Abstract
Background
Lymphatic filariasis and onchocerciasis are disabling and disfiguring neglected tropical diseases of major importance in developing countries. Ivermectin is the drug of choice for mass drug administration programs for the control of onchocerciasis and lymphatic filariasis in areas where the diseases are co-endemic. Although ivermectin paralyzes somatic and pharyngeal muscles in many nematodes, these actions are poorly characterized in adult filariae. We hypothesize that paralysis of pharyngeal pumping by ivermectin in filariae could result in deprivation of essential nutrients, especially iron, inducing a wide range of responses evidenced by altered gene expression, changes in metabolic pathways, and altered developmental states in embryos. Previous studies have shown that ivermectin treatment significantly reduces microfilariae release from females within four days of exposure in vivo, while not markedly affecting adult worms. However, the mechanisms responsible for reduced production of microfilariae are poorly understood.
Methodology/Principal Findings
We analyzed transcriptomic profiles from Brugia malayi adult females, an important model for other filariae, using RNAseq technology after exposure in culture to ivermectin at various concentrations (100 nM, 300 nM and 1 μM) and time points (24, 48, 72 h, and 5 days). Our analysis revealed drug-related changes in expression of genes involved in meiosis, as well as oxidative phosphorylation, which were significantly down-regulated as early as 24 h post-exposure. RNA interference phenotypes of the orthologs of these down-regulated genes in C. elegans include “maternal sterile”, “embryonic lethal”, “larval arrest”, “larval lethal” and “sick”.
Conclusion/Significance
These changes provide insight into the mechanisms involved in ivermectin-induced reduction in microfilaria output and impaired fertility, embryogenesis, and larval development.
Author Summary
Lymphatic filariasis and onchocerciasis are tropical diseases caused by infections with parasitic nematodes. Resulting chronic diseases can be strongly blinding and disfiguring, and contribute to an entrenched cycle of poverty in affected populations. Ivermectin is one of the pivotal drugs used to control these infections. The mechanism of antifilarial action of the drug is incompletely resolved. It kills circulating larval stages (microfilariae), but only reversibly sterilizes adult worms without killing them. Our limited understanding of the involved mechanisms hampers treatment optimization and sustainability of the efficacy of the drug, and investigations into its pharmacology are of paramount importance. Working with Brugia malayi adult females, we employed RNA sequencing and bioinformatics analyses to identify genes for which expression levels changed as a result of exposure to the drug in vitro. Ivermectin exposure altered the expression of genes that are likely to function in the B. malayi female reproductive system even at the lowest concentration tested. Through several biological pathways, genes involved in meiosis were particularly affected. These findings provide some insight into the mechanisms involved in ivermectin-induced reduction in microfilaria output and impaired fertility, embryogenesis, and larval development.
Introduction
Lymphatic filariasis (LF) is caused by the transmission of Wuchereria bancrofti, Brugia malayi, and Brugia timori by infected mosquitos and is endemic in 60 countries, mainly in subtropical and tropical regions, with over 120 million people infected [1]. Onchocerciasis (river blindness) is caused by Onchocerca volvulus and afflicts 25 million people in 24 countries, mainly in Sub-Saharan Africa [2]. Long-running control and elimination programs aim to reduce transmission of both infections by mass drug administration (MDA) of microfilaricidal drugs and to alleviate suffering and disability in onchocerciasis patients [3, 4].
The macrocyclic lactones (ML) are a class of broad-spectrum anthelmintic drugs which include the avermectins and milbemycins (e.g., ivermectin (IVM) and moxidectin, respectively) [5]. Although similar in structure, they differ in pharmacokinetics and dynamics as well as physicochemical properties [6, 7]. These drugs have been extensively used in veterinary medicine for the treatment or prevention of nematode infections and ectoparasite infestations. In human medicine, IVM is one of the three mainstay drugs in the LF global elimination program [8] and is the only drug used in onchocerciasis MDA campaigns [2]. IVM is predominantly microfilaricidal, the killing effect on adults being only moderate and requiring repeated treatment cycles over several years [9].
IVM pseudo-irreversibly activates nematode and arthropod glutamate-gated chloride channels (GluCls), resulting in hyperpolarization of neurons and pharyngeal muscle cells, leading to paralysis of movement and pharyngeal pumping, and ultimately starvation of the worm [10–12]. Treatment results in a dramatic reduction in microfilaria (mf; first-stage larvae (L1) production by adult female filariae, but the mechanisms involved are not understood. GluCl expression in B. malayi mf was localized to a muscle structure surrounding the excretory-secretory (ES) vesicle, suggesting that release of proteins was regulated by GluCls (14). In the same study, IVM exposure resulted in a significant reduction of protein release, possibly preventing the secretion of proteins that allow evasion of the host’s immune system [13]. Recently, the B. malayi GluCl gene avr-14 was found to be highly expressed in reproductive tissues and embryos, suggesting an involvement of GluCls in gamete production and embryogenesis which may account for the sterilizing effect observed in adult filarial worms treated with IVM [14]. Our limited understanding of IVM’s mode of action on adult filarial worms hampers treatment optimization and sustainability of the efficacy of this drug, and investigations into its pharmacology are of paramount importance.
We used adult female B. malayi, a well-characterized model of filarial infections, to study the time-dependent transcriptomic changes induced by exposure to IVM in vitro. We performed two separate studies: 100 nM IVM exposure for up to 72 h, and 300 nM and 1 μM for up to 5 days, to gain insight into the mechanisms which underlie the drug-induced long-term sterility of adult females and subsequent reduction of mf in skin or blood of treated patients. The drug concentrations used in this present study have previously been tested on adult B. malayi females in vitro [15] and have also been tested in C. elegans [7]. The rationale for using the lowest concentration (100 nM) in the first study was to expose worms to a concentration similar to plasma levels detected in humans after administration of a dose typically used in MDA [16–18]. It is recommended that in vitro experimental analyses of drug effects include concentrations higher than the maximal blood/plasma concentration achieved in an animal [19]. We therefore tested higher IVM concentrations in a second phase. These results allowed us to also compare our results with those previously observed in B. malayi at these concentrations.
Materials and Methods
Ethics statement
All animal procedures were approved by the University of Georgia, Institutional Animal Care and Use Committee, and complied with U.S. Department of Agriculture's regulations (USDA Assurance No. A3437-01)
Parasites and study designs
Adult male jirds (Meriones unguiculatus) were infected subcutaneously with ≈400 infective B. malayi larvae (L3). After a minimum of 90 days post-infection (ranging from 3 to 6 months), jirds were euthanized by exposure to CO2 and adult worms were collected from the peritoneal cavity via lavage.
Study design
First study
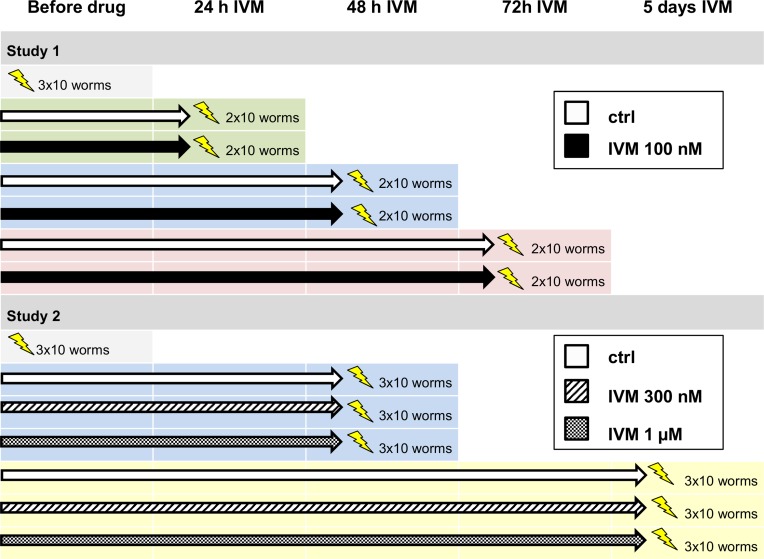
Female worms from three individual jird hosts were randomly assigned to 15 groups (9 untreated and 6 exposed to 100 nM IVM) of 10 worms each. The study design consisted of 4 time points (before drug exposure, 24, 48, and 72 h). Three technical replicates were used before drug exposure and duplicates for the remaining conditions (Fig 1).
Fig 1. Study design.
At each time point, two or three groups of 10 worms were washed, flash-frozen (represented by lightning bolt) and used for RNA extraction. IVM: ivermectin.
Second study
Female worms from three individual jird hosts were randomly assigned to 21 groups (9 untreated, 6 exposed to 300 nM IVM, and 6 exposed to 1 μM IVM) of 10 worms each. The study design consisted of 3 time points (before drug exposure, 48 h and 5 d). Three technical replicates were used for each condition (Fig 1).
For both studies, worms were collected from jirds at the NIH-NIAID Filariasis Research Reagent Resource Center (FR3) at the University of Georgia and separated by sex. Worms were then shipped overnight in separate tubes of 15 ml culture media containing RPMI-1640 (BioWhittaker Classic Cell Culture Media, Lonza Walkersville Inc.) and 1% v/v gentamycin (gentamycin solution, 10 mg/ml Sigma Aldrich Inc.) to McGill University for further analysis. The separate studies were conducted several months apart.
Upon arrival at McGill, the first group (before drug exposure) was thoroughly washed several times in PBS and flash-frozen in liquid N2 for RNA extraction.
Remaining worms were incubated 1 worm per well of a 6-well plate (Costar) containing 6 ml RPMI 1640 (Sigma-Aldrich) supplemented with 10% v/v heat-inactivated fetal bovine serum (Sigma-Aldrich, F1051), 5% penicillin/streptomycin (Sigma–Aldrich P4333) and 2% v/v gentamycin (Gibco, 15750–060). Worms were cultured for another 24, 48, or 72 h (first study) and 48 h or 5 days (second study) at 37°C, 5% CO2 with or without exposure to IVM (Sigma-Aldrich I8898) at 100 nM (study 1), 300 nM or 1 μM (study 2). Control wells contained 0.1% v/v DMSO. On a daily basis, 3 ml of culture media (with or without drug at the different concentrations) in each well was replaced with fresh media.
RNA extraction
RNA extraction was performed as described previously [20]. Briefly, live worms were washed several times with PBS; immotile worms were excluded from the study. RNA was extracted from 5 to 8 worms per group using nuclease-free reagents. On ice, 125 μl 0.1X Tris-EDTA (TE) buffer, pH 8.0 (Ambion, Life Technologies, Burlington, ON) and 375 μl Trizol LS reagent (Ambion) were added to each tube. Worm extracts were obtained by conducting three cycles of flash-freezing in liquid N2 followed by crushing of worms using plastic pestles. One-hundred (100) μl chloroform was added to each sample in a 1.5 ml Eppendorf tube, which was vortexed and incubated 3 min at room temperature. The extracts were transferred to phase-lock gel heavy tubes (5 PRIME, Gaithersburg, MD) and centrifuged at 11,900 x g at 4°C for 15 min for phase separation. The aqueous phase (approximately 150 μl) was transferred to fresh 1.5 ml Eppendorf tubes and mixed with 250 μl ice-cold isopropanol. RNA was precipitated by centrifugation at 12,200 x g at 4°C for 30 min followed by an overnight incubation at -20°C. Pellets were washed twice with 80% EtOH, allowed to dry and resuspended in 50 μl 0.1X TE prior to heating at 55°C for 10 min. Total RNA was purified and concentrated on columns (RNeasy Min-Elute Cleanup Kit, Qiagen, Valencia, CA). Samples were treated with DNase (Ambion DNA-free Kit, Life Technologies, AM1906, Burlington, ON) according to the manufacturer’s instructions to remove contaminating DNA. Assessment of RNA concentration and purity was done by spectrophotometry (NanoDrop 1000, Wilmington, DE) and RNA samples were shipped overnight on dry ice to the NIH-FR3 (Molecular Division) at Smith College (Northampton, MA) for cDNA library preparation and Illumina sequencing. RNA concentration, purity and integrity were precisely measured using the Qubit RNA BR Assay Kit (Life Technologies, Q10210, Burlington, ON) and on an Agilent 2100 Bioanalyzer (Santa Clara, CA).
cDNA library preparation and Illumina sequencing
Messenger RNA was isolated with a NEBNext Poly (A) mRNA Magnetic Isolation Module (New England Biolabs, E7490). The enriched mRNA served as template for cDNA library preparation with a NEBNext Ultra RNA Library Prep Kit Illumina (NEB, E7530) and NEBNext Multiplex Oligos for Illumina (Index Primer 1–12) (NEB, E7600) following the manufacturer’s instructions. Assessment of quality, concentration and size of the cDNA, was performed for each library using a Qubit 2.0 Fluorometer (Life Technologies, Q32866), Qubit dsDNA BR assay kit (Life Technologies, Q32850), High Sensitivity DNA Analysis Kit (Agilent, 5067–4626) and Agilent 2100 Bioanalyzer. cDNA libraries were sequenced on an Illumina MiSeq Platform employing a 150 base pair paired-end NGS setting. Sequencing data were uploaded and stored in BaseSpace (https://basespace.illumina.com) for subsequent analysis.
Data analysis
RNA sequencing analysis
Procedures for data analysis were previously described [20]. Unaligned raw sequencing data (fastq) files were downloaded to the Mason-Galaxy platform hosted at Indiana University (http://galaxy.iu.edu/) [21, 22]. Raw sequence data were first processed using Fastq Groomer (v 1.0.4) to convert file format to Fastq Sanger format and quality scores were verified using FastQC (version 0.52). Based on quality statistics, sequences were trimmed from both the 5’ and 3’ ends using Fastq Quality Trimmer (v 1.0.0) and Trim Galore (v 0.2.8.1). Mapping of gapped reads to the B. malayi reference genome (ftp://ftp.wormbase.org/pub/wormbase/species/b_malayi/sequence/genomic/b_malayi.PRJNA10729.WS243.genomic.fa.gz) was performed with Tophat2 (v 0.6) and RNA sequencing metrics were retrieved using Picard tools (Table 1). Picard alignment summary metrics (http://broadinstitute.github.io/picard/) were generated from BAM files using SAM/BAM Alignment Summary Metrics (version 1.56.0) tool. The read count table was then generated by counting the number of reads per gene feature with HTSeq-count (version 1.0.0) [23] using the union mode to handle reads covering more than one feature and default settings for all other parameters. Differential gene expression analysis between time points and drug concentrations was realized in edgeR (version 3.10.5) [24] through the web interface provided by NetworkAnalyst (http://networkanalyst.ca) [25, 26]. To correct for different library sizes and reduce RNA compositional effects, read counts for gene features were normalized in edgeR using the trimmed mean of M-values (TMM) method [27]. The empirical Bayes method based on weighted conditional maximum likelihood was employed to estimate tagwise (gene-specific) dispersion values [28]. Once negative binomial models were fitted and dispersion values estimated, the exact test was used for pairwise differential expression testing between treated and untreated groups. Significance was set as an experiment-wide false discovery rate (FDR) <0.20 (after the Benjamini-Hochberg method [29]). After filtering, gene lists for each pairwise comparison were imported into Microsoft Excel for further analysis.
Table 1. Picard alignment summary metrics.
Summary of the sequencing and mapping of the data to the B. malayi transcriptome.
| Time point | Concentration | Total # Transcripts | Ave Total # Reads | Ave Total # Reads Mapped | Ave Total # High Quality Reads Mapped | % High Quality Reads Mapped | |
|---|---|---|---|---|---|---|---|
| Study 1 | 24 h | IVM (100 nM) | 10691 | 3537601 | 3537500 | 2990362 | 84.53 |
| Control | 10511 | 2765626 | 2765551 | 2364245 | 85.49 | ||
| 48 h | IVM (100 nM) | 10086 | 2513891 | 2513834 | 2145661 | 85.35 | |
| Control | 10214 | 2957918 | 2957831 | 2519799 | 85.19 | ||
| 72 h | IVM (100 nM) | 10705 | 3875640 | 3874592 | 3380930 | 87.24 | |
| Control | 10428 | 3081545 | 3081499 | 2683543 | 87.09 | ||
| Study 2 | 48 h | IVM (300 nM) | 10087 | 2091222 | 2091198 | 1710598 | 81.80 |
| IVM (1 μM) | 10177 | 2212133 | 2212085 | 1757015 | 79.43 | ||
| Control | 10328 | 2808908 | 2808880 | 2325764 | 82.80 | ||
| 5 d | IVM (300 nM) | 9959 | 1799180 | 1799161 | 1456538 | 80.96 | |
| IVM (1 μM) | 9837 | 1798183 | 1798159 | 1426230 | 79.32 | ||
| Control | 10250 | 2464701 | 2464671 | 2017612 | 81.86 |
Common dispersion, which provides information on the overall variability across the genome for a dataset, and Biological Coefficients of Variation (BCV) were calculated; BCV plots were generated using the edgeR Bioconductor package (version 3.12.0) [24].
Bioinformatics analysis of the sequence data
From lists of retained genes for each comparison, the Wormbase Gene ID was used to retrieve the primary corresponding sequence ID of the gene from Wormbase (http://www.wormbase.org/) and the UniProt accession number [30]. Protein coding sequences from Wormbase were used in BLASTn in order to find the closest B. malayi gene hit and attribute a GenBank ID and annotation to each gene. Gene Ontology (GO) terms [31] were obtained when available from nematode.net (v 4.0; http://nematode.net/NN3_frontpage.cgi) [32, 33]. Enriched GO terms and KEGG/Panther pathways were retrieved and analyzed using WebGestalt (WEB-based Gene SeT AnaLysis Toolkit) [34]. Pathways and GO terms were considered significantly enriched when FDR<0.05. Since many DE genes detected in this study were uncharacterized or not annotated yet had orthologs which could be identified in the Caenorhabditis elegans genome, we used the Uniprot accession numbers of the C. elegans orthologs of B. malayi differentially expressed (DE) genes for enrichment analysis, accepting hits with a minimal E-value of 1*10−20 and using the Blast-based Reciprocal Best Hit (RBH) strategy for identifying orthologs [35, 36]. RNA interference (RNAi) phenotypes associated with the C. elegans orthologs of down-regulated genes at 48 h with 100 nM IVM exposure involved in reproduction, embryo and larval development were retrieved from www.wormbase.org. A hypergeometric test was performed to determine which phenotypes in this dataset were significantly enriched when compared with their overall occurrence in the C. elegans RNAi dataset. P-values were corrected for multiple testing using the Benjamini-Hochberg method (1995) with a level of significance of q<0.05.
Results
Sequencing and mapping
Table 1 displays the total number of transcripts obtained for each sample in both studies and the number of sequence reads for each cDNA library. On average for study 1 (100 nM IVM), 85.82% of the high quality sequence reads were mapped to the B. malayi transcriptome after removal of ambiguous sequence matches (Table 1). Gene mapping fluctuated from 1 to 58487 sequence read(s) per gene. On average for study 2 (300 nM and 1 μM IVM), 81.03% of the high quality sequence reads were mapped to the B. malayi transcriptome after removal of ambiguous sequence matches, and gene mapping fluctuated from 1 to 27507 sequence read(s) per gene (Table 1). In study 1, 93.28–95.79% of reads aligned in pairs to a transcript; in study 2, 90.43–93.52% of reads aligned in pairs.
BCV values ranged from 17 to 27% in the first study and 15 to 35% in the second study (S3 Table). BCV plots show that the BCV values in the first study tended to increase over time and are shown in S1A–S1C Fig. In the second study, the BCV values were higher at the 48 h time point than at 5 days for both IVM concentrations.
We used gene expression levels from untreated (control) worms at each time point as a baseline to identify genes with differential expression after exposure to IVM in each sample. Pairwise comparisons revealed 15 (100 nM, 72 h) to 421 (100 nM, 48 h) DE genes (Table 2, S1 Table). Between 20.59 and 65.15% of DE genes could be assigned GO terms, while similar proportions had a C. elegans ortholog.
Table 2. Numbers of differentially expressed genes over time after IVM exposure in culture.
| 100 nM IVM | 300 nM IVM | 1 μM IVM | |||||
|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 48 h | 5 d | 48 h | 5 d | |
| Total DE genes | 34 | 421 | 15 | 132 | 68 | 147 | 271 |
| Total DE genes up-regulated | 21 | 141 | 2 | 14 | 24 | 136 | 74 |
| Total DE genes down-regulated | 13 | 280 | 13 | 118 | 44 | 11 | 197 |
| Sequences with GO terms* | 7 | 178 | 9 | 86 | 29 | 49 | 160 |
| Sequences with C. elegans ortholog | 7 | 257 | 9 | 92 | 37 | 64 | 186 |
*GO terms were provided by Nematode.net V.4 [33]. IVM: ivermectin; DE: differentially expressed
Transcriptomic profiles of worms exposed to IVM
The greatest overlap of DE genes was seen between the 300 nM and 1 μM groups, with 66 dysregulated genes. When comparing the overlap of DE genes from the first and second study, only 5 genes were found to overlap across the 3 concentrations, with 10 genes in common between the 100 nM and 300 nM groups and 20 genes in common between the 100 nM and 1 μM groups. GO term enrichment showed that 5 of these genes were enriched in “structural molecule activity” molecular function (GO:0005198; FDR = 0.009). Five genes were common to all three IVM treatments (Bm8439 and Bm4605 (two collagens), Bm9776, Bm6450, and Bm9380). Bm9380 is a serpin precursor (Bm-spn-2) and was significantly up-regulated in both studies (100 nM 72 h; 300 nM and 1 μM 5 days; log2 fold-changes 1.37–2.57). Bm9776 encodes a hypothetical protein and was also significantly up-regulated at four time points between both studies (log2 fold-changes 1.06–1.85). Lastly, Bm6450 is annotated as a GTPase activating protein for Arf containing protein and was found to be down-regulated across three different time points between both studies (log2 fold-changes -1.06 to -1.43).
Study 1: Transcriptomic profiles of worms exposed to 100 nM IVM
At 100 nM IVM, 460 DE genes were identified across all time points. Worms exposed to 100 nM IVM for up to 72 h showed the largest change in their transcriptome profiles at 48 h post-exposure (421 DE genes), at which time 67% of those genes were down-regulated (S1 Table, Table 2). Among them, a dynein light chain type 1 family protein, Bm8648, was down-regulated (log2 fold-change: -1.46), as well as Bm14080 (log2 fold-change: -1.51). A cytidine deaminase (Bm4230) was the gene most down-regulated at the 48 h and 72 h time points (log2 fold-change: -5.21 and -6.90, respectively), while the most prominently up-regulated gene in study 1 was Bm8966 (hypothetical protein; log2 fold-change: 6.91; 72 h 100 nM). To cite only a few interesting dysregulated genes at 48 h or 72 h, Bma-nep-1, a neprilysin (Bm7419, log2 fold-change: -3.92), 6 enzymes involved in oxidative phosphorylation (Bm6047, Bm3783, Bm3758, Bm5403, Bm10539, Bm2497; log2 fold-changes between -0.85 and -1.53), and inositol-1 (Bm11145; log2 fold-change: -4.48) were among down-regulated genes. Eight DE genes overlapped across the 24 h and 48 h time points (among them, a kinesin light chain protein 2 (Bm9032), up-regulated at 24 h (log2 fold-change: 2.10) but then significantly down-regulated by 72 h (-3.23 log2 fold-change); only one common DE gene was found at both 48 h and 72 h. One DE gene was shared between 24 h and 72 h (but not with 48 h).
Exposure to 100 nM also resulted in the significant down-regulation (-1.16 to -2.83 log2 fold) of five cuticle collagens at all time points.
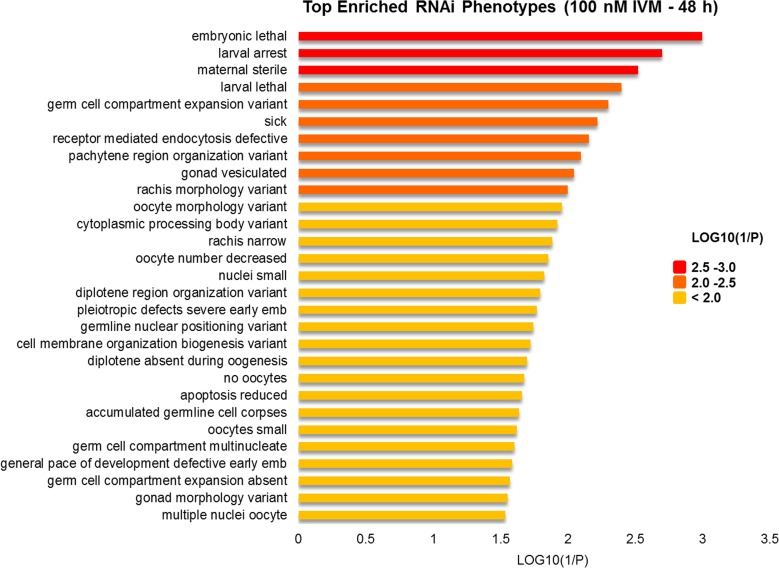
Gene Ontology enrichment analysis of the 100 nM (48 h) dataset revealed significant enrichment of genes with GO terms associated with “growth” (GO:0040007; FDR = 2.03e-08), “locomotion” (GO:0040011; FDR = 2.99e-07) and developmental processes which included “embryo development” (GO:0009790; FDR = 1.07e-06), “embryo development ending in birth or egg hatching” (GO:0009792; FDR = 1.07e-06) and “nematode larval development” (GO:0002119; FDR = 1.07e-06), with 63–91 genes enriched in these categories. Table 3 shows the distribution of the most enriched GO terms at 100 nM IVM exposure for 48 h, for which reproductive processes (reproduction, embryo development and larval development) were statistically overrepresented in the dataset. RNAi phenotypes which were significantly overrepresented in the down-regulated genes dataset compared with their overall abundance in the C. elegans RNAi phenotype dataset in Wormbase are shown in Fig 2 and S4 Table (FDR < 0.05; hypergeometric test). The most commonly observed RNAi phenotype is “embryonic lethal”. Over 81% of down-regulated genes involved in reproduction were associated with an embryonic lethal RNAi phenotype, while 62% were linked to a larval arrest phenotype. At the 24 h time point, no significantly enriched biological processes or molecular functions were noted. “Cytoskeleton” was the greatest enriched cell component term (GO:0005856; FDR = 0.007).
Table 3. Gene Ontology enrichment analysis of top 15 biological processes at 100 nM IVM exposure for 48 h.
| Gene Ontology Term* | GO Identity | Observed | Expected | Reference | Ratio of Enrichment | FDR value** |
|---|---|---|---|---|---|---|
| growth | GO :0040007 | 85 | 45.16 | 2498 | 1.88 | 2.03E-08 |
| locomotion | GO :0040011 | 63 | 30.10 | 1665 | 2.09 | 2.99E-07 |
| embryo development | GO :0009790 | 91 | 55.49 | 3069 | 1.64 | 1.07E-06 |
| embryo development ending in birth or egg hatching | GO :0009792 | 89 | 54.10 | 2992 | 1.65 | 1.07E-06 |
| nematode larval development | GO :0002119 | 72 | 39.00 | 2157 | 1.85 | 1.07E-06 |
| larval development | GO :0002164 | 72 | 39.03 | 2159 | 1.84 | 1.07E-06 |
| post-embryonic development | GO :0009791 | 72 | 39.31 | 2174 | 1.83 | 1.07E-06 |
| establishment of localization | GO :0051234 | 65 | 33.63 | 1860 | 1.93 | 1.07E-06 |
| transport | GO :0006810 | 63 | 32.33 | 1788 | 1.95 | 1.21E-06 |
| regulation of growth | GO :0040008 | 61 | 31.24 | 1728 | 1.95 | 1.96E-06 |
| positive regulation of growth | GO :0045927 | 59 | 29.90 | 1654 | 1.97 | 2.32E-06 |
| positive regulation of growth rate | GO :0040010 | 54 | 26.27 | 1453 | 2.06 | 2.38E-06 |
| regulation of growth rate | GO :0040009 | 54 | 26.31 | 1455 | 2.05 | 2.38E-06 |
| cellular localization | GO :0051641 | 26 | 7.85 | 434 | 3.31 | 2.38E-06 |
| reproduction | GO :0000003 | 80 | 48.00 | 2655 | 1.67 | 3.76E-06 |
*Analysis performed using Web-Based Gene Set Analysis Toolkit (WebGestalt).
**Statistical method: Hypergeometric; p-value adjusted using the Benjamini-Hochberg multiple test adjustment.
Fig 2. Top RNAi phenotypes enriched in reproductive processes after 48 h exposure to 100 nM IVM.
The listed enriched phenotypes obtained were based on Gene Ontology analysis of C. elegans orthologs of DE genes. These phenotypes were significantly overrepresented compared with their overall abundance in the C. elegans RNAi phenotype dataset in Wormbase. Statistical method: Hypergeometric; p-value adjusted using the Benjamini-Hochberg multiple test adjustment (FDR<0.05).
Study 2: Exposure to higher IVM concentrations
In the second study, most changes occurred after 48 h exposure to 300 nM IVM, and only after 5 days when exposed to 1 μM. The expression of 535 genes was dysregulated across several time points and/or concentrations (S1 Table). As the BCVs varied with time and across concentrations, resulting in higher FDRs (as BCVs increased), it was difficult to see a concentration-dependent trend across the DE genes detected in the study and therefore the overlap between DE genes across concentrations and time points was very limited.
At 48 h, only 3 genes overlapped between the groups exposed to 300 nM and 1 μM IVM; at 5 days, 18 genes overlapped between both treatment groups. Among them, 3 genes encoding cuticle collagens (Bm8043, Bm4605, and Bm11024; log2 fold-changes -0.84–-2.23), and others, to cite a few only, coding for a major facilitator superfamily protein (Bm6076; log2 fold-changes -1.1 and -2.21), Bma-zyg-9 (Bm3504; log2 fold-change -1.51 at 48 h and -1.20 at 5 days), which may play a role in meiosis, were down-regulated. Upregulated across concentrations at 5 days was a serpin precursor (Bm9380; log2 fold-changes 1.37 and 2.08).
Transcriptomic profiles of worms exposed to 300 nM IVM
Worms incubated at this concentration showed the largest change after 48 h exposure (132 DE genes). 89% of DE genes were down-regulated at this time. The most strongly down-regulated gene encodes a prominin protein (Bm3187; log2 fold-change: -3.84) predicted to be a 5-transmembrane domain glycoprotein. Other annotated genes of potential interest that were highly down-regulated include IQ calmodulin-binding motif family protein (Bm2777; log2 fold-change: -2.01), Fanconi anemia complementation group D2 protein (Bm6522; log2 fold-change: -2.0), CG7507-PA (Bm5897; log2 fold-change: -1.20), an ortholog of dynein heavy chain-1 in C. elegans, and nucleoporins (Bm6397 and Bm5288; log2 fold-change: -1.19 and -1.91, respectively). The most up-regulated gene was Bm12387 (log2 fold-change: 3.83), annotated as a hypothetical protein. Gene Ontology analysis at this time point showed an enrichment of genes related to “establishment of meiotic spindle orientation” (GO:0051296; FDR = 0.04), “meiotic spindle organization” (GO:0000212; FDR = 0.04) and “establishment of meiotic spindle localization” (GO:0051295; FDR = 0.04) (S2A Table).
By 5 days, the number of DE genes decreased from 132 to 68. Bm11304, a KH domain containing protein, was the most prominently down-regulated gene at this time point and concentration (log2 fold-change: -5.9), while Bm1543, a lectin C-type domain containing protein, was the most up-regulated (log2 fold-change: 5.88). GO enrichment analysis revealed enrichment of genes in molecular processes related to “fatty-acyl-CoA binding” (GO:0000062; FDR = 0.0057), “structural constituent of cuticle” (GO:0042302; FDR = 0.0002), and “structural molecule activity” (GO:0005198; FDR = 0.016) (S2C Table).
Transcriptomic profiles of worms exposed to 1 μM IVM
Exposure to 1 μM IVM resulted in the dysregulation of 147 genes at 48 h and 271 after 5 days. Most strongly down-regulated at 48 h was Bm2880, a tachykinin-like peptide receptor protein (log2 fold-change: -5.51), and a GRIP-domain containing protein, at 5 days (log2 fold-change: -5.71). At both time points, two hypothetical proteins were maximally up-regulated (log2 fold-change: 6.76 and 6.13, respectively). Transcriptomic data from worms exposed to 1 μM IVM for 48 h showed “translation” to be the most significantly enriched GO term (GO:0006412; FDR = 8.87e-06) (S2B Table). After 5 days exposure, down-regulation of a phospholipase C homolog (Bm3716; log2 fold-change: -1.54), and of 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (Bm1768; log2 fold-change: -0.89) was observed. Significantly enriched GO terms also included terms related to “double-strand break repair via nonhomologous end joining” (GO:0006303; FDR = 0.02), “cytokinesis” (GO:0007109; FDR = 0.02), “cytokinetic cell separation” (GO:0000920; FDR = 0.02), “cytokinetic process” (GO:0032506; FDR = 0.003), “cytokinesis after meiosis” (GO:0033206; FDR = 0.01) and “polar body extrusion after meiotic divisions” (GO:0040038; FDR = 0.004). Nine genes were also found enriched in the “collagen and cuticulin-based cuticle development” (GO:0040002; FDR = 0.003) category (S2D Table).
Discussion
We performed time-series studies to assess the effects of exposure to different concentrations of IVM on adult B. malayi females in vitro at the transcriptomic level. Of particular interest was the identification of DE genes involved in reproduction, which could help explain the sterilizing effect of IVM on adult filariae.
Overall BCVs were calculated between samples in both drug studies and were 17–27% in the first study and 15–35% in the second study (S3 Table, S1 Fig), suggesting acceptable biological variability among replicates. Typically, BCVs for datasets from well-controlled studies range from 10% (genetically identical model organisms) to 40% (human data). In a previous study, we reported BCVs to be 14–25% for samples of adult female worms immediately upon extraction from jirds [20]. However, BCVs were slightly higher at the 48 h time point in study 2 (30 and 35%). As BCVs varied across the different concentrations and time points and resulted in higher FDRs as the BCVs increased, we were unable to see obvious trends in concentration- dependence and detected few DE genes that overlapped across the different comparisons. Therefore, our discussion is mainly centered on the analysis of the DE genes detected for each pairwise comparison. We focus on a few processes particularly affected by exposure to IVM in vitro.
IVM significantly reduces the release of mf after 24 h exposure to IVM in vitro and by 4 days, mf output ceases completely at concentrations ≥ 0.15 μg/ml [15]. In the present work, we exposed adult females at a concentration of 100 nM (0.09 μg/ml) for up to 72 h in the first study and 300 nM (0.267 μg/ml) and 1 μM (0.89 μg/ml) for up to 5 days in the second study. Although we did not measure mf output, we suspect that adult females were sterilized at these concentrations based on the previous work. In addition, wsp (Wolbachia surface protein) expression has been shown to decrease significantly with IVM concentrations >0.31 μg/ml (348 nM) [15]. It is therefore possible that some of the observed changes in gene expression might be related to changes in Wolbachia densities; however, we are unable to confirm this as wsp expression was not measured. Wolbachia-related death at these concentrations could have negative implications on reproduction, molting and survival. Interestingly, the Wolbachia surface protein wBm0432 has been shown to interact with several key enzymes involved in the host glycolytic pathway, including aldolase and enolase, and wBm0152 interacts with the host cytoskeletal proteins actin and tubulin [37]. These interactions could have an effect on growth and embryogenesis, effects we observed at the transcriptome level.
IVM dysregulates genes involved in meiosis and calcium signaling surrounding fertilization
Dyneins and kinesins play important roles in mitosis and meiosis [38–40] and both were differentially expressed by exposure to IVM. CG7507-PA (Bm5897), orthologous to dynein heavy chain-1 in C. elegans, was down-regulated after IVM exposure. These molecular motors use ATP to move along microtubules and function in carrying cargo such as vesicles and organelles which are too large to diffuse to other cellular destinations [41]. In C. elegans, dynein is associated with nuclear envelopes, centrosomes and meiotic and mitotic spindle poles [42]. Kinesin-1 and cytoplasmic dynein function together to move the meiotic spindle to the oocyte cortex [39]. During anaphase, the meiotic spindle is attached to the cortex by one pole, allowing selective disposal of half the chromosomes in a polar body [39]. In C. elegans, dynein light chain 1 (dylt-1) is expressed in adult body wall muscle, larval and adult pharynx, posterior intestine and nervous system and is required for normal embryonic and larval viability. The GO term “establishment of meiotic spindle orientation” was enriched in this study. One gene associated with this term was Bm2777 (IQ calmodulin-binding motif family protein), a homolog of aspm-1 in C. elegans and mammalian ASPM (Abnormal Spindles & Primary Microcephaly) [43]. ASPM-1 is required for oocyte meiotic spindle pole assembly and is essential for the meiotic spindles to align orthogonally and in close proximity to the overlying plasma membrane [43]. Another gene that may play a role in meiosis is Bma-zyg-9 (Bm3504), which was down-regulated at the two highest IVM concentrations. In C. elegans, ZYG-9 acts to ensure correct microtubule assembly throughout the cell cycle of early embryos during mitosis, interphase and meiosis [44–46].
A major facilitator superfamily protein (Bm6076), down-regulated at 5 days at the two highest IVM concentrations, is highly expressed in the digestive tract of B. malayi [47]. In C. elegans, RNAi phenotypes associated with the ortholog of Bm6076 include “embryonic lethal”, “asymmetric cell division defective early embryo”, and “spindle pole pulling force variant”, indicating that this gene is probably important for embryo development.
Calcium signaling plays a crucial role in reproduction and fertilization and has been extensively studied in C. elegans [48]. During meiotic maturation of the oocyte, calcium signaling is linked to inositol 1,4,5-triphosphate receptor (ITR-1), N-methyl D-aspartate type glutamate receptor subunit (NMR-1) and UNC-43, which encodes a type II calcium/calmodulin-dependent protein kinase [49]. Phospholipase C catalyzes the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) into diacylglycerol (DAG) and inositol (1,4,5) triphosphate (IP3) [50]. IP3 binds receptors on the endoplasmic reticulum to mobilize Ca2+ into the cytosol. We observed down-regulation of a phospholipase C homolog (Bm3716) and of 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (Bm1768). The latter is an ortholog of egl-8 (egg-laying defective) and is predicted to have phosphatidylinositol phospholipase C activity and Ca2+ ion binding activity. Interestingly, EGL-8 also plays a role in regulating the release of acetylcholine from motor neurons, thus affecting locomotion, and is expressed in all neurons in C. elegans [51]. One of the most down-regulated DE genes we observed was inositol-1 (Bm11145), which is required for the biosynthesis of phosphatidylinositol [52]. Cumulatively, down-regulation of these genes suggests that Ca2+ signaling (via IP3) may be affected following exposure to IVM, with adverse effects on ovulation.
IVM selection on β-tubulin has been reported in O. volvulus and H. contortus; IVM selection changes the frequency of β-tubulin alleles [53]. Microtubules, which are assembled from heterodimers of α- and β-tubulin, drive chromosome motion in mitosis and meiosis [54], and so IVM selection for β-tubulin changes could have downstream effects on meiosis. A recent study in H. contortus has also shown that IVM binds to and alter the tubulin polymerization equilibrium, which can lead to mitotic arrest [55].
A number of other genes that function in fertilization were dysregulated by IVM exposure. Bma-emo-1 (Bm14080), which is involved in ovulation, intracellular protein transport and oocyte growth in C. elegans, was down-regulated. C. elegans emo-1 mutants fail to ovulate after meiotic maturation and oocytes remain trapped in the gonad arm and become endomitotic [56]. Fanconi anemia complementation group D2 protein (Bm6522) was decreased following IVM treatment; deletion of the C. elegans ortholog results in egg-laying defects, precocious oogenesis, and partial defects in fertilization, and may play a role in double-stranded DNA break repair during embryogenesis [57].
IVM exposure dysregulates the expression of genes involved in embryo development
IVM exposure downregulated the expression of several genes involved in RNA transport, with consequences on embryo development. Nuclear pore complexes (NPCs) are highly conserved proteinaceous structures embedded in the nuclear envelope that provide a connection between the nucleus and the cytoplasm [63]. Several genes (Bm6397, Bm9537, Bm5288) orthologous to NPC proteins in C. elegans (npp-13, npp-14 and npp-21 respectively) were down-regulated. Nucleoporins comprise NPCs and may function in transcriptional events, mRNA export, and genome organization [58]. In the nucleoplasm, dynamic nucleoporins may be functioning in the activation of developmental genes [58]. A subclass of nucleoporins plays a role in orienting the mitotic spindle in C. elegans embryos, and RNAi depletion of any of these nucleoporins leads to spindle orientation defects [64]. One of these nucleoporins is NPP-13. NPP-21, an ortholog of human TPR (translocated promoter region), is also involved in embryo development, locomotion and nematode larval development; RNAi knockdown results in embryonic lethal, larval arrest and locomotion variant phenotypes [59]. No obvious RNAi phenotypes have been reported for NPP-14.
Lastly, we observed down-regulation of Bm5608, which is orthologous to mel-46 (maternal effect lethal). In C. elegans, this gene encodes a DEAD-box protein required in the germ line for proper oogenesis and zygotically for post-embryonic development [60].
Pharyngeal pumping
Interestingly, we saw a significant down-regulation of Bma-nep-1 (Bm7419), which has 45.8% identity to nep-1 in C. elegans. Neprilysins are transmembrane zinc-metalloproteases that have roles in cell-to-cell signaling [61]. The expression pattern of nep-1 in C. elegans is limited to pharyngeal cells and a single head neuron; it is an effector of locomotion and pharyngeal pumping [62]. The locomotion of nep-1 knockouts is significantly impaired and RNAi knockdown results in a maternal sterile phenotype [62].
Oxidative phosphorylation
We noted a significant enrichment of dysregulated genes involved in oxidative phosphorylation (100 nM, 48 h). Expression of ubiquinol-cytochrome C reductase (Bm6047), succinate dehydrogenase (Bm3783), ATP synthase epsilon chain (Bm3758), ATP synthase f chain (Bm5403), NADH dehydrogenase subunit 1 (Bm10539) and cytochrome b5-like heme/steroid binding domain containing protein (Bm2497) was down-regulated. In contrast, NADH dehydrogenase subunit 1 was up-regulated at 1 μM (48 h). The down-regulation of these genes may suggest that ATP synthesis is directly or indirectly inhibited by IVM. Abamectin (ABA), another ML in the avermectin family, caused concentration-dependent (5–25 μM) inhibition of the respiratory chain [63]. Reduced expression of ribosomal proteins, histones, and prosaposins [64] are a signature of oxidative stress in C. elegans, but the homologs of these genes were not DE or only marginally so in our study. In nematodes, ATP is obtained by glycolysis or oxidative phosphorylation, using a variety of substrates [65, 66]. Several anthelmintics have been proposed to target helminth oxidative phosphorylation enzymes [67].
Glutamate-gated chloride channels
IVM binding to GluCls induces a slow onset, pseudo-irreversible glutamate-gated current [10, 68, 69]. Given the nature of the drug-receptor association, one can speculate on the necessity for a new cohort of GluCls to be expressed to overcome the effects of IVM. Little is known about the rate of receptor turn-over and synthesis in adult parasitic nematodes, but this scenario could contribute to the prolonged sterilization of adult filariae observed after a single dose of IVM. In this context, it might have been expected that this drug-receptor interaction would lead to increased expression of GluCl genes, which was not observed; expression of the putative receptor for IVM, Bm-avr-14 (Bm1710 isoforms) [10], was not dysregulated following exposure to the drug. GluCl-encoding genes are abundantly expressed in B. malayi female reproductive tissue (among other body parts, such as the pharynx), suggesting that the sterilizing effects of IVM may occur at a very early embryonic stage [14]. In males, GluCl expression in reproductive tissues was less abundant, but was localized to some extent to somatic muscle and the vas deferens, likely controlling body movement as well as sperm cell development and release [14]. Using a high-level IVM-resistant C. elegans strain (the strain DA1316, which has null mutations in three GluCl subunits: avr-14; avr-15; glc-1) [70], no differential expression was noted in the genes encoding the GluCl subunits avr-14 or avr-15, whereas a log fold-change of 0.78 (FDR = 0.06) in glc-1 was observed following treatment with 1.1 μM IVM. Exposure to the drug impaired pharyngeal pumping to some extent in that strain. The authors concluded that IVM induced differential expression of several genes in response to food deprivation, which are likely unrelated to GluCl expression [70].
Effects of in vitro culture on the B. malayi transcriptome
Twenty-nine cuticle collagen genes showed differential expression across both studies, of which 17 were significantly down-regulated following exposure to IVM. Previously, we examined the effect of in vitro cultivation, without anthelmintic, on the transcriptome of B. malayi and found 21 cuticle collagen genes to be significantly DE over time [20]. Thirteen of the DE collagen genes detected in the current work overlapped with dysregulated genes from the previous study (Bm11024, Bm11095, Bm11338, Bm1249, Bm2110, Bm2605, Bm2854, Bm4605, Bm7408, Bm7894, Bm8043, Bm8439 and Bm9021). Bm11024 and Bm11095 were significantly down-regulated in the in vitro cultivation study at 48 h in culture, yet in the present work, we found these genes to be up-regulated at 300 nM IVM for 5 days. Collagen gene dysregulation likely represents a non-specific marker of stress [20, 64]. Scanning electron microscopy has shown that IVM exposure leads to damage of the surface of B. malayi third-stage larvae, inducing the loss of regular cuticular annulations and causing morphological changes in the cuticular surface in the head, body, and tail regions [71]. Previously, we observed the pronounced up-regulation of a serpin precursor (Bm9380), and of a hypothetical protein (Bm337), attributed to in vitro cultivation [20]. In the current study, both genes were up-regulated, but to a much lower extent. This suggests that our untreated control corrects, to some extent, for differential gene expression solely due to life in liquid culture media.
Comparison of the IVM transcriptional response in C. elegans to B. malayi
We compared the IVM transcriptional response in C. elegans to B. malayi to look for common GluCl-independent, IVM-dependent gene expression signatures. The transcriptional response to IVM in C. elegans using whole genome microarray led to the identification of genes involved in early food deprivation response (i.e., fat mobilization) [70]. Using the DA1316 mutant strain (null mutations in three GluCl subunits), which is resistant to the paralytic effects of IVM on the body wall, worms were exposed to 100 nM or 1.1 μM IVM for 4 h. This resulted in the up-regulation of 3 genes and down-regulation of 12 genes in the 100 nM IVM group, with 216 up-regulated and 153 down-regulated in the 1.1 μM group. Comparing our dataset with the C. elegans study showed that only 5 genes overlapped, of which 2 were significantly down-regulated in both studies: 6-pyruvoyl tetrahydropterin synthase (Bm1701) and cytochrome b5-like heme/steroid binding domain containing protein (Bm2497).
Conclusion
We have shown that IVM exposure alters the expression of genes that are likely to function in the B. malayi female reproductive system. Through several biological pathways, genes involved in meiosis were particularly affected. These findings provide some insight into the mechanisms involved in IVM-induced reduction in mf output and impaired fertility, embryogenesis, and larval development and the transcriptomic dataset provides important experimental avenues to pursue the mechanism by which IVM sterilizes worms. This is also the first report of IVM exposure having an effect on oxidative phosphorylation at the transcript level, a metabolic pathway believed to be vital for nematodes.
Supporting Information
The table shows the DE genes at each time point for comparisons between treated groups (100 nM, 300 nM, or 1 μM) and untreated groups at either 24 h, 48 h, 72 h, or 5 d IVM exposure for both studies.
(XLSX)
Tables containing top enriched GO terms at 300 nM IVM exposure for 48 h vs untreated (A), 1 μM IVM exposure for 48 h vs untreated (B), 300 nM IVM exposure for 5 d vs untreated (C), and 1 μM IVM exposure for 5 d vs untreated (D).
(DOCX)
Common dispersion and BCV values were calculated using the edgeR Bioconductor package (Version 3.12.0).
(DOCX)
A hypergeometric test was performed to determine which phenotypes were significantly enriched when compared with their overall occurrence in the C. elegans RNAi dataset in Wormbase. P-values were corrected for multiple testing using Benjamini-Hochberg method (1995) with a level of significance of q<0.05.
(XLSX)
(a-g) Biological coefficient of variation (BCV) vs read abundance (counts per million) for different pairwise comparisons. Tagwise BCV values are plotted against the average log CPM values for each pairwise comparison performed in both studies.
(DOCX)
Data Availability
All sequencing data are available from the GEO database (NCBI) (reference GSE75341) or following the link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75341
Funding Statement
We thank the NIH-NIAID Filariasis Research Reagent Resource Center (FR3) for collaboration in this project (Contract No. HHSN272201000030I-Task D02). We also thank NSERC and the Canada Research Chairs for their support, and acknowledge support from the FRQNT Centre for Host-Parasite Interactions. LT is also grateful to the Swiss National Science foundation (fellowship #PBBSP3_144148; www.snf.ch) and to the Canadian Institutes for Health Research (fellowship #320382). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. Global programme to eliminate lymphatic filariasis: progress report, 2013. Relevé epidémiologique hebdomadaire / Section d'hygiène du Secretariat de la Société des Nations = Weekly epidemiological record / Health Section of the Secretariat of the League of Nations. 2014;89(38):409–18. [PubMed] [Google Scholar]
- 2.WHO. African Programme for Onchocerciasis Control: progress report, 2013–2014. Relevé epidémiologique hebdomadaire / Section d'hygiène du Secretariat de la Société des Nations = Weekly epidemiological record / Health Section of the Secretariat of the League of Nations. 2014;89(49):551–60. [PubMed] [Google Scholar]
- 3.Ottesen EA. The global programme to eliminate lymphatic filariasis. Tropical Medicine & International Health: TM & IH. 2000;5(9):591–4. [DOI] [PubMed] [Google Scholar]
- 4.APOC. Final communiqué of the 11th session of the Joint Action Forum (JAF) of APOC Ouagadougou (Burkina Faso): APOC, Paris, France, 6–9 December 2005. [Google Scholar]
- 5.Chabala JC, Mrozik H, Tolman RL, Eskola P, Lusi A, Peterson LH, et al. Ivermectin, a new broad-spectrum antiparasitic agent. Journal of Medicinal Chemistry. 1980;23(10):1134–6. [DOI] [PubMed] [Google Scholar]
- 6.Prichard R, Menez C, Lespine A. Moxidectin and the avermectins: Consanguinity but not identity. International journal for Parasitology Drugs and Drug Resistance. 2012;2:134–53. 10.1016/j.ijpddr.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ardelli BF, Stitt LE, Tompkins JB, Prichard RK. A comparison of the effects of ivermectin and moxidectin on the nematode Caenorhabditis elegans. Veterinary Parasitology. 2009;165(1–2):96–108. 10.1016/j.vetpar.2009.06.043 [DOI] [PubMed] [Google Scholar]
- 8.Takiguchi Y, Mishima H, Okuda M, Terao M, Aoki A, Fukuda R. Milbemycins, a new family of macrolide antibiotics: fermentation, isolation and physico-chemical properties. The Journal of Antibiotics. 1980;33(10):1120–7. [DOI] [PubMed] [Google Scholar]
- 9.Campbell WC. Ivermectin as an antiparasitic agent for use in humans. Annual Review of Microbiology. 1991;45:445–74. [DOI] [PubMed] [Google Scholar]
- 10.Wolstenholme AJ, Rogers AT. Glutamate-gated chloride channels and the mode of action of the avermectin/milbemycin anthelmintics. Parasitology. 2005;131 Suppl:S85–95. [DOI] [PubMed] [Google Scholar]
- 11.Brownlee DJ, Holden-Dye L, Walker RJ. Actions of the anthelmintic ivermectin on the pharyngeal muscle of the parasitic nematode, Ascaris suum. Parasitology. 1997;115 (Pt 5):553–61. [DOI] [PubMed] [Google Scholar]
- 12.Geary TG, Sims SM, Thomas EM, Vanover L, Davis JP, Winterrowd CA, et al. Haemonchus contortus: ivermectin-induced paralysis of the pharynx. Experimental Parasitology. 1993;77(1):88–96. [DOI] [PubMed] [Google Scholar]
- 13.Moreno Y, Nabhan JF, Solomon J, Mackenzie CD, Geary TG. Ivermectin disrupts the function of the excretory-secretory apparatus in microfilariae of Brugia malayi. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(46):20120–5. 10.1073/pnas.1011983107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li BW, Rush AC, Weil GJ. High level expression of a glutamate-gated chloride channel gene in reproductive tissues of Brugia malayi may explain the sterilizing effect of ivermectin on filarial worms. International Journal for Parasitology Drugs and Drug Resistance. 2014;4(2):71–6. 10.1016/j.ijpddr.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tompkins JB, Stitt LE, Ardelli BF. Brugia malayi: in vitro effects of ivermectin and moxidectin on adults and microfilariae. Experimental Parasitology. 2010;124(4):394–402. 10.1016/j.exppara.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 16.Krishna DR, Klotz U. Determination of ivermectin in human plasma by high-performance liquid chromatography. Arzneimittel-Forschung. 1993;43(5):609–11. [PubMed] [Google Scholar]
- 17.Baraka OZ, Mahmoud BM, Marschke CK, Geary TG, Homeida MM, Williams JF. Ivermectin distribution in the plasma and tissues of patients infected with Onchocerca volvulus. European Journal of Clinical Pharmacology. 1996;50(5):407–10. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez Canga A, Sahagun Prieto AM, Diez Liebana MJ, Fernandez Martinez N, Sierra Vega M, Garcia Vieitez JJ. The pharmacokinetics and interactions of ivermectin in humans—a mini-review. The AAPS Journal. 2008;10(1):42–6. 10.1208/s12248-007-9000-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia L, Liu X. The conduct of drug metabolism studies considered good practice (II): in vitro experiments. Current Drug Metabolism. 2007;8(8):822–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballesteros C, Tritten L, O'Neill M, Burkman E, Zaky WI, Xia J, et al. The effect of in vitro cultivation on the transcriptome of adult Brugia malayi. PLoS Neglected Tropical Diseases 2016;10(1):e0004311 10.1371/journal.pntd.0004311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using Galaxy to perform large-scale interactive data analyses. Current Protocols in Bioinformatics / editoral board, Baxevanis Andreas D [et al. ]. 2007;Chapter 10:Unit 10.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, et al. Galaxy: a web-based genome analysis tool for experimentalists. Current Protocols in Molecular Biology / edited by Ausubel Frederick M [et al. ]. 2010;Chapter 19:Unit 19.0.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England). 2015;31(2):166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England). 2010;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nature Protocols. 2015;10(6):823–44. 10.1038/nprot.2015.052 [DOI] [PubMed] [Google Scholar]
- 26.Xia J, Benner MJ, Hancock RE. NetworkAnalyst—integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Research. 2014;42(Web Server issue):W167–74. 10.1093/nar/gku443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 2010;11(3):R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics (Oxford, England). 2008;9(2):321–32. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 30.Harris TW, Lee R, Schwarz E, Bradnam K, Lawson D, Chen W, et al. WormBase: a cross-species database for comparative genomics. Nucleic Acids Research. 2003;31(1):133–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin J, Abubucker S, Wylie T, Yin Y, Wang Z, Mitreva M. Nematode.net update 2008: improvements enabling more efficient data mining and comparative nematode genomics. Nucleic Acids Research. 2009;37(Database issue):D571–8. 10.1093/nar/gkn744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wylie T, Martin JC, Dante M, Mitreva MD, Clifton SW, Chinwalla A, et al. Nematode.net: a tool for navigating sequences from parasitic and free-living nematodes. Nucleic Acids Research. 2004;32(Database issue):D423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Research. 2005;33(Web Server issue):W741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics (Oxford, England). 2003;19(13):1710–1. [DOI] [PubMed] [Google Scholar]
- 36.Hirsh AE, Fraser HB. Protein dispensability and rate of evolution. Nature. 2001;411(6841):1046–9. [DOI] [PubMed] [Google Scholar]
- 37.Melnikow E, Xu S, Liu J, Bell AJ, Ghedin E, Unnasch TR, et al. A potential role for the interaction of Wolbachia surface proteins with the Brugia malayi glycolytic enzymes and cytoskeleton in maintenance of endosymbiosis. PLoS Negl Trop Dis. 2013;7(4):e2151 10.1371/journal.pntd.0002151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vicente JJ, Wordeman L. Mitosis, microtubule dynamics and the evolution of kinesins. Experimental Cell Research. 2015;334(1):61–9. 10.1016/j.yexcr.2015.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellefson ML, McNally FJ. Kinesin-1 and cytoplasmic dynein act sequentially to move the meiotic spindle to the oocyte cortex in Caenorhabditis elegans. Molecular Biology of the Cell. 2009;20(11):2722–30. 10.1091/mbc.E08-12-1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang HY, Mains PE, McNally FJ. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. The Journal of Cell Biology. 2005;169(3):447–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin MY, Sheng ZH. Regulation of mitochondrial transport in neurons. Experimental Cell Research. 2015;334(1):35–44. 10.1016/j.yexcr.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Rourke SM, Dorfman MD, Carter JC, Bowerman B. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genetics. 2007;3(8):e128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connolly AA, Osterberg V, Christensen S, Price M, Lu C, Chicas-Cruz K, et al. Caenorhabditis elegans oocyte meiotic spindle pole assembly requires microtubule severing and the calponin homology domain protein ASPM-1. Molecular Biology of the Cell. 2014;25(8):1298–311. 10.1091/mbc.E13-11-0687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellanger JM, Carter JC, Phillips JB, Canard C, Bowerman B, Gonczy P. ZYG-9, TAC-1 and ZYG-8 together ensure correct microtubule function throughout the cell cycle of C. elegans embryos. Journal of Cell Science. 2007;120(Pt 16):2963–73. [DOI] [PubMed] [Google Scholar]
- 45.Bellanger JM, Gonczy P. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Current Biology: CB. 2003;13(17):1488–98. [DOI] [PubMed] [Google Scholar]
- 46.Srayko M, Quintin S, Schwager A, Hyman AA. Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Current Biology: CB. 2003;13(17):1506–11. [DOI] [PubMed] [Google Scholar]
- 47.Morris CP, Bennuru S, Kropp LE, Zweben JA, Meng Z, Taylor RT, et al. A proteomic analysis of the body wall, digestive tract, and reproductive tract of Brugia malayi. PLoS Neglected Tropical Diseases 2015;9(9):e0004054 10.1371/journal.pntd.0004054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singaravelu G, Singson A. Calcium signaling surrounding fertilization in the nematode Caenorhabditis elegans. Cell Calcium. 2013;53(1):2–9. 10.1016/j.ceca.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corrigan C, Subramanian R, Miller MA. Eph and NMDA receptors control Ca2+/calmodulin-dependent protein kinase II activation during C. elegans oocyte meiotic maturation. Development (Cambridge, England). 2005;132(23):5225–37. [DOI] [PubMed] [Google Scholar]
- 50.Kariya K, Bui YK, Gao X, Sternberg PW, Kataoka T. Phospholipase C-epsilon regulates ovulation in Caenorhabditis elegans. Developmental Biology. 2004;274(1):201–10. [DOI] [PubMed] [Google Scholar]
- 51.Miller KG, Emerson MD, Rand JB. Goalpha and diacylglycerol kinase negatively regulate the Gqalpha pathway in C. elegans. Neuron. 1999;24(2):323–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen IW, Charalampous CF. Biochemical studies on inositol. IX. D-Inositol 1-phosphate as intermediate in the biosynthesis of inositol from glucose 6-phosphate, and characteristics of two reactions in this biosynthesis. The Journal of Biological Chemistry. 1966;241(10):2194–9. [PubMed] [Google Scholar]
- 53.Eng JK, Blackhall WJ, Osei-Atweneboana MY, Bourguinat C, Galazzo D, Beech RN, et al. Ivermectin selection on beta-tubulin: evidence in Onchocerca volvulus and Haemonchus contortus. Molecular and Biochemical Parasitology. 2006;150(2):229–35. [DOI] [PubMed] [Google Scholar]
- 54.Duro E, Marston AL. From equator to pole: splitting chromosomes in mitosis and meiosis. Genes & Development. 2015;29(2):109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashraf S, Beech RN, Hancock MA, Prichard RK. Ivermectin binds to Haemonchus contortus tubulins and promotes stability of microtubules. International Journal for Parasitology. 2015;45(9–10):647–54. 10.1016/j.ijpara.2015.03.010 [DOI] [PubMed] [Google Scholar]
- 56.Iwasaki K, McCarter J, Francis R, Schedl T. emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. The Journal of Cell Biology. 1996;134(3):699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee KY, Yang I, Park JE, Baek OR, Chung KY, Koo HS. Developmental stage- and DNA damage-specific functions of C. elegans FANCD2. Biochemical and Biophysical Research Communications. 2007;352(2):479–85. [DOI] [PubMed] [Google Scholar]
- 58.Burns LT, Wente SR. From hypothesis to mechanism: uncovering nuclear pore complex links to gene expression. Molecular and Cellular Biology. 2014;34(12):2114–20. 10.1128/MCB.01730-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434(7032):462–9. [DOI] [PubMed] [Google Scholar]
- 60.Minasaki R, Puoti A, Streit A. The DEAD-box protein MEL-46 is required in the germ line of the nematode Caenorhabditis elegans. BMC Developmental Biology. 2009;9:35 10.1186/1471-213X-9-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner AJ, Isaac RE, Coates D. The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology. 2001;23(3):261–9. [DOI] [PubMed] [Google Scholar]
- 62.Spanier B, Sturzenbaum SR, Holden-Dye LM, Baumeister R. Caenorhabditis elegans neprilysin NEP-1: an effector of locomotion and pharyngeal pumping. Journal of Molecular Biology. 2005;352(2):429–37. [DOI] [PubMed] [Google Scholar]
- 63.Castanha Zanoli JC, Maioli MA, Medeiros HC, Mingatto FE. Abamectin affects the bioenergetics of liver mitochondria: A potential mechanism of hepatotoxicity. Toxicology in vitro: an international journal published in association with BIBRA. 2012;26(1):51–6. [DOI] [PubMed] [Google Scholar]
- 64.Shin H, Lee H, Fejes AP, Baillie DL, Koo HS, Jones SJ. Gene expression profiling of oxidative stress response of C. elegans aging defective AMPK mutants using massively parallel transcriptome sequencing. BMC Research Notes. 2011;4:34 10.1186/1756-0500-4-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Behm CA. Metabolism In: The biology of nematodes. 1st ed. London: Francis and Taylor; 2002. [Google Scholar]
- 66.Ramp T, Bachmann R, Kohler P. Respiration and energy conservation in the filarial worm Litomosoides carinii. Molecular and Biochemical Parasitology. 1985;15(1):11–20. [DOI] [PubMed] [Google Scholar]
- 67.Martin RJ, Robertson AP, Bjorn H. Target sites of anthelmintics. Parasitology. 1997;114 Suppl:S111–24. [PubMed] [Google Scholar]
- 68.Yates DM, Wolstenholme AJ. An ivermectin-sensitive glutamate-gated chloride channel subunit from Dirofilaria immitis. International Journal for Parasitology. 2004;34(9):1075–81. [DOI] [PubMed] [Google Scholar]
- 69.Wolstenholme AJ. Glutamate-gated chloride channels. The Journal of Biological Chemistry. 2012;287(48):40232–8. 10.1074/jbc.R112.406280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laing ST, Ivens A, Butler V, Ravikumar SP, Laing R, Woods DJ, et al. The transcriptional response of Caenorhabditis elegans to ivermectin exposure identifies novel genes involved in the response to reduced food intake. PloS One. 2012;7(2):e31367 10.1371/journal.pone.0031367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tippawangkosol P, Choochote W, Na-Bangchang K, Jitpakdi A, Pitasawat B, Riyong D. The in vitro effect of albendazole, ivermectin, diethylcarbamazine, and their combinations against infective third-stage larvae of nocturnally subperiodic Brugia malayi (Narathiwat strain): scanning electron microscopy. Journal of Vector Ecology: Journal of the Society for Vector Ecology. 2004;29(1):101–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table shows the DE genes at each time point for comparisons between treated groups (100 nM, 300 nM, or 1 μM) and untreated groups at either 24 h, 48 h, 72 h, or 5 d IVM exposure for both studies.
(XLSX)
Tables containing top enriched GO terms at 300 nM IVM exposure for 48 h vs untreated (A), 1 μM IVM exposure for 48 h vs untreated (B), 300 nM IVM exposure for 5 d vs untreated (C), and 1 μM IVM exposure for 5 d vs untreated (D).
(DOCX)
Common dispersion and BCV values were calculated using the edgeR Bioconductor package (Version 3.12.0).
(DOCX)
A hypergeometric test was performed to determine which phenotypes were significantly enriched when compared with their overall occurrence in the C. elegans RNAi dataset in Wormbase. P-values were corrected for multiple testing using Benjamini-Hochberg method (1995) with a level of significance of q<0.05.
(XLSX)
(a-g) Biological coefficient of variation (BCV) vs read abundance (counts per million) for different pairwise comparisons. Tagwise BCV values are plotted against the average log CPM values for each pairwise comparison performed in both studies.
(DOCX)
Data Availability Statement
All sequencing data are available from the GEO database (NCBI) (reference GSE75341) or following the link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75341