Abstract
Endosymbiosis is an important evolutionary event for organisms, and there is widespread interest in understanding the evolution of endosymbiosis establishment. Hydra is one of the most suitable organisms for studying the evolution of endosymbiosis. Within the genus Hydra, H. viridissima and H. vulgaris show endosymbiosis with green algae. Previous studies suggested that the endosymbiosis in H. vulgaris took place much more recently than that in H. viridissima, noting that the establishment of the interaction between H. vulgaris and its algae is not as stable as in H. viridissima. To investigate the on-going process of endosymbiosis, we first compared growth and tolerance to starvation in symbiotic and aposymbiotic polyps of both species. The results revealed that symbiotic H. viridissima had a higher growth rate and greater tolerance to starvation than aposymbiotic polyps. By contrast, growth of symbiotic H. vulgaris was identical to that of aposymbiotic polyps, and symbiotic H. vulgaris was less tolerant to starvation. Moreover, our gene expression analysis showed a pattern of differential gene expression in H. viridissima similar to that in other endosymbiotically established organisms, and contrary to that observed in H. vulgaris. We also showed that H. viridissima could cope with oxidative stress that caused damage, such as cell death, in H. vulgaris. These observations support the idea that oxidative stress related genes play an important role in the on-going process of endosymbiosis evolution. The different evolutionary stages of endosymbiosis studied here provide a deeper insight into the evolutionary processes occurring toward a stable endosymbiosis.
Keywords: endosymbiosis, hydra, RNA-seq, differential gene expression, tolerance to starvation, oxidative stress
Introduction
Many cnidarian animals exhibit endosymbiosis with algae (Meyer and Weis 2012), and it is widely known that endosymbiosis has had a massive impact on the survival and lifecycles of cnidarians. One of the most well-studied endosymbiotic systems is that between corals and photosynthetic dinoflagellates (genus Symbiodinium), which is centered on nutrient exchange with dinoflagellates that provide high amounts of photosynthetic products in return for CO2 and (Muller-Parker and D’Elia 1997). Collapse of this endosymbiosis, such as can be induced by environmental stresses, decreases the carbon available for the host and leads to coral death (Brown 1997).
Despite the importance of the endosymbiosis, it remains unclear how the stable endosymbiotic relationships have been established. As coral-dinoflagellate symbiosis has been already present in Triassic era (Stanley and Swart, 1995), it is impossible to elucidate the initial stage of the endosymbiotic relationships.
The genus Hydra is one of the most suitable taxa for studying the evolutionary process of endosymbiosis. Hydras are freshwater cnidarians characterized by the lack of a medusa stage (Kovacevic 2012). In the four species of Hydra maintained at the National Institute of Genetics (NIG, Mishima, Japan), all the six strains of Hydra viridissima Pallas, 1766, exhibit endosymbiosis with green algae belonging to the genus Chlorella. This endosymbiosis is thought to have been initiated in an ancestor of the H. viridissima strains (Kawaida et al. 2013) and is, in fact, considered as a key characteristic of the species (Campbell 1983).
Hydra vulgaris Pallas, 1766, also exhibits endosymbiosis with green algae belonging to the genus Chlorococcum (Kawaida et al. 2013); however, among the several dozens of H. vulgaris strains maintained at the NIG, only two strains (J7 and J10) exhibit endosymbiosis. Previous studies showed that some H. vulgaris group strains can survive in a nonsymbiotic lifestyle even if they are able to form a stable symbiosis with the symbiotic alga (Rahat and Reich 1985; Rahat and Sugiyama 1993). This implies that the endosymbiosis in H. vulgaris is not as stable as in H. viridissima. However, little is known whether the interaction with the algae is different between the two hydra species or not. Previous studies showed that the interaction between H. viridissima and the algae was mutualistic (Muscatine and Lenhoff 1965), but almost nothing is known about the interaction between H. vulgaris and the algae.
Therefore, this study aims to evaluate the endosymbiotic interactions of the two Hydra species with the algae. To investigate these endosymbiotic interactions, we first compared the growth rates and tolerance to starvation in symbiotic and aposymbiotic polyps from which the algae were removed. Next, in order to assess the differences between the interactions at the molecular level, we compared gene expression levels in symbiotic and aposymbiotic polyps. RNA sequencing (RNA-seq) allows for relatively unbiased measurements of transcript expression levels (Wang et al. 2009). This technology also offers the ability to discern aspects of host–symbiont interactions while identifying the genes and pathways regulating those relationships (Meyer et al. 2011). Thus, we conducted differential gene expression analysis between symbiotic and aposymbiotic Hydra spp. using the RNA-seq method. The possible mechanisms underlying a stable endosymbiosis, especially response to oxidative stress by the symbiont, are considered with respect to our results.
Materials and Methods
Hydra spp. Strains and Estimation of Growth and Tolerance to Starvation in Symbiotic and Aposymbiotic Polyps
Endosymbiotic strains of H. viridissima (strain M9) and H. vulgaris (strain J7), stored in the NIG, were used in this study. Polyps were kept in a plastic container filled with Hydra-culture solution (Kawaida et al. 2013) at 18 °C, and growth was estimated by feeding with newly hatched Artemia sp. nauplii three times a week, under a 12 h dark/light cycle (illumination = 2,500 l×). Tolerance to starvation was estimated in nonfed polyps kept in plastic containers; when nonbudding polyps were unable to keep their shape, as observed under the stereomicroscope, they were scored as dead. All polyps were kept under a 12 h dark/light cycle (illumination = 2,500 l×) at 18 °C, and the solution within each container was changed three times per week in both conditions.
RNA Isolation and Sequencing
Total RNA was extracted from intact Hydra individuals, after starvation for 7 days, using a PureLink RNA Mini Kit (Thermo Fisher Scientific Inc., Madison, USA) and following the instructions of the manufacturer. Individuals bearing endosymbiotic algae were disrupted using a μT-12 beads crusher (TAITEC Co., Saitama, Japan). The RNA-integrity number (RIN) of each sample was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA), and only samples with RIN ≥ 9 were used. Total RNA was processed using the TruSeq RNA Library Prep Kit (Illumina Inc., San Diego, USA), following the instructions of the manufacturer, and including a poly-A+ selection step. The indexed libraries produced were then pooled, based on their indices and clustering, and sequenced in an Illumina HiSeq 2000.
De novo Assembly, Functional Annotation, and Reciprocal Best Hit (RBH) Analysis
The de novo assembly of the resulting 101-bp paired-end reads was performed using Trinity (Haas et al. 2013), as implemented in the DNA Data Bank of Japan (DDBJ) Read Annotation Pipeline (Kaminuma et al. 2010; Nagasaki et al. 2013). After removing the de novo assembled contigs shorter than 200 bp, the remaining contigs were compared with those deposited in the UniProtKB/Swiss-Prot database (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gz, last accessed June 16, 2016) and in the nonredundant nucleotide database of the National Centre for Biotechnology (NCBI-NR, ftp://ftp.ncbi.nlm.nih.gov/blast/db/nr.**.tar.gz, last accessed June 16, 2016), using the basic local alignment search tool (BLAST) and an E-value cutoff equal to 1e−5. Gene ontology (GO) terms were then assigned to each contig using the authors’ scripts and a UniProt-GOA file (ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/UNIPROT/gene_association.goa_uniprot.gz, last accessed June 16, 2016).
To identify orthologous genes of H. vulgaris, H. viridissima, Paramecium bursaria, and Ciona varians, an RBH analysis (Moreno-Hagelsieb and Latimer 2008) was conducted using the representative contigs from those remaining after discarding contigs with low expression in these organisms. The contigs of P. bursaria (accession: DRA000907) and C. varians (accession: PRJNA214560) were generated using the method described here from the raw reads deposited in the DDBJ database.
Differential Gene Expression Analysis
Raw reads were mapped to the de novo assembled transcripts of each species using Bowtie2 (Langmead and Salzberg, 2012). Transcript abundance was estimated using eXpress (Roberts and Pachter 2013) and eff_count values were used to determine differential expression (DE). Two biological replicates (1 and 2) were used for each Hydra spp. in both symbiotic and aposymbiotic conditions; normalization and DE analysis between conditions were performed using the iDEGES/edgeR method in TCC R package ver. 1.0.0 (Sun et al. 2013) and a false discovery rate (FDR) ≤ 0.1. Clustering of the organisms with 1,000 approximately unbiased (AU) P values, which is calculated by multiscale bootstrap resampling, and bootstrap probability (BP) replications was performed using the R-package pvclust. Fold-changes in gene expression (determined as log2(symbiotic) − log2(aposymbiotic)) of the orthologous genes were used as the input data as described by Waring et al. (2001), correlation was used as the distance, and the hierarchical clustering was performed using Ward’s method (Murtagh 2014).
GO Enrichment Analysis
GO enrichment analysis was performed for the de novo assembled transcripts of H. viridissima and H. vulgaris using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.7 (Huang et al. 2009). This program performs Fisher’s exact tests to determine the GO terms that are significantly represented among the differentially expressed transcripts in relation to the entire transcriptome. Queries were based on the UniProt ID of de novo annotated assembled references.
Results and Discussion
Change in Growth and Tolerance to Starvation in the Two Hydra spp.
First, growth rates were compared in symbiotic and aposymbiotic polyps of H. viridissima and H. vulgaris. The analysis showed that the growth rate of symbiotic H. viridissima was significantly higher than that of the aposymbiotic state (fig. 1). On one hand, the number of polyps doubled at approximately 4 days of incubation in the symbiotic state and at approximately 28 days of incubation in the aposymbiotic state; the number of polyps was 8.3 ± 4.3 in the symbiotic state and 2.2 ± 0.8 in the aposymbiotic state after 27 days of incubation. On the other hand, the growth rates in symbiotic and aposymbiotic H. vulgaris were almost identical (fig. 1), with the number of individuals (polyps) in both conditions doubling after approximately 10 days. For the symbiotic state, the number of polyps was 3.8 ± 0.8 after 32 days of incubation, and for the aposymbiotic state, it was 3.5 ± 0.8 after 29 days of incubation.
Fig. 1.—
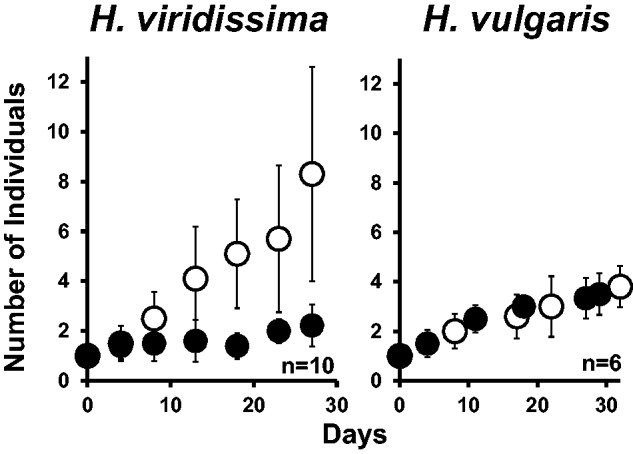
Growth rate of Hydra vulgaris and H. viridissima populations. The hydras were fed three times a week. Open circles correspond to the symbiotic state, and closed circles to the aposymbiotic state. The bars represent standard deviations; n indicates the number of independent experiments conducted.
Considering the tolerance to starvation, on one hand, symbiotic H. viridissima survived for a significantly longer period than aposymbiotic polyps (approximately 45 days vs. approximately 33 days, fig. 2, P < 10−6 by Student’s t-test). In H. vulgaris, on the other hand, aposymbiotic H. vulgaris survived for almost 25 days, whereas symbiotic H. vulgaris survived for a significantly shorter time (approximately 14 days, fig. 2, P < 10−12 by Student’s t-test). Thus, these results support the idea that the endosymbiosis in H. vulgaris is not as stable as in H. viridissima.
Fig. 2.—
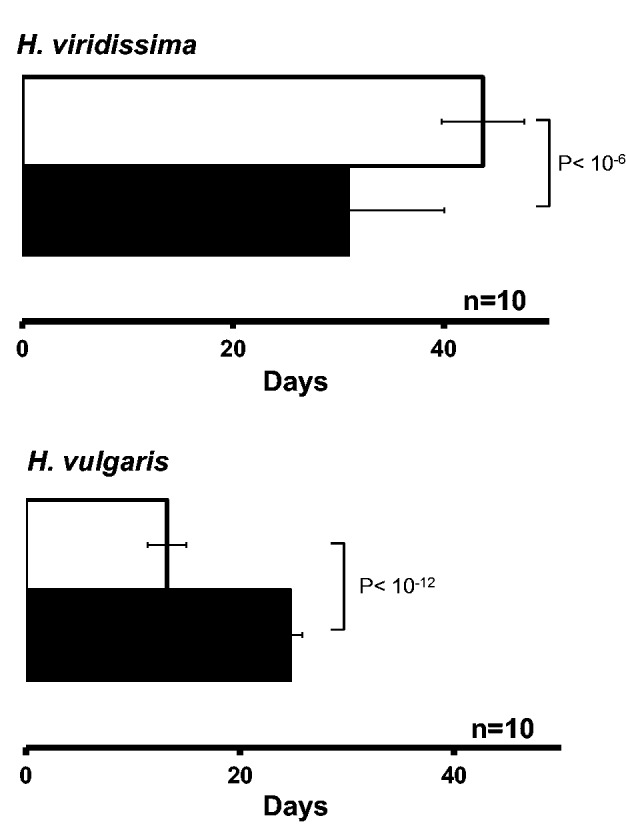
Hydra vulgaris and H. viridissima survival under starvation. The white column represents the symbiotic state and the black column the aposymbiotic state; the bar indicates the standard deviation, n indicates the number of independent experiments conducted.
Sequencing, De Novo Assembly, and Functional Annotation of Hydra spp.
In order to understand the endosymbiotic interaction at the molecular level, the RNA of Hydra spp. after 7 days’ starvation was isolated, and gene expression levels between symbiotic and aposymbiotic state were compared. The RNA-seq yielded a total of 60.7 million pairs of reads containing ∼12.6 Gb in H. viridissima, and a total of 110 million pairs of reads containing ∼20 Gb in H. vulgaris. All reads assembled in Trinity yielded over 100,000 contigs for each species (table 1). In order to choose a representative contig for each gene, the longest transcripts of H. viridissima (72,018 contigs) and H. vulgaris (122,330 contigs) were selected.
Table 1.
Distributions and Annotation Summary of the De Novo Assembled Contigs
| Organism | Contig set type | No. of contigs | N50 | Average GC content (%) | Contigs aligned to |
|
|---|---|---|---|---|---|---|
| UniprotKB (%) | NCBI-NR (%) | |||||
| (E-value ≤ 1e−5) | ||||||
| Hydra viridissima | Whole | 104,912 | 1,621 | 41 | 38,109 (36.3) | 58,368 (55.6) |
| Representative | 72,018 | 1,120 | 43 | 22,933 (31.8) | 34,083 (47.3) | |
| Hydra | 15,114 | 2,166 | 34 | 8,970 (59.3) | 11,380 (75.3) | |
| Hydra vulgaris | Whole | 175,342 | 1,718 | 40 | 42,937 (24.5) | 78,938 (45.0) |
| Representative | 122,330 | 1,102 | 40 | 23,657 (19.3) | 42,625 (34.8) | |
| Hydra | 18,469 | 2,210 | 31 | 9,757 (52.8) | 13,467 (72.9) | |
According to the distribution of the representative contigs within each species (supplementary fig. S1, Supplementary Material online; red dots indicate contigs annotated as cnidarian and green dots indicate contigs annotated as Chlorophyta), the H. viridissima and H. vulgaris contigs also contained contigs derived from endosymbiotic algae (supplementary fig. S1A and C, Supplementary Material online). To exclude these contigs and other contaminants from the analyses, only contigs with more than one fragment per kilo base of contigs per million fragments (FPKM ≥ 1) in all samples were considered. This procedure enabled the elimination of almost all contigs belonging to the endosymbiotic algae (supplementary fig. S1B and D, Supplementary Material online), and yielded 15,144 H. viridissima and 18,469 H. vulgaris “clean” contigs (table 1). After discarding low expression contigs, the proportion of mapped reads decreased by only 1% in the aposymbiotic state, suggesting that most of Hydra spp. contigs were conserved in the process (supplementary table S1, Supplementary Material online). Accordingly, the representative contigs remaining after discarding the low expression contigs were used as the “Hydra contigs” for the differential expression analysis.
After searching for homology against the NCBI-NR (Benson 2015) and the UniProtKB/Swiss-Prot (Magrane and UniProt Consortium 2011) protein databases using BLASTx with an E-value cutoff < 1e−5 (Camacho et al. 2009), contigs were functionally annotated according to protein sequence similarity. A total of 8,970 (59%) reference contigs of H. viridissima matched annotated sequences in the UniProtKB/Swiss-Prot database and 11,380 (75%) matched those in the NCBI-NR database. In H. vulgaris, 9,757 (53%) and 13,467 (73%) reference contigs matched annotated protein sequences in the UniProtKB/Swiss-Prot and NCBI-NR databases, respectively (table 1).
Comparison of Differential Gene Expression Patterns between the Two Hydra spp.
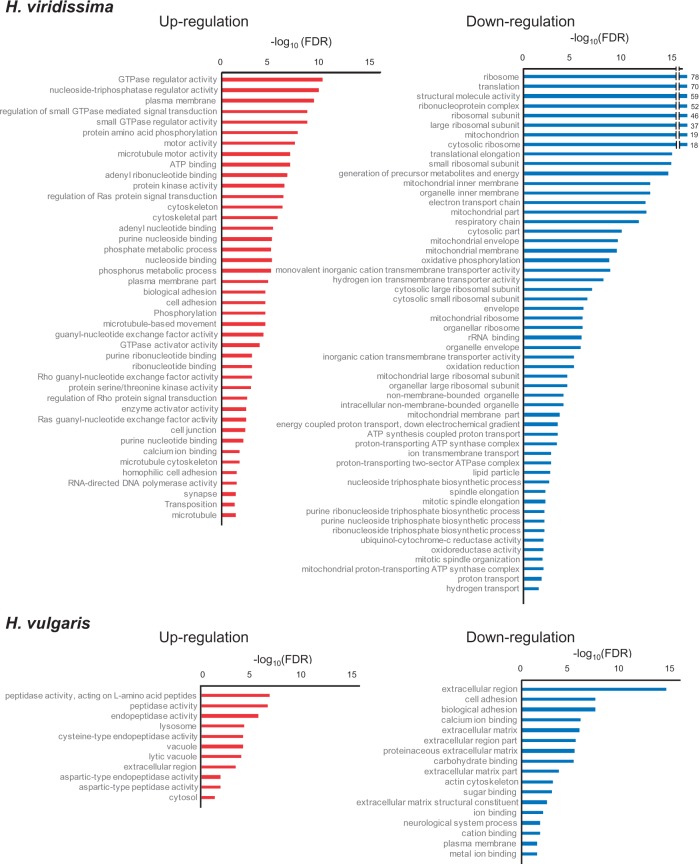
In H. viridissima, 1,890 contigs were upregulated and 2,261 were downregulated in the symbiotic state, while the H. vulgaris symbiotic state had 1,092 upregulated and 1,775 downregulated contigs. Based on these differentially expressed contigs within each species, and using DAVID to analyze Gene Ontology (GO) enrichment, a total of 97 GO terms in H. viridissima (42 upregulated and 55 downregulated) and 28 GO terms in H. vulgaris (11 upregulated and 17 downregulated) were detected as enriched, according to their false discovery rates (FDR ≤ 0.05, fig. 3). No enriched GO terms corresponded to up or downregulated genes in both species.
Fig. 3.—
Enrichment of GO terms in H. viridissima and H. vulgaris genes (FDR ≤ 0.05). GO terms enriched in upregulated and downregulated genes are shown using red and blue bars, respectively. The −log10 (FDR) more than 15 is shown on the right side of the bars.
The reciprocal best hits (RBH) analysis (Moreno-Hagelsieb and Latimer 2008) conducted between the Hydra contigs of the two species yielded 9,934 genes (table 2). Fisher’s exact test revealed that the upregulated genes in H. viridissima tended to be downregulated in H. vulgaris rather than upregulated (P < 10−15), and the downregulated genes in H. viridissima tend to be upregulated in H. vulgaris rather than downregulated (P < 10−5). These results suggested that H. viridissima and H. vulgaris have distinctly different molecular interactions with endosymbiotic algae.
Table 2.
Number of Orthologous Genes Found between Hydra viridissima and H. vulgaris, and Their Regulation in Symbiotic Polyps Relative to Aposymbiotic Polyps
| Total number of orthologous genes |
H. viridissima |
H. vulgaris |
||
|---|---|---|---|---|
| Regulationa,b | Number of genes | Regulation | Number of genes | |
| 9,934 | Up | 1,553 | Up | 65 |
| Non | 1,124 | |||
| Down | 364 | |||
| Non | 6,993 | Up | 390 | |
| Non | 5,935 | |||
| Down | 668 | |||
| Down | 1,388 | Up | 126 | |
| Non | 1,150 | |||
| Down | 112 | |||
In all cases, differential gene expression was significant at a false discovery rate (FDR) ≤ 0.1.
“Up,” significantly upregulated in the symbiotic state; “Non,” not significantly up- or downregulated in the symbiotic state; “Down,” significantly downregulated in the symbiotic state.
Comparison of Differential Gene Expression Patterns with Other Endosymbiotic Organisms
In order to confirm that the differential gene expression patterns between the two Hydra spp. reflect the stability of the endosymbiotic relationship, the differential gene expression patterns obtained in this study were compared with those of other endosymbiotic organisms, P. bursaria and C. varians. Both species exhibit mutualistic relationships with their symbionts (Karakashian 1963; Kodama et al. 2014), and gene expression levels between symbiotic and aposymbiotic states obtained from RNA-seq were compared in previous studies (Riesgo et al. 2014; Lesser 2006). We assembled the reads of the P. bursaria and C. varians and identified orthologous genes in the four organisms by using the RBH analysis. The analysis yielded 1,111 orthologous genes, and a hierarchical cluster analysis was performed using fold-changes in gene expression of these 1,111 orthologous genes (fig. 4). In fig. 4, height represents dissimilarity between clusters, estimated as 1 − (Pearson correlation coefficient). When clusters were positively correlated, height is shown as less than 1. P. bursaria, H. viridissima, and C. varians were grouped into a single cluster with high AU P value and bootstrap value (fig. 4). Thus, the differential gene expression patterns observed in symbiotic H. viridissima were similar to those of other endosymbiotic hosts, but different from those observed in H. vulgaris. This result suggests that these three organisms have a similar type of stable association with their endosymbionts, and that this type of association is not present in H. vulgaris.
Fig. 4.—
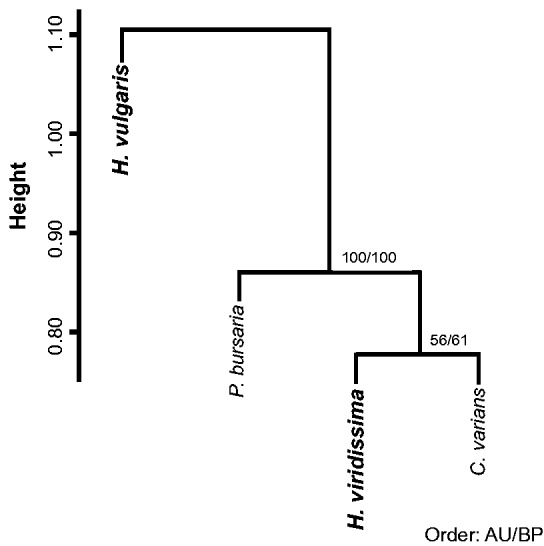
Dendrogram of endosymbiotic hosts based on the differential gene expression patterns. The analysis was conducted in the R package “pvclust” using the Ward’s method as the agglomerative method and Pearson correlations as the distance measure. Height represents dissimilarity between clusters that was calculated as 1 − (Pearson correlation coefficient). Numbers along branches indicate the approximately unbiased P value (AU) and the bootstrap probability (BP).
Possible Mechanisms of Endosymbiotic Interaction
Endosymbionts generate reactive oxygen species (ROS) that cause major cellular damage, including membrane oxidation, protein denaturation, and nucleic acid damage (Marchi et al. 2012). For example, de Vries et al (2015) examined tolerance to starvation of two sea slug species that maintain functional plastids in their cytosol, and concluded that response to ROS mediated by these plastids is a key factor for longer survival. In coral–algal symbiosis, oxidative stress can lead to a breakdown of the symbiosis, and therefore the coral–algal symbiosis has an arsenal of defenses to combat ROS (Roth 2014). As ROS effects are considered to be a major challenge to the host, the possible mechanisms for ROS response of the two Hydra spp. were compared. In H. viridissima downregulated genes, mitochondrion-related GO terms were enriched (fig. 3). Furthermore, most of the transcripts involved in the respiratory chain were significantly downregulated in symbiotic H. viridissima (table 3 and supplementary table S2, Supplementary Material online). Mitochondria generate ROS during the respiratory process (Weis 2008; Lee et al. 2011), and the ROS generated by symbionts and mitochondria have been shown to play a central role in damage to the host, such as in coral bleaching (Dunn et al. 2012). In addition, hyperthermic stress has been shown to induce mitochondrial degradation in cnidarian hosts (Orrenius et al. 1992), which might limit the capacity of the cnidarians to mitigate the effects of ROS generation. Therefore, H. viridissima might respond to ROS by inactivating the respiratory chain process (dashed line indicated as (1) in fig. 5).
Table 3.
Number of Genes Related to Oxidative Stress Response
| Organism | Total | Upregulated | Downregulated | |
|---|---|---|---|---|
| Respiratory chain | H. viridissima | 35 | 0 | 29 |
| H. vulgaris | 34 | 1 | 0 | |
| Peroxidase | H. viridissima | 18 | 2 | 8 |
| H. vulgaris | 20 | 3 | 6 | |
| Polycystin | H. viridissima | 13 | 5 | 0 |
| H. vulgaris | 31 | 1 | 9 | |
| Cadherin | H. viridissima | 10 | 5 | 0 |
| H. vulgaris | 13 | 0 | 8 | |
| Caspase | H. viridissima | 12 | 3 | 0 |
| H. vulgaris | 20 | 7 | 1 | |
| Metalloproteinase | H. viridissima | 5 | 1 | 2 |
| H. vulgaris | 6 | 3 | 1 |
Upregulated; significantly (FDR ≤ 0.1) upregulated in symbiotic state. Downregulated; significantly (FDR ≤ 0.1). We selected the genes that are annotated both in Uniprot and NCBI-NR (E-value < =1e−5).
Fig. 5.—
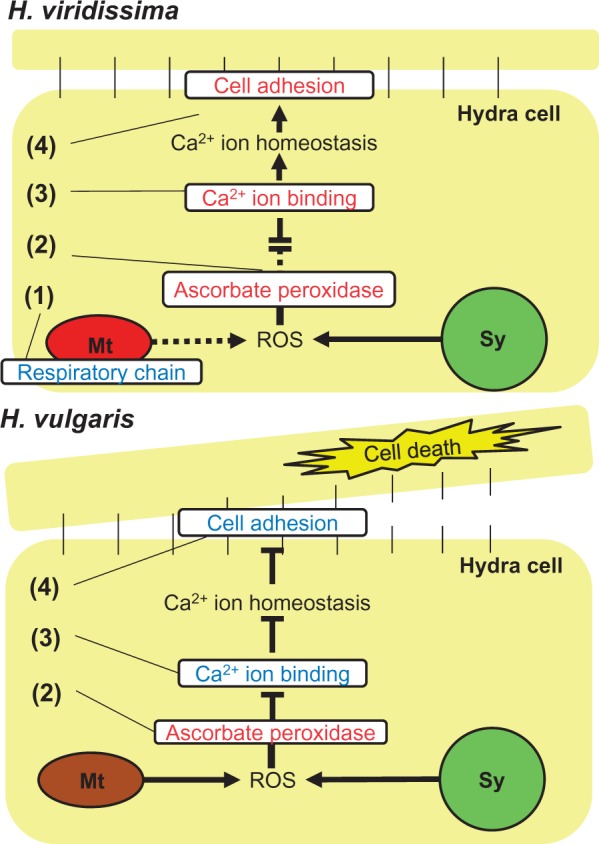
A proposed model of the endosymbiotic interaction between the two Hydra species and algae. The endosymbiotic hydra is exposed to reactive oxygen species (ROS) extracted from both mitochondria (Mt) and the symbiont (Sy). H. viridissima represses ROS by downregulation of respiratory chain genes of mitochondria (indicated by (1)), and respond to the stress by extensive upregulation of ascorbate peroxidase (indicated by (2). Moreover, the disruption of homeostasis is repressed by the upregulation of binding and cell adhesion genes (indicated by (3) and (4)). Although H. vulgaris also responds to the stress by upregualtion of ascorbate peroxidase (indicated by (2)), it still experiences oxidative stress that induces disruption of the homeostasis and cell adhesion (indicated by (3) and (4)), which contributes to the cell death. The genes/systems are shown as upregulated (red) and downregulated (blue).
With regard to the canonical ROS scavenging system, although the GO terms related to peroxidase activity were not enriched in both species, the gene annotated as ascorbate peroxidase were upregulated in the symbiotic state of both species (fig. 6). However, this gene was more dramatically upregulated in H. viridissima than in H. vulgaris (fig. 6). Among the four enzymes responsible for ROS scavenging (superoxide dismutase, ascorbate peroxidase, catalase, and glutathione peroxidase), ascorbate peroxidase only exists in plants (Apel and Hirt 2004). However, Habetha and Bosch (2005) showed that H. viridissima has a plant-related ascorbate peroxidase, which was possibly laterally transferred. The presence of an ascorbate peroxidase gene in both H. vulgaris and H. viridissima, the most distant species in the genus Hydra (Kawaida et al. 2010), suggests that this gene was obtained in the common ancestor of Hydra species. As the gene was more dramatically upregulated in H. viridissima, H. viridissima would scavenge ROS more than H. vulgaris (indicated as (2) in fig. 5).
Fig. 6.—
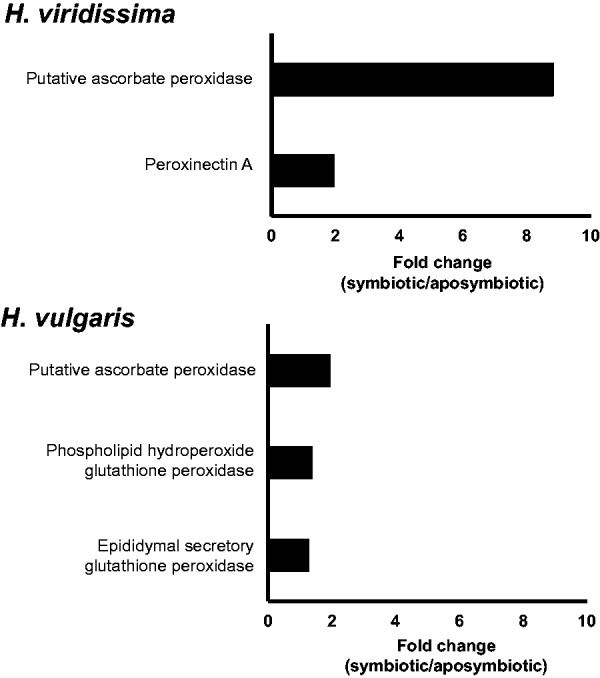
Peroxidase activity related genes significantly (FDR ≤ 0.1) upregulated in symbiotic H. vulgaris and H. viridissima. Supplementary fig. S1, Supplementary Material online. Distribution of contigs based on RNA-seq data. (A) Representative contigs of Hydra vulgaris. (B) H. vulgaris contigs. (C) Representative contigs of H. viridissima. (D) H. viridissima contigs. Contigs that gave the best hits with cnidarian sequences are represented in red, those for Chlorophyta sequences in green, those with Prokaryote sequences in blue, and others in black.
The “Calcium ion binding” GO term was enriched in upregulated H. viridissima genes and downregulated H. vulgaris genes (fig. 3). Oxidative stress can disrupt Ca2+ homeostasis, resulting in an elevation in intracellular Ca2+ (Desalvo et al. 2008), which induces coral bleaching (Koulen et al. 2002). Polycystin, a known intracellular calcium release channel (Montalbetti et al. 2008), which is inhibited by ROS (Hirano et al. 1987), was annotated in both species (table 3 and supplementary table S2, Supplementary Material online). In H. vulgaris, on one hand, the genes related to polycystin tended to be downregulated in the symbiotic state, suggesting that oxidative stress might have increased Ca2+ intracellular concentration in this species. On the other hand, H. viridissima might have managed to keep intracellular Ca2+ homeostasis by upregulating polycystin genes (indicated by (3) in fig. 5).
Disruption of Ca2+ homeostasis affects cell adhesion (Chapman et al. 2010). The GO term “Cell adhesion” was enriched in downregulated H. vulgaris genes and upregulated H. viridissima genes (fig. 3). One of the abundant genes in “Cell adhesion” was cadherin (table 3 and supplementary table S2, Supplementary Material online), which is responsible for cell adhesion, and is protected from proteinases in the presence of Ca2+ ions (Chapman et al. 2010). A previous genome study suggested that Hydra spp. have a classic cadherin that exhibits a highly conserved, bilaterian-type cytoplasmic (CCD) domain (Chapman et al. 2010) and Steinhusen et al. (2001) showed that cadherin was cleaved by caspase 3 and metalloproteinase in apoptotic cells. Some of the caspase 3 and metalloproteinase genes were upregulated in H. vulgaris (table 3 and supplementary table S2, Supplementary Material online), suggesting endosymbiosis might cause apoptosis. Although some caspase 3 and metalloproteinase genes were also upregulated in H. viridissima (table 3 and supplementary table S2, Supplementary Material online), this species might prevent apoptosis by up-regulating cadherin genes (indicated by (4) in fig. 5). These various findings suggest H. viridissima has a well-established mechanism to respond to oxidative stress, whereas H. vulgaris was damaged by oxidative stress, resulting in cell death.
Kawano et al (2004) proposed two models with a focus on ROS tolerance to explain the evolutionary process of stable endosymbiotic relationships. The first model suggested that the host already has mechanisms for ROS response before establishment of the endosymbiosis. The second model proposes that the host first establishes the endosymbiosis and that ROS tolerance emerges later in the host by ROS-driven evolution. The second model implies that endosymbiosis can be a burden for the host especially at the initial stage of the endosymbiotic relationship. In the present study, symbiotic H. vulgaris was shown to be less tolerant to starvation than aposymbiotic polyps, and we suggested that the symbiotic H. vulgaris has not established the mechanisms for ROS responses that are present in H. viridissima. This conclusion is consistent with the second model. Recent studies revealed that mutualism and parasitism have similar evolutionary patterns (Sachs et al. 2011) and that bacterial mutualism evolved from parasitic lineages (Toft and Anderson 2010). Lowe et al. (2016) showed that even beneficial endosymbiosis is costly to the host under certain conditions. Thus, the endosymbiosis, especially the initial stage its establishment, might be a burden for a host such as H. vulgaris, and ROS responses would be one of the key factors towards developing a stable endosymbiotic relationship.
Conclusion
Our study showed that endosymbiotic interaction with algae was significantly different between H. viridissima and H. vulgaris. This difference was evidenced at the physiological and molecular levels: whereas endosymbiosis seemed to be advantageous for H. viridissima survival and its mechanisms were well established, endosymbiosis in H. vulgaris seemed to be disadvantageous ultimately leading to polyp death through cell apoptosis. These results support the conclusion that the endosymbiosis between H. vulgaris and the algae is not yet well established and that it, therefore, lacks the mechanisms for stable endosymbiosis. As the evolution of endosymbiosis in H. vulgaris was more recent than in H. viridissima, the endosymbiotic interaction between H. vulgaris and its algae may still be immature. Thus, understanding this immature endosymbiotic relationship between H. vulgaris and its algae will provide a significant insight into the evolutionary process of endosymbiotic relationship establishment.
Supplementary Material
Supplementary figures S1 and tables S1-S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Ikuko Masujima, Takako Komatsu, and Tae Yamada for helping with Hydra spp. maintenance, and the collaborators of the Beijing Institute for Genomics (BGI) and the Bioscience Core Lab in King Abdullah University of Science and Technology (KAUST) for carrying out sequencing. The authors would also like to thank the ROIS National Institute of Genetics for the use of their NIG supercomputer. This work was supported by the Graduate University for Advanced Studies (SOKENDAI) to M.I. and by KAUST baseline fund to T.G.
Literature Cited
- Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 55:373–399. [DOI] [PubMed] [Google Scholar]
- Benson DA. 2015. GenBank. Nucleic Acids Res. 43:D30–D35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BE. 1997. Coral bleaching: causes and consequences. Coral Reefs 16:S129–S138. [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 1510:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RD. 1983. Identifying Hydra species In: Lenhoff HM, editor. Hydra: research methods. New York: Plenum Press; p. 9–28. [Google Scholar]
- Chapman JA, et al. 2010. The dynamic genome of Hydra. Nature 464:592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desalvo MK, et al. 2008. Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Mol Ecol 17:3952–3971. [DOI] [PubMed] [Google Scholar]
- de Vries J, et al. 2015. Comparison of sister species identifies factors underpinning plastid compatibility in green sea slugs. Proc R Soc Lond B Biol Sci. 282:20142519.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn SR, Pernice M, Green K, Hoegh-Guldberg O, Dove SG. 2012. Thermal stress promotes host mitochondrial degradation in symbiotic cnidarians: are the batteries of the reef going to run out? PLoS One 7:e39024.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habetha M, Bosch TCG. 2005. Symbiotic Hydra express a plant-like peroxidase gene during oogenesis. J Exp Biol. 208:2157–2164. [DOI] [PubMed] [Google Scholar]
- Hirano S, Nose A, Hatta K, Kawakami A, Takeichi M. 1987. Calcium-dependent cell-cell adhesion molecules (cadherins): subclass specificities and possible involvement of actin bundles. J Cell Biol. 105:2501–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. [DOI] [PubMed] [Google Scholar]
- Kaminuma E, et al. 2010. DDBJ launches a new archive database with analytical tools for next-generation sequence data. Nucleic Acids Res. 38:D33–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakashian SJ. 1963. Growth of Paramecium bursaria as influenced by the presence of algal symbionts. Physiol Zool 36:52–68. [Google Scholar]
- Kawaida H, Ohba K, Koutake Y, Shimizu H, Tachida H. 2013. Symbiosis between Hydra and Chlorella: molecular phylogenetic analysis and experimental study provide insight into its origin and evolution. Mol Phylogenet Evol. 66:906–914. [DOI] [PubMed] [Google Scholar]
- Kawaida H, Shimizu H, Fujisawa T, Tachida H, Kobayakawa Y. 2010. Molecular phylogenetic study in genus Hydra. Gene 468:30–40. [DOI] [PubMed] [Google Scholar]
- Kawano T, Kadono T, Kosaka T, Hosoya H. 2004. Green paramecia as an evolutionary winner of oxidative symbiosis: a hypothesis and supportive data. Z Naturforsch C 59:538–542. [DOI] [PubMed] [Google Scholar]
- Kodama Y, et al. 2014. Comparison of gene expression in Paramecium bursaria with and without Chlorella variabilis symbionts. BMC Genomics 15:183.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulen P, et al. 2002. Polycystin-2 is an intracellular calcium release channel. Nat Cell Biol. 4:191–197. [DOI] [PubMed] [Google Scholar]
- Kovacevic G. 2012. Value of the Hydra model system for studying symbiosis. Int J Dev Biol. 56:627–635. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie2. Nat Met 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, et al. 2011. Mitochondrial H2O2 generated from electron transport chain complex I stimulates muscle differentiation. Cell Res. 21:817–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser MP. 2006. Oxidative stress in marine environments: biochemistry and physiological ecology. Ann Rev Physiol. 68:253–278. [DOI] [PubMed] [Google Scholar]
- Lowe CD, Minter EJ, Cameron DD, Brockhurst MA. 2016. Shining a light on exploitative host control in a photosynthetic endosymbiosis. Curr Biol. 26:207–211. [DOI] [PubMed] [Google Scholar]
- Magrane M, Uniprot Consortium. 2011. UniProt Knowledgebase: a hub of integrated protein data. Database 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi S, et al. 2012. Mitochondria-ROS crosstalk in the control of cell death and aging. J Signal Transduct 329635:2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Aglyamova GV, Matz MV. 2011. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-seq procedure. Mol Ecol 20:3599–3616. [DOI] [PubMed] [Google Scholar]
- Meyer E, Weis VM. 2012. Study of cnidarian-algal symbiosis in the “Omics” age. Biol Bull. 223:44–65. [DOI] [PubMed] [Google Scholar]
- Montalbetti N, Cantero MR, Dalghi MG, Cantiello HF. 2008. Reactive oxygen species inhibit polycystin-2 (TRPP2) cation channel activity in term human syncytiotrophoblast. Placenta 6:510–518. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Latimer K. 2008. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24:319–324. [DOI] [PubMed] [Google Scholar]
- Muller-Parker G, D’Elia CF. 1997. Interactions between corals and their symbiotic algae In: Birkeland C, editor. Life and death of coral reefs. New York: Chapman & Hall; p. 96–113. [Google Scholar]
- Murtagh F. 2014. Ward’s hierarchical agglomerative clustering method: which algorithms implement Ward’s criterion? J Classif. 31: 274–295. [Google Scholar]
- Muscatine L, Lenhoff M. 1965. Symbiosis of hydra and algae. II. Effects of limited food and starvation on growth of symbiotic and aposymbiotic hydra. Biol Bull. 123:316–328. [Google Scholar]
- Nagasaki H, et al. 2013. DDBJ read annotation pipeline: a cloud computing-based pipeline for high-throughput analysis of next-generation sequencing data. DNA Res. 20:383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrenius S, Burkitt MJ, Kass GE, Dypbukt JM, Nicotera P. 1992. Calcium ions and oxidative cell injury. Ann Neurol 32:S33–S42. [DOI] [PubMed] [Google Scholar]
- Rahat M, Reich V. 1985. A new alga/hydra symbiosis: Hydra magnipapillata of the 'nonsymbiotic' Vulgaris group hosts a Chlorococcum-like alga. Symbiosis 1:177–184. [Google Scholar]
- Rahat M, Sugiyama T. 1993. The endodermal cells of some “brown” hydra are autonomous in their ability “to host or not to host” symbiotic algae: analysis of Chimera. Endocytobiosis Cell Res. 9:223–231. [Google Scholar]
- Riesgo A, et al. 2014. Transcriptomic analysis of differential host gene expression upon uptake of symbionts: a case study with Symbiodinium and the major bioeroding sponge Cliona varians. BMC Genomics 15:376.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pachter L. 2013. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 10:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS. 2014. The engine of the reef: photobiology of the coral-algal symbiosis. Front Microbiol. 422:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs JL, Essenberg CJ, Turcotte MM. 2011. New paradigms for the evolution of beneficial infections. Trends Ecol Evol. 26:202–209. [DOI] [PubMed] [Google Scholar]
- Stanley GD, Swart PK. 1995. Evolution of the coral-zooxanthenllae symbiosis during the Triassic: a geochemical approach. Paleobiology 21:179–199. [Google Scholar]
- Steinhusen U, et al. 2001. Cleavage and shedding of E-cadherin after induction of apoptosis. J Biol Chem. 276:4972–4980. [DOI] [PubMed] [Google Scholar]
- Sun J, Nishiyama T, Shimizu K, Kadota K. 2013. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 14:219.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft C, Andersson SGE. 2010. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet. 11:465–475. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring JF, Ciurlionis R, Jolly RA, Heindel M, Ulrich RG. 2001. Microarray analysis of hepatotoxins in vitro reveals a correlation between gene expression profiles and mechanisms of toxicity. Toxicol Lett. 120:359–368. [DOI] [PubMed] [Google Scholar]
- Weis VM. 2008. Cellular mechanisms of cnidarian bleaching: stress causes the collapse of symbiosis. J Exp Biol. 211:3059–3066. [DOI] [PubMed] [Google Scholar]