Abstract
Cynomoriaceae, one of the last unplaced families of flowering plants, comprise one or two species or subspecies of root parasites that occur from the Mediterranean to the Gobi Desert. Using Illumina sequencing, we assembled the mitochondrial and plastid genomes as well as some nuclear genes of a Cynomorium specimen from Italy. Selected genes were also obtained by Sanger sequencing from individuals collected in China and Iran, resulting in matrices of 33 mitochondrial, 6 nuclear, and 14 plastid genes and rDNAs enlarged to include a representative angiosperm taxon sampling based on data available in GenBank. We also compiled a new geographic map to discern possible discontinuities in the parasites’ occurrence. Cynomorium has large genomes of 13.70–13.61 (Italy) to 13.95–13.76 pg (China). Its mitochondrial genome consists of up to 49 circular subgenomes and has an overall gene content similar to that of photosynthetic angiosperms, while its plastome retains only 27 of the normally 116 genes. Nuclear, plastid and mitochondrial phylogenies place Cynomoriaceae in Saxifragales, and we found evidence for several horizontal gene transfers from different hosts, as well as intracellular gene transfers.
Keywords: chondriome, Cynomorium, Mediterranean-Irano-Turanian, plastome, parasitic plants, horizontal gene transfer
Introduction
Current phylogenetic systems accept 416 families of flowering plants in 64 orders, with the relationships of most of them known due to molecular phylogenies ( Stevens 2001 onwards; Angiosperm Phylogeny Group, 2009 ; 2016 came out when this study was in revision and includes our results as a personal communication). Among the most difficult to place clades are parasitic plants. They present special challenges because of the deep changes in their genomes that complicate analysis with phylogenetic standard markers and because of the difficulty of obtaining nonhost-contaminated DNA. Horizontal gene transfers (HGTs) between parasites and hosts, resulting in contradictory gene trees, further complicate the task ( Nickrent et al. 1998 , 2004, 2005 ; Barkman et al. 2007 ; Xi et al. 2012 , 2013 ; Zhang et al. 2014 ) and so do high substitution rates in parasite genomes that can lead to long-branch attraction ( Nickrent et al. 1997 ; Barkman et al. 2007 ; Bellot and Renner 2014 ). Phylogenetically long unplaced parasite families include the Lennoaceae (Boraginales) and the Cynomoriaceae ( Stevens 2001 onwards; Angiosperm Phylogeny Group 2009 ). Here, we place the latter, using genomic data from their assembled organellar genomes as well as selected nuclear genes.
Cynomoriaceae comprise one or two species or subspecies, Cynomorium coccineum L. and Cynomorium songaricum Rupr. (= C. coccineum subspecies songaricum (Rupr.) J.Léonard), occurring from the Canary Islands (Lanzarote) through the Mediterranean region to the adjacent Irano-Turanian region, including the Mongolian deserts in western China ( fig. 1 ). The plants grow in rocky or sandy soils, often in saline habitats close to the coast. Their inflorescences are up to 40 cm tall with hundreds of small reddish flowers ( fig. 1 ), and there are no green parts that would carry out photosynthesis. Cynomoriaceae therefore completely rely on water and nutrients from their hosts. Fitting its huge geographic range, Cynomorium parasitizes the roots of plants from many genera and families. In the western part of the genus range, these are Amaranthaceae subfam. Chenopodioideae (usually Atriplex or Salsola ), Plumbaginaceae (e.g., Limonium ), Tamaricaceae ( Tamarix ), Frankeniaceae (all in the Caryophyllales), Cistaceae (Malvales), Fabaceae (Fabales), and Asteraceae (Asterales). In its eastern range (Afghanistan, Mongolia/China), Cynomorium parasitizes Nitraria , a genus of four to five species in Asia and the Mediterranean ( Zhang et al. 2015 ), Peganum harmala L. ( Teryokhin et al. 1975 ; Yang et al. 2012 ; also Nitrariaceae, Sapindales), Tamarix and Reaumuria (both Tamaricaceae), Zygophyllum (Zygophyllaceae), and Salsola ( Chen and Funston 2007 ; Yang et al. 2012 ; Cui et al. 2013 ). Cynomoriaceae may have antioxidant properties ( Zucca et al. 2013 ), and their inflorescences are widely collected as an aphrodisiac throughout the Middle East and in China, where the plant’s conservation status is thought to be critical ( Cui et al. 2013 ).
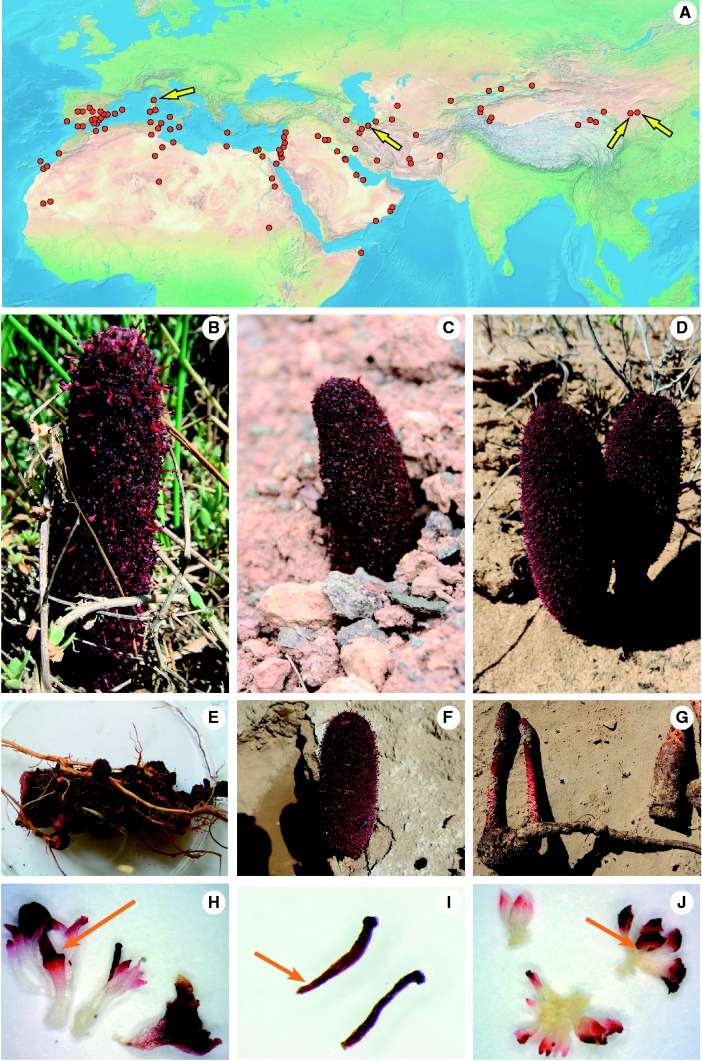
Fig. 1.—
Distribution and habit of Cynomorium . (A) Distribution range of Cynomorium obtained from the map in Hansen (1986) , relevant floras, and 203 GPS coordinates retrieved from the Global Biodiversity Information Facility (GBIF) portal in August 2015. Arrows indicate our own collections (details in table 1). Background map from Naturalearthdata.com. (B–J) Cynomorium plants from Italy (B, E, H), N. Cusimano and C. Cusimano 2, Iran (C and I) Zarre 59621 , and China (D) S.X. Luo 2014 from Tengger Desert; (F, G, J) G. Sun 1, from Gansu. Photos B to G by the respective collectors, photos H to J by N. Cusimano. (B–D) Plants in situ. (E) Part of a rhizome with young inflorescences and connected to the host roots ( Atriplex portulacoides ). (F) A fly ( Musca spec.) visiting Cynomorium in the Tengger Desert. (G) Chinese plant connected to the host root ( Nitraria tangutorum ). (H) Young male and female flowers and a bract, showing a red stamen basis (arrow). (I) Stamen from herbarium material. (J) Male flowers showing white stamen bases (arrow). The color of the stamen basis has been used to differentiate C. coccineum subsp. songaricum from Cynomorium coccineum subsp. coccineum .
The highly reduced flowers and poor preservation (when dried) of the large, fleshy inflorescences in the World’s herbaria have made morphological homology assessment difficult. Tentative inclusion of Cynomorium in Santalales based on the parasitic habit ( Cronquist 1968 ; Takhtajan 1973 ) was not supported by the first mitochondrial ( matR ) and nuclear (small and large rDNA subunits) sequences of Cynomoriaceae, which came from a Spanish population and showed that Cynomorium might belong in the Saxifragales ( Nickrent et al. 2005 ). Sequences of the mitochondrial genes atp1 and cox1 from a specimen from an unknown location placed Cynomorium in the Sapindales, possibly due to a horizontal acquisition from a host ( Barkman et al. 2007 ); the collection location and host are unknown, and the voucher has been lost (dePamphilis CW, personal communication to SSR on September 22, 2015). Surprisingly, the inverted repeat (IR) region sequenced from Chinese material yielded a placement in the Rosaceae, close to Prunus and Fragaria ( Zhang et al. 2009 ). Su et al. (2015 , their supplementary appendix S1) recently summarized these contradictory findings, stressing the problem of HGT and extensive intraindividual variation in plastid rDNA, which they attribute to heteroplasmy in Cynomorium ( García et al. 2004 ).
We here use newly collected material of Cynomoriaceae from populations in China, Iran, and Italy, and Illumina and Sanger sequencing, to obtain 1) a broader sample of gene regions than sequenced in any previous study and 2) a global picture of their copy number and genomic location (whether in the nucleus, plastome, or mitochondrial genome). To calculate the expected genomic coverage (which is essential for interpreting Illumina data), we obtained C value measurements of plants from the western (Italy) and eastern (China) part of the family’s geographic range. We assembled the mitochondrial and chloroplast genomes of an Italian Cynomorium and built angiosperm-wide matrices from different genes to try to circumnavigate the problem of HGT, which can often be detected by comparing topologies from different markers.
Materials and Methods
Collection of Material and DNA Sequencing
Cynomorium specimens were collected in Iran, Italy, and two locations in China, in Ningxia Province and in Gansu Province, both in the Mongolian desert region. Table 1 provides collection locations, herbarium voucher information, and GenBank accession numbers. Total genomic DNA was extracted from fresh material with the DNeasy Plant Maxi Kit (Qiagen) following the manufacturer’s instructions. The DNA of the Italian plant was sent to Genewiz for preparation of five standard paired-end libraries with insert sizes of 200–500 bp, and of one mate-pair library with an insert size of approximately 3.5–4.5 kb. Sequencing was performed on an Illumina HiSeq2500 machine in “Rapid Run Mode.” For Sanger sequencing of selected Chinese and Iranian Cynomorium genes (see below), new primers were designed based on the Italian Cynomorium contigs using Primer3Plus v. 2.3.6 ( Untergasser et al. 2012 ). Supplementary table S1 , Supplementary Material online, shows primer sequences and annealing temperatures. Polymerase chain reaction (PCR) products were purified with the ExoSAP or FastAP clean-up kits (Fermentas Life sciences, St. Leon-Rot, Germany), and sequencing relied on the Big Dye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and an ABI 3130-4 automated capillary sequencer. To confirm the length of the single copy (SC) regions of the plastome, their junctions with the IRs, as well as a low-coverage region in the large single copy (LSC) region (Results), we performed PCR and Sanger resequencing using newly designed primers ( supplementary table S1 , Supplementary Material online), including long-range PCR amplifications using the Q5® High Fidelity DNA Polymerase (New England BioLabs Inc.), following the manufacturer’s protocol.
Table 1.
Collecting Locations, Herbarium Voucher Information, and Genbank Accession Numbers for All Cynomorium Sequences Used in this Study
| Cynomorium Subspecies | Collecting Location | Herbarium Voucher | GenBank# Plastid-Encoded | GenBank# Mitochondrial | GenBank# Nuclear |
|---|---|---|---|---|---|
| coccineum | Israel, year unknown | D. Nickrent 4000 (SIU) | 16S rDNA: U67743 | — | — |
| coccineum | Spain, Cadíz, April 19, 1996 | D. Nickrent 4063 (SIU) | 23S rDNA: AY330869–AY330887 | matR : AY957446 | 18S rDNA: AY957442, 26S rDNA: AY957452 |
| coccineum | Unknown location, April 18, 1996 | J. Hoder s.n. Fide Barkman et al. (2007) , a voucher is in the herbarium PAC (Pennsylvania State University). However, a voucher cannot be found nor information on the collection location (C. dePamphilis, pers. comm. to SSR on September 22, 2015) | matR : EU281095, atp1 : EU280951, cox1 : EU281023 | ||
| coccineum | Iran, Mazandaran Prov., 35 km to Amol, Panjab village, Darli, June 2, 2013 | S. Zarre 59621 (M), 2013 | clpP : KU043173, rpl2 : KU043239, rpl14 : KU043183, rpl16 : KU043226, rps3 : KU043194, rps7 : KU043238, 16S rDNA: KU043211, 23S rDNA: KU043214 | nad5 : KU043231 | |
| coccineum | Iran, same location, May 30, 2014 | S. Zarre 59621 (M), 2014 | clpP : KU043172, rpl2 : KU043240, rpl14 : KU043184, rpl16 : KU043227, rpl36 : KU043188, rps3 : KU043195 rps7 : KU043199, rps11 : KU043202, 4.5S rDNA: KU043216, 5S rDNA: KU043208, 16S rDNA: KU043212, 23S rDNA: KU043236 | nad5 : KU043232 | |
| coccineum | Sardinia, Isola Sinis, May 6, 2014 | N. Cusimano and C. Cusimano 2 (M), Also used for C value measurement and dissection of flowers | clpP : KU043220, rpl2 : KU043223, rpl14 : KU043182, rpl16 : KU043225, rpl36 : KU043187, rps3 : KU043193, rps7 : KU043198, rps11 : KU043201, rps19 : KU043204, 4.5S rDNA: KU043215, 5S rDNA: KU043207, 16S rDNA: KU043210, 23S rDNA: KU043213, ycf2 : KU043218, Complete plastome: KX270752 | atp1 : KU043169, atp6 : KU043163, cob : KU043164, cox1 : KU043219, cox3 : KU043165, matR : KU043179, nad5 : KU043180, rps3: KU043191, Mitochondrial contigs: KX270753 - KX270801 | 18S rDNA: KU043174, 26S rDNA: KU043175, atp1 (nuclear copies): KU043166-8, pepC : KU043177, phyA : KU043178, SMC2 : KU043221 MSH1 : KU043222, |
| songaricum | China, Gansu Prov., Tengger Desert, May 19, 2014 | S.X. Luo 618 (M) | clpP : KU043170, rpl2 : KU043224, rpl14 : KU043186, rpl16 : KU043229, rpl36 : KU043190, rps3 : KU043197 rps19 : KU043206, 23S rDNA: KU043235 | nad5 : KU043181 | |
| songaricum | China, Ningxia Prov., Pingluo, Shizuishan, May 9, 2014 | L. Zhang 1 (M) | clpP : KU043171, rpl2 : KU043241, rpl14 : KU043185, rpl16 : KU043228, rpl36 : KU043189, rps3 : KU043196, rps7 : KU043200, rps11 : KU043203, rps19 : KU043205, 4.5S rDNA: KU043217, 5S rDNA: KU043209, 16S rDNA: KU043234, 23S rDNA: KU043237 | matR : KU043230, nad5 : KU043233, rps3 : KU043192 | |
| songaricum | China, Gansu Prov., Hongshao forest farm, Ganzhou District, near Zhangye, April 24, 2015 | G. Sun 1 (M), used only for C value measurement and dissection of flowers | |||
| songaricum | China, Border of Gansu Prov. and Inner Mongolia | J. Li 5958 (herbarium of Zhejiang University) | IR: FJ895894 - FJ895898 | ||
| songaricum | China, Gansu Prov., Liangucheng, Minqin | J. Li 5941 (herbarium of Zhejiang University) | IR: FJ895885 - FJ895893 | ||
| songaricum | China, Location unknown, no response from authors to repeated emails in September 2015 | Voucher: G. Liu and G. Chen 8611 (vouchering unclear) | matR : JX287337, atp1 : JX287332, cox1 : JX287336 | 18S rDNA: JX287338, 26S rDNA: KJ719256 |
Note .—Herbarium acronyms follow the Index Herbariorum ( http://sciweb.nybg.org/science2/IndexHerbariorum.asp ).
Genome Size Estimation
The C value of two Italian individuals of Cynomorium was measured using flow cytometry with propidium iodide (PI) as the DNA stain and Pisum sativum ‘Kleine Rheinländerin’ as the standard. Fresh material was cochopped together with the standard plant in Otto’s buffer I ( Otto et al. 1981 ). The resulting suspension was filtered (30-μm nylon mesh), RNase treated, and incubated in PI containing Otto’s buffer II. A CyFlow ML flow cytometer (Partec, Muenster, Germany) equipped with a green laser (100 mW, 532 nm, Cobolt Samba, Cobolt, Stockholm, Sweden) was used for the fluorescence measurements, with 5,000 particles measured per run and three runs performed per plant preparation. The C value was calculated according to the formula: 1 C value Object = (mean G1 nuclei fluorescence intensity Object /mean G1 nuclei fluorescence intensity Standard )*1 C value Standard . The peak CV percentages usually were less than 5%. The C value of a Chinese individual was measured in Kunming, also using a Partec CyFlow ML flow cytometer and the method of Temsch et al. (2010) .
Genome Assembly
De novo assembly and scaffolding of the Illumina reads were conducted using CLC Genomic workbench v.7 after adapter trimming and removing bases with poor quality (assembly parameters: similarity fraction = 0.8, length fraction = 0.5, mismatch cost = 2, insertion cost = 3, deletion cost = 3, word size = 45, bubble size = 98, minimum contig length = 1,000). The read depth of each contig and of the final genomes was calculated after remapping the reads showing 100% identity across 100% of their length and removing potential PCR duplicates using the rmdup command of the Samtools suite ( Li et al. 2009 ). Consensus sequences of the assembled contigs/scaffolds were blasted (BLASTn) against 52 plastid genomes to identify plastome fragments. More potential plastid contigs were identified by: 1) blasting the plastid genes of Lindenbergia philippensis (GenBank accession HG530113), several Saxifragales species ( Liquidambar formosa (KC588388) , Paeonia obovata (KJ206533) , Penthorum chinense (JX436155) , Sedum sarmentosum (JX427551)), and Nicotiana undulata (JN563929) against the Cynomorium contig pool using the megablast algorithm, and 2) mapping the reads to the plastid genes of S. sarmentosum . To identify contigs belonging to the mitochondrial genome, we blasted them against the mitochondrial genes of Carica papaya (EU431224), Capsicum annuum (KJ865410), Salvia miltiorrhiza (KF177345), and Malus domestica (NC018554). To identify mitochondrial contigs not carrying any gene, we conducted less stringent BLAST searches (smaller word size, considering lower bitscores) against 28 plant mitochondrial genomes. Many mitochondrial genes were found in more than one copy in Cynomorium , so extra-caution had to be taken to ensure they were assigned to the correct genomic compartment. We used two strategies: 1) the whole contig including the mitochondrial gene region was blasted (BLASTn) against GenBank. If only the gene yielded a hit, we cut it out from the contig and blasted the remaining parts separately; 2) the contig’s read-depth was analyzed, after taking into account possible biases due to GC content (Results). Nuclear copies of plastid or mitochondrial genes are expected to occur at 1–2 orders of magnitude lower coverage than genes residing in the plastome or the chondriome, and this allows distinguishing organelle fragments from potential nuclear copies. Assemblies of both organelle genomes were extended, combined and refined by iterative read remapping using CLC Genomics Workbench v. 8.5.1 ( http://www.clcbio.com ) and Geneious v. 8.1.6 (Biomatters, http://www.geneious.com/ ). Annotations of the plastome and chondriome were performed with DOGMA and GeSeq ( http://dogma.ccbb.utexas.edu , https://chlorobox.mpimp-golm.mpg.de/geseq-app.html ).
Selection of Phylogenetic Markers and Taxon Sampling
We selected markers from all three genomic compartments for an angiosperm-wide taxon sampling, with special focus on sequences from Saxifragales, Rosales, and frequently reported hosts (Sapindales, Caryophyllales). For the plastid gene alignments we added our Cynomorium plastid sequences to a reduced version of the matrix of Ruhfel et al. (2014) , keeping one representative per family of angiosperms. In the plastome of Cynomorium , we found 27 genes of which some were highly degenerated (Results). For reliable alignments and to reduce long-branch attraction, we selected genes that showed ≥70% identity between Cynomorium and a photosynthetic angiosperm ( L. formosa , GenBank accession KC588388). This resulted in 14 alignments of 10 plastid protein-coding genes ( clpP , rpl2 , rpl14 , rpl16 , rpl36 , rps3 , rps7 , rps11 , rps19 , and ycf2 ), and four plastid rDNAs ( rrn4.5 , rrn5 , rrn16 , and rrn23 ).
From 41 mitochondrial genomes in GenBank, we selected 33 genes that satisfy the above-mentioned criteria. As no mitochondrial genome of Sapindales is available so far, we downloaded the Illumina paired read data of Citrus × paradisi × Citrus trifoliata (GenBank accession Nr. SRX374184) and assembled them de novo in the CLC Genomics Workbench with the following parameters: word size: 40; similarity fraction 0.9; length fraction: 0.6. This yielded 893,637 contigs with an N50 of 858 bp, and a total length of 580,244,525 bp. We then blasted 40 mitochondrial genes (not including the tRNAs) of the Rosales species Malus × domestica (GenBank accession NC 018554) against the Citrus contig pool with a maximal E-value of e-20 with the BLASTn tool implemented in Geneious v.8. If the coverage of a recovered contig was between 2,000 and 2,500×, and the rest of the contig yielded plant mitochondrial hits in a BLASTn search in GenBank, it was considered mitochondrial, annotated with the Annotation tool implemented in Geneious v.8, and cross-checked with the ORF finder tool. This yielded 35 mitochondrial genes of Citrus × paradisi × Citrus trifoliata.
From the nuclear genome, we selected four genes ( MSH1 , PEPC , PHYA , and SMC2 ) and the 18S and 26S rDNAs because they could be unambiguously retrieved from the contigs of the Italian Cynomorium and had homologs in GenBank from many other angiosperm orders. We avoided paralogues by choosing low-copy-number genes ( MSH1 and SMC2 ; Zhang et al. 2012 ), and controlled for them by inspecting the gene annotations, reblasting the sequences against GenBank, and also by checking the single-gene trees for characteristic duplicated topologies where paralogues would form two similar clades in the same tree.
All single-gene matrices included at least one gene copy from the Italian Cynomorium obtained by Illumina sequencing, and when a gene was found multiple times in the same or different genomic compartments, all copies were included in the single-gene alignment. In addition, sequences of all plastid genes except ycf2 , and of the mitochondrial matR , nad5 , and rps3 were obtained by Sanger sequencing from our Chinese and Iranian plants and added to the matrices. We also included sequences of Cynomorium from GenBank; accession numbers are given in table 1 for Cynomorium and in supplementary table S2 , Supplementary Material online for other taxa.
DNA Alignments and Phylogenetic Analyses
Mitochondrial genes and ribosomal DNAs were aligned with MAFFT ( Katoh 2013 ), using the Geneious R7 plugin. More variable plastid and nuclear protein-coding regions were aligned based on amino-acid information, using PAL2NAL ( Suyama et al. 2006 ) and MAFFT, or alternatively MACSE ( Ranwez et al. 2011 ). To avoid possible biases in the phylogenetic reconstructions, we removed the 33-bp long coconversion tract of atp1 , the intron of cox1 including the coconversion tract, and the RNA editing sites of all mitochondrial genes (as annotated in the published Arabidopsis thaliana , Brassica napus , and Citrullus lanatus mitochondrial genomes, see supplementary table S2 , Supplementary Material online, for accession numbers).
Matrices of all individual genes were used in maximum likelihood (ML) phylogenetic tree searches conducted in RAxML v.8.2.4 ( Stamatakis 2014 ) with 100 bootstrap replicates. Because most single-gene regions were not sufficiently informative to recover the expected angiosperm topology, the same analyses were also performed using a constrained topology where only Cynomorium was free to move, using the -g option of RAxML and user-designed fully bifurcating trees based on the topology from APG IV. There were no supported conflicts (ML bootstrap support [BS] ≥70%) between unconstrained and constrained trees, so we base further analyses and interpretations on the latter because they provide a consistent angiosperm background. To assess the divergence of the Cynomorium genes we compared the root-to- Cynomorium branch length to the other root-to-tip lengths in each single-gene tree. Finally, we applied to our single-gene matrices the evolutionary placement algorithm ( Berger et al. 2011 ) implemented in RAxML, which uses ML inference to assign a query sequence to the most likely node(s) in a fixed topology. If the probability of the placement is not 1, the probabilities of alternative placements are estimated. The fixed topologies we used were the same as the ones we used to perform the constrained ML tree reconstructions (above).
Analyses of the plastid genes did not reveal statistically supported conflicts, except for the Cynomorium sequences deposited in GenBank by Zhang et al. (2009) . We thus concatenated all plastid genes, keeping these conflicting Cynomorium sequences as separate taxonomic units (Results). The concatenated final plastid matrix contained only plastome-located genes, had a length of 18,104 nt, and included 83 species from 83 families and 42 orders (using the APG IV classification). Concatenation of the nuclear genes (which yielded no statistically supported topological conflicts), resulted in a matrix of 13,723 nucleotides including 388 species from 388 families and 58 orders ( supplementary table S2 , Supplementary Material online). The mitochondrial genes did not show globally conflicting topologies, but their concatenation was less straightforward due to multiple mitochondrion-located Cynomorium copies of the same gene sometimes falling in different orders (Results). We therefore concatenated only those mitochondrial genes that yielded statistically unsupported placements of Cynomorium and that fulfilled the condition that their multiple copies (if any) formed a clade (so that it was meaningful to pick one randomly to concatenate it with other mitochondrial genes). This approach resulted in at least one copy of atp1 , atp6 , atp8 , atp9 , ccmB , cox2 , cox3 , nad1 exon 5, rps3 , rrn18 , and rrn26 sequences not being included in the main concatenated Cynomorium sequence, but being kept as separate taxonomic units. The concatenated matrix comprised 33,271 nt (26 genes in the main Cynomorium sequence) and 46 species from 28 families and 21 orders.
The best partition scheme and evolutionary models for the three concatenated matrices were found with PartitionFinder v. 1.1.1 ( Lanfear et al. 2012 ) in a greedy search of all possible combinations involving single gene and/or codon position partitioning. Phylogenetic inferences were performed on each concatenated matrix using RAxML v.8.1.24 ( Stamatakis 2014 ) through the CIPRES Science Gateway ( Miller et al. 2010 ) with 1,000 bootstrap replicates, and following the best partition scheme found by PartitionFinder, involving 7 (plastid), 9 (nuclear), or 13 (mitochondrial) partitions. Because too many partitions can be problematic for accurate parameters estimation ( Roberts et al. 2009 ) we also ran the same analysis without partitions. The concatenated mitochondrial matrix was run using the angiosperm topology as constraint and allowing only Cynomorium to move.
Neighbor-net splits graph analysis, implemented in SplitsTree ( Huson and Bryant 2006 ), was used to depict the genetic distances, using patristic distances, for a matrix comprising parts of the genes clpP , rpl2 , rpl14 , rpl16 , and rrn23 of several Cynomorium accessions.
Results
Genome Size Estimation and Sequencing Depth
The 1 C values of two Italian plants were 13.70 and 13.61 pg. The 1 C values of three Chinese plants were 13.76, 12.95, and 13.02 pg. Illumina sequencing of one of the Italian plants yielded 1.58 billion reads from the paired-end library and 0.288 billion reads from the mate-pair library (all reads being 150 bp long), corresponding to an expected coverage of 17× before read cleaning. De novo assembly of the remaining 1.68 billion clean reads yielded 1,123,965 contigs and scaffolds with an average size of 3,200 bp representing 3.6 Gbp or approximately 26% of the genome. The N50 was 4,423 bp, and the largest scaffold was 299,171 bp long. We found 53 contigs for the mitochondrial genome that we could assemble as 49 circular subgenomes (below). For the plastome we identified five contigs that we could assemble in one circular molecule; the longest contig was 19,897 bp long, with an average coverage (ac) of 8,609×, and contained genes typically found in the IR; a second contig was identical to a subpart of the latter (3168 bp, ac = 2362×); a third one contained accD and other genes typical of the LSC region (3,244 bp, ac = 1218×), a fourth contig contained clpP exon 3 and rps12 (455 bp, ac = 280×), and the last contig contained a part of ycf1 (1278 bp, ac = 144×). Nuclear contigs have average per-base read depths of 17 ± 18.5× ( table 2 , supplementary fig. S1 A , Supplementary Material online), the average per-base read depth of the mitochondrial genome is 2,772 ± 665×, and that of the plastome is 3,660 ± 3,379. With mate pair reads only, the plastid genome has a more homogeneous coverage, 2.5× higher on average than that of the mitochondrial genome ( table 2 , supplementary fig. S1 A , Supplementary Material online). The few low per-base read depths observed in the mitochondrial genome and the plastid genomes ( supplementary fig. S1 A , Supplementary Material online) are due to the presence of low-complexity regions, especially AT rich regions. This is seen in supplementary figure S1 B and C , Supplementary Material online, which show that the coverage depends on the GC content, with regions having a lower GC content being less covered. This pattern is not due to the restrictive mapping criteria (100% identity of 100% of length) as it is unaffected by less stringent mapping parameters. The GC content of the plastome was 19.5% in noncoding regions, 24% in the ycf genes, 21.5% in suspected pseudogenes, and higher in ribosomal genes (31.9%), tRNAs (47.6%), and rRNAs (50%), and those are followed by variations in per-base read depth, as shown in figure 2 , supplementary figure S1B and S1 C , Supplementary Material online.
Table 2.
Read Depth and GC Content across the Different Genomic Compartments of Cynomorium from a Mapping Using all six Libraries and from a Mapping Using Only the Mate-Pair (MP) Reads (see also supplementary fig. S1 , Supplementary Material online)
|
Read Depth— All Libraries
|
Read Depth— MP Reads
|
GC Content (%)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome | Total Length (bp) | Mean | Median | SD | Mean | Median | SD | Mean | Median | SD | ||
| Mitochondrial | 1,106,389 | 2,772 | 2,827 | 665 | 169 | 167 | 51 | 44 | 44 | 8 | ||
| Plastid | 45,519 | 3,660 | 2,178 | 3,379 | 411 | 428 | 142 | 30 | 31 | 13 | ||
| Nuclear (partial) | 2,991,600 | 17 | 17.25 | 18.5 | 1.6 | 0 | 4 | 38 | 32 | 9 | ||
Note .—Read depth represents the per-base read depth over the whole length of all contigs (1 plastid, 49 mitochondrial, 182 nuclear), except for the first and last 100 bases, the coverage of which could be biased by the necessary stringency of the mapping; GC content was calculated using a sliding window size of 50 bp.
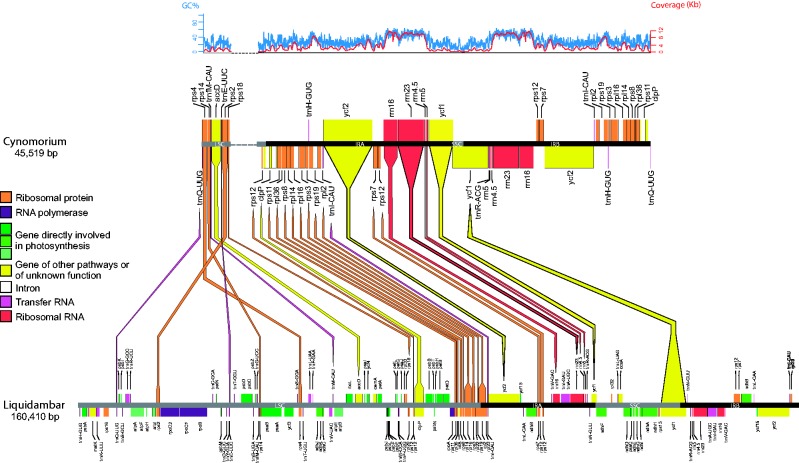
Fig. 2.—
Gene losses and rearrangements in the plastome of Cynomorium compared to that of Liquidambar formosana (GenBank accession KC588388). Colored lines link homologous genes between the two plastomes. The LSC of Cynomorium is divided in two parts (linked by the dotted line) to help visualization. Maps were drawn using OGDraw ( Lohse et al. 2013 ). GC content and coverage of the Cynomorium plastome are depicted in blue and red, respectively. (The pattern is the same when using less stringent mapping parameters; Results.).
Structure and Gene Content of the Organellar Genomes of Cynomorium
The plastome of Cynomorium , presented in figure 2 , has a length of 45,519 bp and resulted from the concatenation of five contigs after iterative extensions. It is divided in two SC regions separated by an IR. A low-complexity (GC = 11%) region between rps18 and rps12 exon 1 had low read-depth (ca. 10×), but Sanger resequencing and/or the Illumina reads supported our assembly, both at this low-coverage region and at the four junctions between the SC and IR regions; in some cases, the sequences obtained by Sanger sequencing were unclear due to mononucleotide stretches ( supplementary fig. S2 , Supplementary Material online).
All genes involved in photosynthesis ( ndh, atp, pet, psa, psb , rbcL ) are missing from the plastome of Cynomorium , which retains a total of 27 genes, namely 14 ribosomal protein genes, clpP, accD , ycf1 , and ycf2 , the four rRNAs, and five tRNAs ( trnE , trnH , trnI , trnfM , and trnQ ). Different from what is observed in the outgroup Liquidambar ( fig. 2 ), the IR makes up the largest part of the plastome, with a length of 2 × 20,136 bp, and comprises most of the genes, starting in clpP intron 2 and ending with a part of ycf1 ( fig. 2 ). The LSC region has a length of 4,066 bp, and contains accD, rps2, rps4, rps12 exon 1, rps18, trnE, trnfM , a part of trnQ (the other part being at the end of the IRb, in clpP intron 2) , clpP exon 3, and a part of clpP intron 2. The small single copy (SSC) region, of 1,190 bp, contains only a part of the ycf1 gene. The genes retained by Cynomorium are collinear with those of Liquidambar except for two rearrangements in the LSC, involving an inversion and displacement of rps14 , and an inversion and displacement of trnE together with rps2 ( fig. 2 ).
Protein-coding genes of the plastome have an open reading frame of at least 80% of the length of the same gene in Liquidambar , except accD , rps18 , ycf1 , and ycf2 , whose lengths are between 53 and 77% of that of Liquidambar ( supplementary table S3 , Supplementary Material online). Cynomorium lost the intron maturase matK but retained four genes containing introns, namely clpP with intron 1 belonging to group II B (gIIb) and intron 2 belonging to group II A (gIIa), rpl2 with one gIIa intron, rpl16 with one gIIb intron, and rps12 , which retained one trans-spliced gIIb intron, but lost the gIIa intron between exon 2 and exon 3 compared to Liquidambar . The rRNA genes share between 75% and 92% identity with Liquidambar along a similar length except for rrn4.5 , which has a deletion of 10 bp. All five tRNAs are able to form a clover-leaf secondary structure according to the tRNAscan-SE webserver ( http://lowelab.ucsc.edu/tRNAscan-SE/ , last accessed on March 29, 2016), although trnQ has a mispairing in its acceptor stem ( supplementary table S3 , Supplementary Material online).
An assembly of the Cynomorium mitochondrial genome comprises 49 circular contigs, ranging between 10,804 and 32,985 bp for a total length of 1,106,389 bp. The contigs have an average GC content of 44.3%, with little difference between coding and noncoding parts. The variation in read depth despite the homogeneous GC content ( table 2 ), as well as our BLAST searches suggest that many parts of the genome are duplicated, and that those repeats can lead to various conformations (possibly also a master genome), depending on the recombination between the circular subgenomes. The mitochondrial subgenomes of Cynomorium are summarized in supplementary fig. S3 , Supplementary Material online; altogether they contain the standard angiosperm mitochondrial gene set with conserved open reading frames, except possibly sdh3 . The main sdh3 sequence lacks a start codon and contains a frameshift, but it may encode a functional protein if its RNA is edited, which requires more investigation.
Insights from Substitution Rates on the Genomic Location of Genes
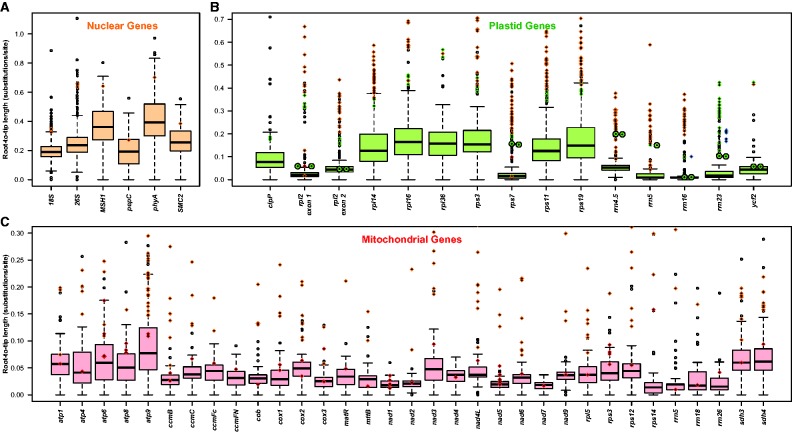
Figure 3 shows the root-to-tip branch length of Cynomorium gene copies relative to those of other angiosperms included in the trees. Average root to tip branch lengths are mostly >0.2 substitutions/site (subst./site) for nuclear genes, >0.1 subst./site for the plastid genes of SC regions, and ≤0.05 subst./site for those of the IR ( fig. 3 A and B), whereas they are <0.1 (and mostly <0.05) subst./site for the mitochondrial genes ( fig. 3 C). For all plastid and all nuclear genes, the branches leading to Cynomorium (orange and green diamonds in fig. 3 A and B) are longer than the average angiosperm branch length, which is not the case for many mitochondrial genes (red diamonds in fig. 3 C). Plastid and mitochondrial gene copies that based on their read-depth are located in the nuclear genome all have higher substitution rates than those assumed to be located in the plastid or mitochondrial genomes (compare orange diamonds and green and red diamonds in fig. 3 B and C). Similarly, plastid gene copies assumed transferred into the mitochondrial genome had a lower substitution rate than those located in the plastome (red diamonds in fig. 3 B). These results fit with the expected differences in substitution rate between the three genomic compartments, indirectly supporting the accuracy of our coverage-based assignments of the genes to the Cynomorium genomic compartments.
Fig. 3.—
Root to tip branch lengths of Cynomorium and other angiosperms in substitutions per site. Boxplots and open circles summarize the branch length distribution of: (A) nuclear, (B) plastid, and (C) mitochondrial genes of photosynthetic plants obtained from the constrained phylogenies in supplementary figures S3–S5 , Supplementary Material online. Black line: median; boxes: upper and lower quartile, including 50% of the data; whiskers: minimum and maximum of the data, provided that their length does not exceed 1.5× the interquartile range; open dots: outliers. Colored diamonds and circles represent, respectively, the branch length of the genes and of their copies found in other genomic compartments of Cynomorium : orange: gene copy located in the nuclear genome; green: gene copy located in the plastid genome; red: gene copy located in the mitochondrial genome; blue: gene copy amplified by Garcia et al. (2004) ; circles: Cynomorium gene copies amplified by Zhang et al. (2009) ; diamonds: all other Cynomorium sequences.
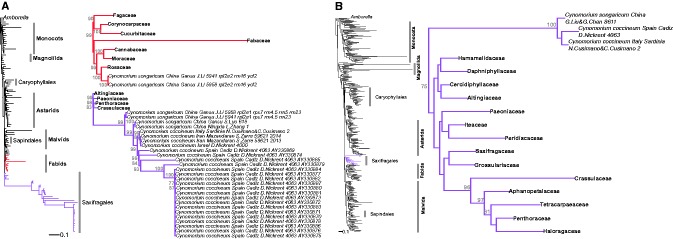
Single Gene Trees and Evolutionary Placement Results Are Congruent
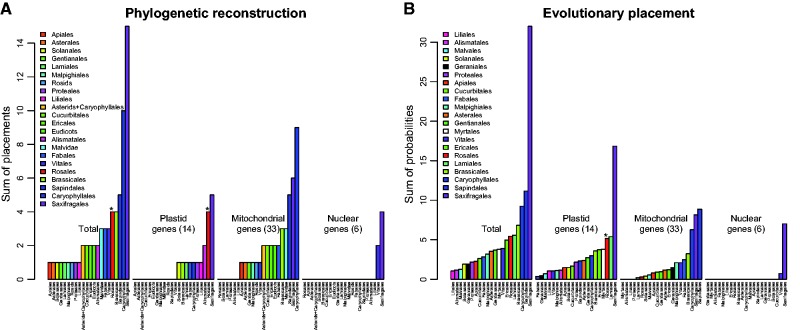
ML searches most commonly placed Cynomorium in the Saxifragales and—with mitochondrial genes—also the host orders Caryophyllales and Sapindales ( fig. 4 A, supplementary figs. S4–S6 and table S4 , Supplementary Material online). The evolutionary placement analyses, which assess the likelihood of alternative phylogenetic placements, yielded the same results as the ML searches ( fig. 4 B and supplementary fig. S7 , Supplementary Material online). Single-gene trees are shown in supplementary figures S4–S6 and table S4 , Supplementary Material online. In three plastid gene trees, our sequences (in green) do not cluster with those amplified by Zhang et al. (2009 ; in blue), which cluster with Rosales (see below), while ours cluster with Saxifragales ( rpl2 exon2, ycf2 with support) or the host order Fabales ( rrn16 , without support). In the other 12 plastid gene trees, all plastid-located plastid genes from this study (green) or others (blue) cluster with each other, and Cynomorium is three times sister to Saxifragales ( rpl2 exon 1, rpl14 and rps3 ) and twice sister to Alismatales ( rps11 , rrn5 ); other placements included Rosales ( rrn23 ) and the host order Caryophyllales ( rrn4.5 ), always without support. Plastid gene copies located in the mitochondrial (red) or nuclear (orange) genomes mostly clustered with those located in the plastome, but in a few cases, they clustered with the host orders Asterales (a mitochondrial and six nuclear copies of rrn5 , and a nuclear copy of rpl2 exon 1), Caryophyllales (a mitochondrial copy of rps7 and a nuclear copy of rps11 ), Fabales (a mitochondrial and a nuclear copy of rrn4.5 ), or Sapindales (three nuclear copies of rrn4.5 ), always without support ( supplementary table S4 and fig. S4 , Supplementary Material online).
Fig. 4.—
Bar plots summarizing the analyses of single gene matrices when the rest of the topology was constrained to fit the currently accepted angiosperm relationships ( Angiosperm Phylogeny Group 2016 ). For simplicity, internally transferred gene copies were not included but their placements can be seen in supplementary figures S4–S6 , Supplementary Material online. (A) Results of ML tree searches. For each clade, the number of genes that placed Cynomorium sequence(s) in the respective clade is shown (details in supplementary figs. S4–S6 and table S4 , Supplementary Material online). (B) Results of evolutionary placement analyses (Materials and Methods). For each clade, the sum of the probabilities across all genes that placed Cynomorium in the respective clade is shown (details in supplementary fig. S7 , Supplementary Material online). For both analyses, results are shown overall and by genomic compartment. The asterisks indicate placements resulting exclusively from the sequences of Zhang et al. (2009) . Numbers of markers for each compartment are indicated in brackets.
Mitochondrial gene copies located in the mitochondrial genome clustered together in 29 out of 37 cases ( supplementary table S4 and fig. S5 , Supplementary Material online). The exceptions are 1) atp6 , where the three copies found clustered with Apiales + Asterales or Cucurbitales, without support; 2) atp8 , where the three copies clustered with Caryophyllales or Sapindales with >80% BS ( supplementary table S4 , Supplementary Material online); 3) atp9 where one copy clustered with Caryophyllales with >80% BS and one copy fell in Fabales without support; 4) ccmB , where one copy fell in Caryophyllales and another in Malvidae without support; 5) cox2 , where one copy placed as sister to asterids and Caryophyllales without support whereas another grouped with Caryophyllales with >80% BS; 6) cox3 where one copy clustered in Caryophyllales with >90% BS and another in Malvidae with >70% BS; 7) nad1 exon 5 where one copy grouped with Caryophyllales and another with Gentianales without support; and 8) rps3 where three Chinese and Italian accessions grouped with Saxifragales with low support whereas one Chinese sequence grouped with Caryophyllales with >90% BS. In the remaining 29 gene trees, Cynomoriaceae usually grouped with Saxifragales ( ccmFc , ccmFN when the long-branched Geraniales were removed from the matrix, matR , mttB , and nad4 ; without support), Brassicales ( nad4L , rps14 , and nad3 when the long-branched Geraniales were removed), or Geraniales ( atp4 , ccmFN , and nad3 ; without support and possibly resulting from long-branch attraction). Other genes gave different placements, notably rosids ( nad7 ) and the host orders Asterales ( rps12 ), Caryophyllales ( atp4 when the long-branched Geraniales were removed, nad2 , and rrn18 —the latter with >70% BS), Fabales ( rrn5 ), or Sapindales ( atp1 with >90% BS, nad1 exon 1, nad6 , and rrn26 with >70% BS), most without strong statistical support. Mitochondrial gene copies located in the nuclear genome mostly clustered with those located in the mitochondrial genome, but in a few cases, they placed somewhere else ( supplementary table S4 , Supplementary Material online), especially in the host orders Asterales and/or Caryophyllales ( atp1 , mttB , nad3 , and nad9 ), and Sapindales (three copies of atp9 and one copy of sdh4 ), usually without support ( supplementary table S4 and fig. S5 , Supplementary Material online).
The six nuclear genes sequenced from different Cynomorium plants always clustered together ( supplementary fig. S6 , Supplementary Material online) and usually grouped with sequences from Saxifragales (18S, 26S, MSH1 , and SMC2 , with between 70% and 100% BS; supplementary table S4 , Supplementary Material online).
Attempts to Reproduce a Placement of Cynomorium in Rosales
Zhang et al. (2009) used primers designed by Dhingra and Folta (2005) for functional chloroplast genomes to PCR-amplify genes from the plastome IR of two Chinese Cynomorium plants. We retrieved their sequences from GenBank and included them in our trees where they fell in two different orders depending on the gene: Rosales ( rpl2 exon 2, and complete ycf2 and rrn16 genes) or Saxifragales ( rpl2 exon 1, rps7 , rrn 4.5 , rrn5 , and rrn23 ; supplementary table S4 and fig. S4 , Supplementary Material online). Our newly generated sequences of all these genes instead clustered in Saxifragales, regardless of their geographic origin (Chinese, Italian, or Iranian), genomic compartment (plastid, mitochondrial, or nuclear copies), or sequencing method (Sanger sequencing with our own primers or with those used by Zhang et al., or Illumina shotgun sequencing). Substitution rates of the copies amplified by Zhang et al. (2009) are also outliers, depending on the gene: their Rosales-like copies had a low substitution rate compared with the higher rate of our Cynomorium copies and their own Saxifragales-like copies (green circles in fig. 3 B). Unspecific amplification of Rosales DNA (either a contamination or a horizontally acquired DNA located in the nuclear or mitochondrial genome) would explain these patterns. When mapped against the Illumina data of the Italian Cynomorium , the primers used by Zhang et al. (2009) to amplify ycf2 and rrn16 do not match the Cynomorium plastome, neither does the reverse primer used to obtain rpl2 -exon 2, whereas the primers used to amplify rps7 , rrn 4.5 , rrn5 , and rrn23 do match. The two primers amplifying rpl2 -exon 1 match the mitochondrial contig in which we also found a copy of this exon. That Zhang et al. (2009) accidentally amplified this copy is supported by the fact that their sequence and our mitochondrial copy cluster together in the tree (in Saxifragales with the plastid copies; supplementary table S4 and fig. S4 , Supplementary Material online) and show the same unusually low substitution rate instead of that of the native plastid-located gene copies (green circles and red diamond in fig. 3 B).
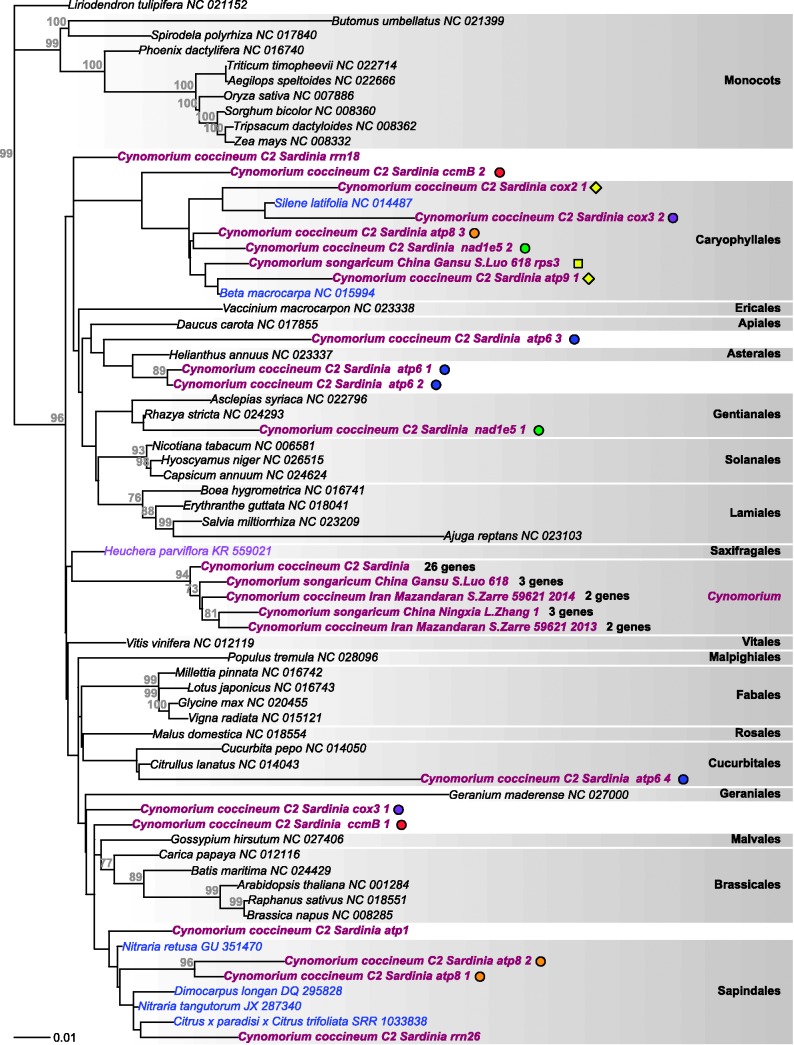
Results from Concatenated Data
The concatenated plastid and nuclear matrices with or without unlinked data partitions yielded topologies that fit with the topology accepted by Angiosperm Phylogeny Group (2016), except for the position of magnoliids (without BS; figs. 5 and 6 ). The concatenated mitochondrial matrix yielded a topology that did not match accepted angiosperm relationships due to lack of signal ( fig. 3 ), and we therefore constrained this topology (Materials and Methods). In the plastid tree ( fig. 5 A), Cynomorium plants newly sequenced for this study as well as previously sequenced plants from Spain (voucher Nickrent 4063 ), Israel ( Nickrent 4000 ), and China (concatenated rps7 , rpl2 exon 1 rrn4.5 , rrn5 , and rrn23 from Zhang et al. 2009 ) fell in Saxifragales (with 99% BS), specifically as sister to Crassulaceae and Penthoraceae (without statistical support). Zhang et al. (2009)rpl2 exon 2, rrn16 , and ycf2 sequences instead grouped with Rosaceae (with 100% BS). Sequences from plants from Israel, Italy, and Spain form a well-supported clade nested in a grade of Chinese accessions, with the most basal being the accessions from Zhang et al. (2009 ; fig. 5 A). In the nuclear tree ( fig. 5 B), Cynomorium is sister to Saxifragales with 75% BS, with the Spanish and Italian sequences closer to each other than to the Chinese sequences. In the mitochondrial tree obtained with a partitioned model ( fig. 6 ), the main Cynomorium clade is sister to Heuchera (Saxifragales), but without support, whereas it is sister to rosids in the unpartitioned analysis, also without support (not shown). The single-gene sequences that were outliers compared with the majority of the mitochondrial genes (see previous section) clustered with the host clades Caryophyllales (Italian atp8 , atp9 , ccmB, cox2 , cox3 , and nad1 exon 5, Chinese rps3 ) or Sapindales (Italian atp1 , atp8 and rrn26 ), or had “intermediate” placements ( ccmB, cox3 , rrn18 ). Different copies of atp6 cluster either with Cucurbitales or Asterales. Removing those sequences did not change the position of the main Cynomorium clade and failed to increase BS (data not shown).
Fig. 5.—
Phylogenetic trees obtained from ML analyses of the concatenated plastid (A) and nuclear (B) genes. Major angiosperm taxa are labeled following Angiosperm Phylogeny Group (2016) , with the orders including Cynomorium in purple (Saxifragales) and red (Fabids), and shown in more detail on the right.
Fig. 6.—
Phylogenetic tree obtained from ML analyses of the concatenated mitochondrial gene matrix. The genus label Cynomorium on the right refers to the placement of 26 genes from the Italian plant. The colored dots mark multiple copies of the respective gene; the two yellow diamonds mark a gene acquired by HGT, of which one native copy was included in the concatenated matrix; the single yellow square marks the rps3 of a Chinese accession (see text).
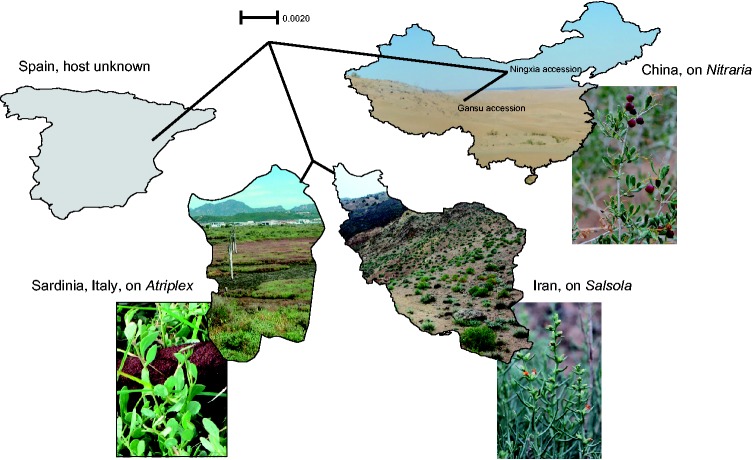
Geography and Genetic Variation within Cynomoriaceae
Figure 7 shows a neighbor net from four plastid protein-coding genes and the plastid 23S rDNA together with the water-stressed, often saline habitats in which the sequenced plants were growing on the hosts shown in the photos. The network reflects the geographical distribution of the sampled populations and is completely tree-like because there is no internal contradiction (no homoplasy) in the data. The great genetic distinctness of the single Spanish sequence is surprising.
Fig. 7.—
Neighbor net obtained from variable plastid regions of Cynomorium from Spain ( Nickrent 4063 ), Italy (N. Cusimano and C. Cusimano 2), Iran ( S. Zarre 59621 ), and China (L. Zhang 1 and S.X. Luo 618). The photos show habitats and hosts of Cynomorium at our collecting sites. The host of the Spanish sample is unknown. Photos by N. Cusimano (Italy and Atriplex ), S.X. Luo (China and Nitraria ), and A. Gröger (Iran and Salsola ).
Discussion
The Mitochondrial and Plastid Genomes of Cynomorium
The Cynomorium mitochondrial genome is only the fourth of any parasitic plant to have been assembled, following the partial genome of Rafflesia lagascae ( Xi et al. 2013 ) and the complete ones of Viscum album and Viscum scurruloideum ( Petersen et al. 2015 , Skippington et al. 2015 ). The organization of the mitochondrial genome of Cynomorium in many sublimons is similar to that found in Silene ( Sloan et al. 2012 ) and V. scurruloideum , with many repeated regions facilitating recombination of the genome and thereby leading to many sublimons. The size and gene content of the chondriome of Cynomorium (and of R. lagascae ) are comparable to that of other angiosperms, while the chondriomes of Viscum have unexpectedly lost all genes from the respiration complex I ( nad genes). Different from Viscum , and to a lesser extent Rafflesia , the substitution rates of Cynomorium mitochondrial genes are of the same order as those of other angiosperms ( fig. 3 C), making it easier to identify ancient HGT events in this lineage (below).
The Cynomorium plastome appears to be circular, and our assembly is probably complete because PCR products of the LSC and SSC are of the expected size, and Sanger sequencing and/or Illumina reads confirmed the IR junctions and the low-coverage region in the LSC region. It contains 27 of the typically 116 angiosperm plastome genes and presents the quadripartite structure observed in many photosynthetic angiosperms, albeit with a very small SC region containing only a piece of ycf1 . In gene content, it is similar to the plastome of Hydnora , another ancient exoholoparasite ( Naumann et al. 2013 , 2016 : about 100 Myr). The 24 genes of the Hydnora plastome are all found in Cynomorium , which in addition retains clpP , trnH , and trnQ , of which trnQ may be a pseudogene. A few genes of Cynomorium are very different in length and identity from the outgroup Liquidambar ( supplementary table S3 , Supplementary Material online), among them accD , which is essential for fatty-acid biosynthesis ( Kode et al. 2005 ) and has the five C-terminal domains shown to be conserved in all known accD sequences ( Lee et al. 2004 ) so that it is probably still functional. The ycf1 and ycf2 genes are also shorter in Cynomorium than in Liquidambar ( supplementary table S3 , Supplementary Material online), but still have large ORFs of 1,273 and 1,761 amino acids, so they could be functional. Length and structure of at least ycf1 are known to vary a lot across land plants ( deVries et al. 2015 ). Their function remains unclear ( deVries et al. 2015 ; Nakai 2015 ), and they are absent from Poales ( Wicke et al. 2011 ) and the non-photosynthetic Sciaphila ( Schelkunov et al. 2015 ), Epipogium ( Lam et al. 2015 ), and Pilostyles ( Bellot and Renner 2016 ). In Hydnora , however, they are expressed ( Naumann et al. 2016 ). Finally, although only half of rps18 of Liquidambar can be aligned to Cynomorium , both lineages conserve the most characteristic domain of this protein.
Four genes contain introns and thus depend on functional splicing machinery, which could present a problem because Cynomorium lost the plastid-encoded maturase matK . However, clpP intron 2, which belongs to group II A, and most of the other introns, which belong to group II B, rely on nuclear-encoded splicing factors ( Zoschke et al. 2010 ; Germain et al. 2013 ). Only rpl2 , which normally depends on matK to splice its group II A intron, may be pseudogenized despite its conserved exons. Interestingly, in Hydnora visseri where matK has also been lost, rpl2 is conserved but does not rely on matK because it lost its intron. In Hydnora longicollis , however, rpl2 is transcribed but is likely a pseudogene ( Naumann et al. 2016 ). A functional loss of rpl2 in Cynomorium would reduce to 16 the minimal set of genes encountered in all exoholoparasites so far examined. The high stem age of Cynomorium and Hydnora and the conservation of 24–27 genes, most of them functional, in their otherwise reduced plastomes contrast with the absent or extremely reduced plastomes of the younger or equally old endoparasites Pilostyles and Rafflesia ( Molina et al. 2014 ; Bellot and Renner 2016 ), and supports a hypothesized difference in plastome function between endo- and exoparasites because the former never have free-living stems.
More generally, the existence of a few plastome genes in Cynomorium and Hydnora that are lost idiosyncratically in other exoparasites ( accD , rps2 , rps18 , rps19 , ycf1, ycf2 ), as well as the retention of clpP and trnH in Cynomorium but not Hydnora , implies either random events or not yet understood lineage-specific selection.
Cynomorium Belongs in Saxifragales and Other Ordinal Placements Are Due to HGT s or Contamination
With sequence data from all three genomic compartments (and from plants representing the family’s range), our study firmly resolves the phylogenetic position of one of the last unplaced angiosperm families ( Stevens 2001 onwards; Angiosperm Phylogeny Group 2009 , 2016 ). That Cynomoriaceae belong in the Saxifragales notably was inferred by Nickrent et al. (2005) a full 10 years ago, based on nuclear 18S and 26S rDNA and the mitochondrial matR from a single plant ( Nickrent 4063 from Spain). The precise placement of Cynomoriaceae within Saxifragales requires denser taxon sampling; Saxifragales comprise 15 families (counting Cynomoriaceae), 117–120 genera, and approximately 2,500 species. Only one of the 55 genes (or their additional copies) obtained from our material placed Cynomorium inside Rosales ( rrn23 , <70% BS), and we suspect that the Rosales placements of the copies of Zhang et al. (2009) were due to sequences obtained with primers designed for functional plastomes, which did not bind to the highly degenerated Cynomorium plastome but instead to contaminant DNA (Results).
We find evidence of HGT, involving both interspecific transfers of mitochondrial genes from host plants into the mitochondrial and the nuclear genomes of Cynomorium , and intracellular transfers of mitochondrial and plastid genes into the nuclear genome. Therefore, we agree with Barkman et al. (2007) interpretation that HGTs from hosts, most likely Nitrariaceae, are the explanation of the grouping in Sapindales obtained from the mitochondrial atp1 and cox1 (including intron) sequences, and we extend this interpretation to the placements of ten other gene copies in Sapindales (two copies of atp8 , rrn26 ) and the other host order Caryophyllales ( atp8 , atp9 , ccmB , cox2 , cox3 , nad1 exon 5, rps3 ). To complicate the picture, some of those genes may have been acquired multiple times from the same or different hosts (e.g., atp8 from Caryophyllales and Sapindales; supplementary table S4 , Supplementary Material online, fig. 6 ). Surprisingly, we could not find native, non-host-like copies of atp1 and atp8 , indicating that host copies of these genes may have replaced the native homologs. Other unexpected phylogenetic placements could be the result of old transfers from unknown or rare hosts such as Asterales (e.g., atp6 ; fig. 6 ), which would further blur phylogenetic reconstructions based on mitochondrial genes.
The inferred extent of HGT places Cynomorium intermediate between the mistletoe V. scurruloideum , in which only the cox1 intron appears horizontally transferred ( Skippington et al. 2015 ), and Rafflesia , which acquired numerous genes from its host ( Tetrastigma , Vitales) or also from unknown past hosts ( Xi et al. 2013 ). Differences in the extent of HGT could be due to the age of a parasitism in a lineage and possibly the type of parasitism: Endoparasites, such as Rafflesia , may be more prone to HGT than exoparasites, such as Viscum and Cynomorium.
Genome Size, Chromosome Numbers, and Geographic History of Cynomorium
The only chromosome count for Cynomorium , obtained from pollen mother cells of a plant parasitizing Tamarix tetragyna Ehrenb. in the lower Jordan Valley, is n = 14, and the karyotype is bimodal ( Pazy et al. 1996 ). For Saxifragales, genome sizes are known for 78 of their 2,500 species, representing 8 of their 15 families with the 1 C values of Cercidiphyllaceae, Daphniphyllaceae, Grossulariaceae, Haloragaceae, Hamamelidaceae, and Saxifragaceae all ≤2.38 pg ( Bennett and Leitch 2012 ). The Cynomoriaceae genomes measured here have 1 C values of approximately 13 pg, similar to those of Crassulaceae and Paeoniaceae (with the single genus Paeonia ), which have genome sizes of 9.1 and 12.05–30.5 pg ( Bennett and Leitch 2012 ).
The Cynomorium neighbor net ( fig. 7 ) illustrates the considerable genetic distances within this lineage, as is expected from populations growing as far apart as Spain, Italy, Iran, and the Mongolian deserts in China ( fig. 1 ). Cynomoriaceae are among the 60 or so seed plant families endemic to the Holarctic ( Takhtajan 1986 ), and Takhtajan considered their single genus ( Cynomorium ) a floristic element of the Tethyan (ancient Mediterranean) subkingdom and the modern Mediterranean region, while stressing that the eastern boundary of the Mediterranean region and its separation from the Irano-Turanian region are difficult to define. This is because the Tethyan flora developed primarily by migration, and the majority of this flora has boreal and eastern Asian origins ( Takhtajan 1986 ; Manafzadeh et al. 2013 ). The Irano-Turanian region includes the Zagros and Alborz mountain ranges of the Iranian plateau, which arose synchronously during the mid-Miocene (references in Manafzadeh et al. 2013 ) and which are part of the Alpine–Himalayan mountain system. Many molecular-biogeographic studies over the past 15 years have inferred east to west expansion of plant lineages from the western Chinese interior towards the Mediterranean (e.g., Zhang et al. 2015 for Nitraria ), and Cynomorium may also have expanded from the Mongolian deserts to Afghanistan and Iran as the Tethys closed and the Arabian Peninsula connected with Eurasia; this would explain the nesting of Iranian and Italian sequences among Chinese ones in the mitochondrial phylogeny ( fig. 6 ). Using the mitochondrial matR sequence of Barkman et al. (EU281095; which correctly placed Cynomorium in Saxifragales, but comes from an unknown collecting site), Naumann et al. (2013) estimated the stem age of Cynomoriaceae as 100 Myr, with a 95% confidence interval of 76–117 Ma. We here refrain from applying a clock-model within Cynomorium , but judging from the within-genus genetic distances ( fig. 7 and supplementary fig. S5 , Supplementary Material online), our sampled populations have been separated for many million years.
Supplementary Material
Supplementary figures S1–S7 and tables S1–S4 are available at Genome Biology and Evolution online ( http://www.gbe.oxfordjournals.org/ ).
Supplementary Material
Acknowledgments
The authors thank Dr M. Silber for her invaluable help in the lab and O. Pérez for help with figures 1 and 2 ; N.C. thanks the “Bayerische Gleichstellungsförderung 2014” (Bavarian equal opportunity program) for a 1-year postdoc stipend.
Literature Cited
- Angiosperm Phylogeny Group . 2009. . An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III . Bot J Linn Soc . 161 : 105 – 121 . [Google Scholar]
- Angiosperm Phylogeny Group . 2016. . An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV . Bot J Linn Soc . 181 : 1 – 20 . [Google Scholar]
- Barkman TJ , et al. . 2007. . Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants . BMC Evol Biol. 7 : 248.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot S, Renner SS. 2014. . Exploring new dating approaches for parasites: the worldwide Apodanthaceae (Cucurbitales) as an example . Mol Phylogenet Evol. 80 : 1 – 10 . [DOI] [PubMed] [Google Scholar]
- Bellot S, Renner SS. 2016. . The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes . Genome Biol Evol. 8 : 189 – 201 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD., Leitch IJ. 2012. . Plant DNA C-values database (release 6.0, Dec 2012). [cited 2016 Jul 9]. Available from: http://data.kew.org/cvalues/ .
- Berger SA, Krompass D, Stamatakis A. 2011. . Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood . Syst Biol. 60 : 291 – 302 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Funston AM. 2007. . Cynomoriaceae . Flora of China 13 : 434 . [Google Scholar]
- Cronquist A. 1968. . The evolution and classification of flowering plants . Boston, USA: : Houghton Mifflin; . [Google Scholar]
- Cui Z , et al. . 2013. . The genus Cynomorium in China: an ethnopharmacological and phytochemical review . J Ethnopharmacol . 147 : 1 – 15 . [DOI] [PubMed] [Google Scholar]
- De Vries J, Sousa FL, Bölter B, Soll J, Gould SB. 2015. . YCF1: a green TIC? Plant Cell 27 : 1827 – 1833 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra A, Folta KM. 2005. . ASAP: amplification, sequencing & annotation of plastomes . BMC Genomics 6 : 176.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García MA, Nicholson EH, Nickrent DL. 2004. . Extensive intraindividual variation in plastid rDNA sequences from the holoparasite Cynomorium coccineum (Cynomoriaceae) . J Mol Evol. 58 : 322 – 332 . [DOI] [PubMed] [Google Scholar]
- Germain A, Hotto AM, Barkan A, Stern DB. 2013. . RNA processing and decay in plastids . Wiley Interdiscip Rev RNA . 4 : 295 – 316 . [DOI] [PubMed] [Google Scholar]
- Hansen B. 1986. . The Balanophoraceae of Continental Africa . Bot Jahrb Für Syst . 106 : 359 – 377 . [Google Scholar]
- Huson DH, Bryant D. 2006. . Application of phylogenetic networks in evolutionary studies . Mol Biol Evol. 23 : 254 – 267 . Software available from www.splitstree.org [DOI] [PubMed] [Google Scholar]
- Katoh S. 2013. . MAFFT multiple sequence alignment software version 7: improvements in performance and usability . Mol Biol Evol. 30 : 772 – 780 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A. 2005. . The tobacco plastid accD gene is essential and is required for leaf development . Plant J. 44 : 237 – 244 . [DOI] [PubMed] [Google Scholar]
- Lam VKY, Soto Gomez M, Graham SW. 2015. . The highly reduced plastome of mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection . Genome Biol Evol. 7 : 2220 – 2236 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. . PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses . Mol Biol Evol. 29 : 1695 – 1701 . [DOI] [PubMed] [Google Scholar]
- Lee SS , et al. . 2004. . Characterization of the plastid-encoded carboxyltransferase subunit ( accD ) gene of potato . Mol Cells . 17 : 422 – 429 . [PubMed] [Google Scholar]
- Li H , et al. . 1000 Genome Project Data Processing Subgroup 2009. . The Sequence alignment/map (SAM) format and SAMtools . Bioinformatics 25 : 2078 – 2079 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. . OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets . Nucleic Acids Res. 1 – 7 . http://ogdraw.mpimp-golm.mpg.de/ , doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manafzadeh S, Salvo G, Conti E. 2013. . A tale of migrations from east to west: the Irano-Turanian floristic region as a source of Mediterranean xerophytes . J Biogeogr. 41 : 366 – 379 . [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. . Creating the CIPRES science gateway for inference of large phylogenetic trees . New Orleans, LA: Gateway Computing Environments Workshop (GCE).p. 1 – 8 . [Google Scholar]
- Molina J , et al. . 2014. . Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) . Mol Biol Evol. 31 : 793 – 803 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M. 2015. . YCF1: a green TIC: response to the de Vries et al. commentary . Plant Cell 27 : 1834 – 1838 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann J , et al. . 2013. . Single-copy nuclear genes place haustorial Hydnoraceae within Piperales and reveal a Cretaceous origin of multiple parasitic angiosperm lineages . PLoS One 8 : e79204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann J , et al. . 2016. . Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae) . Genome Biol Evol. 8 : 345 – 363 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent DL, Ouyang Y, Duff RJ, dePamphilis CW. 1997. . Do nonasterid holoparasitic flowering plants have plastid genomes? Plant Mol Biol. 34 : 717 – 729 . [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Duff RJ, Colwell AE, Wolfe AD, Young ND, Steiner KE, dePamphilis CW , et al. . 1998. . Molecular phylogenetic and evolutionary studies of parasitic plants (Chapter 8). In: Soltis D, Soltis P, Doyle J , editors. Molecular systematics of plants II. DNA sequencing . Boston, MA: : Kluwer Academic Publishers; . p. 211 – 241 . [Google Scholar]
- Nickrent DL, Blarer A, Qiu YL, Vidal-Russell R, Anderson FE. 2004. . Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer . BMC Evol Biol. 4 : 40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent DL, Der JP, Anderson FE. 2005. . Discovery of the photosynthetic relatives of the “Maltese mushroom” Cynomorium. BMC Evol Biol. 5 : 38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Oldiges H, Goehde W, Jain VK. 1981. . Flow cytometric measurement of nuclear DNA content variations as a potential in vivo mutagenicity test . Cytometry 2 : 189 – 191 . [DOI] [PubMed] [Google Scholar]
- Pazy B, Plitmann U, Cohen O. 1996. . Bimodal karyotype in Cynomorium coccineum L. and its systematic implications . Bot J Linn Soc . 120 : 279 – 281 . [Google Scholar]
- Petersen G, Cuenca A, Møller IM, Seberg O. 2015. . Massive gene loss in mistletoe ( Viscum , Viscaceae) mitochondria . Sci Rep . 5 : 17588.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP. . 2011. . MACSE: multiple alignment of coding SEquences accounting for frameshifts and stop codons . PLoS One 6 : e22594 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TE, Sargis EJ, Olson LE. 2009. . Networks, trees, and treeshrews: assessing support and identifying conflict with multiple loci and a problematic root . Syst Biol. 58 : 257 – 270 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. 2014. . From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes . BMC Evol Biol. 14 : 23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelkunov MI , et al. . 2015. . Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol Evol. 7 : 1179 – 1191 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB , et al. . 2012. . Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates . PLoS Biol. 10 : e1001241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E, Barkman TJ, Rice DW, Palmer JD. 2015. . Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes . Proc Natl Acad Sci U S A. 112 : E3515 – E3524 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. . RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies . Bioinformatics . 2014 : 1312 – 1313 . doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens PF. 2001. onwards. Angiosperm Phylogeny Website. Version 12, July 2012 [and more or less continuously updated since] [cited 2016 May].”Available from: http://www.mobot.org/MOBOT/research/APweb/
- Su H-J, Hu J-M, Anderson FE, Der JP, Nickrent D. 2015. . Phylogenetic relationships of Santalales with insights into the origins of holoparasitic Balanophoraceae . Taxon 64 : 491 – 506 . [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. . PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments . Nucleic Acid Res. 34 : W609 – W612 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teryokhin ES, Nikiticheva ZI, Yakovlev MS. 1975. . Development of the seed, endosperm and embryo in Cynomorium songaricum Rupr. (Cynomoriaceae) . Bot Zhurnal . 60 : 153 – 162 . [English translation by A. Shipunov, aided by D. Nickrent]. [Google Scholar]
- Takhtajan A. 1973. . Evolution und Ausbreitung der Blütenpflanzen . Stuttgart: : G. Fisher; . [Google Scholar]
- Takhtajan A. 1986. . Floristic regions of the world . Berkeley: : University of California, Press; . [Google Scholar]
- Temsch EM, Greilhuber J, Krisai R. 2010. . Genome size in liverworts . Preslia 82 : 63 – 80 . [Google Scholar]
- Untergasser A , et al. . 2012. . Primer3—new capabilities and interfaces . Nucleic Acid Res. 40 : E115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, Müller KF, dePamphilis CW, Quandt D. 2011. . The evolution of the plastid chromosome in land plants: gene content, gene order, gene function . Plant Mol Biol. 76 : 273 – 297 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z , et al. . 2012. . Horizontal transfer of expressed genes in a parasitic flowering plant . BMC Genomics 13 : 227.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z , et al. . 2013. . Massive mitochondrial gene transfer in a parasitic flowering plant clade . PLoS Genet. 9 : e1003265.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yi X, Peng M, Zhou Y. 2012. . Stable carbon and nitrogen isotope signatures of root-holoparasitic Cynomorium songaricum and its hosts at the Tibetan plateau and the surrounding Gobi desert in China . Isotopes Environ Health Stud . 2012 : 483 – 493 . doi:10.1080/10256016.2012.680593 [DOI] [PubMed] [Google Scholar]
- Zhang Z-H, Li C-Q, Li J. 2009. . Phylogenetic placement of Cynomorium in Rosales inferred from sequences of the inverted repeat region of the chloroplast genome . J Syst Evol. 47 : 297 – 304 . [Google Scholar]
- Zhang N, Zeng L, Shan H, Ma H. 2012. . Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms . New Phytol . 195 : 923 – 937 . [DOI] [PubMed] [Google Scholar]
- Zhang DL , et al. . 2014. . Root parasitic plant Orobanche aegyptiaca and shoot parasitic plant Cuscuta australis obtained Brassicaceae-specific strictosidine synthase-like genes by horizontal gene transfer . BMC Plant Biol. 14 : 19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ML, Temirbayeva K, Sanderson SC, Chen X. 2015. . Young dispersal of xerophil Nitraria lineages in intercontinental disjunctions of the Old World . Sci Rep . 5 : 13840 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R , et al. . 2010. . An organellar maturase associates with multiple group II introns . Proc Natl Acad Sci U S A. 107 : 3245 – 3250 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucca P , et al. . 2013. . Evaluation of antioxidant potential of “Maltese Mushroom” ( Cynomorium coccineum ) by means of multiple chemical and biological assays . Nutrients 5 : 149 – 161 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.