Abstract
The microbiota of breast milk from Chinese lactating mothers at different stages of lactation was examined in the framework of a Maternal Infant Nutrition Growth (MING) study investigating the dietary habits and breast milk composition in Chinese urban mothers. We used microbiota profiling based on the sequencing of fragments of 16S rRNA gene and specific qPCR for bifidobacteria, lactobacilli and total bacteria to study microbiota of the entire breast milk collected using standard protocol without aseptic cleansing (n = 60), and the microbiota of the milk collected aseptically (n = 30). We have also investigated the impact of the delivery mode and the stage of lactation on the microbiota composition. The microbiota of breast milk was dominated by streptococci and staphylococci for both collection protocols and, in the case of standard collection protocol, Acinetobacter sp. While the predominance of streptococci and staphylococci was consistently reported previously for other populations, the abundance of Acinetobacter sp. was reported only once before in a study where milk collection was done without aseptic cleansing of the breast and rejection of foremilk. Higher bacterial counts were found in the milk collected using standard protocol. Bifidobacteria and lactobacilli were present in few samples with low abundance. We observed no effect of the stage of lactation or the delivery mode on microbiota composition. Methodological and geographical differences likely explain the variability in microbiota composition reported to date.
Introduction
A large number of studies have reported the presence of bacteria in breast milk of healthy lactating women. Early culture studies have showed the consistent presence of commensal bacteria in breast milk of healthy women, but bacteria were seen as a nuisance, complicating the storage and later use of expressed milk e.g. [1–5]. These early studies focused on the detection of potential pathogens and used culture methods that did not support the growth of organisms such as lactobacilli or bifidobacteria. The presence of coagulase-negative staphylococci (usually S. epidermidis), S. aureus, streptococci and Acinetobacter was consistently reported, while other taxa were found more sporadically (enterococci, E. coli, Klebsiella, Moraxella, Pseudomonas). Later studies applied methods allowing the growth of lactobacilli [6] and eventually also bifidobacteria [7–9], and these taxa were detected, but in low numbers (eg. bifidobacteria represented 1.7% and lactobacilli represented 0.4% of all isolated bacteria [8]).
Only recently has it has been increasingly considered that the presence of bacteria in various niches of the human body is not only a physiological norm, but can also be beneficial for the human host [10]. In parallel, the development of technology has permitted the investigation of bacteria in breast milk based on DNA signatures, initially by quantitative PCR targeting specific bacterial groups [11–17]. More recently, several studies have used microbiome profiling based on sequencing of fragments of 16S rRNA gene [8, 15, 16, 18, 19] and shotgun metagenomics consisting in sequencing of the entire bacterial DNA [20, 21]. The reported composition varied widely, suggesting that methodology as well as geographical or ethnical differences could play a role. Some studies observed composition similar to these obtained by culture based studies [16, 18, 21]; however, higher abundance of Pseudomonas than in culture based studies was reported [8, 16, 20, 21]. In one study the predominance of Leuconostoc and Weisella was observed [15]. The above studies were conducted in US or Western European populations. To our knowledge no studies investigating the microbiota of breast milk of lactating mothers were conducted in Asian populations. Most studies which used molecular techniques performed sterile collection, with rejection of the first few millilitres of breast milk. Only a few studies have investigated the microbiota of the entire breast milk collected without cleansing [19, 21] which the suckling infant actually ingests. Although, to our knowledge, no studies have compared the microbiota of the breast milk collected with aseptic cleansing with bacteria present in the entire milk, older studies using culture methods suggested that there were more bacteria in the foremilk [4].
The objective of this cross-sectional study was to investigate the microbiota of breast milk from Chinese lactating mothers during the different stages of lactation, using both aseptic collection protocol used by most studies to date, as well as standard collection protocol to target the breastfeeding-associated microbiota. Our study further substantiates the presence of bacteria in human milk, with a significantly higher number of bacteria identified in the “breastfeeding-associated microbiota” compared to milk obtained under aseptic conditions. We also confirmed the presence of the dominant species such as streptococci, staphylococci, as well as the low abundance of bifidobacteria and lactobacilli.
This study is part of the larger initiative Maternal Infant Nutrition Growth (MING) study.
Methods
Participants
This study was part of MING, a cross-sectional study designed to investigate the dietary and nutritional status of pregnant women, lactating mothers and young children aged from birth up to three years living in urban areas of China. In addition, the human milk composition of Chinese lactating mothers from 3 cities (Beijing, Guangzhou and Suzhou) was characterized in each city. The study was conducted between 2011 and 2012. Two hospitals with maternal and child care units were randomly selected. Furthermore, mothers at lactation period 0 to 240 days were randomly selected based on child registration information. Participants included in the period 0–5 days were recruited at the hospital whereas the other participants were requested by phone to join the study. If participation was dismissed a replacement was found. Response rate was 52%. In the present study of microbiota, only samples from lactating mothers from Beijing were used, and these were limited to three stages of lactation (0–4 days, 5–11 days and 1–2 months after birth). 60 participants chosen at random were included for microbiota analysis with standard collection protocol, and 30 with aseptic protocol, respectively (S1 Fig).
Women between 18–45 years of age who gave birth to a single, healthy, full-term infant and who were exclusively breastfeeding were included in the study. Exclusion criteria included gestational diabetes, hypertension, cardiac diseases, acute communicable diseases and postpartum depression. Lactating women who had nipple or lacteal gland diseases, who had been receiving hormonal therapy during the three months preceding recruitment, or who had insufficient skills to understand study questionnaires were also excluded. The study was conducted according to the guidelines in the Declaration of Helsinki. All of the procedures involving human subjects were approved by the Medical Ethics Research Board of Peking University (No.IRB00001052-11042). Written informed consent was obtained from all subjects participating in the study. The study was also registered in ClinicalTrials.gov with the number identifier NCT01971671.
Data and sample collection
All participants responded to a general questionnaire including socio-economic and lifestyle aspects of the mother. The number of gestational weeks at delivery, delivery method, and the gender of the infant were recorded, the height and weight were measured. None of the participating mothers were treated with antibiotics. The samples were collected between 9:00 am and 11:00 am. The first set of thirty breast milk samples (10 samples/lactation stage collected from 30 mothers) were collected with an aseptic protocol. Sterile gloves were used, the first few drops (0.5–1 ml) were discarded, and the breast was thoroughly cleansed with chlorhexidine solution before manually collecting 3 ml of milk. The second set of sixty breast milk samples (20 samples/lactation stage collected from 60 mothers) were collected from a different group of mothers with standard protocol. Standard protocol did not include any particular cleaning procedure, and it was identical to the one used throughout MING study (S1 Fig, [22], Giuffrida et al in preparation, Affolter et al in preparation, Garcia Rodenas et al submitted). Single full breast milk was sampled with the use of an electric pump (Horigen HNR/X-2108ZB, Xinhe Electrical Apparatuses Co., Ltd), and after securing an aliquot of 10 ml (colostrum) or 40 ml (other lactation stages), the rest of the milk was returned to the mother for feeding to the infant. All the samples were frozen at -80°C before shipping to Lausanne, Switzerland for characterization, and they remained frozen until DNA extraction.
DNA extraction
1 ml of milk was centrifuged for 20 minutes at 8000 g, and the pellets were dissolved in 500 μl of the supernatant. The first set of 30 samples was processed with the DNA Stool Mini kit (QIAGEN, Switzerland) according to the manufacturer’s instructions, with two modifications: the milk samples were heated at 95°C for 5 s and a bead-beating step with Fastprep (MP Biochemicals, USA) was added (30 s, speed 6 with 0.1 mm silica beads). The second set of 60 samples was processed with the Fast DNA SPIN Kit for soil (MO BIO, USA) according to the manufacturer’s instructions.
Quantitative PCR
Quantitative PCR for total Bacteria, total Lactobacillus and total Bifidobacterium were performed as described in Table 1. All assays targeted variable regions of the 16S rRNA gene. The 25 μl PCR mixture contained TaqMan gene expression Master Mix (Applied Biosystems, Switzerland) or SensiMix Plus SYBR (Quantace, Switzerland), 2 μl of DNA extract, primers and Milli-Q water. The amplification reactions were performed in duplicate for each sample using the StepOnePlus Real-Time PCR System (Life Technology Europe, Switzerland). For each of the organisms standard curves were prepared by a tenfold dilutions of DNA from the reference strains within a range of 50 ng to 50 fg for Lactobacillus and Bifidobacterium and a range of 40 ng to 40 fg for total bacteria. The DNA concentration of the reference strains was measured by nanodrop ND-1000 (Nanodrop Technologies, USA). The results were expressed as genomic DNA copy numbers per sample volume (ml), based on the copy number present in the reference strains (Table 1).
Table 1. qPCR conditions.
| Target | Reference strain | Primers 5’– 3’ | Concentration of primers and probes (nM) | Detection limit (CFU/m)l | Cycling conditions | Reference |
|---|---|---|---|---|---|---|
| Total Bacteria | Escherichia coli K12 | For: TCCTACGGGAGGCAGCAGT | 300 | 5.5 x 102 | 95°C 10’(95°C 15”/60°C 60”) 45x | [23] |
| Rev: GGACTACCAGGGTATCTAATCCTGTT | 300 | |||||
| Probe: CGTATTACCGCGGCTGCTGGCAC | 175 | |||||
| Total bifidobacteria Sample set collected with Standard protocol | Bifidobacterium longum NCC3001 | For: CGATGCAACGCAAGAACC | 500 | 1.4 x 103 | 95°C 10’ (95°C 15”/60°C 60”) 50x | This study |
| Rev: ATCTCACGACACGAGCTGAC | 500 | |||||
| Probe: tt(C)gggg(S)ggttcaca | 200 | |||||
| Total bifidobacteria Sample set collected with Aspetic protocol | Bifidobacterium lactis NCC2818 | g-Bifid-F: CTCCTGGAAACGGGTGG | 250 | 1.4 x 102 | 94°C 5’ (94°C 20”/55°C 20”/72°C 50”) 40x | [24] |
| g-Bifid-R: GGTGTTCTTCCCGATATCTACA | 250 | |||||
| Total lactobacilli | Lactobacillus johnsonii NCC533 | For: AGCAGTAGGGAATCTTCCA | 500 | 2.4 x 102 | 95°C 10’ (95°C 15”/58°C 20”/72°C 45”) 40x | [25] |
| Rev: CACCGCTACACATGGAG | 500 |
Microbiota profiling
For the 16S rRNA gene sequencing, the approach proposed by [26] was followed with some modifications. The V4 region was amplified by PCR (for 30μl reaction volume: 6μl of DNA, 1.2μl of each primer at 10 pM, 0.6μl of each dNTP at 2.5 mM, 6μl of Expand High Fidelity PLUS Reaction Buffer, 0.3μl of Expand High Fidelity PLUS Enzyme Blend 5U/μl (Roche Applied Science, Switzerland). Amplification conditions were the following: 94°C for 2 min followed by 30 cycles of 94°C for 30 seconds, 50°C for 30 seconds and 72°C for 30 seconds followed by 7 minutes at 72°C and finally held at 4°C). The primers mapped to the conserved regions flanking V4 and produced an amplicon of 425 bp. In addition, they carried a specific barcode to multiplex the samples for sequencing and the Illumina sequencing adapter. The primers were identical to these used in [26]. After quantification and quality check by capillary electrophoresis (LabChip GX, Perkin Elmer, USA), all the amplicons were pooled in equimolar amount and purified. In cases where the DNA concentration was below the detection limit, all available sample volume was used for pooling. Sequencing was performed on the MiSeq instrument V2.1 (with Hard Coded matrix for data acquisition) paired-end 250 bp (MS-102-2003). Demultiplexing was performed by the MiSeq software (V2.1). Raw sequence data were deposited in the GenBank Short Read Archive (Accession number: SRP077712).
Processing of the sequencing data
Data processing was done with mothur software v.1.32.1 [27]. Paired-end sequences were joined as described [28]. The sequences were aligned to Silva reference alignment and trimmed so that complete overlap was achieved. A preclustering step with 2%-linkage algorithm was performed [29]. Chimeras were removed using uchime algorithm using de novo option [30]. The sequences were classified with RDP classifier (version 9) using 0.8 confidence level. The sequences were clustered into OTUs using 0.03 similarity cut-off value. The distances among sequences were calculated as pairwise uncorrected distance, where gaps were considered a single position, and clustering using average neighbor method. Before calculating beta diversity measures and statistical analyses, the number of sequences per sample was made uniform by random subsampling of 2226 reads from each sample. This eliminated samples that had fewer than 2226 sequencing reads: 8 out of 30 samples from the first data set, and 2 out of 60 samples from the second data set.
Results
Participants’ characteristics
S1 Fig displays the recruitment flowchart from eligibility to sample analysis. Demographics and anthropometry of the participants are described in Table 2. Although there was no specific selection to exclude smokers, there were only few smokers in the investigated population and all mothers in this subset were non-smokers.
Table 2. Maternal and infant characteristics.
| Variable | Aseptic protocol (n = 30) | Standard protocol (n = 60) |
|---|---|---|
| Mother | ||
| Age (years), Mean (SD) | 28.1 (3.6) | 28.2 (3.7) |
| Height (cm), Mean (SD) | 163 (5.1) | 162 (5.4) |
| Weight (kg), Mean (SD) | 63.4 (7.8) | 65.4 (9.5) |
| BMI (kg/m2) | 23.9 (3.3) | 25.1 (3.5) |
| Caesarean delivery, N (%) | 13 (43) | 33 (55) |
| Infant | ||
| Males, N (%) | 13 (48) | 21 (37) |
| Gestational age at birth (weeks), Mean (SD) | 39.5 (1.1) | 39.2 (1.2) |
Quantification of bacteria by qPCR
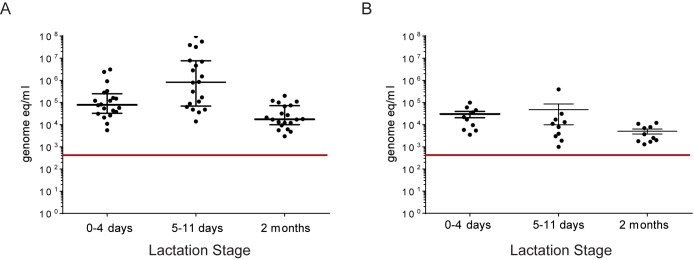
Total bacterial load measured by qPCR (Fig 1) was higher in breast milk collected with standard protocol (median 7.5 x 104 counts/ml) than in samples taken with aseptic precautions (median 7.8 x 103 counts/ml). This difference was significant for samples taken at 5–11 days (Mann-Whitney test, p<0.0001), but not at 0–4 days (Mann-Whitney test, ns) or at 2 months (Mann-Whitney test, ns). Bifidobacteria and lactobacilli were found in only few samples (Table 3). It has to be noted that a different method of detection of bifidobacteria was used for the second set of samples (taken without aseptic precautions), resulting in a lower detection limit.
Fig 1. Quantification of bacteria by qPCR.
Total bacteria counts measured by qPCR in samples collected with standard (A) and aseptic protocol (B) at three lactation stages. Detection limits are shown by red line.
Table 3. Detection of lactobacilli and bifidobacteria in breast milk samples.
Number of positive samples is shown.
| Condition | n | Bifidobacteria | Lactobacilli |
|---|---|---|---|
| Aseptic Protocol | |||
| 0–4 days | 10 | 2 | 2 |
| 5–11 days | 10 | 0 | 1 |
| 2 months | 10 | 0 | 0 |
| Standard protocol | |||
| 0–4 days | 20 | 2 | 0 |
| 5–11 days | 20 | 0 | 0 |
| 2 months | 20 | 1 | 2 |
Microbiota profiling by sequencing of 16S rRNA gene amplicons
The quantity of bacteria in the set of breast milk samples collected with aseptic protocol was mostly below the limit usually considered sufficient for microbiota profiling based on 16S rRNA gene sequencing (106 genome copies/ml) [31]. The majority of samples (23) gave no quantifiable PCR product. Out of 30 samples, only one yielded adequate PCR amplicon and six further samples gave weak products. The samples collected with standard protocol contained more bacterial DNA, and out of 60 samples 17 yielded adequate PCR product, 10 samples yielded weak PCR product, and 33 gave no quantifiable PCR product. Despite the fact that many samples did not reach standard quality requirements for sequencing, all samples were sequenced. The samples which gave good amplification products were compared to those with week amplification products, and no differences were seen (S2 Fig).
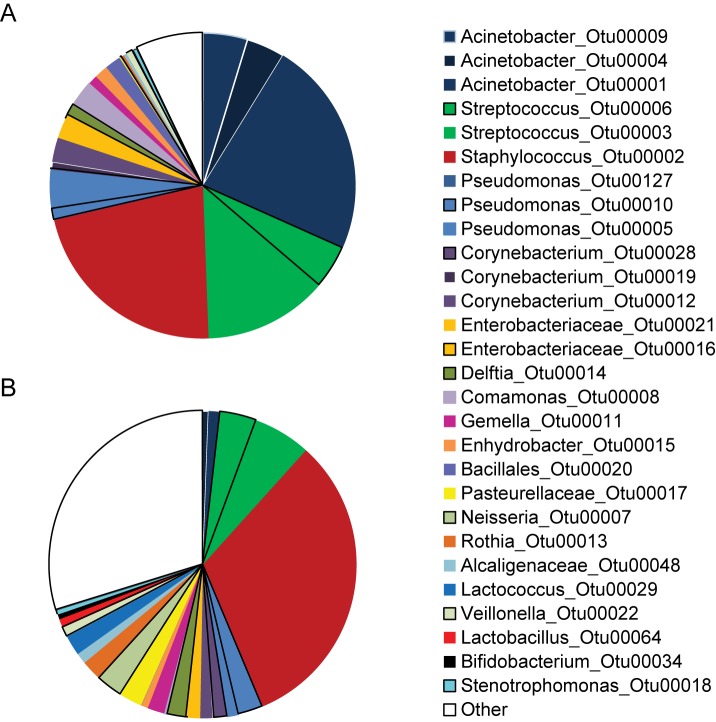
Staphylococci and streptococci were equally abundant in both sets of samples, on average 42% and 40% of total microbiota in samples collected using the aseptic and the standard protocol respectively (Fig 2). High abundance of Acinetobacter sp. (average 32%) was seen in samples collected with standard protocol, but not in samples collected with strict aseptic protocol (average 1.8%).
Fig 2. Average microbiota profiles.
Microbiota composition in samples collected with standard (A) and aseptic protocol (B). Average abundances of OTUs are shown. Sequences belonging to OTUs which contained smaller number of sequences were pooled and are labeled “Other”.
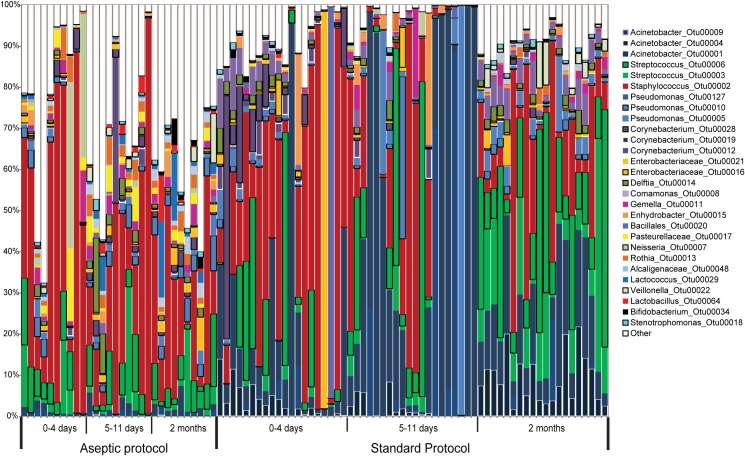
High intra-individual variability in microbiota composition was observed (Fig 3). Low proportion of lactobacilli in the sample set collected with aseptic protocol (0.9%) and standard protocol (0.03%) and bifidobacteria (0.5% and 0.15%, respectively) was consistent with qPCR results (Table 3).
Fig 3. Microbiota profiles of individual subjects.
The composition of breast milk microbiota collected with aseptic and standard protocol at different stages of lactation. The 28 most abundant OTUs are shown. Sequences belonging to OTUs which contained smaller number of sequences were pooled and are labeled “Other”.
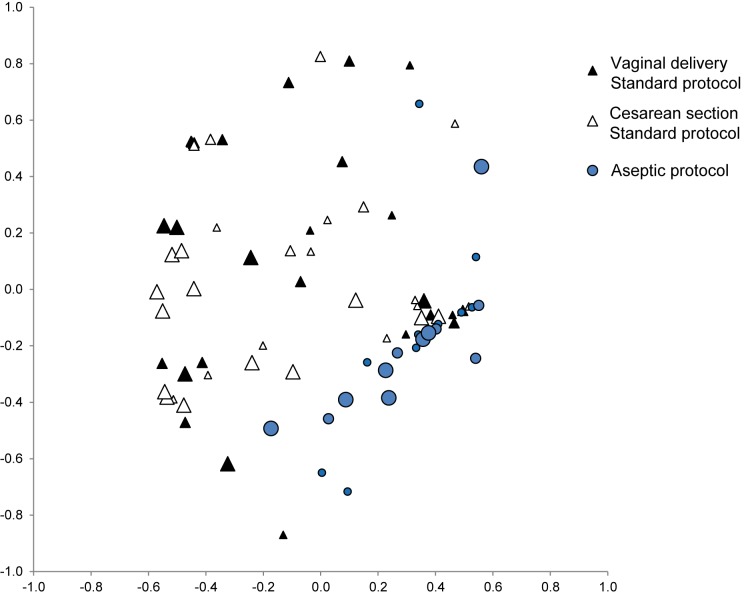
Multivariate NMDS analysis showed significant differences between the samples collected with standard and aseptic protocols (AMOVA, p<0.001), but no differences due to the stage of lactation or the delivery mode (AMOVA, ns), (Fig 4).
Fig 4. Multivariate analysis.
Non-metric multidimensional scaling (NMDS) ordination plot, showing axis 1 and 2. The input data for ordination plots were Yue & Clayton measure of dissimilarity, based on microbiota composition at operational taxonomic unit level. The size of the points represents the time of lactation, the smallest points corresponding to days 0–4 days, the medium to days 5–11, and the largest to month 2.
Discussion
Microbiota profiles
To our knowledge, this is the first report on microbiota composition in a large cohort of Chinese mothers. The microbiota of breast milk from Chinese mothers was dominated by streptococci, staphylococcci and, in the case of standard collection protocol, Acinetobacter sp. The composition of microbiota in breast milk reported previously varied considerably, but the high abundance of staphylococcci and streptococci was perhaps the most consistent feature in the majority of studies which used microbiota profiling (Table 4).
Table 4. Studies with characterization of bacteria present in breast milk by microbiota profiling.
| Predominant taxa | Characterization method | Abundance of bifidobacteria and lactobacilli | N | Population | Collection protocol | Reference |
|---|---|---|---|---|---|---|
| Staphylococcus sp., Streptococcus sp. | Pyrosequencing | “very few sequences” | 16 | US | Cleansing with iodine | [18] |
| Entire breast | ||||||
| Leuconostoc sp. Weisella sp. | Pyrosequencing | No Bifidobacterium or Lactobacillus reported | 18 | Finnish | Cleansing with iodine | [15] |
| Rejection of first drops | ||||||
| Staphylococcus sp., Pseudomonas sp., Streptococcus sp., | Pyrosequencing | Bifidobacterium 1.3% Lactobacillus absent | 7 | Swiss | Cleansing with aseptic soap | [8] |
| Rejection of foremilk | ||||||
| Staphylococcaceae, Streptococcaceae, Pseudomonadaceae | Pyrosequencing | No Bifidobacterium or Lactobacillus reported | 10 | Spanish | Cleansing with chlorhexidine | [36] |
| Rejection of first drops | ||||||
| Acinetobacter sp., Staphylococcus sp. | Ion Torrent | Bifidobacterium 2% Lactobacillus 1.6% | 8 | US | Cleansing with saline | [19] |
| First 10 to 15 ml taken | ||||||
| Staphylococcus sp., Unclassified genus of Enterobacteriaceae, Pseudomonas sp., Streptococcus sp., | Illumina | Bifidobacterium 0.8% Lactobacillus 3% | 39 | USA Caucasian | Cleansing with saline | [37] |
| First 5 to 15 ml taken | ||||||
| Staphylococcus sp., Pseudomonas sp., | Metagenomics | No Bifidobacterium Lactobacillus 0.2% | 10 | US | None | [21] |
| Staphylococcus sp., Pseudomonas sp., | Metagenomics | Bifidobacterium 0.1% Lactobacillus 0.6% | 10 | Spanish | Cleansing with chlorhexidine | [20] |
| Rejection of first drops |
The predominance of staphylococci and in most studies also streptococci was universally observed in early studies based on culture methods [1–5]. These early studies treated the bacterial presence as contamination so naturally focused on the detection of potential pathogens. However, the dominance of staphylococci and streptococci was confirmed more recently when a more diverse range of culture media to cultivate bacteria was used [6, 8, 9].
Our study is consistent with this pattern, suggesting that the microbiota of breast milk from Chinese lactating mothers is similar to that seen at other geographic locations. However, further studies in this and other Asian populations are needed before more definitive conclusions can be drawn.
Only a few studies have investigated the microbiota of the entire breast milk collected without aseptic cleansing, which we propose to call “breastfeeding-associated microbiota” [19]. We considered that breastfeeding-associated microbiota is relevant, as it represents the bacteria ingested by the suckling infant. It is of note that in a study where high abundance of Acinetobacter sp. was reported by [19], milk collection was done without aseptic cleansing of the breast and rejection of foremilk, similarly to the approach we used in the present study (Table 4). Although in our study the experimental methods, such as the extraction protocol, differed between the two sets of samples, we find it unlikely that this was a reason for the predominance of Acinetobacter sp. in samples collected with standard protocol. It remains to be determined whether the high abundance of Acinetobacter sp. is a specific feature of breastfeeding-associated microbiota.
While PCR amplification of samples proved to be difficult, most likely due to low numbers of bacteria, we feel confident that our results reflect the actual composition of microbiota. Indeed we obtained relatively similar results when considering exclusively the samples with good amplification, as indicated by lack of clustering seen in multivariate NMDS analysis. The bacteria were more abundant in the milk collected with standard protocol without rejection of the first drops, which is in agreement with an early study based on bacterial culture [4] where the median number of bacteria decreased ten times, from approximately 103 to 102 CFU per ml, when the first milliliters of breast milk were rejected. In other studies the reported total quantity of live bacteria in breast milk varied, but most studies reported median numbers of 102 to 103, and a range from 101 to 107 CFU per ml of breast milk [1–3, 5–8, 32–35]. One early study suggested that using breast milk pump leads to increased bacterial counts as compared to manual milk expression [1].
In most of the studies, these numbers represent bacteria present inside the breast because the samples were taken with aseptic precautions and elimination of the first few microliters to milliliters of milk. As mentioned above, the numbers present in the entire breast milk appeared higher, but only one early study examined this question directly [4].
Of note, a recent study examined the bacteria present in breast milk tissue of non-lactating women [19]. Low (up to 103CFU/g tissue) numbers of skin commensals were found, suggesting that the presence of live bacteria inside the breast is not limited to the period of breastfeeding.
Abundance of lactobacilli and bifidobacteria
The abundances of bifidobacteria and lactobacilli in microbiota profiles found in our study are in agreement with previously published data (Table 4). Previous studies of breast milk using qPCR detection techniques yielded considerably variable results (summarized in Table 5). In particular, a previous study [15] reported high abundance of bifidobacteria; in addition, bifidobacteria constituted a considerable proportion of the entire bacterial population (Tables 4 and 5). However, the high abundance of bifidobacteria and lactobacilli measured by qPCR was not seen in microbiota profiles [15]. Studies which employed culture techniques rarely reported quantitative results and if reported, low abundance of bifidobacteria and lactobacilli were seen: [8]—1.7% of bifidobacteria and 2.1% lactobacilli; [6]—no culture of bifidobacteria performed and 1.8% of lactobacilli; [7]—0.2% of bifidobacteria and 0.4% of lactobacilli. One study where no sterile cleansing of the breast was performed [9], found higher proportions of lactobacilli and bifidobacteria (5% each of all isolates). A recent study using culture methods [35] found that 40% of samples positive for lactobacilli and 11% were positive for bifidobacteria while the average counts of positive samples were 10 cells per ml for both taxa.
Table 5. Studies with characterization of bacteria present in breast milk by quantitative PCR.
| Abundance of Lactobacilli | % bacterial population1 | Abundance of Bifidobacteria | % bacterial population1 | N | Population | Collection protocol | Reference |
|---|---|---|---|---|---|---|---|
| Not tested | Not tested | 3.9E+2 | Not tested | 20 | Finnish | Not specified | [13] |
| Not tested | Not tested | 1.4E+3 | Not tested | 61 | Finnish | Samples collected after infant has suckled on the breast | [12] |
| Not tested | Not tested | 3.4E+3 | 2.8% | 23 | Spanish | Cleansing with chlorhexidine | [14] |
| Rejection of first drops | |||||||
| 3.7E+3 | <0.01% | 3.6E+3 | <0.01% | 50 | Spanish | Cleansing with chlorhexidine | [11] |
| Rejection of first drops | |||||||
| 9.6E+5 | 197% | 3.3E+5 | 68% | 56 | Finnish | Cleansing with iodine | [15]* |
| Rejection of first drops | |||||||
| 2.7E+4 | 72% | 2.7E+4 | 54% | 18 | Finnish | Cleansing with iodine | [38]* |
| Rejection of first drops | |||||||
| 2.1E+4 | 40% | 1.7+2 | 0.3% | 32 | Spanish | Cleansing with chlorhexidine | [17] |
| 1.8E+4 | 44% | 1.6+2 | 0.4% | 10 | Spanish | Cleansing with chlorhexidine | [36] |
| Rejection of first drops |
1 Based on division of the counts of bifidobacteria and lactobacilli by total bacterial counts
*Different subsets of the same study, NCT00167700
Methodological and geographical differences may explain some of this variability. The low number of bacteria found in breastmilk samples very likely contributes to the variability. The qPCR methods used were initially developed for stool samples, where bacterial loads are five to seven orders of magnitude higher than in breast milk. In the culture based studies, lack of standardization of the details of sample handling such as time to plating and oxygen exposure likely contributed to the variability of results.
The impact of the stage of lactation and the delivery mode
We found no effect of the delivery mode or the stage of lactation on microbiota composition, although this last conclusion is limited to the first 2 months of lactation. Three previous studies evaluated the impact of the stage of lactation on the bacterial counts of total bacteria, lactobacilli or bifidobacteria by qPCR [15, 17, 38] and one of them [17] found that the counts of bifidobacteria increased with the time of lactation, while the remaining studies found no differences. A recent study [34] using culture methods also showed the increase in the counts of bifidobacteria with the time of lactation. The impact of the stage of lactation was also investigated by microbiota profiling [15, 17, 38], but statistical tests to investigate the differences were not reported.
Concerning the impact of the delivery mode, one study [17] found slightly higher counts of total bacteria in colostrum and transition milk of women who delivered by caesarean section, compared to those who delivered vaginally, while this difference was not seen in mature milk. The counts of lactobacilli and bifidobacteria did not differ between the three stages of lactation. It was reported [15] that, apart from slightly higher counts of bifidobacteria in colostrum of women who delivered vaginally (p = 0.05) compared to those who delivered by caesarean section, no differences were seen in the bacterial counts of total bacteria and lactobacilli at three stages of lactation, and in bifidobacteria counts in transition and mature milk. [36] found no differences in the counts of total bacteria, lactobacilli or bifidobacteria between the two delivery modes. Microbiota profiles were described in [15, 36], but no statistical tests to assess the differences between the delivery modes were presented. The differences between the delivery modes were again assessed in a recent study [37] and no significant differences in microbiota composition were found.
Overall, no consistent patterns were found regarding the impact of the delivery mode and the time of lactation on the microbiota of breast milk.
In conclusion our study further substantiates the presence of bacteria in human milk, with a significantly higher number of bacteria identified in the “breastfeeding-associated microbiota” compared to milk obtained under aseptic conditions. We also confirmed the presence of the dominant species such as streptococci, staphylococci, as well as the low abundance of bifidobacteria and lactobacilli.
Supporting Information
(PDF)
Non-metric multidimensional scaling (NMDS) ordination plot, showing axis 1 and 2. The input data for ordination plots were Yue & Clayton measure of dissimilarity, based on microbiota composition at operational taxonomic unit level. The difference on overall microbiota composition between samples that gave good PCR product (PCR pos), and those that gave weak PCR product (PCR neg) was not significant (AMOVA).
(PDF)
Acknowledgments
The authors would like to thank the participants who volunteered for this study, Lawrence Li for project support and guidance, Celia Ning for project management, Qiaoji Li for clinical project management, Yingdong Zheng for sample study calculation and Emilie Ba for data management. Special acknowledgment to Ai Zhao and team at Peking University School of Public Health for recruitment and data collection. Many thanks go to Annick Mercenier for helpful comments on the manuscript and to Francis Foata for deposition of sequencing data in GenBank Sequence Read Archive.
Data Availability
Fastq files containing sequencing data are deposited in the GenBank Short Read Archive (accession number SRP077712).
Funding Statement
OS, GVP, MD, SC, and ST are employees of Nestec Ltd. PD and DM are employees of Nestlé Institute of Health Sciences SA. Nestlé Research Center and Nestlé Nutrition Institute China sponsored the study. We would like to confirm that the funders of the study, Nestlé Research Center and Nestlé Nutrition Institute China, provided support in the form of salaries for authors (OS, GVP, MD, SC, and ST are employees of Nestec Ltd. and PD and DM are employees of Nestlé Institute of Health Sciences SA), but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Liebhaber M, Lewiston NJ, Asquith MT, Sunshine P. Comparison of bacterialcontamination with two methods of human milk collection. The Journal of Pediatrics. 1978;92(2):236–7. 10.1016/S0022-3476(78)80014-6 [DOI] [PubMed] [Google Scholar]
- 2.Eidelman AI, Szilagyi G. Patterns of bacterial colonization of human milk. Obstet Gynecol. 1979;53(5):550–2. [PubMed] [Google Scholar]
- 3.Carroll L, Osman M, Davies DP, McNeish AS. Bacteriology of raw breast milk. The Lancet. 1979;314(8153):1186 10.1016/S0140-6736(79)92409-7 [DOI] [PubMed] [Google Scholar]
- 4.West PA, Hewitt JH, Murphy OM. The Influence of Methods of Collection and Storage on the Bacteriology of Human Milk. J Appl Bacteriol. 1979;46:269–11. [DOI] [PubMed] [Google Scholar]
- 5.El-Mohandes AE, Schatz V, Keiser JF, Jackson BJ. Bacterial contaminants of collected and frozen human milk used in an intensive care nursery. AJIC: American Journal of Infection Control. 1993;21(5):226–30. [DOI] [PubMed] [Google Scholar]
- 6.Heikkilä MP, Saris PEJ. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003;95(3):471–8. [DOI] [PubMed] [Google Scholar]
- 7.Jiménez E, Delgado S, Fernández L, García N, Albújar M, Gómez A, et al. Assessment of the bacterial diversity of human colostrum and screening of staphylococcal and enterococcal populations for potential virulence factors. Res Microbiol. 2008;159(9–10):595–601. 10.1016/j.resmic.2008.09.001 [DOI] [PubMed] [Google Scholar]
- 8.Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. 2013;110(7):1253–62. 10.1017/S0007114513000597 [DOI] [PubMed] [Google Scholar]
- 9.Solís G, de los Reyes-Gavilan CG, Fernández N, Margolles A, Gueimonde M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. 2010;16(3):307–10. 10.1016/j.anaerobe.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 10.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collado MC, Delgado S, Maldonado A, Rodríguez JM. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett Appl Microbiol. 2009;48(5):523–8. 10.1111/j.1472-765X.2009.02567.x [DOI] [PubMed] [Google Scholar]
- 12.Grönlund MM, Gueimonde M, Laitinen K, Kociubinski G, Grönroos T, Salminen S, et al. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin Exp Allergy. 2007;37(12):1764–72. [DOI] [PubMed] [Google Scholar]
- 13.Gueimonde M, Laitinen K, Salminen S, Isolauri E. Breast milk: A source of bifidobacteria for infant gut development and maturation? Neonatology. 2007;92(1):64–6. [DOI] [PubMed] [Google Scholar]
- 14.Martín R, Jiménez E, Heilig H, Fernández L, Marín ML, Zoetendal EG, et al. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol. 2009;75(4):965–9. 10.1128/AEM.02063-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96(3):544–51. 10.3945/ajcn.112.037382 [DOI] [PubMed] [Google Scholar]
- 16.Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC. Impact of mode of delivery on the milk microbiota composition of healthy women. Journal of Developmental Origins of Health and Disease. 2015. 10.1017/S2040174415001397 [DOI] [PubMed] [Google Scholar]
- 17.Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martínez-Costa C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. 2014;34(8):599–605. 10.1038/jp.2014.47 [DOI] [PubMed] [Google Scholar]
- 18.Hunt KM, Preuss J, Nissan C, Davlin CA, Williams JE, Shafii B, et al. Human milk oligosaccharides promote the growth of staphylococci. Appl Environ Microbiol. 2012;78(14):4763–70. 10.1128/AEM.00477-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urbaniak C, McMillan A, Angelini M, Gloor GB, Sumarah M, Burton JP, et al. Effect of chemotherapy on the microbiota and metabolome of human milk, a case report. Microbiome. 2014;2(1):24 10.1186/2049-2618-2-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiménez E, De Andrés J, Manrique M, Pareja-Tobes P, Tobes R, Martínez-Blanch JF, et al. Metagenomic analysis of milk of healthy and mastitis-suffering women. J Hum Lact. 2015;31(3):406–15. 10.1177/0890334415585078 [DOI] [PubMed] [Google Scholar]
- 21.Ward TL, Hosid S, Ioshikhes I, Altosaar I. Human milk metagenome: A functional capacity analysis. BMC Microbiology. 2013;13(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Austin S, De Castro C, Bénet T, Hou Y, Sun H, Thakkar S, et al. Temporal Change of the Content of 10 Oligosaccharides in the Milk of Chinese Urban Mothers. Nutrients. 2016;8(6):346 10.3390/nu8060346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148(1):257–66. [DOI] [PubMed] [Google Scholar]
- 24.Matsuki T, Watanabe K, Fujimoto J, Miyamoto Y, Takada T, Matsumoto K, et al. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl Environ Microbiol. 2002;68(11):5445–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–21. [DOI] [PubMed] [Google Scholar]
- 26.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME Journal. 2012;6(8):1621–4. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–41. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozich J, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–20. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environmental Microbiology. 2010;12(7):1889–98. 10.1111/j.1462-2920.2010.02193.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biesbroek G, Sanders EAM, Roeselers G, Wang X, Caspers MPM, Trzciński K, et al. Deep Sequencing Analyses of Low Density Microbial Communities: Working at the Boundary of Accurate Microbiota Detection. PLoS ONE. 2012;7(3):e32942 10.1371/journal.pone.0032942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albesharat R, Ehrmann MA, Korakli M, Yazaji S, Vogel RF. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst Appl Microbiol. 2011;34(2):148–55. 10.1016/j.syapm.2010.12.001 [DOI] [PubMed] [Google Scholar]
- 33.Martín V, Maldonado-Barragán A, Moles L, Rodriguez-Baños M, Campo RD, Fernández L, et al. Sharing of bacterial strains between breast milk and infant feces. J Hum Lact. 2012;28(1):36–44. 10.1177/0890334411424729 [DOI] [PubMed] [Google Scholar]
- 34.Makino H, Martin R, Ishikawa E, Gawad A, Kubota H, Sakai T, et al. Multilocus sequence typing of bifidobacterial strains from infant's faeces and human milk: are bifidobacteria being sustainably shared during breastfeeding? Benef Microbes. 2015;6(4):563–72. 10.3920/BM2014.0082 . [DOI] [PubMed] [Google Scholar]
- 35.Soto A, Martín V, Jiménez E, Mader I, Rodríguez JM, Fernández L. Lactobacilli and bifidobacteria in human breast milk: Influence of antibiotherapy and other host and clinical factors. J Pediatr Gastroenterol Nutr. 2014;59(1):78–88. 10.1097/MPG.0000000000000347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC. Impact of mode of delivery on the milk microbiota composition of healthy women. Journal of Developmental Origins of Health and Disease. 2016;7(01):54–60. 10.1017/S2040174415001397 [DOI] [PubMed] [Google Scholar]
- 37.Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. 2016;4(1):1–9. 10.1186/s40168-015-0145-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collado MC, Laitinen K, Salminen S, Isolauri E. Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr Res. 2012;72(1):77–85. 10.1038/pr.2012.42 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Non-metric multidimensional scaling (NMDS) ordination plot, showing axis 1 and 2. The input data for ordination plots were Yue & Clayton measure of dissimilarity, based on microbiota composition at operational taxonomic unit level. The difference on overall microbiota composition between samples that gave good PCR product (PCR pos), and those that gave weak PCR product (PCR neg) was not significant (AMOVA).
(PDF)
Data Availability Statement
Fastq files containing sequencing data are deposited in the GenBank Short Read Archive (accession number SRP077712).