Abstract
Halophilic microbes living in hypersaline environments must counteract the detrimental effects of low water activity and salt interference. Some halophilic prokaryotes equilibrate their intracellular osmotic strength with the extracellular milieu by importing inorganic solutes, mainly potassium. These “salt-in” organisms characteristically have proteins that are highly enriched with acidic and hydrophilic residues. In contrast, “salt-out” halophiles accumulate large amounts of organic solutes like amino acids, sugars and polyols, and lack a strong signature of halophilicity in the amino acid composition of cytoplasmic proteins. Studies to date have examined halophilic prokaryotes, yeasts, or algae, thus virtually nothing is known about the molecular adaptations of the other eukaryotic microbes, that is, heterotrophic protists (protozoa), that also thrive in hypersaline habitats. We conducted transcriptomic investigations to unravel the molecular adaptations of two obligately halophilic protists, Halocafeteria seosinensis and Pharyngomonas kirbyi. Their predicted cytoplasmic proteomes showed increased hydrophilicity compared with marine protists. Furthermore, analysis of reconstructed ancestral sequences suggested that, relative to mesophiles, proteins in halophilic protists have undergone fewer substitutions from hydrophilic to hydrophobic residues since divergence from their closest relatives. These results suggest that these halophilic protists have a higher intracellular salt content than marine protists. However, absence of the acidic signature of salt-in microbes suggests that Haloc. seosinensis and P. kirbyi utilize organic osmolytes to maintain osmotic equilibrium. We detected increased expression of enzymes involved in synthesis and transport of organic osmolytes, namely hydroxyectoine and myo-inositol, at maximal salt concentration for growth in Haloc. seosinensis, suggesting possible candidates for these inferred organic osmolytes.
Keywords: ectoine, osmoregulation, hypersaline, salt-in, salt-out, extremophile
Introduction
Extremely halophilic microbes are adapted to hypersaline conditions and require salt concentrations well above those of seawater in order to grow. They typically have optimal reproductive rates at approximately 4× the salinity of seawater (Hauer and Rogerson 2005; Oren 2008). Some halophiles can actually sustain growth in saturating salt concentrations, and some even show their optimal growth in near-saturated media (e.g., Park et al. 2007). Research on extreme halophiles has focused for decades almost entirely on Archaea and Bacteria and has yielded substantial information in terms of adaptations of their proteins and cellular metabolism and physiology (Oren 2002a).
In order to stay hydrated in hypersaline conditions, halophiles equilibrate the osmotic strength of their cytoplasm with the extracellular medium by accumulating solutes, either inorganic or organic. For example, the salt-in haloarchaea accumulate molar concentrations of potassium (Oren 2002b). These have highly acidic proteomes, resulting from an abundance of negatively charged aspartate and glutamate residues and a depletion of positively charged lysine residues at the protein surface (Frolow et al. 1996; Paul et al. 2008). X-ray crystallography studies have demonstrated that this negative net charge is neutralized by water molecules and thus increases protein solubility as well as preventing protein aggregation in hypersaline conditions (Elcock and McCammon 1998; Richard et al. 2000). Halophilic proteins are also depleted of hydrophobic amino acids, and this is often offset by a higher content of borderline hydrophilic–hydrophobic residues such as serine and threonine (Lanyi 1974). High salt increases the hydrophobic effect, and the low hydrophobicity of halophilic proteins possibly allows them to avoid overly rigid folded conformations.
On the other hand, the proteomes of halophiles that use organic solutes as their main osmolytes (salt-out organisms) are not enriched in highly acidic proteins, although they typically produce extracellular proteins that are very acidic and less hydrophobic compared with mesophilic counterparts (e.g., Coronado et al. 2000; Oren et al. 2005). Organic osmolytes are strong “water structure-formers” that are excluded from protein hydration shells, and thus prevent cell desiccation while being compatible with metabolism (Bolen and Baskakov 2001). They are uncharged or zwitterionic low-molecular weight molecules and include some amino acids and their derivatives (glutamate, aspartate, proline, ectoine), polyols (myo-inositol, mannitol, glycerol), sugars (trehalose, sucrose), and betaines (Galinski 1995). To save energy, osmolytes are preferentially imported from the extracellular environment if available, otherwise they are synthesized de novo (Oren 1999). Accumulation of organic osmolytes is not limited to halophiles but rather represents a widespread long-term osmotic balance response in most organisms. In contrast, massive accumulation of inorganic osmolytes is restricted to a few groups, including the Halobacteria (i.e., haloarchaea), the Halanaeorobiales (Rengpipat et al. 1988), Salinibacter ruber (Oren et al. 2002), and Halorhodospira halophila (Deole et al. 2013).
Recently, the view that intracellular salt content is absolutely coupled with proteome acidity has been challenged (Oren 2013). For example, the proteobacterium Halor. halophila employs K+ as its main osmolyte and expresses a highly acidic proteome, but can grow at salinity as low as 3.5%, with intracellular K+ concentrations varying from 0.4 to 2.1 M as salinity ranges from 5% to 35% (Deole et al. 2013). This shows that acidic proteins can function in cytosolic K+ concentrations experienced by mesophiles (e.g., Escherichia coli) and contradicts the hypothesis that halophilic proteins contain an excess of negative surface charges to establish stabilizing interactions with K+. It is suggested instead that the high hydration state of ionized glutamate and aspartate side chains contributes to protein solubility (Deole et al. 2013). The use of a mixture of inorganic and organic osmolytes has also been observed, as in the halophilic archaeon Haladaptatus paucihalophilus which accumulates trehalose and glycine betaine as compatible solutes while keeping a stable intracellular K+ content that is 17× higher than E. coli, but 0.3–0.6× that of Halobacterium salinarum (Youssef et al. 2014). As expected, Hala. paucihalophilus possesses a highly acidic proteome. More puzzling, however, are the cases of the Halanaerobiales Halanaerobium praevalens, “Halanaerobium hydrogeniformans,” and Halothermothrix orenii that accumulate high levels of K+ but apparently do not have an acidic signature in their proteomes (Mavromatis et al. 2009; Elevi Bardavid and Oren 2012). These studies show that proteome acidity is, at best, strongly suggestive of osmoadaptive strategy, rather than a perfect diagnostic.
Knowledge regarding salt adaptation in halophilic and halotolerant eukaryotic microbes is restricted to species of the chlorophycean alga Dunaliella, and certain yeasts, such as Hortaea werneckii, Debaryomyces hansenii, Wallemia ichthyophaga, and Aureobasidium pullulans (Chen and Jiang 2009; Gunde-Cimerman et al. 2009). The yeasts D. hansenii and A. pullulans are considered halotolerant and can grow without salt. In fact, the former grows optimally at salinity around 3–6%, and the latter grows better without added salt (Kogej et al. 2005; Gunde-Cimerman et al. 2009). In contrast, W. ichthyophaga is an extreme halophile that cannot grow without salt and grows optimally at approximately 21–27% salinity (Gunde-Cimerman et al. 2009). These organisms are salt-out strategists that accumulate glycerol as the main compatible solute. Glycerol is one of the cheapest and simplest osmolytes to produce, but is rarely found in other halophiles studied to date, possibly because of its high diffusion rate through standard cell membranes (Oren 1999). Yeasts and Dunaliella have developed mechanisms to increase glycerol retention, potentially lowering permeability by adjusting the membrane sterol content and lipid composition, or cell wall melanin level (Sheffer et al. 1986; Kogej et al. 2006; Gunde-Cimerman et al. 2009). Yeasts also use other polyols such as erythritol, arabitol, and mannitol, with the composition of the compatible solute mix depending on salinity (Hohmann 2002). Nonetheless, the halotolerant yeast D. hansenii has a higher intracellular sodium content than Saccharomyces cerevisiae (Prista et al. 1997), A. pullulans, and H. werneckii (Kogej et al. 2005), suggesting that sodium ions can substantially contribute to osmotic adjustment along with glycerol in this species.
Compared with halophilic archaea, bacteria, the alga Dunaliella, and fungi, virtually nothing is known about halophilic heterotrophic protists, that is, the ecological guild of “protozoa.” Several evolutionarily distinct groups of halophilic protozoa have been recorded in hypersaline environments (Hauer and Rogerson 2005; Foissner et al. 2014; Stoeck et al. 2014; Park and Simpson 2015), and they represent potentially important grazers of prokaryotes in these habitats (Park et al. 2003). Several species show high minimum and optimum salinities for growth under laboratory conditions (Park et al. 2006; Park et al. 2007; Cho et al. 2008; Foissner et al. 2014), and based on this, the salt-in strategy has been speculatively suggested for at least one species (Foissner et al. 2014). Here, we present a molecular examination of two protozoa: Haloc. seosinensis strain EHF34 (Park et al. 2006) and P. kirbyi strain AS12B (Park and Simpson 2011). Both are obligate halophiles that cannot grow at salinities <7.5% under laboratory conditions. The AS12B strain of P. kirbyi grows optimally at a slightly lower salt concentration than Haloc. seosinensis EHF34 (12% vs. 15%, respectively). Concordantly, Haloc. seosinensis EHF34 can still divide in media close to saturation (at 30% salt) whereas P. kirbyi AS12B has a maximum salt concentration for growth around 25% (Park et al. 2006; Park and Simpson 2011).
As a first investigation of the adaptations of these protozoa to high salt conditions, we aimed to gather evidence about their molecular adaptations, and strategy for osmotic adjustment. As they grow in mixed cultures with food prokaryotes, direct measurement of the intracellular content in these protists, although required, is technically challenging. Instead, as a first investigation, we sequenced their transcriptomes and studied the molecular features of their predicted proteomes, assuming that a high level of intracellular salt would lead to the classical acidic signature of salt-in microbes (for example). We also investigated the expression of putative organic osmolyte synthesizers and transporters as a function of salinity in Haloc. seosinensis. We find that the cytoplasmic proteomes of Haloc. seosinensis and P. kirbyi are not highly acidic, although they are significantly more hydrophilic than eukaryotic microbes inhabiting marine environments. At high salt, Haloc. seosinensis upregulated genes related to ectoine hydroxylase, amino acid transporters, and myo-inositol carriers. Collectively, these observations suggest that these halophilic protists exploit organic solutes as major osmolytes, although they potentially also have higher intracellular salt content relative to mesophilic protists.
Materials and Methods
RNA and DNA Extraction and Sequence Generation
RNA was extracted from midexponential-phase cultures of Haloc. seosinensis strain EHF34 (Park et al. 2006) and P. kirbyi strain AS12B (Park and Simpson 2011) grown at 37 °C on a shaker at 50 rpm in salt medium adjusted to desired concentrations after dilution of 30% salt medium (salt proportion as in medium #5 in Park 2012). Haloc. seosinensis was grown in triplicate at 15% and 30% salt, and was fed with Haloferax sp. isolated from one of our cultures. Pharyngomonas kirbyi was grown as flagellated cells at 15% salt and as amoebae at 10% salt, and fed in both cases with Citrobacter sp. As growth controls, cultures grown in parallel with the RNA experiment replicates were not sacrificed for RNA extraction, but kept alive until they reach stationary phase, to ensure proper growth. RNA was extracted using TRIzol following the manufacturer’s instructions (Ambion, Carlsbad, NM). RNA extracts were treated with Turbo DNAse (Ambion) prior to cDNA library preparation using the TruSeq RNA sample preparation kit version 2 (Illumina, San Diego, CA).
Halocafeteria seosinensis’ samples were sequenced on an Illumina HiSeq 2000 platform by Génome Québec generating a total of 188,229,640 100-bp paired-end reads. Pharyngomonas kirbyi samples were sequenced on a MiSeq platform generating 26,040,129 250-bp paired-end reads. Reads were trimmed to remove low-quality sequences and adapter sequences using Trimmomatic 0.30 with a sliding window of 10 nt and a PHRED33 quality threshold of 25 (Bolger et al. 2014), and mapped to genomes of prokaryote species known to be in the culture (Haloferax volcanii DS2—GenBank accession number GCA_000025685.1, Citrobacter freundii 4_7_47CFAA genomic scaffolds—GCA_000238735.1, Salinivibrio costicola subsp. costicola ATCC 33508—GCA_000390145.1) in order to discard contaminant sequences using Stampy 1.0.23 (Lunter and Goodson 2011). After decontamination, reads with k-mer coverage between 6× and 50× were selected using BBNorm (from the BBMap package: http://sourceforge.net/projects/bbmap/, last accessed March 2015) and assembled using Trinity 2.0.2 (Grabherr et al. 2011). Open-reading frames (ORFs) were predicted using TransDecoder (from the Trinity package) and translated to protein sequences. Trinity typically generates many putative isoforms and polymorphic sequences. In order to reduce the redundancy in the data set, ORFs were compared with each other by BLASTP searches (Altschul et al. 1990). For highly similar ORF pairs, we discarded the smallest sequence if the alignment covered >90% of its length and if <5 mismatches were observed. Finally, to remove sequences belonging to unknown prokaryotic contaminants present in the cultures, the nucleotide sequences of ORFs were compared with sequences in the NT database using BLASTN. Sequences having 100-bp-long fragments >90% identical to a prokaryotic sequence were discarded. The generated transcriptomes contained 16,852 ORFs for Haloc. seosinensis and 15,521 ORFs for P. kirbyi.
DNA from Haloc. seosinensis was purified using a salt extraction protocol (Aljanabi and Martinez 1997). Briefly, cells were disrupted by vortexing in lysis buffer and digested with proteinase K (0.2 mg/ml) and 0.01% sodium dodecyl sulfate. DNA was separated from the other organic phases by centrifugation in a supersaturated NaCl solution (3 M) and precipitated with 70% ethanol. A paired-end DNA library (250 bp) was prepared using the Nextera XT kit (Illumina) prior to high-throughput sequencing on a MiSeq platform. In total, 19,726,040 reads were generated and “cleaned” as described for the RNA-derived sequences. Genomic contigs were assembled with MIRA 4.9.5_2 (Chevreux et al. 2004). Genes, including intron/exon boundaries, were delimited by mapping the read sequences obtained from RNA extracts using TopHat2 2.0.13 (Kim et al. 2013) and predicted by Braker 1.1 (Hoff et al. 2016).
Protein Localization Prediction and Calculations of isoelectric point and Grand Average of Hydropathy
Sequences were assumed to encode soluble cytoplasmic proteins if no mitochondrial targeting peptide, signal peptide, or chloroplast transit peptide was predicted by TargetP 1.1 given a reliability class of 1 or 2 (Emanuelsson et al. 2000), and if no transmembrane domain was predicted by TMHMM 2.0 (Krogh et al. 2001). Protein sequences were considered to be secreted if predictions by Phobius 1.01 (Käll et al. 2004), WoLF PSORT 0.2 (Horton et al. 2007), TargetP and SignalP 4.0 (Petersen et al. 2011) all agreed on the extracellular localization of the protein and if no transmembrane domain was predicted by TMHMM. Furthermore, predicted secreted proteins were excluded if they contained the ER retention signals KK, KXK, KDEL, or HDEL at the C-terminus or RR at the N-terminus. Signal peptides of predicted secreted proteins were cleaved prior to further analysis using the cleavage sites predicted by SignalP (Nielsen et al. 1997).
The isoelectric points (pI) of protein sequences were computed by iteratively calculating protein charge at given pH values, using side chain pKa values of charged amino acids (Nelson and Cox 2005), until a neutral charge was obtained. Hydropathicity of protein sequences was determined by calculating the GRand AVerage of hydropathY (GRAVY) score using the Kyte and Doolittle hydrophobicity scale (Kyte and Doolittle 1982). In order to investigate which amino acid had the most impact on the hydrophilicity of the halophilic protist proteomes, we iteratively calculated GRAVY scores by bringing the frequency of amino acids to the average frequency of mesophilic sequences at fastest-evolving sites of the Mandor data set (see below). At each round, the amino acid that contributed the most at bringing the GRAVY scores closest to the mesophilic level was identified and its frequency was kept at this level for further calculation rounds.
Homology Searches
Homologs of Haloc. seosinensis and P. kirbyi proteins were searched for, using the BLAST (Basic Local Alignment Search Tool) algorithm, in the predicted proteomes of 24 protists sequenced during the Marine Microbial Eukaryote Sequencing Project (MMETSP) (supplementary table S5a, Supplementary Material online; Keeling et al. 2014). Taxa were selected if the salinity of the habitat from which they were sampled, or the salinity of the medium in which they were cultured, was close to seawater (i.e., ∼3.5%). Extremophiles (e.g., psychrophiles or acidophiles) were avoided, as were organisms fed with other eukaryotes (due to cross-contamination issues). As a result, taxa in MarProt data sets were not salt-in extreme halophiles, and their proteomes did not contain a molecular signature for halophilicity. Homologous sequences were kept if alignments covered >60% of the smallest sequence examined with identity >25%. Harvested homologs were screened out if they were not predicted to be soluble cytoplasmic proteins given the criteria described above. We refer to these two data sets (one for each halophile) as the Marine Protists comparative data sets (MarProt).
In order to compare halophilic protists with salt-in Archaea, we identified orthologous groups from the eggNOG 4.1 database (Powell et al. 2014) common to 16 species of Halobacteria and Haloc. seosinensis and P. kirbyi through hidden Markov model searches using the hmmscan program of the HMMER package (Eddy 1998). Orthologs with e-evalue <0.00001 were chosen for further comparisons using BLAST. The same coverage and identity thresholds (i.e., >60% of smallest sequence and >25% identity, respectively) used during the MMETSP homolog searches were applied to select sequences for statistical comparison. Transmembrane proteins were excluded as described previously. As a control, sequences in MMETSP taxa homologous to halobacterial genes were also identified and compared.
Putative genes for ectoine/hydroxyectoine synthesis in a wide range of protists were recovered from the complete MMETSP nucleotide data set using TBLASTN, with Haloc. seosinensis EctABCD protein sequences as queries (MMETSP database downloaded in April 2015 from http://data.imicrobe.us). Subject sequences >15% identical to Haloc. seosinensis sequences were compared with nucleotide sequences in the NT database using BLASTN to identify putative prokaryotic contaminants. Sequences >50% identical (without alignment length threshold) to bacterial or archaeal homologs were removed from further analysis. Protein sequences corresponding to the remaining nucleotide sequences (i.e., confirmed to be from protists) were phylogenetically examined as described below.
Phylogenetic Analysis and Ancestral Sequence Inference
We also used a curated “phylogenomic” data set containing 252 house-keeping genes from a broad range of eukaryotes (supplementary table S5b, Supplementary Material online; Brown et al. 2012; Burki et al. 2012), referred as the Mandor data set. Using this data set provided a phylogenetic context to correct for nonindependence among organisms during statistical analyses (see below). For comparison purposes, we only included free-living mesophilic organisms (i.e., no multicellular, symbiotic, parasitic, or thermophilic organisms), and discarded organisms that had <50% of genes represented, with the exception of Dunaliella salina (45% of genes present), as this was the only halophile in the original data set. Sequences orthologous to the 252 genes were obtained from Haloc. seosinensis, P. kirbyi, W. ichthyophaga EXF-994 (GenBank accession number: GCA_000400465.1), D. hansenii CBS767 (GCA_000006445.2), A. pullulans EXF-150 (GCA_000721785.1), and the MarProt taxa using methods described in Brown et al. (2012). Concatenated sequences from each organism were automatically aligned using MAFFT 7.205 (Katoh et al. 2002) and trimmed using BMGE 1.1 (Criscuolo and Gribaldo 2010), resulting in a superalignment containing 110 taxa and 61,522 sites (available upon request to the corresponding author). A maximum-likelihood phylogenetic tree was generated in RAxML 8.1.22 (Stamatakis et al. 2005) using the PROTCAT-LGF model of amino acid substitution and 12 independent starting trees. Support values were calculated from 100 replicates using a rapid bootstrap analysis. Evolutionary rates were computed at each site in the alignment using dist_est (Susko et al. 2003) and the 10,000 fastest-evolving sites were selected as sites that are more likely to change in response to environmental conditions such as salinity.
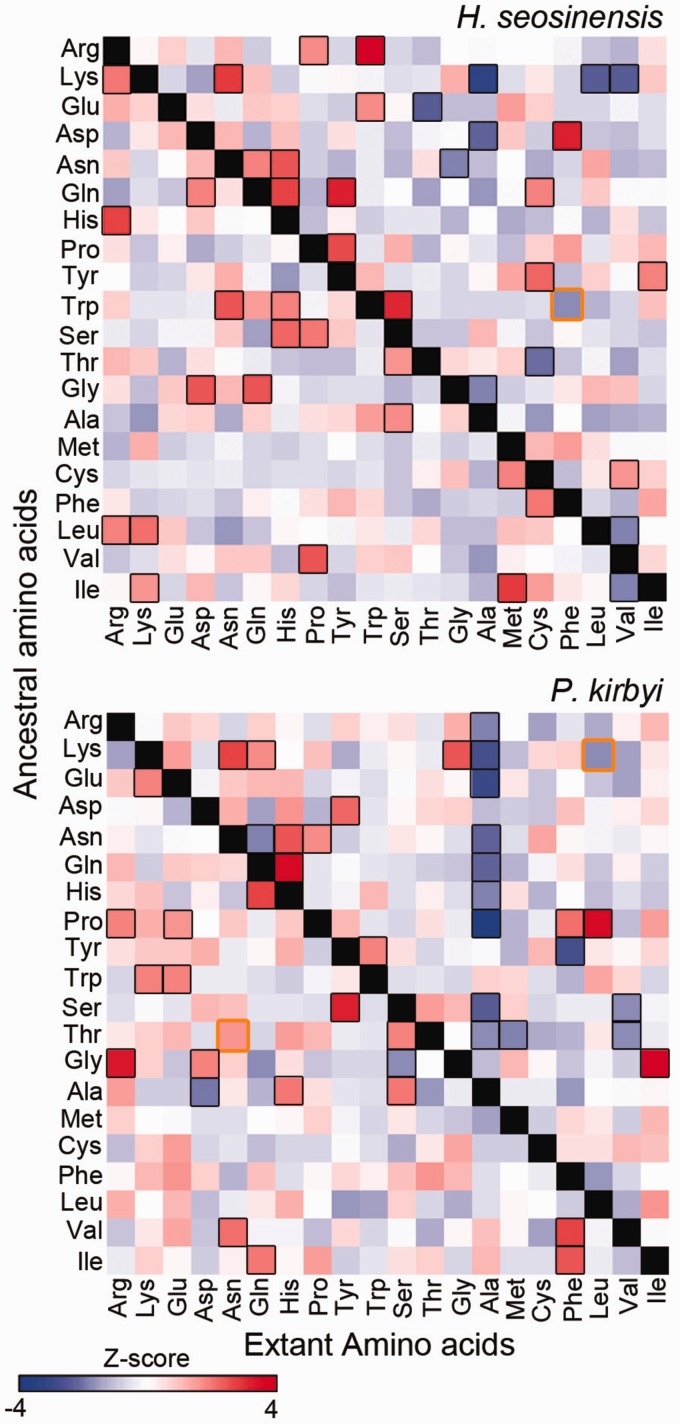
Ancestral sequences were reconstructed with codeml in the PAML package (Yang 2007) using the WAG + Gamma model. Sites for which the probability of a particular ancestral state was >0.9 were kept for further analysis. In order to determine which types of substitutions were significantly enriched in halophiles (from hydrophobic residue to hydrophilic residue or vice versa), we identified substitutions that showed significant variations (P < 0.05) above and below the diagonal of substitution matrices (fig. 4), and analyzed each half of the matrix independently. For each organism, Z-scores for these substitutions were multiplied by the difference between the GRAVY scores of the extant amino acid and of the ancestral amino acid and summed up across the considered half of the substitution matrix. Calculated scores from individual organisms were compared where high scores signify variation patterns in substitutions that led to increased hydrophobicity. Amino acid substitution matrices (fig. 4) were generated using matrix2png (Pavlidis and Noble 2003).
Fig. 4.—
Occurrence of substitutions from predicted ancestral amino acids (on the left of matrices) to extant residues (at bottom of matrices) in halophilic protists at fastest-evolving sites of the Mandor superalignment. Amino acids are ordered from most hydrophilic to most hydrophobic. Each square is colored according to Z-score where blue represents substitutions that occurred less often in halophiles compared with the other taxa and red represents substitutions that occurred more often in halophiles. Boxed squares refer to Z-scores with P < 0.05 (in black P values corrected for multiple testing, in orange P values that were significant prior to correction).
Putative protistan EctABCD sequences were phylogenetically analyzed as described above, except that the PROTGAMMA-LG4X model was used for the maximum-likelihood tree searches. In two instances (EctB in Cafeteria roenbergensis and EctD in Azadinium spinosum), we concatenated partial sequences that likely originated from the same gene. Figures including alignments were generated using AliView 1.17.1 (Larsson 2014).
Statistical Comparisons
Distributions of amino acid frequencies, GRAVY scores, and pI values from Haloc. seosinensis and P. kirbyi were compared against the equivalent data from the marine protists by Mann–Whitney U tests using the scipy package 0.13.3 (Jones et al. 2001). Z-scores for amino acid enrichment analysis and amino acid substitution analysis were calculated as follows:
where p1 is the frequency of the amino acid considered for the halophile, p0 is the amino acid frequency for the mesophiles, p is the amino acid frequency for all organisms, whereas N1 and N0 are the total numbers of amino acids for the halophile and the mesophiles, respectively. GRAVY scores computed from the Mandor data set were compared by standard Z-test. P-values were corrected for multiple testing using the Benjamini–Hochberg method.
Comparisons of the cytoplasmic proteomes and secreted proteomes of salt-out halophilic bacteria were performed as described above, using the protein sequences predicted from the genomes of Actinopolyspora halophila (NZ_AQUI00000000.1), Marinococcus halotolerans (NZ_ATVM00000000.1), Nocardiopsis halophila (NZ_ANAD00000000.1), Virgibacillus alimentarius (NZ_JFBD00000000.1), Chromohalobacter salexigens (NC_007963.1), Halor. halochloris (CP007268.1), and Thiomicrospira halophila (NZ_ARAR00000000.1).
Phylogenetic canonical correlation analysis and phylogenetic principal component analysis were performed using phytools 0.4–31 (Revell 2012) to account for nonindependence of organisms in the Mandor data. Correlations between the habitat salinity and varying protein features (amino acid frequencies, GRAVY and pI) were tested. Salt concentrations were obtained based on the salinity of the environment from which the protists were sampled or the salinity of the medium in which they were commonly maintained (supplementary table S5b, Supplementary Material online).
Protein Tertiary Structure Investigation
In order to investigate the nature of the molecular signature detected in halophilic protists, we modeled in silico the tertiary structure of nine selected proteins from Haloc. seosinensis, and determined the contribution of amino acid substitutions (relative to template sequences) to the overall difference in protein GRAVY scores. Using BLASTP against the Protein Data Bank, we selected sequences that were >60% identical to Haloc. seosinensis sequences and for which the alignment covered >80% of the largest sequence in the pair compared. To avoid noise in the signal of surface residues, we favored soluble monomeric proteins with simple substrates over proteins interacting with nucleic acid (e.g., histones), with lipids (e.g., acyl-CoA dehydrogenase) and with several protein partners that could significantly vary between organisms (e.g., ubiquitin-conjugating enzyme E2 and chaperones).
Tertiary structures were modeled using SWISS-MODEL (Arnold et al. 2006; Biasini et al. 2014), and models with QMEAN4 score <−3 were discarded from the analysis. Primary sequences of each pair (Haloc. seosinensis—template) were aligned using mafft and ambiguously aligned sites were removed manually. Homologous sites where the absolute difference in hydropathy index between substituted amino acids was >1 and where the absolute difference in relative solvent accessibility (RSA) was <5% were considered for further analysis.
The difference in hydrophobicity of compared proteins was examined spatially by calculating the difference in GRAVY (delta-GRAVY) of amino acids at substituted sites, given their RSA, as follows:
where delta-GRAVY is the cumulative difference in GRAVY for each RSA bin (range: 5%) that sums up the difference in hydropathy index (given the hydrophobicity scale of Kyte and Doolittle 1982) between Haloc. seosinensis amino acid (Hh) and the corresponding amino acid in the template sequence (Ht) for each substituted site s divided by the total number of N homologous sites for a maximum of M substitutions. A positive value for delta-GRAVY implies that amino acids in Haloc. seosinensis proteins for this RSA bin contributed to increase the GRAVY score (i.e., this class contributed to increase the hydrophobicity of the Haloc. seosinensis protein) whereas a negative value indicates that they contributed to decrease Haloc. seosinensis GRAVY score (i.e., this class contributed to decrease the hydrophobicity of the Haloc. seosinensis protein).
Differential Expression Assessment
Gene expression at optimal and maximal salt concentrations in Haloc. seosinensis was quantified using RSEM (Li and Dewey 2011). Briefly, forward sequence reads from each replicate were mapped to the Trinity assembly using Bowtie 2 v.2.2.4 (Langmead et al. 2009). Reads mapping to multiple isoforms were assigned proportionally to the number of reads mapping to unique regions of the said isoforms. After removal of ORFs having low read counts in all samples (75th quantile <10 reads), differential expression was assessed using three independent softwares: The empirical Bayesian analysis tool EBSeq following ten iterations (Leng et al. 2013), DESeq2 (Love et al. 2014) and the limma package (Ritchie et al. 2015) after normalization using the Voom method (Law et al. 2014). P values were corrected for multiple testing using the Benjamini–Hochberg method. In the text, we report values from EBSeq and consider differentially expressed ORFs with probability of being differentially expressed >0.95.
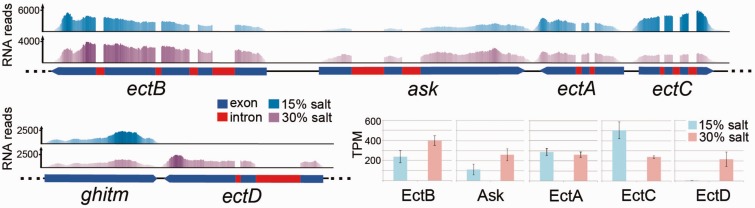
To construct figure 7, RNA read sequences were mapped onto genomic contigs encoding ectoine-related genes using TopHat2 and alignments were visualized using Tablet 1.15.09.01 (Milne et al. 2013).
Fig. 7.—
Genomic context of ectoine/hydroxyectoine synthesis genes in Haloc. seosinensis and their expression levels as a function of external salt concentration expressed in RNA read abundance mapped on genomic contigs and transcripts per million (TPM) as calculated by RSEM. On top, the region between nucleotides 48000 and 56100 on contig c75 contains genes coding for diaminobutyrate aminotransferase (ectB), aspartokinase (ask), diaminobutyrate acetyltransferase (ectA), and ectoine hydroxylase (ectC). On the bottom-left, the region between nucleotides 70000 and 74000 on contig c361 contains genes coding for growth hormone-inducible transmembrane protein (ghitm, shown as a reference) and ectoine hydroxylase (ectD). RNA read abundance is shown on top of genomic contigs on which they were mapped (15% salt in dark blue and 30% salt in purple). On genomic contigs, exons of ORFs are displayed in blue and introns in red. On the bottom-right, RNA transcript abundance (average TPM, error bars indicate 1 SD) for each gene is shown in blue and pink for 15% and 30% salt conditions, respectively.
Results
Cytoplasmic Protein Set
Comparative data sets of diverse marine protistan homologs of Haloc. seosinensis and P. kirbyi sequences were constructed from the MMETSP (Keeling et al. 2014) data set. These are henceforth referred to as the Marine Protists comparative data sets, or “MarProt.” These transcriptomes were used as reference data sets as they are from organisms that thrive in seawater (and not from salt-in extreme halophiles). Predicted cytoplasmic proteins in Haloc. seosinensis and P. kirbyi (1,639 and 1,574 sequences, respectively) had 10,187 and 9,192 homologs, respectively, in MarProt. Applying the homology criteria described in the Materials and Methods section, Haloc. seosinensis and P. kirbyi shared 664 homologous cytoplasmic proteins. As a second approach to compare protein sequences from halophilic protists to nonhalophilic ones we employed a curated “phylogenomic” data set of 252 highly conserved “universal” eukaryotic proteins, “Mandor.” Forty-two proteins from Haloc. seosinensis and 41 from P. kirbyi were in both the MarProt and the Mandor data sets (supplementary table S1, Supplementary Material online).
Cytoplasmic Proteins without Acidic Signature in Halophilic Protists
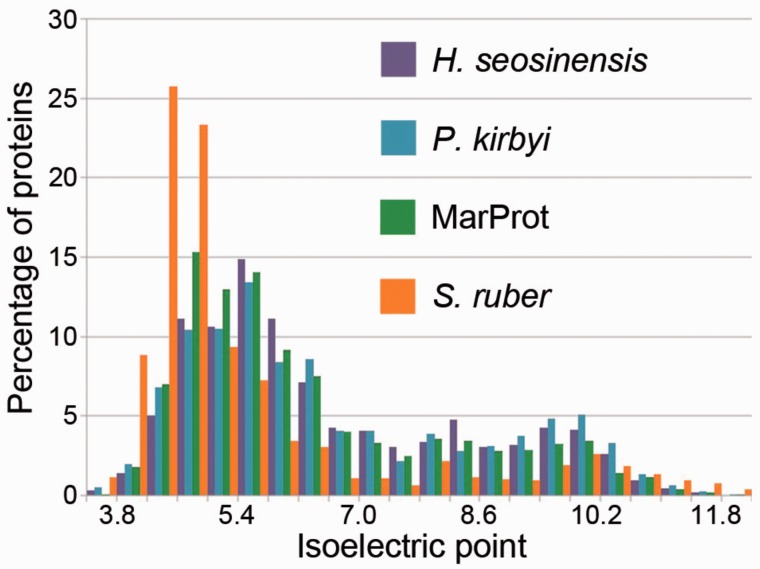
The isoelectric point (pI) distributions of predicted cytoplasmic proteins in Haloc. seosinensis and P. kirbyi were not enriched in acidic proteins as commonly observed in salt-in halophiles (fig. 1 and supplementary fig. S1, Supplementary Material online). The pI distributions of acidic cytoplasmic proteins (pI < 6) in Haloc. seosinensis and P. kirbyi were actually shifted toward basic values as compared with MarProt (Mann–Whitney U test, P < 0.001; supplementary fig. S1, Supplementary Material online). Ranked pI values calculated from amino acids at the fastest-evolving sites in the Mandor alignment also showed that cytoplasmic proteins of halophilic protists were unremarkable in their acidity among this taxonomically broad sample of eukaryotes (rank 31/90 for Haloc. seosinensis and rank 46/90 for P. kirbyi).
Fig. 1.—
Predicted cytoplasmic proteomes of halophilic protists, P. kirbyi and Haloc. seosinensis, are not enriched in acidic proteins as in “salt-in” microbes, represented here by Salinibacter ruber DSM 13855 (GCA_000013045.1). Marine protists sequences come from a concatenation of nonredundant sequences from both MarProt data sets.
Increased Hydrophilicity of Cytoplasmic Proteins in Halophilic Protists
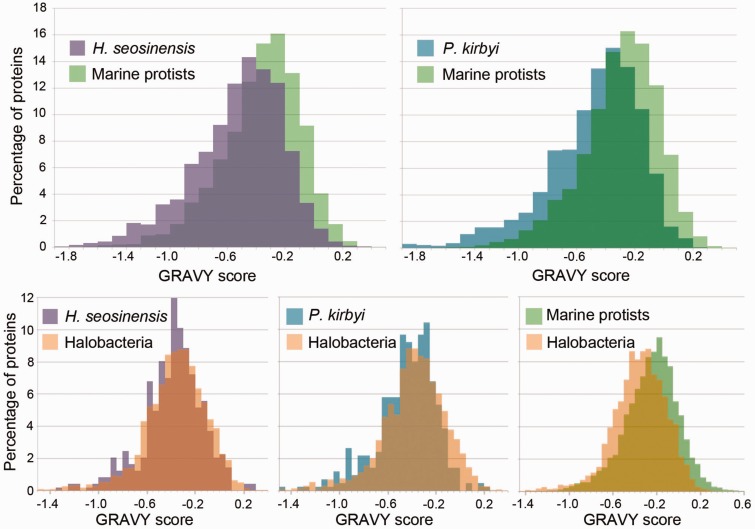
Analyses of GRAVY scores showed that halophilic protists had more hydrophilic cytoplasmic proteomes than typical for marine protists, or mesophilic eukaryotes in general. The distributions of GRAVY scores for Haloc. seosinensis and P. kirbyi were significantly shifted toward more hydrophilic values compared with the distributions of MarProt (Mann–Whitney U test, P < 0.0001; fig. 2). Concordantly, GRAVY scores calculated from the fastest-evolving sites in the Mandor alignment were significantly more hydrophilic for Haloc. seosinensis and P. kirbyi compared with the other taxa (Z-test, P = 0.0005 and 0.0036, respectively). Interestingly, halophilic and halotolerant yeasts did not follow this trend (P > 0.16) whereas the alga Du. salina did (P = 0.009). Furthermore, phylogenetic canonical correlation analysis, which corrects for species nonindependence due to phylogenetic history (Revell and Harrison 2008), indicated that GRAVY scores and the salinity of an organism’s habitat were significantly correlated (Mandor data set; phylogenetic correlation analysis: canonical correlation = 0.39, P = 2.4 × 10−5; supplementary fig. S2, Supplementary Material online). GRAVY scores obtained from the Mandor data set were computed from amino acids at the fastest-evolving sites. These theoretically represent sites in conserved proteins that are not subject to strong selective constraints based on overall protein structure or function, and could respond more easily to environmental conditions, such as salinity. The measured correlation between GRAVY scores calculated at these sites and the habitat salinity supports this assumption.
Fig. 2.—
GRAVY score distributions of predicted cytoplasmic soluble proteins of halophilic protists, marine protists, and Halobacteria. On top, distributions of halophilic protists are significantly shifted toward hydrophilic values compared with homologous proteins from marine protists (Mann–Whitney U test, P < 0.0001). On the bottom, distributions of Haloc. seosinensis and P. kirbyi are slightly shifted toward hydrophilic values compared with the ones of Halobacteria (Whitney, P < 0.02), whereas the distribution for marine protists is significantly shifted toward more hydrophobic values compared with the distribution for Halobacteria (Mann–Whitney U test, P < 0.0001).
Surprisingly, the hydropathy of halophilic protist proteomes was comparable to proteomes of the Halobacteria, salt-in Archaea that typically contain a hydrophilic signature in their proteomes. This was indicated by comparing 561 sequences from Haloc. seosinensis homologous to 3,863 halobacterial sequences, and 478 sequences from P. kirbyi homologous to 3,418 halobacterial sequences. GRAVY distributions of halophilic protists were slightly shifted toward more hydrophilic values compared with those of the Halobacteria (Mann–Whitney U test, P < 0.02; fig. 2). As expected, the distribution of GRAVY scores for 11,529 homologous sequences from the MMETSP marine protists was shifted to be more hydrophobic than the comparable distribution of GRAVY scores for 5,790 sequences from Halobacteria (Mann–Whitney U test, P < 0.0001; fig. 2).
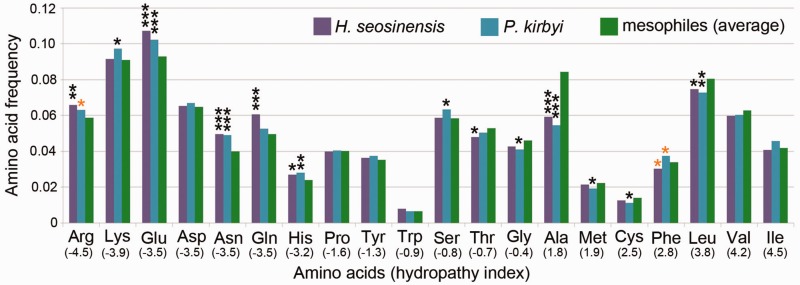
The increased hydrophilicity of the cytoplasmic proteomes of the halophilic protists was due to an overall overrepresentation of polar residues and a general depletion of hydrophobic residues (fig. 3). In both P. kirbyi and Haloc. seosinensis alanine and leucine were significantly underrepresented compared with their frequencies in mesophiles, whereas glutamate, asparagine and histidine were significantly overrepresented. In addition, Haloc. seosinensis cytoplasmic proteins had significantly more arginine and glutamine but less threonine and phenylalanine whereas P. kirbyi had significantly more arginine and glutamine but less cysteine, glycine, and methionine. Cytoplasmic proteins from P. kirbyi were also enriched in phenylalanine, isoleucine, and lysine, but this is potentially a result of the AT-richness of coding sequences in this organism (see below). Interestingly, frequencies of asparagine, histidine and alanine were correlated with habitat salinity (phylogenetic correlation analysis, P < 0.01; supplementary fig. S2, Supplementary Material online). Comparisons of amino acid frequency distributions of halophilic protists to those of MarProt all showed the same general trends of polar residue enrichment and hydrophobic residue depletion (Mann–Whitney U test, P < 0.01, except arginine for P. kirbyi, P = 0.48).
Fig. 3.—
Frequency of amino acids at fastest-evolving sites of the Mandor superalignment. Comparison of values from halophilic protists to the average frequencies of other taxa (free-living microbial eukaryotes not adapted to extreme conditions) in the alignment revealed a general overrepresentation of hydrophilic residues and an underrepresentation of hydrophobic residues (*P < 0.05, **P < 0.01, ***P < 0.001, in black P values corrected for multiple testing, in orange P values that were significant prior to correction). Amino acids are ordered on the x-axis from hydrophilic to hydrophobic residues (hydropathy indices in parentheses).
The contribution of each amino acid to protein GRAVY scores is a function of their frequency and hydropathicity. We therefore evaluated how adjusting each amino acid frequency to a mesophilic level would impact protein GRAVY scores. In both halophiles, these iterative calculations identified alanine as the amino acid most responsible for the low GRAVY scores. In Haloc. seosinensis, the next most important amino acids, in order, were glutamate, glutamine, leucine, arginine, asparagine, valine, phenylalanine, histidine, and isoleucine. In P. kirbyi, the order was alanine, leucine, glutamate, asparagine, lysine, arginine, valine, and histidine. In contrast, when frequencies of the amino acids other than these were adjusted to the mesophilic level, halophiles’ protein GRAVY scores stayed more or less unchanged compared with the average GRAVY of mesophiles.
Phylogenetic canonical correlation analysis of the Mandor data set showed that AT content and GRAVY scores were correlated (canonical correlation = 0.3, P = 0.002; supplementary fig. S2, Supplementary Material online), with the proteomes of GC-rich organisms tending to be more hydrophobic than those of AT-rich organisms. In order to control for a potential influence of AT bias on GRAVY scores, we excluded organisms with AT content <45% and >55% (66 taxa removed) and recalculated the GRAVY scores at fastest-evolving sites. After this data filtering, the extremely hydrophilic GRAVY score of Haloc. seosinensis stood out even more (Z = 4.05, P = 0.00003 after removal of AT-biased organisms compared with Z = 3.29, P = 0.0005 before; supplementary fig. S3, Supplementary Material online). As P. kirbyi was excluded from this previous analysis due to the high AT content of its coding sequences (59% AT on average), we also recalculated GRAVY scores by removing AT-biased sequences from MarProt, retaining 180 sequences from P. kirbyi with AT between 45% and 55% and comparing them to 4,352 homologous sequences from marine protists. Even when AT-biased sequences were removed, the GRAVY score distribution from P. kirbyi was still significantly shifted toward more hydrophilic scores compared with the distribution for MarProt (Mann–Whitney U test, P < 0.00001; supplementary fig. S3, Supplementary Material online).
Next, we aimed to determine which amino acid substitutions had occurred more often in halophiles than in closely related mesophiles, or vice versa, since their divergences from common ancestors. We reconstructed ancestral sequences at internal nodes of a phylogenetic tree inferred from the Mandor data set, and compiled the occurrence of each possible amino acid substitution in the whole alignment for each taxon (tree shown as supplementary fig. S4, Supplementary Material online). Substitutions leading to more hydrophobic residues have occurred less often in the lineages of halophilic protists than in mesophiles (fig. 4). The latter observation is supported by calculations of scores that considered the Z-scores and the difference between GRAVY scores of extant and ancestral amino acids (see Materials and Methods). Halocafeteria seosinensis and P. kirbyi had the lowest scores for substitutions leading to more hydrophobic residues. In particular, substitutions from polar residues to alanine were observed significantly less often in the halophile lineages (adjusted P < 0.05 for substitutions from lysine in both Haloc. seosinensis and P. kirbyi, from arginine, glutamate, asparagine, glutamine, histidine, proline, serine and threonine in P. kirbyi, and from aspartate in Haloc. seosinensis).
Candidate Secreted Proteins in Selected Halophiles
Given the high salinity of the extracellular milieu, secreted proteins in Haloc. seosinensis and P. kirbyi were expected to show the canonical acidic signature of halophilic proteins. However, no acidic signature was detected in proteins predicted to be secreted in Haloc. seosinensis and P. kirbyi (N = 231 and 52 proteins, respectively) when compared with those predicted from the 24 MMETSP protists used for the MarProt data sets (N = 6,584 proteins). The secreted proteins from halophilic protists did show more hydrophilic GRAVY scores, as observed for cytoplasmic proteins (Mann–Whitney U test, P < 0.001).
In order to evaluate the accuracy of the predictions, we searched for homologs of Haloc. seosinensis and P. kirbyi’s predicted secreted proteins in the NR database using BLASTP. Most queries having hits (165 and 30 proteins for Haloc. seosinensis and P. kirbyi, respectively) could be assigned to lysosomal functions (e.g., proteases/cathepsin/carboxypeptidase, beta-N-acetylhexosaminidase, proteins with a saposin domain, physaropepsin) or were homologous to proteins acting at the cell membrane (e.g., phospholipid transfer protein, N-acylsphingosine amidohydrolase), suggesting that these might employ the secretion pathway but were not exported outside the cell. Others were homologous to proteins known to work intracellularly (e.g., dynein). This demonstrates that predicting exoproteins in phagotrophic protists is challenging, and that our analysis probably suffered from this limitation. However, it is notable that some predicted secreted proteins with low pI values (pI < 4.3) were homologous to extracellular proteins, for example, tenascin-like proteins (extracellular matrix proteins), growth factor-binding proteins, and protocadherin (partial hit to extracellular cadherin repeats).
Secretion and retention signals have been intensively studied in yeast (e.g., Vonheijne and Abrahmsen 1989; Gaynor et al. 1994; Conibear and Stevens 1998) and, in contrast to phagotrophic protozoa, yeasts secrete various soluble exoenzymes as part of their osmotrophic lifestyle. Bearing this in mind, as a form of positive control, we also compared predicted exoproteins from the halotolerant/halophilic yeasts D. hansenii and W. ichthyophaga with those of the marine protists in MarProt. The predicted secreted proteins from D. hansenii (103 proteins with median pI of 4.4) and W. ichthyophaga (110 proteins with median pI of 4.3) were significantly more acidic than the inferred exoproteins from marine protists (6,479 proteins with median pI of 4.6, Mann–Whitney U test, P < 0.0008). Therefore, these results support the existence of an acidic signature in the exoproteins of halotolerant/halophilic eukaryotes when such proteins can be confidently identified.
As another control, we compared the predicted secreted proteome to the cytoplasmic proteome in each of several salt-out halophilic bacteria that had optimal salinities for growth between 9% and 18% (i.e., similar to P. kirbyi and Haloc. seosinensis). The species examined were Actinopolyspora halophila, Marinococcus halotolerans, Nocardiopsis halophila, Virgibacillus alimentarius, Chromohalobacter salexigens, Halorhodospira halochloris, and Thiomicrospira halophila. As expected, the secreted proteome was more acidic and hydrophilic than the cytoplasmic proteome in each of these species (Mann–Whitney U test, P < 0.001; supplementary table S2, Supplementary Material online).
Localization of the Hydrophilic Signature Inside Protein Tertiary Structure
In silico predictions of the tertiary structure of proteins in Haloc. seosinensis provided insights into the localization of the hydrophilic signature. The structures of nine proteins were modeled on templates from nonhalophilic organisms (supplementary table S3, Supplementary Material online). All examined Haloc. seosinensis protein sequences were more hydrophilic (i.e., they had lower GRAVY scores) than the corresponding homologous template sequence, except aconitase, which was slightly more hydrophobic (difference in GRAVY score of 0.01).
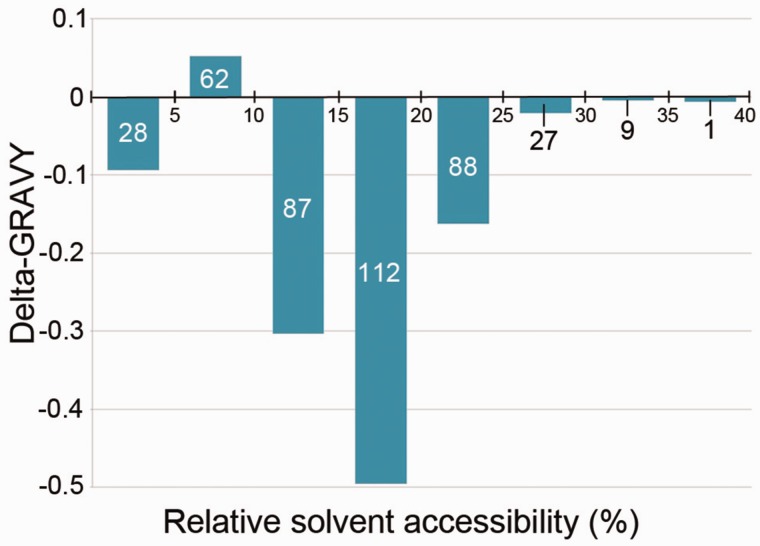
We examined the locations of substitutions that led to an absolute change in amino acid hydropathy index >1 in Haloc. seosinensis proteins compared with the protein sequences of the templates used to generate the structures (fig. 5). Residues that contributed the most to the low hydrophobicity of Haloc. seosinensis proteins had a relative solvent accessibility (RSA) between 10% and 25%. Typically, a threshold of 20% RSA is used to discriminate surface from buried residues (e.g., Chen and Zhou 2005). Our analysis indicated that residues that influenced GRAVY scores the most had RSA values under this threshold. High salt enhances the hydrophobic effect, thus halophilic proteins contain less hydrophobic interactions in order to avoid a too rigid conformation and protein aggregation. A more hydrophilic core in Haloc. seosinensis, relative to template proteins, is in line with this property of halophilic proteins.
Fig. 5.—
Cumulative difference in GRAVY (delta-GRAVY) scores of amino acids at substituted sites in nine Haloc. seosinensis proteins compared with the templates used to model the tertiary structures (y-axis), as a function of their RSA (x-axis). Negative y-axis values indicate that these residues contributed overall to decrease the hydrophobicity of Haloc. seosinensis proteins. The total number of substitutions in each bin is indicated on respective bars.
Expression of Putative Osmolyte Synthesizers/Importers
The lack of a notably acidic proteome suggests that the halophilic protozoa under study might use the salt-out strategy. That would require them to import and/or synthesize organic solutes as a response to elevated salinity. In a differential gene expression analysis of Haloc. seosinensis, proteins whose transcripts were extremely upregulated at high salt included ectoine hydroxylase and transporters for amino acids and myo-inositol. We elected to examine these interesting cases in more detail, using a purely bioinformatic approach.
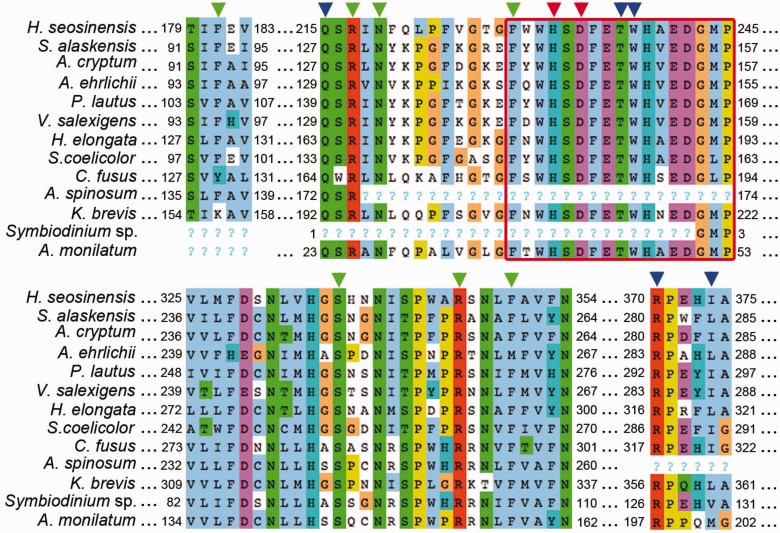
5-hydroxyectoine is one of the osmolytes with the best protein-stabilizing properties (Lippert and Galinski 1992). It is synthesized from ectoine by ectoine hydroxylase (EctD), which belongs to the nonheme-containing iron(II) and 2-oxoglutarate-dependent oxygenases, a ubiquitious and large enzyme superfamily (Schofield and Zhang 1999). Halocafeteria seosinensis expressed transcripts annotated as EctD, and remarkably, these were 227-fold upregulated at high salt in Haloc. seosinensis (table 1). The Haloc. seosinensis EctD sequence contained all the conserved residues involved in binding iron, 2-oxoglurate and 5-hydroxyectoine as well as the ectoine hydroxylase consensus sequence (fig. 6 and supplementary fig. S5, Supplementary Material online; Höppner et al. 2014).
Table 1.
Expression of Genes Potentially Involved in Organic Osmolyte Synthesis and Transport in Halocafeteria seosinensis
| Sequence Name | TPM |
EBSeq |
DESeq2 |
Limma |
Annotation | ||||
|---|---|---|---|---|---|---|---|---|---|
| Opt | Max | PPDE | Post FC | P Value | log2 FC | P Value | log2 FC | ||
| Ectoine biosynthesis | |||||||||
| m.89065 | 289 | 261 | 0.04 | 0.7 | 0.172 | −0.5 | 0.233 | −0.5 | EctA |
| m.89060 | 238 | 397 | 1.0 | 1.3 | 1E−04 | 0.4 | 0.052 | 0.4 | EctB |
| m.89066 | 504 | 239 | 0.7 | 0.4 | 3E−05 | −1.4 | 0.015 | −1.4 | EctC |
| m.14216 | 0.7 | 216 | 1.0 | 220 | NA | 7.4 | 0.001 | 7.8 | Ectoine hydroxylase (EctD) |
| m.89063 | 113 | 259 | 1.0 | 1.8 | 4E−04 | 0.9 | 0.017 | 0.9 | Aspartate kinase |
| Amino acid transport | |||||||||
| m.15646 | 8 | 83 | 1.0 | 7.9 | 2E−59 | 3.0 | 2E−04 | 3.0 | Sodium-amino acid symporter |
| m.82938 | 4 | 38 | 1.0 | 7.1 | 2E−09 | 2.7 | 0.009 | 2.8 | Amino acid transporter |
| m.16444 | 9 | 245 | 1.0 | 22.6 | 2E−28 | 4.3 | 4E−04 | 4.5 | Amino acid transporter |
| m.16489 | 2 | 80 | 1.0 | 29.3 | 8E−50 | 4.7 | 3E−04 | 4.9 | Amino acid transporter |
| m.62883 | 29 | 25 | 1.0 | 0.7 | 2E−05 | −0.6 | 0.022 | −0.6 | Amino acid transporter |
| m.11656 | 16 | 11 | 1.0 | 0.5 | 2E−05 | −0.9 | 0.012 | −0.9 | Amino acid transporter |
| m.18092 | 55 | 36 | 1.0 | 0.5 | 2E−11 | −1.0 | 0.004 | −1.0 | Amino acid transporter |
| m.41609 | 18 | 14 | 1.0 | 0.6 | 4E−06 | −0.7 | 0.014 | −0.6 | Amino acid transporter |
| m.1357 | 48 | 44 | 1.0 | 0.7 | 3E−06 | −0.5 | 0.029 | −0.5 | Amino acid transporter |
| Myo-inositol transport | |||||||||
| m.89752 | 3 | 76 | 1.0 | 18.0 | 3E-44 | 4.1 | 2E-04 | 4.3 | Myo-inositol transporter |
| m.27262 | 5 | 62 | 1.0 | 9.4 | 1E-26 | 3.2 | 4E-04 | 3.4 | Myo-inositol transporter |
| m.25988 | 6 | 14 | 1.0 | 1.9 | 7E-06 | 0.9 | 0.012 | 0.9 | Myo-inositol transporter |
| m.27173 | 2 | 5 | 0.6 | 2.3 | 0.042 | 1.1 | 0.102 | 1.1 | Myo-inositol transporter |
Note.—TPM, average mRNA Transcripts Per Million at optimal (Opt) and at maximal (Max) salt concentrations; PPDE, Posterior Probability of being Differentially Expressed; Post FC, Posterior Fold Change for maximal over optimal salt concentration; EctA, l-2,4-diaminobutyrate acetyltransferase; EctB, l-2,4-diaminobutyrate transaminase; EctC, ectoine synthase. P values calculated by DESeq2 and limma were corrected for multiple testing. (NA: due to low mean normalized count at optimal concentration.)
Fig. 6.—
Alignment of ectoine hydroxylase including sequences of characterized enzymes from Sphingopyxis alaskensis (WP_011543221), for which the crystal structure is available, Acidiphilum cryptum (AER00256), Alkalilimnicola ehrlichii (AER00257), Paenibacillus lautus (ACX67869), Virgibacillus salexigens (AAY29689), Halomonas elongata (WP_013333764) and Streptomyces coelicolor (Q93RV9), and sequences from the protists Haloc. seosinensis, Ceratium fusus (CAMPEP_0172939100), Az. spinosum (concatenation of CAMPEP_0180530970, CAMPEP_0180661784 and CAMPEP_0180535134), Karenia brevis (CAMPEP_0178068410), Symbiodinium sp. (CAMPEP_0169646080) and Alexandrium monilatum (CAMPEP_0175754634). Arrowheads indicate residues involved in binding iron (red), 2-oxoglutarate (green), and 5-hydroxyectoine (blue). The consensus sequence of ectoine hydroxylase (FXWHSDFETWHXEDG-M/L-P) is squared in red. “?” indicates missing data for partial sequences.
5-hydroxyectoine biosynthesis depends on a supply of ectoine, one of the most common osmolytes in halophilic bacteria (Severin et al. 1992). It is possible to speculate that Haloc. seosinensis might import ectoine from food bacteria. This might be the case, however, Haloc. seosinensis also seemed to express all the enzymes necessary for ectoine biosynthesis; diaminobutyrate aminotransferase (EctB), diaminobutyrate acetyltransferase (EctA), and ectoine synthase (EctC) (table 1). Interestingly, these genes are arranged in a cluster on the Haloc. seosinensis genome, similarly to the way the ectoine synthesis operon is encoded on bacterial genomes, including an aspartate kinase (ask) gene (Widderich et al. 2014; fig. 7). A gene positioned at another locus in the Haloc. seosinensis genome encoded a bifunctional aspartate kinase/diaminopimelate decarboxylase enzyme (ORF m.33370), suggesting that the ask gene inside the ectoine synthesis gene cluster might be specialized for ectoine synthesis, with the bifunctional enzyme instead used for lysine biosynthesis, as observed in bacteria (Stöveken et al. 2011). However, phylogenetic analysis of the Haloc. seosinensis Ask sequence (including the aspartokinase cohesion group representative sequences; Lo et al. 2009) did not recover a strongly supported clade made up of Ask sequences specialized for ectoine synthesis (supplementary fig. S6, Supplementary Material online).
Ectoine synthesis also requires the action of aspartate-β-semialdehyde dehydrogenase (ASADH), which is encoded three genes downstream of ectC on the Haloc. seosinensis genome. Except for asadh, all of the ectoine-related genes are closely related to bacterial sequences (supplementary figs. S7–S10, Supplementary Material online). Nonetheless they all contain spliceosomal introns (fig. 7) and thus are truly eukaryotic sequences and do not represent bacterial contamination in our genomic assemblies.
Predicted N-terminal mitochondrial targeting signals were detected in all Haloc. seosinensis ectoine synthesis-related proteins (EctABCD, Ask, and ASADH; see supplementary fig. S5, Supplementary Material online for an example), providing evidence that ectoine synthesis may occur in the mitochondria of Haloc. seosinensis. Ectoine synthesis requires aspartate and glutamate as precursors; these amino acids are synthesized using intermediates of the mitochondrial Krebs cycle (Salway 1999).
Ectoine synthesis is thought to be restricted to bacteria and a few archaea (Widderich et al. 2014 and references therein); however, no systematic search for eukaryotic homologs has been published to our knowledge. We therefore searched for homologs of EctA, B, C, and D in the MMETSP data (all taxa, not just the MarProt subset) as well as the NR and NT databases. We detected candidates for all four genes in a variety of eukaryotes. EctA and EctC were found in transcriptomic data from at least six protists: The amoebozoan Vexillifera sp., the dinoflagellate Az. spinosum, the cryptomonad Goniomonas sp., the ochrophyte stramenopile Bolidomonas pacifica, and in C. roenbergensis, which is a bicosoecid stramenopile and thus quite closely related to Haloc. seosinensis (supplementary figs. S7 and S8, Supplementary Material online). Interestingly, EctA and EctC were encoded on a single transcript (i.e., both proteins appeared to be encoded in a single ORF) in Vexillifera sp. (sequence CAMPEP_0201547320) and Az. spinosum (CAMPEP_0180652276), suggesting that they were expressed as single multifunctional polypeptides. Azadinium spinosum also had EctD encoded on this ORF (i.e., EctA, C and D formed a single ORF). More protists putatively expressed either EctA or EctC, like the amoebozoan Stygamoeba regulata and Stereomyxa ramosa, the ciliate Tiarina fusus, the choanoflagellate Monosiga brevicollis, the diatom Pseudonitzschia fraudulenta, and the oomycetes Albugo candida, Albugo laibachii, Saprolegnia parasitica, Saprolegnia diclina, Phytophthora infestans, Phytophthora sojae, and Plasmospora halstedii. Interestingly, genes closely related to ectC were also detected in the deuterostome animals Branchiostoma floridae and Saccoglossus kowalevskii. In contrast with the wide distribution of EctA and C, only three protists, Goniomonas sp. (strain M), Pelagodinium beii and C. roenbergensis, expressed bona fide EctB candidates. These EctB candidates grouped with bacterial EctB sequences in phylogenetic trees (supplementary fig. S9, Supplementary Material online). Other eukaryotic sequences harvested using Haloc. seosinensis EctB as a query were markedly more distantly related, and were annotated as being involved in amino metabolism: Alanine–glyoxylate aminotransferase, ornithine aminotransferase, putrescine—2-oxoglutarate aminotransferase, 2,2-dialkylglycine decarboxylase, and 4-aminobutyrate aminotransferase (supplementary fig. S9, Supplementary Material online). These latter sequences are unlikely to be orthologs of EctB.
The apparent absence of EctABC homologs in some of the protists that we have investigated must be interpreted with care; putative sequences were mostly identified in the MMETSP data where the transcriptome coverage could be too low in some cases to recover all EctABC genes from individual species, or, alternatively, the genes might just not be expressed in the growth conditions analyzed. Furthermore, many eukaryotic sequences from the MMETSP data set were probably excluded from our analysis due to our stringent filtering of prokaryotic sequences that used a 50% identity cut-off in DNA sequences. However, this analysis demonstrates that ectoine synthesis might well occur in at least a few protists, including in two marine protists that, as in Haloc. seosinensis, seemed to express all ectABC genes. Experimental confirmation is therefore greatly needed to assess the function of these genes in protists.
Regarding ectoine hydroxylase, putative EctD sequences were detected in the dinoflagellate taxa Ceratium fusus, Az. spinosum, Symbiodinium sp., Alexandrium monilatum, Karenia brevis, and Gymnodinium cantenatum (supplementary fig. S10, Supplementary Material online). Where determined, these sequences all included the consensus signature sequence of EctD and the conserved functional residues, except one residue binding 2-oxoglutarate (although some sequences were partial and were missing regions of interest, fig. 6). As for EctC, putative EctD sequences were detected in the animals Branchiostoma floridae and Saccoglossus. Kowalevskii; however, most functional residues were substituted, including the consensus sequence that was not conserved in S. kowalevskii.
In Haloc. seosinensis, ectoine hydroxylase was greatly upregulated at high salt and EctA was not differentially expressed. Predictions for EctB and EctC were not consistent among the three programs used for differential expression analysis (table 1). EctB seemed to be slightly upregulated (30% increase in expression) but limma predicted this to be not significant (adjusted P-value 0.052). Regarding EctC, EBSeq generated a relatively low posterior probability of 0.7 for a 2.5-fold repression at high salt whereas the other two programs returned significant P-values (adjusted P < 0.015).
In principle, import of amino acids with osmoprotectant properties (e.g., glutamate, glycine, proline, aspartate) from food prokaryotes might also contribute to osmotic equilibrium in Haloc. seosinensis. Several genes annotated as amino acid transporters were highly upregulated at high salt (up to 29-fold increase; table 1). In comparison, other amino acid transporter-related genes that were upregulated at optimal salt concentration did not show more than a 2-fold increase in expression (table 1).
A gene related to sodium-neurotransmitter symporters was 8-fold upregulated at high salt (m.15646 in table 1). This family of transporters is well documented in animals. They use a Na+ or Cl− gradient to transport monoamines such as serotonin, dopamine and norepinephrine, and the neurotransmitter ϒ-aminobutyric acid (GABA) and glycine (Torres et al. 2003). They can also potentially transport proline and taurine, two molecules reported as osmoprotectants. Alignment of the Haloc. seosinensis sequence with LeuT (leucine transporter in Aquifex), GlyT1b (glycine transporter in humans), GAT1 (GABA transporter in humans), DAT (dopamine transporter in humans), and SERT (serotonin transporter in humans) showed conservation of functionally relevant residues, including at substrate-binding sites, and at sites involved in coordinating one of the sodium ions (supplementary fig. S11, Supplementary Material online; Yamashita et al. 2005). In the Haloc. seosinensis sequence, amino acids at sites known to interact with the substrate are closer in identity to Glyt1b and GAT1, consistent with the prediction that this transporter in Haloc. seosinensis carries amino acids (with osmoprotectant potential) rather than biogenic amides (neurotransmitters). Expression of several tRNA synthetases and elongation factor genes was modestly repressed at high salt (< 3-fold decrease) suggesting that the inferred increase in amino acid import was probably not coupled to translational activity (supplementary table S4, Supplementary Material online).
Myo-inositol is another well-known compatible solute (Garcia-Perez and Burg 1991; Klages et al. 1999; Majee et al. 2004). Halocafeteria seosinensis expressed four related genes encoding myo-inositol transporters (table 1). Two were highly upregulated at high salt (9- and 18-fold increases in expression) whereas the other two were not differentially expressed. Myo-inositol is also one of the main precursors of phosphatidylinositol (PI), a minor component of the cell membrane, and an important molecule in cell signaling. However, the three enzymes that together convert myo-inositol to PI were actually downregulated at high salt (1.3- to 3.6-fold repression; supplementary table S4, Supplementary Material online), suggesting that the increased gene expression of myo-inositol transporters at high salt was not directed toward lipid biosynthesis.
Discussion
Molecular Signature in Halophilic Protists
We detected an unusual signature in two halophilic protists (specifically phagotrophic protozoa), whereby cytoplasmic proteins were not highly acidic but showed an increased hydrophilicity. We interpret these observations as indicating that the cytosolic concentration of salt was lower than in typical salt-in microbes, while also suggesting a higher cytosolic salt concentration than in marine organisms. In particular, the relative increase in hydrophilicity suggests that cytoplasmic proteins in halophilic protists have evolved to require fewer hydrophobic interactions to remain folded, as one would expect whether the cytosolic salt concentration was higher in these organisms than in mesophiles. As mentioned earlier, although most salt-in strategists have extremely acidic proteomes, whereas salt-out strategists do not, there are several known exceptions (e.g., absence of an acidic signature in the proteomes of the salt-in Halanaerobiales; Elevi Bardavid and Oren 2012). For this reason, the absence of an acidic signature in the predicted proteomes of Haloc. seosinensis and P. kirbyi strongly suggests a broad salt-out strategy, but is not proof of one. Further, experimental work is needed to test this hypothesis.
We also detected the upregulation in high salt conditions of genes potentially involved in organic osmolyte metabolism and transport in Haloc. seosinensis. This supports the idea that organic solutes contribute to osmotic adjustment in these organisms, although the relative importance of inorganic osmolytes remains to be determined. Obtaining direct experimental evidence for organic osmolyte accumulation would be an important avenue for further research, notwithstanding the technical challenges to working with necessarily nonaxenic cultures of small bacterivorous protozoa.
The measured correlation between GRAVY scores and habitat salinity also suggests that the extracellular osmolarity might influence the hydropathicity of the cytoplasmic proteomes of salt-out eukaryotic microbes (taxa in the Mandor data set). Despite active salt expulsion, higher extracellular salt concentration likely leads to a relatively higher salt content that the cell has to manage, at least some of the time. For instance, bacterivores presumably bring salt into the endomembrane system during phagotrophy, consequently increasing the risk of salt intrusion in the cytosol.
Contrary to expectations, most predicted exoproteins in our halophilic protozoa did not show the acidic signature of halophilic proteins. This can probably be explained, to a large extent, by the difficulty of predicting secreted proteins in protists. Even when predictions from multiple programs are combined, as we did in this investigation, the accuracy of prediction is approximately 50% when studying protist sequences (Min 2010). Furthermore, phagotrophs such as Haloc. seosinensis and P. kirbyi do not secrete digestive enzymes outside the cell but rather engulf particles in tightly packed vacuoles. Several proteins predicted to be secreted in halophilic protozoa had functions related to lysosomal activity or to membrane biogenesis, suggesting that they were following the secretory pathway but were not exported outside the cell. This is in contrast with osmotrophs such as fungi and prokaryotes in which a higher number of secreted enzymes is expected. Interestingly, we actually did detect an acidic signature for the halotolerant/halophilic yeast proteins that we predicted to be secreted.
In addition, it is possible that features other than acidity could represent mechanisms of salt adaptation in proteins. The surfaces of acidic halophilic proteins are enriched in negative charges that are thought to interact with water. Protein glycosylation and phosphorylation could also lead to increased negative surface charges and increased solubility. We did not examine such posttranslational modifications and hence we cannot determine whether they play a role in salt adaptation.
Expression of Genes Involved in Organic Osmolyte Metabolism in Haloc. Seosinensis
Analyses of genes that are differentially expressed as a function of salinity in Haloc. seosinensis identified candidates for involvement in biosynthesis and transport of organic osmolytes. At high salt, we detected the upregulation of genes related to hydroxyectoine synthesis and to transporters of myo-inositol and amino acids, potentially glutamate, glycine, proline or taurine. Glycerol, a common osmolyte in yeasts and certain algae, probably does not accumulate in Haloc. seosinensis and P. kirbyi as they did not express glycerol phosphatase or known glycerol transporters. This is consistent with the observation that the addition of glycerol in the medium does not improve growth of Haloc. seosinensis (Park et al. 2006).
The presence of an ectoine hydroxylase with the consensus sequence signature and conserved functional residues, its activation at high salt, and the presence of a bacteria-like ectoine biosynthesis pathway, all suggest that Haloc. seosinensis uses and modifies these organic solutes for osmotic adjustment. This hypothesis is surprising a priori, as ectoine synthesis has not been reported in eukaryotes to our knowledge (e.g., Widderich et al. 2014). Nonetheless, we found modest evidence from available transcriptomes that ectoine biosynthesis may be found in some marine protists, including a close relative of Haloc. seosinensis (C. roenbergensis). However, direct observations of osmolyte accumulation will be necessary to validate these conjectures.
The composition and proportion of accumulated osmolytes commonly vary in microorganisms as a function of growth conditions such as external salinity (García-Estepa et al. 2006), and it is possible that signatures of these differences might be seen at the transcript level. Knowledge on transcriptional regulation of the ectoine operon in bacteria is still fragmented. Studies of a few bacteria showed that transcription is polycistronic, sometimes regulated by EctR, a repressor protein of the MarR family, and is complex as it involves five putative promoters, including one inside the operon (Calderón et al. 2004; Mustakhimov et al. 2010). In Haloc. seosinensis, RNA read-mapping on a genomic contig indicated that four mRNA transcripts were generated from the ectABC-ask cluster of genes (fig. 7), but no EctR or MarR domains (PF01047.17, PF12802.2) were detected in any predicted protein sequences. Our transcriptomic experiment detected the upregulation of ectB, ask, and ectD at high salt, but unexpectedly, no differential expression of ectA and repression of ectC. Regarding ectA, it is possible that repression of activity at low salt occurred through allosteric regulation. This possibility has been discussed in the case of the ectoine biosynthetic enzymes in C. salexigens and Methylomicrobium alcaliphilum (Calderón et al. 2004; Reshetnikov et al. 2005). A higher expression of ectC at optimal salt concentrations suggests that ectoine, the product of this gene, might be an important organic osmolyte at this salinity, whereas hydroxyectoine, generated by ectD, would become relatively more important at extremely high salinity.
Several lines of evidence suggest that hydroxyectoine may be more beneficial than ectoine when salinity gets very high (i.e., in our case 4.7M NaCl). First, hydroxyectoine has superior desiccation protection properties compared with ectoine (Lippert and Galinski 1992). Furthermore, in Brevibacterium sp. JCM6894, accumulation of ectoine starts to plateau or decreases at salinity >2 M whereas hydroxyectoine starts to accumulate at salinity >1.5 M (Nagata et al. 1996, 2008). In Brevibacterium linens, ectoine content also decreases for salinities >2 M NaCl (Bernard et al. 1993). In C. salexigens, although ectoine content increases with salinity ranging from 0.75 to 3 M NaCl, the magnitude of the increase is higher for hydroxyectoine compared with ectoine (13.8- vs. 2.8-fold increase, respectively; García-Estapa et al. 2006). In addition, some strains of Pseudomonas stutzeri preferentially accumulate hydroxyectoine over ectoine (Seip et al. 2011; Stöveken et al. 2011).
Hydroxyectoine was reported to accumulate substantially only during the stationary growth phase in Virgibacillus salexigens, Bacillus clarkii (Bursy et al. 2007), Marinoccocus sp. (Schiraldi et al. 2006), Streptomyces coelicolor (Bursy et al. 2008), and Halomonas elongata (Cánovas et al. 1999). Our growth controls indicated that the Haloc. seosinensis cultures were at midexponential phase at the time of RNA extraction. Therefore, the observed difference in ectoine hydroxylase expression between conditions was not a result of different growth phases. Rather, it suggests that hydroxyectoine contributed to Haloc. seosinensis’ survival in an extreme salt condition (30% ∼ 4.7 M NaCl). Furthermore, in the halophilic proteobacterium C. salexigens, ectoine and hydroxyectoine accumulate together during the exponential phase (García-Estepa et al. 2006) and the same is observed in S. coelicolor when salt is added to the medium (Bursy et al. 2008).
The close phylogenetic relationship of Haloc. seosinensis ectABCD genes with bacterial sequences and the bacteria-like genomic arrangement of ectABC-ask suggest that these genes were acquired by lateral gene transfer from bacteria. The wide but sporadic diversity of protists that putatively express ectoine synthetic genes also suggests that these genes might have afterwards spread horizontally between protists. Explaining this distribution pattern by EctABC being ancestral to all eukaryotes and lost multiple times is less parsimonious. More sampling of eukaryotic sequences will shed light on this by either revealing further patchiness of these genes in eukaryotes or, alternatively, showing a broader distribution within eukaryotes.
Summary
Our results suggest that Haloc. seosinensis and P. kirbyi most probably use organic solutes as the main osmolytes while likely experiencing higher intracellular salt content relative to organisms inhabiting marine environments. This is based on the presence of a hydrophilic signature in cytoplasmic proteins and on the expression pattern of genes potentially involved in organic osmolyte synthesis and transport, namely ectoine/hydroxyectoine, myo-inositol and undetermined amino acids. Importantly, future metabolomic investigation is required to directly measure these osmolytes using nuclear magnetic resonance spectroscopy, mass spectrometry or high performance liquid chromatography. Characterization of enzymatic activity at varying salinities could also help in determining whether cytoplasmic enzymes of halophilic protists function optimally at relatively high salt concentrations.
Supplementary Material
Supplementary figures S1–S11 and tables S1–S5 are available at Genome biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Edward Susko for providing advice on the statistical framework of this study. This work was supported by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada to A.G.B.S. (grant number 298366-2014) and A.J.R. (grant number 227085-2011); and by the Tula Foundation.
Literature Cited
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25:4692–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. [DOI] [PubMed] [Google Scholar]
- Bernard T, et al. 1993. Ectoine accumulation and osmotic regulation in Brevibacterium linens. J Gen Microbiol. 139:129–136. [Google Scholar]
- Biasini M, et al. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42:W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolen DW, Baskakov IV. 2001. The osmophobic effect: natural selection of a thermodynamic force in protein folding. J Mol Biol. 310:955–963. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MW, Kolisko M, Silberman JD, Roger AJ. 2012. Aggregative multicellularity evolved independently in the eukaryotic supergroup Rhizaria. Curr Biol. 22:1123–1127. [DOI] [PubMed] [Google Scholar]
- Burki F, Okamoto N, Pombert JF, Keeling PJ. 2012. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc R Soc Lond [Biol]. 279:2246–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursy J, et al. 2008. Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl Environ Microbiol. 74:7286–7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursy J, Pierik AJ, Pica N, Bremer E. 2007. Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J Biol Chem. 282:31147–31155. [DOI] [PubMed] [Google Scholar]
- Calderón MI, et al. 2004. Complex regulation of the synthesis of the compatible solute ectoine in the halophilic bacterium Chromohalobacter salexigens DSM 3043. Microbiol-SGM. 150:3051–3063. [DOI] [PubMed] [Google Scholar]
- Cánovas D, et al. 1999. Role of N gamma-acetyldiaminobutyrate as an enzyme stabilizer and an intermediate in the biosynthesis of hydroxyectoine. Appl Environ Microbiol. 65:3774–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Jiang J-G. 2009. Osmotic responses of Dunaliella to the changes of salinity. J Cell Physiol. 219:251–258. [DOI] [PubMed] [Google Scholar]
- Chen HL, Zhou HX. 2005. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res. 33:3193–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, et al. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BC, Park JS, Xu KD, Choi JK. 2008. Morphology and molecular phylogeny of Trimyema koreanum n. sp., a ciliate from the hypersaline water of a solar saltern. J Eukaryot Microbiol. 55:417–426. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. 1998. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta. 140:211–230. [DOI] [PubMed] [Google Scholar]
- Coronado MJ, et al. 2000. The alpha-amylase gene amyH of the moderate halophile Halomonas meridiana: cloning and molecular characterization. Microbiology 146:861–868. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10:210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deole R, Challacombe J, Raiford DW, Hoff WD. 2013. An extremely halophilic proteobacterium combines a highly acidic proteome with a low cytoplasmic potassium content. J Biol Chem. 288:581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763. [DOI] [PubMed] [Google Scholar]
- Elcock AH, McCammon JA. 1998. Electrostatic contributions to the stability of halophilic proteins. J Mol Biol. 280:731–748. [DOI] [PubMed] [Google Scholar]
- Elevi Bardavid R, Oren A. 2012. The amino acid composition of proteins from anaerobic halophilic bacteria of the order Halanaerobiales. Extremophiles 16:567–572. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- Foissner W, Jung J-H, Filker S, Rudolph J, Stoeck T. 2014. Morphology, ontogenesis and molecular phylogeny of Platynematum salinarum nov. spec., a new scuticociliate (Ciliophora, Scuticociliatia) from a solar saltern. Eur J Protistol. 50:174–184. [DOI] [PubMed] [Google Scholar]
- Frolow F, Harel M, Sussman JL, Mevarech M, Shoham M. 1996. Insights into protein adaptation to a saturated salt environment from the crystal structure of a halophilic 2Fe-2S ferredoxin. Nat Struct Biol. 3:452–458. [DOI] [PubMed] [Google Scholar]
- Galinski EA. 1995. Osmoadaptation in bacteria. Adv Microb Physiol. 37:272–328. [PubMed] [Google Scholar]
- García-Estepa R, et al. 2006. The ectD gene, which is involved in the synthesis of the compatible solute hydroxyectoine, is essential for thermoprotection of the halophilic bacterium Chromohalobacter salexigens. J Bacteriol. 188:3774–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez A, Burg MB. 1991. Role of organic osmolytes in adaptation of renal cells to high osmolarity. J Membr Biol. 119:1–13. [DOI] [PubMed] [Google Scholar]
- Gaynor EC, Heesen ST, Graham TR, Aebi M, Emr SD. 1994. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 127:653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunde-Cimerman N, Ramos J, Plemenitaš A. 2009. Halotolerant and halophilic fungi. Mycol Res. 113:1231–1241. [DOI] [PubMed] [Google Scholar]
- Hauer G, Rogerson A. 2005. Heterotrophic protozoa from hypersaline environments In: Gunde-Cimerman N, Oren A, Plemenitaš A, editors. Adaptation to life at high salt concentrations in archaea, bacteria, and eukarya. Dordrecht: Springer; p. 519-539. [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. 2016. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32:767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 66:300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höppner A, Widderich N, Lenders M, Bremer E, Smits SHJ. 2014. Crystal structure of the ectoine hydroxylase, a snapshot of the active site. J Biol Chem. 289:29570–29583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, et al. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35:W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E, Oliphant T, Peterson P. 2001. SciPy: Open Source Scientific Tools for Python. [cited 2015 Jan]. Available from: http://www.scipy.org/.
- Käll L, Krogh A, Sonnhammer ELL. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 338:1027–1036. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, et al. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klages K, Boldingh H, Smith GS. 1999. Accumulation of myo-inositol in Actinidia seedlings subjected to salt stress. Ann Bot. 84:521–527. [Google Scholar]
- Kogej T, Gorbushina AA, Gunde-Cimerman N. 2006. Hypersaline conditions induce changes in cell-wall melanization and colony structure in a halophilic and a xerophilic black yeast species of the genus Trimmatostroma. Mycol Res. 110:713–724. [DOI] [PubMed] [Google Scholar]
- Kogej T, Ramos J, Plemenitaš A, Gunde-Cimerman N. 2005. Halophilic fungus Hortaea werneckii and the halotolerant fungus Aureobasidium pullulans maintain low intracellular cation concentrations in hypersaline environments. Appl Environ Microbiol. 71:6600–6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 157:105–132. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi JK. 1974. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol Rev. 38:272–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. 2014. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng N, et al. 2013. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29:1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert K, Galinski EA. 1992. Enzyme stabilization by ectoine-type compatible solutes: protection against heating, freezing and drying. Appl Microbiol Biotechnol. 37:61–65. [Google Scholar]
- Lo C-C, Bonner CA, Xie G, D'Souza M, Jensen RA. 2009. Cohesion group approach for evolutionary analysis of aspartokinase, an enzyme that feeds a branched network of many biochemical pathways. Microbiol Mol Biol Rev. 73:594–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21:936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majee M, et al. 2004. A novel salt-tolerant L-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice - Molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco-conferring salt tolerance phenotype. J Biol Chem. 279:28539–28552. [DOI] [PubMed] [Google Scholar]
- Mavromatis K, et al. 2009. Genome analysis of the anaerobic thermohalophilic bacterium Halothermothrix orenii. PLoS One 4:e4192.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I, et al. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 14:193–202. [DOI] [PubMed] [Google Scholar]
- Min XJ. 2010. Evaluation of computational methods for secreted protein prediction in different eukaryotes. J Proteomics Bioinform. 3:143–147. [Google Scholar]
- Mustakhimov II, Reshetnikov AS, Khmelenina VN, Trotsenko YA. 2010. Regulatory aspects of ectoine biosynthesis in halophilic bacteria. Microbiology 79:583–592. [Google Scholar]
- Nagata S, Adachi K, Sano H. 1996. NMR analyses of compatible solutes in a halotolerant Brevibacterium sp. Microbiology 142:3355–3362. [DOI] [PubMed] [Google Scholar]
- Nagata S, et al. 2008. Efficient cyclic system to yield ectoine using Brevibacterium sp JCM 6894 subjected to osmotic downshock. Biotechnol Bioeng. 99:941–948. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. 2005. Lehninger principles of biochemistry. 4th ed. New York: W. H. Freeman. [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6. [DOI] [PubMed] [Google Scholar]
- Oren A. 1999. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev. 63:334–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. 2002a. Halophilic microorganisms and their environments. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Oren A. 2002b. Intracellular salt concentration and ion metabolism in halophilic microorganisms In: Seckbach J, editor. Halophilic microorganisms and their environments. Dordrecht: Kluwyer Academic Publishers; p. 207–231. [Google Scholar]
- Oren A. 2008. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 4:2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. 2013. Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front Microbiol. 4:315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A, Larimer F, Richardson P, Lapidus A, Csonka LN. 2005. How to be moderately halophilic with broad salt tolerance: clues from the genome of Chromohalobacter salexigens. Extremophiles 9:275–279. [DOI] [PubMed] [Google Scholar]
- Park JS. 2012. Effects of different ion compositions on growth of obligately halophilic protozoan Halocafeteria seosinensis. Extremophiles 16:161–164. [DOI] [PubMed] [Google Scholar]
- Park JS, Cho BC, Simpson AGB. 2006. Halocafeteria seosinensis gen. et sp. nov. (Bicosoecida), a halophilic bacterivorous nanoflagellate isolated from a solar saltern. Extremophiles 10:493–504. [DOI] [PubMed] [Google Scholar]
- Park JS, Kim HJ, Choi DH, Cho BC. 2003. Active flagellates grazing on prokaryotes in high salinity waters of a solar saltern. Aquat Microb Ecol. 33:173–179. [Google Scholar]
- Park JS, Simpson AGB. 2011. Characterization of Pharyngomonas kirbyi (=“Macropharyngomonas halophila” nomen nudum), a very deep-branching, obligately halophilic Heterolobosean. Protist 162:691–709. [DOI] [PubMed] [Google Scholar]
- Park JS, Simpson AGB. 2015. Diversity of heterotrophic protists from extremely hypersaline habitats. Protist 166:422–437. [DOI] [PubMed] [Google Scholar]
- Park JS, Simpson AGB, Lee WJ, Cho BC. 2007. Ultrastructure and phylogenetic placement within Heterolobosea of the previously unclassified, extremely halophilic heterotrophic flagellate Pleurostomum flabellatum (Ruinen 1938). Protist 158:397–413. [DOI] [PubMed] [Google Scholar]
- Paul S, Bag SK, Das S, Harvill ET, Dutta C. 2008. Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 9:R70.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Noble WS. 2003. Matrix2png: a utility for visualizing matrix data. Bioinformatics 19:295–296. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8:785–786. [DOI] [PubMed] [Google Scholar]
- Powell S, et al. 2014. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 42:D231–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prista C, Almagro A, Loureiro-Dias MC, Ramos J. 1997. Physiological basis for the high salt tolerance of Debaryomyces hansenii. Appl Environ Microbiol. 63:4005–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengpipat S, Lowe SE, Zeikus JG. 1988. Effect of extreme salt concentrations on the physiology and biochemistry of Halobacteroides acetoethylicus. J Bacteriol. 170:3065–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshetnikov AS, Mustakhimov II, Khmelenina VN, Trotsenko YA. 2005. Cloning, purification, and characterization of diaminobutyrate acetyltransferase from the halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. Biochem (Mosc). 70:878–883. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3:217–223. [Google Scholar]
- Revell LJ, Harrison AS. 2008. PCCA: a program for phylogenetic canonical correlation analysis. Bioinformatics 24:1018–1020. [DOI] [PubMed] [Google Scholar]
- Richard SB, Madern D, Garcin E, Zaccai G. 2000. Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 Å resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui. Biochemistry 39:992–1000. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, et al. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salway JG. 1999. Metabolism at a Glance. Oxford: Blackwell Science. [Google Scholar]
- Schiraldi C, Maresca C, Catapano A, Galinski EA, De Rosa M. 2006. High-yield cultivation of Marinococcus M52 for production and recovery of hydroxyectoine. Res Microbiol. 157:693–699. [DOI] [PubMed] [Google Scholar]
- Schofield CJ, Zhang ZH. 1999. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr Opin Struct Biol. 9:722–731. [DOI] [PubMed] [Google Scholar]
- Seip B, Galinski EA, Kurz M. 2011. Natural and engineered hydroxyectoine production based on the Pseudomonas stutzeri ectABCD-ask gene cluster. Appl Environ Microbiol. 77:1368–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin J, Wohlfarth A, Galinski EA. 1992. The predominant role of recently discovered tetrahydropyrimidine for the osmoadaptation of halophilic eubacteria. J Gen Microbiol. 138:1629–1638. [Google Scholar]
- Sheffer M, Fried A, Gottlieb HE, Tietz A, Avron M. 1986. Lipid composition of the plasma membrane of the halotolerant alga, Dunaliella salina. Biochim Biophys Acta. 857:165–172. [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463. [DOI] [PubMed] [Google Scholar]
- Stoeck T, et al. 2014. Living at the limits: evidence for microbial eukaryotes thriving under pressure in deep anoxic, hypersaline habitats. Adv Ecol. 2014:532687. [Google Scholar]
- Stöveken N, et al. 2011. A specialized aspartokinase enhances the biosynthesis of the osmoprotectants ectoine and hydroxyectoine in Pseudomonas stutzeri A1501. J Bacteriol. 193:4456–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susko E, Field C, Blouin C, Roger AJ. 2003. Estimation of rates-across-sites distributions in phylogenetic substitution models. Syst Biol. 52:594–603. [DOI] [PubMed] [Google Scholar]
- Torres GE, Gainetdinov RR, Caron MG. 2003. Plasma membrane monoamine transporters: structure, regulation and function. Nature Rev Neurosci. 4:13–25. [DOI] [PubMed] [Google Scholar]
- Vonheijne G, Abrahmsen L. 1989. Species-specific variation in signal peptide design - implications for protein secretion in foreign hosts. FEBS Lett. 244:439–446. [DOI] [PubMed] [Google Scholar]
- Widderich N, et al. 2014. Biochemical properties of ectoine hydroxylases from extremophiles and their wider taxonomic distribution among microorganisms. Plos One 9: e93809e93809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. 2005. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature 437:215–223. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Youssef NH, et al. 2014. Trehalose/2-sulfotrehalose biosynthesis and glycine-betaine uptake are widely spread mechanisms for osmoadaptation in the Halobacteriales. Isme J. 8:636–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.