Abstract
Aim
To evaluate the microbial ecology of the coronal and apical segments of infected root canal systems using a complete sampling technique and next-generation sequencing.
Methodology
The roots of 23 extracted teeth with apical periodontitis were sectioned in half, horizontally, and cryo-pulverized. Bacterial communities were profiled using tagged 454 pyrosequencing of the 16S rDNA hypervariable V5–V6 region.
Results
The sequences were classified into 606 taxa (species or higher taxon), representing 24 bacterial phyla or candidate divisions and one archaeal phylum. Proteobacteria were more abundant in the apical samples (p<0.05), while Actinobacteria were in significantly higher proportions in the coronal samples. The apical samples harbored statistically significantly more taxa than the coronal samples (p=0.01), and showed a higher microbial diversity. Several taxa belonging to fastidious obligate anaerobes were significantly more abundant in the apical segments of the roots compared to their coronal counterparts.
Conclusions
Endodontic infections are more complex than reported previously. The apical part of the root canal system drives the selection of a more diverse and more anaerobe community than the coronal part. The presence of a distinct ecological niche in the apical region explains the difficulty of eradication of the infection, and emphasizes the need that new treatment approaches should be developed.
Keywords: apical, apical periodontitis, coronal, cryo-pulverization, microbiome, pyrosequencing
Introduction
Ever since microorganisms were discovered to be the cause of apical periodontitis (Kakehashi et al. 1965), researchers have explored which microorganisms are involved in this infection of the root canal system. Over 460 bacterial taxa have been associated with infected root canals (Siqueira & Rôças 2009). Although identification techniques have improved, little is known on the interplay of specific microbial communities within the root canal system. The natural habitat of the microorganisms found in infected root canals is expected to be the oral cavity. However, the unique environment of the root canal may also allow transient species, which cannot settle in the mouth, to successfully colonize the root canal. Differences in oxygen and nutrient gradients in the root canal compared to the oral cavity will promote the growth of certain microbiota and inhibit others. In apical periodontitis, the microbiota induces a host response at the point of contact, the apical foramen foremost (Kakehashi et al. 1965). Since the apical microbiota is most adjacent to the inflammation, it is likely to have the greatest influence on its course. Only by a comprehensive understanding of the disease aetiology and pathogenesis, would it be possible to develop effective prevention and treatment strategies. Additionally, the information gathered on the endodontic microbiome in close vicinity of the apical tissue will increase the knowledge on infectious diseases elsewhere in the human body (Siqueira & Rôças 2009).
In the process of determining the endodontic microbiota, the major challenge is sampling the complete root canal system. The root canal configuration does not allow a paper point or file to touch every niche, groove or tubules of the root canal system (Sathorn et al. 2007). Attempts to improve sampling with the use of fluids, files or ultrasonic agitation have not been successful (Möller 1966). The discrepancy between paper point sampling and the actual root canal microbiota was reported more than three decades ago (Akpata 1976). Samples from root canals in extracted human teeth associated with apical periodontitis were obtained using paper points and cultivated. The teeth were then crushed and the tooth particles were also cultivated. Despite negative cultures from three paper point samples per tooth, microorganisms could be cultured from the crushed tooth particles in 7 of the 20 teeth, even after mechanical preparation, irrigation and medication (Akpata 1976).
For decades, identification of a bacterial species has been dependent on its cultivability. This has been a challenge due to elaborate culturing conditions for fastidious bacteria, with more than 50% of the oral bacteria being uncultivable (Paster et al. 2001). Developments in molecular tools have allowed identification of microorganisms independently from their cultivability based on 16S rRNA gene cloning and sequencing. With that method a clone library of each sample is prepared and individual clones are sequenced one by one, using so called traditional Sanger sequencing. This process results in nearly complete 16S rRNA sequences (about 1500 nt) and allows accurate identification of the involved microorganism at strain or species level. In this way novel, uncharacterized species (phylotypes) have been added to the existing knowledge (Munson et al. 2002; Sakamoto et al. 2006). However, this method is elaborate and limited to sequencing the most predominant clones within each sample. With the advent of next-generation sequencing, researchers have developed a high-throughput, deep coverage sequencing tool (Keijser et al. 2008; Voelkerding et al. 2009). Pyrosequencing differs from the traditional Sanger sequencing in that a single pyrosequencing run allows parallel sequencing of over a million sequences. Multiple samples can be distinguished within a sequencing run by adding a unique barcode sequence to the template of each individual sample. Another major difference from the Sanger sequencing is the read length of the obtained sequences. Only a fragment (200 – 500 nt, depending on the sequencing system) of the 16S rRNA can be reliably sequenced by pyrosequencing. Therefore, the data generated by pyrosequencing does not allow as deep taxonomic classification of the obtained sequences as the data obtained by Sanger sequencing. The pyrosequencing method was demonstrated to be superior to the traditional cloning and Sanger sequencing technique in defining microbiota in seven endodontically infected teeth (Li et al. 2010). However, this study was limited by the low number of specimens and, most importantly, by the use of a traditional paper point sampling technique. In the process of determining the entire breadth of endodontic microbiota the sampling method remains the weakest link (Moller 1966, Akpata 1976, Sathorn et al. 2007).
This study aimed at 1) complete sampling of the infected root canal system, including the surrounding dentine, and 2) evaluation of the ecology of the microbiome in the apical and coronal parts of the infected roots at the coverage depth of pyrosequencing.
Materials and methods
The ethical committee of the Academic Centre for Dentistry Amsterdam (ACTA) approved the collection of extracted human teeth. The teeth (premolars and molars) were planned for extraction due to the choice of the patient not to undergo endodontic treatment. The patients agreed that the extracted teeth would be used in this research. The teeth were asymptomatic at the time of extraction and were selected according to the following criteria: 1) a clear periapical radiolucency on the periapical radiographs; 2) no visible exposure of the pulp chamber; 3) no advanced periodontal involvement.
All of the following procedures were performed under strict aseptic conditions, using sterile materials and instruments at all times. After tooth extraction, the outer surface of the tooth was wiped off repeatedly with a piece of gauze soaked in 0.5% sodium hypochlorite. The tooth was then placed in a sodium thiosulfate solution to inactivate the sodium hypochlorite, and decoronated with a diamond disc under saline cooling. The root was cut in 2 halves horizontally (coronal and apical) with the use of another diamond disc, and frozen at −80°C until cryo-pulverization. The samples were cryo-pulverized with the use of a freezer mill (Spex, Certiprep, Metuchen, NJ, USA). The powdered root segments were frozen at −80°C in 5-mL UV-treated RNA stabilization reagent (RNAlater QIAGEN, Hilden, Germany).
To confirm that the procedures were aseptic, a real-time polymerase chain reaction (PCR) using universal 16S rRNA primers (Nadkarni et al. 2002) with conjugated minor groove binders (MGBs) was run on 2 extracted sound teeth with vital pulp before extraction, processed in the same way as the test samples. The amount of 16S rDNA yielded by these samples was below the detection limit and equal to the negative PCR controls.
Samples were transferred to a sterile screw-cap Eppendorf tube with 0.25 ml of lysis buffer (AGOWA mag Mini DNA Isolation Kit, AGOWA, Berlin, Germany). Then 0.3 g zirconium beads (diameter, 0.1 mm; Biospec Products, Bartlesville, OK, USA) and 0.2 ml phenol were added to each sample. The samples were homogenized with a Mini-beadbeater (Biospec Products) for 2 minutes. DNA was extracted with the AGOWA mag Mini DNA Isolation Kit and quantified (Nanodrop ND-1000; NanoDrop Technologies, Montchanin, DE, USA).
PCR amplicon libraries of the small subunit ribosomal RNA gene V5–V6 hypervariable region were generated for the individual samples. PCR was performed using the forward primer 785F (GGATTAGATACCCBRGTAGTC) and the reverse primer 1061R (TCACGRCACGAGCTGACGAC). The primers included the 454 Life Sciences Adapter A (forward primer) and B (reverse primer) fused to the 5’ end of the 16S rRNA bacterial primer sequence and a unique trinucleotide sample identification key. The amplification mix contained 2 units of Pfu Ultra II Fusion HS DNA polymerase and 1× PfuUltra II reaction buffer (Stratagene, La Jolla, CA, USA), 200 µM dNTP PurePeak DNA polymerase Mix (Pierce Nucleic Acid Technologies, Milwaukee, Wl, USA), and 0.2 µM of each primer. After denaturation (94°C; 2 min), 30 cycles were performed that consisted of denaturation (94°C; 30 s), annealing (50°C; 40 s), and extension (72°C; 80 s). The amplicons were purified by means of the MinElute kit (Qiagen, Hilden, Germany). The quality and the size of the amplicons were analyzed on the Agilent 2100 Bioanalyser with the DNA 1000 Chip kit (Agilent Technologies, Santa Clara, CA, USA) and quantified using Nanodrop ND-1000 spectrophotometer. The amplicon libraries were pooled in equimolar amounts and sequenced unidirectionally in the reverse direction (B-adaptor) by means of the Genome Sequencer FLX (GS-FLX) system (Roche, Basel, Switzerland).
GS-FLX sequencing data were processed as previously described (Sogin et al. 2006). In brief, sequences were trimmed by removing primer sequences and low-quality data, sequences that did not have an exact match to the reverse primer, that had an ambiguous base call (N) in the sequence, or that were shorter than 150 nt after trimming. Next, the GAST algorithm (Huse et al. 2008) was used to calculate the percent difference between each unique sequence and its closest match in a database of eubacterial and archaeal V5–V6 sequences from SSU rRNA sequences in the SILVA database (Pruesse et al. 2007). Taxa were assigned to each reference sequence using several sources including Entrez Genome entries, cultured strain identities, SILVA, and the Ribosomal Database Project Classifier (Cole et al. 2005). In cases where reads were equidistant to multiple V5–V6 reference sequences, and/or where identical V5–V6 sequences were derived from longer sequences mapping to different taxa, reads were assigned to the lowest common taxon of at least two-thirds of the sequences. All data were normalized to an equal number of reads per sample and converted to relative proportion data for analysis.
To compare the samples originating from the apical segments with those from the coronal segments of the roots, weighted UniFrac (Lozupone et al. 2007) was used. UniFrac is a method for comparing microbial communities measuring the phylogenetic distance between sets of taxa in a phylogenetic tree as the fraction of the branch length of the tree that leads to descendants from either one environment or the other, but not both. Weighted UniFrac takes into account not only the presence but also the abundance of taxa. The output of the analysis, a distance matrix, was visualized using Principal Coordinate Analysis (PCoA) using open source software QIIME (QIIME version 1.3.0) (Caporaso et al. 2010).
Sequences were clustered into operational taxonomic units (OTUs) at 97% similarity using UCLUST average linkage algorithm (Edgar 2010). Only OTUs with at least 5 reads were included in further analyses. The samples were normalized to an equal number of reads. Then the prevalence of each OTU per sample was calculated. To reduce the effect of sequencing noise and to exclude transient OTUs without potential clinical relevance, an arbitrary “prevalence cutoff” was applied: an OTU was counted as present in a sample if at least 10 reads in the respective sample belonged to this OTU.
Statistical analysis
The number of OTUs and Unifrac distances between the corresponding apical and coronal root segments were compared using a Paired Samples T-test (SPSS, version 17.0). Dominance, Margalef’s richness index and Fisher’s alpha diversity index on taxonomy data were determined using PAST software (Hammer et al. 2001) version 2.05. These outputs were not normally distributed and therefore compared using a Wilcoxon Signed Ranks Test (SPSS).
Results
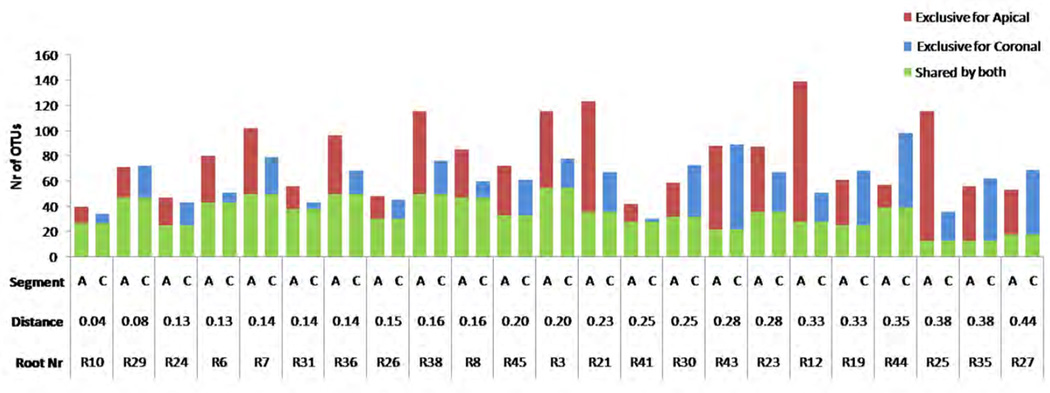
In total, 301298 (73.9%) reads passed quality filtering. The reads (6549 ± 3583 reads/sample) were clustered into 10969 OTUs of which 2168 OTUs contained at least 5 reads. The application of the “prevalence cutoff” resulted in 78 ± 29 OTUs (range 40 – 139 OTUs) per apical segment and 62 ± 18 OTUs (range 30 – 98 OTUs) per coronal root segment. The apical segments harbored significantly more OTUs than the coronal segments of the same root (p=0.01). On average, 34 ± 12 OTUs (range 13 – 55 OTUs) were shared by both segments of the same root, which accounted for 47 ± 17% of the apical and 58 ± 21% of the coronal OTUs of the same root (Figure 1). Of each pair significantly more OTUs were exclusively found in the apical segment than in the coronal part of the root (p=0.01). On average, 44 ± 27 OTUs (range 13 – 111) were exclusive for the apical segment, while 28 ± 17 OTUs (range 2 – 67) were prevalent only in the coronal segment of the same root (Figure 1).
Figure 1.
Prevalence of exclusive and shared OTUs (sequences that cluster within 97% similarity level) by the two root segments of 23 root pairs. Only those OTUs that contained at least 10 reads/sample were counted as present in the corresponding sample. The order of the samples (Root numbers) from left to right corresponds to the increasing difference in phylogenetic distance (Weighted Unifrac) between the corresponding apical and coronal segments. The distances are shown above the respective root numbers. The two segments from R10 were highly similar (Distance=0.04), while those from R27 differed the most (Distance=0.44) among the corresponding pairs of samples.
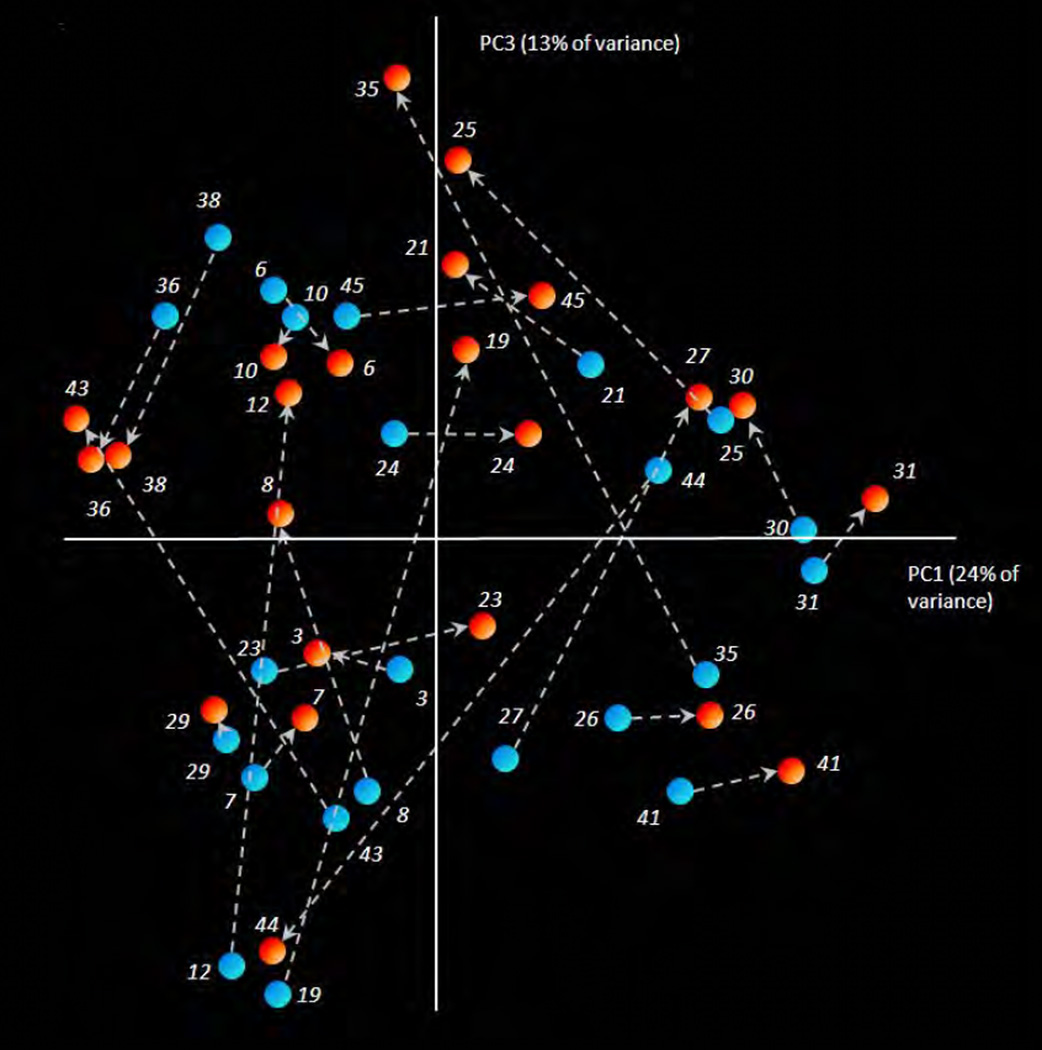
Samples originating from the same root had significantly lower UniFrac distance (0.225 ± 0.11) than the average distance calculated from all samples (0.298 ± 0.04; p<0.001), meaning that two segments from the same roots harbored more similar communities than unrelated roots. However, the data contained a few strong outliers (roots nr. 12, 19, 44, 25, 35 and 27), where the difference between the samples of the same pair was larger than the overall distance (Figure 1, UniFrac distances of each pair are shown above the root numbers). Visualization of the Unifrac analysis on phylogenetic distances among the different samples showed that there was no clear clustering of samples by the segment of the root (Figure 2). Some sub-clustering of the groups of the samples from the apical segments could be observed (Figure 2, apical samples - red dots - of 36, 38 and 43; and 19, 21, 25, 35 and 45, respectively). Additionally, three patterns were observed in the way the microbial communities shifted in their composition when a coronal sample was compared to its corresponding apical sample. Most (70% or 16/23) of the apical samples “shifted” from the coronal samples in the direction of the third coordinate PC3 (Figure 2, arrows connecting the respective pairs of the samples pointing upwards). The position of two samples was not affected by PC3, while in 5 (22%) samples the direction from the coronal to the apical segment was opposite to the majority of the samples (Figure 2, arrows pointing downwards).
Figure 2.
Principal coordinates analysis (PCoA) plot of the phylogenetic distances (Weighted UniFrac) among the samples from the corresponding pairs of segments (red dots – apical; blue dots – coronal segments) from 23 infected roots. Principal coordinates PC1, PC2 (not shown) and PC3 explained 24%, 17% and 13% of the overall variance among the samples, respectively. The sample pairs (apical and coronal) from the same root are connected with dashed lines. Arrows are pointing towards the apical sample of each pair. The pairs of the samples differ in their phylogenetic relatedness from being very adjacent or similar (roots nr 29 and 10) to very distant or phylogenetically different (roots nr 19, 44, 35 and 12). For the majority of the pairs (16 out of 23) the apical sample was positioned in the positive direction of the PC3 axis, with arrow pointing upwards, indicating the common direction of the microbial shift from coronal to apical microbiome in these samples.
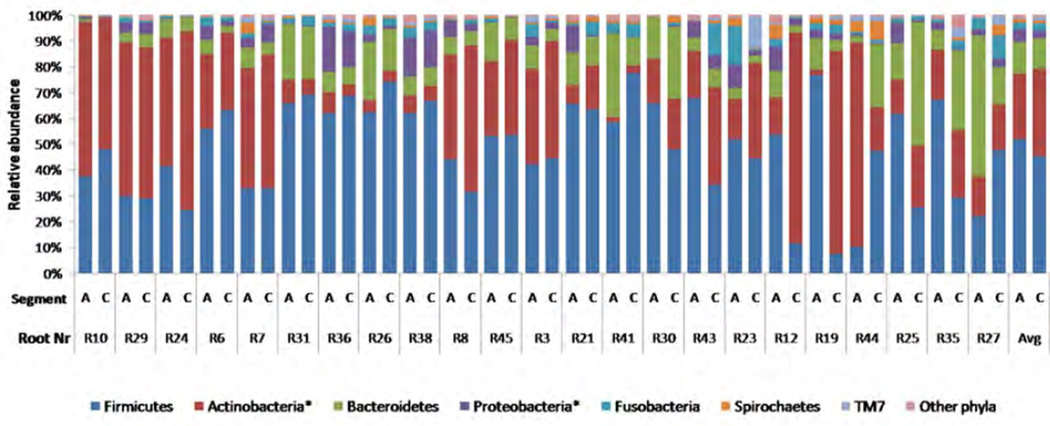
Taxonomical classification of the reads resulted in 606 taxa (species or higher taxon) with approximately 20% of the reads being classified to species level, 72% to genus level (317 genera), 80% to family level, 84% to order level, 97% to class level and 99.7% to phylum level, while the remaining 0.3% reads could not be assigned to any known domain. On average, each sample contained 125 ± 30 taxa (range 70 – 185). Representatives of 24 known bacterial phyla or candidate divisions (Acidobacteria, Actinobacteria, Bacteroidetes, BRC1, Chlamydiae, Cyanobacteria, Deinococcus-Thermus, Firmicutes, Fusobacteria, Nitrospira, OD1, OP11, OP3, OP9, Planctomycetes, Proteobacteria, Spirochaetes, Synergistetes, Tenericutes, TG1, Thermomicrobia, TM7, Verrucomicrobia and WS3) and a single archaeal phylum – Euryarchaeota were detected. Firmicutes and Actinobacteria predominated in most of the samples (Figure 3). When the two types of samples of each root were compared, a significantly higher proportion of Proteobacteria was found apically, and more Actinobacteria – coronally of the same tooth (p<0.05; Figure 3).
Figure 3.
Relative abundance of bacterial phyla per root segment (A; Apical, C; Coronal and Avg; average of all roots) in 23 cryo-pulverized roots. Other phyla represent the sum of the reads belonging to all minor phyla: Thermomicrobia, Cyanobacteria, Synergistes, OD1, Deinococcus Thermus, Chlamydiae, Acidobacteria, OP3, Planctomycetes, OP11, Nitrospira, Tenericutes, Verrucomicrobia, BRC1, Euryarchaeota, OP9 and unclassifiable reads. The order of the roots (Root number) from left to right corresponds to the increasing difference in phylogenetic distance (Weighted Unifrac) between the corresponding apical and coronal segments. The distances are shown in figure 1.
* Statistically significant difference between apical and coronal segments (Wilcoxon Signed Ranks Test, p<0.05).
Of the 317 genera, representatives of the genera Lactobacillus, Actinomyces, Streptococcus and Prevotella were found in all samples, and the genera Fusobacterium, Porphyromonas, Eubacterium, Selenomonas and Corynebacterium were found in all but one sample. The 50 most abundant genera (or higher common taxon if the reads could not be assigned to genus level), ranked by their abundance in the apical segments of the roots, contributed to 93% of the reads obtained from the apical samples (Table 1). Lactobacillus was the most abundant genus in the apical samples (average 14 ± 15%; range 0.05 – 53.7% of all reads/sample) and genus Actinomyces – the most abundant in the coronal samples (average 18 ± 21%; range 0.09 – 64% of all reads/sample). Some with endodontic infections frequently associated genera were found below the top-50 abundance: Enterococcus was ranked number 56 (0.2%, found in 18/23 teeth), Campylobacter ranked number 64 (0.15%, found in 15/23 teeth) and Capnocytophaga ranked number 74 (0.1%, found in 16/23 teeth).
Table 1.
Top 50 prevalent higher taxa (bacterial genera or higher taxon).
| Apical samples§ | Coronal samples | ||||
|---|---|---|---|---|---|
| Phylum | Genus or higher taxon | % Relative abundance (SD) |
N# | % Relative abundance (SD) |
N |
| Firmicutes | Lactobacillus | 14.3 (15.1) | 23 | 9.0 (13.5) | 23 |
| Actinobacteria | Actinomyces | 11.9 (18.8) | 23 | 17.7 (20.8) | 23 |
| Firmicutes | Streptococcus | 8.4 (10.9) | 23 | 5.5 (9.5) | 23 |
| Actinobacteria | class Actinobacteria | 6.9 (13.1) | 23 | 8.8 (16.1) | 23 |
| Bacteroidetes | Prevotella | 6.1 (10.9) | 23 | 6.1 (10.1) | 23 |
| Firmicutes | Parvimonas | 3.4 (4.7) | 21 | 4.1 (8.0) | 23 |
| Firmicutes | Pseudoramibacter | 3.0 (4.9) | 22 | 4.0 (6.1) | 22 |
| Bacteroidetes | order Bacteroidales | 2.7 (3.9) | 23 | 3.7 (5.4) | 22 |
| Firmicutes | fam. Veillonellaceae | 2.5 (2.5) | 23 | 3.2 (2.9) | 23 |
| Fusobacteria | Fusobacterium | 2.0 (2.8) | 23 | 2.2 (2.3) | 22 |
| Firmicutes | Peptostreptococcus | 2.0 (3.9) | 17 | 1.6 (3.3) | 13 |
| Bacteroidetes | Porphyromonas * | 1.8 (1.7) | 23 | 1.1 (1.2) | 22 |
| Firmicutes | fam. Lachnospiraceae * | 1.6 (1.4) | 23 | 0.7 (0.7) | 23 |
| Firmicutes | Mogibacterium | 1.6 (2.8) | 21 | 2.2 (3.6) | 20 |
| Firmicutes | Eubacterium | 1.5 (2.2) | 23 | 2.3 (2.8) | 22 |
| Actinobacteria | Propionibacterium | 1.3 (2.0) | 22 | 1.2 (1.8) | 22 |
| Spirochaetes |
Treponema fam. |
1.3 (1.6) | 22 | 1.2 (1.6) | 22 |
| Firmicutes | Syntrophomonadaceae | 1.2 (1.8) | 20 | 1.6 (2.4) | 19 |
| Firmicutes | Selenomonas | 1.2 (1.4) | 22 | 1.5 (2.3) | 23 |
| Firmicutes | Anaerovorax | 1.1 (1.4) | 22 | 2.0 (1.9) | 22 |
| Actinobacteria | Parascardovia | 0.9 (1.8) | 12 | 0.7 (2.0) | 12 |
| Firmicutes | Filifactor | 0.9 (2.1) | 17 | 1.5 (4.6) | 16 |
| Proteobacteria | Neisseria | 0.9 (2.5) | 21 | 0.8 (2.4) | 15 |
| Firmicutes |
Oribacterium fam. |
0.9 (1.3) | 20 | 0.7 (1.3) | 21 |
| Firmicutes | Peptostreptococcaceae | 0.8 (1.3) | 22 | 0.4 (0.4) | 22 |
| Firmicutes | Uncl. Firmicutes | 0.8 (1.8) | 22 | 0.7 (1.2) | 18 |
| Firmicutes | Moryella | 0.7 (2.1) | 15 | 0.3 (0.4) | 17 |
| Actinobacteria | Corynebacterium | 0.7 (1.0) | 22 | 1.3 (3.5) | 23 |
| Bacteroidetes | fam. Prevotellaceae * | 0.7 (0.7) | 22 | 0.2 (0.3) | 22 |
| Proteobacteria | Enhydrobacter * | 0.7 (0.7) | 17 | 0.2 (0.3) | 17 |
| Firmicutes | Anoxybacillus * | 0.6 (0.8) | 17 | 0.2 (0.5) | 14 |
| Firmicutes | fam. Ruminococcaceae * | 0.6 (0.8) | 21 | 0.2 (0.3) | 21 |
| Firmicutes | fam. Erysipelotrichaceae | 0.6 (0.9) | 20 | 0.5 (1.4) | 18 |
| Fusobacteria | Leptotrichia | 0.5 (0.8) | 21 | 0.5 (1.2) | 21 |
| Actinobacteria | Collinsella * | 0.5 (0.7) | 19 | 0.2 (0.3) | 20 |
| TM7 | TM7 | 0.5 (0.7) | 22 | 1.3 (2.6) | 22 |
| Firmicutes | Granulicatella | 0.5 (1.5) | 15 | 0.7 (2.0) | 11 |
| Bacteroidetes | Bacteroides * | 0.5 (0.9) | 19 | 0.1 (0.2) | 18 |
| Actinobacteria | Bifidobacterium | 0.4 (0.7) | 13 | 0.2 (0.3) | 13 |
| Unclassified reads | 0.4 (0.5) | 23 | 0.3 (0.4) | 22 | |
| Firmicutes | Faecalibacterium * | 0.4 (0.4) | 17 | 0.1 (0.2) | 14 |
| Firmicutes | Dialister | 0.4 (0.7) | 13 | 0.2 (0.4) | 15 |
| Actinobacteria | Actinobaculum | 0.4 (0.8) | 15 | 1.2 (3.2) | 16 |
| Actinobacteria | fam. Bifidobacteriaceae | 0.4 (0.9) | 15 | 0.1 (0.4) | 13 |
| Bacteroidetes | Uncl. Bacteroidetes | 0.3 (1.4) | 9 | 0.2 (0.6) | 9 |
| Actinobacteria | fam. Coriobacteriaceae | 0.3 (0.7) | 17 | 0.5 (1.5) | 19 |
| Proteobacteria | Desulfobulbus | 0.3 (0.8) | 12 | 0.3 (0.5) | 15 |
| Actinobacteria | Rothia * | 0.3 (0.7) | 15 | 0.2 (0.5) | 12 |
| Proteobacteria | Shigella * | 0.3 (0.3) | 17 | 0.05 (0.1) | 12 |
| Actinobacteria | Olsenella | 0.2 (0.7) | 10 | 0.2 (0.5) | 11 |
| Proteobacteria | Acidovorax | 0.2 (0.3) | 18 | 0.1 (0.3) | 15 |
- The data is sorted according to the relative abundance in the apical samples
- Number of samples with the respective taxon
- Denotes significantly higher proportion in apical samples than in coronal samples (Wilcoxon Signed Ranks Test, p<0.05)
Eleven of the top-50 genera were at significantly higher proportions (p < 0.05) in the apical segment of the root compared to its coronal part (Table 1). Of these eleven taxa eight belonged to known obligate anaerobes, and three to facultative anaerobes. The anaerobes belonged to the Bacteroidia class of the Bacteroidetes phylum (genus Porphyromonas, genus Bacteroides and Prevotellaceae family), to the Clostridia (genus Faecalibacterium, Ruminococcaceae and Lachnospiraceae family) and the Bacilli class (genus Anoxybacillus) of the Firmicutes phylum, and the Actinobacteria class of the Actinobacteria phylum (genus Collinsella). Two gamma-proteobacteria class members of the phylum Proteobacteria (genus Enhydrobacter and Shigella), as well as genus Rothia from the phylum and class Actinobacteria belong to facultative anaerobe taxa. Of all genera only genus Anaerovorax was significantly more abundant in the coronal samples than in the apical samples (p < 0.05). Eighty nine of all genera were found exclusively in the apical samples, most of which were present at a very low prevalence (present in 1 to 2 samples) and at a low abundance (see supporting information). Four of these genera, Anaerococcus (phylum Firmicutes), Brevibacterium (phylum Actinobacteria), Rhodopseudomonas and Mesorhizobium (Proteobacteria), however, were found in at least four apical samples and contributed to 0.01 – 0.2% of the reads/sample. Forty seven of all genera were exclusive for the coronal samples, all being at a low prevalence (1 – 3 samples) and all but one at a very low abundance. In this single coronal sample (Root nr 35) 5% of the reads belonged to the genus Jonquetella (phylum Synergistetes).
All 606 taxa (final taxon to species level) were used to perform diversity analyses. The apical samples harbored statistically significantly more taxa (132 ± 32) than the coronal samples (117 ± 27; p<0.05). There was no difference in the ecological dominance of taxa between apical and coronal samples (Dominance Index, p>0.05). Three of the 23 apical root segments, however, contained a very high proportion (40 – 60%) of either genus Actinomyces (root nr 44: 70% of the reads), Lactobacillus (root nr 35: 50% of the reads) or Prevotella (root nr 27: 47% of the reads). Seven of the 23 coronal samples were predominated by either genus Actinomyces (root nr 12: 43% of the reads; root nr 19: 54% of the reads; root nr 8: 48% of the reads; root nr 3: 38% of the reads), genus Prevotella (root nr 25: 45% of the reads), genus Lactobacillus (root nr 6: 46% of the reads) or unclassified representatives of the class Actinobacteria (root nr 24: 67% of the reads). The apical samples had a higher species richness (Margalef’s Richness Index, p<0.05) and a higher microbial diversity than the coronal samples (Fisher’s alpha, p<0.05). In other words, the apical microbiota contained more species, most of which were present in greater relative abundance apically compared to the coronal part of the root.
Discussion
This study compares, for the first time, the complexity of the microbiome of the infected root canal system including the surrounding dentine at the depth of pyrosequencing in different parts of the root. The results demonstrate that the apical segments of the roots are distinct ecological niches and harbour a more complex microbiome than their coronal counterparts.
The most complete microbiological sampling method was used, cryo-pulverization of the root segments, that has been applied previously in the assessment of endodontic microbial communities (Alves et al. 2009; Rôças et al. 2010). To exclude microorganisms of carious tissue and from periodontal origin, the crowns were removed, and the root surfaces were disinfected. The amount of bacterial 16S rDNA from the control samples (sound teeth processed the same way as the test samples), was below the detection limit and equal to the negative PCR controls. The only, but very relevant, limitation of this sampling technique is of ethical and practical nature, as analysis requires extraction of the tooth. However, it was now possible to identify the entire microbial population in the root canal system and dentinal tubules, which would have remained undetected using paper points.
The study identified 606 bacterial taxa at a species or higher taxonomic level belonging to 317 genera from 24 bacterial and one archaeal phylum. Although, this is noticeably higher than the previously reported 468 taxa (Siqueira & Rôças 2009) and 179 bacterial genera (Li et al. 2010), only 20% of the reads in the present study could be classified to species level. An OTU-approach was used where sequences were clustered at a 97% similarity level. These clusters could belong to the same species- or strain-level phylotypes. The OTU-approach resulted in 10969 OTUs of which 2168 OTUs contained at least 5 reads. This indicates that the overall diversity within the root canals at the species level may be considerably higher than currently reported. Other organisms, such as fungi (Baumgartner et al. 2000) or viruses (Sabeti et al. 2003) not assessed in the current study, may also contribute to the diversity of the infection of the root canal system and the inflamed periapical tissue. The 16S rRNA hypervariable region was used to profile bacteria, while fungi and viruses lack the 16S rRNA gene. Additional targets, e.g. fungal internal transcribed spacer (ITS) regions (Ghannoum et al. 2010) and clustered regularly interspaced short palindromic repeats (CRISPRs) (Pride et al. 2011) could be used to target fungi and bacteriophages (viruses infecting bacteria), respectively. Different targets could be combined in future studies to assess the overall diversity.
In line with previous reports (Munson et al. 2002, Saito et al. 2006, Sakamoto et al. 2006), Firmicutes was the most predominant phylum in our samples (48% of the reads). The second largest phylum in the data set was Actinobacteria (30%), followed by Bacteroidetes (12%). This, however, does not agree with the pyrosequencing results of Li et al. (2010), where phylum Bacteroidetes dominated the data with nearly 60% of the reads, followed by a much lower proportion of Firmicutes (20%) and Actinobacteria (5%). This difference could be due to the paper point sampling technique used (Sathorn et al. 2007) and the limited sample size (7 teeth) compared to 46 root segments from 23 cryo-pulverized teeth in this study. The most likely reason, however, is due to differences in DNA extraction technique. Li et al. (2010) used Qiagen lysis buffer and proteinase K treatment (QIAamp DNA mini kit, Qiagen, Valencia, CA, USA), which has been shown to underestimate Gram-positive bacteria compared to more easily lyzed Gram-negatives (Nadkarni et al. 2009). In the experience of the authors (unpublished findings) this particular DNA extraction method, if used according to the manufacturer’s instructions, yields lower sample diversity than if an additional bead beating step is added to enhance rupture of more resilient Gram-positive cells. The method that was used in the current study involves a bead beating step and has been shown to be highly efficient (Keijser et al. 2007).
Most of the abundant taxa in this study represent the genera known to play a prominent role in endodontic infections (Siqueira & Rôças 2009). However, the majority of the 606 taxa were at a low abundance and thus their clinical relevance may be questioned. For instance, only three reads originating from two coronal samples were classified as Archaea, phylum Euryarchaeota, genus Metanoregula. Archaea have been detected in the oral cavity (Alves et al. 2009; Siqueira & Rôças 2009) but this low prevalence and abundance in the present samples suggests that the root canal system is not a preferred niche for these microorganisms.
The specific aim of this study was to compare the apical and coronal parts of the same infected roots. As expected, there was significantly higher phylogenetic relatedness (lower Unifrac distance) between the root segments originating from the same root than among unrelated roots. Interestingly, in six (26%) of the 23 roots, the microbial community of the apical segment differed more from the coronal segment (higher Unifrac distance) than the average difference among all roots. This suggests that different ecological factors have contributed to shaping the community within individual roots. This is in line with the DGGE-profiling results from similarly obtained samples (Alves et al. 2009) where high inter- and intra-individual variability among the sample profiles was observed.
Several taxa, eight of which were obligate anaerobes, were present in significantly higher proportions in the apical than in the coronal segments of the root. Two of the strictly anaerobe taxa that were more abundant in the apical segments, belonged to genus Faecalibacterium (Firmicutes, Ruminococcaceae family) and to unclassified members of the Ruminococcaceae family. Ruminococcaceae are gut commensals and have only recently been associated with endodontic infection in a sample from a single individual (Ribeiro et al. 2011). This group of microorganisms was present in 22 of 23 root pairs, ranging from 0 (both segments of the root nr 24) to 6% (apical segment of the root 21) of the reads. Much higher prevalence of these organisms in the present study could be due to the more efficient sampling by cryo-pulverization than by the paper point approach used by Ribeiro et al. (2011) and due to the more sequences obtained per sample (6549 reads/sample in the present study versus 40.7 clones/sample by Ribeiro et al. 2011), since the number of species detected in a sample is affected strongly by the number of sequences analyzed (Schloss & Handelsman 2005). In a study on cultivable microbiota in previously filled roots with failed endodontic treatment, no association between the root segment and the isolated bacterial strains could be found (Adib et al. 2004). This study differed from the present one not only in methodology (culturing versus pyrosequencing and paperpoint sampling versus cryo-pulverization) but also by the use of root-filled teeth as opposed to primary infections. Another study (Rôças et al. 2010) targeting 28 bacterial taxa in the checkerboard panel did report a difference in prevalence of several taxa between the apical and coronal segments of the infected roots: streptococci were found more often in the coronal part and Prevotella baroniae, Tannerella forsythia and Fusobacterium nucleatum more often in the apical segment of the root.
An important and previously unreported finding of this study is the observation of a microbial shift within the root towards increased microbial complexity. This contradicts with the previously mentioned DGGE-profiling study (Alves et al. 2009) where no difference in diversity (the number of DGGE bands) between the apical and the coronal parts of the same roots was found. This could be due to the difference in methodology used. Pyrosequencing allows much deeper resolution than DGGE profiling (a species is visible as a DGGE band if it constitutes at least 1% of the total community; Muyzer et al. 1993; Murray et al. 1996). When the apical and coronal sample data in the present study (Table 1) were compared, only one taxon (genus Porphyromonas) that was found at a significantly higher abundance in the apical segments, constituted more than 1% of the sample (1.4 ± 1.5%, range 0 – 7.7% of reads). All other taxa that were found at a higher abundance in the apical samples would have been missed using the DGGE approach, and may explain the disagreement with the above-mentioned study (Alves et al. 2009).
Intuitively one may expect the opposite – higher diversity at the entrance of the root canal due to the connection with the oral cavity. An hypothesis to explain the higher diversity at the apical segments of the roots is as follows. The coronal part of the root is rich in nutrients due to influx from the oral cavity. This selects for fast growing, best-adapted microorganisms, and may result in communities with relatively low diversity, as has been shown in modeling studies on ecology of biofilms (Kassen et al. 2000, Pham et al. 2009). Although the potential communication with the oral cavity provides the influx of new microbial species, these species will have to outcompete the existing community which has already been selected as the most fit for this environment. The more apical parts of the root, particularly the complex network of canals in the apical delta, may be limited in fresh nutrients from the oral cavity. However, the surrounding debris of decomposed pulp, the dead bacteria and a potential exudate from periapical inflammatory tissue provide a protein rich and complex nutrient-profile. To metabolize such complex compounds microorganisms are relatively more dependent on their inter-species interactions in the food chain. This cooperative process and interdependency precludes the overgrowth of few species (Kreft 2004). As a result, diverse slow-growing and fastidious organisms may be able to establish in the apical community.
Slow growth of the apical microorganisms has an important clinical implication – this together with the complex anatomy of the apex, might lead to the difficulty in eradicating the infection. It has been shown that the sensitivity of bacterial cells to antimicrobials increases with increasing growth rate (Evans et al. 1991) and slow growth is known to be one of the biocide resistance mechanisms in biofilms (Mah & O'Toole 2001). This emphasizes the need for more research in potent antimicrobial therapies and cleaning protocols for the whole root canal system.
One major shortcoming of DNA-based studies is the inability to discriminate dead from live microorganisms. Instead, all genetic material, including damaged and nonviable cells is assessed. To overcome this limitation, several approaches are available, unfortunately each with their own limitations. Assessment of messenger RNA (mRNA) instead of the total RNA (rRNA, tRNA and mRNA) allows targeting the cells that are active and thus vital (Sheridan et al. 1998). For this the mRNA is transcribed into its complementary DNA using reverse transcriptase PCR. A major technical challenge is the low relative abundance (1–5%) of mRNAs in total cellular RNA and selective removal of rRNA in mixed communities (He et al. 2010). Another more recently described solution to the problem is the use of photoactivated DNA-intercalating agent such as ethidium monoazide (Nocker& Camper 2006, Soejima et al. 2007) or propidium monoazide (Loozen et al. 2011), which irreversibly binds DNA of damaged cells and precludes this part of DNA from being amplified in the subsequent PCR reaction. It is easy to use and seems promising for specifically targeted species. However, the first results on different taxa show that optimal assay conditions, such as concentrations of the compounds and light exposure time, might depend upon the targeted bacterial species (Loozen et al. 2011) and thus may not be suitable for microbiome studies such as the present one. On the other hand, it has been shown that microbial enzymes (e.g., deoxyribonucleases) contribute to rapid degradation of DNA in non-sterile environments (Josephson et al. 1993). Recent study on the persistence of dead-cell bacterial DNA in ex vivo root canals showed that, after 24 months, DNA of heat-inactivated Enterococcus faecalis could still be PCR-amplified, while in the presence of nucleases, the decomposition of DNA occurred within 21 days if the DNA was still cell-bound and already within 30 minutes if the DNA was freely available (Brundin et al. 2010). With the knowledge of this recycling of material from dead cells within complex communities, such as samples used in the present study, and the prevalence cutoff used (minimum of 10 reads/sample per OTU), there is no evidence to suggest that the results have overestimated the diversity of the microbiome of the root canal system.
Conclusions
A combination of sampling by cryo-pulverization with profiling by 454 pyrosequencing revealed that the endodontic microbiome is more complex than previously described. Separate assessment of the apical and the coronal parts of the same teeth revealed that the apical microbiota was more diverse than its coronal counterpart. This supports the presence of a distinct ecological niche adjacent to the periapical inflammation. These data will contribute to a further understanding of the disease etiology and pathogenesis, which should result in more effective prevention and treatment strategies.
Acknowledgments
The authors would like to thank Prof. Dr. Ü.K. Akal Aktaş and Prof. Dr. F. Tulga Öz for their help with the collection of the extracted teeth, and H.W. van Essen and N. Bravenboer for their technical assistance. Dr. S.M. Huse was supported by a subcontract to Mitchell Sogin from the NIH Human Microbiome 1UH2DK083993-01 awarded to the University of Michigan.
References
- Adib V, Spratt D, Ng Y-l, Gulabivala K. Cultivable microbial flora associated with persistent periapical disease and coronal leakage after root canal treatment: a preliminary study. International Endodontic Journal. 2004;37:542–551. doi: 10.1111/j.1365-2591.2004.00840.x. [DOI] [PubMed] [Google Scholar]
- Akpata E. Effect of endodontic procedures on the population of viable microorganisms in the infected root canal. Journal of Endodontics. 1976;2:369–373. doi: 10.1016/S0099-2399(76)80099-4. [DOI] [PubMed] [Google Scholar]
- Alves FR, Siqueira JFJ, Carmo FL, et al. Bacterial community profiling of cryogenically ground samples from the apical and coronal root segments of teeth with apical periodontitis. Journal of Endodontics. 2009;35:486–492. doi: 10.1016/j.joen.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Baumgartner JC, Watts CM, Xia T. Occurrence of Candida albicans in infections of endodontic origin. Journal of Endodontics. 2000;26:695–698. doi: 10.1097/00004770-200012000-00003. [DOI] [PubMed] [Google Scholar]
- Brundin M, Figdor D, Roth C, Davies JK, Sundqvist G, Sjögren U. Persistence of dead-cell bacterial DNA in ex vivo root canals and influence of nucleases on DNA decay in vitro. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontology. 2010;110:789–794. doi: 10.1016/j.tripleo.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J, Chai B, Farris R, et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Research. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Evans DJ, Allison DG, Brown MR, Gilbert P. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. Journal of Antimicrobial Chemotherapy. 1991;27:177–184. doi: 10.1093/jac/27.2.177. [DOI] [PubMed] [Google Scholar]
- Ghannoum MA, Jurevic RJ, Mukherjee PK, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathogens. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer O, Harper D, Ryan P. PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:1–9. [Google Scholar]
- He S, Wurtzel O, Singh K, et al. Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nature Methods. 2010;7:807–812. doi: 10.1038/nmeth.1507. [DOI] [PubMed] [Google Scholar]
- Huse S, Dethlefsen L, Huber J, Mark Welch D, Relman D, Sogin M. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genetics. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson KL, Gerba CP, Pepper IL. Polymerase chain reaction detection of nonviable bacterial pathogens. Applied and Environmental Microbiology. 1993;59:3513–3515. doi: 10.1128/aem.59.10.3513-3515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakehashi S, Stanley H, Fitzgerald R. The effects of surgical exposures of dental pulps in germ-free and conventional laboratory rats. Oral Surgery Oral Medicine Oral Pathology. 1965;20:340–349. doi: 10.1016/0030-4220(65)90166-0. [DOI] [PubMed] [Google Scholar]
- Kassen R, Buckling A, Bell G, Rainey PB. Diversity peaks at intermediate productivity in a laboratory microcosm. Nature. 2000;406:508–512. doi: 10.1038/35020060. [DOI] [PubMed] [Google Scholar]
- Keijser B, ter Beek A, Rauwerda H, et al. Analysis of temporal gene expression during Bacillus subtilis spore germination and outgrowth. Journal of Bacteriology. 2007;189:3624–3634. doi: 10.1128/JB.01736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijser BJ, Zaura E, Huse SM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. Journal of Dental Research. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- Kreft JU. Biofilms promote altruism. Microbiology. 2004;150:2751–2760. doi: 10.1099/mic.0.26829-0. [DOI] [PubMed] [Google Scholar]
- Li L, Hsiao W, Nandakumar R, et al. Analyzing endodontic infections by deep coverage pyrosequencing. Journal of Dental Research. 2010;89:980–984. doi: 10.1177/0022034510370026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loozen G, Boon N, Pauwels M, Quirynen M, Teughels W. Live/dead real-time polymerase chain reaction to assess new therapies against dental plaque-related pathologies. Molecular Oral Microbiology. 2011;26:253–261. doi: 10.1111/j.2041-1014.2011.00615.x. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Applied and Environmental Microbiology. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends in Microbiology. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Möller AJR. Microbiological examination of root canals and periapical tissues of human teeth (PhD Thesis) Göteborg, Sweden: University of Göteborg; 1966. [PubMed] [Google Scholar]
- Munson M, Pitt-Ford T, Chong B, Weightman A, Wade W. Molecular and cultural analysis of the microflora associated with endodontic infections. Journal of Dental Research. 2002;81:761–766. doi: 10.1177/0810761. [DOI] [PubMed] [Google Scholar]
- Murray AE, Hollibaugh JT, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Applied and Environmental Microbiology. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and Environmental Microbiology. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni M, Martin F, Hunter N, Jacques N. Methods for optimizing DNA extraction before quantifying oral bacterial numbers by real-time PCR. FEMS Microbiology Letters. 2009;296:45–51. doi: 10.1111/j.1574-6968.2009.01629.x. [DOI] [PubMed] [Google Scholar]
- Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- Nocker A, Camper AK. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Applied and Environmental Microbiology. 2006;72:1997–2004. doi: 10.1128/AEM.72.3.1997-2004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. Journal of Bacteriology. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham LC, van Spanning RJ, Roling WF, et al. Effects of probiotic Lactobacillus salivarius W24 on the compositional stability of oral microbial communities. Archives of Oral Biology. 2009;54:132–137. doi: 10.1016/j.archoralbio.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Pride DT, Sun CL, Salzman J, et al. Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects overtime. Genome Research. 2011;21:126–136. doi: 10.1101/gr.111732.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, et al. SUVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Research. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AC, Matarazzo F, Faveri M, Zezell DM, Mayer MPA. Exploring bacterial diversity of endodontic microbiota by cloning and sequencing 16S rRNA. Journal of Endodontics. 2011;37:922–926. doi: 10.1016/j.joen.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Röças IN, Alves FR, Santos AL, Rosado AS, Siqueira JFJ. Apical root canal microbiota as determined by reversecapture checkerboard analysis of cryogenically ground root samples from teeth with apical periodontitis. Journal of Endodontics. 2010;36:1617–1621. doi: 10.1016/j.joen.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Sabeti M, Valles Y, Nowzari H, Simon JH, Kermani-Arab V, Slots J. Cytomegalovirus and Epstein-Barr virus DNA transcription in endodontic symptomatic lesions. Oral Microbiology and Immunology. 2003;18:104–108. doi: 10.1034/j.1399-302x.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- Saito D, de Toledo Leonardo R, Rodriques J, Tsai S, Höfling J, Gonçalves R. Identification of bacteria in endodontic infections by sequence analysis of 16S rDNA clone libraries. Journal of Medical Microbiology. 2006;55:101–107. doi: 10.1099/jmm.0.46212-0. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Röças IN, Siqueira JFJ, Benno Y. Molecular analysis of bacteria in asymptomatic and symptomatic endodontic infections. Oral Microbiology and Immunology. 2006;21:112–122. doi: 10.1111/j.1399-302X.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- Sathorn C, Parashos P, Messer HH. How useful is root canal culturing in predicting treatment outcome? Journal of Endodontics. 2007;33:220–225. doi: 10.1016/j.joen.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Applied and Environmental Microbiology. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan GE, Masters Cl, Shallcross JA, MacKey BM. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Applied and Environmental Microbiology. 1998;64:1313–1318. doi: 10.1128/aem.64.4.1313-1318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira JJ, Röças IN. Diversity of endodontic microbiota revisited. Journal of Dental Research. 2009;88:969–981. doi: 10.1177/0022034509346549. [DOI] [PubMed] [Google Scholar]
- Soejima T, lida K, Qin T, et al. Photoactivated ethidium monoazide directly cleaves bacterial DNA and is applied to PCR for discrimination of live and dead bacteria. Microbiology and Immunology. 2007;51:763–775. doi: 10.1111/j.1348-0421.2007.tb03966.x. [DOI] [PubMed] [Google Scholar]
- Sogin M, Morrison H, JA H, et al. Microbial diversity in the deep sea and the underexplored "rare biosphere". Proceedings of the National Academy of Sciences U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkerding KV, Dames SA, Durtschl JD. Next-Generation Sequencing: from basic research to diagnostics. Clinical Chemistry. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]