Abstract
Loss of introns in plant mitochondrial genes is commonly explained by retroprocessing. Under this model, an mRNA is reverse transcribed and integrated back into the genome, simultaneously affecting the contents of introns and edited sites. To evaluate the extent to which retroprocessing explains intron loss, we analyzed patterns of intron content and predicted RNA editing for whole mitochondrial genomes of 30 species in the monocot order Alismatales. In this group, we found an unusually high degree of variation in the intron content, even expanding the hitherto known variation among angiosperms. Some species have lost some two-third of the cis-spliced introns. We found a strong correlation between intron content and editing frequency, and detected 27 events in which intron loss is consistent with the presence of nucleotides in an edited state, supporting retroprocessing. However, we also detected seven cases of intron loss not readily being explained by retroprocession. Our analyses are also not consistent with the entire length of a fully processed cDNA copy being integrated into the genome, but instead indicate that retroprocessing usually occurs for only part of the gene. In some cases, several rounds of retroprocessing may explain intron loss in genes completely devoid of introns. A number of taxa retroprocessing seem to be very common and a possibly ongoing process. It affects the entire mitochondrial genome.
Keywords: intron loss, retroprocessing, RNA editing, mitochondrial genome, Alismatales
Introduction
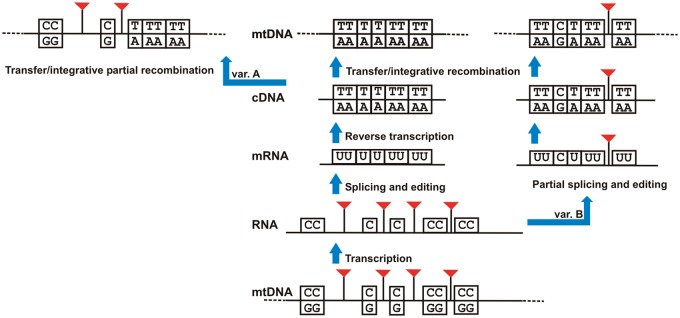
Over evolutionary time intron loss has occurred in virtually all organisms with spliced genes (Joly et al. 2001; Lambowitz and Zimmerly 2004; Odom and Herrin 2013). However, the mechanisms underlying intron loss remain poorly understood. Currently, the most widely accepted mechanism of intron loss is a reverse transcriptase (RT)-mediated model (also called the cDNA-mediated model by some authors, e.g. Odom and Herrin 2013, or simply retroprocessing). Under this model, a processed (spliced) mRNA is reverse transcribed and the resulting intron-free cDNA integrates into the genome by homologous recombination (fig. 1) (Fink 1987; Derr and Strathern 1993; Mourier and Jeffares 2003; Cohen et al. 2012).
Fig. 1.—
Integration of mitochondrial gene sequences through a cDNA intermediate. The central panel shows a gene with four introns and eight edited sites. Following splicing and RNA editing, the mRNA sequence is reverse transcribed and through homologous recombination it replaces the original gene completely. Variation A shows how partial recombination replaces only part of the original sequence. Variation B shows how partial RNA editing and incomplete splicing followed by retrotranscription and homologous recombination may produce a gene where some edited sites and introns remain. The figure is modified from Henze and Martin (2001).
Although reverse transcription has been widely invoked in discussions of gene evolution, reports of cDNA production in vivo are extremely rare (Derr and Strathern 1993; Dhellin et al. 1997; Odom and Herrin 2013). Indirect evolutionary evidence supporting the RT-mediated model of intron loss has been found, however, in different groups. One of the predictions of this model is a positional bias of intron loss towards the 3′-end of genes, consistent with the fact that reverse transcription starts at the 3′-end and that the reverse transcriptase tends to dissociate prematurely from the template, without reaching the mRNA 5′-end (Zhang et al. 2010). This 3′-bias has been found in some organisms, but not in others (Mourier and Jeffares 2003). A second piece of evidence comes from an apparent association between losses of multiple adjacent introns and a higher rate of intron-less pseudogene production (that may also be created by intron-less cDNAs), observations that are both consistent with a RT-mediated intron loss model. As in the first case, empirical evidence is inconclusive, with some groups showing a clear tendency to lose adjacent introns, and not others, reviewed by Zhu and Niu (2013a, 2013b).
Intron content varies greatly among plant mitochondria. Comparative analyses of different plant lineages indicate a high rate of intron gain and loss early in land-plant evolution, with subsequent stabilization in descendant lineages (Mower et al. 2012). In angiosperms, most of the variation in mitochondrial intron content is not caused by intron loss, but by an overall higher frequency of gene loss and presumably transfer to the nucleus. A number of lineage-specific intron losses have, however, been detected, and multiple losses were recently found in the genus Geranium (Park et al. 2015). In addition, a few genes (e.g., nad1 and cox2) seem to have a very dynamic evolutionary history, with frequent intron loss among angiosperm groups (Gugerli et al. 2001; Joly et al. 2001; Hepburn et al. 2012).
Comparative analyses of mitochondrial genomes seem to indicate that some genes/lineages are particularly labile with respect to intron content. The few cases in which intron loss have been studied in plant mitochondrial genomes do suggest the presence of a RT-mediated mechanism, although a model of intron loss through horizontal gene transfer and gene conversion has been proposed recently as being a more likely explanation of intron loss in angiosperm mitochondria (Hepburn et al. 2012), thus questioning the extent of involvement of retroprocessing in intron loss in this organelle.
Plant mitochondrial transcripts are subject to frequent RNA editing. Thus, besides from producing intron-less genes an RT-mediated mechanism has been used to explain loss of RNA editing in plant mitochondrial (and plastid) genes (Geiss et al. 1994; Bowe and dePamphilis 1996; Petersen et al. 2006; Ran et al. 2010; Grewe et al. 2011; Cuenca et al. 2012). Reverse transcription of a processed (edited) mRNA produces a cDNA copy that differs in its edited sites from the original gene, and recombination with a cDNA converts the genomic sequence from one that requires editing to one that does not (fig. 1) (Odom and Herrin 2013). Thus, a positive correlation between loss of introns and loss of edited sites supports the TR-mediated model of intron loss.
Evidence that retroprocessing is acting in plant mitochondria comes mostly from the lycophyte Isoetes engelmannii (which represents the oldest extant split of vascular plants), where simultaneous absence of RNA editing and introns was found in three gene sequences (cox2, nad1, and nad4; Grewe et al. 2011). Similarly, the rps3 gene in some conifers is also intron-less and associated with a loss of editing at numerous sites (Ran et al. 2010), and the entire mitochondrial genome of Welwitschia has experienced a massive loss of both edited sites and cis-spliced introns (Guo et al. 2016). Within the angiosperms RT-mediated intron loss has also been proposed but examples are less well documented (e.g., Park et al. 2015), and the RT-mediated model has been rejected in a recent study evaluating the mechanism of intron loss in the cox2 gene in Magnolia (Hepburn et al. 2012).
All studies addressing intron loss in plant mitochondria share the same weakness, as they are based on only one gene (or intron) in a single species, or on a small sample of a few closely related species. In this paper, we present a detailed study of intron loss for 22 plant mitochondrial introns (out of the 19–25 introns present in the typical angiosperm mitochondrial genome) in 31 species representing all 14 families of the order Alismatales (Monocotyledons). This order was chosen because we previously observed some correlation between intron presence/absence and the occurrence of edited sites (Cuenca et al. 2012). Remarkably, in the current study, we have detected an unusually high degree of variation in the intron content among members of this group, spanning all the variation known in the mitochondrial genomes of all other angiosperms. Some families within the group display a typical angiosperm intron content, whereas others reveal different degrees of intron loss, including the most intron-poor angiosperm species found so far. In this study, we present evidence that an RT-mediated model is able to explain most cases of intron loss within Alismatales.
Materials and Methods
Plant Material, Sequencing, and Assembly
We included mitochondrial genome data for 31 species within the Alismatales, 29 of which are reported in this paper for first time; two represent previously published mitochondrial genomes, Spirodela polyrhiza (Wang et al. 2012) and Butomus umbellatus (Cuenca et al. 2013). Our taxon sampling includes members for all families within the order.
About 454 Titanium series libraries were constructed for mitochondrial DNA of three taxa: Zostera marina, Stratiotes aloides, and Potamogeton natans following the protocol of Cuenca et al. (2013). Library construction and sequencing was carried out at the National High-throughput DNA Sequencing Centre at the University of Copenhagen in a quarter of plate for each. About 454 reads were assembled in Newbler v. 2.3 using default parameters. Twenty-six Illumina libraries were obtained from the MonAToL project (see Ross et al. 2016 for details). Details about taxon sampling can be found in Petersen et al. (2016) and Ross et al. (2016).
Local databases were created with all assembled contigs. Contigs including mitochondrial genes were identified by BLAST using as a template all the cDNAs extracted from the mitochondrial genome of B. umbellatus (NC021399), S. polyrhiza (NC017840), and Liriodendron tulipifera (KC821969), and annotated manually in Geneious ver. 6.1 (Kearse et al. 2012). In addition, Illumina reads were mapped against the Butomus and the Liriodendron mitochondrial genome using Bowtie ver. 7.1.0. This was useful where complete genes and/or introns were not present in the assembled contigs.
To construct a phylogeny including representatives of other angiosperm groups, mitochondrial protein-coding genes where extracted from 26 published complete mitochondrial genomes. Gene matrices were constructed for each protein-coding gene and aligned based on translations using MUSCLE (Edgar 2004).
Estimation of Edited Sites
To empirically identify edited sites, we compared DNA sequences with available transcriptome data from seven members of Alismatales: Posidonia australis, Sagittaria latifolia, Triglochin maritima, and Pistia stratiotes, all obtained from the MonAToL project (http://www.botany.wisc.edu/monatol/welcome.htm), and S. polyrhiza (SRR497624), Elodea nuttalli (ERR062145, ERR062146, and ERR062147), and Z. marina (SRX021650) downloaded from the SRA database. Transcriptome data were mapped against our taxon specific DNA reference sequences. Whenever a C to T change was found in a taxon, this position was annotated as being edited in the matrix. As we do not have transcriptome data for all taxa and all genes (nad4L is missing from all transcriptomes), this strategy does not identify all edited sites (especially for taxa outside Alismatales). Thus, we complemented the results from empirical observations by using the program Prep-Mt (Mower 2009) to further predict presence of edited sites within the dataset.
Phylogenetic Analyses
In all phylogenetic analyses, the edited sites were excluded. Maximum-likelihood (ML) trees were obtained for each individual gene as well as for a concatenated matrix including all 20 mitochondrial genes. ML trees were obtained using the program RAxML (ver. 7.2.8) with the GTR + gamma model, and 1,000 replicates of rapid bootstrapping (Stamatakis et al. 2008). The ML tree obtained from the concatenated matrix was used for character mapping as well as for correlation analyses.
Testing the Correlation between the Numbers of Edited Sites and the Presence of Introns
We used a model of continuous random walk implemented in the program BayesTraits v. 2 (Meade and Pagel 2014) to test the correlation between numbers of edited sites and presence of introns. This program uses Markov chain Monte Carlo (MCMC) methods to derive posterior distributions and ML to obtain point estimates of the parameters of statistical models, and the values of traits at ancestral nodes of phylogenies. BayesTraits allows the analysis of continuous characters using a generalized least squares (GLS) approach where the non-independence among the species is accounted for by reference to a matrix of the expected covariance among species. This matrix is derived from the phylogenetic tree. The model estimates the covariance of changes between pairs of traits, to test for correlation. In a MCMC framework, tests of likelihood often rely on Bayes factors. The logic of Bayes factors is similar to a likelihood test, but instead of comparing the ML between two models, the harmonic means of the likelihoods are compared (Meade and Pagel 2014). In this context, a Bayes factor > 5 is considered strong evidence for rejecting the model with fewer parameters, which in this case is one of the independence between characters (presence/absence of introns vs. number of edited RNA sites).
The analyses were done in three different ways: (a) genome-based analyses, comparing frequency of edited sites in the genes that usually include introns in angiosperm mitochondria with the proportion of introns (both cis- and trans-spliced) present in each taxa compared with the ancestor of monocots + eudicots; (b) gene-based analyses, where correlation between edited site frequency and intron content was calculated for each individual gene; and (c) intron-based analyses, where the correlation was tested for each intron coded as a binary character (absence/presence) against the number of edited sites in the two adjacent exons. As there are no differences in gene length among taxa (the few gap columns in the alignments where not included in the analyses), the total number of edited sites is essentially reflecting the editing frequency at both adjacent exons. The ccmFc gene was not included in the analyses partly because we were not able to empirically determine edited sites using transcriptome data, and partly due to alignment problems within Alismatales, making assignment of homologous edited sites dubious. Genes coding for ribosomal proteins were also excluded because rpl2 and rps10 are only rarely present in Alismatales, and rps3 seems to be degenerated in some species of this group (data not shown).
For all correlations, we used a MCMC of 1,100,000 iterations with a burn-in of 10,000 iterations, uniform priors, and a single tree; the ML tree obtained from the mitochondrial data excluding edited sites (see above). Bayes factors were obtained by comparing the harmonic mean from an analysis where both characters are allowed to change independently, against the harmonic mean from an analysis where both characters are forced to evolve in a correlated manner.
Mapping of Edited Sites and Presence of Introns
Intron loss was mapped on the ML mitochondrial tree using Dollo optimization, which is based on the assumption that a complex character that has been lost during evolution of a particular lineage cannot be regained (Rogozin et al. 2006). Similarly, Dollo optimization was used to identify branches where major changes in editing frequency occurred (somewhat arbitrarily considering a major drop to have occurred when the number of edited sites is reduced to less than 50% of the edited sites present in the most recent common ancestor, MRCA).
In the mapping analysis, we considered a simultaneous loss of intron(s) and edited sites as evidence of a RT-meditated model of intron loss, if (a) the edited sites are adjacent to the intron position and (b) the edited sites lost constitute at least 50% of the edited sites present in at least one of the flanking exons in the intron-bearing MRCA.
Results
Intron Presence is Highly Variable within Alismatales with Several Introns Lost in Parallel
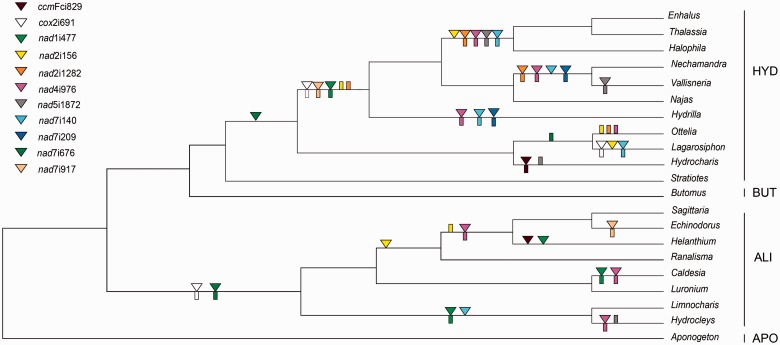
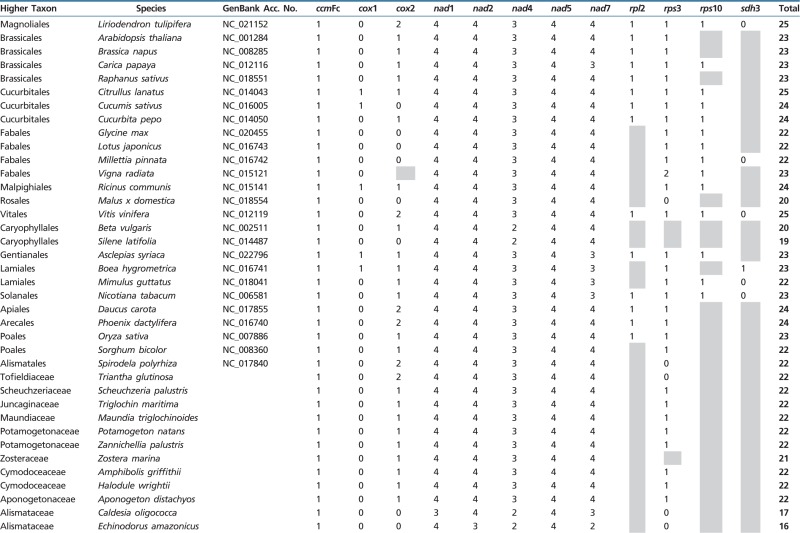
The mitochondrial genome of flowering plants typically contains between 19 and 25 introns (table 1). However, the present study shows that there is more variation in intron content within the order Alismatales than among all other angiosperms. Intron content within the sampled Alismatales ranged from 12 to 22 introns. In particular, some members of families Alismataceae and Hydrocharitaceae are exceptionally depauperate in cis-spliced mitochondrial introns (table 1). Mapping the presence/absence of introns onto a phylogeny including 31 members of Alismatales reveals that most cis-spliced introns were lost in parallel in these two families (table 1 and fig. 2). The most extreme cases of intron loss are found in the Hydrocharitaceae, where only six cis-spliced introns are retained in Enhalus, Thalassia and Halophila. In contrast, no trans-spliced intron was found to be missing within Alismatales.
Table 1.
Mitochondrial Intron Content in Different Angiosperm Groups. Cells in Grey Indicate Gene Absence. The Total Number of Introns is Given for Whole Mitochondrial Genomes, and Plant Species are Sorted by Order. GenBank Data from Complete Mitogenomes Harvested March 2015. For Species of Alismatales Gene Sequences are Deposited in GenBank Under Accession nos. KU642045–KU642059, KU642204–KU642232, KX254028–KX254102, KX272887–KX272933 and KX363594–KX363635.
 |
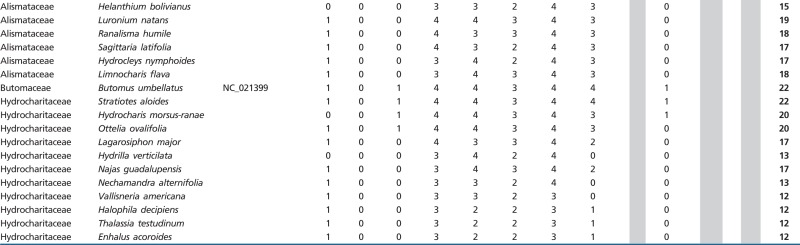 |
Fig. 2.—
Mapping of intron loss in Alismatales. Triangles represent an intron loss, and rectangles represent a loss >50% of edited sites in the adjacent exons to the intron indicated. Colors represent different introns, as specified in the figure. For simplicity, Alismatales families other than Aponogetonaceae (APO), Alismataceae (ALI), Butomaceae (BUT), and Hydrocharitaceae (HYD) are not shown in the tree.
Five introns (nad4i976, nad7i676, cox2i373, cox2i691, and nad1i477) previously reported as lacking in some angiosperms (Knoop et al. 2011) are also all lacking in some members of Alismatales. We also found that additional introns previously considered universally present in the seed plants are lacking in some Alismatales (ccmFci829, nad2i156, nad2i1282, nad5i1872, nad7i140, nad7i209, and nad7i917). Surprisingly, five of these otherwise universally present introns were lost not only once but also several times within the group (fig. 2).
Strong Correlation between Intron Loss and Removal of Edited Sites Supports Retroprocessing
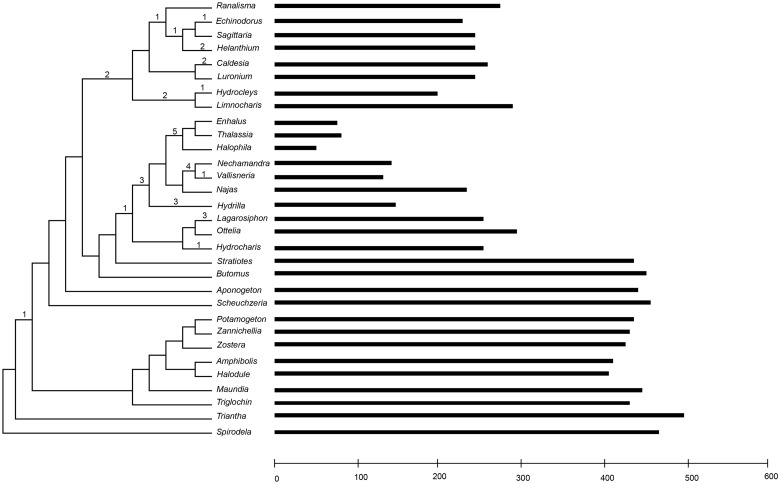
To test whether a RT-mediated model can explain intron loss in Alismatales, we can test for correlation between the occurrence of introns and edited sites. Under this model, intronless genes that also contain the edited nucleotide state will be produced, eliminating the need for editing in the retroprocessed gene. In this study, we used a combination of empirical evidence (transcriptome data) and computational methods (Prep-Mt, Mower 2009) to predict edited sites for all mitochondrial genes. Approximately 85% of the edited sites are supported by empirical data. In figure 3, the location of introns and edited sites in the nad7 gene is shown for the all taxa of Alismatales and similar maps of the remaining genes are included as supplementary material (supplementary figs. S2–S6, Supplementary Material online). Our results clearly show a near 10-fold difference in editing frequency among taxa within Alismatales, ranging from 496 sites in 21 mitochondrial genes in Triantha to only 51 sites in Halophila (fig. 4 and supplementary table S1, Supplementary Material online).
Fig. 3.—
Distribution of introns and edited sites in the nad7 gene in members of Alismatales. Rectangles represent exons and black triangles represent cis-spliced introns. Vertical lines inside exons represent edited sites. Horizontal lines joining exons indicate the lack of introns.
Fig. 4.—
Number of edited sites in 21 mitochondrial genes in members of the order Alismatales. Horizontal bars indicate number of edited sites for each taxon in the phylogeny. Values above branches in the phylogenetic tree indicate number of cis-spliced introns lost in that branch, as obtained from our mapping analyses. The phylogenetic tree has been pruned from the ML tree of 46 angiosperm taxa based on 20 mitochondrial genes.
When correlations are evaluated using total number of introns (with the exception of ccmFci829 and introns present in ribosomal genes; see Materials and Methods section) against the number of edited sites in the intron-bearing genes, a Bayes factor of 18 is obtained, indicating very strong evidence for correlation between intron presence and editing frequency. This result indicates that species with low intron content also have a low editing frequency, but is silent concerning the processes affecting individual genes. Therefore, we also performed gene-by-gene correlations, finding a strong correlation for nad7 and cox2 and, to a lesser extent, for nad1 and nad5 (table 2). In general, genes carrying trans-spliced introns showed a lower correlation between intron presence and editing status (nad1, nad2, and nad5). This is hardly surprising, because it is not clear whether it is possible to lose trans-spliced introns, once they are established.
Table 2.
Correlation between Intron Numbers and Edited Sites Frequencies for Individual Genes. Bayes Factors (BF) Were Obtained Using BayesTraits and by Comparing Harmonical Means Between a Model Where Both Characters are Independent Against a More Complex Model Where Both Characters Evolve in a Correlated Manner. In this Context, a BF > 5 is Considered to be Evidence of Correlation, and BF > 10 as Strong Evidence of Correlation (Meade and Pagel 2014)
| cox2 | nad1 | nad2 | nad4 | nad5 | nad7 | all | |
|---|---|---|---|---|---|---|---|
| BF | 13.2 | 8.1 | 3.3 | 5.8 | 4.4 | 18 | 18 |
To remove the confounding signal created by trans-spliced introns, and to test whether cis-spliced introns in a single gene could have been removed by a RT-mediated mechanism, we tested for correlation between intron presence/absence and the number of edited sites in the two adjacent exons. Strong correlation was found in seven of the 11 cases of intron loss tested here (table 3). Within the same gene, heterogeneity in the correlation between intron presence and editing frequency was found. For example, in cox2, the lack of cox2i373 is independent of editing frequencies (not supporting a RT-mediated model for intron removal), whereas the lack of cox2i691 shows a very strong correlation with editing status in the adjacent exons (supporting retroprocessing). The same phenomenon is seen in nad7, where nad7i209 seems to have been removed by a different mechanism than the adjacent intron nad7i676.
Table 3.
Correlation between Intron Presence and Edited Sites Frequencies in Adjacent Exons. Bayes Factors (BF) Were Obtained Using BayesTraits and by Comparing Harmonical Means Between a Model Where Both Characters are Independent Against a More Complex Model Where Both Characters Evolve in a Correlated Manner. In this Context, a BF > 5 is Considered to be Evidence of Correlation, and BF > 10 as Strong Evidence of Correlation (Meade and Pagel 2014)
| cox2i373 | cox2i691 | nad1i477 | nad2i156 | nad2i1282 | nad4i976 | nad5i1872 | nad7i140 | nad7i209 | nad7i676 | nad7i917 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BF | 0.4 | 30.8 | 21.9 | 0.8 | 13.5 | 13.8 | 12.9 | 6.7 | 1 | 26.8 | 16.2 |
Character Mapping Reveals 27 Cases Where Edited Sites and Introns Where Lost Simultaneously
To obtain a detailed picture of the frequency of retroprocessing in Alismatales, we optimized all instances of intron loss and reduced editing frequency in the adjacent exons on a phylogeny of the group (fig. 2). As intron loss occurs only in Alismataceae and Hydrocharitaceae, we show only the clade comprising these families plus Butomus (Butomaceae), plus Aponogeton (Aponogetonaceae) as the sister group (fig. 2). The complete phylogeny including all taxa used in this study is shown in supplementary fig. S1 (Supplementary Materials online). We used Dollo optimization for the loss of introns, meaning than once an intron is lost it cannot be regained (McPherson et al. 2004; Rogozin et al. 2006). Dollo optimization does not strictly apply to editing frequencies, as a gain of edited sites is not an uncommon event, although it occurs at a significantly lower rate than loss. However, it has been shown that the gain of edited sites follows a scattered pattern in the mitochondrial genome (Petersen et al. 2006; Cuenca et al. 2010), making the likelihood of gaining a series of edited sites in adjacent positions very small.
Our results show that the history of intron loss in the two families is complex (detailed descriptions of the behavior of each individual gene may be found in supplementary material, Supplementary Materials online). When all evidence is considered, we found 27 instances of intron loss associated with loss of edited sites in one or both the adjacent exons (table 4), supporting a RT-mediated model for intron removal. However, in seven instances, introns were removed without affecting the adjacent edited sites, and a different mechanism may need to be invoked to explain intron loss (table 5).
Table 4.
Cases of Intron Loss in Alismatales Supporting the RT-Mediated Model
| Intron | Group | ES lost/totala | ES 3b | ES 5c | Retroprocessing max size (nt)d | Retroprocessing min size (nt)e |
|---|---|---|---|---|---|---|
| ccmFci829 | Hydrocharis | 15/18 | 9 | 6 | 680 | 412 |
| cox2i691 | Alismataceae | 8/12 | 6 | 2 | 226 | 162 |
| cox2i691 | Lagarosiphon | 12/12 | 10 | 2 | 499 | 399 |
| cox2i691 | Hydrilla-Enhalus | 9/12 | 8 | 1 | 281 | 265 |
| nad1i477 | Hydrocleys-Limnocharis | 7/7 | 1 | 6 | 273 | 249 |
| nad1i477 | Hydrilla-Enhalus | 5/5 | 1 | 4 | 273 | 231 |
| nad1i477 | Caldesia | 3/5 | 1 | 2 | 183 | 63 |
| nad1i477 | Helanthium | 2/5 | 0 | 2 | 273 | 63 |
| nad2i1282 | Nechamandra-Vallisneria | 6/6 | 1 | 5 | 754 | 206 |
| nad2i1282 | Halophila-Enhalus | 6/6 | 1 | 5 | 754 | 206 |
| nad4i976 | Helanthium-Echinodorus | 28/30 | 12 | 18 | 910 | 788 |
| nad4i976 | Caldesia | 18/26 | 2 | 16 | 410 | 270 |
| nad4i976 | Hydrocleys | 21/30 | 10 | 11 | 574 | 532 |
| nad4i976 | Halophila-Enhalus | 5/5 | n.a | 5 | 936 | >421f |
| nad4i976 | Nechamandra-Vallisneria | 4/5 | n.a | 4 | 909 | >396f |
| nad4i976 | Hydrilla | 4/5 | n.a | 4 | 909 | >396f |
| nad5i1872 | Halophila-Enhalus | 13/13 | 7 | 6 | 541 | 467 |
| nad5i1872 | Vallisneria | 7/13 | 1 | 6 | 367 | 94 |
| nad7i140 | Lagarosiphon | 5/5 | 5 | n.a | 212 | >143f |
| nad7i140 | Hydrilla | 5/6 | 3 | 2 | 1143 | >207f |
| nad7i140 | Halophila-Enhalus | 6/6 | 4 | 2 | 210 | 171 |
| nad7i140 | Nechamandra-Vallisneria | 2/6 | 0 | 2 | 1098 | >75f |
| nad7i209 | Hydrilla | 2/2 | 2 | n.a | 1143 | >68f |
| nad7i209 | Nechamandra-Vallisneria | 2/2 | 2 | n.a | 1098 | >68f |
| nad7i676 | Alismataceae | 18/19 | 12 | 6 | 520 | 499 |
| nad7i917 | Hydrilla-Enhalus | 9/13 | 5 | 4 | 793 | 244 |
| nad7i917 | Echinodorus | 5/6 | 1 | 3 | 716 | 189 |
Edited sites lost in the area homologous to the two adjacent exons/Edited sites present in the intron-bearing MRCA of the group in question.
Edited sites lost in a continuous stretch downstream the area of intron loss.
Edited sites lost in a continuous stretch upstream the area of intron loss.
Calculated as the stretch between the last edited site present in the MRCA upstream of the region of possible retroprocessing and the first edited site downstream that region.
Calculated as the stretch between the most upstream and the most downstream edited site lost by retroprocessing event.
No edited sites are present in one of the exons in the intron-bearing MRCA, impeding estimate the minimum stretch of the retroprocessing in that exon. A minimum stretch was only calculated for one exon, indicated by the symbol >.
Table 5.
Cases of Intron Loss in Alismatales that Do Not Support the RT-Mediated Model
| Intron | Group | ES lost/totala | Distance 5’(nt)b | Distance 3’(nt)b |
|---|---|---|---|---|
| ccmFci829 | Helanthium | 0/18 | 337 | 366 |
| cox2i373 | Alismatidsc | 0/14 | 105 | 59 |
| nad2i156 | Ranalisma-Sagittaria | 3/12 | 9 | 69 |
| nad2i156 | Lagarosiphon | 0/18 | 9 | 60 |
| nad2i156 | Hydrilla-Enhalus | 0/3 | 9 | 201 |
| nad7i140 | Hydrocleys-Limnocharis | 0/7 | 7 | 56 |
| nad7i676 | Hydrocharitaceae excl. Stratiotes | 3/15 | 448 | 45 |
Edited sites lost in the area homologous to the two adjacent exons/Edited sites present in the intron-bearing MRCA of the group in question.
Distance from the closest edited sites to the position of intron in other taxa.
Refers to the order Alismatales except for families Araceae and Tofieldiaceae.
Furthermore, we found cases where editing frequency dropped without affecting intron presence. In at least 10 instances, this reduction in the number of edited sites occurred adjacent to a trans-spliced intron, most commonly in exons 2 and 3 of nad2 (fig. 3 and supplementary figs. S2–S6, Supplementary Materials online). However, to our knowledge, no trans-spliced intron has been reported missing, either in Alismatales or in any other angiosperm. There are also a number of cases where edited sites have been lost, but the cis-spliced intron remains, particularly within nad4 (fig. 2 and supplementary fig. S5, Supplementary Materials online). In some of these examples, introns have been lost subsequently.
Phylogenetic Position of Najas
One potential source of error in our character mapping is the position of Najas in the phylogeny. The position of Najas in a clade with other members of the Hydrocharitaceae is strongly supported here, but is in conflict with the position suggested by a total evidence analysis and data from the plastid genome (Petersen et al. 2016; Ross et al. 2016). In some genes, Najas shows a different pattern of editing and intron presence than the other members of the clade, and this may inflate the estimated number of retroprocessing events if Najas is misplaced, as a number of parallel retroprocessing events in nad2, nad4, and nad7 need to be inferred under the Dollo scheme to explain the observed pattern (fig. 2). An alternative hypothesis would be that Najas actually gained introns and/or edited sites, which seems highly implausible. In order to evaluate whether Najas could have gained edited sites and an intron by horizontal gene transfer, we tested recombination in the nad7 gene using the software 3seq (Boni et al. 2007). This analysis indicates 13 cases of chimeric sequences in our dataset, one in Najas with a minimum recombinant region of 183 nucleotides spanning exons 1 and 2 (data not shown). However, this result is a direct consequence of the number of edited sites, and when the analysis is repeated using a matrix with edited sites excluded, no sequence triplet was found significant for recombination (data not shown). Similarly, no chimeric sequences were found when the nad2 gene was analyzed edited sites excluded, indicating that our results here are not affected by possible horizontal transfer, which may occur in the mitochondria of other plant groups.
Extent of Retroprocessing in the Mitochondrial Genome—Are Other Genes Affected?
If retroprocessing occurred at high rates in members of Alismatales, the effect would obviously not be limited to intron-bearing genes. To investigate whether this is the case, we determined edited sites for all mitochondrial genes in Alismatales, and mapped their history on the phylogeny. Ribosomal genes were not included in the analysis because most of them are lacking in Alismatales, and the few that are present show evidence of pseudogenization in some species (data not shown).
As with intron-bearing genes, many other genes seem to have lost their edited sites in members of Alismataceae and Hydrocharitaceae (fig. 4 and supplementary table S1, Supplementary Material online). This is in contrast with the more typical pattern of editing in the remaining 12 families in Alismatales. In fact, our analyses indicate that some members of Hydrocharitaceae have the lowest reported numbers of edited sites in all angiosperms.
Discussion
Although intron loss is known in a few mitochondrial genes in some angiosperms groups (e.g., Silene), massive loss of mitochondrial introns has hitherto only been reported from Geranium and species of Viscum, where the loss in the latter is associated with complete gene loss (Park et al. 2015; Petersen et al. 2015; Skippington et al. 2015). With regard to the number of introns retained in the mitochondrial genome, our data are comparable with those from Geranium, where only nine introns (five trans-spliced, four cis-spliced) were found (Park et al. 2015). The loss of introns seems to be associated with a reduction in numbers of edited sites. Among non-angiosperm seed plants, a similar case has been reported from Welwitschia with 10 introns (seven trans-spliced, three cis-spliced) retained and coupled with a strongly reduced number of edited sites (Guo et al. 2016). Although both cases may represent examples of RT-mediated intron loss, this was not an aspect explored in any of the two studies.
In the Alismatales, we observe that members of both Alismataceae and Hydrocharitaceae have experienced massive losses of introns, too. But an intron-poor mitochondrial genome is not an ancestral condition in either of these families; rather losses occurred several times independently in nested groups within each family. Our results from detailed analyses of correlation between intron loss and loss of edited sites support an RT-mediated model, which can explain many (but not all) of the cases of intron loss in Alismatales. Our results also point to retroprocessing as having been particularly active in the families Alismataceae and Hydrocharitaceae, but not in the other members of Alismatales. However, the biological reasons for this difference among clades remain unknown. The RT-mediated model cannot readily, however, explain all instances of intron loss, and in the more some cases, as for example, in cox2, more than one mechanism of intron loss may need to be invoked to explain the loss of adjacent introns within a gene. An alternative model of intron loss is genomic deletion, but in this case, the intron will usually be cut imprecisely, altering the intron–exon borders. Although imperfect intron cuts altering the intron–exon border by many amino acids would not be seen, as they presumably should be removed by selection, cuts causing indels of only one or a few amino acids may very well be conserved. However, no evidence of such imprecise intron cuts was found here for any of the intron losses where the RT-mediated model is not supported. Additionally, it has been proposed that introns can be lost by local gene conversion after an intron-lacking exogenous gene copy is integrated in the mitochondrial genome by horizontal transfer (Hepburn et al. 2012).
We found only one case, involving atp9 in Tofieldia, where a gene is duplicated and one of the copies clearly is of exogenous origin (data not shown). However, this gene does not possess introns, and, therefore, has not been included in our analyses of intron loss. Although small gene fragments of obviously exogenous origin were found in our Illumina libraries, it is not clear whether they are integrated into the mitochondrial genome (usually these gene fragments have very low coverage, suggesting that they could represent contamination or nuclear copies). In any case, none of these gene fragments spans more than a small piece of a single exon, and no evidence of recombination between these small pieces and the native gene was found. Therefore, we are inclined to believe that horizontal transfer is not causing intron loss in Alismatales.
Trans-Spliced Introns
No trans-spliced intron has been reported missing in Alismatales or any other angiosperm, raising questions about how retroprocessing and/or gene conversion might affect this kind of intron. One possible explanation lies in the manner in which processed genes (or parts thereof) are reinserted into the genome. This is generally believed to require homologous recombination, which is active and frequent in the plant mitochondrial genome (Arrieta-Montiel et al. 2009; Davila et al. 2011). In order to remove the intron in a cis-spliced gene by homologous recombination, crossovers must take place on either side of the intron (Fink 1987), which is clearly not possible in a trans-spliced intron. However, we did find sequences flanking trans-spliced introns that seem to have experienced retroprocessing, as inferred by the simultaneous loss of a considerable number of edited sites (table 6). Examples of this include six sites upstream from nad1i394 lost in Alismataceae and in Nechamandra and Vallisneria, five sites downstream from nad1i728 in Hydrilla, 13 edited sites upstream from nad2i709 in some Hydrocharitaceae, nine edited sites upstream from nad2i709 in Sagittaria and Echinodorus, 14 sites upstream from nad5i1477 in some members of Hydrocharitaceae, and seven sites upstream from nad5i1477 in Hydrocleys. Given the large number of adjacent edited sites removed simultaneously, we believe that these cases support a localized variant of the RT-mediated model (see below).
Table 6.
Areas Close to a Trans-Spliced Intron Showing a Significant Reduction of Edited Sites
| Trans-spliced intron | Group | ES lost/ES totala | Positionb |
|---|---|---|---|
| nad1i394 | Vallisneria-Nechamandra | 6/7 | Upstream |
| nad1i394 | Alismataceae | 6/7 | Upstream |
| nad1i728 | Hydrilla | 10/12 | Downstream |
| nad2i542 | Ottelia | 13/15 | Upstream |
| nad2i542 | Ottelia | 3/3 | Downstream |
| nad2i542 | Hydrilla-Enhalus | 14/15 | Upstream |
| nad2i542 | Sagittaria | 8/9 | Upstream |
| nad2i542 | Helanthium | 8/9 | Upstream |
| nad5i1455 | Hydrilla-Enhalus | 16/21 | Upstream |
| nad5i1477 | Hydrocleys | 7/8 | Downstream |
Edited sites lost in the two adjacent exons/edited sites present in the intron-bearing MRCA of the group in question.
Refers to the position of the exon showing reduced number of edited sites respect to the trans-spliced intron.
Variants of the RT-Mediated Model
Although our data provide evidence supporting a RT-mediated model in 27 cases of intron loss, there are variants of the model that should be considered. The most straightforward scenario involves the insertion of a complete and fully edited gene copy, in which case all introns and all edited sites will be removed simultaneously (fig. 1). This leads to the processed copy either replacing the native gene copy, or the co-existence of both copies in the genome. We did not find evidence of coexistence of a full-length processed paralog and the unprocessed gene in the mitochondrial genome of any of the Alismatales representatives examined here (or in any other angiosperm studied so far). It has been proposed that if homologous recombination is responsible for integration of a cDNA copy into the genome, the only site for recombination would be the gene copy that originally produced the transcript (Fink 1987), making the co-existence of both copies highly unlikely. In addition, plant mitochondrial genomes tend to retain a single allele per gene, as has been shown in the case of hybrids, cybrids (Sanchez-Puerta et al. 2015), and mtDNA that acquired mitochondrial genes by horizontal transfer (Bergthorsson et al. 2004; Mower et al. 2010). This may indicate that even in the rare case in which a processed paralog is inserted in a different location than that of the original gene copy, as has been shown for a short fragment of nad1 in some Alismatales (Cuenca et al. 2012), one of the copies will rapidly degenerate.
In several species of Hydrocharitaceae, we found instances (as in nad7 and cox2) that might initially be interpreted as if a native copy had been replaced by a completely processed paralog, as the whole gene has lost its introns and edited sites. However, a more detailed analysis shows that intron removal has been a gradual process, with adjacent introns being lost at different times in the evolution of the group, pointing instead to several rounds of retroprocessing (see below) or, as in the case of cox2, for intron loss mediated by a mechanism different than the RT-model (cox2i373), followed by one or more rounds of retroprocessing (removal of cox2i691, plus removal of the edited sites).
A second variant of the RT-mediated model also involves retroprocessing with a fully edited copy, but as a localized event, affecting only part of the original gene (fig. 1 var. A). In most cases of reported retroprocessing, the pattern of edited site loss is consistent with this model (Geiss et al. 1994; Itchoda et al. 2002; Grewe et al. 2009; Ran et al. 2010; Sloan et al. 2010). This model is also the best supported by our data, with affected sequence regions ranging in size from a few bases to approximately 1 kb (table 4), or a whole gene fragment flanked by two trans-spliced introns, as the case of the nad1 exons 2 and 3. In mammalian nuclear genomes, conversion events between duplicated sequences are frequently short (< 500 bp, average close to 200 bp) (McGrath et al. 2009), so it is possible that localized retroprocessing may explain at least some of the seven cases of intron loss where adjacent edited sites are not removed if the inserted cDNA fragments are simply confined to regions between but not including edited sites. The maximum possible length of the potentially inserted fragments (164–703 bp) would make four cases easily explained by localized retroprocessing, whereas the remaining three cases with fragments being very short (63–78 bp) may be more doubtful (table 5).
A third variant also suggests that retroprocessing can occur with a partially processed cDNA copy, but in addition, the time sequence in which RNA editing and splicing occur becomes relevant (fig. 1, var. B). If editing occurs before splicing, there will be more cases of genes having no edited sites, but a conserved number of introns, whereas if splicing occurs before, edited sites will tend to remain when introns are removed. Although we know that not all sites of RNA editing are edited simultaneously, it has been shown in other angiosperms that most of the editing activity occurs before or in parallel with intron splicing (Castandet et al. 2010; Hepburn et al. 2012), indicating that it is possible to find intron-bearing processed copies (as we have seen mostly in the nad4 gene, see below). In contrast, although RNA editing seems to be needed in order to facilitate splicing, it has been shown that full RNA editing is not a prerequisite for splicing (Castandet et al. 2010). In fact, the editing of some few sites seems to be dependent of intron splicing, for example, the presence of intron inhibits RNA editing in some sites, such as the edited site closest to the intron–exon borders in nad7 (Li-Pook-Than et al. 2007). This indicates that depending of the maturation stage of the precursor RNA when it is retrotranscribed, the resulting cDNA copies could either conserve introns, or some few edited sites.
As mentioned above, particularly in nad4, we found a number of cases where editing has been lost, but the intron remains (even though in most cases the intron is subsequently lost in the evolution of the group, fig. 2). Besides this model where recombination occurs with a partially processed copy, alternative scenarios of intron presence in what looks like a fully processed copy (or part of it) should be considered.
Why Are Some Group II Introns Present in Genes That Otherwise Seem to Be Retroprocessed?
Theoretically, a retroprocessed gene copy could gain an intron by insertion. This scenario, however, is highly improbable and calls for a mobile intron able to invade an intron-free gene copy. Although some group II introns may be autocatalytic, a homing mechanism for these introns seems to be lost in seed plant mitochondria (Ahlert et al. 2006; Bonen 2012).
A second possibility is that recombination between the gene and the cDNA copy is strongly suppressed upstream of the intron, as recombination involving both exons is needed in order to remove the intron. This has been suggested to occur in other groups (e.g., Chlamydomonas; Newman et al. 1992; Odom and Herrin 2013). However, this again does not explain cases in which the adjacent upstream intron is removed, because the suppression in recombination in an exon will, in principle, affect both adjacent introns. To complicate the situation further, it has been shown that some group II introns are needed for the correct splicing of other group II introns (Petersen et al. 2011), affecting the splicing order and the intron retention through evolutionary time. In a recent study, it was found that the plastid ycf3 intron 1 is needed for the correct splicing of ycf3 intron 2. Plants that have lost the ycf3 intron 1 have a low fitness, caused by the defective splicing of ycf3 intron 2. However, in plants where the ycf3 intron 2 is lost first, the ycf3 intron 1 is no longer needed and is readily lost (Petersen et al. 2011).
As mentioned above, in some of the cases of initial intron retention, the intron in question has been lost subsequently. This observation is consistent with inferred events among members of the Caryophyllales, where the loss of editing preceded the loss of the rps3 intron in Amaranthaceae and Achatocarpaceae (Kagami et al. 2012). These authors suggested that intron presence might have an impact on gene expression, affecting particularly the ability to correctly edit the transcript. This inference may be supported by the fact that mRNAs of modified mitochondrial genes from which introns have been removed are less frequently edited than RNAs transcribed from the original intron-bearing genes (Castandet et al. 2010). In accordance with this, there is evidence for differential status between un-spliced and spliced RNA in the cox2, nad4, and nad7 genes of several plant species (Sutton et al. 1991; Li-Pook-Than et al. 2007). Following this hypothesis, in a gene that has lost RNA edited sites, the presence of introns would become superfluous, and their subsequent loss inconsequential to gene expression. If so, the removal of edited sites is a precondition for intron loss, which may be an alternative explanation for the correlation between lack of editing and lack of intron. However, a mechanism of intron loss under these proposed scenarios still needs to be determined.
In summary, in the present study, we show strong evidence for an RT-mediated process as the most plausible model to explain intron loss in the Alismatales and possibly in many other plant groups. Our results do not support horizontal gene transfer followed by gene conversion as a mechanism of intron loss, thus questioning the generality of this model. However, as only very few groups of plants have been investigated, it would be useful to study intron loss in a wider taxonomic context.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online (http://gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Danish Council for Independent Research | Natural Sciences Grant no. 12-126713 to O.S. and G.P., and NSERC (Natural Sciences and Engineering Research Council of Canada) Discovery Grant to S.W.G., and US National Science Foundation Grants DEB 0830036 to S.W.G and DEB 0830020 to J.I.D.
Literature Cited
- Ahlert D, Piepenburg K, Kudla J, Bock R. 2006. Evolutionary origin of a plant mitochondrial group II intron from a reverse transcriptase/maturase-encoding ancestor. J. Plant Res. 119:363–371. [DOI] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, Shedge V, Davila J, Christensen AC, Mackenzie SA. 2009. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 183:1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD. 2004. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc. Natl. Acad. Sci. U S A. 101:17747–17752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L. 2012. Evolution of mitochondrial introns in plants and photosynthetic microbes In: Marechal-Drouard L, editor. Mitochondrial genome evolution. London: Academic Press, Elsevier Science; p. 155–186. [Google Scholar]
- Boni M, Posada D, Feldman M. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowe LM, dePamphilis CW. 1996. Effects of RNA editing and gene processing on phylogenetic reconstruction. Mol. Biol. Evol. 13:1159–1166. [DOI] [PubMed] [Google Scholar]
- Castandet B, Choury D, Begu D, Jordana X, Araya A. 2010. Intron RNA editing is essential for splicing in plant mitochondria. Nucl. Acids Res. 38:7112–7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NE, Shen R, Carmel L. 2012. The role of reverse transcriptase in intron gain and loss mechanisms. Mol. Biol. Evol. 29:179–186. [DOI] [PubMed] [Google Scholar]
- Cuenca A, Petersen G, Seberg O. 2013. The complete sequence of the mitochondrial genome of Butomus umbellatus a member of an early branching lineage of monocotyledons. PLoS One 8:e61552.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca A, Petersen G, Seberg O, Davis J, Stevenson D. 2010. Are substitution rates and RNA editing correlated? BMC Evol. Biol. 10:349.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca A, Petersen G, Seberg O, Jahren AH. 2012. Genes and processed paralogs co-exist in plant mitochondria. J. Mol. Evol. 74:158–169. [DOI] [PubMed] [Google Scholar]
- Davila JI, et al. 2011. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 9:64.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr LK, Strathern JN. 1993. A role for reverse transcripts in gene conversion. Nature 361:170–173. [DOI] [PubMed] [Google Scholar]
- Dhellin O, Maestre J, Heidmann T. 1997. Functional differences between the human LINE retrotransposon and retroviral reverse transcriptases for in vivo mRNA reverse transcription. EMBO. J. 16:6590–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink GR. 1987. Pseudogenes in yeast. Cell 49:5–6. [DOI] [PubMed] [Google Scholar]
- Geiss KT, Abbas GM, Makaroff CA. 1994. Intron loss from the NADH dehydrogenase subunit 4 gene of lettuce mitochondrial DNA: evidence for homologous recombination of a cDNA intermediate. Mol. Gen. Genet. 243:97–105. [DOI] [PubMed] [Google Scholar]
- Grewe F, et al. 2011. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucl. Acids Res. 39:2890–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe F, Viehoever P, Weisshaar B, Knoop V. 2009. A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucl. Acids Res. 37:5093–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugerli F, et al. 2001. The evolutionary split of Pinaceae from other conifers: evidence from an intron loss and a multigene phylogeny. Mol. Phylogenet. Evol. 21:167–175. [DOI] [PubMed] [Google Scholar]
- Guo W, et al. 2016. Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol. Biol. Evol. 33:1448–1460. [DOI] [PubMed] [Google Scholar]
- Henze K, Martin W. 2001. How do mitochondrial genes get into the nucleus? Trends Genet. 17:383–387. [DOI] [PubMed] [Google Scholar]
- Hepburn NJ, Schmidt DW, Mower JP. 2012. Loss of two introns from the Magnolia tripetala mitochondrial cox2 gene implicates horizontal gene transfer and gene conversion as a novel mechanism of intron loss. Mol. Biol. Evol. 29:3111–3120. [DOI] [PubMed] [Google Scholar]
- Itchoda N, Nishizawa S, Nagano H, Kubo T, Mikami T. 2002. The sugar beet mitochondrial nad4 gene: an intron loss and its phylogenetic implication in the Caryophyllales. Theor. Appl. Genet. 104:209–213. [DOI] [PubMed] [Google Scholar]
- Joly S, Brouillet L, Bruneau A. 2001. Phylogenetic implications of the multiple losses of the mitochondrial coxII.i3 intron in the angiosperms. Int. J. Plant Sci. 162:359–373. [Google Scholar]
- Kagami H, Nagano H, Takahashi Y, Mikami T, Kubo T. 2012. Is RNA editing implicated in group II intron survival in the angiosperm mitochondrial genome? Genome 55:75–79. [DOI] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V, Volkmar U, Hecht J, Grewe F. 2011. Mitochondrial genome evolution in the plant lineage In: Kempken F, editor. Plant mitochondria. New York (NY: ): Springer Science+Business Media; p. 3–29. [Google Scholar]
- Lambowitz AM, Zimmerly S. 2004. Mobile group II introns. Annu. Rev. Genet. 38:1–35. [DOI] [PubMed] [Google Scholar]
- Li-Pook-Than J, et al. 2007. Relationship between RNA splicing and exon editing near intron junctions in wheat mitochondria. Physiol. Plant 129:23–33. [Google Scholar]
- McGrath CL, Casola C, Hahn MW. 2009. Minimal effect of ectopic gene conversion among recent duplicates in four mammalian genomes. Genetics 182:615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson MA, Fay ME, Chase MW, Graham SW. 2004. Parallel loss of a slowly evolving intron from two closely related families in asparagales. Syst. Botany 29:296–307. [Google Scholar]
- Meade A, Pagel M. 2014. BayesTrait V2 manual. Reading Evolutionary Biology Group. Available from: http://www.evolution.rdg.ac.uk/BayesTraits.html.
- Mourier T, Jeffares DC. 2003. Eukaryotic intron loss. Science 300:1393.. [DOI] [PubMed] [Google Scholar]
- Mower JP. 2009. The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucl. Acids Res. 37:W253–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Sloan D, Alverson AJ. 2012. Plant mitochondrial genome diversity: the genomics revolution In: Wendel JF, editor. Plant genome diversity, vol. I. Wien: Spinger-Verlag; p. 123–144. [Google Scholar]
- Mower JP, et al. 2010. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol. 8:150.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SM, Harris EF, Johnson AM, Boynton JE, Gillham NW. 1992. Nonrandom distribution of chloroplast recombination events in Chlamydomonas reinhardtii: evidence for a hotspot and an adjacent cold region. Genetics 132:1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom OW, Herrin DL. 2013. Reverse transcription of spliced psbA mRNA in Chlamydomonas spp. and its possible role in evolutionary intron loss. Mol. Biol. Evol. 30:2666–2675. [DOI] [PubMed] [Google Scholar]
- Park S, et al. 2015. Dynamic evolution of Geranium mitochondrial genomes through multiple horizontal and intracellular gene transfers. New Phytol. 208:570–583. [DOI] [PubMed] [Google Scholar]
- Petersen G, Cuenca A, Møller IA, Seberg O. 2015. Massive gene loss in mistletoe (Viscum, Viscaceae) mitochondria. Sci. Rep. 5:17588.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G, et al. 2016. Phylogeny of the Alismatales (Monocotyledons) and the relationship of Acorus (Acorales?). Cladistics 32: DOI: 10.1111/cla.12120 [DOI] [PubMed] [Google Scholar]
- Petersen G, Seberg O, Davis JI, Stevenson DW. 2006. RNA editing and phylogenetic reconstruction in two monocot mitochondrial genes. Taxon 55:871–886. [Google Scholar]
- Petersen K, Schöttler MA, Karcher D, Thiele W, Bock R. 2011. Elimination of a group II intron from a plastid gene causes a mutant phenotype. Nucl. Acids Res. 39:5181–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran J-H, Gao H, Wang X-Q. 2010. Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer II and further evidence for the phylogeny of gymnosperms. Mol. Phylogenet. Evol. 54:136–149. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Babenko VN, Koonin EV. 2006. Dollo parsimony and the reconstruction of genome evolution In: Albert VA, editor. Parsimony, phylogeny, and genomics. Oxford: Oxford University Press; p. 240. [Google Scholar]
- Ross TG, et al. 2016. Plastid phylogenomics and molecular evolution of Alismatales. Cladistics 32: DOI: 10.1111/cla.12133 [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Zubko MK, Palmer JD. 2015. Homologous recombination and retention of a single form of most genes shape the highly chimeric mitochondrial genome of a cybrid plant. New Phytol. 206:381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E, Barkman TJ, Rice DW, Palmer JD. 2015. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl. Acad. Sci. U S A. 112:E3515–E3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR. 2010. Extensive loss of RNA editing sites in rapidly evolving Silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A fast bootstrapping algorithm for the RAxML web-servers. Syst. Biol. 57:13.. [DOI] [PubMed] [Google Scholar]
- Sutton CA, Conklin PL, Pruitt KD, Hanson MR. 1991. Editing of pre-mRNAs can occur before cis- and trans-splicing in Petunia mitochondria. Mol. Cell. Biol. 11:4274–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WQ, Wu YR, Messing J. 2012. The mitochondrial genome of an aquatic plant, Spirodela polyrhiza. PLoS One 7(10):e46747.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L-Y, Yang Y-F, Niu D-K. 2010. Evaluation of models of the mechanisms underlying intron loss and gain in Aspergillus fungi. J. Mol. Evol. 71:364–373. [DOI] [PubMed] [Google Scholar]
- Zhu T, Niu DK. 2013a. Mechanisms of intron loss and gain in the fission yeast Schizosaccharomyces. PLoS One 8(4):e61683.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Niu DK. 2013b. Frequency of intron loss correlates with processed pseudogene abundance: a novel strategy to test the reverse transcribed model. BMC Biol. 11:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.