Abstract
Background
Simazine is a triazine herbicide used worldwide in both agricultural and non-agricultural fields that is frequently detected in surface water and groundwater. Due to its widespread use, an increasing amount of research has focused on the potentially serious environmental and health risks.
Material/Methods
We used Western blotting and real-time quantitative PCR to analyze the effects of simazine on dopamine neuronal development-related factors in MN9D dopaminergic cells.
Results
The expression of tyrosine hydroxylase (TH) mRNA was significantly increased after treatment with 300 and 600 μmol L−1 simazine after 24 and 48 h. Levels of nuclear-related receptor 1 (Nurr1) mRNA after 24- and 48-h exposure were decreased with 50 μmol L−1 simazine, but increased with 600 μmol L−1 simazine. Significant increases in TH and Nurr1 protein were observed in all simazine-treated groups at 24 and 48 h. The expression of neurogenin 2 and LIM homeobox transcription factor 1 beta (Lmx1b) mRNA were significantly increased after exposure to 600 μmol L−1 simazine for 48 h, while the expression of wingless-type MMTV integration site family member 1 (Wnt1) mRNA was increased by all doses of simazine.
Conclusions
Simazine may have an impact on TH in MN9D cells through 2 mechanisms; one mechanism is through the Lmx1a/Ngn2 pathway, and the other mechanism is through the Lmx1b-pitx3/Wnt1-Nurr1 pathway. These 2 pathways likely do not operate in isolation, but rather together, during the cellular response to simazine exposure.
MeSH Keywords: Central Nervous System, Dopamine, Simazine
Background
Simazine (6-chloro-N, N’-diethyl-1, 3, 5-triazine-2, 4-diamine) is a member of the triazine family, which also includes other herbicides such as atrazine and propazine. Simazine has been used worldwide in both agricultural and non-agricultural fields to inhibit photosynthesis in broadleaf weeds, grasses, and algae in farm ponds. Simazine has become a mainstream herbicide because it is highly efficient, has broad-spectrum activity, and is economical. Due to its widespread use, an increasing amount of research has focused on the potentially serious environmental and health risks as a result of simazine toxicity.
Exposure routes for simazine include: ingestion through polluted drinking water, absorption through skin contact, and inhalation from during occupational exposure [1]. Due to its slight solubility in water, and relative non-volatility, simazine can partition into the organic phase where it is easily degraded through photolysis. Simazine is also one of the most frequently detected pesticides in surface water and groundwater as a result of herbicide runoff [2]. Available data have demonstrated that atrazine, a close homologue of simazine, results in toxicological responses that include effects on antioxidant mechanisms, behavior, mammary gland development, and neuroendocrine systems [3–8]. In addition, simazine has demonstrated toxicity in reproductive and immune systems and has also been reported to have ecological impacts on the environment [9–13]. However, data on the neurotoxicity of simazine are sparse. In a previous study, simazine suppressed PC12 cell growth via regulation of the tyrosine hydroxylase (TH)-mediated synthesis of dopamine (DA) and the release of noradrenaline (NE) [14].
In the present study, the effects of 24- or 48-h simazine exposure on the changes in developmental and differentiation transcription factors were evaluated in MN9D cells at the mRNA and protein level as markers of potential neurotoxicity.
Material and Methods
Chemicals and cells culture
Simazine (98% pure) was a gift from ZhongShan Chemical Co., Ltd. (Zhejiang, China). Other chemicals were purchased from Amersco (USA) unless otherwise indicated.
Simazine was finely ground to smaller particles in an agate mortar and then dissolved in DMSO (0.5%) as the stock solution. The stock solution was diluted with culture medium to the required concentration according to the experiments.
The MN9D cell line (number: 0514A077) was purchased from ShangHai FoleiFort Biological Technology Development Co., Ltd. The cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco BRL), supplemented with 10% (v/v) heat-inactivated fetal calf serum (Hyclone), 100 units mL−1 penicillin, and 100 units mL−1 streptomycin, in an incubator with 5% CO2 at 37°C. Cells were cultured in 10-cm dishes, harvested at 90–95% confluence, and RNA or protein was isolated for further analysis.
Simazine treatment
The MN9D cells were collected in the logarithmic phase, cell concentration was adjusted to approximately 1×104 cells mL−1, and cells were plated in 96-well plates with 90 μl of DMEM per well. The experimental groups included 0.5% DMSO (solvent control group) and simazine at concentrations of 0 (negative control group), 50, 300, and 600 μmol L−1. During simazine treatment, cell viability was determined every 12 h for 72 h using a colorimetric MTT reduction assay [15,16]. The optical density of the dissolved grains was assessed at 490 nm (TECAN-sunrise). Cell survival in each group is expressed as a percentage rate. The changes in cell morphology were observed under an inverted microscope every 24 h for 72 h.
Total RNA extraction and real-time quantitative PCR
Total RNA from MN9D cells was isolated using Trizol reagent following the manufacturer’s instructions. The concentration of RNA was determined with an ND-2000C spectrophotometer (NanoDrop, Thermo Scientific, USA). Synthesis of cDNA was performed with 1 μg of total RNA using a PrimeScript® RT with gDNA eraser reagent kit (TaKaRa Biotechnology Co., Ltd., Dalian, China) following the manufacturer’s instructions. The PCR primers were designed and synthesized by Generay Biotechnology Co., Ltd. (Shanghai, China) (Table 1).
Table 1.
Sequences of primers used for PCR amplification.
| Target | Primer | Primer sequence | Size (bp) | Accession number (Genbank) |
|---|---|---|---|---|
| TH | Forward | 5′ ccg cac att tgc cca gtt c 3′ | 148 | NM_009377.1 |
| Reverse | 5′ tgc acc gta agc ctt cag ctc 3′ | |||
|
| ||||
| Nurr1 | Forward | 5′ gac ggg ctg gat tcc caa tag 3′ | 157 | NM_001139509.1 |
| Reverse | 5′ cct tga ggc gag gac cca ta 3′ | |||
|
| ||||
| Lmx1b | Forward | 5′ gga agg tcc gag aga cat tgg 3′ | 125 | NM_010725.2 |
| Reverse | 5′ ctg ctc ctg ctg ttg ctg gtg t 3′ | |||
|
| ||||
| Lmx1a | Forward | 5′ cag tgt gcc tcc tgc aaa gag 3′ | 137 | NM_033652.5 |
| Reverse | 5′ aac tca ttg ggc gca atg g 3′ | |||
|
| ||||
| Wnt1 | Forward | 5′ gct gtg cga gag tgc aaa tgg 3′ | 133 | NM_021279.4 |
| Reverse | 5′ cgg agg tga ttg cga aga tg 3′ | |||
|
| ||||
| Ngn2 | Forward | 5′ aag aag acc cgc agg ctc aag 3′ | 158 | NM_009718.2 |
| Reverse | 5′ cag atg taa ttg tgg gcg aag c 3′ | |||
|
| ||||
| Pitx3 | Forward | 5′ tgt cgt tat cgg acg cag g 3′ | 139 | NM_008852.4 |
| Reverse | 5′ tgc ttc ttc ttc aga gag ccg tc 3′ | |||
|
| ||||
| β-actin | Forward | 5′ tga gag gga aat cgt gcg aga c 3′ | 149 | NM_007393.3 |
| Reverse | 5′ gct cgt tgc caa tag tga tga cc 3′ | |||
TH – tyrosine hydroxylase; Nurr1 – nuclear receptor related-1 protein; Lmx1a/Lmx1b – LIM homeobox transcription factor 1 alpha/1 beta; Wnt1 – Wingless type MMTV integration site family member 1; Ngn2 – neurogenin 2; Pitx3 – paired-like homeodomain transcription factor 3.
The cDNA was amplified using an ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). A relative quantitative real-time PCR was chosen using the SYBR Green method (SYBR® Premix Ex TaqTM II, TaKaRa Biotechnology Co., Ltd., Dalian, China). The cycling conditions included denaturation at 95°C for 5 s, 40 cycles of annealing at 58°C for 34 s, and 30s of extension at 72°C for 30 s. The cycle at which sample fluorescence reached threshold was defined as the threshold cycle (Ct). The results were are expressed as the relative expression ratio calculated based upon the real-time PCR efficiency (E) and ΔCt. The ΔCt for each gene (target or reference) was calculated by subtracting the Ct number of the target sample from that of the control sample. As shown in Equation 1, the ratio of target gene expression in treatment versus control can be derived from the ratio between target gene efficiency (E target) to the power of target ΔCt (ΔCt target) and reference gene efficiency (E reference) to the power of reference ΔCt (ΔCt reference) (Equation 1) [17].
| 1 |
Western blot analysis
Protein was extracted from MN9D cells using RIPA lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China). Cells were lysed on ice for 2 h in lysis buffer and then centrifuged at 12, 000 rpm for 10 min at 4°C. The supernatant was collected and a BCA protein assay kit (Beyotime Institute of Biotechnology) was used to determine the protein concentration. Equal amounts of total protein (80 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 1% bovine serum albumin (BSA) for 1 h at room temperature. Membranes were stripped and incubated overnight with rabbit polyclonal antibodies against TH (Abcam, ab41528) and nuclear-related receptor 1 (Nurr1; Abcam, ab93332) at a 1:500 dilution in blocking buffer (ZSGB Biotechnology, Peking, China) at 4°C. After washing 3 times with Tris-buffered saline containing 0.1% Tween-20 (TBS-T) at room temperature, the membranes were incubated at room temperature for 1 h with alkaline phosphatase goat anti-rabbit IgG secondary antibody at a 1:1000 dilution in blocking buffer. Membranes were washed with TBS 3 times at room temperature and then incubated with Western blue stabilized substrate for alkaline phosphatase (Promega, USA) for 5 min. Relative expression (%) was calculated based upon protein density using the Chemi Analysis System (Bioshine, Shanghai, China). Sample blot density was normalized to β-actin (Immunoway Biotechnology, USA) at 1:500 dilution in blocking buffer.
Statistical analysis
Data analysis was performed using SAS version 9.1.3 (Cary, NC, USA). Factorial design analysis of variance was performed for all data using SAS software. The SNK-q test was used to compare between the control and test groups. Differences were considered significant when P values less than 0.01 were obtained.
Results
Effects of simazine on the viability of MN9D cells
The viability of MN9D cells after simazine treatment for 60 and 72 h was significantly decreased when compared to the negative control group (P<0.01). When compared to the solvent (0.5% DMSO) control group, cell viability was also significantly decreased in a dose-dependent manner (P<0.01) (Figure 1).
Figure 1.
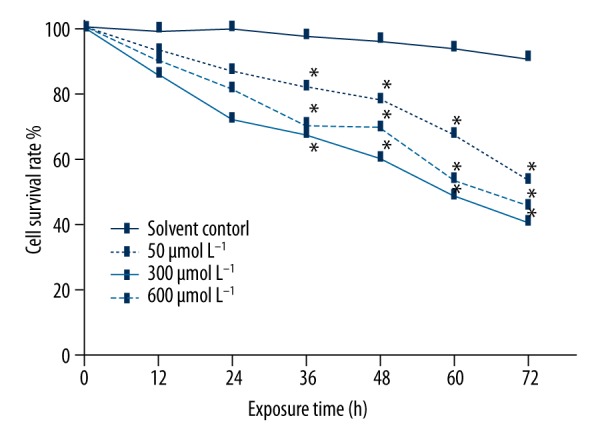
Effects of simazine on the viability in MN9D cells. Cell viability was significantly reduced; * P<0.01, compared to the solvent control group.
Expression of TH, Nurr1, Ngn2, Lmx1a, Lmx1b, Wnt-1, and Pitx3 genes in MN9D cells treated with simazine
There were no significant differences between the solvent control group and the negative control group in TH, Nurr1, Ngn2, Lmx1a, or Lmx1b mRNA expression after 24 or 48 h. Simazine treatment significantly increased the expression of TH, Nurr1, Ngn2, Lmx1b, and Wnt1 genes in MN9D cells in a dose-dependent manner (P<0.01) (Figures 2, 3).
Figure 2.
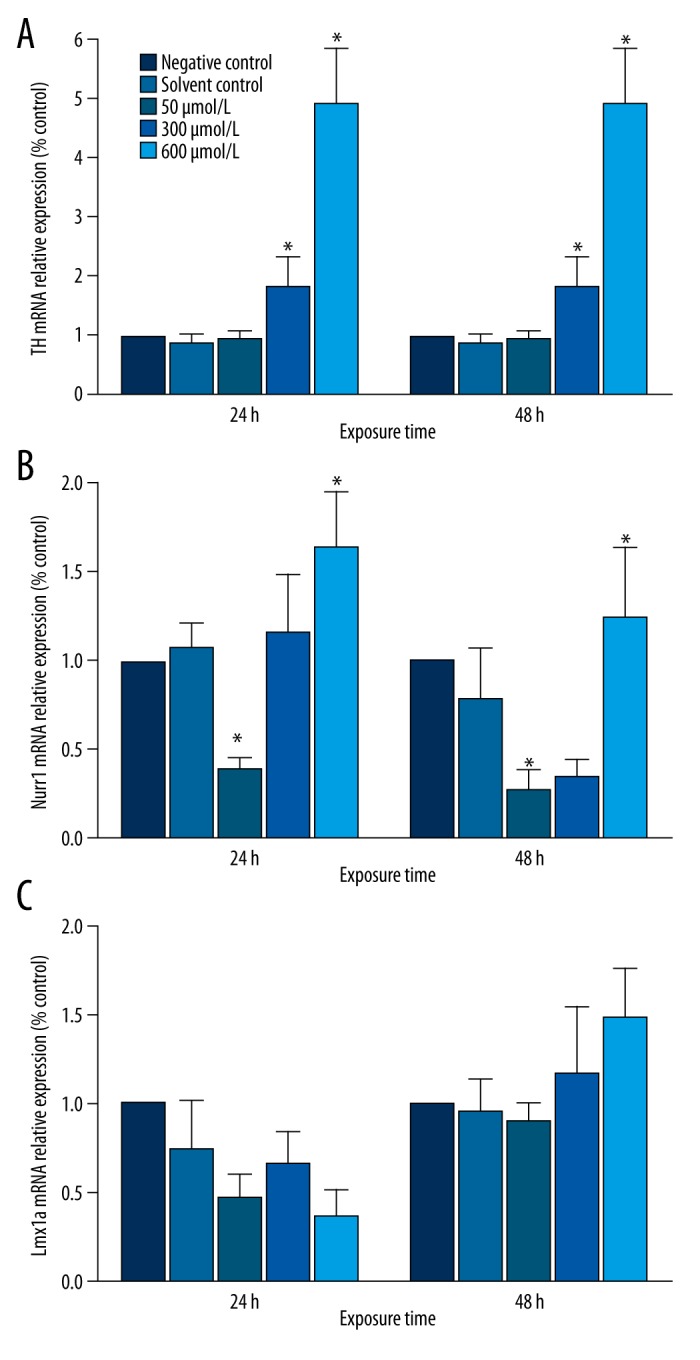
Effects of simazine on the mRNA levels for TH, Nurr1, and Lmx1a in MN9D cells after exposure for 24 or 48 h (mean expression relative to control ±SE; * P<0.01, compared to the solvent control group).
Figure 3.
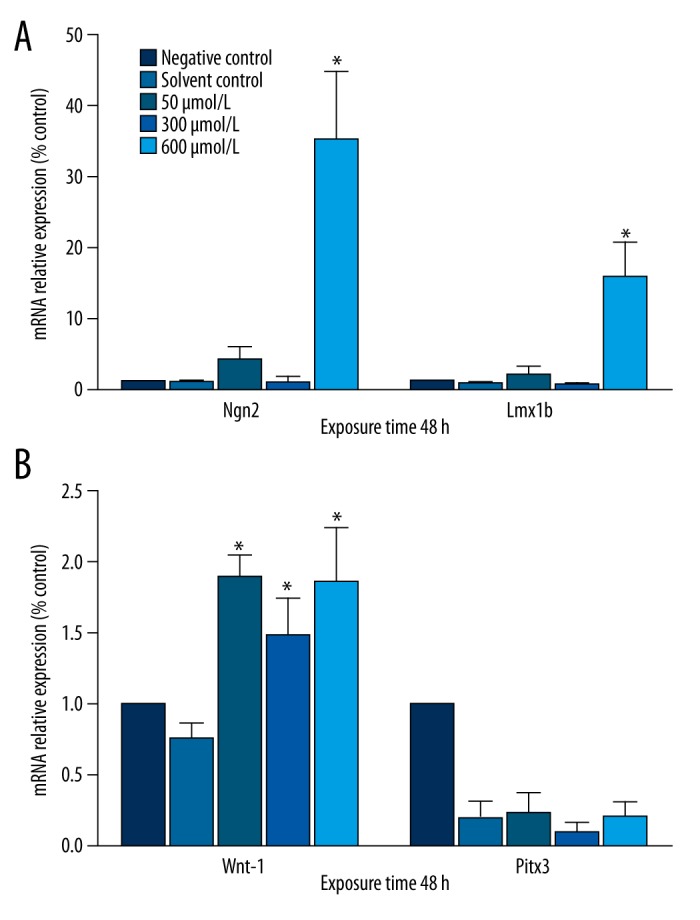
Effects of simazine on the mRNA levels for Wnt1, Pitx3, Ngn2, and Lmx1b in MN9D cells after exposure for 48 h (mean expression relative to control ±SE; * P<0.01, compared to the solvent control group).
Expression of TH and Nurr1 protein in MN9D cells treated with simazine
After 24 and 48 h of exposure, simazine significantly increased the expression of TH and Nurr1 protein in MN9D cells in comparison with the control groups (P<0.01) (Figures 4–6).
Figure 4.
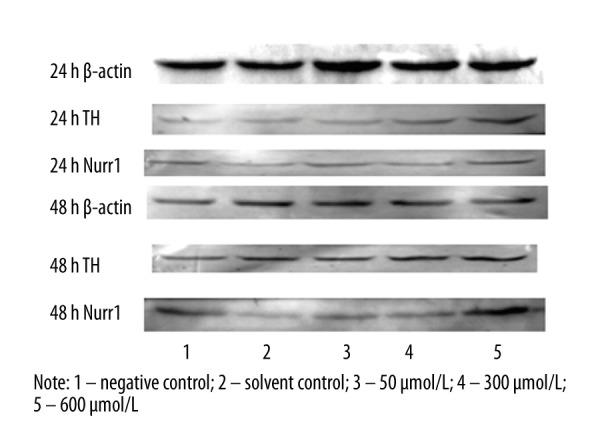
The expression of TH and Nurr1 protein (relative to β-actin) in MN9D cells after exposure for 24 or 48 h.
Figure 5.
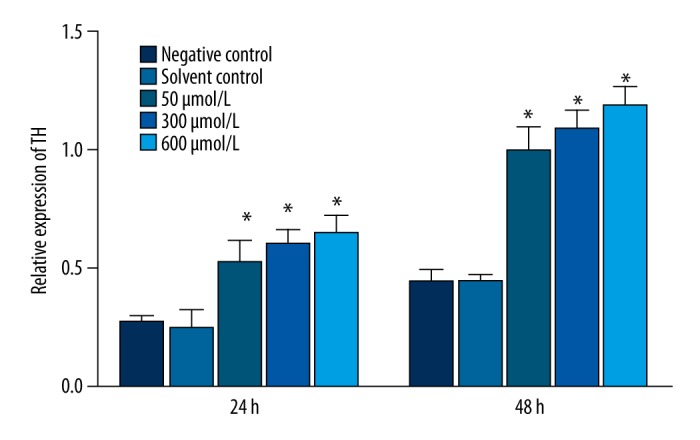
Effect of simazine on the expression of TH protein (relative to β-actin) in MN9D cells after exposure for 24 or 48 h (mean relative expression ±SE; * P<0.01, compared to the solvent control group).
Figure 6.
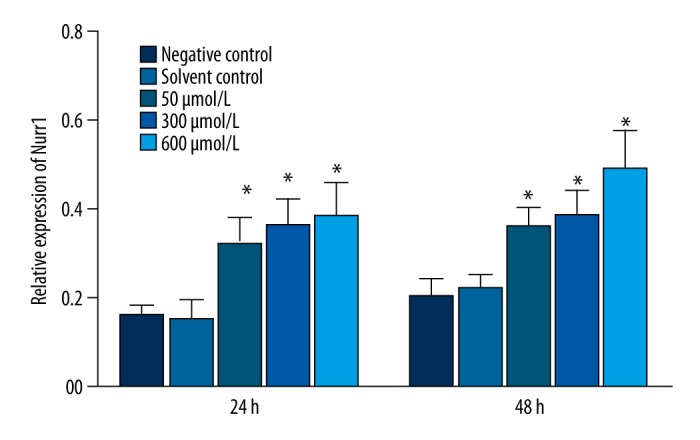
Effects of simazine on the expression of Nurr1 protein (relative to β-actin) in MN9D cells after exposure for 24 or 48 h (mean relative expression ±SE; * P<0.01, compared to the solvent control group).
Discussion
The MN9D cell is a murine mesencephalon-derived dopaminergic neuronal cell line, derived from a fusion of embryonic mesencephalic neurons of mouse origin and N18TG cells, that can synthesize, release, and take up dopamine [18]. The MN9D cell is commonly used as an experimental model of DA neurons because there are a number of important similarities between these two 2 cell types [19–21]. The MN9D cell has also been used to study the mechanisms and potential therapeutics relevant to the loss of DA neurons in Parkinson’s disease [22,23]. After exposure to simazine, MN9D cells displayed irregular cell morphology and an increased number of dead cells. Our findings demonstrate that simazine has effects on MN9D cell growth and proliferation, indicating dopaminergic neurotoxicity or induced dopaminergic neuronal injury.
In the mammalian central nervous system, the dopamine system has 3 main projection pathways: the nigrostriatal pathway, which plays a key role in the control of voluntary movement; the mesolimbic dopamine pathway, which is associated with schizophrenia and drug addiction; and the mesocortical dopamine pathway, which is associated with emotion and motivation [24,25]. Due to the involvement of the dopaminergic neurons of the midbrain in the various advanced features of the brain, such as motor control, emotion, motivation, learning, and memory, an in-depth understanding of the roles of dopaminergic neurons in developmental processes is helpful for the study of the mechanisms of contaminant exposure.
TH is the rate-limiting enzyme in DA synthesis and is a specific marker for DA neurons. Nurr1 protein is a transcription factor, mainly found in the substantia nigra and ventral tegmental area [26], that is also essential for the occurrence, migration, and survival of DA neurons. Deletions of the Nurr1 gene affect the normal development of DA neurons in the developing brain, leading to TH and DA neurotransmitter deficiency. The expression of the TH gene can also be promoted by Nurr1 [27,28]. In our study, a low dose of simazine (50 μmol L−1) inhibited TH expression in MN9D cells after 24- and 48- h exposure. However, in MN9D cells exposed to high doses of simazine (300 or 600 μmol L−1), levels of TH were significantly increased when compared to the control groups. The expression of Nurr1, which is in the same genetic pathway as TH, was significantly reduced in the low-dose simazine exposure, and then significantly increased in the high-dose simazine exposure. The expression of TH and Nurr1 protein was also significantly increased when MN9D cells were treated with all doses of simazine. These findings suggest that the increasing expression of TH in the MN9D cells was due to enhanced expression of Nurr1.
The development of DA neurons is a very complex process, requiring coordination and interaction between external and internal factors. It includes early neural induction, precursor cell fate decisions, differentiated cell migration, and building neural projections [29,30]. During early embryonic development, some neuroepithelial cells in the ventral midbrain are gradually transformed into dopamine progenitor cells under the influence of a number of signaling molecules and transcription factors. Some studies have confirmed that a number of transcription factors play key roles in the decision of the fate of dopamine neurons [31]. Later in development, dopamine progenitor cells differentiate into mature cells with some cells gradually exiting the cell cycle and migrating to their destination. During the process of migration, TH and neurotransmitter expression and synthesis begin. Many transcription factors and cell cycle regulatory proteins are involved in this process [32,33]. These proteins include LIM homeobox transcription factor 1 alpha (Lmx1a), LIM homeobox transcription factor 1 beta (Lmx1b), wingless-type MMTV integration site family member1 (Wnt1), neurogenin2 (Ngn2), and paired-like homeodomain transcription factor3 (Pitx3).
As an important determinant of dopamine neurons, Lmx1a can regulate the expression of Ngn2 to produce mature midbrain dopamine neurons [34,35]. The transcription factor, Ngn2, has important functions in cell fate determination, differentiation, and developmental processes of the nervous system [36]. This transcription factor is necessary for the differentiation and development of dopamine neurons in mice and can promote the formation of immature Nurr1+ dopamine neurons, while also playing a role in their late maturation. In the current study, the levels of Lmx1a were decreased after simazine exposure for 24 h and increased after 48 h, but these changes were not statistically significant when compared with the solvent control group. After a 48-h exposure to a high dose of simazine, the expression of Ngn2 was significantly increased to a level that was 35 times higher than in the solvent control group. The formation of Nurr1+ in immature dopamine neurons is promoted by Ngn2, so we speculate that the expression of Ngn2 may be one of the mechanisms by which levels of TH and Nurr1 were increased following treatment with simazine.
The glycoprotein, Wnt1, can regulate cell proliferation, differentiation, and determine cell fate [37]. The proliferation and differentiation of Nurr1-positive precursor cells in the ventral midbrain from cultured dopamine neural precursor cells is promoted by Wnt1 [38]. Lmx1b is aA homeobox transcription factor playing important roles in the development of limbs, kidneys, and the central nervous system, andLmx1b, is also constitutively expressed in dopamine early progenitor cells and mature dopaminergic neurons. The effects of Lmx1b on dopamine neurons are primarily observed in relatively mature dopaminergic neurons, where it promotes and maintains the survival and proliferation of dopaminergic neurons by regulating the expression of Wnt1 [39,40]. In the current study, the expression of Lmx1b in the high-dose simazine exposure group and the expression of Wnt1 in all of the treatment groups were significantly increased after 48 h. This demonstrates that a certain dose of simazine can upregulate the expression of Nurr1 and further promote the expression of TH through the regulation of Lmx1a, Lmx1b, and Wnt1. However, further study is needed to investigate the neurotoxic effects of simazine on MN9D cells.
Conclusions
This study demonstrates that simazine induces neurotoxicity resulting in dopaminergic damage in MN9D cells. Exposure to 300 and 600 μmol L−1 simazine for 24 and 48 h resulted in the upregulation of the expression of TH in MN9D cells. The effects of simazine on several other factors involved in the process of dopamine precursor cell differentiation into mature dopaminergic neurons were also determined. Simazine may have an impact on TH in MN9D cells through 2 mechanisms: one mechanism is through the Lmx1a/Ngn2 pathway, and the other mechanism is through the Lmx1b-pitx3/Wnt1-Nurr1 pathway. These 2 pathways likely do not operate in isolation, but rather together, during the cellular response to simazine exposure.
Acknowledgements
We are grateful to Jiaren Liu for critically revising the manuscript.
Footnotes
Source of support: This study was supported by the National Natural Sciences Foundation of China (Grant number: 81072332)
Conflict of interest statement
We declare that we have no conflicts of interest.
References
- 1.Riviere JE, Brooks JD. Predicting skin permeability from complex chemical mixtures: Dependency of quantitative structure permeation relationships on biology of skin model used. Toxicol Sci. 2011;119(1):224–32. doi: 10.1093/toxsci/kfq317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salasnich L, Sattin F. Charge-exchange processes between excited helium and fully stripped ions. Phys Rev A. 1995;51(5):4281–83. doi: 10.1103/physreva.51.4281. [DOI] [PubMed] [Google Scholar]
- 3.Belloni V, Dessi-Fulgheri F, Zaccaroni M, et al. Early exposure to low doses of atrazine affects behavior in juvenile and adult CD1 mice. Toxicology. 2011;279(1–3):19–26. doi: 10.1016/j.tox.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Hovey RC, Coder PS, Wolf JC, et al. Quantitative assessment of mammary gland development in female Long Evans rats following in utero exposure to atrazine. Toxicol Sci. 2011;119(2):380–90. doi: 10.1093/toxsci/kfq337. [DOI] [PubMed] [Google Scholar]
- 5.Rohr JR, McCoy KA. A qualitative meta-analysis reveals consistent effects of atrazine on freshwater fish and amphibians. Environ Health Perspect. 2010;118(1):20–32. doi: 10.1289/ehp.0901164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abarikwu SO, Adesiyan AC, Oyeloja TO, et al. Changes in sperm characteristics and induction of oxidative stress in the testis and epididymis of experimental rats by a herbicide, atrazine. Arch Environ Contam Toxicol. 2010;58(3):874–82. doi: 10.1007/s00244-009-9371-2. [DOI] [PubMed] [Google Scholar]
- 7.Foradori CD, Hinds LR, Hanneman WH, Handa RJ. Effects of atrazine and its withdrawal on gonadotropin-releasing hormone neuroendocrine function in the adult female Wistar rat. Biol Reprod. 2009;81(6):1099–105. doi: 10.1095/biolreprod.109.077453. [DOI] [PubMed] [Google Scholar]
- 8.Cooper RL, Stoker TE, Tyrey L, et al. Atrazine disrupts the hypothalamic control of pituitary-ovarian function. Toxicol Sci. 2000;53(2):297–307. doi: 10.1093/toxsci/53.2.297. [DOI] [PubMed] [Google Scholar]
- 9.Park HO, Bae J. Disturbed relaxin signaling pathway and testicular dysfunction in mouse offspring upon maternal exposure to simazine. PloS One. 2012;7(9):e44856. doi: 10.1371/journal.pone.0044856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren R, Sun DJ, Yan H, et al. Oral exposure to the herbicide simazine induces mouse spleen immunotoxicity and immune cell apoptosis. Toxicol Pathol. 2013;41(1):63–72. doi: 10.1177/0192623312452488. [DOI] [PubMed] [Google Scholar]
- 11.Morgante V, Flores C, Fadic X, et al. Influence of microorganisms and leaching on simazine attenuation in an agricultural soil. J Environ Manage. 2012;95(Suppl):S300–5. doi: 10.1016/j.jenvman.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 12.Wilson PC, Wilson SB. Toxicity of the herbicides bromacil and simazine to the aquatic macrophyte, Vallisneria americana Michx. Environ Toxicol Chem. 2010;29(1):201–11. doi: 10.1002/etc.22. [DOI] [PubMed] [Google Scholar]
- 13.Gunasekara AS, Troiano J, Goh KS, Tjeerdema RS. Chemistry and fate of simazine. Rev Environ Contam Tox. 2007;189:1–23. doi: 10.1007/978-0-387-35368-5_1. [DOI] [PubMed] [Google Scholar]
- 14.Das PC, McElroy WK, Cooper RL. Differential modulation of catecholamines by chlorotriazine herbicides in pheochromocytoma (PC12) cells in vitro. Toxicol Sci. 2000;56(2):324–31. doi: 10.1093/toxsci/56.2.324. [DOI] [PubMed] [Google Scholar]
- 15.Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119(2):203–10. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 16.Oh YJ, Uhland-Smith A, Kim JE, O’Malley KL. Regions outside of the Bcl-2 homology domains, BH1 and BH2 protect a dopaminergic neuronal cell line from staurosporine-induced cell death. Brain Res Mol Brain Res. 1997;51(1–2):133–42. doi: 10.1016/s0169-328x(97)00229-5. [DOI] [PubMed] [Google Scholar]
- 17.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Chen HZ, Chen FF, et al. Global microRNA expression profiling reveals differential expression of target genes in 6-hydroxydopamine-injured MN9D cells. Neuromolecular Med. 2013;15(3):593–604. doi: 10.1007/s12017-013-8244-z. [DOI] [PubMed] [Google Scholar]
- 19.Choi HK, Won LA, Kontur PJ, et al. Immortalization of embryonic mesencephalic dopaminergic neurons by somatic cell fusion. Brain Res. 1991;552(1):67–76. doi: 10.1016/0006-8993(91)90661-e. [DOI] [PubMed] [Google Scholar]
- 20.Rick CE, Ebert A, Virag T, et al. Differentiated dopaminergic MN9D cells only partially recapitulate the electrophysiological properties of midbrain dopaminergic neurons. Dev Neurosci. 2006;28(6):528–37. doi: 10.1159/000095115. [DOI] [PubMed] [Google Scholar]
- 21.Choi HK, Won L, Roback JD, et al. Specific modulation of dopamine expression in neuronal hybrid cells by primary cells from different brain regions. Proc Natl Acad Sci USA. 1992;89(19):8943–47. doi: 10.1073/pnas.89.19.8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen CX, Huang SY, Zhang L, Liu YJ. Synaptophysin enhances the neuroprotection of VMAT2 in MPP+-induced toxicity in MN9D cells. Neurobiol Dis. 2005;19(3):419–26. doi: 10.1016/j.nbd.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Holtz WA, Turetzky JM, O’Malley KL. Microarray expression profiling identifies early signaling transcripts associated with 6-OHDA-induced dopaminergic cell death. Antioxid Redox Signal. 2005;7(5–6):639–48. doi: 10.1089/ars.2005.7.639. [DOI] [PubMed] [Google Scholar]
- 24.Bjorklund A, Dunnett SB. Dopamine neuron systems in the brain: An update. Trends Neurosci. 2007;30(5):194–202. doi: 10.1016/j.tins.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Van den Heuvel DM, Pasterkamp RJ. Getting connected in the dopamine system. Prog Neurobiol. 2008;85(1):75–93. doi: 10.1016/j.pneurobio.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Zetterstrom RH, Solomin L, Jansson L, et al. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276(5310):248–50. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 27.Sakurada K, Ohshima-Sakurada M, Palmer TD, Gage FH. Nurr1, an orphan nuclear receptor, is a transcriptional activator of endogenous tyrosine hydroxylase in neural progenitor cells derived from the adult brain. Development. 1999;126(18):4017–26. doi: 10.1242/dev.126.18.4017. [DOI] [PubMed] [Google Scholar]
- 28.Castro DS, Hermanson E, Joseph B, et al. Induction of cell cycle arrest and morphological differentiation by Nurr1 and retinoids in dopamine MN9D cells. J Biol Chem. 2001;276(46):43277–84. doi: 10.1074/jbc.M107013200. [DOI] [PubMed] [Google Scholar]
- 29.Riddle R, Pollock JD. Making connections: the development of mesencephalic dopaminergic neurons. Brain Res Dev Brain Res. 2003;147(1–2):3–21. doi: 10.1016/j.devbrainres.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Simon HH, Bhatt L, Gherbassi D, et al. Midbrain dopaminergic neurons: determination of their developmental fate by transcription factors. Ann NY Acad Sci. 2003;991:36–47. [PubMed] [Google Scholar]
- 31.Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3(7):531–41. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- 32.Simon HH, Thuret S, Alberi L. Midbrain dopaminergic neurons: control of their cell fate by the engrailed transcription factors. Cell Tissue Res. 2004;318(1):53–61. doi: 10.1007/s00441-004-0973-8. [DOI] [PubMed] [Google Scholar]
- 33.Smits SM, Burbach JP, Smidt MP. Developmental origin and fate of meso-diencephalic dopamine neurons. Prog Neurobiol. 2006;78(1):1–16. doi: 10.1016/j.pneurobio.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Andersson E, Tryggvason U, Deng Q, et al. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124(2):393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 35.Prakash N, Wurst W. Genetic networks controlling the development of midbrain dopaminergic neurons. J Physiol. 2006;575(Pt 2):403–10. doi: 10.1113/jphysiol.2006.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kele J, Simplicio N, Ferri AL, et al. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development. 2006;133(3):495–505. doi: 10.1242/dev.02223. [DOI] [PubMed] [Google Scholar]
- 37.Wurst W, Prakash N. Wnt1-regulated genetic networks in midbrain dopaminergic neuron development. J Mol Cell Biol. 2014;6(1):34–41. doi: 10.1093/jmcb/mjt046. [DOI] [PubMed] [Google Scholar]
- 38.Castelo-Branco G, Wagner J, Rodriguez FJ, et al. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc Natl Acad Sci USA. 2003;100(22):12747–52. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsunaga E, Katahira T, Nakamura H. Role of Lmx1b and Wnt1 in mesencephalon and metencephalon development. Development. 2002;129(22):5269–77. doi: 10.1242/dev.129.22.5269. [DOI] [PubMed] [Google Scholar]
- 40.Prakash N, Brodski C, Naserke T, et al. A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development. 2006;133(1):89–98. doi: 10.1242/dev.02181. [DOI] [PubMed] [Google Scholar]


