Abstract
Meiosis is a highly regulated process by which genetic information is transmitted through sexual reproduction. It encompasses unique mechanisms that do not occur in vegetative cells, producing a distinct, well-regulated meiotic transcriptome. During vegetative growth, many meiotic genes are constitutively transcribed, but most of the resulting mRNAs are rapidly eliminated by the Mmi1-MTREC (Mtl1-Red1 core) complex. While Mmi1-MTREC targets premature meiotic RNAs for degradation by the nuclear 3′–5′ exoribonuclease exosome during mitotic growth, its role in meiotic gene expression during meiosis is not known. Here, we report that Red5, an essential MTREC component, interacts with pFal1, an ortholog of eukaryotic translation initiation factor eIF4aIII in the fission yeast Schizosaccharomyces pombe. In mammals, together with MAGO (Mnh1), Rnps1, and Y14, elF4AIII (pFal1) forms the core of the exon junction complex (EJC), which is essential for transcriptional surveillance and localization of mature mRNAs. In fission yeast, two EJC orthologs, pFal1 and Mnh1, are functionally connected with MTREC, specifically in the process of meiotic gene expression during meiosis. Although pFal1 interacts with Mnh1, Y14, and Rnps1, its association with Mnh1 is not disrupted upon loss of Y14 or Rnps1. Mutations of Red1, Red5, pFal1, or Mnh1 produce severe meiotic defects; the abundance of meiotic transcripts during meiosis decreases; and mRNA maturation processes such as splicing are impaired. Since studying meiosis in mammalian germline cells is difficult, our findings in fission yeast may help to define the general mechanisms involved in accurate meiotic gene expression in higher eukaryotes.
Keywords: MTREC, elF4AIII, MAGO, meiosis, splicing, exon junction complex
INTRODUCTION
Proper meiosis ensures that sexually reproducing organisms accurately transmit genetic information to the next generation. While nearly all diploid eukaryotic cells contain the genetic information required to undergo meiosis, only a few very specialized cells or cells exposed to particular environmental stimuli will enter meiosis, suggesting that the switch from mitosis to meiosis is precisely regulated by distinct transcriptional programs. Indeed, transcriptome analysis revealed that a massive transcriptional shift is required to enter the meiotic phase (Mata et al. 2002; Harigaya and Yamamoto 2007). In the fission yeast Schizosaccharomyces pombe (S. pombe), expression of ∼25% of the genome increases at least fourfold during meiosis (Mata et al. 2002; Chalmel et al. 2007).
During mitotic growth, most meiotic genes are silenced. Aberrant expression of meiotic genes results in cell-cycle defects in yeast and is a hallmark of some human tumors (Harigaya and Yamamoto 2007; Fratta et al. 2011). In fact, many cancers are detected by the presence of meiotic proteins. For example, the cancer/testis (CT) antigens represent a large family of meiotic proteins that are normally expressed in the spermatocytes of the testis but are often detected in cancerous tumors as well (Simpson et al. 2005). Hence, ensuring that meiotic genes remain silenced during vegetative growth is important. In fission yeast, many meiotic genes are constitutively transcribed during both vegetative growth and meiosis (Chen et al. 2011). However, in vegetative cells, their transcripts are rapidly eliminated (Harigaya et al. 2006; Harigaya and Yamamoto 2007). The best understood mechanism that selectively eliminates the meiotic transcripts during vegetative growth is the Mmi1 pathway (Harigaya et al. 2006; Chen et al. 2011). This pathway specifically targets transcripts containing the determinant of selective removal (DSR) sequence, which consists of hexameric U(U/C)AAAC repeats (Yamashita et al. 2012). Mmi1 is a sequence-dependent RNA-binding protein (Chen et al. 2011) that binds DSR-containing transcripts. Biochemical analysis has shown that Mmi1 associates with Erh1 protein to form EMC (Mmi1-Erh1 complex) (Sugiyama et al. 2016), and recruits the Red1-Mtl1-containing MTREC complex (Lee et al. 2013; Egan et al. 2014). MTREC subsequently recruits the exosome, a 3′–5′ exoribonuclease complex, to degrade these Mmi1-associated meiotic mRNAs in the nucleus during vegetative growth (Lee et al. 2013; Egan et al. 2014; Zhou et al. 2015).
The exosome is an evolutionarily conserved protein complex that functions as one of the main RNA-degradation systems in eukaryotes (Houseley et al. 2006). RNA elimination by the exosome requires 3′-processing factors including the poly(A) polymerase Pla1 and the poly(A)-binding protein Pab2, both of which are associated with MTREC (St-Andre et al. 2010; Yamamoto 2010; Egan et al. 2014). Red5, which was found to interact with Pab2 and function in RNA degradation (Sugiyama et al. 2013), copurified with MTREC in another study, and appears to be one of its core subunits (Egan et al. 2014; Zhou et al. 2015). Red5 and the other MTREC subunits Red1, Mtl1, and Iss10 are all required for degradative function (Egan et al. 2014; Zhou et al. 2015). MTREC is the S. pombe ortholog of the human NEXT complex (Lee et al. 2013; Zhou et al. 2015), suggesting a similar conserved mechanism for RNA degradation between S. pombe and higher eukaryotes (Lubas et al. 2011; Andersen et al. 2013). In addition to MTREC-associated RNA degradation, Mmi1 restricts splicing and RNA 3′ processing during mitotic growth (Chen et al. 2011). In cells with functional Mmi1-DSR degradation systems, meiotic RNAs remain unspliced and unpolyadenylated during vegetative growth, leading to their rapid degradation (Chen et al. 2011). During meiosis, or in mutants lacking Mmi1, meiotic transcripts are stabilized and are processed into fully mature mRNAs (Chen et al. 2011).
Similar to RNA polymerase II (RNAP II) mitotic transcripts, meiotic mRNAs are processed from longer precursors and packed into mRNA/protein particles (mRNPs) before being transported to the cytoplasm for translation. mRNP formation requires 5′ capping, splicing, polyadenylation at the 3′ end, and loading of mRNA packaging and export factors. These processes are precisely regulated, which may allow for multiple gene products such as the alternative splicing common in metazoans (Maniatis and Tasic 2002). In fission yeast, although ∼47% of genes contain introns, no evidence of alternative splicing that results in a single gene coding for multiple proteins has been found (Wood et al. 2002; Rhind et al. 2011). It was believed that regulatory splicing in fission yeast is limited to splice versus do-not-splice determinations (Engebrecht et al. 1991; Chen et al. 2011). About 10% of known meiotic transcripts contain introns, are spliced only during meiosis, and remain unspliced during vegetative growth (Averbeck et al. 2005). In addition, their splicing is concurrent with 3′ end maturation, and polyadenylated RNAs are not detected during vegetative growth (Potter et al. 2012). However, the exact mechanism of how specific splicing events are determined and controlled to enable meiotic transcripts to mature remains unknown (Wilhelm et al. 2008).
Mmi1-DSR-mediated degradation is inactivated during meiosis, thus Mmi1-regulated meiotic genes are efficiently expressed (Yamamoto 2010). This process relies on the coordinated action of the long noncoding RNA meiRNA and the protein Mei2 (Watanabe and Yamamoto 1994; Yamamoto 2010). During meiosis, Mei2 is phosphorylated and binds to meiRNA, forming a complex that can efficiently sequester Mmi1 from binding to DSR-containing transcripts, thereby inactivating MTREC-mediated degradation (Watanabe and Yamamoto 1994; Harigaya et al. 2006; Ding et al. 2012). During vegetative growth, Mmi1 colocalizes with Red1, a core subunit of MTREC. While Mmi1 is sequestered by meiRNA during meiosis, Red1 does not colocalize with meiRNA foci (Mei2 dot) during meiotic sequestration (Sugiyama and Sugioka-Sugiyama 2011), calling into question the function of MTREC during meiosis.
Here we show that Spac1f5.10 (pFal1, the S. pombe translation-initiation factor Four A Like and ortholog of eIF4AIII) interacts with the MTREC subunit Red5 to regulate the crucial splicing of meiotic transcripts during meiosis. pFal1 is a predicted Asp-Glu-Ala-Asp (DEAD)-box ATP-dependent RNA helicase (Li et al. 1999; Caruthers et al. 2000). eIF4A family proteins contain prototypical DEAD-box domains (Li et al. 1999; Lu et al. 2014). Of the three major isoforms in yeast and mammals, eIF4AI and eIF4AII are found primarily in the cytoplasm and function similarly in translational initiation (Tuteja et al. 2008; Lu et al. 2014). In contrast, eIF4AIII is a third, functionally distinct eIF4A protein that localizes mainly to the nucleus (Li et al. 1999). Like all DEAD-box family proteins, eIF4AIII exhibits nucleic-acid binding activity (Cordin et al. 2006). In mammals, eIF4AIII is a member of the exon junction complex (EJC), acting as a molecular clamp mediated by its sequence-independent nucleic-acid binding activity (Shibuya et al. 2004). It remains associated with RNA after splicing to serve as a binding platform for the other EJC proteins (Shibuya et al. 2004). Essentially, its stable binding works as a placeholder, marking RNA species that have completed their splicing (Le Hir et al. 2001; Chan et al. 2004). Constitutive binding of the EJC to RNA molecules enables recruitment of other proteins for various downstream RNA metabolic processes (Shibuya et al. 2004; Lu et al. 2014). S. pombe contains putative orthologs of the EJC components eIF4aIII, MAGO, Y14, and RNPS1, but lacks MLN51 (Wen and Brogna 2010).
We demonstrate that accurate meiotic gene expression during meiosis requires EJC orthologs pFal1 and Mnh1, and MTREC members Red5 and Red1. Loss of any of these proteins causes defective sporulation in diploid cells accompanied by an impairment of meiotic gene splicing. Red5 interacts with pFal1, which is associated with the other S. pombe orthologs of EJC, including Mnh1, Y14, and Rnps1. Unlike mammalian EJC, which cannot be assembled without Y14, we found that in S. pombe, the interaction between pFal1 and Mnh1 remains even in the absence of Y14 or Rnps1. Although additional evidence is necessary to fully address whether these EJC orthologs form an EJC-like complex in fission yeast, our findings pinpoint a cooperative role of elF4aIII, MAGO, MTREC, and the exosome in regulating the maturation of meiotic transcripts when meiosis is induced.
RESULTS
Cells without pFal1 remain viable, but display severe growth defects
In budding yeast, the eIF4AIII ortholog Fal1 is an essential protein with reported roles in rRNA processing (Kressler et al. 1997; Alexandrov et al. 2011). In S. pombe, prior to this study, at least one attempt to delete spac1f5.10+ (S. pombe Fal1, pFal1) failed (Wen and Brogna 2010), leading the authors to assume that pFal1 is essential for cell viability like its orthologs in Drosophila melanogaster and S. cerevisiae (Kressler et al. 1997; Palacios et al. 2004). In contrast, a genome-wide deletion analysis obtained viable pfal1Δ cells (Kim et al. 2010; Hayles et al. 2013). The strain library is commercially available, although we were not able to confirm the deletion of pfal1+ from it using standard genotyping methods. Therefore, the precise function of pFal1 in S. pombe remains obscure.
To study the role of pFal1, we generated pfal1Δ cells by replacing the entire open-reading frame of pfal1+ with an antibiotic-selectable marker through homologous recombination. To assay growth, cells were serially diluted 10-fold, plated on rich media (YEA), and incubated at different temperatures for 3 d. While viable, pfal1Δ showed a strong growth defect at all three temperatures tested (Fig. 1A). This phenotype is distinct from the deletion of budding yeast Fal1, which is lethal, suggesting a possible divergence in function between the two orthologs.
FIGURE 1.
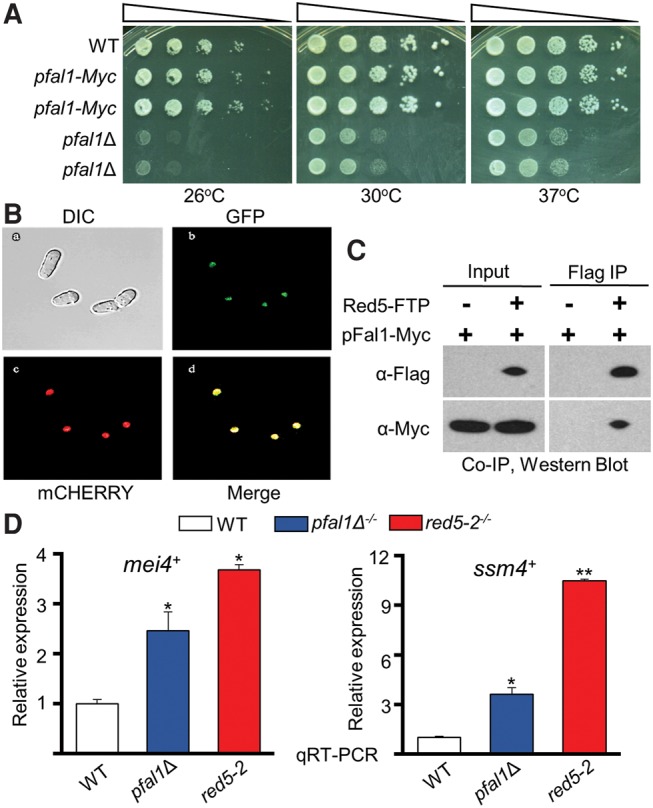
pFal1 localizes to a chromatin-rich region in the nucleus and interacts with Red5. (A) pfal1Δ haploid cells have a severe growth defect. Cells were serially diluted 10-fold, plated on rich media (YEA), and incubated at the indicated temperatures for 3 d. (B) pFal1-GFP colocalizes with Hta1-mCherry in confocal images of live pombe cells. (C) pFal1 interacts with Red5 as determined by Flag Co-IP and Western blot using antibodies as indicated. Immunoprecipitated samples were treated with 20 µg of RNase A/H mixture and 33 µg of DNase I before the final wash. (D) qRT-PCR analysis shows increased mei4+ and ssm4+ transcripts relative to act1+ normalized to wild type (WT = 1), during vegetative growth in diploid cells containing red5-2 or pfal1Δ. Asterisks denote a significant difference comparing each sample with wild type, (*) P ≤ 0.05 and (**) P ≤ 0.005.
pFal1 localizes to a distinct nuclear compartment
In S. cerevisiae, Fal1 primarily localizes in the nucleolus where it is required for the maturation of 40S ribosomal RNA (Kressler et al. 1997). To analyze the cellular localization of pFal1 in S. pombe, we created a green fluorescent protein (GFP)-tagged allele of pfal1+. This allele was combined with a mCHERRY-tagged allele of the histone protein hta1+. Visualization of a fluorescently marked histone protein allows the detection of chromatin-containing nuclear compartments. When imaged via confocal microscopy, pFal1-GFP colocalized with Hta1-mCHERRY (Fig. 1B), indicating that in S. pombe, pFal1 localizes to chromatin-containing regions of the nucleus and is not restricted to the nucleolus. The viability of pfal1Δ cells and its different pattern of nuclear localization compared to Fal1 further indicate that pFal1 may play roles distinct from those of Fal1.
pFal1 interacts with Red5
pFal1 has previously been implicated in association with Red5, a component of the MTREC complex involved in meiotic mRNA elimination, although the function of pFal1 was not studied (Egan et al. 2014; Zhou et al. 2015). We confirmed this interaction using coimmunoprecipitation (Co-IP) followed by Western blot (Fig. 1C). To detect pFal1 by Western blot using a commercially available antibody, we added a Myc epitope tag to the carboxyl terminus of pFal1 to generate a pfal1-Myc allele. Unlike pfal1Δ, the pfal1-Myc allele is fully functional, and cells carrying pFal1-Myc have no growth defect at any temperatures examined by 10-fold serial dilution assay on rich media (Fig. 1A). The pfal1-Myc allele was further combined with a tagged Red5 allele, red5-FTP. FTP is a tandem affinity epitope tag containing 3× Flag and a protein A motif, separated by a Tobacco Etch Virus (TEV) protease cleavage site. We performed Co-IP by precipitating Red5-FTP with IgG-sepharose followed by Western blot using anti-Myc antibody. Interaction of Red5 and pFal1 was confirmed as pFal1-Myc and was detected in the Red5-FTP precipitated sample, but not in the mock control (Fig. 1C). Notably, the interaction was not sensitive to RNase/DNase treatment, indicating that it is mediated through protein–protein association.
Loss of pFal1 impairs elimination of meiotic transcripts during vegetative growth
Red5 is a subunit of the MTREC complex that targets DSR-containing meiotic transcripts for degradation during mitotic growth (Lee et al. 2013; Egan et al. 2014). Red5 is essential. Conditional loss of function of Red5 (red5-2) reduces the efficacy of the Mmi1-DSR system, resulting in the accumulation of meiosis-specific transcripts during vegetative growth at restrictive conditions (Sugiyama et al. 2013; Egan et al. 2014). The interaction between pFal1 and Red5 indicates that pFal1 may also contribute to meiotic gene elimination during vegetative growth. To test this possibility, we compared the abundance of meiotic transcripts in pfal1Δ cells with that of red5-2, the same allele used in previous studies (Sugiyama et al. 2013). We chose to study mei4+ and ssm4+ transcripts because they both contain a DSR region (Chen et al. 2011). These transcripts are known targets of Mmi1 binding and are subsequently degraded during vegetative growth (Yamashita et al. 2012). In agreement with previous findings, we observed an increase in mei4+ and ssm4+ transcripts during vegetative growth in cells containing red5-2 (Fig. 1D). Cells without pfal1 accumulate intermediate levels of these transcripts between wild type and red5-2 (Fig. 1D). These results suggest that pFal1 plays a role in the degradation of mei4+ and ssm4+ during vegetative growth, although the effect is less pronounced than that of red5-2.
Meiotic defect(s) of pfal1 mutants
Much attention has been focused on the function of MTREC/Red5 during vegetative growth, in which the complex works together with Mmi1 to facilitate the degradation and silencing of meiotic transcripts (Lee et al. 2013; Egan et al. 2014). During meiosis/sporulation, Mmi1 is sequestered by Mei2-meiRNA so that meiotic RNAs are shielded from Mmi1-mediated RNA degradation and are thereby stabilized and translated into functional proteins important for the progression of meiosis (Yamamoto 2010). It has been shown that Red1, the core component of MTREC, does not colocalize with Mmi1 during meiosis, suggesting that MTREC may possess functions distinct from its Mmi1-DSR-associated role in meiosis (Sugiyama and Sugioka-Sugiyama 2011). In addition, defective MTREC, such as that of the red5 mutant, reduces sporulation efficiency, suggesting an essential role for MTREC in meiosis (Sugiyama et al. 2013). Yet, the exact roles of MTREC during this process have not been explored.
Since pFal1 interacts with Red5, we next sought to determine whether pfal1 mutants also exhibit a sporulation defect. Diploid cells are advantageous because they allow direct analysis of meiosis independent of the mating and conjugation steps. To induce meiosis, diploid cells were first grown in rich media to mid-log phase, then transferred to SPA sporulation media and grown for an additional 24 h before being imaged. The efficiency of meiosis was then measured by analysis of the sporulation profile.
More than 70% of wild-type diploid cells efficiently sporulate, producing tetrads of haploid spores contained in asci following meiotic induction. Diploid red5-2 cells show severe sporulation defects, with <1% of cells producing asci. Diploid pfal1Δ−/− cells show a decreased sporulation efficiency compared to wild-type cells (Fig. 2B). Less than 5% of pfal1Δ−/− cells sporulated, and ∼20% form misshapen asci on SPA medium (Fig. 2C), suggesting a meiotic defect.
FIGURE 2.
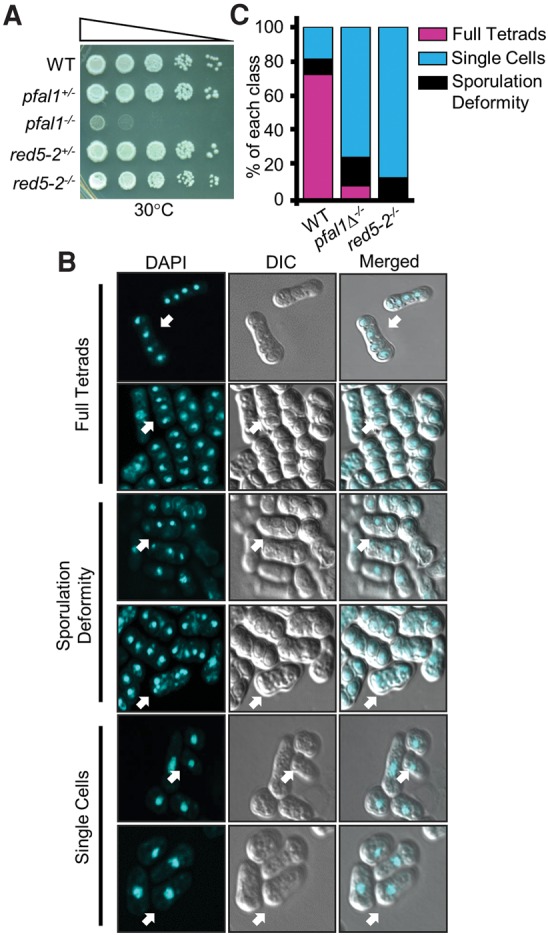
Loss of pFal1 results in defective sporulation. (A) Diploid pfal1Δ−/− cells show a growth defect similar to that in haploid pfal1Δ cells. Cells were serially diluted 10-fold, plated on rich media (YEA), and incubated at 30°C for 3 d. (B) Sporulated cells were visualized by DAPI staining and confocal microscopy. Images are presented in three classes: full tetrads (four nuclei), deformed tetrads (three or more than four nuclei), and single cells (one nucleus). Arrowheads indicate representative cells in each class. (C) Imaged cells with indicated genotypes were counted and categorized as defined in B. Bars represent the percent of each class versus the total cells.
Defective splicing of meiotic genes in pfal1 and red5 mutants
To investigate the molecular basis behind the meiotic defect in diploid pfal1Δ and red5-2 cells, we assayed the abundance and splicing patterns of meiotic transcripts during meiosis, when these genes are expressed at high levels. Since MTREC is involved in the recognition and degradation of unspliced or abnormally spliced RNAs during vegetative growth (Zhou et al. 2015), and pFal1 is the S. pombe homolog of eIF4aIII, a member of the EJC in mammals with a putative role in abnormal splicing-mediated decay (Shibuya et al. 2004), we analyzed meiotic transcripts of rec8+. Rec8 is a component of the meiotic cohesin complex and involved in linear element formation, chromosome pairing, and sister-chromatid cohesion during meiosis (Molnar et al. 1995). rec8+ is an early meiotic gene that is barely spliced and ultimately degraded by the exosome during vegetative growth, but is known to be regulated by splicing during meiosis (Averbeck et al. 2005). To assay the spliced RNAs of rec8+ during meiosis by qRT-PCR, we designed one primer of each pair to span an exon–exon junction so only the cDNA produced from spliced RNA species would be analyzed (Fig. 3A, top). qRT-PCR results show a large increase in rec8+ transcript levels during meiosis in wild-type cells, but no increase in pfal1Δ and red5-2 mutants (Fig. 3A). To determine whether this phenotype was specific to meiotic genes, we included a constitutively spliced gene, idh2+, as a control. idh2+ codes for isocitrate dehydrogenase (NAD+) subunit 2, and is constitutively expressed in vegetative and meiotic cells (Marguerat et al. 2012). As expected, the relative abundance of the idh2+ transcript is not increased in meiotic diploid pfal1Δ, red5-2, or red1Δ cells (Fig. 3A). These results demonstrate that pFal1, Red5, and Red1 are required to generate spliced rec8+ mRNA during starvation-induced meiosis.
FIGURE 3.
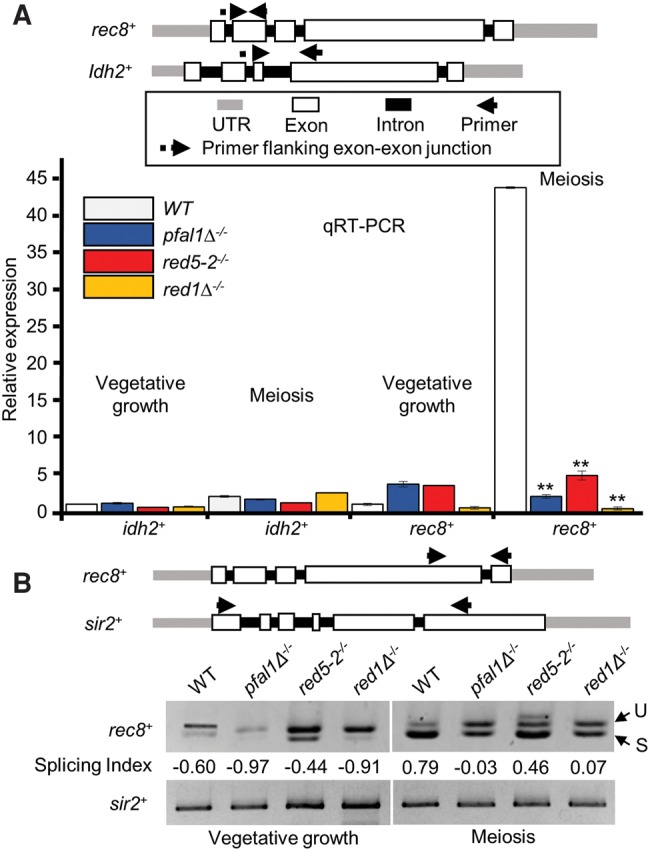
Both pFal1 and Red5 are essential for meiotic splicing of rec8+ transcripts. (A) qRT-PCR using primers flanking the exon–exon junction of an early meiotic gene (rec8+) and a nonmeiotic gene (idh2+) of indicated strains during mitosis (vegetative growth) or meiosis. Expression was normalized against act1+ and reported relative to vegetative wild-type values (WT = 1). (B) Semiquantitative RT-PCR with primers spanning two adjacent exons of rec8+ or multiple introns of sir2+. The ratios of spliced to unspliced forms of rec8+ transcripts were calculated using the splicing index. (U) Unspliced, (S) spliced.
Next, we wanted to determine whether the decrease in meiotic transcripts observed in pfal1Δ, red5-2, and red1Δ mutants was specific to meiosis, or whether these genes were affected during vegetative growth as well. As part of the MTREC complex, Red5 helps to degrade DSR-containing meiotic genes during vegetative growth. Since rec8+ RNA levels are affected by mutation of mmi1 (Averbeck et al. 2005; Chen et al. 2011), we investigated whether partial loss of Red5 function or complete loss of Red1 would result in the aberrant accumulation of rec8+ during vegetative growth. qRT- PCR results show that the loss of pfal1+ or red5+ moderately affected the level of transcript in rec8+ during vegetative growth (Fig. 3A), indicating that rec8+ is regulated by the Mmi1-MTREC system as suggested previously (Chen et al. 2011). The RNA level of the nonmeiotic, intron-containing gene idh2+ was not affected in wild-type, pfal1Δ, or red5-2 mutant cells. These results show that the decreased abundance of spliced rec8+ transcripts in pfal1Δ or red5-2 mutants relative to wild type is specific to meiosis, suggesting a meiotic role in regulatory splicing for pFal1 and Red5.
To further investigate this meiotic role of pFal1 and Red5, we assayed the amount of variant rec8+ transcripts, including unspliced and spliced isoforms, by semiquantitative RT-PCR with primers spanning two adjacent introns of rec8+. A nonmeiotic intron-containing gene, sir2+, is used as a control for total RNA. Like idh2+, sir2+ is constitutively expressed in both vegetative and in meiotic cells (Alper et al. 2013). Although we performed RT-PCR using the primers flanking the multiple introns of sir2+, we consistently detected only one band corresponding to the completely spliced form of sir2+ mRNA, suggesting that efficient splicing of the transcript occurs in each mutant tested. In contrast, we could clearly visualize the spliced versus unspliced form of rec8+ transcripts (Fig. 3B). To quantitatively evaluate the splicing efficiency, we calculated the splicing index, which is defined by band intensity [(spliced − unspliced)/(spliced + unspliced)]. The value, ranging from −1 (fully unspliced) to 1 (fully spliced), is normalized to total cDNA of each sample. During vegetative growth, all rec8+ transcripts show a negative splicing index, indicating that they are not spliced efficiently in both wild-type and mutant cells during mitosis (Fig. 3B). When meiosis is induced, however, rec8+ transcripts from wild-type cells show a splicing index value of 0.79, indicating a shift toward active splicing. Consistent with the meiotic defects, this shift did not occur efficiently in diploid homozygous pfal1 and red1 mutants, reflected by a decreased splicing index (Fig. 3). Partial loss of function of Red5 (red5-2) also exhibits a moderate reduction of the splicing index compared to wild-type cells during meiosis. These data indicate that pFal1, Red5, and Red1 are required for the efficient splicing of meiotic genes during meiosis.
pFal1 interacts with the other S. pombe EJC orthologs
The roles of pFal1 and Red5 in the splicing of meiotic genes during meiosis prompted us to study the roles of their associated proteins. pFal1 is the ortholog of eIF4AIII, which is a core component of the EJC in Drosophila and mammals (Tange et al. 2004). In addition to pFal1, S. pombe orthologs of EJC core members were identified, including Y14, MAGO (Mnh1), and RNPS1 (systematic IDs SPAC23A1.09, SPBC3B9.08c, and SPBC13G1.14c, respectively). These orthologs are not required for splicing-dependent nonsense-mediated decay (NMD) (Wen and Brogna 2010), raising the question whether a mammalian-like EJC is assembled in S. pombe. To study this question, we generated strains expressing functional epitope-tagged Mnh1, Y14, or Rnps1, and investigated their interactions by Co-IP (Fig. 4A–D). We can easily detect interaction between pFal1-Myc and Mnh1-FTP (Fig. 4A). We also observed interactions of pFal1-Myc with Rnps1-GFP and Y14-HA (Fig. 4B,D). Notably, the interaction between pFal1-Myc and Mnh1-FTP is preserved in the absence of Y14 or Rnps1 (Fig. 4A,C). In addition, pFal1-Myc coimmunoprecipitated with Rnps1-GFP with or without Y14 (Fig. 4B). In higher eukaryotes, MAGO and Y14 form a stable dimer, and without Y14, a mammalian-like EJC cannot be formed (Lau et al. 2003; Shi and Xu 2003). Our results suggest that fission yeast does not assemble a mammalian-like EJC, which is not surprising since fission yeast does not have the ortholog of the EJC stabilizing protein MLN51, and its Y14 lacks the well-conserved amino-terminus domain found in other organisms (Wen and Brogna 2010).
FIGURE 4.
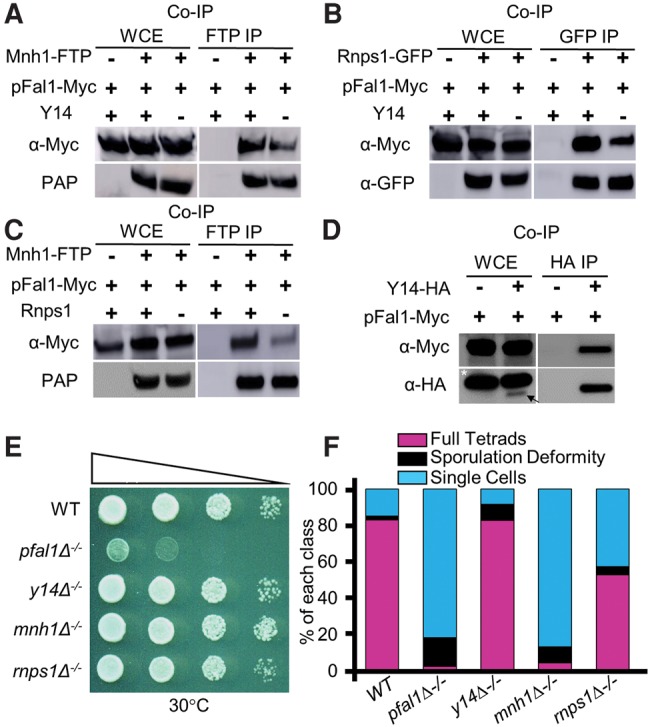
While the other EJC orthologs interact with pFal1, only Mnh1 shows a meiotic defect. (A) pFal1 interacts with Mnh1 with or without Y14. (B) pFal1 interacts with Rnps1 with or without Y14. (C) pFal1 interacts with Mnh1 with or without Rnps1. (D) pFal1 interacts with Y14. All Co-IPs were performed simultaneously using the appropriate antibodies as indicated. (*) Nonspecific binding background bands. Arrow indicates desired Y14-HA product. (E) Diploid cells with indicated genotypes were diluted 10-fold, spotted on rich medium, and grown at 30°C for 3 d. (F) Loss of either pFal1 or Mnh1 is associated with decreased sporulation efficiency, as calculated in 2C.
Mnh1/MAGO is indispensable for producing functional meiotic transcripts during meiosis
Since the other core EJC orthologs are associated with pFal1, it is possible that these proteins in S. pombe have a meiotic defect and are also involved in meiotic mRNA splicing. To investigate this, we generated deletion mutants of Y14, Mnh1, and Rnps1. Surprisingly, cells lacking these genes showed no obvious growth defects at 30°C, unlike pfal1Δ−/− (Fig. 4E), suggesting that pFal1 performs additional functions essential for cell growth. While deletion of y14 or rnps1 moderately decreases sporulation efficiency, loss of mnh1 causes severe sporulation defects (Fig. 4F), indicating an essential role of Mnh1 in meiosis.
We next investigated whether Y14, Mhn1, or Rnps1 play roles in meiotic gene splicing, similar to pFal1. Using qRT-PCR, we found a reduction in spliced meiotic transcripts of rec8+ in mnhΔ1−/− but not y14Δ−/− or rnps1Δ−/− (Fig. 5A), consistent with the degree of defective sporulation seen in these mutants. We also analyzed unspliced versus spliced transcripts of rec8+ in diploid wild-type and EJC ortholog mutants by semiquantitative RT-PCR (Fig. 5B). The splicing index of rec8+ significantly decreased in pfal1Δ−/− and mnh1Δ−/− homozygous diploid cells, while no reduction was observed in y14Δ−/− and rnps1−/− cells. These results indicate that in S. pombe, pFal1 and Mnh1 are required for maturation of meiotic RNAs.
FIGURE 5.
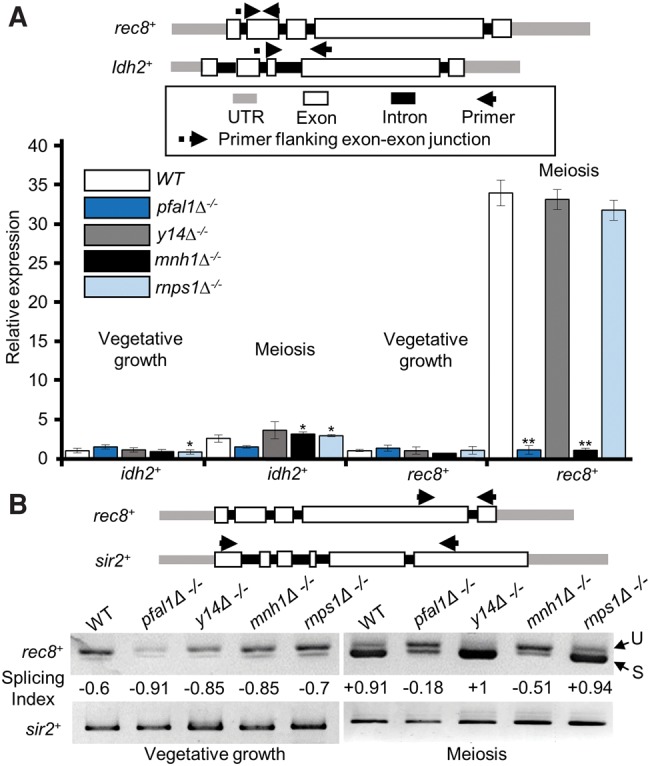
Mnh1, but not Y14 or Rnps1, is required for meiotic splicing of rec8+ transcripts. (A) The spliced rec8+ transcripts were analyzed by qRT-PCR using primers flanking the exon–exon junction of rec8+ and idh2+ in the indicated strains during vegetative growth or meiosis. Expression was normalized against act1+, and values reported are relative to vegetative wild type (WT = 1). (B) Semiquantitative RT-PCR with primers spanning two adjacent exons of rec8+ (top). The splicing efficiencies of rec8+ transcripts at the indicated strains were calculated using the splicing index. (U) Unspliced, (S) spliced. Asterisks denote significant difference comparing each sample with wild type within the same group (meiosis or vegetative growth), (*) P ≤ 0.05 and (**) P ≤ 0.005.
MTREC and pFal1 function in ensuring proper levels of meiotic transcripts during meiosis
During meiosis, Mmi1-DSR mediated degradation must be inactivated to ensure the expression of meiotic transcripts. Although Mmi1 is sequestered by meiRNA and Mei2 (Mei2 dot), it is not clear whether MTREC has any role in the processing of meiotic transcripts, ensuring their accurate maturation and quality control. While investigating the splicing of rec8+ transcripts in meiosis, we consistently obtained fewer RT-PCR products in pfal1Δ−/− cells (Figs. 3B, 5B). These results prompted us to investigate whether rec8+ transcripts generated in MTREC and the pfal1 mutants might be abnormal and thereby targeted for fast elimination. To answer this question, we used primers within one exon of rec8+ or idh2+ to measure the total RNA levels by qRT-PCR in wild-type and mutant cells lacking functional MTREC or EJC orthologs. In addition to the splicing defect, the total meiotic RNA level of rec8+ was significantly lower without either functional pFal1, Mnh1 (Fig. 6A), or MTREC components (Fig. 6B). These data suggest that pFal1, Mnh1, and MTREC ensure stable expression of meiotic genes during meiosis. The decreased RNA level of rec8+ could be a result of degradation of the transcripts due to defective splicing and/or 3′ formation. It is possible that pFal1/Mnh1 and MTREC can link exosome targeting to meiotic genes through RNA-processing factors such as those involved in splicing.
FIGURE 6.
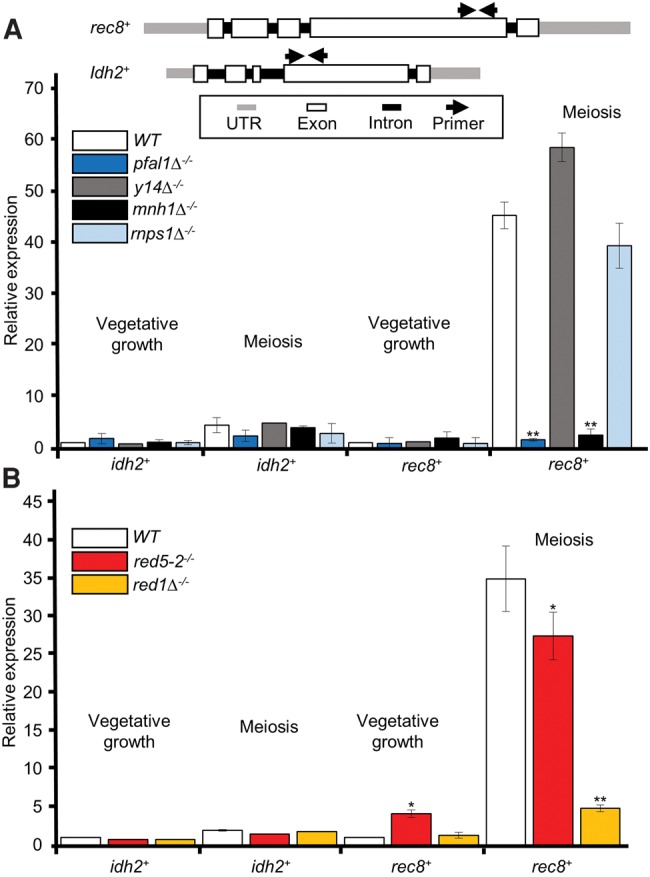
Total rec8+-transcript levels are lower in cells carrying either MTREC or EJC mutations. (A,B) Relative expression using primers within the same exon of rec8+ and idh2+ in response to deletion of EJC orthologs (A) or MTREC members (B). Expression was normalized against act1+, and values are reported relative to vegetative wild type (WT = 1). Asterisks denote significant difference comparing each sample with wild type within the same group (meiosis or vegetative growth), (*) P ≤ 0.05 and (**) P ≤ 0.005.
DISCUSSION
eIF4AIII has been studied primarily in mammalian tissue culture and Drosophila, where it functions in diverse RNA-processing pathways, including splicing, export, and degradation (Palacios et al. 2004; Shibuya et al. 2004; Malone et al. 2014). In S. pombe, the function of its ortholog had not been explored. In this study, we identified and investigated the function of S. pombe pFal1, a previously uncharacterized DEAD-box RNA helicase that plays a crucial role in the dynamic switching of meiotic gene expression during reproductive transition. We found that pFal1 is primarily localized in the chromatin-rich nuclear compartment and is required for the regulatory splicing of meiotic transcripts. Not only does it associate with MTREC via interaction with Red5, but it also associates with the other identified orthologs of EJC members Mnh1, Y14, and Rnps1. Diploid cells with defective MTREC, pFal1, or Mnh1 cannot sporulate efficiently, probably due to defective expression of meiotic transcripts, such as rec8+. Our results identify the functional coordination between MTREC, elF4aIII, and MAGO in ensuring the rapid processing and maturation of meiotic transcripts as an essential step for the proper execution of the meiotic program.
Is there an assembled EJC in fission yeast?
In mammals, the EJC mediates splicing and nonsense-mediated decay (NMD), a translation-linked process that degrades mRNAs with premature translation termination codons (PTCs) (Shibuya et al. 2004; Le Hir and Andersen 2008). Current evidence argues against the formation of a mammalian-like EJC in fission yeast. First, open reading frames (ORFs) in fission yeast contain very few introns, so NMD probably occurs independent of splicing (Wen and Brogna 2010). Indeed, EJC orthologs are not required for NMD, and the presence of an intron in the mature transcript stimulates NMD regardless of its position relative to the PTC (Wen and Brogna 2010). Second, our study shows that EJC orthologs play distinct roles during meiosis; only pFal1 and Mnh1 exhibit meiotic defects correlated with defective splicing of meiotic transcripts (Figs. 4, 5). Third, pFal1 associates with Mnh1 without Y14 or Rnps1 (Fig. 4A,C), suggesting that pFal1 and Mnh1 form a separate complex, which interacts with MTREC. Additional biochemical analysis, such as cosedimentation, is necessary to fully address whether an EJC-like complex is formed in fission yeast.
The potential role of the pFal1 in regulatory splicing
Transcriptome-wide analyses discovered unexpected levels of mRNA diversity, which is crucial for understanding the regulation of eukaryotic gene expression. Regulatory splicing helps to increase the complexity of mRNA isoforms, resulting in extensive proteome diversity. It can also introduce PTCs that induce RNA elimination by NMD (Kalsotra and Cooper 2011). Regulatory splicing contributes to key developmental processes such as sex determination in insects (Kalsotra and Cooper 2011), production of functionally distinct peptide hormones in mammals (Amara et al. 1982), and the transition of the meiotic developmental program in budding yeast (Engebrecht et al. 1991). In budding yeast, only 5% of genes contain at least one intron. In fission yeast, however, nearly 50% of genes contain an intron, and almost half of those contain multiple introns, indicating that splicing must be prevalent in this organism (Wood et al. 2002; Rhind et al. 2011). It was believed that regulatory splicing in both yeasts is focused on decisions of splicing versus not splicing (Engebrecht et al. 1991; Chen et al. 2011). A recent study in fission yeast suggests that mRNA levels can be regulated under different survival conditions by recruiting Mmi1 to “decay-promoting” introns (Kilchert et al. 2015). Some evidence also supports the presence of alternative splicing in fission yeast (Awan et al. 2013); however, it is not known whether functional proteins are produced as a result of exon skipping.
In this study, we found that only a very small proportion of rec8+ transcripts are spliced during vegetative growth; however, splicing is efficient and common during meiosis (Fig. 3). This observation is in agreement with a role for regulatory splicing in the switch to the meiotic developmental program. Without pFal1, rec8+ is not efficiently spliced during meiosis, resulting in a significantly different profile of band intensity compared to that of wild-type cells. In addition to a role for the pFal1 in splicing, we found that the total RNA of rec8+ decreases when pFal1 and Mnh1 are mutated (Fig. 6), suggesting that unspliced transcripts are most likely targeted for degradation by the MTREC–exosome pathway. We attempted to combine pfal1 or mnh1 mutant with a deletion of Rrp6 (rrp6Δ), the core catalytic subunit of the nuclear exosome, but recovered no double mutants, suggesting a synthetic lethality between these EJC orthologs and the exosome (data not shown). This genetic analysis implies that pFal1-Mnh1 and the exosome have overlapping functions essential for survival. It is likely that MTREC links the function of these proteins with the exosome.
Multifaceted functions of MTREC in mitotic growth and meiosis
To suppress meiosis in vegetative fission yeast, Mmi1 binds to DSR-containing meiotic transcripts and cooperates with MTREC to selectively degrade them (Yamanaka et al. 2010). During meiosis, relocalization of Mmi1 to the Mei2 dot allows meiotic transcripts to accumulate and be translated into meiosis-specific proteins (Yamanaka et al. 2010). The core component of MTREC, Red1, is colocalized with Mmi1 during vegetative growth but does not move with it to mei2 dot during meiosis (Sugiyama and Sugioka-Sugiyama 2011). This difference suggests that MTREC and Mmi1 are independently regulated during meiosis (Sugiyama and Sugioka-Sugiyama 2011). Although it is known that MTREC is crucial for DSR-mediated mRNA decay and interacts with splicing factors (Lee et al. 2013; Zhou et al. 2015), its exact role in meiosis was not known. Our results demonstrate its specific role in regulatory splicing of meiotic genes and support a positive role during meiosis in contrast with its role in the DSR-mediated degradation of meiotic RNAs during vegetative growth. MTREC may facilitate the maturation of meiotic transcripts by recruiting RNA-processing factors, including splicing and 3′ end formation factors. This hypothesis is supported by the observation that when rec8+ transcripts are not properly spliced during meiosis following the loss of pFal1, the abundance of rec8+ transcripts decreases (Figs. 3B, 5B, 6). Since Red5 is associated with cleavage and poly(A) machineries (Lee et al. 2013; Egan et al. 2014; Zhou et al. 2015), the protein–protein interaction between pFal1 and Red5 may provide evidence of a physical link between splicing and 3′ end processing factors (Fig. 7). The pFal1-Mnh1 may bind upstream of the exon–exon junction where it interacts with Red5 to recruit other processing factors including cleavage and poly(A) machinery. The interactions between these factors ensure their close proximity to the site of meiotic RNA transcription. Governing accurate meiotic gene expression, MTREC performs two seemingly opposite roles: DSR-mediated degradation of meiotic transcripts in mitosis and processing and maturation of meiotic transcripts in meiosis.
FIGURE 7.
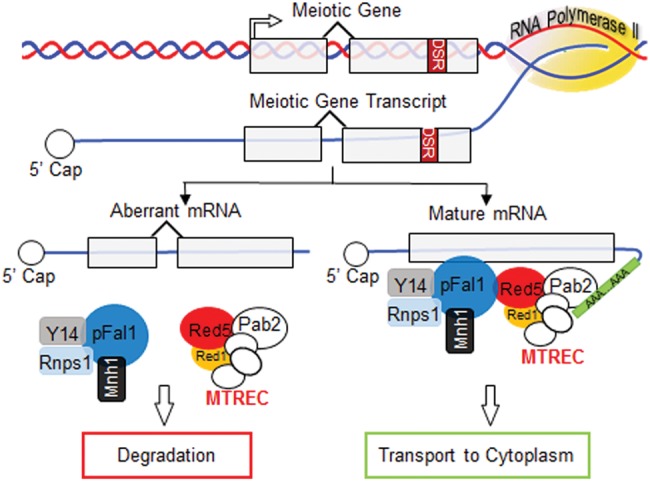
Meiotic roles for the multifaceted MTREC complex, transcript processing, and facilitation of their maturation. Abnormal meiotic transcripts generated in the absence of functional MTREC and some EJC orthologs are targeted for degradation. Physical interactions between MTREC and the EJC ensure their close proximity to accurately govern the regulation of meiotic gene expression.
Meiosis is an immensely complex process that requires the concerted effort of myriad gene products (Yamanaka et al. 2010). Elucidating the processes that control the expression of meiotic genes is necessary to understand eukaryotic sexual reproduction. Considering that MTREC and the EJC orthologs are conserved from yeast to humans (Wen and Brogna 2010; Zhou et al. 2015), our results may provide insights that are broadly applicable across eukaryotes into various RNA-processing pathways essential for meiosis.
MATERIALS AND METHODS
Strains and cell culture
Both the Red5-FLAG (ts) and Red5-FTP strains were generously provided by Dr. Tamás Fischer, Heidelberg University Biochemistry Center. All other strains were created using standard methods described previously (Bahler et al. 1998). Standard methods of S. pombe culture were followed as previously described (Moreno et al. 1991; Forsburg 2003). Vegetative cells were maintained at 30°C in YEA media. Diploid cells were created by inducing conjugation of cells of complementary h+ and Smt0 mating types. Conjugation was induced by plating the mixture of two yeast strains of opposite mating type on SPA media at 26°C for 18 h. Using strains containing the intragenically complementary ade6-M210 and ade6-M216 alleles allowed for selection for diploid cells on media lacking adenine (Szankasi et al. 1988). S. pombe cells proliferate and remain as diploids if starvation is interrupted shortly after conjugation. To this end, newly formed diploid cells were transferred into YEA media at 30°C. They grew in liquid YEA media until mid-log phase when OD at 595 nm reached 0.3–0.5. They were then harvested by centrifugation at 1000 RCF, washed with sterile double-distilled H2O, and resuspended in the same amount of SPA sporulation media. Around 1OD cells were taken at this point and counted as time 0. The rest of the cells were incubated in SPA sporulation media at 30°C for 6 h before harvest. A list of strains used in this study is available online: http://college.wfu.edu/biology/people/faculty/zhang/zhang-lab/zhang-lab-links/.
Quantifying sporulation efficiency
After sporulation, 5 µL of each culture was placed on a glass coverslip and heat-fixed at ∼70°C on a ceramic hotplate. The coverslip with attached cells was placed onto a drop of DAPI mounting media (Southern Biotech) and then imaged using a confocal microscope. Sporulation efficiency was measured using the cell counter Image J plugin as previously described (Tucker et al. 2016). Cells with one or two nuclei were considered to be growing vegetatively; cells with four nuclei were considered to form a full tetrad; and cells with any other number of nuclei were considered to have a sporulation deformity. Efficiency was reported as the percentage ratio of each group versus the total counted cells.
qRT-PCR analysis of meiotic gene expression
Total RNA was isolated from cells during vegetative growth and 6 h after placement in SPA sporulation media using the Masterpure Yeast RNA Purification Kit (Epicentre). cDNA was synthesized using M-MLV reverse transcriptase (Promega) according to the manufacturer's specifications and primed with Oligo dT18 (Fisher). The analysis of meiotic gene expression was performed by qPCR using primers specific to the mature mRNA. This was accomplished by designing one primer of each pair so that 3–7 bases on its 3′ end spanned an exon–exon junction to ensure that only spliced transcripts were detected. At least two biological repeats were performed for all experiments. A Student's t-test was performed (two-tailed distribution) in order to compare each of the indicated sample values with wild-type values of the same group (mitosis or meiosis). Error bars represent standard error (s.e.). Asterisks denote the level of significance, (*) P ≤ 0.05 and (**) P ≤ 0.005 (Figs. 1, 3A, 5A, and 6).
Semiquantitative analysis of meiotic transcripts
Semiquantitative PCR was used to examine the abundance of spliced transcripts relative to unspliced transcripts. cDNA was amplified by PCR using primers spanning at least one intron. The resulting amplicons were separated by 1% agarose electrophoresis, stained with SYBR green at a 1:10,000 dilution in TBE, and imaged on an Amersham Imager 600 RGB (GE Healthcare). Band intensity was quantified using ImageJ software, and the splicing index of band intensity [(spliced − unspliced)/(spliced + unspliced)] was calculated.
Coimmunoprecipitation (Co-IP) and Western blotting
Coimmunoprecipitation experiments were carried out as described previously (Zhang et al. 2008; Gerace and Moazed 2015). All strains were grown simultaneously in 150 mL cultures to an equal OD595 ≈ 1.5. Pelleted cells were washed with 1× PBS and 0.5 g pellets were saved. Whole-cell lysate (WCE) was obtained through bead beating (425–600 µm Glass Beads, Sigma-Aldrich) and tandem spinning followed by incubation with 2 μL Benzonase (25 unit/µL, EMD Millipore) for 2 h at 16°C. Protein A/G Agarose (Roche) coupled with the appropriate antibody were used to immunoprecipitate Myc-, HA-, and GFP-tagged proteins. FTP-tagged proteins were pulled down directly with IgG Sepharose (GE Healthcare). IP samples were incubated on an end-over-end rotator for 2 h at 4°C before being washed and eluted with 1× SDS Sample Buffer. Immunoprecipitated fractions and the equivalent of 2%–5% of the input extracts were analyzed by Western blot using anti-Flag (M2, Sigma), anti-Myc (A14, Santa Cruz), PAP (Sigma), anti-HA (Y11, Roche), or anti-GFP (Roche) antibodies.
ACKNOWLEDGMENTS
We thank Shiv Grewal and Tamas Fischer for sharing strains. We thank Glen Marrs for help with confocal image acquisition and for comments on the manuscript. We also thank Jeff Muday for figure preparation. This work is supported by the National Institutes of Health, National Cancer Institute, grant R21 (RG1200), the Pilot Research Award (K.Z.); Wake Forest University, grant U01737, the Science Research Award (K.Z.); Wake Forest University, D00641; and by the Graduate Student Fellowship, The Center for Molecular Communication and Signaling, Wake Forest University, U01740 (J.F.T.; graduate student receiver).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.055608.115.
Freely available online through the RNA Open Access option.
REFERENCES
- Alexandrov A, Colognori D, Steitz JA. 2011. Human eIF4AIII interacts with an eIF4G-like partner, NOM1, revealing an evolutionarily conserved function outside the exon junction complex. Genes Dev 25: 1078–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper BJ, Job G, Yadav RK, Shanker S, Lowe BR, Partridge JF. 2013. Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast. EMBO J 32: 2321–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara SG, Jonas V, Rosenfeld MG, Ong ES, Evans RM. 1982. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature 298: 240–244. [DOI] [PubMed] [Google Scholar]
- Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, et al. 2013. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol 20: 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J. 2005. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol Cell 18: 491–498. [DOI] [PubMed] [Google Scholar]
- Awan AR, Manfredo A, Pleiss JA. 2013. Lariat sequencing in a unicellular yeast identifies regulated alternative splicing of exons that are evolutionarily conserved with humans. Proc Natl Acad Sci 110: 12762–12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Caruthers JM, Johnson ER, McKay DB. 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci 97: 13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel F, Rolland AD, Niederhauser-Wiederkehr C, Chung SS, Demougin P, Gattiker A, Moore J, Patard JJ, Wolgemuth DJ, Jegou B, et al. 2007. The conserved transcriptome in human and rodent male gametogenesis. Proc Natl Acad Sci 104: 8346–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, Dreyfuss G. 2004. eIF4A3 is a novel component of the exon junction complex. RNA 10: 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Futcher B, Leatherwood J. 2011. The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PLoS One 6: e26804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367: 17–37. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Okamasa K, Yamane M, Tsutsumi C, Haraguchi T, Yamamoto M, Hiraoka Y. 2012. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science 336: 732–736. [DOI] [PubMed] [Google Scholar]
- Egan ED, Braun CR, Gygi SP, Moazed D. 2014. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. RNA 20: 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht JA, Voelkel-Meiman K, Roeder GS. 1991. Meiosis-specific RNA splicing in yeast. Cell 66: 1257–1268. [DOI] [PubMed] [Google Scholar]
- Forsburg SL. 2003. Growth and manipulation of S. pombe. Curr Protoc Mol Biol 64: 13.16.1–13.16.17. [DOI] [PubMed] [Google Scholar]
- Fratta E, Coral S, Covre A, Parisi G, Colizzi F, Danielli R, Nicolay HJ, Sigalotti L, Maio M. 2011. The biology of cancer testis antigens: putative function, regulation and therapeutic potential. Mol Oncol 5: 164–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace E, Moazed D. 2015. Affinity purification of protein complexes using TAP tags. Methods Enzymol 559: 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Yamamoto M. 2007. Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res 15: 523–537. [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. 2006. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50. [DOI] [PubMed] [Google Scholar]
- Hayles J, Wood V, Jeffery L, Hoe KL, Kim DU, Park HO, Salas-Pino S, Heichinger C, Nurse P. 2013. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol 3: 130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. 2011. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 12: 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C, Wittmann S, Passoni M, Shah S, Granneman S, Vasiljeva L. 2015. Regulation of mRNA levels by decay-promoting introns that recruit the exosome specificity factor Mmi1. Cell Rep 13: 2504–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 28: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, de la Cruz J, Rojo M, Linder P. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 17: 7283–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Diem MD, Dreyfuss G, Van Duyne GD. 2003. Structure of the Y14-Magoh core of the exon junction complex. Curr Biol 13: 933–941. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Andersen GR. 2008. Structural insights into the exon junction complex. Curr Opin Struct Biol 18: 112–119. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NN, Chalamcharla VR, Reyes-Turcu F, Mehta S, Zofall M, Balachandran V, Dhakshnamoorthy J, Taneja N, Yamanaka S, Zhou M, et al. 2013. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell 155: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Imataka H, Morino S, Rogers GW Jr, Richter-Cook NJ, Merrick WC, Sonenberg N. 1999. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol Cell Biol 19: 7336–7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu WT, Wilczynska A, Smith E, Bushell M. 2014. The diverse roles of the eIF4A family: you are the company you keep. Biochem Soc Trans 42: 166–172. [DOI] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43: 624–637. [DOI] [PubMed] [Google Scholar]
- Malone CD, Mestdagh C, Akhtar J, Kreim N, Deinhard P, Sachidanandam R, Treisman J, Roignant JY. 2014. The exon junction complex controls transposable element activity by ensuring faithful splicing of the piwi transcript. Genes Dev 28: 1786–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418: 236–243. [DOI] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bahler J. 2012. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151: 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147. [DOI] [PubMed] [Google Scholar]
- Molnar M, Bahler J, Sipiczki M, Kohli J. 1995. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics 141: 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E. 2004. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753–757. [DOI] [PubMed] [Google Scholar]
- Potter K, Cremona N, Sunder S, Wise JA. 2012. A dominant role for meiosis-specific 3′ RNA processing in controlling expression of a fission yeast cyclin gene. RNA 18: 1408–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xu RM. 2003. Crystal structure of the Drosophila Mago nashi–Y14 complex. Genes Dev 17: 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Sonenberg N, Moore MJ. 2004. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol 11: 346–351. [DOI] [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. 2005. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 5: 615–625. [DOI] [PubMed] [Google Scholar]
- St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F. 2010. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J Biol Chem 285: 27859–27868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Sugioka-Sugiyama R. 2011. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J 30: 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Wanatabe N, Kitahata E, Tani T, Sugioka-Sugiyama R. 2013. Red5 and three nuclear pore components are essential for efficient suppression of specific mRNAs during vegetative growth of fission yeast. Nucleic Acids Res 41: 6674–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Thillainadesan G, Chalamcharla VR, Meng Z, Balachandran V, Dhakshnamoorthy J, Zhou M, Grewal SI. 2016. Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol Cell 61: 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P, Heyer WD, Schuchert P, Kohli J. 1988. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination host spot allele ade6–M26. J Mol Biol 204: 917–925. [DOI] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. 2004. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol 16: 279–284. [DOI] [PubMed] [Google Scholar]
- Tucker JF, Ohle C, Schermann G, Bendrin K, Zhang W, Fischer T, Zhang K. 2016. A novel epigenetic silencing pathway involving the highly conserved 5′-3′ exoribonuclease Dhp1/Rat1/Xrn2 in Schizosaccharomyces pombe. PLoS Genet 12: e1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N, Vashisht AA, Tuteja R. 2008. Translation initiation factor 4A: a prototype member of dead-box protein family. Physiol Mol Biol Plants 14: 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M. 1994. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78: 487–498. [DOI] [PubMed] [Google Scholar]
- Wen J, Brogna S. 2010. Splicing-dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO J 29: 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm E, Pellay FX, Benecke A, Bell B. 2008. Determining the impact of alternative splicing events on transcriptome dynamics. BMC Res Notes 1: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Yamamoto M. 2010. The selective elimination of messenger RNA underlies the mitosis-meiosis switch in fission yeast. Proc Jpn Acad Ser B Phys Biol Sci 86: 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. 2010. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J 29: 2173–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Shichino Y, Tanaka H, Hiriart E, Touat-Todeschini L, Vavasseur A, Ding DQ, Hiraoka Y, Verdel A, Yamamoto M. 2012. Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Biol 2: 120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu J, Schermann G, Ohle C, Bendrin K, Sugioka-Sugiyama R, Sugiyama T, Fischer T. 2015. The fission yeast MTREC complex targets CUTs and unspliced pre-mRNAs to the nuclear exosome. Nat Commun 6: 7050. [DOI] [PMC free article] [PubMed] [Google Scholar]


