. Proc Natl Acad Sci 100: 10824–10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Liu Y, Li L, Wang B, Qiu Y. 2010. The complete mitochondrial genome sequence of the hornwort Phaeoceros laevis: retention of many ancient pseudogenes and conservative evolution of mitochondrial genomes in hornworts. Curr Genet 56: 53–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.