Abstract
Beetles represent the largest insect order and they display extreme morphological, ecological and behavioral diversity, which makes them ideal models for evolutionary studies. Here, we present the draft genome of the scarab beetle Oryctes borbonicus, which has a more basal phylogenetic position than the two previously sequenced pest species Tribolium castaneum and Dendroctonus ponderosae providing the potential for sequence polarization. Oryctes borbonicus is endemic to La Réunion, an island located in the Indian Ocean, and is the host of the nematode Pristionchus pacificus, a well-established model organism for integrative evolutionary biology. At 518 Mb, the O. borbonicus genome is substantially larger and encodes more genes than T. castaneum and D. ponderosae. We found that only 25% of the predicted genes of O. borbonicus are conserved as single copy genes across the nine investigated insect genomes, suggesting substantial gene turnover within insects. Even within beetles, up to 21% of genes are restricted to only one species, whereas most other genes have undergone lineage-specific duplications and losses. We illustrate lineage-specific duplications using detailed phylogenetic analysis of two gene families. This study serves as a reference point for insect/coleopteran genomics, although its original motivation was to find evidence for potential horizontal gene transfer (HGT) between O. borbonicus and P. pacificus. The latter was previously shown to be the recipient of multiple horizontally transferred genes including some genes from insect donors. However, our study failed to provide any clear evidence for additional HGTs between the two species.
Keywords: Coleoptera, Scarabaeoidea, cytochrome P450, glutathione S-transferase
Introduction
Insects are one of the most diverse and successful animal groups on earth. More than one million species have been documented and there are probably 10 times more waiting to be described (Scherkenbeck and Zdobinsky 2009). Within insects, the Coleoptera (beetles) is the most evolutionarily successful order (Hunt et al. 2007) containing over 400,000 described species (Hammond 1992) demonstrating extraordinary morphological, ecological and behavioral diversity (Crowson 1960). However, despite the enormous size of this group, only few beetles have had their genomes sequenced so far, the red flour beetle Tribolium castaneum (Richards et al. 2008), the mountain pine beetle Dendroctonus ponderosae, a major forest pest (Keeling et al. 2013), the coffee berry borer Hypothenemus hampei (Vega et al. 2015) and the social beetle Nicrophorus vespilloides (Cunningham et al. 2015).
The Scarabaeoidea (=Lamellicornia) represent a major superfamily within the Coleoptera consisting mostly of phytophagous beetles. Some species are notorious agricultural pests such as the cockchafers, whereas others are considered useful because of their contribution to the decompositon of livestock manure (Scholtz and Grebennikov 2005). Besides their enormous ecological significance, scarab beetles are also noted for their impressive phenotypic plasticity, which gives rise to a great variety of sexually dimorphic horns and antlers, whose size depends on the nutritional status of the animal (Moczek 2009; Lavine et al. 2015).
Within Scarabaeoidea, the genus Oryctes, family Dynastidae (Endrödi 1985) comprises of 43 often tropical species (Bedford 1976; Dechambre 1983). Some species are pests of palm trees like the most widespread Oryctes rhinoceros (Ohler 1999), which can do considerable harm on coconut farms (Bedford 1980). In contrast to O. rhinoceros and its close relative O. monoceros, the majority of Oryctes species have root-feeding grub stages and have undergone enormous speciation in Africa, Southeast Asia and the Indian Ocean (Dechambre 1983). One such beetle species is Oryctes borbonicus (Dechambre 1982), a species that is endemic to La Réunion Island, a biodiversity hotspot located in the Indian Ocean (Myers et al. 2000). Oryctes borbonicus is included in the subgenus Insuloryctes placed together with endemics from Mauritius (O. tarandus and O. chevrolatii) and Rodriguez (O. minor) (Dechambre 1982). The endemic scarab beetles of Mauritius might be extinguished due to the usage of a baculovirus to control the palm tree pest species O. rhinoceros and O. monoceros. Therefore, it has not been possible to get DNA of the putative sister species to construct a phylogeny of the genus. The biology of O. borbonicus is largely unknown. In its distribution area are no palm trees growing, so in contrast to many other Oryctes species, O. borbonicus does not use palms as food source on La Réunion. The larvae of O. borbonicus have not yet been found in nature, but all the area where O. borbonicus can be caught is covered with grassland. Therefore, we assume that grubs feed on roots of different grasses. Also, breeding experiments suggest that the development of the grubs can be estimated to be ∼3 years (personal communication Jacques Rochat).
The nematode Pristionchus pacificus is known to be associated with O. borbonicus (Herrmann et al. 2006). P. pacificus is a nematode model organism used in evolutionary biology for studies that aim to integrate developmental biology, ecology and population genetics (Sommer and McGaughran 2013; Sommer 2015). Pristionchus was shown to have a necromenic association with scarab beetles. After invading the beetles as developmentally arrested dauer stages, dauer larvae wait for the death of the beetle to continue their development, feeding on the microbes growing on the carcass (Weller et al. 2010; Ragsdale et al. 2015). Pristionchus pacificus had its genome originally sequenced a decade ago, the results of which revealed a surprisingly complex genomic composition encoding >25,000 protein coding genes (Dieterich et al. 2008). Surprisingly, these studies revealed a substantial amount of horizontal gene transfer (HGT) into the nematode genome, most likely from different donors. For example, the P. pacificus genome contains seven cellulase genes that are originally of microbial origin but have duplicated and diversified within the genus Pristionchus (Dieterich et al. 2008; Mayer et al. 2011; Schuster and Sommer 2012). It also contains diapausins, which are thought to be beetle-derived (Dieterich et al. 2008; Rödelsperger and Sommer 2011). Further analysis based on various homology searches and codon usage suggested that insects might have been the donors of additional Pristionchus gene acquisitions by HGT, which could be confirmed by phylogenetic analysis for a family of retrotransposons (Rödelsperger and Sommer 2011). HGT between insects and nematodes appears very likely, given the close entomophilic association of Pristionchus species and other genera of the family Diplogastridae (Kanzaki and Giblin-Davis 2015). Furthermore, roughly one-third of the P. pacificus gene predictions do not have homologs in other nematodes indicating the presence of so-called orphan (or taxonomically restricted) genes (Borchert et al. 2010; Rödelsperger et al. 2013). Therefore, sequencing the genome of the scarab beetle O. borbonicus will provide the opportunity to search for HGT events, which could not be detected previously due to the scarcity of beetle genomes.
In this study, we present the draft genome of O. borbonicus as a resource for the insect genomics community. In a first general characterization of the O. borbonicus genome we show that, in spite of its basal phylogenetic position relative to the two other sequenced beetles, O. borbonicus has undergone a substantial amount of lineage-specific gene expansions and losses. We were able to further confirm this fact in a detailed phylogenetic analysis of two large gene families, the Glutathione S-Transferases (GST) and Cytochrome P450 (CYP) gene families, respectively. At the same time, the analysis of the O. borbonicus genome revealed no further evidence for HGT between beetles and Pristionchus nematodes.
Materials and Methods
Genome Sequencing
High-Molecular Weight DNA Extraction
One male Oryctes borbonicus beetle was dissected under sterile conditions. The thoracal flight muscles were prepared for DNA extraction with the Qiagen genomic DNA extraction kit and Qiagen genomic tip columns (Qiagen, Hamburg, Germany) following the manufacturer’s instructions. DNA quality and integrity was determined with a NanoDrop ND 1000 spectrometer (PeqLab, Erlangen, Germany) and by gel electrophoresis. The high molecular weight DNA was used to generate mate-pair libraries. Paired-end libraries were prepared with DNA extracted from two legs of the same beetle with the epicenter DNA extraction kit (Epicentre, Madison) following the manufacturer’s instructions.
RNA Extraction
Two legs from the same individual beetle were frozen in liquid nitrogen and ground to a fine powder using mortar and pestle. The powder was directly transferred into Tri Reagent® (Sigma-Aldrich, Munich, Germany) and RNA was extracted with the Direct-zol™ RNA MiniPrep (Zymo, Freiburg, Germany) following the manufacturer’s protocol. RNA quality was determined with a NanoDrop ND 1000 spectrometer (PeqLab, Erlangen, Germany) and RNA integrity was checked with the Agilent RNA Nano Chip Assay (Agilent, Santa Clara). Only RNA samples of high quality (OD 260/280 > 2 and OD 260/230 > 1.8) and high integrity were used for library preparation.
DNA Sequencing
Two paired-end libraries with an average insert size of 450 and 500 bp and four mate-pair libraries of 3, 5, 8 and 11 kb were prepared using Illumina’s paired-end and Nextera mate-pair kits, respectively (Illumina, Eindhoven, Netherlands) following the manufacturer’s protocols. For the paired-end libraries a starting amount of 100 ng of DNA was sheared with a Covaris S2 ultrasonicator (Covaris Inc. Woburn, San Diego). The mate-pair libraries were prepared from 4 μg DNA. Both the paired-end libraries (2 plex) and the mate-pair libraries (5 plex) were each run on a single 101 bp paired-end flow cell lane of an Illumina HiSeq 2000 Sequencer (Illumina, Eindhoven, Netherlands).
Overlap Library
The overlap library was created using the Illumina Nano kit (Illumina, Eindhoven, Netherlands) following the manufacturer’s protocol. The average size of the library was ∼450 bp. Subsequent sequencing was performed on the MiSeq platform with the reagent Kit v2, with 2×250 bp read length (Illumina, Eindhoven, Netherlands).
RNA Sequencing
The high quality RNA obtained from two beetle legs was used to prepare two independent libraries following Illumina’s TruSeq RNA library preparation protocol (Illumina, Eindhoven, Netherlands). Both libraries (2 plex) were sequenced together on a single 101-bp pair-end flow cell lane of an Illumina HiSeq 2000 Sequencer.
Genome Assembly
Genomic paired and mate-pair reads were quality trimmed and filtered as described in Rödelsperger et al. 2014. Genomic paired-end libraries were used to generate a first draft assembly with the Velvet assembler (Version 1.2.10) with a k-mer size of 41 (Zerbino and Birney 2008). In a second step, we used SSPACE (version 2.0) with default settings for scaffolding using the mate-pair data (Boetzer et al. 2011). Intrascaffold gaps were closed using the Gapclosing module (version 1.10) of the SOAP package. For removal of contamination, we ran a BLASTn search against the NCBI nt database, searching for hits with sequence identity >95% over a length of 100 bp. After taxonomic inspection of significant hits, the contaminating contigs (mostly of fungal origin) were removed from the assembly. The final assembly comprised 150,243 scaffolds spanning 518 Mb (494 Mb excluding gaps) with an N50 of 105 kb. The largest scaffold spans 1.1 Mb. The genome-wide GC content was 34.4%. To obtain independent estimates of genome size and repeat content we used the software jellyfish (version 1.1.4) to generate k-mer spectra of original raw sequencing data (Marçais and Kingsford 2011).
Gene and Repeat Annotation
Repeats were identified using the RepeatModeler and RepeatMasker pipeline, which identified 154 Mb (29.2%) as repetitive (∼4.5% LINE elements, ∼10% DNA elements, and ∼14.4% unclassified). Transcriptomic reads were aligned to the Oryctes borbonicus assembly using TopHat (v2.0.3), and reference-guided transcriptome assemblies were generated using the software Cufflinks (v2.0.1). Transcriptome assemblies were used to train the gene finder AUGUSTUS (v2.6.1) (Stanke et al. 2006), which predicted 23,278 protein coding genes in the repeat masked assembly. To compare gene predictions with a purely evidence-based set of gene annotations, we made use of the MAKER2 pipeline (version 2.31.8; Holt and Yandell 2011). MAKER2 was run once on the repeat masked genome using only de novo assembled transcripts (Trinity version: trinityrnaseq_r20140413p1; Grabherr et al. 2011) and protein homology data from D. ponderosae and T. castaneum. Thus, no implicit call of ab initio gene finders was done for this set of gene annotations.
Protein domains were annotated using the hmmsearch program of the HMMER3 package and the included PFAM profiles. For analysis of core eukaryotic genes, we downloaded a set of 458 profile HMMs, which were searched against the database of predicted proteins using hmmsearch (evalue < 0.001) (CEGMA database, Parra et al. 2007).
Comparative Genomic Data
We downloaded protein sequences of two more beetles and of seven representative species from seven different insect orders from Ensembl Metazoa release 25. This data set comprised 13,457 protein sequences from Dendroctonus ponderosae (Coleoptera), 16,526 sequences from Tribolium castaneum (Coleoptera), 30,362 protein sequences representing 13,918 genes from Drosophila melanogaster (Diptera), 15,314 sequences from Apis mellifera (Hymenoptera), 15,441 sequences from Rhodnius prolixus (Hemiptera), 12,829 sequences from Heliconius melpomene (Lepidoptera), 10,788 sequences from Pediculus humanus (Phthiraptera), 14,610 sequences from Zootermopsis nevadensis (Isoptera). In the case of D. melanogaster, we used the longest isoform per gene for further analysis. Conserved synteny between beetle genomes was detected by means of CYNTENATOR software (Rödelsperger and Dieterich 2010), which computes gene order alignments using a phylogenetic scoring function. This approach identified conserved syntenic blocks between O. borbonicus and the T. castaneum genome spanning the 42 Mb of the T. castaneum genome and 39 Mb of conserved syntenic blocks between O. borbonicus and D. ponderosae. Conserved synteny was used to identify contigs that are likely of X-chromososomal origin (supplementary fig. S3, Supplementary Material online). For the CYP family, manually curated sequences, including those from the moth Bombyx mori, were retrieved from the website of David Nelson (http://drnelson.uthsc.edu/biblioC.html, last accessed June 6, 2016), and experimentally characterized sequences of other beetles (Ips paraconfusus, Phyllopertha diversa, and Leptinotarsa decemlineata) were retrieved from Genbank. Artifactual fusions and fissions in protein predictions were manually corrected as described previously (Markov et al. 2015), with further expert polishing by D. Nelson regarding CYPs, and are provided as a supplementary dataset, Supplementary Material online.
Ortholog Clustering and Phylogenetic Analysis
In order to identify orthologous clusters we used orthomcl (Li et al. 2003) software with default settings. We identified 2,355 1:1 orthologs from 14,748 orthomcl clusters using a custom perl script. We generated multiple sequence alignments for each 1:1 ortholog cluster using Clustal Omega (Sievers et al. 2011). After model testing with ProtTest 3 (Darriba et al. 2011) 2,355 individual gene trees and one tree based on all concatenated alignments were constructed using RAxML (version 8.1.20, Stamatakis 2014). Concatenation as well as looking at the most frequently reconstructed gene trees, resulted in the same species tree topology (fig. 1A). The analysis of the GST and CYP gene families was done by aligning sequences using MUSCLE (Edgar 2004). Conserved sites were manually selected using Seaview (Gouy et al. 2010). Phylogenetic trees were inferred using PhyML (Guindon and Gascuel 2003) with the LG + G substitution model. Branch support values were assessed using the approximate likelihood ratio test (Anisimova and Gascuel 2006).
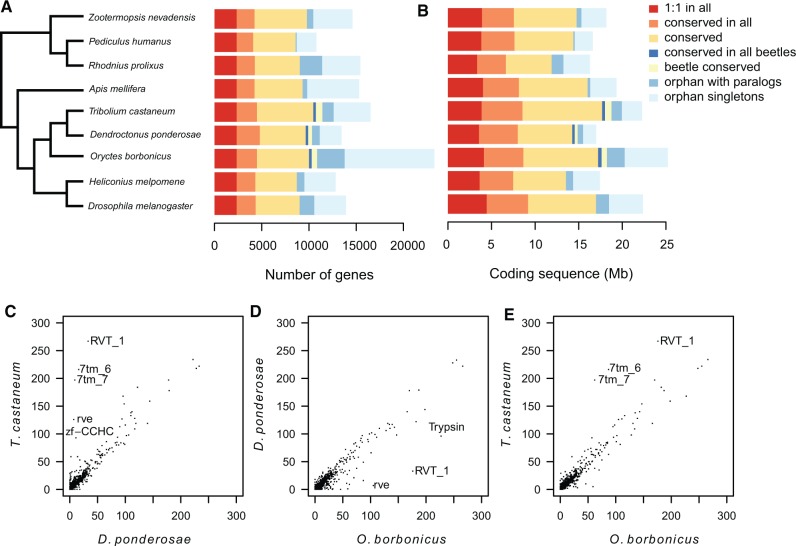
Fig. 1.—
Conserved and lineage-specific patterns of gene content evolution. (A) Schematic phylogeny of investigated insect genomes (Hunt et al. 2007; Trautwein 2012) and distribution of genes in different orthology classes. (B) Amount of coding sequence in different orthology classes. (C–E) Protein domain (PFAM) count comparison between all three beetle genomes. Large protein domain families that show the most extreme differences in gene counts are labeled in each comparison.
Results
Draft Genome Assembly of Oryctes borbonicus
From a sampling trip to La Réunion island, of which the primary goal was to find more beetle associated nematode strains, we set two male beetle specimen aside that should provide enough material to sequence the genome and transcriptomes. Our original intention was to use only data from one specimen for the assembly, but since additional sampling trips were not an option, we collected a second individual as backup.
We used state-of-the-art sequencing technology including mate-pair, paired-end and overlap libraries to assemble a draft genome of O. borbonicus. We initially planned to generate an assembly using data from a single individual with the Allpath-LG assembler. However, without knowing the actual genome size, we found that the coverage of the overlap library was insufficient to generate an ab initio assembly by Allpath-LG (supplementary fig. S1, Supplementary Material online). We therefore followed an alternative approach, that used data from a single individual for the initial assembly and data from both individuals for scaffolding and gap-closing (see Materials and Methods). Our final assembly has a size of 518 Mb (table 1), which is substantially larger than the two previously sequenced beetle genomes (Tribolium castaneum has 230 Mb and Dendroctonus ponderosae 208 Mb). Calculation of genome size based on the total amount of sequence data, the expected coverage and the size of the X-chromosomes (as inferred by coverage and synteny analysis), results in a similar estimate of roughly 500 Mb. High quality reads without any uncalled bases were realigned against the final assembly in order to assess the completeness of the genome assembly. This showed that >99% of reads could be placed onto the assembly, indicating that the assembly covers almost all of the raw sequencing data. However, we find evidence that unresolved repeats caused some problems in the assembly resulting in high rates of ambiguous base calls and a heterogenous coverage profile (supplementary figs. S2–S4, Supplementary Material online). The fraction of repetitive sequences in the final assembly was estimated to be 29%, which matches the range of 17–34% found in the two previously sequenced beetle genomes (Richards et al. 2008; Keeling et al. 2013). In addition, analyzing the k-mer spectrum of raw sequence data (supplementary fig. S5, Supplementary Material online), we find that ∼70% of sequence data is represented by k-mers that have at most the expected coverage (30×). About 23,278 protein-coding genes were predicted using the AUGUSTUS gene finder after training with RNA-seq data. Again, this number of protein-coding genes is substantially larger than the values reported for the two previously sequenced beetle genomes (N = 13,457 for D. ponderosae and N = 16,526 for T. castaneum).
Table 1.
Genome statistics
| Genome Assembly |
De Novo Transcriptome | Evidence-Based Annotation | Gene Prediction | ||
|---|---|---|---|---|---|
| Scaffolds | Contigs | Trinity | MAKER2 | AUGUSTUS | |
| Total size (Mb) | 517.9 | 426.3 | 27.2 | 16.6 | 25.2 |
| N sequences | 150,243 | 30,471 | 18,177 | 20,504 | 23,278 |
| Largest (kb) | 1,101 | 457 | 16 | 32.4 | 41 |
| Smallest (kb) | 0.1 | 0.1 | 0.5 | <0.1 | <0.1 |
| N50 (kb) | 104.8 | 33.1 | 2.0 | 1.5 | 1.8 |
| N sequences (length>N50) | 1,365 | 3,590 | 3,915 | 3,168 | 3,712 |
Note.—Assembly features such as size and number of sequences were collected for the raw Contig assembly, scaffolded genome, and three types of gene annotations. Please note that arbitrary minimum cutoffs were used by the different programs.
We used the MAKER2 pipeline (version 2.31.8; Holt and Yandell 2011) to generate a set of 20,504 evidence-based gene annotations based on de novo assembled transcripts and protein sequences from other insects (Grabherr et al. 2011; Holt and Yandell 2011). These annotations were used to further evaluate the AUGUSTUS predictions showing that ∼81% of evidence-based exons overlap predicted exons. Second, we tested for the representation of core eukaryotic genes in the predicted gene set. We used the definition of core eukaryotic genes as provided by the CEGMA database (Parra et al. 2007) and found that 445 (97%) of the 458 core eukaryotic gene profiles are represented in the predicted proteome of O. borbonicus and the best hit covered in median 92% (interquartile range: 73–98%) of the query profile. Third, we generated our “own” core insect gene data set by assembling homologous sequences into orthologous clusters (Li et al. 2003) in order to identify genes that are presented as 1:1 orthologs in all three beetle-, as well as in the six nonbeetle insect genomes (see Materials and Methods). This approach identified 2,355 genes that have a 1:1 relationship in all nine insect genomes. We used these orthologous clusters to test whether O. borbonicus genes show a tendency to represent incomplete or partial predictions by comparing their size with the size of the corresponding genes in the other eight insect genomes. As indicated in supplementary figure S6A and B, Supplementary Material online, O. borbonicus gene predictions show a similar size range relative to Drosophila as the gene sets of the other sequenced insect genomes.
Next, to assess the degree of completely missing gene predictions, we counted outlier orthology clusters representing those genes that have a 1:1 orthology relationship in all but one genome (supplementary fig. S6C, Supplementary Material online). The number of outliers was in a range of a few dozen for all nine genomes, again supporting the notion that at least in highly conserved regions most if not all of the sequenced genomes are of comparable quality. In summary, these results underpin the high quality of gene predictions in O. borbonicus relative to the eight other insect genomes (supplementary fig. S6, Supplementary Material online).
The considerable differences in genome size and gene number between O. borbonicus and the two previously sequenced beetle motivated us to investigate further to what extent these differences are manifested across different gene classes. We made the surprising finding that 41% of O. borbonicus gene predictions represent orphan singleton genes, i.e. genes that lack sequence similarity in any other of the tested insect genomes as well as in the O. borbonicus genome itself (as opposed to orphans with intra-species paralogs) (supplementary figs. S1A and B and S6, Supplementary Material online). This is in strong contrast to the two previously sequenced beetle genomes, for which we found 17% of genes to be orphan singletons in D. ponderosae and 24% in T. castaneum. Genes can be classified as orphan genes due to multiple technical as well as biological reasons. Technical reasons are the lack of phylogenetic resolution of sampled genomes, but also artifactual gene predictions. Consistent with a previous analysis of orphan genes in insects (Wissler et al. 2013), orphan singletons are relatively short, their total coding sequence does not correspond to gene number (fig. 1B) and they show lesser evidence of expression (supplementary fig. S7, Supplementary Material online). Biologically, orphan genes may very well represent truly functional lineage-specific genes.
To further support that at least a fraction of identified orphan genes represent truly functional sequences, we investigated to what extent, orphan singletons show evidence of protein domains (PFAM, e-value < 0.001), weak homologies in other insects (BLASTP e-value < 0.001), or have expression evidence (FPKM >1). In total, we found that 4,057 (43%) of orphan singletons fulfill at least one of the three mentioned criteria suggesting that large portions of these predictions are real genes. However, further insight into the origins of orphan genes can only be gained on the basis of much broader phylogenetic sampling and also broader transcriptome data. As this is a future project, the remaining part of our analysis will focus on genes with homologs in other beetle and insect genomes—a gene set that might be of greatest value for current comparative genomic analyses.
Distribution of Conserved, Beetle-Specific and Orphan Genes
We used the identified orthologous clusters to compare the following: (1) conserved genes, with detectable homologs across different insect orders, (2) beetle-specific genes that are found in at least two of the beetle genomes but not in any other insect, and (3) orphan genes, which are restricted to one specific lineage. In order to obtain more robust estimates of the relative size of different gene classes, we excluded orphan singletons from this analysis, as it is unclear to what extent this class includes artifactual gene predictions. Figure 1A and B shows the fine scale distribution of different homology classes across all nine insect genomes. As mentioned above, we originally identified 2,355 orthologous clusters, whose genes were predicted as 1:1 orthologs in all nine species. These 1:1 orthologs make up between 17% and 25% of the gene repertoire in these different insects, indicating that the majority of gene families have undergone lineage-specific gene birth and death events. The first three categories in figure 1A and B define conserved genes that are found across all analyzed insect orders. It is important to note that this gene set is by far the largest class. Focusing only on the beetle genomes, the fraction of conserved genes ranges from 73% for O. borbonicus to 87% for D. ponderosae. At the same time, the second most abundant group of genes are orphan genes (orphan with paralogs), which constitute between 7% of genes in D. ponderosae and 21% in O. borbonicus. Finally, only a minor fraction of genes (6–8%) are specific to and conserved only within beetles (with homologs in at least one other beetle genome), suggesting that the generation of novel genes has not strongly contributed to the evolution of the order Coleoptera.
Lineage-Specific Patterns in Beetle Genome Evolution
Our prior analyses indicated that a substantial fraction of genes are either conserved among insects or highly specific to individual lineages (fig. 1A and B). Therefore, one major benefit of the O. borbonicus genome is its position as an outgroup relative to the two previously sequenced beetle genomes (Hunt et al. 2007). In order to confirm previous analyses, we reconstructed phylogenetic trees based on the 2,355 orthologous gene clusters showing that the most frequently predicted tree topologies are fully consistent with the tree shown in figure 1. Similarly, the reconstruction of a genome-wide species phylogeny based on the concatenation of the alignments (supermatrix approach) of all 2,355 orthologous clusters revealed that O. borbonicus indeed represents an outgroup to the two previously sequenced beetles.
To further characterize lineage-specific patterns in the evolution of beetle genomes, we first screened for drastic changes in the size of gene families. In the ideal case, detailed phylogenetic analyses (see below) would be necessary to reconstruct the exact evolutionary history of duplication and gene loss events. However, comparisons of approximate gene family sizes provide a proxy to prioritize candidate gene families for more detailed investigation. Here, we defined gene families based on the presence of a certain protein PFAM domain and performed all three pair-wise comparisons of the distributions of gene family sizes (fig. 1C–E). In the comparison between T. castaneum and D. ponderosae, reverse transcriptases (PF00078) and seven transmembrane proteins (PF02949 and PF08395) are present in much higher number in T. castaneum (fig. 1C). In the absence of an outgroup it would remain unclear whether this result is caused by an expansion in the T. castaneum lineage or by a loss in the D. ponderosae lineage. The comparison with O. borbonicus as outgroup (fig. 1E) shows higher numbers for both gene families in T. castaneum, suggesting that these families have undergone a lineage-specific expansion in the branch leading to T. castaneum. It should be noted however, that the number of genes encoding reverse transcriptases is also moderately larger in O. borbonicus relative to D. ponderosae (fig. 1D). We interpret this finding as evidence that a second expansion in the lineage leading to O. borbonicus is the most likely scenario to explain this increase, because we are not aware of a mechanism that would lead to a lineage-specific loss of transposable elements. Nonetheless, detailed phylogenetic analyses are ultimately needed to confirm this second expansion. While expansions of reverse transcriptases and retroviral integrases (PF00665) in the O. borbonicus and T. castaneum lineages could point to recent activity of retroviruses and retrotransposons, the expansion of seven transmembrane proteins could be due to a variety of different events. Proteins of the seven transmembrane families are involved in many essential signaling pathways, for example as receptors for neuropeptides. These short proteins have been shown to be crucial not only for developmental processes, such as molting and diapause (Verlinden et al. 2015), but also play important roles in the maintenance of homeostasis of metabolites and proteins, in the regulation of water balance and in several behaviors (Scherkenbeck and Zdobinsky 2009; van Hiel et al. 2010).
Accelerated Gene Turnover in the D. ponderosae Lineage
To quantify the rate of gene turnover across the beetle phylogeny, we identified orthologous clusters with beetle-specific gene losses or expansions. In addition, we mapped these events onto specific branches in the beetle phylogeny and compared these numbers with the estimations of simulated gene losses and gains that were randomly placed onto the beetle phylogeny. For this analysis, we restricted ourselves to orthologous clusters with one member in H. melpomene and D. melanogaster, respectively, and screened for beetle-specific losses and gains. For example, the presence of a member of such an orthologous cluster in O. borbonicus but its absence in the two other beetles would suggest that the ortholog was lost in the common ancestor of T. castaneum and D. ponderosae. Following this methodology, we identified 320 clusters with O. borbonicus-specific losses and 220 with expansions, 281 losses and 459 expansions in D. ponderosae, 52 losses and 74 expansions in T. castaneum, and 48 losses and 34 expansions in the ancestor of T. castaneum and D. ponderosae, respectively.
We then simulated 10,000 evolutionary scenarios, randomly placing gene losses and gains onto branches of the phylogeny with probabilities proportional to the branch lengths as derived from our supermatrix tree. The most striking patterns discovered in these analyses, are the strong depletions of gene losses and gains in the T. castaneum lineage (P < 10 − 4) with a simultaneously increased number of duplications in the D. ponderosae lineage (P < 10 − 4). This result is particularly interesting, as D. ponderosae did not show any large expansions of specific gene families in our previous analysis based on protein domains (fig. 1C and D), suggesting that both approaches reveal rather complementary patterns of gene family evolution. In summary, our analysis has shown lineage-specific expansions and losses as a recurrent trend in beetles. However, more detailed phylogenetic analysis is needed to confirm these trends in individual gene families. We thus focus our final analysis on the GST and CYP gene families, which are important molecular players in insect physiology.
Oryctes-Specific Expansions in the Ancient GST Sigma and Theta Families
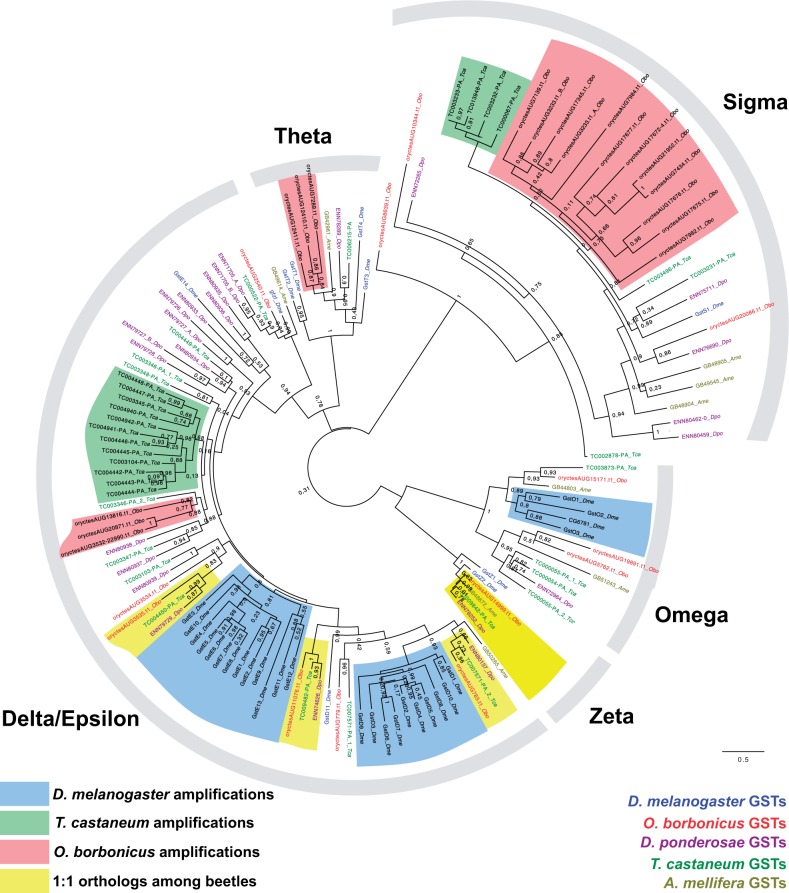
GSTs are members of an ancient gene family, present in both bacterial and eukaryotic organisms. Insect GSTs can be divided into two major groups, the cytosolic and the microsomal GST genes. The cytosolic group further divides into six classes, the Delta, Epsilon, Sigma, Omega, Theta, and Zeta classes, respectively (Friedman et al. 2011). The Delta and Epsilon classes are insect-specific, and comprise enzymes that metabolize pesticides (Enayati et al. 2005; Che-Mendoza et al. 2009). However, some of these GST genes are also regulating the metabolism of endogenous hormonal compounds (Enya et al. 2015), whereas others have nonenzymatic functions (Sheehan et al. 2001). Consistent with previous reports (Shi et al. 2012; Keeling et al. 2013), we find that the Delta/Epsilon GSTs are highly expanded in T. castaneum independent of D. melanogaster (fig. 2 and supplementary fig. S8, Supplementary Material online). The overall number of cytosolic GSTs is similar across the three beetle species (36 in T. castaneum, 28 in D. ponderosae, and 30 in O. borbonicus). However, the addition of the O. borbonicus genome suggests that only 3 out of the 30 cytosolic GSTs are 1:1 orthologs among beetles (highlighted in fig. 2). Interestingly, the biggest expansion regarding Oryctes GSTs occurs in the Sigma class, which comprises enzymes involved in the synthesis of prostaglandins in vertebrates and nematodes (Sheehan et al. 2001), and also in Bombyx mori (Yamamoto et al. 2013). Furthermore, our analysis suggests that four of the biggest expansions are located in the same genomic cluster. A second smaller Oryctes-specific expansion, limited to three paralogs, is found in the theta class and a third one concerns three genes in the Delta-Epsilon family.
Fig. 2.—
A maximum-likelihood tree of beetle cytosolic GSTs. The tree is rooted with sequences from Drosophila and Apis, and was calculated under the LG+G model. A linear version is available in supplementary figure S8, Supplementary Material online.
Ten Independent Oryctes-Specific Expansion Events across the CYP Family
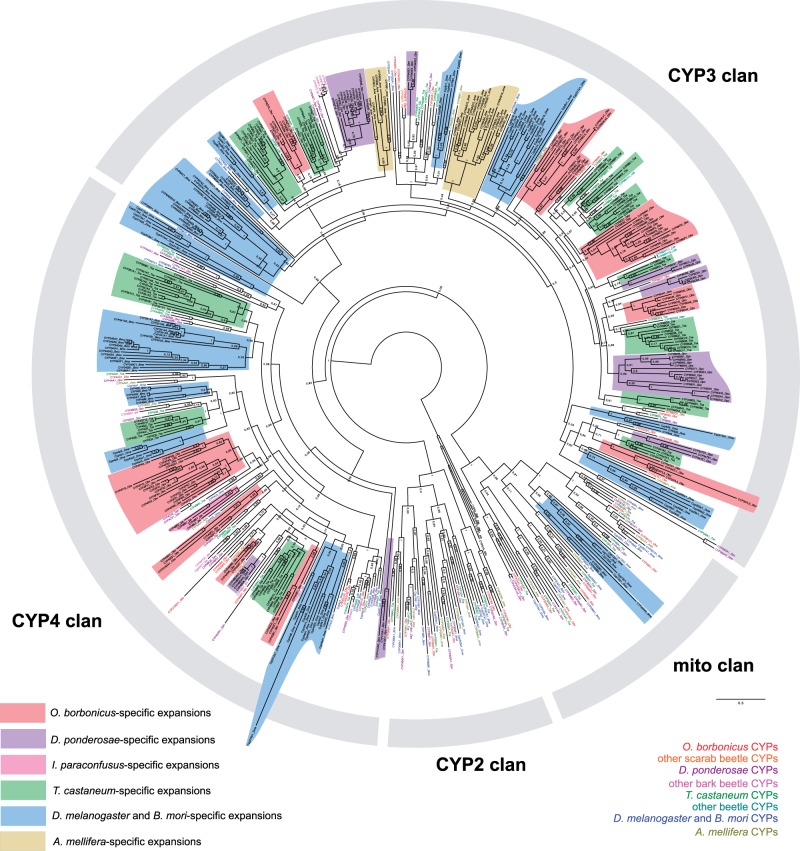
The CYP family is present in most aerobic eukaryotes, and also in some bacteria (Nelson et al. 2013). In insects, the CYP family divides into four major clans, clan 2, clan 3, clan 4 and the mitochondrial or mito clan, respectively (Feyereisen 2006). We found a total of 115 CYPs in O. borbonicus: 6 in clan 2, 62 in clan 3, 39 in clan 4, and 8 in the mito clan (fig. 3 and supplementary fig. S9, Supplementary Material online). Interestingly, clan 2 and the mito clan are enriched in 1:1 orthologs across insects and many of these genes are suggested to be involved in the biosynthesis or the degradation of the molting hormone ecdysone. Also, one of them is involved in the final step of juvenile hormone synthesis. Interestingly, the mito clan also comprises the xenobiotic-metabolizing CYP12 gene from dipterans, which just like the Oryctes-specific expansion of Sigma GSTs, represents a case of lineage-specific expansion in a part of the tree that is otherwise conserved across insects.
Fig. 3.—
A maximum-likelihood tree of insect CYPs. The tree was calculated under the LG+G model, and is rooted by CYP51 sequences. A linear version of the tree is available in supplementary figure S9, Supplementary Material online.
Clan 4 comprises some fatty-acid hydroxylating enzymes (Nelson et al. 2013), but also enzymes involved in the synthesis or degradation of pheromones in other beetles (Qiu et al. 2012). For example, individual sequences from another scarab beetle, Phyllopertha diversa (Maïbèche-Coisne et al. 2004) and the bark beetle Ips paraconfusus (Huber et al. 2007), indicate that duplication events are widespread among species and higher taxa, confirming the notion that pheromone-synthesizing genes evolve rapidly (Liénard et al. 2008). It is also interesting to note that clan 4 provides the most striking example of Oryctes-specific gene expansions. Specifically, in the group termed CYP4C3 (supplementary fig. S9, Supplementary Material online), comprising a single gene from D. melanogaster (CYP4C3), A. mellifera (CYP4AV1) and T. castaneum (CYP4BM1), D. ponderosae has two duplicates. In addition, the partial data for I. paraconfusus suggest the existence of at least four paralogs. In contrast, the number rises to 19 for O. borbonicus, suggesting extremely high gene birth, and potentially death rates, consistent with previous reports in insects (Feyereisen 2006).
Clan 3 indicates an additional example of high numbers of lineage-specific clusters, including genes that are involved in insecticide degradation in at least two beetles (Zhu et al. 2010; Zimmer et al. 2014) and pheromone synthesis in bark beetles (Sandstrom et al. 2006; Song et al. 2014). Again, there are many O. borbonicus-specific amplifications including one with 16 genes (supplementary fig. S9, Supplementary Material online). Interestingly, the same pattern of lineage-specific expansions has also been observed in other insects (Richards et al. 2008).
Absence of Additional HGT Events from Beetles to Nematodes
Our initial motivation to study the genome of O. borbonicus goes back to previous studies of the P. pacificus genome, which characterized a number of HGTs from different donor organisms including insects (Dieterich et al. 2008; Mayer et al. 2011; Rödelsperger and Sommer 2011) and showed that roughly one-third of P. pacificus genes, so called orphan genes, do not have homologs in other nematodes (Borchert et al. 2010; Rödelsperger et al. 2013). Thus, the analysis of the genome of the scarab beetle O. borbonicus, a well established host of P. pacificus, provided the unique opportunity to search for other HGTs that had so far missed detection due to the scarcity of beetle genomes.
We obtained previously identified candidate gene sets for HGT (Rödelsperger and Sommer 2011) on the basis of BLAST analysis using nematode and insect data and used them to screen for homologs in O. borbonicus. However, based on BLASTp and tBLASTn searches, followed by multiple alignment and phylogenetic analysis, we could neither identify members of the Diapausin family in the O. borbonicus genome, which had been previously proposed to be transferred from beetles (Dieterich et al. 2008; Rödelsperger and Sommer 2011), nor could we find any other convincing candidates for HGT from the beetle to the nematode. Similarly, previously identified horizontally transferred retrotransposons did not show higher similarity to O. borbonicus sequences than to sequences from Lepidopterans (Rödelsperger and Sommer 2011). These results suggest that the horizontal transfer events could date back to a time before the Pristionchus—scarab beetle association.
Discussion
Beetles represent the largest insect order but in comparison to flies and ants they are largely underrepresented at the level of genome sequencing analysis (Drosophila 12 genomes consortium 2007; Roux et al. 2014). In this study, we have sequenced and annotated the genome of the scarab beetle O. borbonicus, which was proposed to be a basal representative in comparison to two previously sequenced genomes of T. castaneum (Richards et al. 2008) and D. ponderosae (Keeling et al. 2013), and might therefore be of importance to polarize genomic patterns between the two pest species. Please note that during the last phase of finalizing this manuscript, the genomes of Nicrophorus vespilloides and Hypothenemus hampei were published (Cunningham et al. 2015; Vega et al. 2015), which unfortunately we were not able to include in our study without redoing all analyses.
The O. borbonicus genome is 518 Mb in size and thus substantially larger, encoding a higher number of predicted genes than other sequenced beetle genomes. While we find a higher ratio of orphan genes in O. borbonicus (genes that lack sequence homology in any of the eight other insect genomes that are analyzed in this study), further data will be needed to quantify, to what extent orphan singleton genes represents a true biological phenomenon. For our overall insect genome analysis, we have excluded singleton orphan genes to ensure that they do not affect any of the conclusions drawn from these analyses. Overall, our comparative genomic analyses show that while many gene families are conserved across multiple insect orders, some have undergone lineage-specific gene duplications and losses. Even among beetles, various different approaches (comparing protein domain counts, screening for branches with increased rates of duplications in the beetle phylogeny, detailed phylogenetic analyses of individual gene families) show a common trend of substantial gene turnover among genomes. In this context it is important to note that even in gene families that do not show any striking differences in gene family size (fig. 1C–E) we can detect numerous large gene expansions using detailed phylogenetic analysis. This highlights the importance of detailed phylogenetic analyses of manually curated data sets to ensure the robustness of duplication patterns in genome evolution (Markov et al. 2015). Furthermore, we would like to add, that although heterozygosity in the sequenced beetle individual can lead to nearly identical duplicates, our general analysis of genome quality (supplementary fig. S4, Supplementary Material online) suggests that the assembler rather has the tendency to overcompress and merge repetitive regions. Thus, we conclude that the identified gene expansions in the O. borbonicus genome are indeed real.
Despite the lack of further evidence for HGTs, the O. borbonicus genome is of particular interest because of the study of the association of Pristionchus nematodes with scarab beetles. Previous work has shown that Pristionchus species and P. pacificus strains show specificity in their response to the pheromones of their beetle hosts (Hong and Sommer 2006; McGaughran et al. 2013; Cinkornpumin et al. 2014). Nonhydroxylated fatty acid ester derivatives pheromones were identified in four species from the Oryctes genus, that all are coconut pest species (Said et al. 2015). If the same kind of molecules would be active as pheromones in O. borbonicus, some Cytochrome P450s may be involved in their inactivation, as they do for molecules from other chemical classes in the scarab beetle Phyllopertha diversa (Maïbèche-Cosne et al. 2004). However, a detailed phenotypic interpretation of genomic patterns is hampered by the lack of functional data and also by the low phylogenetic resolution. Thus, more genomic and functional studies will be needed to better characterize the interaction between nematodes and beetles.
Supplementary Material
Supplementary figures S1-S9, table S1 and dataset are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was funded by the Max Planck Society. We would like to thank the Office National des Forêts and the Parc National de La Réunion, for supplying collection permits. Jacque Rochat and Sophie Gasnier (Insectarium de La Réunion) for their help with beetle collection. Finally, we thank Metta Riebesell for language corrections and David Nelson for naming and further polishing of Oryctes CYP sequences.
Literature Cited
- Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 55:539–552. [DOI] [PubMed] [Google Scholar]
- Bedford GO. 1976. Observations on the biology and ecology of Oryctes rhinoceros and Scapanes australis (Coleoptera: Scarabaeidae: Dynastinae): pests of coconut palms in Melanesia. J Aust Ent Soc. 15:241–2510. [Google Scholar]
- Bedford GO. 1980. Biology, ecology, and control of palm rhinoceros beetles. Ann Rev Entomol. 25:309–339. [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. [DOI] [PubMed] [Google Scholar]
- Borchert N, et al. 2010. Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res. 20:837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che-Mendoza A, et al. 2015. Long-lasting insecticide-treated house screens and targeted treatment of productive breeding-sites for dengue vector control in Acapulco, Mexico. Trans R Soc Trop Med Hyg. 109:106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinkornpumin JK, et al. 2014. A host beetle pheromone regulates development and behavior in the nematode Pristionchus pacificus. elife 3: doi:10.7554/elife.03229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowson RA. 1960. The phylogeny of coleoptera. Annu Rev Entomol. 5:111. [Google Scholar]
- Cunningham CB, et al. 2015. The genome and methylome of a beetle with complex social behavior, Nicrophorus vespilloides (Coleoptera: Silphidae). Genome Biol Evol. 7: 3383–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechambre RP. 1983. Contribution to the phylogenetic study of the Oryctes of the madagascan region; use of the propygidial stridulatory stria (Col. Dynastidae). J Bull Soc Ent Fr. 88:436–448. [Google Scholar]
- Dieterich C, et al. 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet. 40:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 genomes consortium 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Edgar R. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrödi S, editor. 1985. The dynastinae of the world. Bosten: Dordrecht. [Google Scholar]
- Enayati AA, Ranson H, Hemingway J. 2005. Insect glutathione transferases and insecticide resistance. Insect Mol Biol. 14:3–8. [DOI] [PubMed] [Google Scholar]
- Enya S, et al. 2015. The silkworm glutathione S-transferase gene noppera-bo is required for ecdysteroid biosynthesis and larval development. Insect Biochem Mol Biol. 61:1–7. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. 2006. Evolution of insect P450. Biochem Soc Trans. 34:1252–1255. [DOI] [PubMed] [Google Scholar]
- Friedman R. 2011. Genomic organization of the glutathione S-transferase family in insects. Mol Phylogenet Evol. 61:924–932. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27:221–224. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Hammond PM. 1992. Species inventory In: Groombridge B, editor. Global biodiversity, status of the earth’s living resources. London: Chapman and Hall; p. 17–39. [Google Scholar]
- Herrmann M, Mayer WE, Sommer RJ. 2006. Nematodes of the genus Pristionchus are closely associated with scarab beetles and the Colorado potato beetle in western Europe. Zoology 109:96–108. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong RL, Sommer RJ. 2006. Chemoattraction in Pristionchus nematodes and implications for insect recognition. Curr Biol. 16:2359–2365. [DOI] [PubMed] [Google Scholar]
- Huber DPW, Erickson ML, Leutenegger CM, Bohlmann J, Seybold SJ. 2007. Isolation and extreme sex-specific expression of cytochrome P450 genes in the bark beetle, Ips paraconfusus, following feeding on the phloem of host ponderosa pine, Pinus ponderosa. Insect Mol Biol. 16:335–349. [DOI] [PubMed] [Google Scholar]
- Hunt T, et al. 2007. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318:1913–1916. [DOI] [PubMed] [Google Scholar]
- Kanzaki N, Giblin-Davis RM. 2015. Diplogastrid systematics and phylogeny In: Sommer RJ, editor. Pristionchus pacificus: a nematode model for comparative and evolutionary biology. Leiden-Bosten: Brill; p. 43–76. [Google Scholar]
- Keeling CI, et al. 2013. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 14:R27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine L, Gotoh H, Brent CS, Dworkin I, Emlen DJ. 2015. Exaggerated trait growth in insects. Annu Rev Entomol. 60:453–472. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liénard MA, Strandh M, Hedenström E, Johansson T, Löfstedt C. 2008. Key biosynthetic gene subfamily recruited for pheromone production prior to the extensive radiation of Lepidoptera. BMC Evol Biol. 8:270.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maïbèche-Coisne M, Nikonov AA, Ishida Y, Jacquin-Joly E, Leal WS. 2004. Pheromone anosmia in a scarab beetle induced by in vivo inhibition of a pheromone-degrading enzyme. Proc Natl Acad Sci. 101: 11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov GV, Baskaran P, Sommer RJ. 2015. The same or not the same: lineage-specific gene expansions and homology relationships in multigene families in nematodes. J Mol Evol. 80:18–36. [DOI] [PubMed] [Google Scholar]
- Mayer WE, Schuster LN, Bartelmes G, Dieterich C, Sommer RJ. 2011. Horizontal gene transfer of microbial cellulases into nematode genomes is associated with functional assimilation and gene turnover. BMC Evol Biol. 11:13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughran A, Morgan K, Sommer RJ. 2013. Natural variation in chemosensation: lessons from an island nematode. Ecol Evol. 3:5209–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczek AP. 2009. On the origins of novelty and diversity in development and evolution: a case study on beetle horns. Cold Spring Harb Symp Quant Biol. 74:289–296. [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403:853–858. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Goldstone JV, Stegeman JJ. 2013. The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Philos Trans R Soc Lond B Biol Sci. 368:20120474.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler JG, editor. 1999. Modern coconut management: palm cultivation and products. Bourton: Practical Action. [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- Qiu Y, et al. 2012. An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc Natl Acad Sci. 109: 14858–14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale EJ, Kanzaki N, Herrmann M. 2015. Taxonomy and natural history: the genus Pristionchus In: Sommer RJ, editor. Pristionchus pacificus: a nematode model for comparative and evolutionary biology. Leiden-Bosten: Brill; p. 77–120. [Google Scholar]
- Richards S, et al. 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452:949–955. [DOI] [PubMed] [Google Scholar]
- Rödelsperger C, Dieterich C. 2010. CYNTENATOR: progressive gene order alignment of 17 vertebrate genomes. PLoS One 5:e8861.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C, Sommer RJ. 2011. Computational archaeology of the Pristionchus pacificus genome reveals evidence of horizontal gene transfers from insects. BMC Evol Biol. 11:239.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C, Streit A, Sommer RJ. 2013. Structure, function and evolution of the nematode genome In: eLS. Chichester: John Wiley & Sons, Ltd. doi: 10.1002/9780470015902.a0024603. [Google Scholar]
- Rödelsperger C, et al. 2014. Characterization of genetic diversity in the nematode Pristionchus pacificus from population-scale resequencing data. Genetics 196:1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux J, et al. 2014. Patterns of positive selection in seven ant genomes. Mol Biol Evol. 31:1661–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said I, et al. 2015. Identification of the aggregation pheromone of the date palm root borer Oryctes agamemnon. J Chem Ecol. 41:446–457. [DOI] [PubMed] [Google Scholar]
- Sandstrom P, Welch WH, Blomquist GJ, Tittiger C. 2006. Functional expression of a bark beetle cytochrome P450 that hydroxylates myrcene to ipsdienol. Insect Biochem Mol Biol. 36:835–845. [DOI] [PubMed] [Google Scholar]
- Scherkenbeck J, Zdobinsky T. 2009. Insect neuropeptides: structures, chemical modifications and potential for insect control. Bioorg. Med. Chem. 17:4071–4084. [DOI] [PubMed] [Google Scholar]
- Scholtz CH, Grebennikov VV. 2005. Scarabaeoidea Latreille, 1802 In: Beutel RG, Leschen R, editors. Coleoptera, Beetles. vol. 1: morphology and systematics (Archostemata, Adephaga, Myxophaga, Polyphaga partim). Berlin: de Gruyter Olenbourg. [Google Scholar]
- Schuster LN, Sommer RJ. 2012. Expressional and functional variation of horizontally acquired cellulases in the nematode Pristionchus pacificus. Gene 506:274–282. [DOI] [PubMed] [Google Scholar]
- Sheehan D, Meade G, Foley VM, Dowd CA. 2001. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 360:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, et al. 2012. Glutathione S-transferase (GST) genes in the red flour beetle, Tribolium castaneum, and comparative analysis with five additional insects. Genomics 100:327–335. [DOI] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ, McGaughran A. 2013. The nematode Pristionchus pacificus as a model system for integrative studies in evolutionary biology. Mol Ecol. 22:2380–2393. [DOI] [PubMed] [Google Scholar]
- Sommer RJ. 2015. Nematoda In: Wanninger A, editor. Invertebrate evo-devo. Wien: Springer; p. 15–34. [Google Scholar]
- Song M, et al. 2014. Exo-Brevicomin biosynthetic pathway enzymes from the mountain pine beetle, Dendroctonus ponderosae. Insect Biochem Mol Biol. 53:73–80. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Tzvetkova A, Morgenstern B. 2006. AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 7:S11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hiel MB, et al. 2010. Neuropeptide receptors as possible targets for development of insect pest control agents. Adv Exp Med Biol. 692:211–226. [DOI] [PubMed] [Google Scholar]
- Vega FE, et al. 2015. Draft genome of the most devastating insect pest of coffee worldwide: the coffee berry borer Hypothenemus hampei. Sci Rep. 5:12525.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinden H, et al. 2015. The pleiotropic allatoregulatory neuropeptides and their receptors: a mini-review. J Insect Physiol. 80:2–14. [DOI] [PubMed] [Google Scholar]
- Weller AM, Mayer WE, Rae R, Sommer RJ. 2010. Quantitative assessment of the nematode fauna present on Geotrupes dung beetles reveals species-rich communities with a heterogeneous distribution. J Parasitol. 96:525–531. [DOI] [PubMed] [Google Scholar]
- Wissler L, Gadau J, Simola DF, Helmkampf M, Bornberg-Bauer E. 2013. Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 5:439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, et al. 2013. Crystal structure of a Bombyx mori sigma-class glutathione transferase exhibiting prostaglandin E synthase activity. Biochim Biophys Acta. 1830:3711–3718. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer CT, et al. 2014. Molecular and functional characterization of CYP6BQ23, a cytochrome P450 conferring resistance to pyrethroids in European populations of pollen beetle, Meligethes aeneus. Insect Biochem Mol Biol. 45:18–29. [DOI] [PubMed] [Google Scholar]
- Zhu F, et al. 2010. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc Natl Acad Sci. 107:8557–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.