Abstract
Membrane proteins are designed to fold and function in a lipid membrane, yet folding experiments within a native membrane environment are challenging to design. Here we show that single-molecule forced unfolding experiments can be adapted to study helical membrane protein folding under native-like bicelle conditions. Applying force using magnetic tweezers, we find that a transmembrane helix protein, Escherichia coli rhomboid protease GlpG, unfolds in a highly cooperative manner, largely unraveling as one physical unit in response to mechanical tension above 25 pN. Considerable hysteresis is observed, with refolding occurring only at forces below 5 pN. Characterizing the energy landscape reveals only modest thermodynamic stability (ΔG = 6.5 kBT) but a large unfolding barrier (21.3 kBT) that can maintain the protein in a folded state for long periods of time (t1/2 ~3.5 h). The observed energy landscape may have evolved to limit the existence of troublesome partially unfolded states and impart rigidity to the structure.
Helical membrane protein folding can be broken down into two major stages1,2. The first stage is initial insertion of transmembrane helices, which appears to be largely governed by the water-membrane partitioning of free energy3. In the second stage, the protein completes folding to its final native structure. Thus, once insertion occurs, membrane protein folding and unfolding occurs within the membrane. Ideally, studies of the second stage of folding would be performed in a membrane environment, yet folding studies require a means for altering the energy landscape to favor the unfolded state, which is hard to achieve in a membrane. One method, called steric trapping, drives unfolding by using a protein that binds preferentially to the unfolded state4,5. Atomic force microscopy (AFM) has been extensively used to study forced unfolding of membrane proteins from bilayers6–9. The AFM studies, however, apply force parallel to the membrane normal so that the proteins are physically pulled out of the membrane. To study the more physiological process of folding within a membrane, it is necessary to apply force along the membrane plane.
Here we developed a new method to observe the forced unfolding and refolding behavior of a single membrane protein within a lipid bilayer environment. By adapting techniques pioneered for soluble protein folding10,11, we hold a single membrane protein in a magnetic trap and provide a bilayer environment for the protein using bicelles, self-assembled bilayer discs wrapped by detergent molecules12–14. We use this magnetic trapping strategy to study folding and unfolding of a helical membrane protein, GlpG. GlpG is an E. coli rhomboid intramembrane protease that has six transmembrane α-helices15–18 and cleaves other transmembrane substrates in a lipid bilayer19–23. Previously reported extensive bulk equilibrium and kinetic folding studies on GlpG mutants in detergent provide a useful comparison24,25. Because GlpG has an even number of helices, the pulling direction is exactly defined along the membrane plane when the N and C termini of GlpG are pulled.
We found a remarkably high degree of cooperativity and a high barrier to unfolding, so large forces were required to unfold the protein at an appreciable rate. To see refolding at a measurable rate, we must return to much lower forces. Thus, we were unable to observe reversible folding directly. Nevertheless, we could construct a putative energy landscape by extrapolating the observed folding and unfolding rates to zero force. We found that GlpG is held close to its native state by a deep energy well near the folded conformation. The energy landscape is ideal for preventing the formation of misfolded states both during insertion and after the protein is synthesized.
RESULTS
Cooperative unfolding and refolding of GlpG in bicelles
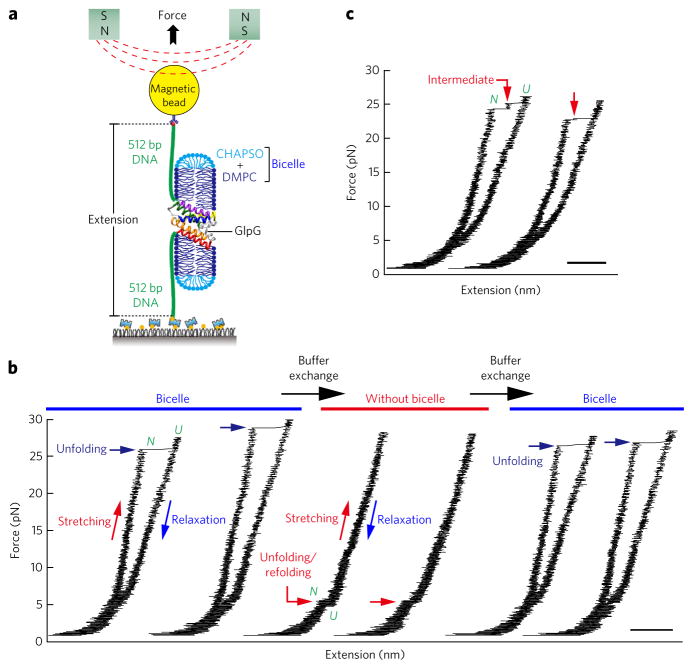
Single GlpG proteins were covalently linked to two DNA handles (512 base pairs each) at the N- and C-terminal ends10,26,27 (Fig. 1a and Supplementary Results, Supplementary Fig. 1). One DNA handle was anchored to a PEG-coated surface via biotin-avidin binding, and the other handle was attached to a magnetic bead. As a pair of magnets approaches, the magnetic bead experiences increasing force of up to tens of pN, which is then delivered to the tweezed GlpG protein27–33. The change in the bead height (i.e., the extension value) as a result of the force application can be measured (Fig. 1a). With this experimental scheme, we are able to apply tension in a direction vertical to the membrane normal vector, allowing the GlpG protein to unfold and refold within the lipid membrane (Supplementary Fig. 2). The experiment is free from nonspecific interactions with the surface because the DNA handles completely separate the GlpG protein from the surface.
Figure 1. Cooperative unfolding and refolding of GlpG in bicelles.
(a) Schematic of the single-molecule magnetic tweezers experiment for studying unfolding and refolding of a single GlpG protein. (b) Representative force-extension curves in each buffer condition. After several cycles of unfolding and refolding in bicelles (left), the bicelles were removed and the unfolding and refolding cycles were repeated (middle). In the buffer condition without bicelles, a very small amount of CHAPSO (0.0038%) was added to prevent nonspecific binding. After up to tens of pulling cycles, the bicelle condition could be restored by another round of microfluidic buffer exchange (right), and the unfolding behavior seen previously in bicelles was fully restored. (c) Representative force-extension curves showing multiple-step unfolding of single GlpG proteins. In b and c, scale bar represents 50 nm.
Gradual pulling experiments with GlpG, in which the force was slowly increased as the magnets approached the sample at a constant speed (0.1 mm s−1, corresponding to an average force-loading rate of ~0.5 pN s−1), revealed a high degree of unfolding cooperativity (Fig. 1b). The GlpG protein remained intact until the magnetic force was increased to ~25 pN and then showed one abrupt unfolding event with a step size of 40 nm (Fig. 1b). The observed 40-nm increase was very close to the expected value when a fully folded GlpG was unfolded to a completely unstructured polypeptide at 25 pN (Supplementary Fig. 3). Thus, although unfolding was initiated in a bicelle environment, driving unfolding with a reasonable probability required such high force that the entire protein ultimately unraveled. Our observation suggests highly cooperative unfolding of the entire GlpG protein.
We observed a large refolding hysteresis. The unfolded GlpG protein only refolded when we decreased the force to a few pN so the unfolding and refolding cycle had a force gap of more than 20 pN (Fig. 1b). At these low forces, the transmembrane helical structure could be restored before refolding, allowing refolding within a protein–bicelle complex (Supplementary Fig. 3). The unfolding and refolding cycle could be repeated up to tens of times in a very reproducible manner, indicating that, in spite of the hysteresis, the protein completely refolded using our experimental setup.
The experimental system was remarkably robust. We removed the bicelles by buffer exchange, leaving the hydrophobic polypeptide in an aqueous environment (Fig. 1b and Supplementary Fig. 4). Under these conditions, we saw a single unfolding event at low (5 pN) forces, and the large hysteresis completely vanished (Fig. 1b and Supplementary Fig. 5d). When the bicelle condition was restored by another round of buffer exchange, however, the unfolding and refolding behavior of the GlpG protein was fully restored (Fig. 1b). Moreover, addition of detergent molecules alone, instead of bicelles, substantially decreased the unfolding force and made its distribution much more heterogeneous (Supplementary Fig. 5). These observations indicate that the bicelle condition has a crucial role in the cooperative unfolding and refolding of GlpG and also point to the advantage of the single-molecule tethering approach for studying the folding of membrane proteins that are so prone to irreversible aggregation.
Intermediates in C- to N-terminal unfolding of GlpG
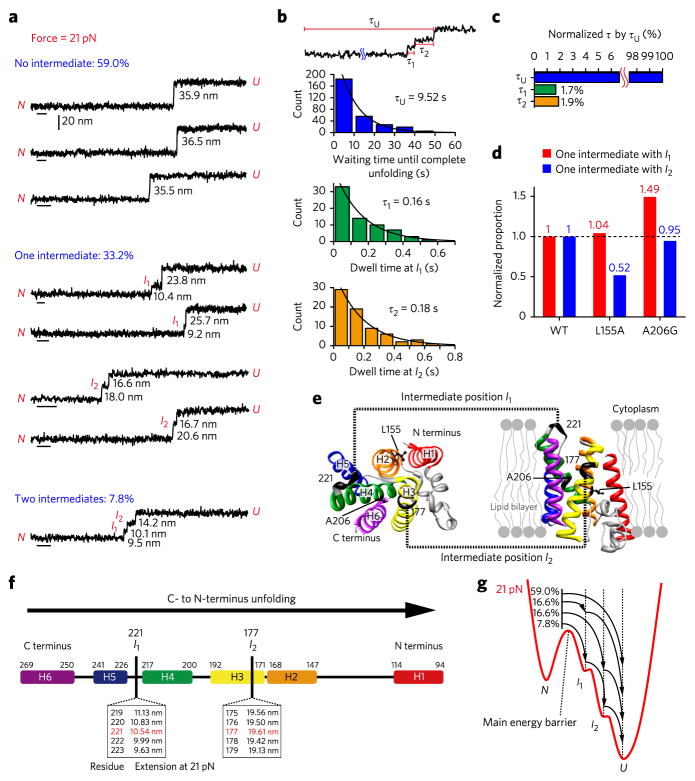
Although unfolding of GlpG was essentially a cooperative process, we noticed transient intermediates, or pauses, during unfolding (Fig. 1c). We sought to identify where these unfolding pauses occurred. In the pulling experiment shown in Figure 1, however, where mechanical tension was gradually increased, the unfolding events stochastically occurred at different force levels, which precluded direct comparison of observed step sizes. We therefore designed a ‘force-jump’ experiment where the magnetic force was rapidly increased and maintained at a predetermined value27 (Fig. 2). In such force-jump experiments, unfolding of GlpG was induced at a constant force level, and the observed extension increases could be pooled together to elucidate the structure of unfolding intermediates34–36.
Figure 2. C- to N-terminus unfolding of single GlpG with two intermediates.
(a) Representative extension traces at 21 pN for unfolding events (n = 295) with no intermediates (59.0%), one intermediate (33.2%) and two intermediates (7.8%). Statistics of unfolding step sizes are in Supplementary Figure 6. Scale bars, 1 s. (b) Dwell time analysis (n = 295). τU is the waiting time until complete unfolding (blue), and τ1 and τ2 are the dwell times in the intermediate states I1 (green) and I2 (yellow). (c) Dwell times in the intermediates normalized by τU. (d) Comparison of the normalized proportion of unfolding patterns with one intermediate between the WT and the L155A and A206G mutants. The normalized proportion is defined by P(X)/P(WT), where P(X) means the proportion of each unfolding pattern in the total number of traces for X = WT (n = 295), L155A (n = 81) or A206G (n = 97). The histograms for one intermediate with I1 or I2 are shown in red and blue, respectively. (e) GlpG structure showing the intermediate positions I1 and I2 (black). The mutation sites Leu155 and Ala206 are shown in ball-and-stick representation. Left, cytoplasmic view; right, side view showing a lipid bilayer. (f) Schematic diagram showing the mapping of Gaussian peak values to the intermediate residue positions. Arrow indicates unfolding direction. (g) Conceptual folding energy landscape at 21 pN. The arrows denote the structural transitions among the native state (N), the intermediate states (I1 and I2) and the unfolded state (U).
When we employed force jumps to 21 pN, we were able to observe four different patterns in the unfolding of single GlpG proteins (Fig. 2a and Supplementary Fig. 6). In about 60% of the total unfolding trajectories (n = 295), no intermediates were resolved with our time resolution. In one-third of the trajectories, one unfolding intermediate was detected. The extension distribution of these intermediates showed two Gaussian peaks with one peak at ~10 nm (I1) and the other at ~20 nm (I2). In 7.8% of the trajectories, we observed two intermediates. These two intermediates almost exactly overlap with the I1 and I2 intermediates observed for the one-intermediate cases, suggesting that I1 and I2 do not represent two independent pathways but two intermediates along one unfolding pathway (Supplementary Fig. 6). Finally, we measured the dwell times in I1 and I2 (τ1 and τ2) and compared these dwell times with the total unfolding time, τU, which was the time elapsed between the force jump to 21 pN and the moment of complete unfolding (Fig. 2b). Both τ1 and τ2 were <2% of τU, quantitatively showing that the unfolding process of GlpG had essentially one rate-limiting step and paused only briefly in the I1 and I2 intermediate states after the rate-limiting step (Fig. 2c). In fact, no intermediates were observed in 60% of the unfolding trajectories with our current time resolution. This dwell-time analysis quantitatively illustrates again that the intermediates are very transient compared to the total unfolding; thus, the unfolding of the entire GlpG protein is highly cooperative.
We reasoned that the observed unfolding is a unidirectional process beginning at either the N or C terminus (Supplementary Fig. 7). To determine the directionality of the unfolding process, we examined the unfolding of GlpGL155A and GlpGA206G (refs. 24,25), whose N- and C-terminal parts are respectively destabilized by their mutation (Fig. 2d,e and Supplementary Fig. 8). For the L155A mutant, the observation probability of the I2 intermediate was selectively reduced compared to the wild type (WT) (Fig. 2d and Supplementary Fig. 8). Thus, this N-terminal mutation lowered the stability of the region that was unfolded in the I2 to U step, and the corresponding unfolding step (from I2 to U) became accelerated. In contrast, for the A206G mutant, the observation probability of the I1 intermediate was selectively increased (Fig. 2d and Supplementary Fig. 8). Thus, the C-terminal mutation of A206G accelerated the N to I1 step by lowering the stability of the corresponding region. These observations collectively suggest that mechanical unfolding of GlpG starts at the C terminus and propagates toward the N-terminal end.
We next pinpointed the residues comprising the unfolding intermediates. Using the Marko-Siggia formulation of the worm-like chain model (Supplementary Fig. 9), the I1 intermediate was found to extend to approximately residue 221, which corresponds to the unfolding of helices 5 and 6 (Fig. 2e,f). The I2 intermediate was found to extend until approximately position 177, corresponding to the unfolding of helices 3 and 4. These results suggest that mechanical unfolding of GlpG at high forces takes place in units of helical hairpins, two helices at a time.
Characterization of folding and unfolding kinetics
Our observations of the unfolding and refolding of GlpG collectively point to the existence of one main energy barrier that separates the folded and unfolded states (Fig. 2g), with minor energy barriers separating the I1 and I2 intermediates located between the primary energy barrier and the unfolded U state. Crossing of the main energy barrier becomes the ratelimiting step for unfolding, and, once crossed, the unfolding process only briefly pauses in the I1 and I2 states.
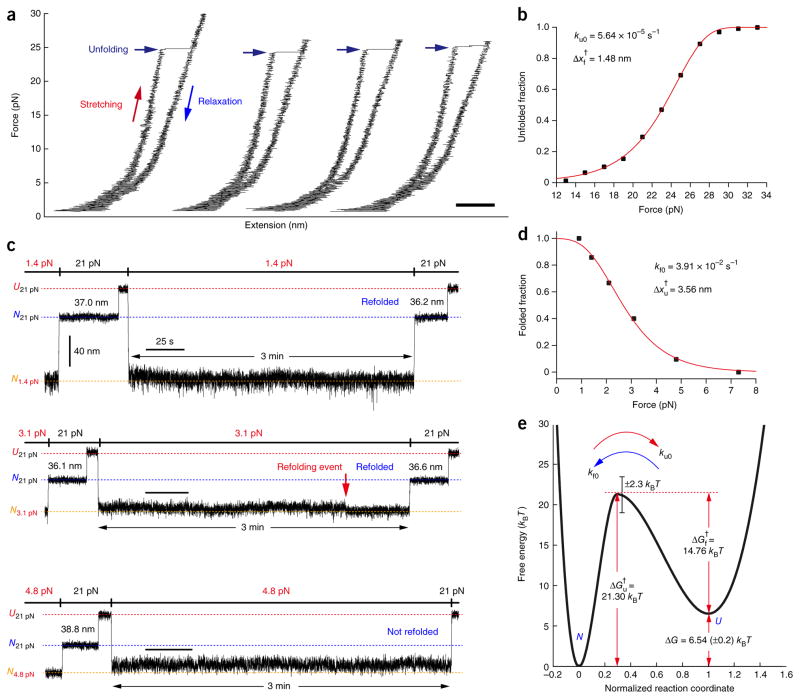
To characterize the main unfolding energy barrier in a quantitative way, we studied the unfolding and refolding kinetics. We first revisited the gradual pulling experiments of Figure 1 (Fig. 3a). As noted above, each unfolding event stochastically occurred at a different force level, meaning that we could study the unfolded fraction as a function of force (Fig. 3b). Fitting this unfolding probability (Online Methods) yielded a kinetic rate for GlpG unfolding at zero tension (ku0) of 5.64 × 10−5 s−1 and a distance from the folded state to the transition state (Δxf†) of 1.48 nm. To characterize the opposite side of the energy barrier, we repeated the refolding experiments but varied the force levels during refolding (Fig. 3c). After waiting 3 min, we checked the folding status by pulling the GlpG protein at 21 pN to determine whether the extension reflected the U or N state. We studied the folded fraction (within the given 3 min) as a function of mechanical tension. By extrapolation, we estimated the kinetic rate for folding at zero tension (kf0) to be 3.91 × 10−2 s−1 and the distance from the unfolded state to the transition state (Δxu†) to be 3.56 nm (Fig. 3d and Online Methods). These data reporting unfolding and refolding kinetics as a function of force are analogous to chevron plots in bulk membrane protein folding studies in detergent that report kinetic parameters as a function of denaturant concentration. Our reaction coordinate (x) is conceptually a thermally averaged end-to-end distance of GlpG measured at zero force along the pulling direction, and the distance (Δx) indicates how x changes37,38. Thus, the mechanical tension and distance to the transition state are analogous to the denaturant concentration and its denaturant power (reflected in the m values) used in the bulk folding studies, but the mechanical parameters have direct physical implications.
Figure 3. Folding energy landscape of GlpG.
(a) Representative gradual pulling experiments measuring the unfolding force of single GlpG proteins. Scale bar, 50 nm. (b) Unfolded fraction versus force (n = 233) from which the zero-force unfolding rate (ku0) and the distance from the native state to the transition state (Δxf†) were obtained. (c) Representative extension traces in force-cycle experiment for obtaining the folding kinetics. After unfolding at 21 pN, the force was lowered back to 0.9–7.3 pN and maintained for 3 min. The extent of refolding was then determined by restoring the 21-pN force and comparing the observed extension with the extensions observed for the native and unfolded states (N21 pN and U21 pN are shown as blue and red dashed lines, respectively). (d) Folded fraction versus force (n = 125), which was used to obtain the folding kinetic rate at zero force (kf0) and the distance from the unfolded state to the transition state (Δxu†). (e) Putative folding energy landscape of GlpG. The energy difference between the native state and the unfolded state (ΔG) and the energy barriers (ΔGu†, ΔGf†) are denoted with red arrows. The error of the ΔG represents s.e.m., and the error of the energy barriers represent the error of the frequency factor kw (Online Methods).
To test whether the kinetic rates we determined were affected by the specific bicelle conditions, we repeated the unfolding measurements at different lipid/detergent ratios and temperature conditions (Supplementary Fig. 10 and Supplementary Tables 1 and 2). When the lipid/detergent ratio was increased from 2.2:1 to 2.8:1, the kinetic rates and the distance to the transition state were largely unaffected (with only a 0.6 kBT difference; Supplementary Table 1), indicating that the edge effects of the detergent belt surrounding the bicelle structure were negligible. The gel phase melting temperature of 1,2- dimyristoyl-sn-glycero-3-phosphocholine (DMPC)/1,2-diheptanoylsn- glycero-3-phosphocholine (DHPC) bicelles (analogous to our DMPC/3-((3-cholamidopropyl)dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO) bicelles) is 21 °C (ref. 39), which is close to the temperature (22 °C) used in our experiments, so we tested whether increasing the temperature would have any effect. When we increased the measurement temperature up to 25 °C, however, we did not see any obvious change in the kinetic rates or the transition state distance (Supplementary Table 2). Thus, our results do not seem highly sensitive to small changes in the bicelle conditions.
Folding energy landscape of GlpG
In characterizing the unfolding and refolding kinetics, we needed to use different force ranges (Fig. 3b,d) because of the large hysteresis observed in the unfolding and refolding cycle of GlpG (Fig. 1). Nevertheless, we believe that we can reconstruct an energy landscape at zero force within bicelles by extrapolation if we assume that the transition state for the unfolding induced by high force levels is the same as that of the refolding pathway observed at low force levels. We believe this is a reasonable assumption because: (i) We do not see discontinuities in the unfolding and refolding rates as a function of force that would imply a change in pathway (Fig. 3b,d). The two force regions used for unfolding (13–33 pN) and refolding (1–7 pN) are separated only by 6 pN. (ii) Even though the GlpG–bicelle complex must ultimately become highly distorted as GlpG is unraveled to an unstructured polypeptide at high forces, unfolding is initiated within the bicelle structure, and the distance to the transition state is only 1.5 nm (Figs. 1b and 3b). Thus, unfolding rates reflect unfolding within a bicelle. (iii) Helical structure is restored at low forces, indicating that refolding occurs again within the protein–bicelle complex (Supplementary Fig. 3). (iv) Finally, we find that our measured thermodynamic values (ΔG and ΔΔG) for the WT and mutants are largely consistent with the values from the bulk equilibrium unfolding experiments described below (Supplementary Table 3).
With the assumption we made above, we constructed a putative folding energy landscape of GlpG (Fig. 3e and Table 1). The ratio of the unfolding and folding rates at zero force led to an unfolding free energy of ΔG = −kBT × ln(ku0/kf0) = 6.54 kBT. Bulk SDS unfolding experiments report ΔG values from 7.08 kBT to 13.88 kBT (refs. 24,25), which is in reasonable agreement considering the completely different methods for driving unfolding, the different environments and the uncertain extrapolations in SDS unfolding studies5 (Supplementary Table 3). Our measured refolding rate of 3.91 × 10−2 s−1 is very similar to the rate measured in the detergent refolding experiments (2.7 × 10−2 s−1), a parameter that does not require much extrapolation. The main discrepancy occurs in the unfolding rates (1.0 × 10−7 s−1 versus 5.64 × 10−5 s−1), but this involves a large extrapolation. Using the Kramer equation (Online Methods), the height of the energy barrier encountered during GlpG folding (ΔGf†) was estimated to be 14.76 kBT, rendering the folding process slow (t1/2 ~18s). We also mapped the transition state onto the normalized reaction coordinate x/(Δxf†+Δxu†), where the Δxf† and Δxu† are respectively from the folded and unfolded state to the transition state. Notably, the transition state turned out to be much closer to the native state than to the unfolded state (i.e., βf ≡ Δxf†/(Δxf†+Δxu†) = 0.29), consistent with our observations that the six transmembrane helices were tightly coupled and essentially worked as one unit when GlpG was folded and unfolded.
Table 1.
Summary of kinetic and thermodynamic properties of WT and mutant GlpG
| Δxf† (nm) | Δxu† (nm) | βf | ku0 (s−1) | kf0 (s−1) | ΔGu† (kBT) | ΔGf† (kBT) | ΔG (kBT) | |
|---|---|---|---|---|---|---|---|---|
| WT | 1.48 (0.03) | 3.56 (0.24) | 0.29 (0.02) | 5.64 (0.91) × 10−5 | 3.91 (0.54) × 10−2 | 21.30 (2.30) | 14.75 (2.30) | 6.54 (0.21) |
| L155A | 1.26 (0.09) | 2.99 (0.34) | 0.29 (0.03) | 1.73 (0.81) × 10−4 | 1.37 (0.18) × 10−2 | 20.18 (2.30) | 15.80 (2.30) | 4.37 (0.48) |
| A206G | 1.68 (0.10) | 3.57 (0.17) | 0.32 (0.02) | 1.17 (0.44) × 10−4 | 2.42 (0.19) × 10−2 | 20.57 (2.30) | 15.23 (2.30) | 5.33 (0.38) |
Numbers in parentheses indicate error. For ΔGu† and ΔGf†, error represents the error of the frequency factor kw (Online Methods). All other error values represent s.e.m.
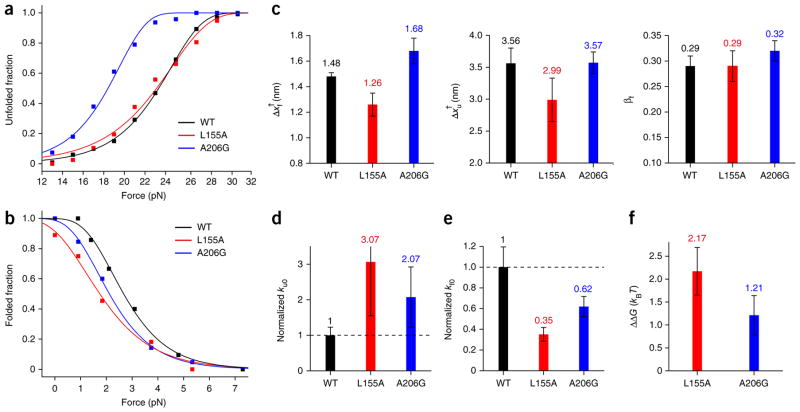
We also studied how the L155A and A206G mutations affected the energy landscape (Fig. 4 and Table 1). As in our previous study of unfolding patterns (Fig. 2d), kinetic measurements of the two mutants revealed detailed changes in the unfolding and refolding probabilities as a function of force (Fig. 4a,b). These data reconfirm that our measurements do not simply measure disruption and association of bicelle complexes but rather reflect subtle differences in the energy landscapes of the mutants. At the same time, however, the general shapes of the energy landscape were essentially preserved for the two mutants. The position of the transition state (βf ) remained at the normalized distance of 0.3, close to the native folded state (Fig. 4c). The unfolding and the refolding rates were modestly changed for the two mutants. The unfolding rates at zero force (ku0) were increased by a factor of two or three, corresponding to lowering of the unfolding energy barrier (ΔGu †) by ~1 kBT (Fig. 4d and Table 1). Although the force values reaching 50% unfolding were similar to those of the L155A mutant and the WT (Fig. 4a), the difference in the unfolding curve slopes gave a smaller Δxf † for the L155A mutant (Fig. 4c), which in turn led to a higher ku0 (Fig. 4d). The refolding rates were decreased by almost the same factors, indicating that the refolding energy barrier was increased as much as the unfolding energy barrier was decreased (Fig. 4e and Table 1). The calculated destabilizing extents, ΔΔG (calculated as ΔGWT − ΔGmutant), were thus in the range of 1–2 kBT (Fig. 4f), consistent with the values obtained from the bulk SDS unfolding experiments24,25 (Supplementary Table 3). Thus, our method of reconstructing energy landscapes is sensitive to modest changes in the intrinsic stability of GlpG.
Figure 4. Comparison of kinetic and thermodynamic properties between WT and mutant GlpG.
(a,b) Unfolded fraction (a) and folded fraction (b) as a function of force for the WT and mutant GlpG proteins. The total number of unfolding and refolding events are n = 233 and n = 125 for WT; n = 77 and n = 58 for the L155A mutant; and n = 95 and n = 87 for the A206G mutant. Fitting the data (Online Methods) yields kinetic rates for unfolding and folding at zero tension (ku0 and kf0) and distances from the folded (and unfolded) state to the transition state (Δxf† and Δxu†). (c) Comparison of the distance values (left, Δxf†; middle, Δxu†) and the transition state positions (right, βf) of the WT and the mutants. (d,e) Comparison of the unfolding rate (d) and refolding rate (e) for the WT and mutant proteins, normalized to the WT rate. (f) The change in unfolding free energy of the mutants relative to the WT observed in forced unfolding experiments (ΔΔG = ΔGWT − ΔGmutant). All error bars represent s.e.m.
DISCUSSION
Overall, the primary features of the folding energy landscape for GlpG we observe are (i) high cooperativity, (ii) low thermodynamic stability, (iii) a high kinetic barrier and (iv) a transition state that is structurally closer to the folded state than the unfolded state. As there is still limited information on the folding of large helical membrane proteins, particularly under native conditions, it is unclear how common these characteristics of GlpG folding will be for membrane proteins in general.
In contrast to what we see for GlpG, the transition states found with SDS-driven unfolding of bacteriorhodopsin, DsbB and even GlpG are all placed closer to the unfolded state than the folded state25,40,41. It is possible that the difference simply reflects folding and unfolding in the more native-like bicelle. In contrast, it could reflect different requirements for structural flexibility. The close proximity of the energy barrier to the folded state would imply high local curvature of the energy landscape around the folded state, which could impart structural rigidity to GlpG.
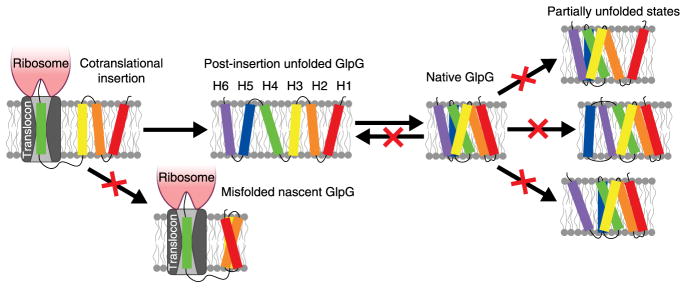
The high degree of cooperativity in mechanical unfolding for a helical membrane protein was surprising to us. Individual transmembrane helices are stable within a bilayer1, so we expected that helices could be pulled off one at a time. Instead, the six transmembrane helices largely behave as one unfolding unit. The folding of bacteriorhodopsin and GlpG also seems to be highly cooperative when studied by SDS unfolding25,40, so it is possible that this is a common property of membrane proteins. Although cooperativity is not theoretically required, there may also be evolutionary pressure favoring cooperativity in membrane proteins as in soluble proteins42. Cooperativity would prevent the formation of structure before complete insertion, thereby limiting the development of stable, albeit incorrect partial structures before the entire protein is available for structure formation (Fig. 5). Once formed, these misfolded structures might be difficult to unravel. Thus, it makes evolutionary sense to select an energy function that requires the protein to wait until complete insertion before adopting a stable structure. There is evidence for some structure formation during biological insertion43,44 and for preferred folding from the N terminus25,45, but it is unclear whether partially inserted states can generate stable enough structures to direct folding.
Figure 5. How the folding energy landscape of GlpG may prevent dangerous misfolded states.
Cooperativity can limit the formation of stable off-pathway structures before completion of translation. The high kinetic barrier near the folded state prevents folded GlpG from returning to the unfolded state on a biologically relevant time scale, imparts rigidity and limits the existence of partially unfolded states that might be prone to inappropriate interactions.
A high kinetic barrier for unfolding, as signified by our observation of cooperative unfolding, represents another mechanism for limiting the existence of aggregation-prone unfolded states (Fig. 5). Although GlpG is not very thermodynamically stable, once folded, the 3.5-h unfolding half-life (because of the high ΔGu † of 21.30 kBT) implies that an E. coli cell will rarely see a GlpG unfold on the time scale of a cell division. Very slow unfolding has also been observed for both diacylglycerol kinase and bacteriorhodopsin5,40,46. It is not known how slowly DsbB unfolds under native conditions, but it refolds from an SDS-denatured state on a similar time scale to bacteriorhodopsin and GlpG47. Slow folding may reflect a rugged energy landscape for membrane proteins48.
Like soluble proteins, it is likely that there will be wide variation in the folding behavior of membrane proteins. We need to see more examples of well-characterized folding landscapes under native conditions to learn about structural correlations with folding properties. The approach described here may now allow us to expand our analysis of membrane protein folding to more proteins.
ONLINE METHODS
Protein expression and purification
The membrane domain of the E. coli GlpG gene (residues 87–276) was amplified from the genome of E. coli XL1-Blue strain by PCR. The PCR primers included codons to add cysteine residues at both the N and C termini. The amplification primers were:
FWD: 5′-GGAAAGAGCTCTGTGCCGCCTTGCGTGAACGCG-3′
REV: 5′-CCCTTAAGCTTTTAACATTTTCGTTTTCGCGCATTGAGCG-3′.
The amplified gene was cloned into the pTrcHisB vector at the SacI/HindIII restriction sites, thereby adding an N-terminal His6 tag. The natural cysteine at position 104 was changed to alanine, and the N-terminal cysteine was shifted two residues away from the N terminus using site-directed mutagenesis with PfuUltra II Fusion HS DNA polymerase (Agilent Technologies) to give the following protein construct, GlpGCys-TM-Cys, which includes residues 89–276 of E. coli GlpG between the two cysteines.
MGHHHHHHELAACLRERAGPVTWVMMIAAVVVFIAMQILGDQE VMLWLAWPFDPTLKFEFWRYFTHALMHFSLMHILFNLLWWWYLGG AVEKRLGSGKLIVITLISALLSGYVQQKFSGPWFGGLSGVVYALMGY VWLRGERDPQSGIYLQRGLIIFALIWIVAGWFDLFGMSMANGAH IAGLAVGLAMAFVD SLNARKRKC.
GlpGCys-TM-Cys/L155A and GlpGCys-TM-Cys/A206G were created by site-directed mutagenesis of Leu155 to alanine (residue position 80 in our construct) and Ala206 to glycine (residue position 131 in our construct).
The GlpG protein constructs in BL21-Gold (DE3) were grown in LB medium at 37 °C and induced with 0.5 mM IPTG at 0.7 OD600, and cells were harvested after an additional 3 h incubation. Cells were resuspended in 50 mM Tris-HCl, 1 mM EDTA, 1 mM PMSF, 1 mM DTT and 2 μg/ml DNAse I (pH 8.0) and lysed by two passes through an Avestin EmulsiFlex-C3 at 15,000 psi. Cell debris was removed by centrifugation at 25,000g for 15 min. The membrane fraction was collected by centrifugation of the supernatant at 100,000g for 90 min at 4 °C. The pellet was resuspended in 25 mM Tris-HCl, 1.25% n-decyl-β-D-maltopyranoside (DM, Affymetrix) and 1 mM TCEP (pH 8.0) (5 ml per liter of culture) with the aid of a dounce homogenizer. Membranes were further solubilized with gentle rotation for 45 min at room temperature. The soluble fraction was collected after centrifugation at 100,000g for 30 min at 4 °C. 4 M NaCl and 5 M imidazole were added to the supernatant to a final concentration of 300 mM and 10 mM, respectively. The supernatant was incubated for 1 h at 4 °C with Ni-NTA (0.5 ml resin per liter of culture) that had been preequilibrated with 20 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole, 0.2% DM and 1 mM TCEP (pH 8.0). The resin was packed into a column by gravity and, after collecting the flow-through, washed with 5 column volumes of 10 mM and 30 mM imidazole before eluting with 300 mM imidazole in 20 mM Tris-HCl, 300 mM NaCl, 0.2% DM and 1 mM TCEP (pH 8.0). Elution fractions containing protein (detected by absorbance at 280 nm) were pooled, concentrated to 3 ml using a 10,000 MWCO Amicon Ultra centrifugal filter (Millipore) and buffer exchanged into 25 mM Tris-HCl, 0.2% DM, 1 mM TCEP (pH 8.0) using an Econo-Pac 10DG column (BioRad). The protein was then passed over a 1-ml HiTrap Q HP ion exchange column (GE Healthcare Life Sciences) equilibrated with 25 mM Tris-HCl, 0.2% DM, 1 mM TCEP (pH 8.0). The flow-through was collected and bound to Ni-NTA (0.5 ml resin per liter of culture) equilibrated with 20 mM Tris-HCl, 150 mM NaCl, 0.2% DM and 1 mM TCEP (pH 7.5). The resin was packed into a column by gravity and washed with 0.2% DM before washing with 10 column volumes of 0.5% n-dodecyl-β-D-maltopyranoside (DDM) to exchange GlpG into DDM micelles. The resin was washed with 0.1% DDM to return to a low concentration of detergent and eluted with 300 mM imidazole. Protein-containing fractions of 1 ml volume were pooled and exchanged into 50 mM Tris-HCl, 150 mM NaCl and 0.1% DDM (pH 7.5) with an Econo-Pac 10DG desalting column to remove TCEP and imidazole. Aliquots of purified GlpGCys-TM-Cys and GlpGCys-TM-Cys/L155A were flash frozen in liquid nitrogen and stored at −80 °C. Fresh aliquots were used for activity assays and 2,2′-dithiodipyridine (DTDP, Sigma-Aldrich) derivatization.
The GlpG substrate, SN-Spitz, was a modified version of SNGpATM, which contains staphylococcal nuclease fused to the transmembrane segment of glycophorin A transmembrane domain and a C-terminal His tag4. To convert SNGpATM into a GlpG substrate, the transmembrane segment was modified by Quickchange mutagenesis to include the sequence of Spitz (ASIASGA), which is a known cleavage site for E. coli GlpG49.
MATSTKKLHKEPATLIKAIDGDTVKLMYKGQPMTFRLLLVDT PETKHPKKGVEKYGPEASAFTKKMVENAKKIEVEFDKGQRTDKYG RGLAYIYADGKMVNEALVRQGLAKVAYVYKPNNTHEQHLRKSE AQAK KEKLNIWSEDNADSGPERVQLAHHFSEPGASIASGAVMAGVIGTI LLISYGIRRLIKKLEHHHHHH.
SN-Spitz was expressed and purified in DDM as previously described for SN-GpA4.
GlpG activity in detergent and bicelles
Bicelles for activity assays were composed of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 3-((3-cholamidopropyl)dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO, Avanti Polar Lipids) and prepared as described12,50,51. To prepare a stock solution of GlpGCys-TM-Cys in bicelles, 53 μM GlpGCys-TM-Cys in 50 mM Tris-HCl, 150 mM NaCl, 0.1% DDM (pH 7.5) was mixed 16.5:1 with 35% DMPC/CHAPSO (2.8:1, w/w) to give a final concentration of 50 μM GlpGCys-TM-Cys in 2% DMPC/CHAPSO. The mixture was then incubated on ice for 30 min followed by 2 h at room temperature before use. We also prepared a stock solution of GlpGCys-TM-Cys in detergent which contained 50 μM GlpGCys-TM-Cys in 50 mM Tris-HCl, 150 mM NaCl, 0.1% DDM (pH 7.5). A stock solution of SN-Spitz was prepared containing 200 μM SN-Spitz in 50 mM Tris-HCl, 150 mM NaCl and 0.1% DDM (pH 7.5). Reactions were initiated by adding 1.6 μl of the GlpGCys-TM-Cys stock, 1 μl of the SN-Spitz stock,17.4 μl of 50 mM Tris-HCl and 150 mM NaCl and then were incubated at 37 °C for 18 h. Reactions were stopped by adding 10 μl of 4× SDS sample buffer and heating for 10 min at 65 °C. The cleaved product was visualized by SDS-PAGE using a 4–12% NuPAGE BisTris gradient gel (Life Technologies) run in MES SDS running buffer (Supplementary Fig. 1a).
DNA handle attachment to bicelle-incorporated protein
The bicelle stock was made with DMPC lipid (Avanti Polar Lipids) and CHAPSO detergent (Sigma-Aldrich or Affymetrix) in the deionized water12,51. The molar ratio of lipid to detergent ranged from 2.2:1 to 2.8:1 with a final bicelle concentration of 8.8%. To dissolve the lipid and detergent, cycles of cooling on ice, brief vortexing, freezing at −80 °C, brief heating to 33 °C and vortexing were performed. Finally a quick spin at 4 °C with a tabletop centrifuge helped remove any remaining powders. The bicelle stock solution was stored at −80 °C.
Purified GlpGCys-TM-Cys was derivatized with DTDP by mixing 250 μl of 27 μM GlpGCys-TM-Cys in 50 mM Tris-HCl, 150 mM NaCl, 0.1% DDM (pH 7.5) with 20 μl of 67 mM DTDP dissolved in acetonitrile for a final concentration of 25 μM GlpG and 5 mM DTDP. The reaction was incubated on a rotator for 1 h at room temperature. Unreacted DTDP was removed using a BioRad Econo-Pac 10DG desalting column equilibrated with 50 mM Tris-HCl, 150 mM NaCl, 0.1% DDM (pH 7.5). Protein was collected in 250-μl fractions. The peak fractions were pooled and concentrated to 200 μl of 26 μM DTDP-derivatized GlpG. Complete labeling of GlpG was verified by LC/MS as described using an orifice potential of 90 V (ref. 52). We added the bicelle stock solution to the DTDP-derivatized GlpG in a 1:4 (v/v) ratio while keeping it on ice and gently pipetted the contents up and down until the solution became clear and homogenous12,51. We incubated the mixture on ice for 1 h to allow for complete reconstitution of GlpG into bicelles and kept the bicelle-GlpG mixture on ice until it was used in the next step.
Two types of 512-bp DNA (biotin- and digoxigenin-modified handles) were PCR-amplified using a λDNA template, the forward primer CATGTGGGTGACG CGAAA with a 5′ thiol–modified C6 S-S and the reverse primer TCGCCACCATCATTTCCA with either 5′ biotin or 5′ digoxigenin modification (each 0.4 ml and 2 ml). The thiol modifications of the PCR products were activated by adding 100 mM DTT final concentration and incubating for 1 h at 37 °C. The products were purified using a PCR purification column, eluted into 50 mM Tris-HCl (pH 7.5) and concentrated to 3–10 μM in a final volume of ~30 μl using a 10K Amicon centrifugal filter (Millipore). The DNA handles were stored at −80 °C.
To maximize the likelihood of two different handles attaching to a GlpG protein, the handles were attached sequentially, maintaining the bicelle concentration as 0.5–2% during the whole attachment reaction. First, about 20-fold excess of the protein (14 μM) was reacted with the biotin-modified DNA handle (0.8 μM) in 40 mM Tris-HCl, 80 mM NaCl, 1.3% bicelle (pH 7.5) for >12 h at room temperature. Repetitive buffer exchange into 50 mM Tris-HCl, 100 mM NaCl, 2% bicelle (pH 7.5) were then performed using a 100K Amicon centrifugal filter. Then the GlpG attached to the biotin-modified handle (0.2 μM) was reacted with about 40-fold excess of digoxigenin-modified DNA handle (7.5 μM) in 50 mM Tris-HCl, 150 mM NaCl and 0.5–1% bicelle (pH 7.5) for >20 h at room temperature. The bicelle-incorporated GlpG covalently linked to the respective DNA handles (bicelle-GlpG-DNA) was diluted tenfold with 50 mM Tris-HCl, 150 mM NaCl and 1.3% bicelle (pH 7.5) and stored at −80 °C. The bicelle-GlpG-DNA sample was analyzed by 6% SDS-PAGE stained with SYBR Safe DNA Gel Stain (Invitrogen) (Supplementary Fig. 1b).
Single-molecule magnetic tweezers experiment
A single-molecule magnetic tweezers apparatus was built on an inverted microscope (Olympus, IX73) as previously described27–33, in which force can be easily controlled by changing the vertical distance of a pair of magnets from the sample. The imaging room of the magnetic tweezers was maintained at constant humidity and constant temperature (22 °C) to prevent an undesirable bicelle phase transition at higher temperature. The sample chamber was a ~20 μl volume channel, constructed by putting together a 24 × 40 mm cleaned coverslip and 24 × 50 mm polyethylene glycol-coated coverslip with double-sided tape. The bicelle-GlpG-DNA sample was injected into the sample channel and then attached to the bottom coverslip by biotin-neutravidin binding and to 2.8-μm magnetic beads by dig-antidig binding. The buffer condition in the sample channel was 50 mM Tris-HCl, 150 mM NaCl, 1.3% bicelle (pH 7.5). We can exchange various buffer solutions by capillary force into the channel. By approaching the pair of magnets to the experiment sample, we can apply a few to tens of pN force to the single GlpG protein and then measure the change of extension, i.e., the end-to-end distance in the bicelle-GlpG-DNA molecule (Fig. 1a). The extension change is obtained from the change of diffraction patterns of attached magnetic bead captured in 60 Hz CCD camera (JAI). We corrected vertical drifts of microscope stage by maintaining the vertical position of a nonmagnetic reference bead immobilized directly on the bottom surface every 500 ms.
In the gradual pulling experiments (Fig. 1 and Fig. 3a), the force-loading rate at every moment is far below 1 pN s−1, which is near equilibrium condition during protein unfolding and folding. In the force-jump and force-cycle experiments (Fig. 2 and Fig. 3c), the unfolding step sizes were measured as the difference between arithmetic mean values of extensions over the appropriate intervals before and after the unfolding event and then statistically analyzed as Gaussian distributions by collecting them (further analysis is described below). We can assess the relevant error in the step-size measurement (σstep) with the equation , which illustrates that σstep is a s.e.m. because the s.d. of extension trace (σi, σf) is divided by the number of data points (Ni, Nf). Because the fluctuation of extension traces is typically less than 5 nm and we include more than 300 data points for each measurement, σstep is less than ~0.4 nm, indicating that we can estimate the step size with an accuracy down to the level of a few Å27.
Extension analysis for finding intermediate positions
To map the extension values measured in the force-jump experiment to the corresponding residue positions (Fig. 2), we analyzed the expected extension when a GlpG protein unfolds from the native state to specific residues27 (Supplementary Fig. 9). The total extension is described as the sum of three terms:
| (1) |
where xp,n is the extension expected for the nth helix or linker that has lost its secondary structure (i.e., an unstructured polypeptide) as calculated by the Marko-Siggia formula; xh,n is the extension for the nth helix as calculated by Kessler-Rabin formula53; and Δd is the axial length change of the tertiary structure between DNA handles calculated from the GlpG structure information15–18.
The extension for unfolded polypeptide (xp) is obtained using the Marko-Siggia formula of the worm-like chain (WLC) model:
| (2) |
where F is the applied tension; kBT is the thermal energy; Lp is the contour length of polypeptide, which is the number of unfolded residues (Np) times the average residue step size (lp) of 0.36 nm (ref. 6); and Pp is the persistence length of polypeptide (measured as 0.39 nm; described in the next section). Equation (2) can be applied when the contour length of a polymer is much greater (by at least a factor of five) than its persistence length. Therefore, μ-helices are not well described by equation (2) because the persistence length of the helices (tens of nm) is greater than the contour lengths of each helix (a few nm). Thus the extension for the helical part (xh) is estimated by the Kessler-Rabin formula (KR model), which is applicable for any arbitrary ratio between persistence length and contour length:
| (3) |
where f = F/kBT, , Lh is the contour length of helix that is the number of residues in the helix (Nh) times the average helical rise per amino acid (lh) of 0.16 nm, and Ph is the persistence length of helix (measured as 9.17 nm; described in next section). Finally, from the GlpG structure information, the axial length change of Δd can be obtained as Δd = d − d0, where d is the axial width of the partially folded structure up to specific residue and d0 is the axial width of the fully folded structure (Supplementary Fig. 9a).
In analyzing the Gaussian peaks of the extension distribution measured in the force-jump experiment (Fig. 2), equation (1) can be reduced to
| (4) |
because we observe that a helix-coil transition occurs at about 18 pN; thus, all of the helices can be assumed to be unraveled upon unfolding at the 21-pN force used. Thus, with equations (2) and (4), we calculated the expected extension value for a GlpG protein unfolding up to a specific residue position (Supplementary Fig. 9b). In this calculation, we compared two versions of extension estimation from two different GlpG structures: one in detergent condition15 and the other in lipid bilayer condition18. The difference in protein structures is reflected in the structural factor Δd in equations (1) and (4), but we did not see any obvious difference between the two estimations (Supplementary Fig. 9b).
Helix-coil transition
By pooling the unfolding data (unfolding force Fu and step size ΔLu) from all traces in the gradual pulling experiments (Supplementary Fig. 3a), we produced a scatter plot showing the unfolding force against the step size (Supplementary Fig. 3b). When the six transmembrane helices are completely unraveled to polypeptide coils upon unfolding, the data points are expected to be distributed along the line of equation (2) (WLC model; Supplementary Fig. 3b). We observed a definite deviation from the WLC model below 20 pN, which indicates that the helix-coil transition in the corresponding force range (Supplementary Fig. 3b,c). This is further supported by the observation that in the force-jump experiment (Fig. 2a and Supplementary Fig. 6), the observed step size of ~35 nm corresponds to unfolding of GlpG to the completely unstructured state with no α-helical content (Supplementary Fig. 9b).
To obtain the persistence length of the polypeptide (Pp), we fitted the data only for the region over 20 pN with the WLC model,
| (5) |
which is derived from equations (2) and (4) with d = 0 because there is no tertiary structure (Supplementary Fig. 3b). The average residue step size (lp) of 0.36 nm was used6. The persistence length determined for the GlpG polypeptide (Pp = 0.39 nm) is consistent with what was reported for a similar helical membrane protein (Pp = 0.4 nm) (ref. 6).
For the persistence length of helix (Ph), we fitted the data for the region below 17 pN with the WLC-KR model
| (6) |
(Supplementary Fig. 3b). Equation (6) is derived from the simplified equation (1), x ≈ xp + 6·xh − d0, and equation (3). The extension for loop regions between helices (xp) is calculated by equation (2). The estimated persistence length for helices of GlpG (Ph = 9.17 nm) is broadly consistent with the known value for a helix in a coiled coil (Ph = 25 nm) (ref. 54). The fact that the persistence length for the coiled coil is somewhat larger seems reasonable because of the tight association of two helices in the coiled coil.
Quantitative analysis of the folding energy landscape
To obtain a quantitative picture for the folding energy landscape, we measured unfolding and folding kinetics. We used the gradual pulling experiment to obtain the unfolded fraction (U) as a function of force (F) (Figs. 3a,b and 4a), from which we determined the kinetic rate constant for unfolding at zero force (ku0) and the distance from the native state to the transition state (Δxf†) (Table 1). To this end, we used the following equation,
| (7) |
where kBT is the thermal energy and A is the proportional constant of dF/dt = AF. The constant A is determined from the data of force calibration with magnet heights, which is approximated as a single exponential function in the analyzed force range. Equation (7) is derived from the first-order rate equation, dN/dt = −kuN, and the Bell equation, ku = ku0 exp(−FΔxf†/kBT), where N represents the folded fraction and ku represents the unfolding kinetic rate at a given force. Equation (7) can be derived from
For folding kinetics, we performed refolding experiments at lower forces ranging from 0 pN to 8 pN (Figs. 3c,d and 4b). At these forces, the thermal noise is too high to detect the individual refolding events. Hence after unfolding at 21 pN, we lowered the force to specified levels, waited for 3 min and increased the force to 21 pN to see whether GlpG was refolded during the 3-min waiting time (Fig. 3c). From the folding probability (N) as a function of force (Figs. 3d and 4b), we measured the kinetic rate for folding at zero force (kf0) and the distance from the unfolded state to the transition state (Δxu†; Table 1). The fitting equation
| (8) |
in which Δt is the waiting time at specific force for refolding, is likewise derived from the first-order rate equation dU/dt = −kfU, and the Bell equation, kf = kf0 exp(−FΔxu† / kBT), where kf is the folding kinetic rate at arbitrary force. The formula derivation is developed as
at constant force.
From the kinetic rate constants (ku0, kf0), we obtained the unfolding free energy (ΔG) and the kinetic energy barriers (ΔGu†, ΔGf†) (Fig. 3e and Table 1) by the Kramer equation
| (9) |
| (10) |
| (11) |
where kw is the frequency factor in the range of 104–106 s−1 (refs. 55–60), which is why the energy barriers are measured with an error of 2.3 kBT. The unfolding free energy (ΔG) indicating the thermodynamic stability of protein is more reliably measured with an error of 0.2 kBT because it is obtained only from the ratio of ku0 to kf0, regardless of the frequency factor.
Supplementary Material
Acknowledgments
This work was supported by the National Creative Research Initiative Program (Center for Single-Molecule Systems Biology to T.-Y.Y.) funded by the National Research Foundation of Korea and Marine Biotechnology Program (20150220 to T.-Y.Y.) funded by the Ministry of Oceans and Fisheries of Korea, and supported by US National Institutes of Health grant 2R01GM063919 to J.U.B.
Footnotes
Author contributions
D.M., R.E.J., J.U.B. and T.-Y.Y. designed the experiments. R.E.J. expressed and purified proteins. D.M. prepared the DNA-protein hybrid sample and performed the magnetic tweezers experiments. All of the authors analyzed the data and contributed to writing of the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Engelman DM, et al. Membrane protein folding: beyond the two stage model. FEBS Lett. 2003;555:122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 2.Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 3.White SH, von Heijne G. How translocons select transmembrane helices. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- 4.Hong H, Blois TM, Cao Z, Bowie JU. Method to measure strong protein-protein interactions in lipid bilayers using a steric trap. Proc Natl Acad Sci USA. 2010;107:19802–19807. doi: 10.1073/pnas.1010348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang YC, Bowie JU. Measuring membrane protein stability under native conditions. Proc Natl Acad Sci USA. 2014;111:219–224. doi: 10.1073/pnas.1318576111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oesterhelt F, et al. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 7.Kedrov A, Janovjak H, Sapra KT, Muller DJ. Deciphering molecular interactions of native membrane proteins by single-molecule force spectroscopy. Annu Rev Biophys Biomol Struct. 2007;36:233–260. doi: 10.1146/annurev.biophys.36.040306.132640. [DOI] [PubMed] [Google Scholar]
- 8.Engel A, Gaub HE. Structure and mechanics of membrane proteins. Annu Rev Biochem. 2008;77:127–148. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- 9.Zocher M, et al. Single-molecule force spectroscopy from nanodiscs: an assay to quantify folding, stability, and interactions of native membrane proteins. ACS Nano. 2012;6:961–971. doi: 10.1021/nn204624p. [DOI] [PubMed] [Google Scholar]
- 10.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 11.Shank EA, Cecconi C, Dill JW, Marqusee S, Bustamante C. The folding cooperativity of a protein is controlled by its chain topology. Nature. 2010;465:637–640. doi: 10.1038/nature09021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 13.Joh NH, et al. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453:1266–1270. doi: 10.1038/nature06977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dürr UH, Gildenberg M, Ramamoorthy A. The magic of bicelles lights up membrane protein structure. Chem Rev. 2012;112:6054–6074. doi: 10.1021/cr300061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YC, Zhang YJ, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–183. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 16.Wu ZR, et al. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 17.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci USA. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinothkumar KR. Structure of rhomboid protease in a lipid environment. J Mol Biol. 2011;407:232–247. doi: 10.1016/j.jmb.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemberg MK, Freeman M. Cutting proteins within lipid bilayers: rhomboid structure and mechanism. Mol Cell. 2007;28:930–940. doi: 10.1016/j.molcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Freeman M. Rhomboid proteases and their biological functions. Annu Rev Genet. 2008;42:191–210. doi: 10.1146/annurev.genet.42.110807.091628. [DOI] [PubMed] [Google Scholar]
- 21.Ha Y, Akiyama Y, Xue Y. Structure and mechanism of rhomboid protease. J Biol Chem. 2013;288:15430–15436. doi: 10.1074/jbc.R112.422378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinothkumar KR, Freeman M. Intramembrane proteolysis by rhomboids: catalytic mechanisms and regulatory principles. Curr Opin Struct Biol. 2013;23:851–858. doi: 10.1016/j.sbi.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 23.Lemberg MK. Sampling the membrane: function of rhomboid-family proteins. Trends Cell Biol. 2013;23:210–217. doi: 10.1016/j.tcb.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Baker RP, Urban S. Architectural and thermodynamic principles underlying intramembrane protease function. Nat Chem Biol. 2012;8:759–768. doi: 10.1038/nchembio.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paslawski W, et al. Cooperative folding of a polytopic α-helical membrane protein involves a compact N-terminal nucleus and nonnative loops. Proc Natl Acad Sci USA. 2015;112:7978–7983. doi: 10.1073/pnas.1424751112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cecconi C, Shank EA, Marqusee S, Bustamante C. In: DNA Nanotechnology: Methods and Protocols. 1. Zuccheri G, Samorì B, editors. Vol. 749. Humana Press; 2011. pp. 255–271. [Google Scholar]
- 27.Min D, et al. Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism. Nat Commun. 2013;4:1705–1714. doi: 10.1038/ncomms2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bae W, et al. Programmed folding of DNA origami structures through single-molecule force control. Nat Commun. 2014;5:5654–5661. doi: 10.1038/ncomms6654. [DOI] [PubMed] [Google Scholar]
- 29.Gosse C, Croquette V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saleh OA, Allemand JF, Croquette V, Bensimon D. Single-molecule manipulation measurements of DNA transport proteins. ChemPhysChem. 2005;6:813–818. doi: 10.1002/cphc.200400635. [DOI] [PubMed] [Google Scholar]
- 31.Kim K, Saleh OA. A high-resolution magnetic tweezer for single-molecule measurements. Nucleic Acids Res. 2009;37:136–142. doi: 10.1093/nar/gkp725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipfert J, Hao XM, Dekker NH. Quantitative modeling and optimization of magnetic tweezers. Biophys J. 2009;96:5040–5049. doi: 10.1016/j.bpj.2009.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Vlaminck I, Dekker C. Recent advances in magnetic tweezers. Annu Rev Biophys. 2012;41:453–472. doi: 10.1146/annurev-biophys-122311-100544. [DOI] [PubMed] [Google Scholar]
- 34.Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alegre-Cebollada J, Kosuri P, Rivas-Pardo JA, Fernandez JM. Direct observation of disulfide isomerization in a single protein. Nat Chem. 2011;3:882–887. doi: 10.1038/nchem.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stigler J, Ziegler F, Gieseke A, Gebhardt JCM, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 37.Carrion-Vazquez M, et al. Mechanical and chemical unfolding of a single protein: A comparison. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 39.Beaugrand M, et al. Lipid concentration and molar ratio boundaries for the use of isotropic bicelles. Langmuir. 2014;30:6162–6170. doi: 10.1021/la5004353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curnow P, Booth PJ. Combined kinetic and thermodynamic analysis of α-helical membrane protein unfolding. Proc Natl Acad Sci USA. 2007;104:18970–18975. doi: 10.1073/pnas.0705067104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otzen DE. Mapping the folding pathway of the transmembrane protein DsbB by protein engineering. Protein Eng Des Sel. 2011;24:139–149. doi: 10.1093/protein/gzq079. [DOI] [PubMed] [Google Scholar]
- 42.Watters AL, et al. The highly cooperative folding of small naturally occurring proteins is likely the result of natural selection. Cell. 2007;128:613–624. doi: 10.1016/j.cell.2006.12.042. [DOI] [PubMed] [Google Scholar]
- 43.Cymer F, von Heijne G, White SH. Mechanisms of integral membrane protein insertion and folding. J Mol Biol. 2015;427:999–1022. doi: 10.1016/j.jmb.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim SJ, Skach WR. Mechanisms of CFTR folding at the endoplasmic reticulum. Front Pharmacol. 2012;3:201. doi: 10.3389/fphar.2012.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curnow P, et al. Stable folding core in the folding transition state of an α-helical integral membrane protein. Proc Natl Acad Sci USA. 2011;108:14133–14138. doi: 10.1073/pnas.1012594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jefferson RE, Blois TM, Bowie JU. Membrane proteins can have high kinetic stability. J Am Chem Soc. 2013;135:15183–15190. doi: 10.1021/ja407232b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otzen DE. Folding of DsbB in mixed micelles: a kinetic analysis of the stability of a bacterial membrane protein. J Mol Biol. 2003;330:641–649. doi: 10.1016/s0022-2836(03)00624-7. [DOI] [PubMed] [Google Scholar]
- 48.Kim BL, Schafer NP, Wolynes PG. Predictive energy landscapes for folding α-helical transmembrane proteins. Proc Natl Acad Sci USA. 2014;111:11031–11036. doi: 10.1073/pnas.1410529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urban S, Wolfe MS. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc Natl Acad Sci USA. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faham S, Ujwal R, Abramson J, Bowie JU. In: Membrane Protein Crystallization. 1. DeLucas LJ, editor. Vol. 63. Academic Press; 2009. pp. 109–125. [Google Scholar]
- 51.Ujwal R, Bowie JU. Crystallizing membrane proteins using lipidic bicelles. Methods. 2011;55:337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitelegge JP, et al. Toward the bilayer proteome, electrospray ionizationmass spectrometry of large, intact transmembrane proteins. Proc Natl Acad Sci USA. 1999;96:10695–10698. doi: 10.1073/pnas.96.19.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kessler DA, Rabin Y. Distribution functions for filaments under tension. J Chem Phys. 2004;121:1155–1164. doi: 10.1063/1.1760743. [DOI] [PubMed] [Google Scholar]
- 54.Schwaiger I, Sattler C, Hostetter DR, Rief M. The myosin coiled-coil is a truly elastic protein structure. Nat Mater. 2002;1:232–235. doi: 10.1038/nmat776. [DOI] [PubMed] [Google Scholar]
- 55.Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 56.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 57.Rhoades E, Cohen M, Schuler B, Haran G. Two-state folding observed in individual protein molecules. J Am Chem Soc. 2004;126:14686–14687. doi: 10.1021/ja046209k. [DOI] [PubMed] [Google Scholar]
- 58.Kubelka J, Hofrichter J, Eaton WA. The protein folding ‘speed limit’. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 59.Chung HS, Louis JM, Eaton WA. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc Natl Acad Sci USA. 2009;106:11837–11844. doi: 10.1073/pnas.0901178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gebhardt JCM, Bornschlogla T, Rief M. Full distance-resolved folding energy landscape of one single protein molecule. Proc Natl Acad Sci USA. 2010;107:2013–2018. doi: 10.1073/pnas.0909854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.