Graphical abstract
Keywords: Isothermal loop-mediated amplification, SYTO16, Real time LAMP, Visceral leishmaniasis, Leishmania donovani
Highlights
-
•
Three systems of loop-mediated isothermal amplification (LAMP) were developed for diagnosing leishmaniasis.
-
•
The green nucleic acid stain, SYTO-16 was adapted for monitoring the reactions in real-time.
-
•
The LAMP assays proved highly sensitive detecting >100Fg DNA/reaction.
-
•
Leishmania DNA was detected in a significant number of asymptomatic individuals living in endemic areas.
Abstract
Visceral leishmaniasis (VL), one of the most important neglected tropical diseases, is caused by Leishmania donovani eukaryotic protozoan parasite of the genus Leishmania, the disease is prevalent mainly in the Indian sub-continent, East Africa and Brazil. VL can be diagnosed by PCR amplifying ITS1 and/or kDNA genes. The current study involved the optimization of Loop-mediated isothermal amplification (LAMP) for the detection of Leishmania DNA in human blood or tissue samples. Three LAMP systems were developed; in two of those the primers were designed based on shared regions of the ITS1 gene among different Leishmania species, while the primers for the third LAMP system were derived from a newly identified repeated region in the Leishmania genome. The LAMP tests were shown to be sufficiently sensitive to detect 0.1 pg of DNA from most Leishmania species. The green nucleic acid stain SYTO16, was used here for the first time to allow real-time monitoring of LAMP amplification. The advantage of real time-LAMP using SYTO 16 over end-point LAMP product detection is discussed. The efficacy of the real time-LAMP tests for detecting Leishmania DNA in dried blood samples from volunteers living in endemic areas, was compared with that of qRT-kDNA PCR.
1. Introduction
The leishmaniasis are a group of diseases, caused by eukaryotic protozoan parasites of the genus Leishmania, transmitted by blood-sucking phlebotomine sand flies. Leishmaniasis affects 12 million men, women and children in 88 countries around the world, over 350 million humans are at risk, and over 2 million new cases emerge every year (Alvar et al., 2012, Ready, 2014, WHO/TDR, 2010). Visceral leishmaniasis (VL), the most serious form of the disease, occurs mainly in the Indian sub-continent, East Africa and Brazil. Although some 400,000 people are affected by VL annually with up to 40,000 deaths per year, the disease is still ranked among the neglected tropical diseases (Alvar et al., 2012, Desjeux, 2004, Ready, 2014).
VL can be diagnosed by microscopic examination of stained, splenic or bone marrow biopsy material (Chappuis et al., 2007, WHO/TDR, 2010). Serological methods with moderate sensitivity based on direct agglutination test (DAT) and enzyme-linked immune-sorbent assays (ELISA) are also used (Boelaert et al., 2008, ter Horst et al., 2009). In addition, several other methods based on in vitro DNA amplification of different repeated Leishmania genes, using the polymerase chain reaction (PCR), were also adapted for detection of parasites in tissue samples. The most commonly targeted genes are the ribosomal ITS1 and the kinetoplast minicircle genes (kDNA) (el Tai et al., 2000, Nicolas et al., 2002, Rodgers et al., 1990, van Eys et al., 1992). These PCR assays have been developed for the detection of Leishmania DNA in a variety of clinical samples such as skin biopsies and smears, bone marrow and lymph node aspirates as well as peripheral blood. To date, there is no generally-accepted gold standard for Leishmania identification (Miller et al., 2014). Van der Auwera and Dujardin (2015), gave a comprehensive literature analysis for the different available and accepted methods used for Leishmania species detection in clinical and epidemiological studies.
Rapid, minimally invasive diagnostic test for cutaneous leishmaniasis (CL) would constitute a valuable tool, especially when large numbers of patients are to be tested in a short time and under field conditions. In the case of VL, prevalent asymptomatic infections in endemic regions may serve as parasite reservoirs for infecting vectors (Adler et al., 1966, Ali and Ashford, 1994, Sharma et al., 2000). Thus, a rapid and simple field-test to determine putative reservoir potential of L. donovani, would comprise a valuable tool for curtailing disease transmission.
Loop-mediated isothermal amplification (LAMP) was first introduced by Notomi et al. in 2000. The technique is considered a highly sensitive and rapid method for DNA amplification at a constant temperature (60–65 °C). LAMP can amplify a few copies of DNA to 109 molecules in about one hour under isothermal conditions The methodology relies on auto-cycling strand-displacement DNA synthesis using Bacillus stearothermophilus (Bst) DNA polymerase, and the use of 4 specially designed primers that enable the production of amplification products having a stem and loop structure. LAMP has proved its usefulness for the detection of many infectious agents including disease–causing parasites (Abbasi et al., 2013, Aryan et al., 2010, Notomi et al., 2000, Poon et al., 2006, Salant et al., 2012).In recent years several studies have utilized LAMP for detecting Leishmania DNA in tissue and blood of infected humans as well as the sand fly vectors (Khan et al., 2012, Nzelu et al., 2014, Takagi et al., 2009, Verma et al., 2013). The first LAMP test for diagnosis of infection was developed by Takagi et al., who used specific primers for L. donovani that were based on kDNA sequences and achieved an amplification sensitivity of 1fg per reaction (Takagi et al., 2009). Later on, this test was used by other researchers to validate VL infections from patients' blood samples (Khan et al., 2012, Verma et al., 2013).
We designed three sets of primers for LAMP DNA amplification appropriate for the detection of most Leishmania species. Two sets of primers derive from shared regions of the Leishmania ITS1 sequences and the third set was obtained from a newly-identified repeat region known to have shared sequences in L. donovani and L. major. Results showed that the sequence is also shared by other Leishmania species. The described method employs SYTO-16, a nucleic acid stain found to be suitable for real-time LAMP detection by fluorescence. Thus, our assay avoids the necessity for end-point analysis, be it using green fluorescence (SYBR Green I), turbidity (spectrophotometry measurement of absorbance at 400 nm) or agarose gel electrophoresis.
2. Materials and methods
2.1. Ethical concerns
Informed consent was sought from all the adults recruited for the study. Consent for inclusion of young children, was obtained from parents or guardians. Study procedures were approved by the ethical review committees at the Medical Faculty, Addis Ababa University and the National Research Ethics Review Committee (NRERC) at the Ethiopian Ministry of Science and Technology.
2.2. Samples
Genomic DNA was obtained from the following Leishmania reference strains: L. donovani (MHOM/SD/62/IS), L. major (MHOM/IL/2014/LRC-L1671), L. tropica (MHOM/IL/2011/LRC-L1558), L. aethiopica (MHOM/ET/72/L100), L. infantum chagasi (MHOM/CR/199?/LRC-L744), L. mexicana (MNYC/BZ/62/M 379), L. amazonensis (MHOM/BR/73/M2269), L. braziliensis (MHOM/BR/75/M-4037), L. panamensis (MHOM/CO/86/UA126), L. b. guyanensis (MHOM/BR/75/M4147). Human finger-prick blood samples were collected from 44 individuals living in a village in the Tahatay Adiabo district of north Ethiopia. From these 44 samples; 9 individuals were previously treated VL cases residing in different households. The rest of the samples were taken from volunteers living nearby.
2.3. DNA extraction
Total genomic DNA was purified using phenol-based DNA extraction method (28). Leishmania promastigotes or ∼20 μl dried blood (blotted in two 6 mm [dia.] 3MM filter paper punches) were incubated at 60 °C for two hours in a 1.5 ml micro-centrifuge tube containing 200 μl of lysis buffer (50 mM NaCl, 10 mM EDTA, 50 mM Tris-HCl pH 7.4, 1% triton X-100, and 200 μg/ml of proteinase K). This was followed by extraction with equal volumes of TE-saturated phenol (pH 8), and then precipitating the DNA with ethanol. The extracted lyophilized DNA was resuspended in 50 μl DNAse/RNase-free, double distilled water and stored at −20 °C until further use.
2.4. LAMP primer design
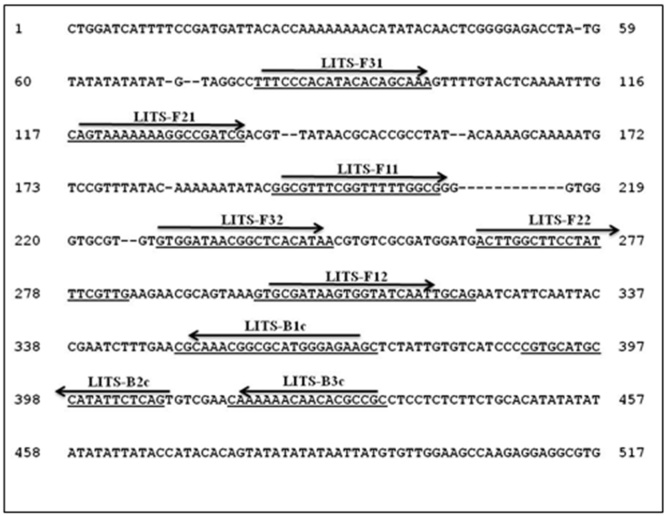
Three different sets of LAMP primers (external primers: F3 and B3, internal primers: FIP and BIP) were designed; the sequences for two sets were based on shared ITS1 DNA sequences of different Leishmania species. The third set of primers was based on L. donovani repetitive sequence as elaborated in the “results” section. All primers were designed manually taking into consideration the required melting temperatures, and the distances between all the primers. Table 1 shows the nucleotide sequences of the newly designed LAMP primers, and Fig. 1 presents their location within the conserved Leishmania ITS1 gene.
Table 1.
DNA sequences of the primers designed for the three LAMP systems. Two targeting the ITS1 gene (See Fig. 1) and one targeting the L. donovanirepeat DNA region.
| LAMP System | Primer Name | Primer sequence |
|---|---|---|
| LITS-LAMP1 | LITSF3.1 | TTTCCCACATACACAGCAAA |
| LITSB3.3 | GCGGCGTGTTGTTTTTTG | |
| LITSFIP1 | CGCCAAAAACCGAAACGCCTTTTCAGTAAAAAAAGGCCGATCG | |
| LITSBIP3 | CGCAAACGGCGCATGGGAGAAGCTTTTCTGAGAATATGGCATGCACG | |
| LITS-LAMP2 | LITSF3.2 | GTGGATAACGGCTCACATAA |
| LITSB33 | GCGGCGTGTTGTTTTTTG | |
| LITSFIP2 | CTGCAATTGATACCACTTATCGCACTTTTACTTGGCTTCCTATTTCGTTG | |
| LITSBIP3 | CGCAAACGGCGCATGGGAGAAGCTTTTCTGAGAATATGGCATGCACG | |
| L151-LAMP | L151F3 | GATGAGAAGCTCACGGAG |
| L151B3 | ATCCTCCTCCTCGTCTTC | |
| L151FIP | CGGGTACGTGAGTCCGTATTTTGTCTGCCAGTCAACAAGA | |
| L151BIP | GAAGCTCATGATCGAGAAGGAGTTTTTTCGTCTGATGCGTTGCT |
Fig. 1.
DNA sequence of the L. donovaniITS1 gene, showing the location of the LAMP primers on the sequences shared by different Leishmania species.
2.5. LAMP assays
LAMP reactions were performed in 25 μl enzyme reaction buffer (20 mM Tris-HCl [pH 8.8], 10 mM KCl, 10 mM [NH4]2 SO4, 8 mM MgSO4, and 1% Tween 20).; that contains 40pmoles FIP and BIP; 5pmoles F3 and B3 outer primers; 8 units of Bst-WarmStart DNA polymerase (New England Bio-labs Inc., MA, USA);0.2 mM dNTPs mixture; 0.8 M Betaine; 2 μM SYTO-16 green fluorescence nucleic acid stain (Thermo-Fisher Scientific, Grand Island, NY).The reaction was carried out at 65 °C for 2 h in a real time PCR thermocycler (Rotor-Gene 6000, Qiagene, Hilden, Germany) with fluorescence data acquisition at one minute intervals.
2.6. Detection of LAMP
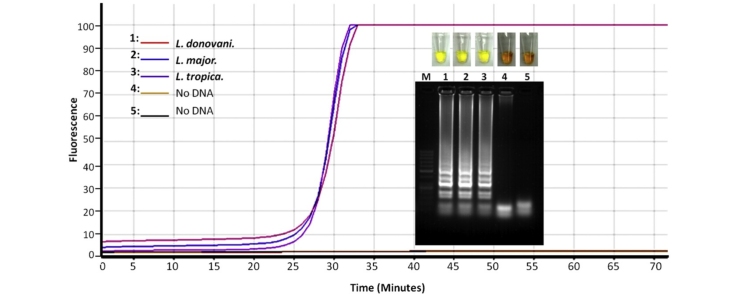
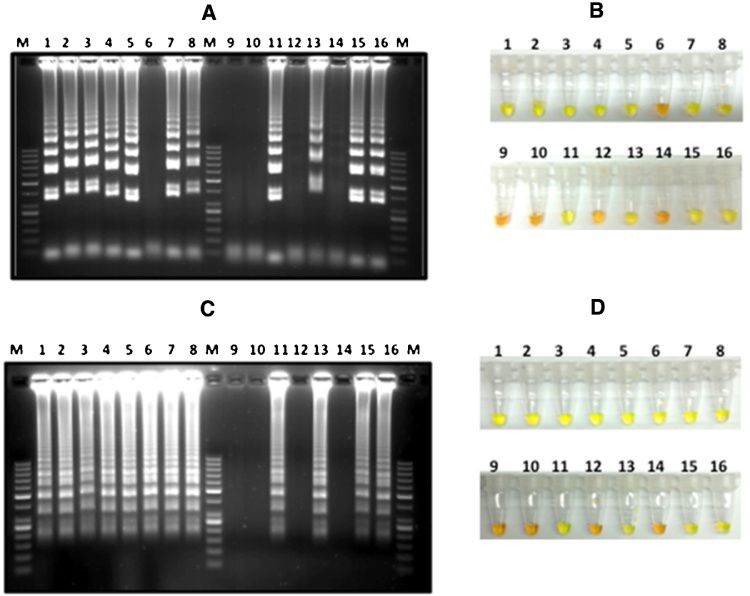
DNA amplification was assessed using two methods. The SYTO®16 green-fluorescent nucleic acid stains are cell-permeant nucleic acid stains that show intense fluorescence enhancement upon binding nucleic acids. SYTO-16 (2 μM, Excitation/Emission, 488/518 nM) was used for real-time detection and quantitation of the DNA accumulating in the reaction tubes.The second method employed end-point quantification of DNA by adding SYBR Green-I(>1 μM,Thermo-Fisher Scientific, Grand Island, NY). SYBR Green-I detects the presence of LAMP-DNA immediately causing the color of the reaction to change from orange to green (Fig. 2). To verify the results, LAMP–DNA was electrophoresed and viewed on standard agarose gels stained with ethidium-bromide (Fig. 2).
Fig. 2.
Agarose gel electrophoresis analysis and SYBR Green I end point detection of LITS-LAMP1 (A,B), and LITS-LAMP2 (C,D). The analysis shows LAMP DNA amplification products of different concentrations amplified from different amounts of L. donovani template DNA, (1) 0.1 ng, (2) 0.01 ng, (3) 1 pg, (4) 0.1 pg. And the amplification of 1 ng template DNA of each of the following Leishmania species: (5) L. major, (6) L. aethiopica, (7) L. tropica, (8) L. infantum chagasi. (9) and (10) No DNA [control], (11–16) Select blood samples obtained by finger prick from volunteers in North Ethiopia. (M) DNA size marker.
2.7. Quantitative real-time kinetoplast DNA PCR (qRT-kDNA PCR)
The concentration of Leishmania parasites in blood was estimated as described previously (Abbasi et al., 2013). Briefly, kDNA amplification by qPCR was achieved usingt he kDNA minicircle-specific primers JW11 (CCTATTTTACACCAACCCCCAGT) and JW12 (GGGTAGGGGCGTTCTGCGAAA) (Nicolas et al., 2002). The qPCR reaction was performed using the Absolute Blue qPCR SYBR Green mix kit (Thermo scientific, Surrey, UK), and the amplified DNA was detected and quantified by a real time PCR thermocycler (Rotor-Gene 6000, Qiagene, Hilden, Germany).
3. Results
3.1. Selection of Leishmania LAMP primers
ITS1 Sequences from different Leishmania species selected from GenBank, were aligned in order to identify the DNA sequences shared by all Leishmania species and, thus, suitable for LAMP primers. Based on comprehensive ITS1 alignments; two external forward (F3) primers (LITSF3.1 and LITSF3.2), and one backward (LITSB3) primer were identified. In addition, two forward internal primers(LITSFIP1 and LITSFIP2) and one backward inner primer (LITSBIP) were designed. These primers were used to build two Leishmania ITS1LAMP systems (LITS-LAMP1 and LITS-LAMP2). Where; LITS-LAMP1 spans 353 bp, and LITS-LAMP2 covers 204 bp (Table 1, Fig. 1).
The selected primers for the third Leishmania LAMP system (L151) (Table 1) were based on a L. donovani repetitive DNA sequence (458-bp) identified previously (Abbasi, unpublished data). The repeat sequence forms part of conserved L. donovani gene segment (GenBank accession no. XM-003858114), and it also displays a high degree of homology to a segment in the L. major chromosome 4 (GenBank accession no. FR796400), but no known homology with other Leishmania species.
3.2. Optimization of the detection of real-time LAMP DNA amplification
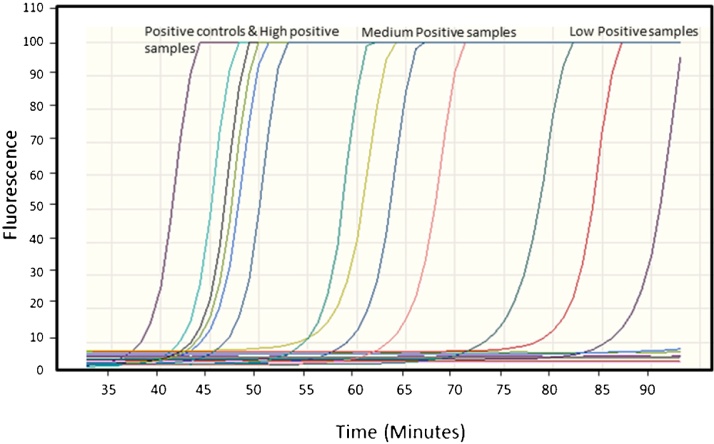
A comparison between SYBR Green I and SYTO-16 fluorescent nucleic acid showed that SYBR Green-I at concentrations of 1–5 μM inhibited LAMP amplification almost completely. Real-time detection of DNA amplification was possible only at concentrations below 1 μM. However, at these extremely low concentrations, fluorescence intensity was lower than 50% of the intensity required for detection (data not shown) severely limiting its efficacy. On the other hand, SYTO-16 (2 μM) allowed optimal real time visualization of the LAMP products. Fig. 3, shows real time detection of LAMP DNA amplification detected on a qRT-PCR thermocycler using SYTO-16.
Fig. 3.
Monitoring of LAMP amplification from different Leishmania DNA control samples and from DNA extracted finger pricks samples. The analysis shows the DNA amplification detection in real time by measuring the increasing fluorescence of DNA binding to SYTO-16 dye.
3.3. LAMP sensitivity and specificity
LAMP was performed using different concentrations of L. donovani template DNA (0.1 ng, 0.01 ng, 1 pg, and 100fg) and 1 ng DNA from additional Leishmania species from the Old World and from Latin America (L. major, L. tropica, L. aethiopica, L. [infantum] chagasi, L. mexicana, L. amazonensis, L. braziliensis, L. panamensis, and L. guyanensis). LITS-LAMP1/2 and L151, amplified L. donovani DNA at the lowest concentration tested (100fg, Fig. 2), similar levels of sensitivity were also obtained with L. major template DNA. While LITS-LAMP2 and L151 amplified DNA from all the Leishmania species tested, LITS-LAMP1 failed to amplify DNA from L. aethiopica (Fig. 2).
3.4. Samples analysis
The three LAMP systems were compared for their ability to detect Leishmania DNA in finger prick blood collected from 44 individuals living in a VL endemic area in Ethiopia. LAMP results were compared with those obtained by qRT-kDNA PCR. All volunteers were asymptomatic at the time of sampling although some had succumbed to VL and were treated in the past (Table 2, column 2). Two samples (number 22 and 36) showed positive results with all the three LAMP systems as well as the qRT-kDNA PCR. The strength of LAMP positivity was indicated by the time (in minutes) needed for obtaining fluorescence signals caused by the binding of SYTO-16 to the amplified DNA. This parameter is known as take-off time, and the shorter time means relatively more template DNA in the sample. The take-off times of LITS-LAMP1, LITS-LAMP2, and L151-LAMP using 0.1 ng L. donovani template DNA were 42, 40, and 54 min respectively. Takeoff times longer than 95 min for LITS-LAMP1 and LITS-LAMP2, and 110 min for L151-LAMP were considered negative. Note sample number 22; that showed the highest parasite number by kDNA-qPCR, also showed an early take-off time in the three LAMP systems (Fig. 3).
Table 2.
Parasite concentrations (per ml) in finger prick blood of volunteers from northern Ethiopia estimated by qRT-kDNA PCR. Results are compared with the corresponding results obtained by the different LAMP assays. Positive LAMP results are designated by the take-off time in mins (see materials and methods).
| Sample number | History of VL | Parasite/ml qRT-kDNA-PCR |
Estimated take-off time of LAMP amplification (min) |
||
|---|---|---|---|---|---|
| LITS- LAMP1 | LITS- LAMP2 | L151 LAMP | |||
| 1 | + | 0 | – | – | 61 |
| 2 | – | 0 | – | – | – |
| 3 | – | 0 | – | – | – |
| 4 | – | 0 | – | – | – |
| 5 | – | 0 | – | – | – |
| 6 | + | 0 | – | – | 67 |
| 7 | – | 0 | – | – | – |
| 8 | – | 0 | – | – | – |
| 9 | – | 0 | – | – | – |
| 10 | – | 0 | – | – | – |
| 11 | + | 18 | 69 | 57 | – |
| 12 | – | 0 | – | – | – |
| 13 | – | 0 | – | – | – |
| 14 | – | 0 | – | – | – |
| 15 | – | 0 | – | – | – |
| 16 | + | 0 | – | – | – |
| 17 | – | 0 | – | – | – |
| 18 | – | 0 | – | – | – |
| 19 | – | 0 | – | – | – |
| 20 | – | 3 | – | – | – |
| 21 | + | 11 | – | 78 | 63 |
| 22 | – | 595 | 59 | 54 | 52 |
| 23 | – | 0 | – | – | 86 |
| 24 | – | 59 | – | 85 | 65 |
| 25 | – | 0 | – | – | – |
| 26 | + | 0 | – | – | – |
| 27 | – | 0 | – | 72 | 86 |
| 28 | – | 0 | – | 62 | 65 |
| 29 | – | 0 | – | 48 | – |
| 30 | – | 0 | – | 48 | 103 |
| 31 | + | 0 | – | 41 | 34 |
| 32 | – | 0 | – | 57 | – |
| 33 | – | 0 | – | 47 | – |
| 34 | – | 9 | – | – | – |
| 35 | – | 0 | – | – | – |
| 36 | + | 2 | 75 | 41 | 34 |
| 37 | – | 0 | – | 59 | – |
| 38 | – | 1 | – | 43 | – |
| 39 | – | 0 | – | – | – |
| 40 | + | 0 | 56 | 44 | 58 |
| 41 | – | 0 | 60 | 54 | – |
| 42 | – | 23 | – | 42 | 52 |
| 43 | – | 0 | 60 | 53 | – |
| 44 | – | 0 | – | 45 | – |
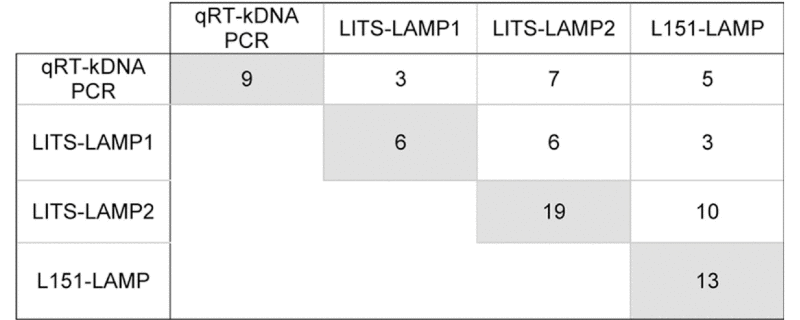
Of the nine previously-treated VL cases; five and six individuals were positive by LITS-LAMP2 and L151 LAMP respectively. Only three of those were positive by qRT-kDNA PCR. Of the nine samples positive by qRT-kDNA PCR, three, seven and 5 were also positive by LITS-LAMP1, LITS-LAMP2, and L151 LAMP respectively (Table 3). Looking at the results of the shared positives between kDNA-PCR and LITS-LAMP2 (that was 7/9), the other two samples that were negative by LITS-LAMP2 were found to have 3, and 9 parasites/ml, based on previous validation of kDNA-PCR results it was calculated that 41.7% of the estimated parasite number with less than 10 parasites/ml could be false positives (Abbasi et al., 2013).The LAMP system that detected the highest number of positive individuals was LITS-LAMP2 (19 positives from 44) followed by L151-LAMP that detected 13 including 10 positive individuals whom were also detected by LITS-LAMP2 (Table 3).
Table 3.
A four by four matrix depicting the relationships between the different LAMP systems and the qRT-kDNA PCR results among the 44 individuals tested. Total numbers detected by each system are in the shaded cells. Note higher sensitivity of the LITS LAMP2 system compared with the PCR as well as the other two LAMP systems.
 |
4. Discussion
In this study we evaluated the efficacy of several newly developed LAMP assays for detecting low concentrations of Leishmania DNA in biological samples. The main advantages of LAMP over conventional and qRT-PCR, are the extremely high sensitivity (detection of 1fg target DNA per reaction (Aryan et al., 2010, Notomi et al., 2000)), the speed with which the assays can be completed, and the simple, relatively inexpensive equipment required. These traits make LAMP assays readily adaptable to field conditions (Hamburger et al., 2013, Njiru, 2012). The three LAMP systems we describe here, detected very low concentrations (>100fg) of L. donovani DNA. This is about 104 more sensitive than conventional ITS1-PCR (el Tai et al., 2000), and about 103 more sensitive than kDNA-PCR (Nicolas et al., 2002). While high sensitivity is certainly an asset, it can also be a liability, necessitating special care to avoid contamination during DNA preparation and mixing of the reaction components in order to avoid false positive results. Thus, it is preferable not to open the tubes at the end of the LAMP reaction for endpoint analysis by SYBR Green I or agarose gel electrophoresis (Kubota and Jenkins, 2015). This can be achieved by adding assimilating primers; one fluorescent and one quenching to the reaction mixture. These primers fluoresce only once they are assimilated into the LAMP product, thereby, demonstrating a positive LAMP reaction (Kubota and Jenkins, 2015).
We selected a direct nucleic acid stain (SYTO-16) to follow the LAMP reactions in real time and avoid the necessity to open the reaction tubes. Selection of a suitable fluorescent dye to be used in real time LAMP detection is crucial (Watts et al., 2014). Previous studies clearly showed that SYBR green I, the most commonly used DNA fluorescent dye, inhibits LAMP reactions (Salant, 2013). The use of 1 μM SYTO-16 dye enabled real time monitoring of LAMP at a low concentration with no inhibitory effect. The importance of developing a LAMP test is its use for point of care nucleic acid based diagnosis, so the advantage of incorporating such low concentration of SYTO-16 dye, renders such tests inexpensive, beside it is possible to use a portable real time machine for monitoring the LAMP amplification (Kubota and Jenkins, 2015, Seyrig et al., 2015).
The shortest takeoff times (35–45 min) were those of the positive controls (Fig. 3). The positive test samples took-off during some 30 min thereafter, while any signals that appeared after 95 min were considered false positives since, at that stage, many of the negative controls with no target DNA or known non-endemic negative human DNA also started to fluoresce. In fact, real time monitoring showed that LAMP reactions produced false positive signals if allowed to proceed for 95 min or longer (data not shown). Similarly, in conventional PCR, amplification occurring after 35 cycles is frequently attributable to spurious priming and considered false positive. Thus, when performing endpoint analyses, LAMP reactions need to be calibrated for maximum incubation with minimal false positive results by running positive control samples with ascending concentrations of template DNA, for different lengths of time before adding SYBR Green I. This illustrates the main advantage of real time LAMP using SYTO-16 over end-point evaluation.
Previous studies using L. donovani-specific LAMP assays showed a sensitivity of 90.7% in one study (Khan et al., 2012), and 96.4% in another study using the same LAMP system. Both studies were performed on samples from clinically-confirmed VL cases (Verma et al., 2013). The diagnostic LAMP assays described previously were based on a primer sets designed specifically for the detection of low concentrations (1fg) of kinetoplast mini-circle DNA of L. donovani in blood and tissue samples (Takagi et al., 2009, Verma et al., 2013). Another LAMP amplification system was developed based on the Leishmania 18S rRNA gene for the detection of Leishmania DNA in cutaneous lesions tissue spotted on FTA card. Malachite green was used for end point detection (Chaouch et al., 2013). The use of a reverse transcriptase LAMP assay based on the 18S ribosomal RNA for the diagnosis of visceral leishmaniasis and cutaneous leishmaniasis was also possible with a less sensitivity than direct detection of DNA by LAMP assay (Adams et al., 2010). In contrast with the previously developed LAMP tests, the LAMP assays we describe were designed to amplify DNA from as many Leishmania species as possible. Once adapted to run on a field LAMP apparatus, our general LAMP assays may be useful for epidemiological field surveys rather than for diagnostic purposes, even when more than one species of Leishmania species is endemic (Hamburger et al., 2013, Njiru, 2012).
In VL foci, a large proportion of asymptomatic persons are frequently infected with L. donovani or L. infantum (Abbasi et al., 2013, Topno et al., 2010, Ukil et al., 2011). For example in an endemic focus in Bihar, India, the rate of sero-conversion among healthy individuals residing in households with a history of VL was 62%, and the disease conversion rate in PCR positive subjects was higher than sero- positivity (Ukil et al., 2011). In another epidemiological study from Nepal, Anti-Leishmania antibodies were found in 40/416 (9.6%) persons without VL and the risk factor for VL upon having a VL case in the neighbourhood was higher (Ostyn et al., 2015), the same group were able to detect Leishmania DNA in 5% of the individuals using Leishmania SSU-rDNA PCR. We focused on, asymptomatic volunteers; nine of whom were previously-treated VL cases. The numbers of previously-treated cases shown to be infected were 9/44 by qRT-kDNA PCR, 6/44 by LITS-LAMP1, 19/44 by LITS-LAMP2, and 13/44 by L151-LAMP. Thus, the LAMP tests indicate the presence of parasite DNA in the blood of some asymptomatic individuals. Identifying asymptomatic cases based on immunological or molecular analysis among household members of individuals with previously confirmed VL cases can classify this group as being at risk of infection. In different studies the use of molecular methods for Leishmania detection identified higher percentages of asymptomatic cases (Alborzi et al., 2008). In addition, blood PCR or LAMP can not conclusively prove the presence of live Leishmania parasites. Positive results may be due to parasite DNA circulating in the blood having leached out from infected skin or internal organs (Banu et al., 2016). Therefore, follow-up monitoring for clinical signs is crucial for validating the effectiveness of such sensitive methods for identification of asymptomatic cases and future prevention of disease.
Control of vector-borne diseases calls for the accurate appraisal of the hosts’ potential for infecting the arthropod vectors. Diagnosis of host infectiousness is optimally achieved by determining the infection rates of insectary-reared vectors that had fed on infected human volunteers (xenodiagnosis). Unfortunately, xenodiagnosis for leishmaniasis is an intricate operation encumbered by technical, logistical and ethical hurdles. We are currently working on replacement technologies based on skin microbiopsies that will be analyzed in the field using Leishmania specific LAMP to identify those persons that serve as reservoir hosts for propagating VL in the community and treat them.
Acknowledgement
This study was supported by the Bill and Melinda Gates Foundation Global Health Program (grant number OPPGH5336).
Contributor Information
Ibrahim Abbasi, Email: ibrahima@ekmd.huji.ac.il.
Oscar D. Kirstein, Email: oscark@ekmd.huji.ac.il.
Asrat Hailu, Email: hailu_a2004@yahoo.com.
Alon Warburg, Email: alonw@ekmd.huji.ac.il.
References
- Abbasi I., Aramin S., Hailu A., Shiferaw W., Kassahun A., Belay S., Jaffe C., Warburg A. Evaluation of PCR procedures for detecting and quantifying Leishmania donovani DNA in large numbers of dried human blood samples from a visceral leishmaniasis focus in Northern Ethiopia. BMC Infect. Dis. 2013;13:153. doi: 10.1186/1471-2334-13-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams E.R., Schoone G.J., Ageed A.F., Safi S.E., Schallig H.D. Development of a reverse transcriptase loop-mediated isothermal amplification (LAMP) assay for the sensitive detection of Leishmania parasites in clinical samples. Am. J. Trop. Med. Hyg. 2010;82:591–596. doi: 10.4269/ajtmh.2010.09-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler S., Foner A., Montiglio B. The relationship between human and animal strains of Leishmania from the Sudan. Trans. R. Soc. Trop. Med. Hyg. 1966;60:380–386. doi: 10.1016/0035-9203(66)90305-1. [DOI] [PubMed] [Google Scholar]
- Alborzi A., Pourabbas B., Shahian F., Mardaneh J., Pouladfar G.R., Ziyaeyan M. Detection of Leishmania infantum kinetoplast DNA in the whole blood of asymptomatic individuals by PCR-ELISA and comparison with other infection markers in endemic areas, southern Iran. Am. J. Trop. Med. Hyg. 2008;79:839–842. [PubMed] [Google Scholar]
- Ali A., Ashford R.W. Visceral leishmaniasis in Ethiopia. IV. Prevalence, incidence and relation of infection to disease in an endemic area. Ann. Trop. Med. Parasitol. 1994;88:289–293. doi: 10.1080/00034983.1994.11812869. [DOI] [PubMed] [Google Scholar]
- Alvar J., Velez I.D., Bern C., Herrero M., Desjeux P., Cano J., Jannin J., den Boer M. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryan E., Makvandi M., Farajzadeh A., Huygen K., Bifani P., Mousavi S.L., Fateh A., Jelodar A., Gouya M.M., Romano M. A novel and more sensitive loop-mediated isothermal amplification assay targeting IS6110 for detection of mycobacterium tuberculosis complex. Microbiol. Res. 2010;165:211–220. doi: 10.1016/j.micres.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Banu S.S., Meyer W., Ahmed B.N., Kim R., Lee R. Detection of Leishmania donovani in peripheral blood of asymptomatic individuals in contact with patients with visceral leishmaniasis. Trans. R. Soc. Trop. Med. Hyg. 2016;110:286–293. doi: 10.1093/trstmh/trw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelaert M., El-Safi S., Hailu A., Mukhtar M., Rijal S., Sundar S., Wasunna M., Aseffa A., Mbui J., Menten J., Desjeux P., Peeling R.W. Diagnostic tests for kala-azar: a multi-centre study of the freeze-dried DAT, rK39 strip test and KAtex in East Africa and the Indian subcontinent. Trans. R. Soc. Trop. Med. Hyg. 2008;102:32–40. doi: 10.1016/j.trstmh.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Chaouch M., Mhadhbi M., Adams E.R., Schoone G.J., Limam S., Gharbi Z., Darghouth M.A., Guizani I., BenAbderrazak S. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of Leishmania infantum in canine leishmaniasis based on cysteine protease B genes. Vet. Parasitol. 2013;198:78–84. doi: 10.1016/j.vetpar.2013.07.038. [DOI] [PubMed] [Google Scholar]
- Chappuis F., Sundar S., Hailu A., Ghalib H., Rijal S., Peeling R.W., Alvar J., Boelaert M. Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat. Rev. Microbiol. 2007;5:873–882. doi: 10.1038/nrmicro1748. [DOI] [PubMed] [Google Scholar]
- Desjeux P. Leishmaniasis: current situation and new perspectives. Comp. Immunol. Microbiol. Infect. Dis. 2004;27:305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Hamburger J., Abbasi I., Kariuki C., Wanjala A., Mzungu E., Mungai P., Muchiri E., King C.H. Evaluation of loop-mediated isothermal amplification suitable for molecular monitoring of schistosome-infected snails in field laboratories. Am. J. Trop. Med. Hyg. 2013;88:344–351. doi: 10.4269/ajtmh.2012.12-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.G., Bhaskar K.R., Salam M.A., Akther T., Pluschke G., Mondal D. Diagnostic accuracy of loop-mediated isothermal amplification (LAMP) for detection of Leishmania DNA in buffy coat from visceral leishmaniasis patients. Parasit. Vectors. 2012;5:280. doi: 10.1186/1756-3305-5-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota R., Jenkins D.M. Real-time duplex applications of loop-mediated AMPlification (LAMP) by assimilating probes. Int. J. Mol. Sci. 2015;16:4786–4799. doi: 10.3390/ijms16034786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E., Warburg A., Novikov I., Hailu A., Volf P., Seblova V., Huppert A. Quantifying the contribution of hosts with different parasite concentrations to the transmission of visceral leishmaniasis in Ethiopia. PLoS Negl. Trop. Dis. 2014;8:e3288. doi: 10.1371/journal.pntd.0003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas L., Milon G., Prina E. Rapid differentiation of Old World Leishmania species by LightCycler polymerase chain reaction and melting curve analysis. J. Microbiol. Methods. 2002;51:295–299. doi: 10.1016/s0167-7012(02)00099-4. [DOI] [PubMed] [Google Scholar]
- Njiru Z.K. Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Negl. Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzelu C.O., Gomez E.A., Caceres A.G., Sakurai T., Martini-Robles L., Uezato H., Mimori T., Katakura K., Hashiguchi Y., Kato H. Development of a loop-mediated isothermal amplification method for rapid mass-screening of sand flies for Leishmania infection. Acta Trop. 2014;132:1–6. doi: 10.1016/j.actatropica.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Ostyn B., Uranw S., Bhattarai N.R., Das M.L., Rai K., Tersago K., Pokhrel Y., Durnez L., Marasini B., Van der Auwera G., Dujardin J.C., Coosemans M., Argaw D., Boelaert M., Rijal S. Transmission of Leishmania donovani in the hills of Eastern Nepal, an outbreak investigation in Okhaldhunga and Bhojpur districts. PLoS Negl. Trop. Dis. 2015;9:e0003966. doi: 10.1371/journal.pntd.0003966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Wong B.W., Ma E.H., Chan K.H., Chow L.M., Abeyewickreme W., Tangpukdee N., Yuen K.Y., Guan Y., Looareesuwan S., Peiris J.S. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin. Chem. 2006;52:303–306. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- Ready P.D. Epidemiology of visceral leishmaniasis. Clin. Epidemiol. 2014;6:147–154. doi: 10.2147/CLEP.S44267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers M.R., Popper S.J., Wirth D.F. Amplification of kinetoplast DNA as a tool in the detection and diagnosis of Leishmania. Exp. Parasitol. 1990;71:267–275. doi: 10.1016/0014-4894(90)90031-7. [DOI] [PubMed] [Google Scholar]
- Salant H., Abbasi I., Hamburger J. The development of a loop-mediated isothermal amplification method (LAMP) for Echinococcus granulosis coprodetection. Am. J. Trop. Med. Hyg. 2012;87:883–887. doi: 10.4269/ajtmh.2012.12-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salant H. The development of a loop-mediated isothermal amplification method (LAMP) for echinococcus granulosis coprodetection (vol. 87, pg 883, 2012) Am. J. Trop. Med. Hyg. 2013;88 doi: 10.4269/ajtmh.2012.12-0184. 606–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyrig G., Stedtfeld R.D., Tourlousse D.M., Ahmad F., Towery K., Cupples A.M., Tiedje J.M., Hashsham S.A. Selection of fluorescent DNA dyes for real-time LAMP with portable and simple optics. J. Microbiol. Methods. 2015;119:223–227. doi: 10.1016/j.mimet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Sharma M.C., Gupta A.K., Das V.N., Verma N., Kumar N., Saran R., Kar S.K. Leishmania donovani in blood smears of asymptomatic persons. Acta Trop. 2000;76:195–196. doi: 10.1016/s0001-706x(00)00068-1. [DOI] [PubMed] [Google Scholar]
- Takagi H., Itoh M., Islam M.Z., Razzaque A., Ekram A.R., Hashighuchi Y., Noiri E., Kimura E. Sensitive, specific, and rapid detection of Leishmania donovani DNA by loop-mediated isothermal amplification. Am. J. Trop. Med. Hyg. 2009;81:578–582. doi: 10.4269/ajtmh.2009.09-0145. [DOI] [PubMed] [Google Scholar]
- Topno R.K., Das V.N., Ranjan A., Pandey K., Singh D., Kumar N., Siddiqui N.A., Singh V.P., Kesari S., Kumar N., Bimal S., Kumar A.J., Meena C., Kumar R., Das P. Asymptomatic infection with visceral leishmaniasis in a disease-endemic area in Bihar, India. Am. J. Trop. Med. Hyg. 2010;83:502–506. doi: 10.4269/ajtmh.2010.09-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukil A., Kar S., Srivastav S., Ghosh K., Das P.K. Curative effect of 18beta-glycyrrhetinic acid in experimental visceral leishmaniasis depends on phosphatase-dependent modulation of cellular MAP kinases. PLoS One. 2011;6:e29062. doi: 10.1371/journal.pone.0029062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G., Dujardin J.C. Species typing in dermal leishmaniasis. Clini. Microbiol. Rev. 2015;28:265–294. doi: 10.1128/CMR.00104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Avishek K., Sharma V., Negi N.S., Ramesh V., Salotra P. Application of loop-mediated isothermal amplification assay for the sensitive and rapid diagnosis of visceral leishmaniasis and post-kala-azar dermal leishmaniasis. Diagn. Microbiol. Infect. Dis. 2013;75:390–395. doi: 10.1016/j.diagmicrobio.2013.01.011. [DOI] [PubMed] [Google Scholar]
- WHO/TDR, Control of the Leishmaniasis: Report of a Meeting of the WHO Expert Committee on the Control of Leishmaniases; World Health Oganization, Geneva, March 22–26; 2010. [Google Scholar]
- Watts M.R., James G., Sultana Y., Ginn A.N., Outhred A.C., Kong F.R., Verweij J.J., Iredell J.R., Chen S.C.A., Lee R. A Loop-mediated isothermal amplification (LAMP) assay for Strongyloides stercoralis in stool that uses a visual detection method with SYTO-82 fluorescent dye. Am. J. Trop. Med. Hyg. 2014;90:306–311. doi: 10.4269/ajtmh.13-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Tai N.O., Osman O.F., el Fari M., Presber W., Schonian G. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans. R. Soc. Trop. Med. Hyg. 2000;94:575–579. doi: 10.1016/s0035-9203(00)90093-2. [DOI] [PubMed] [Google Scholar]
- ter Horst R., Tefera T., Assefa G., Ebrahim A.Z., Davidson R.N., Ritmeijer K. Field evaluation of rK39 test and direct agglutination test for diagnosis of visceral Leishmaniasis in a population with high prevalence of human immunodeficiency virus in Ethiopia. Am. J. Trop. Med. Hyg. 2009;80:929–934. [PubMed] [Google Scholar]
- van Eys G.J., Schoone G.J., Kroon N.C., Ebeling S.B. Sequence analysis of small subunit ribosomal RNA genes and its use for detection and identification of Leishmania parasites. Mol. Biochem. Parasitol. 1992;51:133–142. doi: 10.1016/0166-6851(92)90208-2. [DOI] [PubMed] [Google Scholar]