Abstract
Collembola (springtails) are detritivorous hexapods that inhabit the soil and its litter layer. The ecology of the springtail Orchesella cincta is extensively studied in the context of adaptation to anthropogenically disturbed areas. Here, we present a draft genome of an O. cincta reference strain with an estimated size of 286.8 Mbp, containing 20,249 genes. In total, 446 gene families are expanded and 1,169 gene families evolved specific to this lineage. Besides these gene families involved in general biological processes, we observe gene clusters participating in xenobiotic biotransformation. Furthermore, we identified 253 cases of horizontal gene transfer (HGT). Although the largest percentage of them originated from bacteria (37.5%), we observe an unusually high percentage (30.4%) of such genes of fungal origin. The majority of foreign genes are involved in carbohydrate metabolism and cellulose degradation. Moreover, some foreign genes (e.g., bacillopeptidases) expanded after HGT. We hypothesize that horizontally transferred genes could be advantageous for food processing in a soil environment that is full of decaying organic material. Finally, we identified several lineage-specific genes, expanded gene families, and horizontally transferred genes, associated with altered gene expression as a consequence of genetic adaptation to metal stress. This suggests that these genome features may be preadaptations allowing natural selection to act on. In conclusion, this genome study provides a solid foundation for further analysis of evolutionary mechanisms of adaptation to environmental stressors.
Keywords: Collembola, springtails, de novo genome assembly, gene family expansions, horizontal gene transfer, heavy metal tolerance
Introduction
The soil environment is home to a species-rich community of invertebrates which, in interaction with microorganisms and responding to specific physical and chemical soil factors, contribute to essential ecosystem services of the soil, such as organic matter degradation, nutrient cycling, disease antagonism, soil fertility and even human health (Wall et al. 2015). Despite the great ecological relevance of the soil community, our insight into molecular processes underlying the ecology and evolution of soil invertebrates is largely lagging behind. For some free-living nematode species genome assemblies have been reported (Dieterich et al. 2008; Hillier et al. 2005), while for Collembola a more limited molecular database is available (Timmermans et al. 2007; Faddeeva et al. 2015). Collembolans are an extremely abundant and species-rich component of the soil invertebrate community. Moreover, several species are susceptible to the effects of soil contamination and are often used for soil toxicity testing. An internationally standardized soil toxicity test has been developed for the species Folsomia candida (ISO 1999; Organisation for Economic Co-operation and Development 2009). In this article, we report the first draft genome of a soil-living collembolan, Orchesella cincta. We analyze the genome with the aim to shed light on genomic signatures of adaptation to the soil environment.
Orchesella cincta (fig. 1) is a member of the hexapod subclass Collembola (springtails), a group that shares the most recent common ancestor with the insects (Misof et al. 2014). It is an obligate sexually reproducing species with a diploid genome (2n = 12) (Hemmer 1990). The animals hatch from eggs deposited on the substrate and grow to an adult of about 6-mm length over a period of 6 weeks. During growth at 20 °C they molt about every 6 days but in contrast to insects, molting continues during adult life. O. cincta populations can be easily sampled in the field; its life-cycle and population structure have been studied extensively (Van Straalen 1985; Van der Wurff et al. 2003). At the same time, it is a suitable species for laboratory experiments, including gene knock-out and molecular studies of development (Konopova and Akam 2014).
Fig. 1.—
Orchesella cincta. Photo by Jan van Duinen. Both male and female O. cincta animals are on average 6 mm long and contain three thoracal- and six abdominal segments. Their brown-black slender body is covered with hair. They have a fully developed furca that is used for jumping and ventral tube, which is involved in osmoregulation. The mandibula of O. cincta is a chewing type with presented molar plate.
Interestingly, several previous studies provided evidence of adaptive evolution of stress tolerance to long-term metal pollution in populations living in metal-contaminated areas (Posthuma et al. 1993; Roelofs et al. 2007, 2009). The tolerant phenotype comprises increased excretion efficiency of heavy metals upon shedding its gut epithelium during molting, which is associated with an earlier time of reproduction and less growth reduction upon cadmium exposure (Posthuma et al. 1993). At the molecular level, adaptive evolution of metal tolerance is partly explained by transcriptional regulation of genes essential for metal detoxification. Among them are metallothionein, diapausin, and proteins involved in the stress-activated protein kinase pathway (Roelofs et al. 2009). For instance, it has been shown that metallothionein (mt), a protein that chelates heavy metals due to its high binding efficiency, is constitutively overexpressed in metal-tolerant O. cincta populations (Roelofs et al. 2007, 2009). This has demonstrated that heavy metal soil contamination can act as a selective pressure favoring phenotypes with high mt expression and causing an increased frequency of highly inducible mt promotor alleles in tolerant populations (Janssens et al. 2008). However, the mechanism by which such altered transcriptional responses can be selected still remains to be elucidated. Generating a genome sequence will be a valuable step in further studying the microevolution of stress responses in the metal-tolerant O. cincta populations. In a broader context, the genome sequence of a collembolan could provide more information on the evolution of this intriguing group of organisms and their adaptation to soil factors.
In this study, we describe the genome content of O. cincta. Furthermore, we examine gene families that have undergone significant expansions as well as genes and gene families that evolved in the O. cincta genome after horizontal gene transfer (HGT) events. Homologs of some of these genes and gene families were identified in previous studies to be involved in the evolution of the metal tolerance in O. cincta populations living in metal-contaminated areas (Roelofs et al. 2007, 2009). We discuss the functional significance of these genes in the light of the adaptive evolution of metal tolerance.
Materials and Methods
Sample Preparation and Sequencing
Orchesella cincta (Collembola, Entomobryidae) was cultured for many generations in the laboratories of the Department of Ecological Science, Vrije University Amsterdam. It is commonly raised in mass culture on twigs overgrown with green algae. The original population was sampled from the forest Roggebotzand, The Netherlands. Over the years, the stock population was regularly amended with new infusions from the same field site and occasionally from other forests in the Netherlands.
For Illumina sequencing, DNA was isolated from one single female of the 8th generation of an inbred line using the Promega SV genomic DNA purification system with some modifications described before (Roelofs et al. 2006). A paired-end O. cincta genome library was sequenced with 2 × 100 bp read lengths on the Illumina HiSeq2000 platform at the Leiden Genome Technology Center, resulting in overlapping forward and reverse reads. To improve the contiguity of the genome, we additionally generated long Pacific Bioscience (PacBio) reads. For this, 40 animals (males and females) were pooled and crushed in 300 µl Nuclei Lysis Solution (Promega). Genomic DNA was isolated using the Promega SV genomic DNA purification system as described by the manufacturer. An additional cleanup step was performed using phenol chloroform isoamyl alcohol extraction, followed by isopropanol precipitation described before (Sterenborg and Roelofs 2003). Finally, the DNA pellet was dissolved in H2O at a final concentration of 90 µg/ml DNA, and was sheared with Covaris G-tube and 15–20 kb fragments. These were sequenced using 12 single molecule real time cells on the PacBio RS II platform according to the manufacturer’s protocol at the Leiden Genome Technology Center.
Assembly and Pre-Processing
Raw Illumina sequencing data were pre-processed in the Trimmomatic tool v.0.32 (Bolger et al. 2014) with the following parameters: leading:20; trailing:20; slidingwindow:4:20; headcrop:10; minlen:20. Adapters were removed with the cutadapt tool v.1.8.1 (Martin 2011). We used the Blobology scripts (Kumar et al. 2013) and Kraken tool v.0.10.5 (Wood and Salzberg 2014) to identify and remove prokaryote, viral, human, yeast and fungal reads from the data. Sequencing errors in the raw reads were corrected with the SGA tool (Simpson and Durbin 2012) and duplicates were removed with the fastq-mcf tool (Aronesty 2011). Based on the cleaned Illumina reads we estimated the genome size with the BGI method (Liu et al. 2013).
SparseAssembler v.1beta (Ye et al. 2011) was applied (NodeCoveTh = 1, EdgeCovTh = 1, k = 21, g = 15, PathCovTh = 100) to build contigs based on Illumina data and the DBG2OLC tool (Ye et al. 2014) was used (KmerCovTh = 2, AdaptiveTh = 0.004, MinOverlap = 10, RemoveChimera = 1, k = 17) to complement the Illumina assembly with long PacBio reads. After the polishing step, we performed further scaffolding and gap filling using PBJelly v. 2.2.0 (English et al. 2012). Finally, we used HaploMerger (release: 20120810) (Huang et al. 2012) to discard all duplicate heterozygous contigs and to define the reference haploid assembly. The final polishing was done with the Pbdagcon (c = 2, m = 200) (Chin et al. 2013) and with the Pilon software (–fix bases, –diploid) (Walker et al. 2014). Finally, we used VecScreen against the UniVec database to remove the remaining vector contamination. The completeness of the genome was estimated with the core eukaryotic genes in the CEGMA pipeline v.2.4 (Parra et al. 2007) and with the core arthropods gene set in the BUSCO pipeline v.1.1 (Simão et al. 2015). De novo transcripts, generated in a previous study (Faddeeva et al. 2015), were mapped to the genome scaffolds using the isoblat tool v.3.0 with default parameters (Ryan 2013).
In addition, the mitochondrial genome was assembled with MITObim v.1.6 (Hahn et al. 2013) using the mitochondrial genome of another springtail Orchesella villosa (Carapelli et al. 2007) as reference. Annotation was performed with MITOS (Bernt et al. 2013) annotation web-tool. Further manual curation was performed based on read alignment.
Annotation
Structural genome annotation was performed in MAKER2 v.2.31.8 (Holt and Yandell 2011). Ab initio gene predictions were produced with SNAP (version 2006-07-28) (Korf 2004), Augustus v.3.1.0 (Stanke and Morgenstern 2005) and GeneMark (Lomsadze et al. 2005). Augustus and Genemark were trained with BRAKER1 (Hoff et al. 2015) and SNAP using two iterative runs of MAKER2 (est2genome = 1 and protein2genome = 1). Cleaned lllumina paired-end RNA-seq data (SRR935330, Faddeeva et al. 2015) aligned with the TopHat2 from Tuxedo package (read-mismatches = 8, read-gap-length = 4, read-edit-dist = 8) (Trapnell et al. 2012). O. cincta transcripts (Faddeeva et al. 2015) and proteomes of Daphnia pulex, Tribolium castaneum, Pediculus humanus, Acyrthosiphon pisum, and Drosophila melanogaster from the Ensembl Genomes database were used as evidence for the annotation. We used the de novo repeat library constructed in RepeatModeler (Smit and Hubley 2014) and the RepBase database (Jurka et al. 2005) for repeat identification in the RepeatMasker tool (Tarailo‐Graovac and Chen 2009). We applied BlastP (Basic Local Alignment Search Tool) (Altschul et al. 1990) with an E-value threshold of 0.1 against SwissProt and TrEMBL databases to assign homology-based gene functions. We performed an InterProScan (Quevillon et al. 2005) search against the Superfamily (Wilson et al. 2009) and Pfam (Finn et al. 2014) protein databases to identify protein domains. GO annotations were assigned to protein sequences in the Blast2GO suite v.3.1 based on InterPro domains and a BlastP against Swiss-Prot database (with an E-value threshold of 1e−5).
Ortholog Clustering and Gene Family Analysis
In order to assess gene family size evolution in O. cincta we first predicted orthologs clusters shared between the insects T. castaneum, P. humanus, A. pisum, D. melanogaster and Aedes aegypti, together with the centipede Strigamia maritima, the arachnids Ixodes scapularis, and Tetranychus urticae, the branchiopod D. pulex, the nematode Caenorhabditis elegans and the Entognatha O. cincta in OrthoMCL version 1.4 (Li et al. 2003). We used the E-value threshold cut-off of 1e−5 and a matching percentage of ≥50%. The method proposed by Cao et al. (2013) was applied to identify significant expansions based on z-scores in O. cincta. For gene families represented by at least three species besides O. cincta, we calculated z-score as: (the gene number for each family − the average gene number of the family from all species)/the standard deviation of gene numbers of the family from all species. The families with z-scores ≥2 were considered as significantly expanded.
Horizontal Gene Transfer
HGT analysis was performed by calculating the HGT index h based on the protocol described by Crisp et al. (2015) with a number of modifications. Two gene categories were defined instead of three: native genes and potential cases of HGT. Genes with an h score <30, or with an h score ≥30 and best nonmetazoan bit score <100 were considered native genes. A gene was considered a candidate case of HGT if h ≥ 30 and if the best nonmetazoan bit score was ≥100. To confirm integration in O. cincta’s genome, a genome linkage test was performed by inspecting the mapping of HGT candidates to genomic scaffolds ascertaining that (1) native genes were present on the same scaffold and (2) the alignments of the PacBio long reads confirmed the linkage of the foreign gene with the native DNA. The HGT candidates with best metazoan bit score <50 that passed the genome linkage test were considered as confirmed cases of HGT; those with best metazoan bit score ≥50 were subjected to phylogenetic validation to verify their nonmetazoan origin. For the phylogenetic validation, we performed BlastP of HGT candidates against the following Uniprot databases: Fungi, Metazoa (excluding Arthropoda), Bacteria, Archaea, Plants, Arthropoda, and Protists with an E-value threshold of 1e−5. The top five hits for each database were aligned with the HGT candidate from O. cincta with the Muscle tool (Edgar 2004). Alignments were trimmed using TrimAl (Capella-Gutiérrez et al. 2009) with the gt = 0.6 option. We used PhyML (Guindon and Gascuel 2003) to build trees with aLRT SH branch support. HGT was confirmed when metazoan sequences were absent in the monophyletic group formed by the HGT candidate and the potential donor taxa. Subsequently, we performed a GO enrichment analysis for the genes that were confirmed to be horizontally acquired with the elim algorithm in the topGO package (Alexa et al. 2006) in R (v.3.2.2); p values <0.05 were considered significantly enriched. Furthermore, we determined whether there is a difference in the proportion of horizontally transferred genes that encode enzymes (genes with EC annotation) compared with native genes using a chi-squared test. Finally, we performed a BlastN search of genes that showed an interaction effect between cadmium exposure and population in a previous microarray study (Roelofs et al. 2009) against expanded, lineage-specific and HGT gene families with an E-value threshold of 1e−5. Finally, a chi-square test was performed to verify whether genes linked to metal tolerance are significantly overrepresented among expanded gene families (proportional to nonexpanded gene families), lineage-specific gene families (proportional to nonlineage-specific gene families), and among HGT genes (proportional to native genes).
Results
Orchesella cincta Genome
We have assembled the draft genome of O. cincta with an estimated genome size of 283.8 Mbp. This is in concordance with previously reported O. cincta haploid genome size based on flow cytometry of sperm cells (Janssens 2008); in this study two peaks were identified corresponding to sizes of 217 and 270 Mbp, the smaller peak most likely indicating the absence of one sex chromosome. Genome assembly was performed using 9.8 Giga basepairs (Gbp) of high-quality paired-end corrected Illumina HiSeq2000 reads (22.6 Gbp before correction) and 6.6 Gbp PacBio long reads (with average read size 3,808.6 kbp). The draft assembly comprises 9,402 scaffolds with a total sequence length of 286.8 Mbp, an N50 of 65.9 kbp with a maximum sequence length of 807.1 kbp and GC content of 36.8% (table 1). Moreover, 184,664 repeats were identified, representing 15% of the genome. Largest categories are represented by simple repeats (51.6% of total amount of repeats), repeats that could not be classified (25.2%), low complexity repeats (6.1%) and Gypsy LTR retrotransposons (3.5%) (supplementary table S1, Supplementary Material online).
Table 1.
Orchesella cincta Genome Properties
| Assembly | |
|---|---|
| Total sequences | 9,402 |
| Total bases (Mbp) | 286.8 |
| Min sequence length (bp) | 340 |
| Max sequence length (kbp) | 807.1 |
| N50 length (kbp) | 65.9 |
| GC % | 36.8 |
| N % | 0.01 |
| Structural annotation | |
| Genes | 20,249 |
| Mean gene length (bp) | 2,990.7 |
| Exon (%) | 12.6 |
| Intron (%) | 8.5 |
| Repeats (%) | 15.0 |
| Functional annotation | |
| Swiss | 14,909 |
| TrEMBL | 15,954 |
| InterPro | 14,565 |
| GO | 14,438 |
| EC | 4,550 |
| Validation | |
| CEGMA complete | 231 (93.1%) |
| GEGMA partial | 246 (99.2%) |
| CEGMA 458 set | 446 (97.4%) |
Validation using CEGMA scores indicated that the gene content is adequately covered by this assembly: 246 out of 248 (99.2%) and 446 out of 458 (97.4%) core eukaryotic CEGMA genes are present in the O. cincta genome. Furthermore, 30,970 (95.5%) de novo transcripts generated in a previous study (Faddeeva et al. 2015), could be mapped to the assembled scaffolds.
In this assembly, we predicted 20,249 genes of which 88.6% were supported by RNA-Seq data with FPKM values more than 0.5. This resulted in a gene density of 70.6 genes per Mbp, and the coding regions cover 12.6% of the genome. Interpro domains could be retrieved for 71.9% of protein-coding genes. Among all proteins in O. cincta, 3,905 sequences (19.3%) do not show any similarity with proteins in SwissProt or TrEMBL databases. Gene ontology terms (GO) histogram (level 2) provides a general overview of biological processes (BPs) among the predicted protein sequences (supplementary fig. S1, Supplementary Material online). Also, a subset of 4,550 (22.5%) proteins is supported by Enzyme codes (EC). Plotting these codes onto metabolic pathways with iPATH 2.0 (Yamada et al. 2011) indicates that most of the essential metabolic pathways are present in the dataset (supplementary fig. S2, Supplementary Material online).
In addition, we reconstructed the complete 15.7 kbp mitochondrial genome (supplementary table S2, Supplementary Material online). The mitochondrial genome shows the well-known A + T bias (72.5%), which is lower than in other insect species but higher when compared to other arthropods (Nardi et al. 2001). The A + T content in O. cincta is slightly higher than in O. villosa (72.2%). A BlastN search (dc-megablast) of O. cincta against O. villosa shows a sequence identity of 75.2% and 75.8% alignment coverage, suggesting a large sequence variation in the genus. However, both size and gene content seem to be similar to the previously sequenced collembolan mitochondrial genomes: 13 protein-coding genes were identified, as well as 22 tRNAs and two rRNAs (Nardi et al. 2001; Carapelli et al. 2007).
Orthologs Clustering and Gene Family Analysis
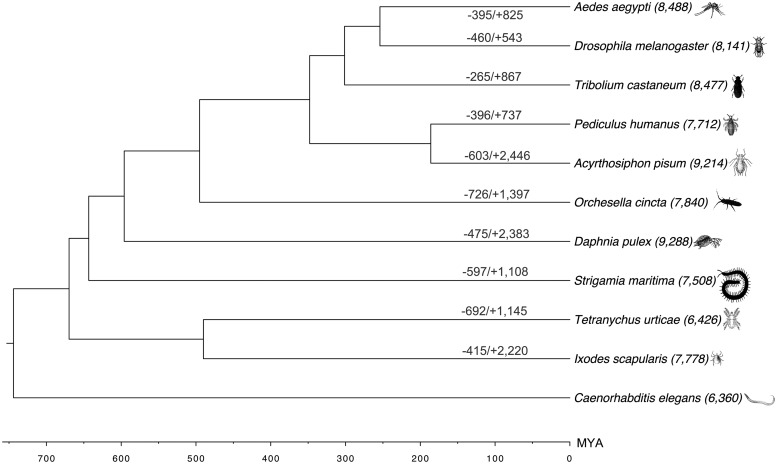
Orthologous gene clusters among 11 species with well-characterized genomes were compared to O. cincta gene to assess gene family loss and gain events (fig. 2). In total, we identified 20,513 orthologous clusters in the 11 species (supplementary table S3, Supplementary Material online), of which 2,416 were shared among all 11 species (fig. 2). From 20,249 predicted O. cincta proteins, orthology analysis clustered 14,378 protein sequences (71%) into 7,840 orthologous groups whereas 5,871 (29%) sequences remained unclassified. We found that 1,169 clusters were lineage-specific for O. cincta (supplementary table S4, Supplementary Material online).
Fig. 2.—
Gene gain and loss analysis in O. cincta genome. The species tree was built by using species distances from TimeTree database (Hedges et al. 2006) Common Taxonomy Tree at NCBI. The divergence time between species is marked in million years. A total number of gene families, gene family gain (+) and loss (−) are indicated. The loss of a gene family is identified in species when a gene family exists in the neighboring branch and the out-group, but not in itself. The gene families in most recent common ancestor were defined as (1) those shared by two direct descendants and (2) shared by each of the direct descendants and the out-group (Cao et al. 2013).
Gene Family Expansions and Metal Tolerance
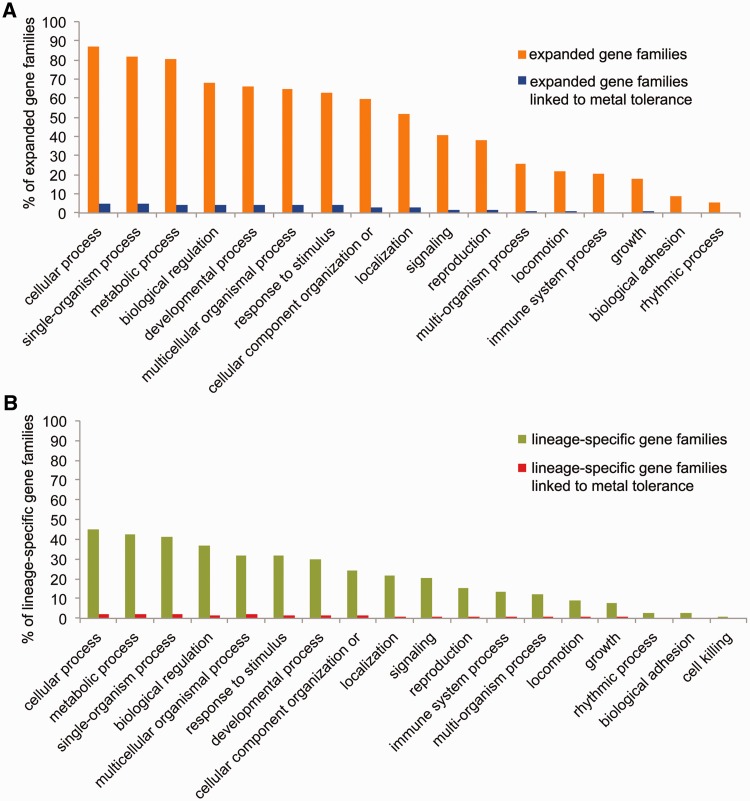
We identified 446 cases of gene family expansion in O. cincta (supplementary table S5, Supplementary Material online). The percentage of GO BPs among expanded gene clusters is represented relative to a total number of expanded gene families in the O. cincta genome in figure 3A. The top ten largest expanded gene families in O. cincta (with 15–80 genes) include two orthologous clusters of cytochrome P450s/monooxygenases (part of xenobiotic metabolism phase I), gene families involved in lipid metabolism (O-acyltransferases, lipases), transposition (AC transposases), sugar transport (solute carrier 35 proteins), CAP protein family (Golgi-associated plant pathogenesis-related proteins), immune system processes (lysosome membrane proteins), and signaling (FMRFamide receptor proteins). Previously, we showed that O. cincta populations living at metal-contaminated field sites evolved tolerance to nonessential heavy metals. This tolerance phenotype was associated with altered expression of genes involved in stress response, signaling and immune response (Roelofs et al. 2007, 2009). It is interesting to know whether genes that were previously linked to the tolerance phenotype based on transcript abundance (microarray- and, Q-PCR analysis), could be identified among the clusters that show gene family expansion. Indeed, we identified 25 expanded gene clusters that could be linked to metal tolerance (supplementary table S5, Supplementary Material online). However, the chi-square test did not confirm a significant enrichment of metal-tolerant genes among expanded gene families when compared to nonexpanded gene families (P = 0.14). Among the O. cincta clusters represented by at least ten genes, cytochrome P450s, lipases, proteins with a chitin-binding domain and NADP-dependent oxidoreductases (oxidation-reduction) were identified (associated GO BPs are summarized in fig. 3A). Cytochrome P450s as well as ATP-binding cassette (ABC) transporters participate in xenobiotic metabolism and have undergone substantial gene family expansion in O. cincta (supplementary table S5, Supplementary Material online). Previously, we have shown that a cDNA homologous to immune response-related cd36 receptor protein croquemort was highly inducible by cadmium exposure and showed high constitutive expression in tolerant animals (Roelofs et al. 2007). This cDNA matches with a gene family that contains two lineage-specific members in the O. cincta genome. Moreover, a highly related protein family with similar annotation (cd36, croquemort) is expanded up to eight members.
Fig. 3.—
(A) The proportion of expanded gene families (orange) and expanded gene families linked to metal tolerance (blue) annotated with gene ontology BPs relative to the total number of expanded gene families in O. cincta. (B) The proportion of lineage-specific gene clusters (green) and lineage-specific gene clusters associated with metal tolerance (red) annotated with gene ontology BPs relative to the total number of lineage-specific gene families in O. cincta.
Lineage-Specific Families
We identified 1,169 O. cincta lineage-specific gene families. The top ten of largest gene families include ortholog clusters containing 34–71 genes in O. cincta, that are mainly involved in ubiquitination (BTB/POZ domain-containing proteins, Speckle-type POZ proteins, E3 ubiquitin-protein ligases), transport (SEC14-like proteins), proteolysis (bacillopeptidases), chitin metabolism (putative chitinases), immune system process (C-type lectins), oxidation-reduction (glucose dehydrogenases, DBH-like monooxygenases) as well as CUB domain containing proteins (fig. 3B). Among other lineage-specific gene clusters with at least five genes, we also observed genes involved in the general stress response (coding for heat-shock protein 70s), phase I xenobiotic metabolism (cytochrome p450s and carboxylesterases), phase II xenobiotic metabolism (glutathione S-transferases), and cellular antioxidants (catalases). Again, we identified 77 gene clusters supported by transcripts that show a population-specific gene expression pattern upon cadmium treatment (supplementary table S5, Supplementary Material online, fig. 3B), and thus may potentially be a preadaptation that allowed metal tolerance to evolve in O. cincta populations living at metal-contaminated sites. Noteworthy, the chi-square test indicates that O. cincta lineage-specific gene families are significantly enriched with genes involved in metal tolerance when compared to gene clusters shared with other animals (P = 1.71e−05). Among them, gene families that are represented by at least ten genes include proteins involved in proteolysis (e.g., bacillopeptidases), chitin metabolism (chitinases), oxidation-reduction (e.g., DBH-like monooxygenases, glucose dehydrogenases), calcium-dependent phospholipid binding (C2-domain containing proteins) and lipid metabolism (putative fatty acid elongation proteins). Finally, we identified a lineage-specific gene family of ABC transporters and diapausins (antifungal enzymes), for which the differential gene expression pattern was previously shown to be associated with the metal-tolerant phenotype (Roelofs et al. 2009).
Horizontal Gene Transfer
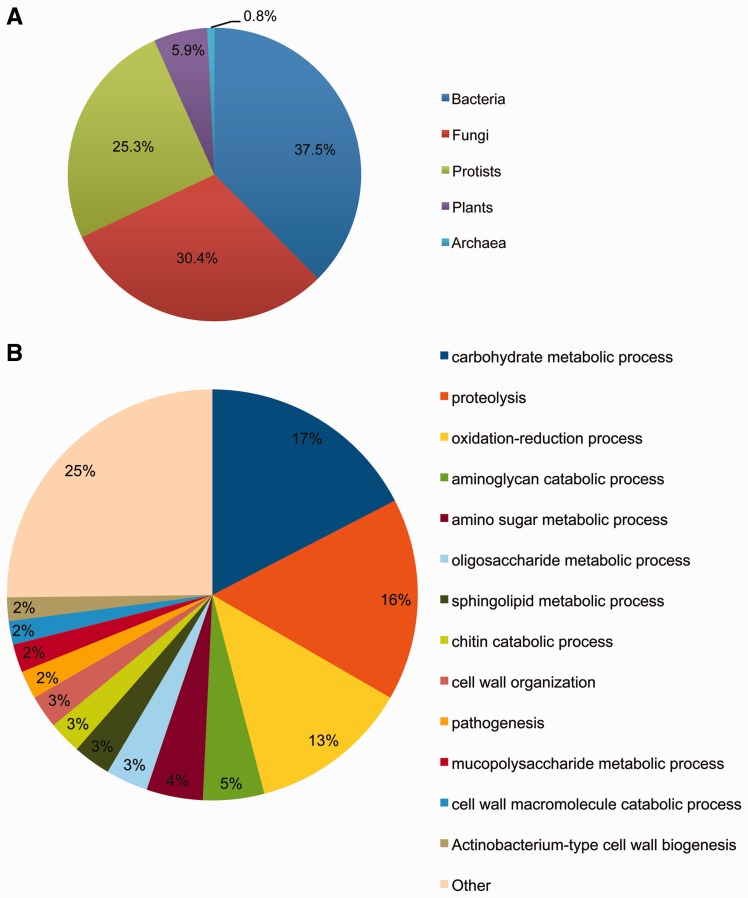
HGT analysis was conducted by calculating h-scores for all 20,249 genes in O. cincta. As expected, most of the genes (19,876) were classified as native, but 373 were classified as potential HGT candidates. The subsequent genome linkage test revealed that 336 genes were located on a genomic scaffold next to native genes. The remaining 37 genes were not linked to native genes and were discarded from further analysis. The low metazoan bit score of 83 candidates directly confirmed their status as horizontally transferred genes (Crisp et al. 2015). The remaining 253 candidates were subjected to a phylogenetic test. From these, 170 genes (67.2%) passed the test (Supplementary file S1 online) and were confirmed as horizontally transferred. In total, 253 genes were identified to have evolved in the O. cincta genome after HGT (supplementary table S6, Supplementary Material online), which constitutes ∼1.2% of all genes in O. cincta. Figure 4A shows the origin of these foreign genes. Bacterial origin is suggested for 37.5% of the HGT genes while 30.4% seems to originate from a fungal source.
Fig. 4.—
(A) Origin of foreign genes in O. cincta. The figure indicates what percentage of foreign genes in O. cincta originates from each of the donor groups. (B) Gene ontology terms associated with foreign genes. The figure shows the percentage of GO terms associated with foreign genes in O. cincta. The group ‘other’ includes categories represented by less than five genes (see supplementary table S7, Supplementary Material online) and accounts for 25% of the genes.
GO enrichment analysis demonstrates that 40 BPs and 35 molecular functions are enriched in the set of horizontally transferred genes (supplementary table S7, supplementary Material online) when compared with the complete set of predicted genes in O. cincta. The largest functional categories (fig. 4B) include genes involved in carbohydrate metabolism (47 genes), proteolysis (43 genes), and oxidation-reduction processes (34 genes). Most genes in the ‘proteolysis’ category are annotated as bacillopeptidases. In the ‘oxidation-reduction’ category, genes are identified to be involved in amine metabolism, response to oxidative stress, purine biosynthesis, detoxification of lignin-derived products, fatty acid biosynthesis, and aromatic compound degradation. The top five significant GO BP terms are related to chitin catabolism, cell wall macromolecule catabolism, actinobacterium-like cell wall biogenesis, carbohydrate metabolism and nitrogen utilization. The category ‘cell wall catabolism’ is represented by chitinases, genes involved in cell separation, and genes that break down peptidoglycan (the main component of bacterial cell walls). The category ‘carbohydrate metabolism’ also includes many genes that participate in the degradation of components of the cell walls of plants, bacteria or fungi (chitinase, arabinosidase, lysozyme, and beta-glucanase). Interestingly, 36 HGT genes (14.2%) show homology to transcripts that were previously associated with metal stress and evolution of the metal tolerance (Roelofs et al. 2009) (supplementary table S7, Supplementary Material online). The chi-square test reveals a significant enrichment of genes involved in metal tolerance among the HGT genes when compared to native genes (P = 3.65e−31). Finally, the bacillopeptidase gene family seems to have undergone expansion after HGT (see section above on gene family expansion).
Discussion
Here, we present the first genome of a collembolan species, O. cincta. Collembola belong to a primitive group of monophyletic hexapods that shares the most recent common ancestor with all insects (Misof et al. 2014). We show that both genome size and gene content are within the range expected for arthropods and do not seem to deviate from the values observed in other arthropod species used in the comparative analysis. O. cincta seems to have lost a substantial number of gene families when compared with other analyzed arthropods (fig. 2). At the same time, O. cincta shows a high level of lineage-specific gene gains when compared with other hexapods. This may indicate a substantial level of gene turn over, comparable with previous observations in the genomes of Diptera (Tautz and Domazet-Loso 2011). Around 19.3% of translated genes in O. cincta do not show homology to any other organisms, which is substantially less than in fast evolving genomes like those of D. pulex, where 36% proteins have no homologs (Colbourne et al. 2011). Daphnia is currently, the most closely related species to springtails with an available high quality annotated genome (Misof et al. 2014). Additional genome information is needed for Collembola genomes, as well as Diplura and Protura genomes, to more precisely estimate the percentage of lineage-specific genes because such estimation depends strongly on the phylogenetic distance with more related organisms.
Collembola are typically soil-dwelling animals. More specifically, O. cincta thrives in the soil litter layer, a habitat rich in plant decaying material, fungal and bacterial activity. High levels of secondary metabolites accumulate in this environment. For instance, a decay of lignocellulose gives rise to potentially toxic lignin polymers (phenols) and small organic acids (Van der Pol et al. 2014). Such compounds are known to have toxic properties and are even used as bio-control agents against insects (Broza et al. 2001). Broza et al. (2001) showed that Collembola are nonsusceptible to such compounds, suggesting that they have well-evolved detoxification systems to facilitate this. Enzymes involved in xenobiotic metabolism are known to be involved in detoxification of plant- and microbial toxins and their evolution in insects is associated with insecticide resistance (Feyereisen 1999). Such xenobiotic biotransformation can be subdivided into three phases. During phase I, cytochrome P450s facilitate water solubility of the compounds, which therefore become more reactive. In phase II glutathione S-transferases catalyze the synthesis of glutathione needed for conjugation of the reactive metabolite. In phase III ABC-transporters recognize these conjugates and export them outside the cell. Orchesella’s most expanded gene families comprise the phase I enzymes, cytochrome P450s. However, also, several phase II and phase III proteins are among expanded or lineage-specific gene families, for example, carboxylesterases, ABC transporters and glutathione S-transferases. These gene families, which are responsible for metabolic resistance to insecticides, have also undergone considerable expansion in the genome of Anopheles gambiae (Ranson et al. 2002). Further studies should elucidate whether these gene family expansions have evolved in O. cincta in order to facilitate living in the soil litter layer.
Orchesella cincta exerts some remarkable features with regard to genetic adaptation to stress from the environment. Several previous studies (Roelofs et al. 2007, 2009, 2010) provide evidence for microevolution of the metal tolerance in populations living in areas contaminated with heavy metals (cadmium and lead). A growing body of evidence points towards transcriptional regulatory evolution facilitating the constitutive overexpression of genes important in stress response, metal detoxification, immune response, signaling and chromatin remodeling (Roelofs et al. 2007, 2009, 2010). Here, we identified that some of these genes, altered in expression pattern, are also expanded in the genome. For instance, diapausins could provide protection against cadmium uptake as follows. The peptide is known to exert antifungal activity by blocking Ca2+ channels (Kouno et al. 2007). Since toxic heavy metals, such as cadmium, are actively taken up by Ca2+ channels, we have previously (Roelofs et al. 2009) suggested that blocking these channels could prevent uptake of this toxic metal aiding in the metal-tolerant phenotype.
Previously, we provided evidence that specific variants in the promotor of the metal detoxifying protein metallothionein (mt) contribute to an over-expression phenotype in cadmium tolerant populations (Janssens et al. 2007). The genome of Orchesella contains only one copy of mt (Sterenborg and Roelofs 2003), suggesting that cis-regulatory evolution is the most plausible explanation for constitutive overexpression of this gene in metal tolerant animals. However, the current study identified expanded gene families that are associated with metal tolerance. This suggests that amplification of genes could be a preadaptation, by which selection can act on so that gene expression can be induced by the coordinated expression of paralogs. This was, for instance, shown in D. pulex (Colbourne et al. 2011; Shaw et al. 2007). Whether amplified gene families could also contribute to metal stress adaptation in O. cincta, can only be investigated when we re-sequence tolerant genotypes both at genome as well as at transcriptome level. So, further investigation should be done to understand this mechanism.
Although HGT for a long time was thought to be more common among prokaryotes, many cases of HGT in eukaryotes have now been described (Chapman et al. 2010; Boschetti et al. 2012; Crisp et al. 2015). Some invertebrates seem to have exceptionally high levels of foreign genes in their genomes. For example, 9.5% of the transcribed sequences in the bdelloid rotifer Adineta vaga originated from HGT (Boschetti et al. 2012). The O. cincta genome contains a substantially lower number of foreign genes (1.2%). Still, this percentage is higher compared to Hydra magnipappillata (0.4%) (Chapman et al. 2010), Caenorhabditis spp. (0.4%), Drosophila spp. (0.1%) and primates (0.3%) (Crisp et al. 2015). The majority of foreign genes in O. cincta (37.5%) originate from bacteria (fig. 4A), which is similar to Caenorhabditis (46.2%) and bdelloid rotifers (59%) (Crisp et al. 2015). In Drosophila and primates, instead, most HGT events came from protists (46.5% and 57.6%) (Crisp et al. 2015), that are also important sources of HGT genes in O. cincta (25.3%). Notably, in O. cincta, we observe a high number of HGT genes (30.4%) with a fungal origin (fig. 4A). Also in the case of bdelloid rotifers fungi represent a high share (23%) of the foreign genes (Boschetti et al. 2012), however in other species fungi contribute less to HGT—only 9.9–11.9% of foreign genes in Drosophila, Caenorhabditis, and primates (Crisp et al. 2015). HGT genes in O. cincta participate mostly in cell wall degradation and carbohydrate metabolism. It is tempting to speculate that the capacity to acquire control over cell wall breakdown provides a selective advantage in the soil litter layer that is particularly rich in plant cell wall degradation products. It may provide and important food source to thrive in such habitat. Of course, this needs to be underpinned by experimental evidence.
An interesting observation is that we identified significantly more (P = 9.04e−15) EC among horizontally transferred genes (42.7%) than among native genes (22.2%). This is in contrasts to informational genes (i.e., genes involved in transcription, translation, and related processes), which were not observed within the HGT gene set. This result is in accordance with the complexity hypothesis that suggests that operational genes (genes involved in metabolism/genes encoding for enzymes) are more likely to be successfully integrated into a host genome following a HGT event than informational genes (Jain et al. 1999). Crisp et al. (2015) indeed suggest that the transfer of metabolic genes contributes to biochemical diversification during animal evolution. It is possible that the acquisition of foreign genes has conferred new functions that played a role in adapting to the chemical environment of the soil litter layer (Moran and Jarvik 2010).
Finally, we also checked whether HGT genes are linked to the metal tolerance phenotype in O. cincta populations living in metal-contaminated areas. Again, we used microarray data of cadmium exposure to an O. cincta sensitive and tolerant population from Roelofs et al. (2009) to identify HGT genes that show significant population-specific expression under metal stress conditions and therefore contribute to the tolerant phenotype. This analysis suggests that a significant number of horizontally transferred genes not only retained their activity after transfer but could also be a target of selection to evolve adaptation to metal stress.
Conclusion
Here, we present the first genome of a collembolan. This work builds a foundation for further comparative genomics of springtails and yields new insights into the evolution of Collembola. The analysis of gene families suggests that expanded and lineage-specific gene clusters are mostly involved in general BPs, however, some of these clusters were shown to be involved in xenobiotic biotransformation pathway and biotic response (immune response, antifungal activity by blocking Ca2+ channels). We speculate that some expanded gene families facilitate successful occupation of the soil litter layer. We also identified horizontally transferred genes in O. cincta genome. Most of them originate from bacteria, but we also identified an exceptionally high number of foreign genes derived from a fungal source. Interestingly, these horizontally transferred genes are mostly involved in carbohydrate metabolism, which may also be beneficial for life in an environment rich in decaying organic matter. Subsets of expanded gene families, as well as HGT genes, could be linked to heavy metal tolerance in O. cincta. This suggests that HGT, an evolution of novel genes, and gene duplication could be a preadaptation facilitating an evolution of metal stress tolerance in populations living at metal-contaminated sites.
Supplementary Material
Supplementary figures S1 and S2, tables S1–S7 and file S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Dr. Romain Studer for constructing the species tree for the gene family evolution analysis, Janine Mariën for genomic DNA isolation, Dr. Ken Kraaijeveld for advice on genome assembly, Peter H. Neleman for help with python scripts, Prof. Dr. Johan den Dunnen for facilitating next-generation sequencing at the LGTC, Dr. Tjalf de Boer for help with GO enrichment analysis, Ana I. Belo and Mima Malcicka for help with text editing. We thank Mathijs Kattenberg and the SURFsara support team for assistance with the calculations on the SURFsara LISA cluster and SURFsara cloud. We also thank Roland den Hollander and the VU “IT for Research” support team for assistance with website hosting. This research was financed by grant F08.001.03 from the BE-BASIC foundation. Dick Roelofs receives additional funding from EU FP7 program Sustainable Nanotechnologies (SUN), grant agreement number 604305.
Literature Cited
- Alexa A, Rahnenfuhrer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22:1600–1607. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Aronesty E. 2011. ea-utils: Command-line tools for processing biological sequencing data. Durham (NC: ): Expression Analysis. [Google Scholar]
- Bernt M, et al. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschetti C, et al. 2012. Biochemical diversification through foreign gene expression in bdelloid rotifers. PLoS Genet. 8:e1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broza M, Pereira RM, Stimac JL. 2001. The nonsusceptibility of soil Collembola to insect pathogens and their potential as scavengers of microbial pesticides. Pedobiologia 45:523–534. [Google Scholar]
- Cao Z, et al. 2013. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat Commun. 4:2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapelli A, Liò P, Nardi F, Van der Wath E, Frati F. 2007. Phylogenetic analysis of mitochondrial protein coding genes confirms the reciprocal paraphyly of Hexapoda and Crustacea. BMC Evol Biol. 7:S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JA, et al. 2010. The dynamic genome of Hydra. Nature 464:592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C-S, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, et al. 2011. The ecoresponsive genome of Daphnia pulex. Science 331:555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Common Taxonomy Tree [Internet]. [cited 2016 Jun 9]. Available from: http://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi.
- Crisp A, Boschetti C, Perry M, Tunnacliffe A, Micklem G. 2015. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 16:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C, et al. 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet. 40:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7:e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensembl Genomes [Internet]. [cited 2016 Jun 9]. Available from: http://ensemblgenomes.org.
- Faddeeva A, et al. 2015. Collembolan transcriptomes highlight molecular evolution of hexapods and provide clues on the adaptation to terrestrial life. PLoS One 10:e0130600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyereisen R. 1999. Insect P450 enzymes. Annu Rev Entomol. 44:507–533. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22:2971–2972. [DOI] [PubMed] [Google Scholar]
- Hemmer W. 1990. Karyotype differentiation and chromosomal variability in springtails (Collembola, Insecta). Biol Fert Soils 9:119–125. [Google Scholar]
- Hillier LW, et al. 2005. Genomics in C. elegans: so many genes, such a little worm. Genome Res. 15:1651–1660. [DOI] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. 2015. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32:767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, et al. 2012. HaploMerger: reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res. 22:1581–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISO 1999. 11267: Soil Quality. Inhibition of Reproduction of Collembola (Folsomia candida) by Soil Pollutants. International Organization for Standardization. Geneva, Switzerland.
- Jain R, Rivera MC, Lake JA. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci. 96:3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens TK, et al. 2007. Recombinational micro-evolution of functionally different metallothionein promoter alleles from Orchesella cincta. BMC Evol Biol. 7:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens TK, et al. 2008. Comparative population analysis of metallothionein promoter alleles suggests stress-induced microevolution in the field. Environ Sci Technol. 42:3873–3878. [DOI] [PubMed] [Google Scholar]
- Janssens TKS. 2008. The role of transcriptional regulation in the micro-evolution of heavy metal tolerance in Orchesella cincta (Collembola). Amsterdam: Vrije University Amsterdam. [Google Scholar]
- Jurka J, et al. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110:462–467. [DOI] [PubMed] [Google Scholar]
- Konopova B, Akam M. 2014. The Hox genes Ultrabithorax and abdominal-A specify three different types of abdominal appendage in the springtail Orchesella cincta (Collembola). EvoDevo. 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouno T, et al. 2007. The structure of a novel insect peptide explains its Ca2+ channel blocking and antifungal activities. Biochemistry 46:13733–13741. [DOI] [PubMed] [Google Scholar]
- Kumar S, Jones M, Koutsovoulos G, Clarke M, Blaxter M. 2013. Blobology: exploring raw genome data for contaminants, symbionts and parasites using taxon-annotated GC-coverage plots. Front Genet. 4:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, et al. 2013. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv:1308.2012.
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33:6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12. [Google Scholar]
- Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763–767. [DOI] [PubMed] [Google Scholar]
- Moran NA, Jarvik T. 2010. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 328:624–627. [DOI] [PubMed] [Google Scholar]
- Nardi F, Carapelli A, Fanciulli PP, Dallai R, Frati F. 2001. The complete mitochondrial DNA sequence of the basal hexapod Tetrodontophora bielanensis: evidence for heteroplasmy and tRNA translocations. Mol Biol Evol. 18:1293–1304. [DOI] [PubMed] [Google Scholar]
- Organisation for Economic Co-operation and Development O. 2009. Test No. 232: Collembolan Reproduction Test in Soil: OECD Publishing.
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- pbdagcon [Internet]. [cited 2016 Jun 9]. Available from: https://github.com/PacificBiosciences/pbdagcon.
- Posthuma L, Hogervorst RF, Joosse EN, Van Straalen NM. 1993. Genetic variation and covariation for characteristics associated with cadmium tolerance in natural populations of the springtail Orchesella cincta (L.). Evolution 619–631. [DOI] [PubMed] [Google Scholar]
- Quevillon E, et al. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson H, et al. 2002. Evolution of supergene families associated with insecticide resistance. Science 298:179–181. [DOI] [PubMed] [Google Scholar]
- Roelofs D, et al. 2009. Adaptive differences in gene expression associated with heavy metal tolerance in the soil arthropod Orchesella cincta. Mol Ecol. 18:3227–3239. [DOI] [PubMed] [Google Scholar]
- Roelofs D, Marien J, Van Straalen NM. 2007. Differential gene expression profiles associated with heavy metal tolerance in the soil insect Orchesella cincta. Insect Biochem Mol Biol. 37:287–295. [DOI] [PubMed] [Google Scholar]
- Roelofs D, Morgan J, Stürzenbaum S. 2010. The significance of genome-wide transcriptional regulation in the evolution of stress tolerance. Evol Ecol. 24:527–539. [Google Scholar]
- Roelofs D, Overhein L, De Boer M, Janssens T, Van Straalen N. 2006. Additive genetic variation of transcriptional regulation: metallothionein expression in the soil insect Orchesella cincta. Heredity 96:85–92. [DOI] [PubMed] [Google Scholar]
- Ryan JF. 2013. Baa. pl: a tool to evaluate de novo genome assemblies with RNA transcripts. arXiv preprint arXiv:1309.2087.
- Shaw JR, et al. 2007. Gene response profiles for Daphnia pulex exposed to the environmental stressor cadmium reveals novel crustacean metallothioneins. BMC Genomics 8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–2112. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Durbin R. 2012. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 22:549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R. 2014. RepeatModeler [Internet]. [cited 2016 Jun 9]. Available from: http://www.repeatmasker.org/RepeatModeler.html.
- Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33:W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterenborg I, Roelofs D. 2003. Field-selected cadmium tolerance in the springtail Orchesella cincta is correlated with increased metallothionein mRNA expression. Insect Biochem Mol Biol. 33:741–747. [DOI] [PubMed] [Google Scholar]
- Tarailo‐Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinfo. 4(10):11–14. [DOI] [PubMed] [Google Scholar]
- Tautz D, Domazet-Loso T. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12:692–702. [DOI] [PubMed] [Google Scholar]
- Timmermans MJ, et al. 2007. Collembase: a repository for springtail genomics and soil quality assessment. BMC Genomics 8:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniVec database UniVec & UniVec_Core databases [Internet]. [cited 2016 Jun 9]. Available from: ftp://ftp.ncbi.nih.gov/pub/UniVec/
- Van der Pol EC, Bakker RR, Baets P, Eggink G. 2014. By-products resulting from lignocellulose pretreatment and their inhibitory effect on fermentations for (bio) chemicals and fuels. Appl Microbiol Biotechnol. 98:9579–9593. [DOI] [PubMed] [Google Scholar]
- Van der Wurff A, Isaaks J, Ernsting G, Van Straalen N. 2003. Population substructures in the soil invertebrate Orchesella cincta, as revealed by microsatellite and TE‐AFLP markers. Mol Ecol. 12:1349–1359. [DOI] [PubMed] [Google Scholar]
- Van Straalen NM. 1985. Comparative demography of forest floor Collembola populations. Oikos 253–265. [Google Scholar]
- Vecscreen [Internet]. [cited 2016 Jun 9]. Available from: http://www.ncbi.nlm.nih.gov/tools/vecscreen/.
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall DH, Nielsen UN, Six J. 2015. Soil biodiversity and human health. Nature 528:69–76. [DOI] [PubMed] [Google Scholar]
- Wilson D, et al. 2009. SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37:D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P. 2011. iPath2. 0: interactive pathway explorer. Nucleic Acids Res. 39:W412–W415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, et al. 2014. DBG2OLC: efficient assembly of large genomes using the compressed overlap graph. arXiv preprint arXiv:1410.2801.
- Ye C, Ma ZS, Cannon CH, Pop M, Yu DW. 2011. SparseAssembler: de novo assembly with the Sparse de Bruijn Graph. arXiv preprint arXiv:1106.2603.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.