Abstract
De novo genes, which originate from ancestral nongenic sequences, are one of the most important sources of protein-coding genes. This origination process is crucial for the adaptation of organisms. However, how de novo genes arise and become fixed in a population or species remains largely unknown. Here, we identified 782 de novo genes from the model plant Arabidopsis thaliana and divided them into three types based on the availability of translational evidence, transcriptional evidence, and neither transcriptional nor translational evidence for their origin. Importantly, by integrating multiple types of omics data, including data from genomes, epigenomes, transcriptomes, and translatomes, we found that epigenetic modifications (DNA methylation and histone modification) play an important role in the origination process of de novo genes. Intriguingly, using the transcriptomes and methylomes from the same population of 84 accessions, we found that de novo genes that are transcribed in approximately half of the total accessions within the population are highly methylated, with lower levels of transcription than those transcribed at other frequencies within the population. We hypothesized that, during the origin of de novo gene alleles, those neutralized to low expression states via DNA methylation have relatively high probabilities of spreading and becoming fixed in a population. Our results highlight the process underlying the origin of de novo genes at the population level, as well as the importance of DNA methylation in this process.
Keywords: de novo genes, DNA methylation, origin, process, protein-coding genes
Introduction
De novo genes originate from nongenic sequences. This mechanism is extremely important for the divergence and adaptation of organisms (Tautz and Domazet-Loso 2011; Long, VanKuren, et al. 2013; Tautz 2014; McLysaght and Guerzoni 2015). Many studies have examined the origin of de novo genes in various species, such as humans (Knowles and McLysaght 2009; Wu, Irwin, et al. 2011; Guerzoni and McLysaght 2016), fruit flies (Levine et al. 2006; Begun et al. 2007; Chen et al. 2007, 2010; Zhou et al. 2008; Reinhardt et al. 2013), primates (Toll-Riera et al. 2009; Xie et al. 2012), yeast (Cai et al. 2008), and plants (Campbell et al. 2007; Yang et al. 2009; Lin et al. 2010; Donoghue et al. 2011; Guo 2013; Hoen and Bureau 2015). De novo genes share various characteristics, such as shorter lengths, lower expression levels, and more highly diversified sequences compared to other genes in the genome. Recent studies in yeast and fruit flies have been particularly insightful. In yeast, de novo gene origination is considered more prevalent than gene duplication (Carvunis et al. 2012). In fruit flies, natural selection plays an important role in the spread of de novo genes (Zhao et al. 2014), and there is a balance between gene gain and loss (Palmieri et al. 2014).
Although several characteristics of de novo genes are well understood, the process by which they originate and become fixed in a population is still largely unknown (Light et al. 2014; Neme and Tautz 2014; Tautz 2014; Zhao et al. 2014; Schlotterer 2015). The difficulties in elucidating this process can be roughly attributed to two major factors. First, most previous studies have used interspecific comparative genomics methods. However, evolutionary processes can be clarified at a much higher resolution at the population level (Lynch 2007), including the origin of de novo genes (Schlotterer 2015). Second, regardless of whether an “ORF sequence” or the acquisition of “transcriptional ability” is the first step in de novo gene origination, de novo genes must arise in a stepwise manner (Carvunis et al. 2012; Light et al. 2014), and transcription and translation are two necessary stages in the evolution of a protein-coding gene (Knowles and McLysaght 2009; Xie et al. 2012; Reinhardt et al. 2013; Schlotterer 2015). Therefore, de novo gene origination is an evolutionary process involving the transition from a nontranscribed and noncoding intergenic sequence to a transcribed and translated coding sequence (CDS) and from being present at low frequency to high frequency in a population before eventually becoming fixed. Accordingly, comparisons of gene features at various evolutionary stages with regard to allele frequencies may reflect and even clarify the process and mechanism by which de novo genes arise.
Here, we focused on the origin of de novo genes in the model plant Arabidopsis thaliana to address the following fundamental questions: 1) How do de novo gene alleles originate? 2) How are these genes maintained? 3) How do they spread and become fixed in a population or species? Based on multiple omics data from the same population, our results indicate that DNA methylation plays a crucial role during the origin of de novo genes in a population.
Results
Identification and Classification of De Novo Genes in A. thaliana
We detected de novo genes based on the similarity of protein sequences of the A. thaliana reference genome (ecotype Col-0) to those of 64 other available genomes from green plants (62 species in total, supplementary table S1, Supplementary Material online). Genes lacking homologues in any other species were regarded as candidate de novo genes. In total, 1,135 candidate de novo genes were found in A. thaliana (see the flowchart in supplementary fig. S1, Supplementary Material online). After excluding candidates with homologues in other species in the NCBI nonredundancy protein database (266 genes), genes with gaps (27 genes) or missing gene models (60 genes) at the syntenic regions in the genomes of two closely related species (Arabidopsis lyrata and Capsella rubella), we identified 782 lineage-specific genes (2.9% of the 27,206 annotated protein-coding genes) (supplementary table S2, Supplementary Material online). In total, 87% (683 genes) of these genes have been identified as Arabidopsis-specific genes in previous studies (Lin et al. 2010; Donoghue et al. 2011; Arendsee et al. 2014) (supplementary fig. S2, Supplementary Material online), suggesting that lineage-specific genes represent a stable gene set that remained after we added many additional genome sequences in the present study. In addition, none of the 782 genes contain conserved protein domains based on sequence similarity searches against the Pfam protein families database (Finn et al. 2016), and none were derived from frameshifts of existing gene sequences or horizontal gene transfer based on similarity searches of public sequence data. Thus, the 782 genes identified in this study have likely originated de novo in the A. thaliana genome and can therefore be regarded as de novo genes. Indeed, comparisons between the CDSs of A. thaliana de novo genes and their syntenic sequences in its close relatives, A. lyrata and C. rubella, support their de novo origin (e.g., AT4G05071 in supplementary fig. S3, Supplementary Material online).
The de novo genes could be divided into three groups based on their transcriptional and translational states in Col-0 (Data 1-19 in supplementary table S3, Supplementary Material online): 145 de novo genes with translational evidence (hereafter referred to as TL type); 239 with transcriptional evidence only (TC type); and 398 without transcriptional or translational evidence (TN type). Although we collected genome-wide expression data from several tissues (leaves, flowers, and roots) and under different conditions (normal, hypoxia, and dark), half of the identified de novo genes belong to the TN type. It is possible that some TN-type genes are artefacts resulting from gene model prediction; however, some might be expressed under other environmental conditions or in other tissues or developmental stages beyond those investigated in the reported studies. Thus, TN-type genes were included in subsequent analysis as well. In addition, 24,018 intergenic open reading frames (ORFs) longer than 90 nt were predicted from intergenic regions of the Col-0 genome without masking either repeats or transposable element (TE) sequences, which were used as randomly selected intergenic regions to compare with de novo genes and common genes (26,424 annotated protein-coding genes other than the identified de novo genes).
Epigenetic Modification Plays an Important Role in De Novo Gene Origin
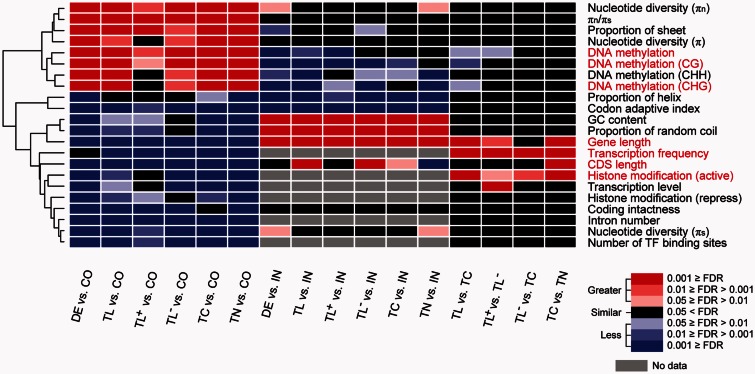
To investigate important factors affecting de novo gene origin, we compared many gene features among de novo genes, common genes and intergenic ORFs. Consistent with previous studies in yeast (Carvunis et al. 2012; Abrusan 2013) and fruit flies (Palmieri et al. 2014), our analyses based on the A. thaliana Col-0 reference genome indicated that de novo genes are generally shorter than common genes and have fewer introns, lower GC contents and codon adaptation indexes (CAI), fewer transcription factor binding sites (TFBS), lower transcription levels (see supplementary text, Supplementary Material online for details), higher β-sheet contents and higher polymorphisms, and are under weaker selective constraint compared to common genes (FDR < 0.05; fig. 1; statistics in supplementary tables S4 and S5, Supplementary Material online). Among these gene features, TFBS and selective constraint are important factors in de novo gene origination (Carvunis et al. 2012; Zhao et al. 2014).
Fig. 1.—
Significance levels for comparisons of gene features. DE, de novo genes (782 genes); CO, common genes (26,474 genes); IN, intergenic ORFs (24,018 ORFs); TL, de novo genes with translational evidence (145 genes); TC, de novo genes with only transcriptional evidence (239 genes); TN, de novo genes without expression evidence (398 genes); TL+, TL genes with detected Poly(A) tails (65 genes); TL−, TL genes without detected Poly(A) tails (80 genes). πn (πs), polymorphism level in nonsynonymous (or synonymous) sites. CG, CHG, and CHH indicate DNA methylation in different contexts (H indicates A, C or T). Active histone modifications are histone markers H3K4me3, H3K36me3, and H3K9Ac in Data 24 of supplementary table S3, Supplementary Material online, and repressive histone modification is marker H3K27me3. P values were transformed into FDR values in the multiple comparisons. Five gene features with significant differences among the three types of de novo genes (TL, TC, and TN) are marked red. Detailed comparisons are shown in supplementary table S5, Supplementary Material online.
Beyond these well-studied gene features, we further evaluated the methylation levels (Schmitz et al. 2013) and histone modification patterns (Luo et al. 2013) (Data 24 and 25 in supplementary table S3, Supplementary Material online) in Col-0, as well as the intact coding region frequency (Cfreq, the percentage of accessions with intact coding regions in the population for a gene) and transcription frequency (Tfreq, the percentage of accessions with transcriptional evidence in the population for a gene) across the 84 previously resequenced accessions (data in supplementary table S6, Supplementary Material online). Compared to common genes, de novo genes are highly methylated in all methylation contexts (CG, CHG, and CHH contexts; H indicates A, C or T) in their genic regions, in addition to having fewer active histone marks and lower Cfreq (fig. 1). Thus, epigenetic modification (DNA methylation and histone modification) may be another important factor affecting the origin of de novo genes.
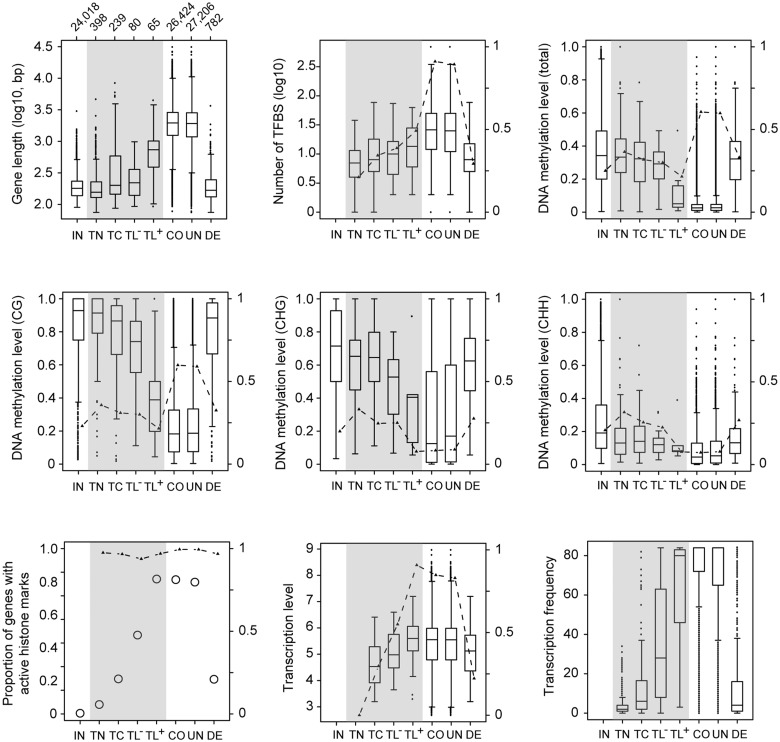
Except for the gene features of CAI, the nucleotide diversity and the proportion of the sequence present in α-helices, comparisons of all other available gene features suggested that genes in the de novo gene set are in an intermediate state between intergenic ORFs and common genes (e.g., de novo genes are shorter than common genes but longer than intergenic ORFs, and the DNA methylation levels are higher in de novo genes than in common genes but lower than in intergenic ORFs; figs. 1 and 2, see “DE vs. CO” and “DE vs. IN”, and supplementary fig. S4, Supplementary Material online). These results indicate that de novo genes are at an evolutionary stage representing the transition from intergenic regions to mature genes and that the de novo genes identified in this study are likely “proto-genes”, as proposed in yeast (Carvunis et al. 2012).
Fig. 2.—
Gene features with significant differences between de novo gene types. Numbers above the top left panel indicate the total numbers of genes or ORFs used in the comparisons. The y-axis on the right side of the boxplot and dashed lines indicate the proportion of ORFs or genes used in the analysis. IN, intergenic ORFs; TN, de novo genes without expression evidence; TC, de novo genes with only transcriptional evidence; TL−, TL genes without detected Poly(A) tails; TL+, TL genes with detected poly(A) tails; CO, common genes; UN, unigenes; DE, de novo genes. Grey background indicates different types of de novo genes. Data 2 in supplementary table S3, Supplementary Material online were used in the comparison of transcription levels. DNA methylation levels were estimated in genic regions. CG, CHG, and CHH indicate DNA methylation in different contexts (H indicates A, C or T). Active histone markers are H3K4me3, H3K36me3, and H3K9Ac in Data 24 of supplementary table S3, Supplementary Material online.
Among the different types of de novo genes (TL, TC, and TN), five gene features were significantly different in at least one of the three-way comparisons, including gene length, CDS length, DNA methylation level (total level and levels in the CG and CHG contexts, respectively), active histone marks, and Tfreq, (FDR < 0.05; fig. 1, see “TL vs. TC” and “TC vs. TN”). This result once again demonstrates the importance of DNA methylation and histone marks in the origination of de novo genes.
Among the three types of de novo genes, TL-type genes are more similar to common genes than the other types (see fig. 2 and supplementary table S4, Supplementary Material online for details). Interestingly, more TL-type genes have poly(A) tails (45%) than TC (6%) or TN (2%) genes based on data from three independent studies on the genome-wide polyadenylation landscape in A. thaliana Col-0 (Wu, Liu, et al. 2011; Sherstnev et al. 2012; Subtelny et al. 2014) (Data 20–22 in supplementary table S3, Supplementary Material online). Given that polyadenylation at the 3′ end of a primary RNA transcript is important for translation initiation, which is a more mature feature of a protein-coding gene (Wells et al. 1998), TL-type genes could be divided into two groups: TL type with poly(A) tails (hereafter referred to as TL+) and TL type without poly(A) tails (TL−). We found that TL+genes are more similar to common genes than TL− genes, as the former share six features with common genes, while the latter share only four features (fig. 1, see “TL+vs. CO” and “TL− vs. CO”). This finding suggests that TL+ genes are much more mature than other de novo genes. More importantly, the DNA methylation levels and histone modification patterns are significantly different between TL+and TL− genes (FDR < 0.05; fig. 1, see “TL+vs. TL−”). Overall, epigenetic modification may play an important role during the origination of de novo genes.
De Novo Genes are Highly Methylated and Stably Inherited across Generations
Epigenetic modification among de novo genes has rarely been studied, except for one study in A. thaliana based on methylation data for the reference line Col-0 (Lin et al. 2010), which demonstrated that lineage-specific genes are more highly methylated than evolutionarily conserved genes. Therefore, we were interested in investigating the importance of DNA methylation during the process of de novo gene origination.
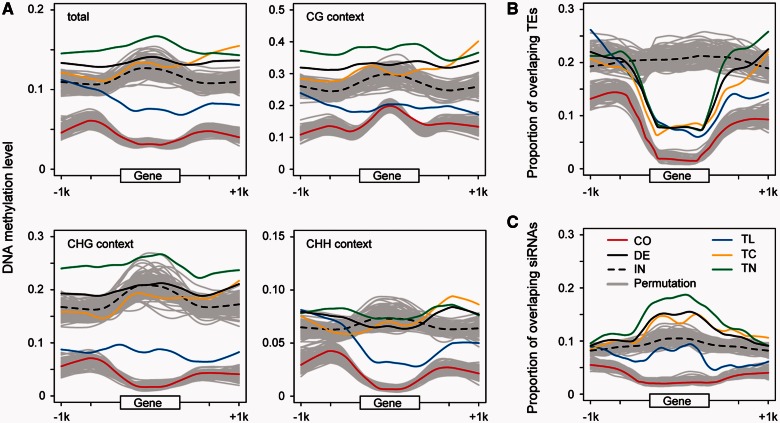
We found that the DNA methylation levels of de novo genes share two major attributes. First, the total DNA methylation levels of de novo genes were significantly higher than those of common genes in either their genic (mean values, 0.11 vs. 0.03; Wilcoxon test, P < 1 × 10−15) or flanking regions (0.11 vs. 0.05 in the upstream 1 kb, and 0.12 vs. 0.04 in the downstream 1 kb; all P < 1 × 10−8) (fig. 3A; see supplementary table S7, Supplementary Material online for details). This pattern held true in different methylation contexts (fig. 3A). Second, in the genic regions of de novo genes, the total DNA methylation level of TL-type genes (0.06) was lower than that of TC- and TN-type genes (0.11 and 0.14, Wilcoxon test, all P < 0.05), which was true in different DNA methylation contexts (fig. 3A). Furthermore, in their upstream regions, the methylation levels of TN genes were the highest, followed by TC and TL types, especially in the CG context (TN, 0.31; TC, 0.27; TL, 0.21; Wilcoxon test, all P < 0.05) (fig. 3A). In the downstream regions, TC and TN genes are significantly more highly methylated than TL genes (Wilcoxon test, all P < 0.05). In summary, de novo genes and their flanking sequences are more highly methylated than common genes, and TL-type genes are less methylated than the other types of de novo genes in either their genic or flanking regions. Accordingly, since the DNA methylation patterns of de novo genes are correlated with their transcription levels (see supplementary text, Supplementary Material online for details), the transcription levels of de novo genes are lower than those of common genes; the transcription levels of the TC type are lower than those of the TL type, and no expression activity was detected in the TN type in this study.
Fig. 3.—
DNA methylation levels and the proportion of overlapping TEs and siRNAs in genic and flanking regions. Genic regions were normalized to a length of 1 kb. The upper and lower grey curves in each panel are the resampling results for intergenic ORFs and 27,206 unigenes (782 loci resampled 100 times), respectively. (A) DNA methylation levels. Detailed statistics are listed in supplementary table S7, Supplementary Material online. (B) Proportions of overlapping TEs. (C) Proportions of overlapping siRNAs. CO, common genes; DE, de novo genes; IN, intergenic ORFs; TL, de novo genes with translational evidence; TC, de novo genes with only transcriptional evidence; TN, de novo genes without expression evidence.
We were also interested in determining whether the DNA methylation patterns of de novo genes can be stably inherited. To investigate the heredity of DNA methylation in de novo genes, we analyzed different methylation positions (DMPs) from 30-generation accumulation lines of Col-0 (Becker et al. 2011). Compared to those of common genes, much fewer sites of de novo genes changed their methylation states after 30 generations (0.6% vs. 1.8%; Fisher’s exact test, P < 2.2 × 10−16). The proportions were even lower in TC- and TN-type genes (both 0.5%) compared to TL-type genes (0.8%; Fisher’s exact test, all P < 0.01). These results suggest that the methylation states of de novo genes are under selective constraint, and their expression products may be deleterious to the organism, especially those of TC- and TN-type genes.
In addition to DNA methylation, histone modification appears to be important to the evolution of de novo genes. More TL-type genes (44%) are associated with the active histone modifications H3K4me3, H3K36me3, and H3K9Ac in Col-0 than either TC- (15%) or TN- (4%) type genes (Fisher’s exact test, all P < 1 × 10−8) (supplementary fig. S5, Supplementary Material online), which is consistent with their transcription patterns, as TL-type genes are highly transcribed. Therefore, variations in epigenetic modification may play an important role in the transcriptional regulation (see supplementary text, Supplementary Material online for details) and evolutionary processes of de novo genes. The effect is stable in the genome and can be inherited, at least for DNA methylation.
Enriched TEs and siRNAs Resulting in High DNA Methylation Levels in De Novo Genes
TEs are usually considered to be a major recruiter of DNA methylation. Compared to common genes, the proportions of TEs in de novo genes were higher in either genic (mean values, 0.08 vs. 0.02) or flanking (0.19 vs. 0.10) regions (Wilcoxon test, all P < 1 × 10−5) (fig. 3B). Furthermore, TEs overlapping with de novo genes were more highly methylated than TEs located in common genes (mean values of methylation levels, 0.37 vs. 0.25; Wilcoxon test, P < 1 × 10−8). Thus, the high proportions and high methylation states of the overlapping TEs likely increase the methylation levels of de novo genes. However, the proportions of TEs were similar in genic regions of all three types of de novo genes (TL, 0.07; TC, 0.08; TN, 0.08; Wilcoxon test for TE-overlapped genes, all P > 0.05). Thus, the proportion of TEs by itself could explain the higher methylation levels of de novo genes compared to common genes, but not the variations in methylation within de novo genes.
To understand the causes of DNA methylation variations among different types of de novo genes, we investigated the distribution patterns of 24-nt siRNAs, a key factor in RNA-directed DNA methylation (RdDM) (Data 27 in supplementary table S3, Supplementary Material online). Compared to common genes, more genic (mean values, 0.15 vs. 0.02) and flanking (0.10 vs. 0.04) regions of de novo genes are targeted by siRNAs (fig. 3C; Wilcoxon test, all P < 1 × 10−23), which probably accounts for their high DNA methylation levels. In contrast to the proportions of overlap with TEs, which are similar among TL, TC, and TN genes (fig. 3B), the proportion of siRNA-targeted regions in the genic regions was significantly higher in TN (0.18) and TC (0.14) genes than in TL (0.08) genes (fig. 3C; Wilcoxon test for siRNA-overlapped genes, all P < 0.001) (fig. 3C). This pattern held true when we used siRNA depth as a measure (mean depths in TN, TC, and TL are 0.95, 0.66, and 0.31, respectively; Wilcoxon test for siRNA-overlapped genes, all P < 0.05; supplementary fig. S6, Supplementary Material online). In the genic regions, the DNA methylation levels of siRNA-targeted genes were significantly higher than those of genes not targeted by siRNA (mean values, 0.07 vs. 0.01 for common genes; 0.30 vs. 0.01 for de novo genes; see supplementary table S8, Supplementary Material online for details). The higher proportion of siRNA-targeted regions might be a direct cause of the increased methylation levels of de novo genes compared to common genes, as well as the DNA methylation variations among de novo genes.
In de novo genes, ∼8% of genic regions overlap with TEs; however ∼15% of genic regions overlap with siRNAs (fig. 3B and C ). Apparently, half of the siRNA-targeted regions do not overlap with TEs, suggesting that at least half of these siRNA-targeted regions may not be targeted by siRNAs produced by TEs. Since the presence of an antisense transcript complementary to the sense transcript is another source of siRNAs (Zilberman et al. 2007; Luo et al. 2013), using published antisense transcript data (Luo et al. 2013), we found that the proportion of genes with antisense transcripts was significantly higher in TC de novo genes than in common genes with only transcriptional evidence (16% vs. 6%; Fisher’s exact test, P < 1 × 10−6), while the proportion was similar between TL de novo genes (21%) and common genes with translational evidence (21%). These results suggest that the high levels of antisense transcripts of TC de novo genes may be responsible for their higher siRNA levels.
Frequencies of De Novo Gene Alleles are Correlated to Their DNA Methylation Levels
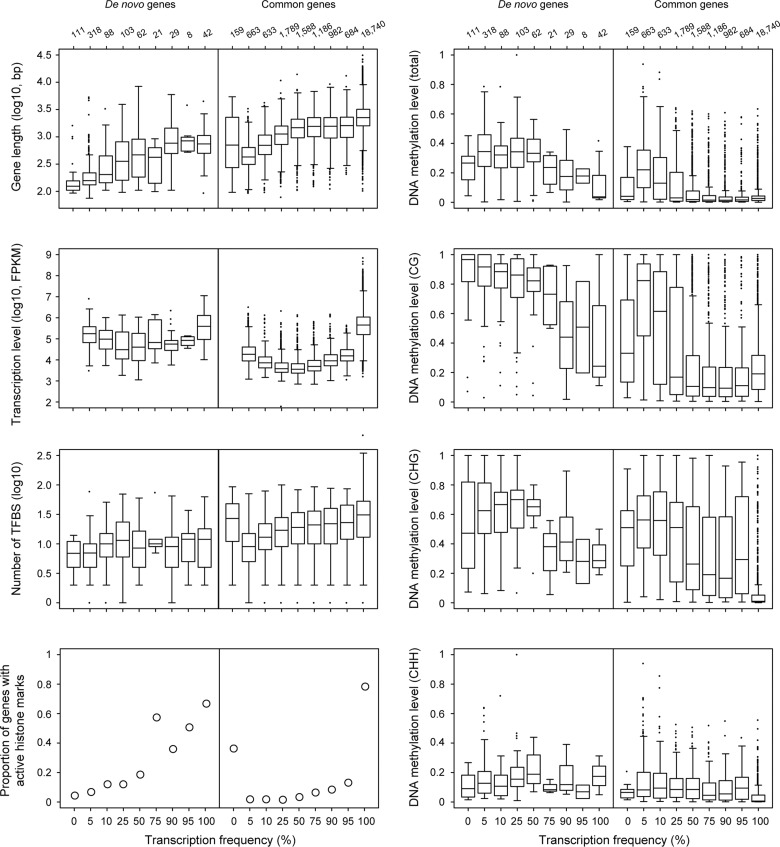
De novo gene origination is a process in which a transcribed and translated DNA segment arises from an intergenic region. However, from the perspective of population genetics, the origin of a de novo gene is the fixation process of a de novo gene allele in a population. Here, we used the frequency of transcribed alleles (Tfreq) as an indicator of whether a de novo gene is fixed in a population to investigate the factors affecting the fixation process of de novo genes. As demonstrated in figure 1, the gene length, DNA methylation level, histone modification pattern, transcription level, and Tfreq differ significantly among de novo gene groups, which most likely correlates with the evolutionary process of de novo gene origin from intergenic sequences to common genes.
To clarify which gene feature is the principal factor that correlates with the spread of de novo gene alleles in a population, we characterized all gene features shown in figure 1 with regard to the Tfreq at the population level (fig. 4 shows gene features with significant differences in fig. 1 and the number of TFBS; for the other gene features, see supplementary fig. S7, Supplementary Material online). For both de novo and common genes, the loci with higher Tfreq values generally have greater gene lengths, more TFBS, and higher proportions of active histone markers but lower DNA methylation levels. Intriguingly, for both de novo and common genes, the transcription levels were negatively correlated with the Tfreq when this value was lower than 50% but positively correlated with Tfreq when this value was higher than 50% (supplementary table S9, Supplementary Material online). Thus, we performed correlation analysis for each gene feature with Tfreq bins of ≤50% and >50% separately (see Methods for details).
Fig. 4.—
Distribution of gene features with significant differences among de novo gene types with respect to Tfreq. Data 2 in supplementary table S3, Supplementary Material online were used in the comparison of transcription levels. DNA methylation levels were estimated in genic regions. Numbers above the top panel indicate the total numbers of genes used in the comparisons. CG, CHG, and CHH are DNA methylation in different contexts (H indicates A, C or T). Active histone markers are H3K4me3, H3K36me3, and H3K9Ac in Data 24 of supplementary table S3, Supplementary Material online.
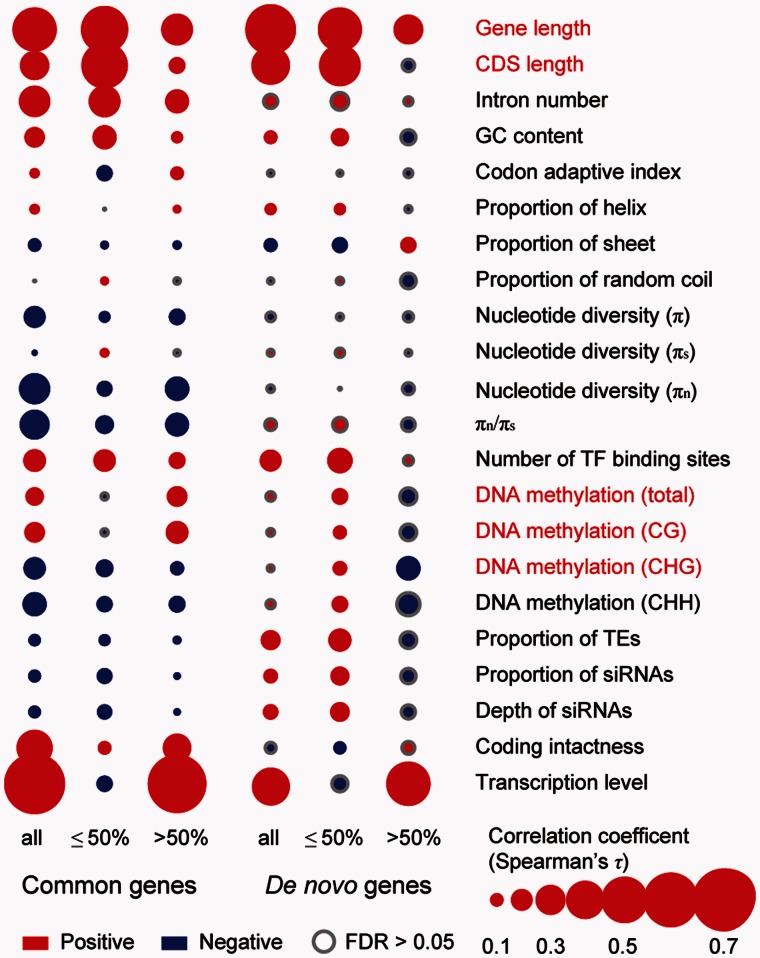
The gene features of gene length, transcription level and DNA methylation, which significantly differed among de novo genes in the reference line Col-0 (fig. 1), were significantly correlated with the Tfreq for both de novo genes and common genes (fig. 5). However, between de novo genes and common genes, the correlation pattern of DNA methylation differed. For common genes, the correlation patterns between DNA methylation and Tfreq were the same for all genes or genes with Tfreq ≤ 50% or >50%; when Tfreq increased, common genes were highly methylated in the CG context but had lower methylation levels in the CHG and CHH contexts. However, for de novo genes, both the total DNA methylation levels and methylation levels in the CG, CHG or CHH context were positively correlated with Tfreq when Tfreq ≤ 50%, but negatively correlated with Tfreq when Tfreq > 50% (the same pattern was observed for the proportion of TEs and siRNAs).
Fig. 5.—
Correlation between each gene feature and Tfreq for de novo genes and common genes. all, all de novo genes or all common genes. ≤50% (>50%), de novo genes or common genes with Tfreq ≤ 50% (Tfreq > 50%). Grey dot indicates correlation coefficient lower than 0.01. πn (πs), polymorphism level in nonsynonymous (or synonymous) sites. The names of gene features marked in red in figure 1 are also marked in red in this figure. Data 2 in supplementary table S3, Supplementary Material online were used in transcription level comparisons. DNA methylation levels were estimated in genic regions. CG, CHG, and CHH are DNA methylation in different contexts (H indicates A, C or T). P values were transformed into FDR values in the multiple comparisons.
Therefore, the de novo gene alleles at intermediate Tfreq are highly methylated in all methylation contexts, which may result in the low transcription levels of de novo genes at intermediate Tfreq. Given that the RNA sequences or peptides derived from de novo genes are likely deleterious for the organisms, one way for de novo genes at a low Tfreq to spread in the population is for their deleterious effects to be relieved by reducing their expression levels through increased DNA methylation. The increased DNA methylation levels could largely result from the increased proportions of overlapping TEs and siRNAs for de novo genes at intermediate Tfreq (fig. 5). Most likely, a newly originated de novo allele transcribed at low Tfreq is deleterious. The repressed Tlevel produced by increased DNA methylation could prevent the de novo allele from becoming extinct. This allele would ultimately be fixed in the population with an increased Tlevel under positive selection if the allele was favorable.
Discussion
The origin of de novo genes is a fundamental biological question (Carvunis et al. 2012; Abrusan 2013; Arendsee et al. 2014; Light et al. 2014; Neme and Tautz 2014; Wu et al. 2014; Cui et al. 2015). The genome contains many small ORFs with expression evidence in intergenic regions, some of which have important phenotypic effects (Hanada et al. 2007, 2013). Most previous studies based on comparative genomics at the interspecific level indicate that, after de novo genes arose, they gradually became longer, more highly expressed and more conserved in their CDSs. In this study, to understand the process of de novo gene origination, we performed intraspecific analysis. We divided the 782 de novo genes into different groups based on whether they are transcribed, translated, translated with detected poly(A) tails or none of these. The results suggest that TL+genes are more similar to mature genes, followed by TL− genes, TC genes, and TN genes.
We used the distribution of transcribed alleles among the 84 resequenced accessions (supplementary fig. S8, Supplementary Material online shows the distribution of the 84 A. thaliana accessions, and see Methods for details) to investigate the geographic spreading of de novo genes, finding that the proportion of widely transcribed de novo genes (transcribed both in Western Europe and Central Asia accessions) is lowest for TN, followed by TC, TL−, TL+, and common genes (supplementary fig. S9, Supplementary Material online). The geographic spread of de novo genes may be affected by selection, demographic history, expression variability or even the age of the gene. The widespread distribution of a transcribed gene generally indicates a relatively early origin rather than a recent origin. The later scenario requires strong selection and high gene flow between geographically isolated populations. In fact, A. thaliana is a globally distributed species, and accessions from Western Europe and Central Asia have been isolated for a long period of time without substantial gene flow (Cao et al. 2011). Under the general hypothesis that genes transcribed in both Western Europe and Central Asia accessions could be regarded as “older” than genes transcribed only in accessions from either Western Europe or Central Asia, the proportion of “older” genes could serve as a proxy for the age of each type of gene in total. Thus, our analysis of geographic spreading suggests that TL+ genes are much “older” than other de novo genes and that TN-type genes are the “youngest”.
Based on the gene features of de novo genes and their predicted ages, it appears that, in general, de novo genes originated from sequences (with or without an ORF) lacking transcriptional ability (e.g., intergenic ORFs and TN type in this study), which first acquired this ability (with or without an ORF; e.g., TC type), followed by translational ability (e.g., TL− genes) and were ultimately modified to become mature protein-coding genes (e.g., TL+genes) (supplementary fig. S10, Supplementary Material online). However, acquiring an ORF sequence is not necessarily the first step in de novo gene birth, and de novo genes originating from different types of genes (TN type representing ORF sequences, TC type representing noncoding RNAs) could vary in terms of half-life times. Thus, we cannot rule out the possibility that de novo genes emerging with one or more mature gene features may have a longer lifetime in the genome than the others (e.g., genes with poly(A) tails may persist for a longer time than genes without poly(A) tails) and are therefore likely detected as “older” genes than those originating without mature gene features.
While much is known about the gene features of de novo genes, much less is known about the evolutionary forces behind de novo gene birth. One must address this question at the population level using a population genetics approach (Schlotterer 2015). However, only a few studies about de novo genes have been performed at the population level, which utilized a small sample size (Palmieri et al. 2014; Zhao et al. 2014). The availability of multiple omics data sets from the same population of A. thaliana, with a large sample size and numerous additional omics data sets as well, provided us with the great opportunity to investigate this question for the first time at the multiple omics data set level.
Based on the results of our intraspecific analysis, we propose that enhanced DNA methylation induced by TEs, siRNAs, and antisense transcripts could improve the probability of survival of de novo alleles, as well as their spread in the population, by repressing the transcriptional activities of de novo gene alleles. This “neutralisation” effect could help maintain the coding potential and expression activity of de novo genes (supplementary fig. S11, Supplementary Material online). Given that the epimutation rate is at least four orders of magnitude higher than the DNA mutation rate (Ossowski et al. 2010; Becker et al. 2011; Schmitz et al. 2011), and that DNA methylation can recover in even a single generation (e.g., methylation reprograming in generative cells) (Slotkin et al. 2009; Calarco et al. 2012), it is possible that de novo gene alleles that are initially deleterious but favorable in a new environment could recover their transcriptional activities via demethylation.
The regulatory effects of DNA methylation are conserved among species to some extent (Takuno and Gaut 2013; Seymour et al. 2014). Therefore, it would be interesting to explore whether the same correlation between the allele frequencies of de novo genes and their DNA methylation levels exists in other species during the process of de novo gene birth. The “neutralisation by DNA methylation” model could help us understand the evolutionary forces through which a large number of transcription units, such as small ORFs, are maintained in the genome, and it emphasizes the importance of epimutations in evolution.
Materials and Methods
Data Sources
The published genome-wide protein-coding gene sequences of 65 green plants, including A. thaliana (TAIR10 annotation), were downloaded and used to identify A. thaliana de novo genes (supplementary table S1, Supplementary Material online). Information regarding the published data, including 13 transcriptomes, six translatomes (genome-wide ribosome footprinting data), three polyadenylation data sets, one DNase hypersensitive sites (DHs) data set, one histone modification data set, one small interfering RNA sequences (siRNAs) data set, and one antisense transcript data set for A. thaliana reference accession Col-0, is listed in supplementary table S3, Supplementary Material online. The TFBS data for A. thaliana genes were downloaded from the Arabidopsis Gene Regulatory Information Server (Yilmaz et al. 2011). Genome sequences, DNA methylation data (leaf tissue), and transcriptome information (leaf tissue) for 84 A. thaliana accessions as well as DNA methylation data from flower tissues of seven of these accessions were downloaded from the Arabidopsis 1001 Genome Database (http://1001genomes.org/data/Salk/, last accessed July 14, 2016) and the NCBI GEO database (GSE43857 and GSE43858) (Schmitz et al. 2013) (supplementary table S6, Supplementary Material online). The methylation data set for Arabidopsis accumulation lines was downloaded from the NCBI GEO database (GSE36844, GSE36845, and GSE47490) and analyzed as previously described (Becker et al. 2011).
De Novo Gene Identification
For genes with alternative transcript isoforms, only the longest CDS of each gene was included in the analyses. In total, 27,206 A. thaliana genes, which are referred to as unigenes, were used in this study. An “all to all” blastp search (Altschul et al. 1997) was performed for protein sequences from the 65 genomes using an E value threshold of 1 × 10−5. All protein sequences were clustered into separate groups using the Markov Cluster Algorithm (Enright et al. 2002) (mcl-12-135, I = 2, S = 6) based on the blastp E values. Thus, genes in groups that only contained A. thaliana genes were extracted as candidate de novo genes for the A. thaliana reference accession Col-0. Using the protein sequences of these genes as queries, PSI-blast (E < 1 × 10−5) searches were performed against the NCBI nonredundancy protein database (NCBI nr database excluding sequences from A. thaliana and nonplant species, as of February 2016) to exclude the candidates with homologues in plants without complete genome sequences. In addition, these genes were searched against the PlantGDB-assembled unique transcripts database (PUTs database excluding sequences from A. thaliana, as of November 2013) using tblastn (E < 1 × 10−5) to exclude candidates derived from frameshift mutations of existing genes. To eliminate possible horizontal gene transfer from nonplant species, all candidates were searched against the NCBI nonredundancy protein database (NCBI nr database excluding green plants, as of February 2016) using PSI-blast (E < 1 × 10−5) and the criterion that the length of the hit must be at least half that of the query sequence.
Gene synteny relationships among A. thaliana, A. lyrata, and C. rubella were estimated using MCScanX software (Wang et al. 2012) based on E values in the above “all against all” blastp analysis and gene model annotation information extracted from the same source used to download their protein-coding gene sequences (supplementary table S1, Supplementary Material online). To reduce the effects of gaps and missing gene annotations in genomes of closely related species on the identification of de novo genes in A. thaliana, de novo gene candidates with sequencing gaps in synteny regions between A. thaliana and A. lyrata and lacking similar sequences (E value < 1 × 10−10) in other regions of the A. lyrata genome were excluded. Furthermore, EXONERATE 2.2.0 (Slater and Birney 2005) (the alignment option is “protein2genome:bestfit”) was used to find missing gene models in A. lyrata and C. rubella sequences that were syntenic to or most similar to A. thaliana de novo gene candidates. The intactness of the coding region for each missing gene sequence was evaluated using an inhouse Perl script. Sequences with complete start and stop codons, no premature stop codons and limited numbers of frame-shifted codons compared to the A. thaliana gene model were defined as complete CDSs. If the total length of the refined CDS of A. lyrata and/or C. rubella was longer than 90% that of the A. thaliana candidate, and the codon-shift length was less than 10% that of the candidate, the sequence was considered to be a missing gene that may be homologous to the corresponding A. thaliana candidate and was thus excluded from the candidate list.
Intergenic ORFs and Common Genes
Genomic regions except for protein-coding genes, noncoding RNA (including tRNA, rRNA, miRNA, snRNA, snoRNA, and other RNA), pseudogenes and TE genes annotated in TAIR10 GFF file were considered to be intergenic regions. The longest single-exon ORF with a complete start and stop codon in each intergenic region in either the sense or antisense strand was predicted (if there was overlap between the sense and antisense ORF in the same genomic region, only the longest was kept), and ORFs longer than 90 nt were considered to be intergenic ORFs for comparisons with de novo genes and common genes. Unigenes, except for de novo genes, were considered to be common genes in this study.
Classification of de novo Genes and Calculation of Gene Features
Transcriptome and translatome data (Data 1–19 produced from A. thaliana Col-0) were used as transcriptional or translational evidence for de novo genes, which were mapped to the TAIR10 reference genome in previous studies (supplementary table S3, Supplementary Material online). When the fragments/reads per kilobase per million mapped fragments/reads (FPKM/RPKM) value for a gene was equal to or greater than 1 in at least one of the expression data sets, or greater than 0 in at least two of the data sets, it was classified as expressed. Thus, de novo genes of A. thaliana Col-0 were divided into three types based on their expression states: genes with translational evidence (TL type, FPKM/RPKM ≥ 1 in at least one translatome data set or FPKM/RPKM > 0 in at least two translatome data sets), genes with transcriptional activities only (TC type, FPKM/RPKM ≥ 1 in at least one transcriptome data set or FPKM/RPKM > 0 in at least two transcriptome data sets) and genes without expression evidence (TN type, the other de novo genes excluding TL and TC types).
The CAI was calculated for A. thaliana unigenes and intergenic ORFs using CodonW 1.4.2 (http://codonw.sourceforge.net/, last accessed July 14, 2016). Protein secondary structures (α-helices, β-sheets, and random coils) were predicted using PSSpred V2 (Y. Zhang, http://zhanglab.ccmb.med.umich.edu/PSSpred/, last accessed July 14, 2016). GC contents were measured in CDS regions. The single nucleotide polymorphism matrix and information regarding unmapped regions of the 84 accessions (Schmitz et al. 2013) were used for polymorphism analysis, in which missing data were controlled in the calculation according to a previous method (Long, Rabanal, et al. 2013). Nucleotide diversities (π) were measured in genic regions. Polymorphism in nonsynonymous (or synonymous) sites of each gene, πn (πs), was computed as the mean of all pairwise substitution rates in nonsynonymous (or synonymous) sites, Ka (Ks), among transcribed alleles from the 84 accessions. The substitution rates were estimated using the “yn00” program in PAML v4.7 with default parameters (Yang 2007). Coding intactness analysis for each gene among accessions was performed following the method used to detect missing gene models described in previous sections. As intraspecific analysis was performed in the study, a strict standard was used (genes with no changes in the states of start and stop codons and less than 10% codon-shift length compared to Col-0 gene models were treated as intact).
DNA Methylation, TEs and siRNAs
The DNA methylation level was calculated using the ratio of the total number of sequenced methylated sites to the total number of sequenced cytosine sites for a given length of sequence. DNA methylation levels at symmetric (CG and CHG, where H indicates A, C or T) and asymmetric (CHH) sites as well as total covered cytosine sites were computed. The proportion of overlapping TEs or siRNAs represents the percentage of genic or flanking region that overlapped with TEs or that was targeted by siRNAs. Information about the TEs in the A. thaliana reference genome was extracted from the TAIR database (http://www.arabidopsis.org/, last accessed July 14, 2016). The 24-nt siRNA sequences were mapped to the A. thaliana reference genome (TAIR10) using blastn without mismatches or gaps. The siRNA sequences that mapped to multiple locations were considered to overlap with all of these locations. The siRNA depth was computed for each genic and flanking region for different types of genes according to a previous method, in which siRNAs with multiple targeted locations were weighted by the number of mapping locations (Hollister et al. 2011). Permutations of DNA methylation levels and the proportion of TEs and siRNAs were performed 100 times for common genes and intergenic ORFs, respectively. Each time, 782 common genes or intergenic ORFs were randomly extracted from all common genes or intergenic ORFs without replication.
Transcription Frequency and Prediction of Origination Times of De Novo Genes
The transcriptomes of the 84 A. thaliana accessions were used to classify the de novo genes and common genes into nine frequency groups based on their transcription frequencies (Tfreq): 0% (111 de novo genes and 159 common genes), 5% (318, 663), 10% (88, 633), 25% (103, 1,789), 50% (62, 1,588), 75% (21, 1,186), 90% (29, 982), 95% (8, 684), and 100% (42, 18,740), representing genes transcribed in 0, 1–4, 5–8, 9–21, 22–42, 43–63, 64–76, 77–80 or 81–84 accessions, respectively. The transcription levels (Data 2 in supplementary table S3, Supplementary Material online) of de novo genes between nearby groups were compared, exhibiting an order of 5% > 10% > 25% ≈ 50% < 75% ≈ 90% ≈ 95% < 100% (“>” indicates that the transcription level of genes in the group on the left side is higher than that on the right side, “<” indicates that of the left side is lower than that of the right side and “≈” indicates that the difference between nearby groups is not significant; Wilcoxon test, P < 0.05), whereas that of common genes was 5% > 10% > 25% > 50% < 75% < 90% < 95% < 100% (P < 1 × 10−4). Thus, a transcription frequency of 50% was considered to be the dividing point.
To estimate the geographic spread of the de novo genes, 65 accessions (excluding accessions located in the United Kingdom, North America, and Japan in the 84 previously resequenced genomes) distributed in Eurasia were divided into three groups: 48 accessions located west of 15°E (group 1), 12 accessions located between 15°E and 60°E (group 2), and five accessions located east of 60°E (group 3) (supplementary fig. S8 and supplementary table S6, Supplementary Material online). The numbers of de novo genes with transcriptional evidence in accessions from both groups 1 and 3 for each type of de novo gene were counted. The same strategy was used to analyse common genes.
Statistical Analysis
Statistical analyses were performed using R (http://www.r-project.org/, last accessed July 14, 2016). P values were transformed into false discovery rate (FDR) values when comparison times in multiple tests were larger than ten, and the other P values were corrected using Bonferroni correction.
Supplementary Material
Supplementary figures S1–S11 and tables S1–S10 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We are indebted to the Arabidopsis community for producing so many valuable and informative data sets and resources, which made this study possible. We would like to thank Brandon S. Gaut and Detlef Weigel for their valuable comments about this work. We thank the anonymous reviewers for taking the time to help us greatly improve the manuscript. This work was supported by the National Natural Science Foundation of China (91231104, 31222006, and 31470331) and the 100 Talents Program of the Chinese Academy of Sciences.
Literature Cited
- Abrusan G. 2013. Integration of new genes into cellular networks, and their structural maturation. Genetics 195:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendsee ZW, Li L, Wurtele ES.. 2014. Coming of age: orphan genes in plants. Trends Plant Sci. 19:698–708. [DOI] [PubMed] [Google Scholar]
- Becker C, et al. 2011. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480:245–249. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA, Kern AD, Jones CD.. 2007. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics 176:1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhao R, Jiang H, Wang W.. 2008. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics 179:487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, et al. 2012. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, et al. 2007. Identification and characterization of lineage-specific genes within the Poaceae. Plant Physiol. 145:1311–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, et al. 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 43:956–963. [DOI] [PubMed] [Google Scholar]
- Carvunis AR, et al. 2012. Proto-genes and de novo gene birth. Nature 487:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M.. 2010. New genes in Drosophila quickly become essential. Science 330:1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ST, Cheng HC, Barbash DA, Yang HP.. 2007. Evolution of hydra, a recently evolved testis-expressed gene with nine alternative first exons in Drosophila melanogaster. PLoS Genet. 3:e107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, et al. 2015. Young genes out of the male: an insight from evolutionary age analysis of the pollen transcriptome. Mol Plant. 8:935–945. [DOI] [PubMed] [Google Scholar]
- Donoghue MT, Keshavaiah C, Swamidatta SH, Spillane C.. 2011. Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol Biol. 11:47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA.. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerzoni D, McLysaght A.. 2016. De novo genes arise at a slow but steady rate along the primate lineage and have been subject to incomplete lineage sorting. Genome Biol Evol. 8:1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YL. 2013. Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 73:941–951. [DOI] [PubMed] [Google Scholar]
- Hanada K, et al. 2013. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci U S A. 110:2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH.. 2007. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 17:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen DR, Bureau TE.. 2015. Discovery of novel genes derived from transposable elements using integrative genomic analysis. Mol Biol Evol. 32:1487–1506. [DOI] [PubMed] [Google Scholar]
- Hollister JD, et al. 2011. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci U S A. 108:2322–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DG, McLysaght A.. 2009. Recent de novo origin of human protein-coding genes. Genome Res. 19:1752–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ.. 2006. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci U S A. 103:9935–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light S, Basile W, Elofsson A.. 2014. Orphans and new gene origination, a structural and evolutionary perspective. Curr Opin Struct Biol. 26:73–83. [DOI] [PubMed] [Google Scholar]
- Lin H, et al. 2010. Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evol Biol. 10:41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, VanKuren NW, Chen S, Vibranovski MD.. 2013. New gene evolution: little did we know. Annu Rev Genet. 47:307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, et al. 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 45:884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, et al. 2013. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. 73:77–90. [DOI] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome architecture. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- McLysaght A, Guerzoni D.. 2015. New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos Trans R Soc Lond B Biol Sci. 370:20140332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neme R, Tautz D.. 2014. Evolution: dynamics of de novo gene emergence. Curr Biol. 24:R238–R240. [DOI] [PubMed] [Google Scholar]
- Ossowski S, et al. 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327:92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri N, Kosiol C, Schlotterer C.. 2014. The life cycle of Drosophila orphan genes. Elife. 3:e01311.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt JA, et al. 2013. De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously non-coding sequences. PLoS Genet. 9:e1003860.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotterer C. 2015. Genes from scratch—the evolutionary fate of de novo genes. Trends Genet. 31:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, et al. 2011. Transgenerational epigenetic instability is a source of novel methylation variants. Science 334:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, et al. 2013. Patterns of population epigenomic diversity. Nature 495:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour DK, Koenig D, Hagmann J, Becker C, Weigel D.. 2014. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 10:e1004785.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstnev A, et al. 2012. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol. 19:845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, et al. 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136:461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP.. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Gaut BS.. 2013. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci U S A. 110:1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. 2014. The discovery of de novo gene evolution. Perspect Biol Med. 57:149–161. [DOI] [PubMed] [Google Scholar]
- Tautz D, Domazet-Loso T.. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12:692–702. [DOI] [PubMed] [Google Scholar]
- Toll-Riera M, et al. 2009. Origin of primate orphan genes: a comparative genomics approach. Mol Biol Evol. 26:603–612. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB.. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 2:135–140. [DOI] [PubMed] [Google Scholar]
- Wu DD, et al. 2014. “Out of pollen” hypothesis for origin of new genes in flowering plants: study from Arabidopsis thaliana. Genome Biol Evol. 6:2822–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DD, Irwin DM, Zhang YP.. 2011. De novo origin of human protein-coding genes. PLoS Genet. 7:e1002379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, et al. 2011. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci U S A. 108:12533–12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, et al. 2012. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 8:e1002942.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Jawdy S, Tschaplinski TJ, Tuskan GA.. 2009. Genome-wide identification of lineage-specific genes in Arabidopsis, Oryza and Populus. Genomics 93:473–480. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yilmaz A, et al. 2011. AGRIS: the Arabidopsis gene regulatory information server, an update. Nucleic Acids Res. 39:D1118–D1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Saelao P, Jones CD, Begun DJ.. 2014. Origin and spread of de novo genes in Drosophila melanogaster populations. Science 343:769–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, et al. 2008. On the origin of new genes in Drosophila. Genome Res. 18:1446–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S.. 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 39:61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.