Abstract
The luminous bacterial symbionts of anomalopid flashlight fish are thought to be obligately dependent on their hosts for growth and share several aspects of genome evolution with unrelated obligate symbionts, including genome reduction. However, in contrast to most obligate bacteria, anomalopid symbionts have an active environmental phase that may be important for symbiont transmission. Here we investigated patterns of evolution between anomalopid symbionts compared with patterns in free-living relatives and unrelated obligate symbionts to determine if trends common to obligate symbionts are also found in anomalopid symbionts. Two symbionts, “Candidatus Photodesmus katoptron” and “Candidatus Photodesmus blepharus,” have genomes that are highly similar in gene content and order, suggesting genome stasis similar to ancient obligate symbionts present in insect lineages. This genome stasis exists in spite of the symbiont’s inferred ability to recombine, which is frequently lacking in obligate symbionts with stable genomes. Additionally, we used genome comparisons and tests of selection to infer which genes may be particularly important for the symbiont’s ecology compared with relatives. In keeping with obligate dependence, substitution patterns suggest that most symbiont genes are experiencing relaxed purifying selection compared with relatives. However, genes involved in motility and carbon storage, which are likely to be used outside the host, appear to be under increased purifying selection. Two chemoreceptor chemotaxis genes are retained by both species and show high conservation with amino acid sensing genes, suggesting that the bacteria may actively seek out hosts using chemotaxis toward amino acids, which the symbionts are not able to synthesize.
Keywords: Photodesmus, bioluminescent symbiosis, genome reduction, symbiont transmission, genome stability
Introduction
Most bacterial symbionts whose growth occurs mainly within their hosts (referred to here as being obligately dependent on hosts for growth, or simply obligately dependent) are vertically transmitted to new host generations and are frequently physically restricted to intracellular habitats (Moran et al. 2008; Bright and Bulgheresi 2010; Sachs et al. 2011). The bacterium “Candidatus Photodesmus katoptron” (Gammaproteobacteria: Vibrionaceae), the luminous symbiont of the anomalopid flashlight fish Anomalops katoptron (Beryciformes: Anomalopidae), has many genomic features in common with unrelated obligately dependent symbionts, such as insect endosymbionts, including genome reduction, gene content similarities, and an elevated evolutionary rate (Hendry and Dunlap 2011; Hendry et al. 2014). In contrast to endosymbionts, however, anomalopid symbionts are extracellular and have an active environmental phase (Kessel 1977; Haygood et al. 1984; Haygood 1993). “Ca. Photodesmus katoptron” has lost genes for metabolism of carbohydrates other than glucose and for synthesis of most amino acids and is therefore unlikely to establish free-living populations in the environment (Hendry et al. 2014). However, symbionts are continually released into seawater, where they remain luminous for a few to several hours and are motile (Kessel, 1977; Haygood et al. 1984; Haygood, 1993; Hendry and Dunlap, 2014). It is not known how long these cells persist or how they are acquired by new hosts, as anomalopid symbionts are unculturable and difficult to study, but it is clear that the lifestyle of these bacteria is unusual compared with most known obligate symbionts and free-living relatives (Herring and Morin 1978; Hastings and Nealson 1981; Haygood and Distel 1993).
No indication of direct parent to offspring vertical transmission of anomalopid symbionts has been found, but genomic and behavioral evidence suggests that the symbionts may be transmitted pseudovertically, from adults to larval fish within a population, after release into the environment (Haygood 1993; Haygood and Distel 1993; Hendry et al. 2014). The two fish hosts from this study, A. katoptron and Photoblepharon palpebratus, co-occur for much of their range (Mccosker and Rosenblatt 1987), but appear to consistently harbor specific symbiont species, “Ca. Photodesmus katoptron” and “Ca. Photodesmus blepharus,” respectively (Haygood and Distel 1993; Hendry and Dunlap 2014). These fishes, which are strictly nocturnal, can sometimes be found at night together in groups near the ocean surface (Morin et al. 1975; Wolfe and Haygood 1991; Hendry and Dunlap 2014). When schooling in groups, the fish use bacterially produced light from two under-eye light organs to hunt prey and avoid predators (Harvey 1922; Morin et al. 1975; Herring and Morin 1978). It has been hypothesized that aggregation of adults, and possibly larval fish, particularly in caves and crevices during the day, has facilitated the convergent evolution of genome reduction and obligate host dependence in anomalopid symbionts, which are nested within a bacterial family containing many free-living, facultatively symbiotic, and host-associated bacteria with horizontal transmission (Haygood 1993; Hendry and Dunlap 2011; Hendry et al. 2014).
In addition to genomic reduction and the loss of redundant genes, findings from physically host-restricted, vertically transmitted symbionts have shown that population bottlenecks between host generations and small effective population sizes increase the effect of genetic drift and cause higher rates of evolution in symbiont populations compared with those of free-living relatives. Selection may also be less effective in symbiont populations, as suggested by fixation rates of deleterious mutations (Moran 1996; Wernegreen and Moran 1999; Woolfit and Bromham 2003; Wernegreen and Funk 2004). Although selection in obligate symbionts is thought to be relatively weak, accumulation of deleterious mutations can be costly. Genes that have changed function or are important for an obligately symbiotic lifestyle may be under increased purifying selection in symbionts compared with orthologs of these genes in free-living relatives (Fares et al. 2002; Toft and Fares 2008). Despite this high rate of evolutionary change, some ancient symbiont lineages are thought to eventually have become relatively static in gene content and genome structure. For example, the aphid symbiont Buchnera aphidicola has codiverged with diverse aphid hosts, diverging at the nucleotide sequence level but retaining a highly conserved gene content and order (Tamas et al. 2002; McCutcheon and Moran 2012). Several causes for this pattern have been suggested, such as limited potential for recombination, loss of mobile genetic elements, or increased selection on the remaining genes to conserve their function (McCutcheon and Moran 2012). The loss of recombination ability has been presented as a likely cause of genome stability (McCutcheon and Moran 2012), but conflicting patterns have been found in different symbionts (Sloan and Moran 2013). Ideally more data from multiple unrelated symbiont lineages could possibly help resolve this question.
The independent evolution of obligate host dependence in anomalopid symbionts presents an opportunity to confirm broad evolutionary patterns of symbiont evolution, such as genome stasis and reduced selection, within a distinct system from commonly studied insect symbionts. Furthermore, it may be possible to connect gene content and evolutionary patterns within anomalopid symbionts to their lifestyle of obligate host dependence with an active environmental phase. For instance, by investigating patterns of gene retention in an obligately dependent stinkbug symbiont with environmental transmission, Kenyon et al. (2015) were able to infer which genes were likely to be involved in environmental persistence of the symbiont. In the A. katoptron symbiont, genes involved in chemotaxis and motility, which are typically lost in obligate symbionts, have been largely retained, suggesting that they are important for the ecology of the bacterium and might be involved in finding new hosts, as seen in other obligate and facultative symbionts (Toh et al. 2006; Mandel et al. 2012; Hendry and Dunlap 2014; Hendry et al. 2014). To infer evolutionary patterns among anomalopid symbionts, we generated a de novo genome sequence for a second anomalopid symbiont, “Ca. Photodesmus blepharus” for comparison with a second de novo sequenced A. katoptron symbiont from the same location. Additionally, we used these comparisons to infer which types of genes may be important for the anomalopid symbiont’s symbiotic lifestyle or environmental persistence.
Materials and Methods
Material
Symbiont DNA was obtained from anomalopid flashlight fish species A. katoptron and P. palpebratus collected in coastal waters in the Republic of Vanuatu in 2011 and DNA was extracted as in Hendry and Dunlap (2011). Bacteria were released from light organs by squeezing into sterile buffered artificial seawater and bacterial cells were pelleted by centrifugation and used for DNA extraction. For the P. palpebratus symbiont, four specimens (Ppalp.1 to Ppalp.4) were collected and DNA from one light organ of each specimen was combined for sequencing. For the A. katoptron symbiont, eight specimens (Akat.10 to Akat.18) were combined. The genome sequences resulting from these samples are technically metagenomic; however, anomalopid symbionts are monoclonal within a host and show little genetic polymorphism between hosts (Hendry et al. 2014; Hendry and Dunlap 2011). Previous work, including polymerase chain reaction (PCR) amplification and sequencing of multiple loci from different hosts at one location (Hendry and Dunlap 2011) and comparisons of whole genome sequences from individuals across a wide geographic range, substantiate this pattern (Hendry 2012). Pooled symbiont genome samples from the A. katoptron and Photoblepharon blepharus individuals used in this study were found to have single nucleotide polymorphisms (SNPs) at a rate of approximately 0.21/kilobase and 0.55/kilobase, respectively (supplementary table S1), making their genomic diversity similar to bacteria considered genetically monomorphic (Achtman 2008) (supplementary fig. S1). Furthermore, symbiont samples from across the geographic range of A. katoptron were found to have only 10 SNPs in 9,591 bases (PCR amplified from 16 coding genes, four rRNA and tRNA genes, and five noncoding spacer regions) (Hendry 2012). A similar pattern is found in some insect endosymbionts (Funk et al. 2001; Abbot and Moran 2002). For consistency, as it is the convention in insect systems for obligate symbiont genomes to be generated using low diversity DNA from multiple host individuals, we will refer to the sequences reported here as genomes rather than metagenomes.
Genome Sequencing and Annotation
Sequencing was done at the University of Michigan DNA Sequencing core on an Illumina HiSeq 2000 machine. Illumina reads of 100 bp in length were assembled in Velvet 1.1.06 (Zerbino and Birney 2008). For each species, six independent assemblies with a minimum read coverage of 45x were generated using subsets of the total reads. At this coverage minimum no host sequence contigs (based on BLAST) were recovered in the assembly. The SeqMan Pro 9.05 software by DNASTAR was then used to combine contigs from the assemblies to check for sequence consistency and to generate consensus sequences. Glimmer 3.02 (Delcher et al. 2007) was used to predict open reading frames (ORFs) within both assemblies, and ORFs were annotated by BLAST comparison with the Swiss-Prot and UniRef 90 databases (December 2011 releases). Predicted ORFs were thrown out if they were <100 amino acids in length and had <40% identity to database protein sequences. Predicted ORFs with <100 amino acids but >40% identity over at least 50% of the length of orthologs were retained. Predicted ORFs were categorized as pseudogenes if they were above this threshold but contained premature stop codons (a stop codon resulting in a gene <66% the length of orthologs). Homologs between the two symbiont genomes were identified with reciprocal best BLAST comparisons and the same identity and length cutoffs described above.
Both assemblies consist of several large (19,935–452,950 bp in length for “Ca. Photodesmus katoptron” and 10,794–244,971 bp in length for “Ca. Photodesmus blepharus”) contigs containing protein coding genes (nine contigs in “Ca. Photodesmus katoptron” and 19 in “Ca. Photodesmus blepharus”) and smaller contigs containing tRNA and rRNA genes. Additionally, in both assemblies, plasmid sequences were recovered. These were determined to be circular in the assembly but this was not confirmed with PCR. The high read depth (average coverage = 181× for “Ca. Photodesmus katoptron” and 197× for “Ca. Photodesmus blepharus”) indicates that each genome is likely fully represented in each assembly. Furthermore, the assembly of the “Ca. Photodesmus katoptron” genome presented here is similar in number of genes to the previously reported complete assembly (916 genes vs. 903, respectively).
Molecular Analysis
For comparisons of synteny, the predicted genome annotations of each symbiont were analyzed with the SynMap tool in CoGe using default parameters (Lyons et al. 2008). We also performed analyses on the following Vibrio relative pairs: Vibrio splendidus ATCC 33789 and Vibrio mimicus MB-451, Vibrio vulnificus CMCP6 and Vibrio harveyi 1DA3, Vibrio cholerae O395 and Vibrio parahaemolyticus RIMD 2210633, V. harveyi 1DA3 and V. mimicus MB-451, V. vulnificus CMCP6 and V. parahaemolyticus RIMD 2210633, and V. splendidus ATCC 33789 and V. cholerae O395. These taxa pairs were chosen because they are of similar phylogenetic distance apart at neutral loci as the two “Ca. Photodesmus” symbionts (Hendry and Dunlap, 2011). We then summed the total length of all syntenic blocks for each comparison and divided by the total genome size. To test a molecular clock hypothesis and investigate evolutionary rate in the anomalopid symbionts compared with free-living relatives, we used Tajima’s relative rate test (Tajima 1993) implemented in Mega7 (Kumar et al. 2016). Seven housekeeping gene loci with conserved function were chosen as in Hendry and Dunlap (2014) and concatenated for 7,285 bp. Sequences from one anomalopid symbiont and a free-living close relative (V. splendidus ATCC 33789) were compared with a free-living outgroup species (Aliivibrio fischeri ATCC 7744T). This test should tend to give a conservative assessment of relative rates as it will underestimate multiple substitutions, particularly in fast-evolving taxa.
To produce estimates of changes in purifying selection compared with relatives, SynMap was used to identify orthologous coding loci and estimate dN and dS (nonsynonymous and synonymous substitution) values. We used default parameters with the exception that the maximum distance between matches was increased to 4,000 genes to maximize the number of matches and any duplicate matches were thrown out. For substitution estimates, SynMap implements the default likelihood-based (codeml) package adapted from the program PAML (Yang 2007). This analysis was run for the anomalopid symbionts as well as the pairs of taxa listed above for comparison of synteny. Comparisons between “Ca. Photodesmus katoptron” and the six relative genomes were used to identify homologs, and only loci identified in both symbionts and at least three of the relative pairs were used in analysis (720 loci in total).
To identify changes in substitution patterns compared with relatives, we determined the ratio of nonsynonymous to synonymous substitutions (ω) for the symbiont pair (ωPk-Pp) and each relative pair. A value of change in purifying selection in the symbionts compared with relatives, R, was estimated by dividing each ratio (R = ωPk-Pp/ωrelative1-relative2). In these analyses, a value of R > 1 indicates that the symbiont locus is experiencing relaxed selective constraints compared with non-symbiotic relatives, R = 1 suggests no change in selection, and R < 1 indicates increased purifying selection compared with relatives. For each locus we determined the mean R value for symbionts compared with the three to six possible pairs of relatives and used the range of values to identify loci for which the mean was greater or less than 1 by two standard deviations. Loci with mean R values significantly >1 were considered to be under relaxed selection and those with values significantly <1 were considered to be under increased purifying selection compared with relatives. We note that this is likely to be a conservative estimate of increased purifying selection, as saturation at synonymous sites, which is common in obligate symbionts, would tend to inflate ωPk-Pp and therefore increase R (Moran 1996; Toft and Fares 2008).
Analysis of Methyl-Accepting Chemotaxis Protein Genes
We sought to determine the possible function of the methyl-accepting chemotaxis protein (MCP) genes present in the anomalopid symbiont genomes, as these genes are responsible for sensing chemoattractants and could therefore be used to find hosts. All MCP genes found in anomalopid symbionts and their free-living relatives V. harveyi 1DA3, V. splendidus ATCC 33789, Vibrio orientalis CIP102891, V. cholerae O395, V. vulnificus CMCP6, V. parahaemolyticus RIMD 2210633, Photobacterium profundum SS9, A. fischeri MJ11, and A. fischeri ZF211 were identified in the IMG DOE database (Markowitz et al. 2012) and aligned by protein sequence in ClustalW2 (Larkin et al. 2007). Only genes with >40% sequence identity to anomalopid symbiont MCP genes, over at least 40% of the gene length, were included. A Bayesian tree was generated in Mr. Bayes (Huelsenbeck and Ronquist 2001) using a fixed rate amino acid model run for 200,000 generations. We found that some anomalopid symbiont MCP genes show sequence similarity to MCP genes known to sense amino acids, and so we sought to verify this by searching for ligand binding sequences. Each of the MCP sequences included in the tree were searched for a conserved amino acid ligand binding sequence using the sequence identified in V. vulnificus (Nishiyama et al. 2012) and was considered to have the conserved binding site if the sequence matched at 9/17 amino acids.
Results and Discussion
Genome Features
At 1.11 Mb (megabases), the de novo sequenced genome of “Ca. Photodesmus blepharus,” the second anomalopid symbiont genome to be sequenced, is reduced by approximately 80% compared with the average genome size of free-living relatives (Hendry et al. 2014). The genome assembly yielded 33 contigs, including two circular plasmids, with an average coverage depth of 197×. The assembly appears to be fragmented at multi-copy loci such as rRNA genes. The genome is predicted to contain 984 functional genes, 923 protein coding genes, 46 tRNA genes, and 15 rRNA genes in 5 operons (based on coverage depth compared with single-copy genes), and 23 pseudogenes. The predicted ORFs include genes for all pathways considered necessary for life and predicted to indicate completeness of genome coverage in the previously sequenced anomalopid symbiont genome; the “Ca. Photodesmus blepharus” assembly therefore appears to fully represent the genomic content of this bacterium (Raes et al. 2007; Hendry et al. 2014).
The genome of “Ca. Photodesmus katoptron” from Vanuatu is highly similar to the previously sequenced and annotated genome for this symbiont species from the Philippines (Hendry et al. 2014). The genome assembly yielded 18 contigs including one circular plasmid. This assembly is the same size (1 Mb) and contains a similar number of predicted functional genes, 916 compared with 903, as in the previously published assembly of “Ca. Photodesmus katoptron.” These include 873 protein coding genes, 35 tRNA genes, 15 rRNA genes in 5 operons, and 13 pseudogenes. The gene differences between genome assemblies for “Ca. Photodesmus katoptron” are mostly copy number disparities and likely result from differences in the methods and sequencing technology used rather than meaningful geographic differences. This new assembly does not change previous conclusions involving genome reduction or metabolic capabilities in this species.
Genome Similarity between Symbionts
The “Ca. Photodesmus blepharus” genome, like “Ca. Photodesmus katoptron,” is severely reduced in amino acid synthesis and energy metabolism genes. The “Ca. Photodesmus blepharus” genome has only four of the many amino acid synthesis genes missing from the “Ca. Photodesmus katoptron” genome, and it lacks one amino acid synthesis gene present in the “Ca. Photodesmus katoptron” genome (supplementary tables S1 and S2). Both symbionts appear to be unable to synthesize most amino acids and we surmise therefore that they acquire amino acids from their host fishes. Similarly, “Ca. Photodesmus blepharus” appears to be restricted to glucose as a carbon/energy source and has only two genes involved in energy metabolism not found in the “Ca. Photodesmus katoptron” genome (supplementary table S1). Together these findings suggest that both symbionts are obligately dependent on their hosts for growth.
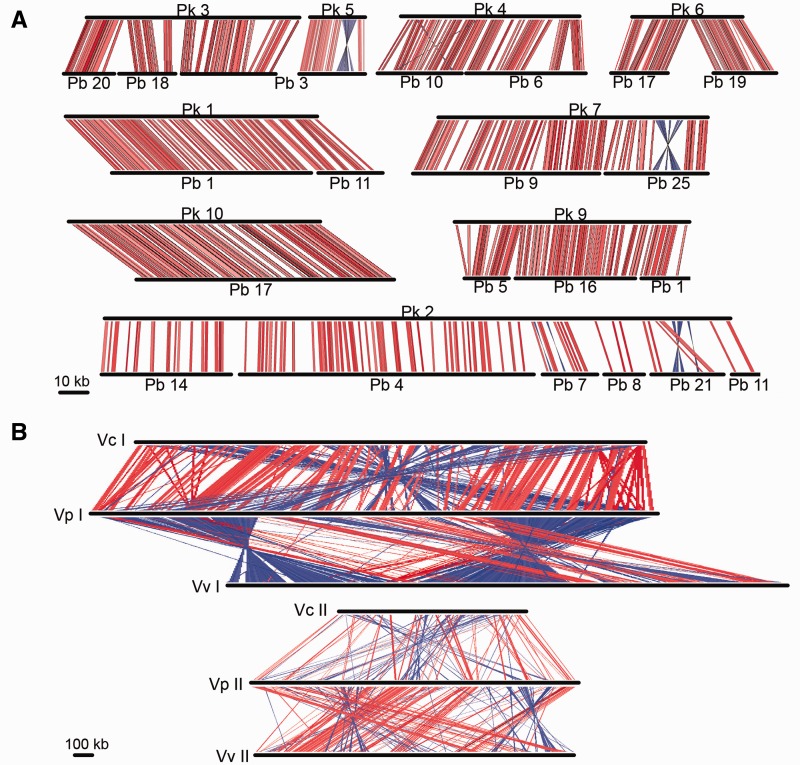
The gene content of the two anomalopid symbiont genomes is highly similar. The symbionts share orthologs for 834 protein coding genes (90.4% and 96.3% of “Ca. Photodesmus blepharus” and “Ca. Photodesmus katoptron” gene content, respectively). Furthermore, although these genomes are not closed, the assembled contigs are generally similar in gene order (fig. 1). The two genomes share more syntenic blocks, areas of chromosomes with homology and shared gene order, relative to genome size, than is typical of Vibrio genomes. “Ca. Photodesmus katoptron” and “Ca. Photodesmus blepharus” share 79.3% and 71.4% of their genomes in syntenic blocks compared with six pairs of Vibrio relatives, which share an average of 50.1% of genomic sequence in syntenic blocks (values range from 37.1% to 61.1%). Genome reduction in the symbionts’ common ancestor and subsequent low rates of genomic rearrangement could account for this pattern. The high similarity in gene content and gene order is consistent with genome stasis, as found in long-term intracellular symbionts such as Buchnera, which appears to have a pattern of genome stability over 50 million years of evolution (Tamas et al. 2002; McCutcheon and Moran 2012).
Fig. 1.—
Genome similarity between (A) the anomalopid symbionts, “Ca. Photodesmus katoptron” (Pk) and “Ca. Photodesmus blepharus” (Pb), and (B) free-living relatives V. cholerae N16961 (Vc), V. parahaemolyticus RIMD 2210633 (Vc), and V. vulnificus CMCP6 (Vv). Alignments were determined by the Artemis Comparison Tool (Carver et al. 2005). Each connecting line represents an area of high nucleotide sequence similarity; red lines indicate alignment of two positive strands and blue lines represent similarity between positive and negative strands. Homology was determined with an e value cutoff of 1.0. Chromosome segments in (A) are labeled by contig number and in (B) are labeled by chromosome number.
Genome Divergence between Symbionts
Although the gene content of the genomes of “Ca. Photodesmus katoptron” and “Ca. Photodesmus blepharus” is more similar than is typical of closely related free-living Vibrionaceae species, at the nucleotide sequence level the two symbionts appear to be evolving at a faster rate than free-living close relatives. The anomalopid symbionts are both evolving significantly faster than a free-living close relative (χ2 (1) = 295.79, P < 0.00001; χ2 (1) = 285.59, P < 0.00001 for “Ca. Photodesmus katoptron” and “Ca. Photodesmus blepharus,” respectively), compared with a distant free-living relative outgroup taxon in Tajima’s relative rate test (Tajima 1993). These results support the previous finding that “Ca. Photodesmus” is evolving at a faster rate than is typical for Vibrionaceae species (Hendry and Dunlap 2011), and are consistent with high levels of genetic drift owing to an obligate host association.
The “Ca. Photodesmus blepharus” genome contains 89 unique, species-specific genes and the “Ca. Photodesmus katoptron” genome contains 32 unique genes (supplementary tables S2 and S3). The majority of these genes are also present in other Vibrionaceae genomes, which suggests that they have been retained in one symbiont species but not the other. The species-specific genes represent multiple functional categories and may be the result of stochastic gene loss differences during genome reduction since the split of the two species, or they could relate to ecological differences. In “Ca. Photodesmus blepharus,” all plasmid genes (15 total between both plasmids, excluding the parA and repA plasmid replication genes) are unique to that symbiont, whereas “Ca. Photodesmus katoptron” has 4 out of 11 unique plasmid genes. These unique plasmid genes may contribute to phenotypic differences between the symbionts; however, they are largely of unknown function and it is therefore difficult to infer their purpose. We focus here on genes for which entire pathways or multiple genes involved in a function have been differentially retained between the symbionts, as these genes are less likely to have been retained by chance.
A significant number of unique “Ca. Photodesmus blepharus” genes (22 genes; 24.7% of unique “Ca. Photodesmus blepharus” genes compared with 16.7% of the total “Ca. Photodesmus blepharus” gene content, z-ratio proportion test P = 0.0324) are involved in functions that may be more important outside of the host than within. These functions include the synthesis of cell wall components, chemotaxis and motility, and the metabolism of glycogen (supplementary table S2). Cell wall components, including cell surface proteins, and chemotaxis and motility genes were previously hypothesized to be necessary for an environmental phase for anomalopid symbionts, as genes of these kinds are frequently lost in host-restricted symbionts (Hendry et al. 2014). Genes required for metabolism of glycogen, primarily a carbon storage molecule, are also typically lost in obligate bacteria, presumably because the host supply of carbon is steady (Henrissat et al. 2002). Electron micrographs of anomalopid symbionts have identified what appear to be poly-3-hydroxybutyrate (PHB) granules, another carbon storage molecule, inside cells (Kessel 1977). This observation leads to the hypothesis that carbon storage during host association may prepare the bacteria to survive outside the host where carbohydrate sources may not be plentiful (Haygood 1993). Genes required for PHB synthesis (phaB and phaC) are found in the genomes of both anomalopid symbionts, which indicates that synthesis of PHB is likely in both species. Because “Ca. Photodesmus blepharus” may be able to synthesize and break down glycogen for carbon storage, in addition to PHB, this species may be better able to survive in seawater than “Ca. Photodesmus katoptron,” possibly explaining the retention of these genes in the genome. Consistent with this possibility, cells of the P. palpebratus symbiont remain luminous for longer periods in seawater after release from the host (Hendry and Dunlap 2014). Regulatory genes, including those typically involved in regulating luminescence, are also relatively highly represented among unique “Ca. Photodesmus blepharus” genes, possibly because more regulatory genes may be needed for additional pathways in the slightly larger “Ca. Photodesmus blepharus” genome.
Loci under Selection
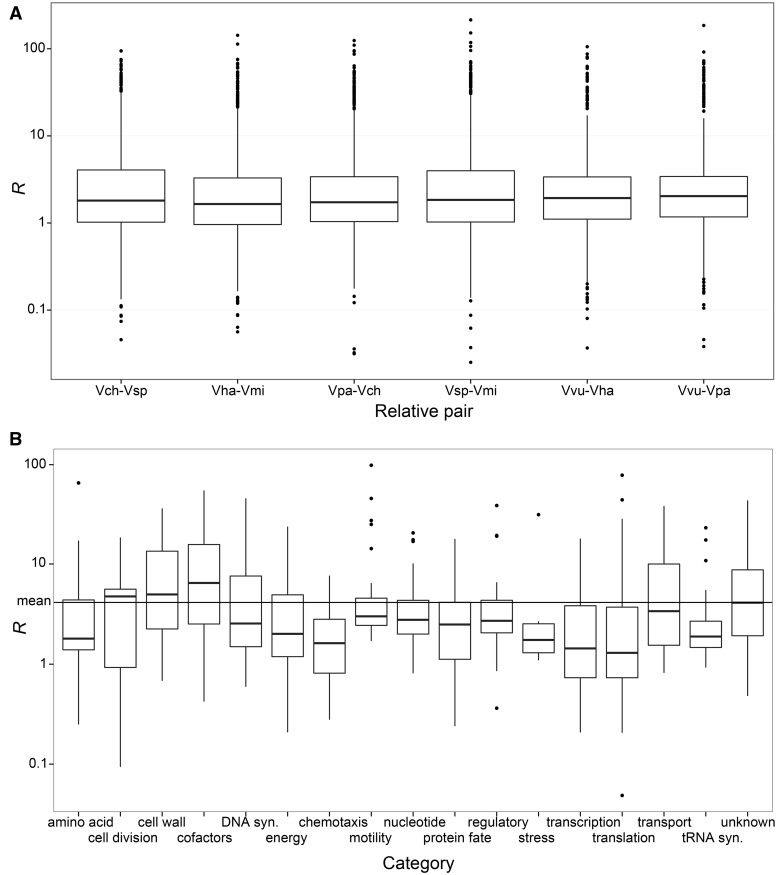
To identify those genes that might be needed for the anomalopid symbiont’s unusual lifestyle, we performed genome wide tests for purifying selection. We focused on testing for loci with signatures of increased purifying selection compared with free-living relatives, as these loci are likely to be important for the bacteria and possibly related to the switch to an obligate symbiotic lifestyle. Following the methods of Toft and Fares (2008), we compared ratios of dN with dS between “Ca. Photodesmus katoptron” and “Ca. Photodesmus blepharus” with six pairs of relatives to obtain a measure of change in purifying selection (R) in symbiont genes compared with orthologs in free-living relatives. The genome-wide R values determined for each symbiont locus were similar regardless of the relative pair used for comparison (fig. 2A). We therefore used the range of comparisons for each locus to determine which genes showed a significant increase or decrease in purifying selection for symbiont genes compared with values seen for the same genes in relatives. Genes that had an R value less than two standard deviations from no change compared with relatives were classified as having increased purifying selection compared with orthologs, whereas genes with an R value greater than two standard deviations from no change were classified as having decreased purifying selection compared with orthologs. These terms will be used throughout to refer to genes with significant changes in selection. This test for purifying selection in the symbiont lineage is predicted to be conservative as it focuses only on increased selection compared with relatives.
Fig. 2.—
Boxplots for R values indicating changes in purifying selection in anomalopid symbiont loci compared with orthologs in free-living relatives. (A) R values for each free-living relative comparison. (B) R values for each locus averaged across relative pair comparisons and divided into functional categories. The mean value of R across all loci is shown with a line and values < 1 indicate increased purifying selection.
Many genes, 49% of those analyzed (352 genes), showed a significant decrease in purifying selection compared with relatives, as has been shown in previous tests of selection on the genomes of other obligate symbionts (Toft and Fares 2008). Additionally, 40% of genes (285) showed no change and only 11% of genes (82) were found to be under increased purifying selection in the symbionts compared with orthologous genes in free-living relatives (supplementary table S4). Of the genes under increased purifying selection, >90% fall into eight functional categories: protein synthesis, energy metabolism, protein fate, motility, cell division, transcription, DNA synthesis and repair, and cell wall synthesis and structures (supplementary table S4). In each of the categories of cell division, transcription, and protein synthesis, >25% of the genes were found to be under increased purifying selection compared with relatives (fig. 2B), a significantly higher percentage compared with genes with no change in or decreased selection (supplementary table S4). These categories are some of the least reduced in anomalopid symbionts compared with relatives (Hendry et al. 2014), and an inverse, although not significant, relationship exists between the percentage of gene reduction and the average R value in each category. This inverse relationship indicates that functional categories with less gene loss tend to have greater increases in purifying selection in the symbionts (supplementary fig. S2). It is possible that genes in those categories most important for cell function are the least likely to be lost and also that, if such genes are lost, perhaps eliminating some redundancy in function, the remaining genes become more functionally constrained and under increased selection compared with relatives with larger genomes. This possibility is consistent with the idea that selection against the loss of necessary genes may partly explain genome stasis in endosymbionts (Tamas et al. 2002).
We were particularly interested in selection on genes involved in chemotaxis and motility. The functions of these genes might differ in the anomalopid symbionts compared with relatives given the symbiont’s potentially limited environmental phase. The median R values for genes involved in chemotaxis and regulation of the flagellar motor is lower than the total median value of R, indicating that a higher proportion of these genes is experiencing increased purifying selection compared with relatives than other genes in the symbiont genomes (fig. 2B). Six of these genes, cheR (involved in chemotaxis), an ortholog of the V. cholerae gene mlp24 (coding for a MCP), and fliG (a motor switch regulator), as well as flagellar genes (flgE, flgJ, and flaD) were found to be under increased purifying selection compared with orthologs in free-living relatives. These genes might therefore be ecologically important for the symbionts. In particular, the MCP gene might be used by symbionts to find hosts, as these genes code for membrane-bound proteins that sense chemical attractants and repellants and lead to a motility response.
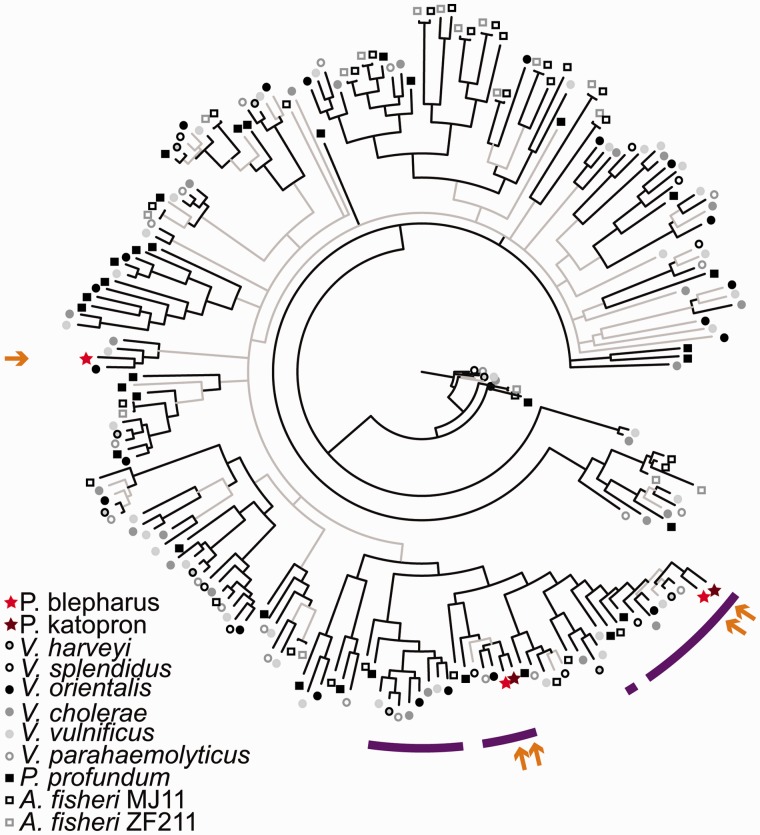
Members of Vibrionaceae typically have large numbers of MCP genes (V. cholerae, for instance, has 45 and A. fischeri has 43), which are thought to respond to a diverse array of chemicals (Nishiyama et al. 2012; Brennan et al. 2013). In contrast, however, only two MCP genes have been retained in “Ca. Photodesmus katoptron” and three in “Ca. Photodesmus blepharus.” Two of the MCP genes are present in the genomes of both symbionts; they are highly similar in amino acid sequence and have protein sequences that are most closely related to Vibrionaceae MCPs thought to be involved in binding environmental amino acids (fig. 3). Mutation experiments and chemotaxis assays have found that MCPs that bind and respond to amino acids in the environment share a ligand-binding sequence motif that is highly conserved across Gammaproteobacteria, including Vibrionaceae species (A. fischeri and V. cholerae) (Taguchi et al. 1997; Glekas et al. 2010; Nishiyama et al. 2012; Brennan et al. 2013); this motif is also present in the shared MCP gene sequences of the anomalopid symbionts (fig. 4).
Fig. 3.—
Bayesian tree based on amino acids sequences for all MCP genes found in anomalopid symbionts and the free-living relatives V. harveyi 1DA3, V. splendidus ATCC 33789, V. orientalis CIP102891, V. cholerae O395, V. vulnificus CMCP6, V. parahaemolyticus RIMD 2210633, P. profundum SS9, A. fischeri MJ11, and A. fischeri ZF211. Genes with >40% sequence identity to anomalopid MCP genes, over 40% of the gene length, were included. Branches shown in black have ≥95% posterior probability and branches with less support are in gray. Arrows indicate location of anomalopid symbiont sequences and purple bars denote sequences with a conserved amino acid ligand binding sequence, suggesting that they may respond to amino acids in the environment (Taguchi et al. 1997; Glekas et al. 2010; Nishiyama et al. 2012; Brennan et al. 2013).
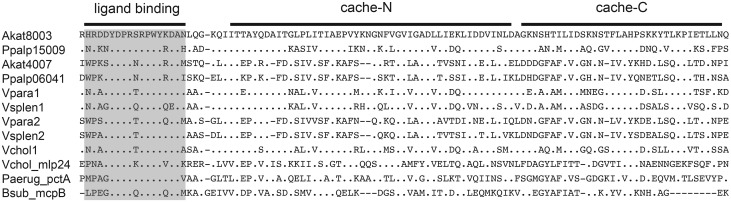
Fig. 4.—
Alignment of the portion of MCP gene amino acid sequence implicated in amino acid ligand binding (ligand binding motif) and signal transduction (Cache domains) in chemotaxis proteins. Sequences are shown for anomalopid symbiont genes, some orthologs from close relatives (V. parahaemolyticus, V. splendidus, V. cholerae sequences in fig. 3), and four orthologs with experimentally demonstrated amino acid binding and response capabilities (Vchol_mlp24: V. cholerae, Afisc_vfcA: A. fischeri, Paeru_pctA: Pseudomonas aeruginosa, and Bsubt_mcpB: Bacillus subtilus). Sites with identical amino acids compared with the Akat8003 sequence are shown with a period. The conserved sequence shown to bind to amino acids is highlighted.
With the exception of the reduction in copy number for MCP genes, both anomalopid symbiont genomes have retained the full complement of chemotaxis and motility genes found in some free-living relatives (Hendry et al. 2014), including all genes necessary for flagellum synthesis and function and all chemotaxis genes necessary for signal transduction from MCPs to the flagella (cheW, cheA, cheY, cheZ, cheB, and cheR). The conservation of these pathways indicates that chemotaxis using the retained MCP genes could be functional for the bacteria. The high sequence similarity of anomalopid MCP genes with probable amino-acid sensing MCPs and the conserved amino-acid binding motif suggests that these “Ca. Photodesmus” proteins could bind and respond to environmental amino acids as chemoattractants. Because the symbionts cannot synthesize most amino acids (this study, Hendry et al. 2014), they must acquire them from the environment or fish hosts. Therefore the ability to sense and move toward amino acids could be important for the bacteria, explaining why these MCP genes, but not others, have been retained. In this case, the anomalopid symbionts would seem to show a narrowing of chemotactic sensory abilities, perhaps in keeping with a restricted lifestyle as a symbiont. However, it is also possible that these few MCP genes have been retained by chance or some unknown function.
Conclusions
The genome conservation observed here is similar to the genome stasis observed in ancient obligate symbionts (Tamas et al. 2002; McCutcheon and Moran 2012). The age of the split between the fish family Anomalopidae and relatives is not known, but divergence of the host order Beryciformes from other teleost fishes has been estimated as approximately 100 million years ago (Santini et al. 2009). The association between anomalopids and their symbionts therefore could have existed for a similar length of time as the Buchnera association, though it is also possible that the rate of reduction in anomalopid symbionts was relatively fast and the observed stability is therefore more recent. We note that the anomalopid symbionts have retained many genes necessary for DNA recombination (table 1), as well as multiple copies of repetitive genes such as rRNA operons, which are frequently lost in intracellular symbionts (Tamas et al. 2002; McCutcheon and Moran 2012). The retention of these genes could increase the likelihood of recombination and genomic rearrangements compared with Buchnera, yet “Ca. Photodesmus” genomes seem to be similarly stable. This result is consistent with patterns found in whitefly symbionts showing that recombination ability and genome stability are not necessarily related (Sloan and Moran 2013). It is possible that genomic stasis is beneficial to some symbionts with significant genome reduction, perhaps because the loss of additional genes would be highly deleterious to both bacteria and hosts, and that genomic stability is selected for in anomalopid symbionts.
Table 1.
Recombination-related genes found in anomalopid symbionts, V. harveyi, and B. aphidicola
| Locus | P. katopron | P. blepharus | V. harveyi | Buchnera |
|---|---|---|---|---|
| recA | X | X | X | |
| recB | X | X | X | X |
| recC | X | X | X | X |
| recD | X | X | X | X |
| recF | X | X | X | |
| recG | X | X | X | |
| recJ | X | X | X | |
| recO | X | X | X | |
| recQ | X | X | ||
| recR | X | X | X | |
| ruvA | X | X | X | |
| ruvB | X | X | X | |
| ruvC | X | X | X | |
| xerC | X | X | ||
| xerD | X |
We hypothesized that loci experiencing increased purifying selection in the anomalopid symbionts compared with orthologs in free-living relatives were likely to have either shifted function or to be increasingly functionally constrained. The fact that many of these genes were from functional categories with low levels of gene reduction, and therefore are presumed to be essential for cell growth, suggests that these loci may be more functionally constrained in the symbionts rather than that they have changed in function. Intriguingly, a number of chemotaxis and motility genes appear to be under increased purifying selection compared with relatives, highlighting their likely importance to the symbionts. It is difficult to know if these genes have shifted function or are simply more important in a new ecological context. The two MCP genes retained in both anomalopid symbionts are conserved with relatives, suggesting that their function has not changed, but the increased purifying selection on one of these genes in the symbionts indicates that their function may be important. To our knowledge, the genomes of only a few obligate symbionts with reduced genomes, the aphid symbiont Buchnera, the tsetse fly symbionts Wigglesworthia glossinidia and Sodalis glossinidius, and the rice weevil symbiont Sodalis pierantonius have retained genes involved in motility. Buchnera is non-motile and uses flagellar apparatus genes for protein secretion, whereas flagella may be involved in host transmission in the tsetse fly symbionts (Akman et al. 2002; Maezawa et al. 2006; Toh et al. 2006; Toft and Fares 2008; Rio et al. 2012). Sodalis glossinidius has retained some chemotaxis genes (cheW and cheZ) but no MCP genes (Toh et al. 2006), and the recently reduced genome of S. pierantonius contains predicted flagellar and chemotaxis genes, but many of these, including all MCP genes, are pseudogenes (Oakeson et al. 2014). Although anomalopid symbionts released from hosts are known to remain luminous for at least a few hours in seawater, it is not known how long they persist or what the fate of environmental cells is. However, it is unlikely that pathways used only outside the host would be retained and maintained by purifying selection if release from the host was a dead end and these cells did not go on to colonize new hosts. The increased purifying selection compared with relatives on the chemotaxis genes retained in anomalopid symbionts is therefore further support for the hypothesis that an extra-host phase is important for symbiont ecology and possibly transmission between hosts
As these analyses show, Vibronaceae members typically have many MCP genes, including one to four copies that appear to respond to amino acids (fig. 4). Because the anomalopid symbionts appear to be unable to synthesize most amino acids, the ability to detect them in the environment could be highly beneficial. That the symbionts have retained these genes and lost most other MCP genes suggests that the bacteria are not generally responding to nutrients (e.g. glucose) in the environment, but specifically to amino acids. It seems unlikely that this ability would be beneficial inside the host light organ, where the bacteria are too densely packed for motility or chemotaxis to be effective (Kessel 1977; Haygood 1993). More likely is that the bacteria sense and respond to amino acids as a way to find new host fish. According to this scenario, bacteria released from adult light organs persist in the local environment and are tactic toward amino acids released by aposymbiotic larvae of the fish. The use of amino acids as a means to identify host cells has been described for the pathogen V. cholerae. The V. cholerae ortholog of one of the MCP genes found in the anomalopid symbionts, mlp24, binds and responds to amino acids and is involved in host cell attachment and subsequent toxin secretion (Lee et al. 2001; Nishiyama et al. 2012). The ancestral pathway to sense environmental amino acids may have become tied to host identification in “Ca. Photodesmus.”
Supplementary Material
Supplementary figures S1 and S2 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgment
We thank Grant Norton of Sustainable Reef Suppliers, Brad Remmer of Sea Dwelling Creatures and Fish Doctors Aquarium of Ypsilanti, Michigan for providing assistance in obtaining fish specimens. We also thank Michael Sheehan for discussions on analysis, Alison Gould for help processing samples, and Mario Fares for thoughts on calculating changes in purifying selection. Departmental Block Grant awards from the University of Michigan Department of Ecology and Evolutionary Biology funded this work.
Literature Cited
- Abbot P, Moran NA. 2002. Extremely low levels of genetic polymorphism in endosymbionts (Buchnera) of aphids (Pemphigus). Mol. Ecol. 11:2649–2660. doi: 10.1046/j.1365-294X.2002.01646.x. [DOI] [PubMed] [Google Scholar]
- Achtman M. 2008. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32: 402–407. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- Brennan CA, DeLoney-Marino CR, Mandel MJ. 2013. Chemoreceptor VfcA mediates amino acid chemotaxis in Vibrio fischeri. Appl. Environ. Microbiol. 79:1889–1896. doi: 10.1128/AEM.03794-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright M, Bulgheresi S. 2010. A complex journey: transmission of microbial symbionts. Nat. Rev. Microbiol. 8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, Rajandream M A, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21: 3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 23 673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares MA, Barrio E, Sabater-Muñoz B, Moya A. 2002. The evolution of the heat-shock protein GroEL from Buchnera, the primary endosymbiont of aphids, is governed by positive selection. Mol. Biol. Evol. 19:1162–1170. [DOI] [PubMed] [Google Scholar]
- Funk DJ, Wernegreen JJ, Moran NA. 2001. Intraspecific variation in symbiont genomes: bottlenecks and the aphid-Buchnera association. Genetics 157:477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glekas GD, Foster RM, Cates JR, Estrella JA, Wawrzyniak MJ, Rao CV, Ordal GW. 2010. A PAS domain binds asparagine in the chemotaxis receptor McpB in Bacillus subtilis. J. Biol. Chem. 285: 1870–1878. doi: 10.1074/jbc.M109.072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey EN. 1922. The production of light by the fishes Photoblepharon and Anomalops. Washington (DC): Carnegie Institution of Washington. [Google Scholar]
- Hastings JW, Nealson KH. 1981. The symbiotic luminous bacteria In: Starr MP, Stolp H, Truper HG, Balows A, Schlegel HG, editors. The prokaryotes: a handbook on habitats, isolate, and identification of bacteria. Berlin: Springer-Verlag; pp. 1332–1345. [Google Scholar]
- Haygood MG. 1993. Light organ symbioses in fishes. Crit. Rev. Microbiol. 19:191–216. doi: 10.3109/10408419309113529. [DOI] [PubMed] [Google Scholar]
- Haygood MG, Distel DL. 1993. Bioluminescent symbionts of flashlight fishes and deep-sea anglerfishes form unique lineages related to the genus Vibrio. Nature 363:154–156. doi: 10.1038/363154a0. [DOI] [PubMed] [Google Scholar]
- Haygood MG, Tebo BM, Nealson KH. 1984. Luminous bacteria of a monocentrid fish (Monocentris japonicus) and two anomalopid fishes (Photoblepharon palpebratus and Kryptophanaron alfredi): population sizes and growth within the light organs, and rates of release into the seawater. Mar. Biol. 78:249–254. doi: 10.1007/BF00393010. [Google Scholar]
- Hendry TA. 2012. Genome reduction and evolution in an obligate luminous symbiont. Ann Arbor (MI): University of Michigan Library; Available from: http://hdl.handle.net/2027.42/94077. [Google Scholar]
- Hendry TA, Dunlap PV. 2014. Phylogenetic divergence between the obligate luminous symbionts of flashlight fishes demonstrates specificity of bacteria to host genera. Environ. Microbiol. Rep. 6:331–338. doi: 10.1111/1758-2229.12135. [DOI] [PubMed] [Google Scholar]
- Hendry TA, Dunlap PV. 2011. The uncultured luminous symbiont of Anomalops katoptron (Beryciformes: Anomalopidae) represents a new bacterial genus. Mol. Phylogenet. Evol. 61:834–843. doi: 10.1016/j.ympev.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Hendry TA, de Wet JR, Dunlap PV. 2014. Genomic signatures of obligate host dependence in the luminous bacterial symbiont of a vertebrate. Environ. Microbiol. 16:2611–2622. doi: 10.1111/1462-2920.12302. [DOI] [PubMed] [Google Scholar]
- Henrissat B, Deleury E, Coutinho PM. 2002. Glycogen metabolism loss: a common marker of parasitic behaviour in bacteria? Trends Genet. 18:437–440. doi: 10.1016/S0168-9525(02)02734-8. [DOI] [PubMed] [Google Scholar]
- Herring PJ, Morin JG. 1978. Bioluminescence in fishes In: Herring PJ, editor. Bioluminescence in action. London: Academic Press; pp. 273–329. [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. [DOI] [PubMed] [Google Scholar]
- Kenyon LJ, Meulia T, Sabree ZL. 2015. Habitat visualization and genomic analysis of ‘Candidatus Pantoea carbekii,’ the primary symbiont of the brown marmorated stink bug. Genome Biol. Evol. 7:620–635. doi: 10.1093/gbe/evv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel M. 1977. The ultrastructure of the relationship between the luminous organ of the teleost fish Photoblepharon palpebratus and its symbiotic bacteria. Cytobiologie 15:145–158. [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 30:2725–2729. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee SH, Butler SM, Camilli A. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. 98: 6889–6894. doi: 10.1073/pnas.111581598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, Freeling M. 2008. The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the rosids. Trop. Plant Biol. 1:181–190. doi: 10.1007/s12042-008-9017-y. [Google Scholar]
- Mandel MJ, et al. 2012. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl. Environ. Microbiol. 78:4620–4626. doi: 10.1128/AEM.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, et al. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40:D115–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccosker J, Rosenblatt R. 1987. Notes on the biology, taxonomy, and distribution of flashlight fishes (beryciformes, Anomalopidae). Jpn. J. Ichthyol. 34:157–164. [Google Scholar]
- McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- Maezawa K, Shigenobu S, Taniguchi H, Kubo T, Aizawa S, Morioka M. 2006. Hundreds of flagellar basal bodies cover the cell surface of the endosymbiotic bacterium Buchnera aphidicola sp. strain APS. J. Bacteriol. 188: 6539–6543. doi: 10.1128/JB.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. U S A. 93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable dacterial symbionts. Annu. Rev. Genet. 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Morin JG, et al. 1975. Light for all reasons: versatility in the behavioral repertoire of the flashlight fish. Science 190:74–76. doi: 10.1126/science.190.4209.74. [Google Scholar]
- Nishiyama S, et al. 2012. Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect. Immunol. 80:3170–3178. doi: 10.1128/IAI.00039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeson KF, et al. 2014. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol. Evol. 6:76–93. doi: 10.1093/gbe/evt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. 2007. Prediction of effective genome size in metagenomic samples. Genome Biol. 8:R10. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio RVM, Symula RE, Wang J, Lohs C, Wu Y, Snyder AK, et al. 2012. Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: Glossinidae) obligate symbiont Wigglesworthia. Mbio 3: e00240. doi: 10.1128/mBio.00240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs JL, Essenberg CJ, Turcotte MM. 2011. New paradigms for the evolution of beneficial infections. Trends Ecol. Evol. 26:202–209. doi: 10.1016/j.tree.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Santini F, Harmon LJ, Carnevale G, Alfaro ME. 2009. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol. Biol. 9: 194. doi: 10.1186/1471-2148-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Moran NA. 2013. The evolution of genomic instability in the obligate endosymbionts of whiteflies. Genome Biol. Evol. 5:783–793. doi: 10.1093/gbe/evt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. 1997. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 143: 3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- Tajima F. 1993. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas I, et al. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- Toft C, Fares MA. 2008. The evolution of the flagellar assembly pathway in endosymbiotic bacterial genomes. Mol. Biol. Evol. 25:2069–2076. doi: 10.1093/molbev/msn153. [DOI] [PubMed] [Google Scholar]
- Toh H, et al. 2006. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ, Funk DJ. 2004. Mutation exposed: a neutral explanation for extreme base composition of an endosymbiont genome. J. Mol. Evol. 59:849–858. doi: 10.1007/s00239-003-0192-z. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ, Moran NA. 1999. Evidence for genetic drift in endosymbionts (Buchnera): analyses of protein-coding genes. Mol. Biol. Evol. 16:83–97. [DOI] [PubMed] [Google Scholar]
- Wolfe C, Haygood M. 1991. Restriction fragment length polymorphism analysis reveals high levels of genetic divergence among the light organ symbionts of flashlight fish. Biol. Bull. 181:135–143. doi: 10.2307/1542496. [DOI] [PubMed] [Google Scholar]
- Woolfit M, Bromham L. 2003. Increased rates of sequence evolution in endosymbiotic bacteria and fungi with small effective population sizes. Mol. Biol. Evol. 20:1545–1555. doi: 10.1093/molbev/msg167. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.